Abstract
Background:
Cadmium (Cd) is a ubiquitous environmental toxicant that can accumulate in the placenta during pregnancy, where it may impair placental function and affect fetal development.
Objectives:
We aimed to investigate Cd-associated variations in placental DNA methylation (DNAM) and associations with gene expression; we also aimed to identify novel pathways involved in Cd-associated reproductive toxicity.
Methods:
Using placental DNAM and Cd concentrations in the New Hampshire Birth Cohort Study (NHBCS, ) and the Rhode Island Child Health Study (RICHS, ), we performed an epigenome-wide association study (EWAS) between Cd and DNAM, adjusting for tissue heterogeneity using a reference-free method. Cohort-specific results were aggregated via inverse variance weighted fixed effects meta-analysis, and variably methylated CpGs were associated with gene expression. We then performed functional enrichment analysis and tests for associations between gene expression and birth size metrics.
Results:
We identified 17 Cd-associated differentially methylated CpG sites with meta-analysis , two of which were within a 5% false discovery rate (FDR). DNAM levels at 9 of the 17 loci were associated with increased expression of 6 genes (5% FDR): TNFAIP2, EXOC3L4, GAS7, SREBF1, ACOT7, and RORA. Higher placental expression of TNFAIP2 and ACOT7 and lower expression of RORA were associated with lower birth weight z-scores ().
Conclusion:
Cd-associated differential DNAM and corresponding DNAM-expression associations were observed at loci involved in inflammatory signaling and cell growth. The expression levels of genes involved in inflammatory signaling (TNFAIP2, ACOT7, and RORA) were also associated with birth weight, suggesting a role for inflammatory processes in Cd-associated reproductive toxicity. https://doi.org/10.1289/EHP2192
Introduction
Cadmium (Cd) is a toxic heavy metal that is released into the environment from mining and industrial processes, the application of phosphate fertilizers, and fossil fuel combustion; it accumulates in soils and is readily taken up by plants (ATSDR 2012). Human exposure primarily occurs via consumption of contaminated foods or through using tobacco products (Järup and Åkesson 2009). Cadmium serves no biological role in humans and can cause severe health consequences such as kidney damage, bone and joint problems, and various cancers (ATSDR 2012). Trace exposures to cadmium may also have toxic effects on pregnant mothers and on the developing fetus (Thompson and Bannigan 2008).
Recent epidemiologic studies have observed associations between Cd concentrations in maternal and/or fetal tissues with restricted fetal growth and pregnancy complications (Al-Saleh et al. 2014, 2015, Kippler et al. 2012b, 2013; Laine et al. 2015; Menai et al. 2012; Wang et al. 2016a). The toxic mechanisms that underlie these associations are not clear, although Cd is known to induce oxidative stress, interfere with cell-cycle regulation, alter apoptotic signaling at the cellular level (Valko et al. 2016), and to act as an endocrine disruptor (Jiménez-Ortega et al. 2012). Additionally, the placenta provides a partial barrier against maternal–fetal Cd transfer, resulting in Cd accumulation in the placenta throughout pregnancy, although its accumulation within the placenta is likely not without consequence (Gundacker and Hengstschläger 2012). Animal models and in vitro studies have suggested that maternal Cd exposure during pregnancy can perturb maternal–fetal nutrient and waste transfer (Kippler et al. 2010; Wang et al. 2016b), increase oxidative stress in placental tissue (Wang et al. 2012), interfere with placental glucocorticoid synthesis (Wang et al. 2014), and alter trophoblast proliferation and apoptosis (Erboga and Kanter 2016). Thus, the placenta appears to be a critical target tissue for the toxic effects of Cd during human pregnancies.
We hypothesize that various Cd-associated perturbations to placental functions are likely linked to the selective expression or repression of specific biological pathways. Similarly, maternal Cd exposure during pregnancy has been associated with variations in DNA methylation (DNAM), an epigenetic mechanism that regulates gene expression potential, in maternal and fetal blood samples (Vilahur et al. 2015). To date, only one study has investigated associations between placental Cd and DNAM throughout the placental genome in humans. Mohanty et al (2015) explored Cd-associated DNAM in a small study of 24 placentae, identifying multiple differentially methylated loci nearby or within genes involved in cell damage, angiogenesis, cell differentiation, and organ development; these associations differed by fetal sex (Mohanty et al. 2015). Studies with larger samples using recent methodological advances to estimate and adjust for tissue heterogeneity are needed to understand how Cd exposure influences the placental epigenome of human pregnancies and whether such variations are related to fetal growth restriction.
We conducted a study of the associations between placental Cd and placental DNAM throughout the epigenome across two well-characterized independent cohorts while examining the potential impact of tissue heterogeneity on these associations. Secondarily, we investigated whether the top hits from our analysis were enriched for biological pathways consistent with findings from other studies; whether differentially methylated loci were associated with the expression levels of nearby genes; and whether these variations in expression were associated with standardized measures of birth weight, birth length, and head circumference.
Methods
New Hampshire Birth Cohort Study
This study included mother–infant pairs from the New Hampshire Birth Cohort Study (NHBCS), an ongoing birth cohort initiated in 2009. Women who were currently pregnant, were between 18 and 45 y of age, were receiving prenatal care from one of the study clinics in New Hampshire, who reported that the primary source of drinking water at their residence was from an unregulated well, and who had resided in the same household since their previous menstrual period with no plans to move before delivery were enrolled in the cohort. All participants provided written informed consent in accordance with the requirements of the Institutional Review Board (IRB) of Dartmouth College. The sample for this study consisted of NHBCS participants who were recruited between February 2012 and September 2013, whose placenta were sampled to conduct genetic and epigenetic assays (). Placental gross measures such as placental diameter (cm) and placental weight (g) were collected immediately after delivery. Interviewer-administered questionnaires and medical record abstraction were utilized to collect sociodemographic, lifestyle, and anthropometric data.
Rhode Island Child Health Study
The Rhode Island Child Health Study (RICHS) enrolled mother–infant pairs with nonpathologic pregnancies at the Women & Infants Hospital in Providence, Rhode Island, between September 2010 and February 2013. Exclusion criteria consisted of mothers being of age or having life-threatening conditions, pregnancies with gestational time , or infants with congenital/chromosomal abnormalities. All protocols were approved by the institutional review boards at the Women & Infants Hospital of Rhode Island and Dartmouth College, and all participants provided written informed consent. Infants who were born small for gestational age [ birth weight (BW) percentile] or large for gestational age ( BW percentile) were oversampled; then, infants who were adequate for gestational age (between the 10th and 90th BW percentiles) that matched on gestational age and maternal age were coincidentally enrolled. This study included all mother–infant pairs for whom placental metal concentrations and placental DNAM arrays had been conducted (). Interviewer-administered questionnaires were utilized to collect sociodemographic and lifestyle data; anthropometric and medical history data were obtained via structured medical records review. Samples with complete covariate, exposure, and outcome data were utilized for this study.
Anthropometric Measures
Birth weight (g), head circumference (cm), and birth length (cm) were abstracted from medical records. z-Scores were calculated for each, standardized by gestational time and infant sex, via Fenton growth curves (Fenton and Kim 2013). We classified infants with standardized BW percentiles below the 10th percentile as small for gestational age (SGA), those above the 90th percentile as large for gestational age (LGA), and those between the 10th and 90th percentile as adequate for gestational age (AGA).
Potential Covariates
Highest achieved maternal education level was self-reported and was defined as those with education beyond the high school level versus those who did not go beyond high school. Maternal smoking during pregnancy was defined as any self-reported smoking during any point of pregnancy versus those who reported no smoking during pregnancy. Fetal sex was abstracted from medical records. Samples with complete covariate, placental Cd, and placental DNAM data were utilized for this study.
Sample Collection
Placenta samples from RICHS and NHBCS were biopsied from the fetal side adjacent to the cord insertion site within 2 h of delivery, and maternal decidua was removed. Samples were placed in RNAlater (Life Technologies) and then frozen at . Both RNA and DNA were extracted (Norgen Biotek) and quantified using a Qubit Flourometer (Life Technologies) and were subsequently stored at . Both cohorts followed the same sampling protocol.
DNA Methylation, Quality Control, and Normalization
Both cohorts measured DNAM via Illumina Infinium HumanMethylation450K BeadArray (Illumina) at the University of Minnesota Genomics Center. Bisulfite modification was performed with an EZ Methylation kit (Zymo Research, Irvine, CA), samples were randomized across multiple batches, batch variables were recorded to allow for batch-effect corrections, and data were assembled using BeadStudio (Illumina). The raw array data are available via the NCBI Gene Expression Omnibus (GEO) for NHBCS and RICHS via accession numbers GSE71678 and GSE75248, respectively. For quality control (QC), we excluded probes with poor detection p-values (), measuring DNAM at X- and Y-linked CpG sites, with a single nucleotide polymorphism (SNP) within 10 bp of the target CpG or single base extension (SBE) (and allelic frequency ), or cross-hybridizing to multiple genomic regions (Chen et al. 2013). Background correction, dye bias, and functional normalization were performed via the minfi package (Fortin et al. 2014), and standardization across probe types was performed via beta mixture quantile (BMIQ) normalization (Teschendorff et al. 2013). Batch effects were corrected for using empirical Bayes via the combat function in R (Johnson et al. 2007). We examined whether principal components from the methylation array were associated with batch variables and found no indications of remaining batch effects after this correction. We also confirmed that batch variables were not associated with continuous [analysis of variance (ANOVA) p-values: , ] or dichotomous Cd ( p-values: , ) in either cohort. DNAM data were reported as , representing the proportion of methylated alleles for each individual CpG site. For the epigenome-wide association (EWAS) models described below we utilized M-values, logit-transformation of the beta values, which better approximate a normal distribution (Du et al. 2010).
RNA Sequencing, QC, and Normalization
Transcriptome-wide sequencing was performed on 200 placental samples from RICHS. We used an RNeasy Mini Kit (Qiagen) to isolate total RNA, which was then stored at until analysis. We quantified RNA using a Nanodrop Spectrophoto-meter (Thermo Scientific), assessed RNA integrity via Agilent Bioanalyzer (Agilent), removed ribosomal RNA with a Ribo-Zero Kit (Huang et al. 2011), converted to cDNA using random hexamers (Thermo Scientific), and performed transcriptome-wide RNA sequencing via the HiSeq 2500 platform (Illumina) (Bentley et al. 2008). Raw reads have been deposited at the NCBI sequence read archive (SRP095910). QC was performed in FastQC; then, reads were mapped to the human reference genome (h19) using the Spliced Transcripts Alignment to a Reference (STAR) aligner. Low-expressed genes were excluded, and read counts were adjusted for GC content (Risso et al. 2011) and then normalized via the trimmed mean of M-values (TMM) (Robinson et al. 2010); final data are normalized counts per million (logCPM) reads.
Cadmium Quantification
Trace element concentrations were quantified in RICHS and NHBCS placental samples at the Dartmouth Trace Elements Analysis Core using inductively coupled plasma mass spectrometry (ICP-MS); details of the processing are described elsewhere (Punshon et al. 2016). None of the RICHS samples had nondetectable Cd concentrations, and only four of the NHBCS samples received nondetects, which were assigned a value equal to the 0.5th percentile of detectable values. Owing to the skewed nature of the Cd concentration distributions and to the infeasibility of exploring modeling assumptions for all EWAS models, we dichotomized cadmium concentrations by the cohort-specific median concentrations (NHBCS ) and (RICHS ) for the EWAS analysis. Cd concentrations were -transformed for follow-up analyses of significant findings, which utilized continuous data.
Estimation of Placental Tissue Heterogeneity
We addressed the potential issue of confounding due to DNAM-associated tissue heterogeneity via the RefFreeEWAS package in R (Houseman et al. 2016). This method utilized nonnegative matrix factorization to estimate the proportions of putative cellular mixtures and has been demonstrated to yield reliable estimates of the constituent cell types and the underlying methylomes that define them (Houseman et al. 2016). Using the 10,000 CpGs with the greatest variability in DNAM, we identified four and five putative constituent cell mixtures in the RICHS and NHBCS cohorts, respectively, and then used the full set of CpGs for each cohort (NHBCS and RICHS ) to estimate the relative proportions of each putative cell type per placental sample. We examined whether the 10,000 CpGs with the largest variance across putative cell-type-specific DNAM values were enriched for gene ontology (GO) terms or Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways via the gometh function in the missMethyl package in R (Phipson et al. 2015). This method adjusts for the potential bias introduced by some genes having a greater probability of being included in the gene set because they have greater numbers of CpGs on the 450K array. We examined the consistency of the CpGs with high cell-type-specific DNAM as well as the consistency in the top 100 GO terms and KEGG pathways associated with those highly variable CpGs.
Epigenome-Wide Association Study
Robust linear regressions (rlm) were used to estimate the associations between CpG-specific DNAM levels and high versus low placental concentrations of Cd. We produced two covariate-adjusted models for each CpG within each cohort, first regressing M-values on dichotomized Cd concentrations (cohort-specific median as the reference) while adjusting for infant sex, maternal smoking during pregnancy, and highest achieved maternal education level. Second, we produced the same models while also adjusting for the estimated proportions of putative placental cell types. Genomic inflation was examined via the genomic inflation factor (), which was calculated using the genABEL package in R (Aulchenko et al. 2007). We selected the 250 CpGs with the smallest Cd-associated meta-analysis p-values and assessed these for functional enrichment with GO terms and KEGG pathways as described above. Statistical significance for enrichment was determined at a 5% false discovery rate (FDR).
Meta-Analyses
We used METAL to meta-analyze the cohort-specific Cd-associations via inverse variance weighted fixed effects models, and we evaluated interstudy heterogeneity via Cochran's Q-test . (Willer et al. 2010). We produced FDR-adjusted p-values using the qvalue package in R. Statistical significance was determined at an FDR of 5%. Volcano plots were produced to visualize the overall magnitude and direction of associations, and Manhattan plots were produced using the qqman package to visualize the genomic distribution of significant associations. Plots for specific regions of the genome were visualized using the coMET package (Martin et al. 2015). Top hits from the study were also evaluated for dose–response relationships via modeling associations between -transformed continuous Cd and DNAM, using linear mixed models, allowing for random intercepts by study. Plots of FDR-significant hits utilized DNAM as the dependent variable for more intuitive interpretation.
Expression Quantitative Trait Methylation Analysis
We then tested whether any of the Cd-associated CpGs () could be expression quantitative trait methylation (eQTM) loci within the 200 RICHS samples, 85 of which overlap with the samples used in the EWAS. We tested for cis-gene–CpG associations (within of the target CpG site) by regressing gene expression levels () on CpG using robust linear models. Statistical significance for eQTM associations was determined at a 5% FDR.
Associations with Fetal Growth
Genes that were identified as potential eQTMs at a 5% FDR threshold were then tested for associations between birth metrics and gene expression using Kendal correlations, a nonparametric correlation test that allows for ties. Because birth metric z-scores were standardized by fetal sex and by weeks of gestation, no additional adjustments were made to these association tests.
Replication of Previous Studies and Sex-Specific Effects
We then investigated whether we could reproduce the results from a previous study of sex-specific associations between placental Cd and placental DNAM (Mohanty et al. 2015). Because multiple studies of various fetal and maternal tissues have suggested that associations between Cd and DNAM may differ by fetal sex (Kippler et al. 2013; Mohanty et al. 2015; Virani et al. 2016), we also screened the CpGs from our study with meta-analysis for interactions with fetal sex. For these analyses, we utilized linear mixed models with DNAM M-values as the outcome, allowing for random intercepts by study, and we included an interaction term between DNAM levels and fetal sex while adjusting for maternal education and maternal smoking. We utilized the Wald test to produce p-values and determined statistically significant interactions at a p-value threshold of 0.05. Any models producing interaction were then rerun stratified by fetal sex.
Results
The EWAS for this study included 343 mother–infant pairs drawn from the New Hampshire Birth Cohort (NHBCS) and 141 mother–infant pairs from the Rhode Island Child Health Study (RICHS). The most notable differences between the NHBCS and RICHS samples (Table 1) were with regard to racial diversity and birth size. Because RICHS was over-sampled for LGA and SGA infants by design, we observed a higher proportion of SGA and LGA infants compared with NHBCS. Placental samples from the RICHS cohort also had slightly higher median Cd concentrations () than samples from the NHBCS cohort (), and concentrations were approximately log-normally distributed in both cohorts (see Figure S1).
Table 1.
Maternal and offspring characteristics for the NHBCS () and RICHS () samples.
| Maternal characteristics | NHBCSa | RICHSa |
|---|---|---|
| Smoking during pregnancy | ||
| Any | 21 (6.1%) | 19 (13.5%) |
| None | 322 (93.9%) | 122 (86.5%) |
| Educational attainment | ||
| High school or less | 37 (10.8%) | 34 (24.1%) |
| More than high school | 306 (89.2%) | 107 (75.9%) |
| Ethnicity | ||
| Not white | 6 (1.7%) | 36 (25.5%) |
| White | 337 (98.3%) | 105 (74.5%) |
| Infant characteristics | ||
| Gestational age (weeks) | 38.95 (1.54) | 39.33 (1.00) |
| Birth weight (g)b | 3435.14 (510.95) | 3623.09 (716.03) |
| Size for gestational ageb | ||
| SGA | 15 (4.4%) | 21 (14.9%) |
| AGA | 292 (86.1%) | 68 (48.2%) |
| LGA | 32 (9.4%) | 52 (36.9%) |
| Sex | ||
| Female | 159 (46.4%) | 68 (48.2%) |
| Male | 184 (53.6%) | 73 (51.8%) |
| Placental Cd (ng/g)c | ||
| Overall | 3.13 (2.61) | 4.37 (2.71) |
| Below cohort-specific median | 2.07 (1.05) | 2.78 (1.35) |
| Above cohort-specific median | 4.68 (2.32) | 5.49 (2.44) |
Note: AGA, adequate for gestational age; Cd, cadmium; LGA, large for gestational age; NHBCS, New Hampshire Birth Cohort Study; RICHS, Rhode Island Child Health Study; SGA, small for gestational age.
Means and standard deviations are presented for continuous variables, whereas frequencies and percentages are presented for categorical variables.
Four samples were missing for birth weight and SGA/AGA/LGA classifications in the NHBCS; all other continuous and categorical variables had no missing values.
Medians and interquartile ranges (IQRs) presented owing to skewed distributions of Cd concentrations.
We regressed DNAM M-values (at 408,367 CpGs in NHBCS and 397,040 CpGs in RICHS) on dichotomized placental Cd concentrations (median split) while adjusting for fetal sex, maternal smoking during pregnancy, and maternal education level in each cohort. We then combined parameter estimates and p-values via inverse variance weighted fixed effects meta-analysis (390,583 CpGs overlapped between the cohorts), which yielded three loci with Cd associations at a 5% FDR and 18 additional loci with (Table 2), in which Cd was predominantly associated with hypermethylation.
Table 2.
Results from cohort-specific and meta-analyses of Cd-associated DNA methylation, for probes with meta-analysis ; adjusted for maternal smoking during pregnancy, maternal education, and fetal sex.
| CpG annotations | Meta-analysis | NHBCS () | RICHS () | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG ID | Ch. | Genomic position | Gene | Regulatory feature | p-Value | FDR | p-Value | p-Value | |||
| cg16034168 | 1 | 6336711 | ACOT7 | Body | 0.39 | 0.107 | 0.48 | 0.31 | |||
| cg18384460 | 1 | 63771428 | - | - | 0.35 | 0.107 | 0.43 | 0.25 | |||
| cg08705382 | 1 | 230322024 | GALNT2 | Body | 0.34 | 0.151 | 0.34 | 0.35 | |||
| cg26262055 | 4 | 25090618 | - | - | 0.22 | 0.151 | 0.29 | 0.21 | |||
| cg26653990 | 5 | 72510846 | - | - | 0.23 | 0.151 | 0.25 | 0.19 | |||
| cg00766220 | 5 | 72598865 | - | - | 0.14 | 0.147 | 0.13 | 0.14 | |||
| cg09391066 | 6 | 3163814 | - | - | 0.19 | 0.151 | 0.21 | 0.18 | |||
| cg03797906 | 7 | 28994696 | TRIL | 3′UTR; Body | 0.17 | 0.151 | 0.21 | 0.14 | |||
| cg11157440 | 8 | 19316005 | CSGALNACT1 | Body | 0.29 | 0.151 | 0.28 | 0.30 | |||
| cg21437157 | 8 | 27346608 | - | - | 0.23 | 0.147 | 0.19 | 0.33 | |||
| cg05888894 | 8 | 119107030 | EXT1 | Body | 0.33 | 0.046 | 0.33 | 0.33 | |||
| cg03509329 | 14 | 24520788 | LRRC16B | TSS1500 | 0.15 | 0.151 | 0.19 | 0.12 | |||
| cg15345741 | 14 | 103674333 | - | - | 0.28 | 0.046 | 0.28 | 0.29 | |||
| cg17232357 | 15 | 67012832 | SMAD6 | Body | 0.107 | ||||||
| cg12642651 | 17 | 9861712 | GAS7 | Body | 0.38 | 0.151 | 0.27 | 0.48 | |||
| cg03762242 | 17 | 9940004 | GAS7 | Body; 5′UTR | 0.26 | 0.142 | 0.26 | 0.26 | |||
| cg16768966 | 17 | 9940227 | GAS7 | Body; TSS200 | 0.37 | 0.046 | 0.34 | 0.40 | |||
| cg15285733 | 17 | 10039537 | GAS7 | Body | 0.17 | 0.062 | 0.16 | 0.18 | |||
| cg02393525 | 17 | 40687925 | NAGLU | TSS200 | 0.23 | 0.151 | 0.16 | 0.31 | |||
| cg06707369 | 19 | 36400425 | TYROBP | TSS1500 | 0.17 | 0.151 | 0.19 | 0.15 | |||
| cg11707084 | 19 | 48345263 | CRX | 3′UTR | 0.23 | 0.151 | 0.25 | 0.19 | |||
Note: Ch., chromosome; CpG, cytosine-phosphate-guanine methylation site; FDR, false discovery rate adjusted p-value; NHBCS, New Hampshire Birth Cohort; RICHS, Rhode Island Child Health Study; TSS200, within 200 bp of the transcription start site; TSS1500, within 1,500 bp of the transcription start site; UTR, untranslated region.
Cell-specific DNAM is a well-recognized confounder in epigenetic epidemiology, leading many studies to estimate the proportions of constituent cell types from the DNAM array data (Houseman et al. 2012) and to adjust for these proportions in their regression models. However, estimation of cell-type proportions requires access to cell-type-specific methylomes for the individual cells that constitute the heterogenous tissue under study, and no reference methylomes currently exist for placental tissue. Thus, we used a reference-free method that deconvoluted the major axes of variation in DNAM, which are likely associated with tissue heterogeneity, to estimate the proportions of the putative constituent cell types (PCTs) in our placental samples (Houseman et al. 2016). With this method, we estimated five and four PCTs in NHBCS and RICHS, respectively. We then tested whether the 10,000 CpGs with the greatest variability in PCT-specific DNAM were enriched for biologically relevant GO terms or KEGG pathways, and we checked for consistency between the two cohorts. We observed 31.6% overlap among these 10,000 PCT-defining CpGs and strong concordance in the biological functions of the top 100 GO terms (60% overlap) and KEGG pathways (76% overlap) across the two cohorts. The top 10 KEGG pathways from both cohorts included metabolic pathways, neuroactive ligand–receptor interaction, axon guidance, calcium signaling, olfactory transduction, cell adhesion molecules (CAMs), and cAMP signaling (see Excel Tables S1 and S2; see also Figure S2).
We then tested whether the distribution of these PCTs varied with placental Cd concentrations. Higher Cd was associated with increased proportions of PCT-1 (3.4% difference, T-test ) and PCT-2 (4.7% difference, T-test ) in NHBCS, whereas higher Cd was associated with increased proportions of PCT-2 (7.3% difference, T-test ) in RICHS. Because these associations may confound the relationships between Cd and locus-specific DNAM, we then reproduced the cohort-specific multivariable models and meta-analysis while also adjusting for the estimated PCT pro-portions. These adjustments substantially reduced inflation in NHBCS ( before adjustment and after adjustment), and only modestly increased inflation in RICHS ( before adjustment and after adjustment). Furthermore, the PCT adjustments reduced the proportion of small heterogeneity p-values () in the meta-analysis from 7.9% in the non-PCT-adjusted models to 4.6% in the PCT-adjusted models. (See Excel Table S3 and Excel Table S4 for full results from all cohort-specific and meta-analyses).
We compared the meta-analysis parameter estimates and p-values before and after PCT adjustment across all CpGs via plots, LOESS curves, and Spearman correlations. We found that overall, the parameter estimates tended to become attenuated by PCT adjustment (see Figure S3) and produced fewer small p-values (see Figure S4), indicating an overall confounding by estimated cellular heterogeneity. For example, the parameter estimates for our top three hits from the original meta-analysis (cg15345741, cg16768966, and cg05888894) were reduced by 11%, 16%, and 45%, respectively, after adjustment for PCTs, indicating that cell mixture likely explained much of the Cd-associated variation for cg05888894, but only a modest proportion of the Cd-DNAM associations at cg15345741 and cg16768966. Additionally, the coefficients for cg15345741 and cg16768966 retained highly significant p-values () after PCT adjustment. Despite a tendency towards attenuation, the estimated effects before and after adjustment were highly correlated (, ), although p-values were only moderately correlated (, ). Overall, higher Cd was more commonly associated with higher DNAM, and our suggestive and FDR-significant hits were spread throughout the genome (Figure 1).
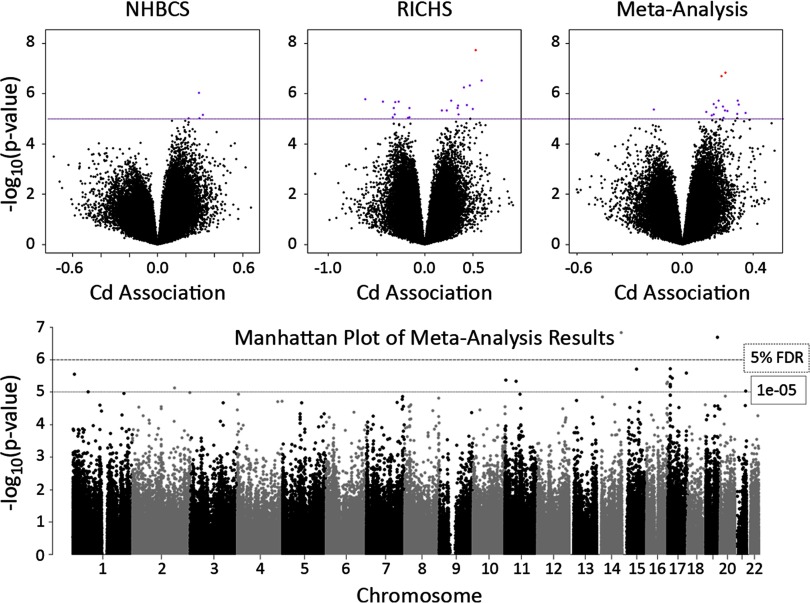
Figure 1.
Volcano plots from each cohort-specific epigenome-wide association study (EWAS) and the meta-analysis, and Manhattan plot of meta-analysis results. All models adjusted for fetal sex, maternal smoking during pregnancy, mother’s highest achieved education level, and putative placental cell types; dashed line represents the false discovery rate (FDR) 5% threshold; dotted line represents suggestive association ().
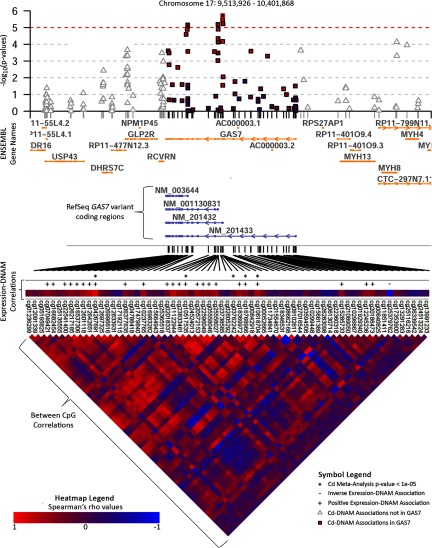
The PCT-adjusted top hit from NHBCS was cg20656525 on chromosome 14, downstream of the genes encoding TNF alpha induced protein 2 (TNFAIP2) and exocyst complex component 3 like 4 (EXOC3L4), whereas the top hit from RICHS was cg07708653 on chromosome 1, within the body of the 4-hydroxyphenylpyruvate dioxygenase like (HPDL) gene. The PCT-adjusted meta-analysis yielded 17 CpGs with Cd-association (Table 3), two of which were significant at a 5% FDR (cg15345741 and cg11707084). Both of the FDR-significant CpGs also had significant linear associations with log-Cd (cg15345741 , ; cg11707084 , ) in linear mixed models. For these two CpGs, we regressed DNAM over dichotomized Cd, continuous Cd, and log-transformed Cd, using the pooled data adjusted for the same covariates and an indicator for study to visualize the approximate percent change in DNAM associated with Cd (Figure 2). The meta-analyses again revealed a clear trend towards higher DNAM levels associated with higher Cd concentrations; 94% of Cd-associated loci with had positive beta coefficients. Of note, 41% of CpGs with suggestive p-values () were on chromosome 17, five of which were within the growth arrest specific 7 (GAS7) gene. Given the high proportion of GAS7 CpGs among the top hits from the meta-analysis, we explored the comethylation structure within GAS7 and all Cd-DNAM associations within and around this gene. The strongest Cd-associated DNAM sites tended to congregate around the first exons of three of the four different GAS7 variants (GAS7-a, -b, and -d) and exhibited a strong comethylation structure as shown by strong positive Spearman correlation p-values (Figure 3).
Table 3.
Results from cohort-specific and meta-analyses of cadmium-associated DNA methylation, for probes with meta-analysis ; adjusted for maternal smoking during pregnancy, maternal education, fetal sex, and estimated tissue heterogeneity.
| CpG annotations | Meta-analysis | NHBCS () | RICHS () | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CpG ID | Ch. | Genomic position | Gene | Regulatory feature | p-Value | FDR | p-Value | p-Value | |||
| cg16034168a | 1 | 6336711 | ACOT7 | Body | 0.32 | 0.171 | 0.32 | 0.32 | |||
| cg24000528 | 2 | 177013630 | MIR10B | TSS1500 | 0.16 | 0.182 | 0.18 | 0.16 | |||
| cg24696183 | 11 | 2891360 | KCNQ1DN | Body | 0.171 | ||||||
| cg23193177 | 11 | 45717613 | - | - | 0.24 | 0.171 | 0.22 | 0.27 | |||
| cg15345741a | 14 | 103674333 | - | - | 0.25 | 0.04 | 0.25 | 0.24 | |||
| cg06100161 | 15 | 60987894 | RORA | Body | 0.20 | 0.171 | 0.12 | 0.28 | |||
| cg03917020 | 16 | 85285139 | - | - | 0.14 | 0.174 | 0.13 | 0.14 | |||
| cg02600679 | 16 | 88832700 | PIEZO1 | Body | 0.25 | 0.171 | 0.17 | 0.34 | |||
| cg04267691 | 17 | 9862752 | GAS7 | Body; 5'UTR | 0.18 | 0.178 | 0.12 | 0.24 | |||
| cg10517535 | 17 | 9929698 | GAS7 | Body; TSS200 | 0.31 | 0.177 | 0.16 | 0.59 | |||
| cg03762242a | 17 | 9940004 | GAS7 | Body; 5'UTR | 0.23 | 0.171 | 0.20 | 0.26 | |||
| cg16768966a | 17 | 9940227 | GAS7 | Body; TSS200 | 0.31 | 0.171 | 0.23 | 0.42 | |||
| cg10618704 | 17 | 9940970 | GAS7 | TSS1500; Body | 0.36 | 0.174 | 0.24 | 0.54 | |||
| cg11851174 | 17 | 17712609 | RAI1 | Body | 0.19 | 0.171 | 0.20 | 0.18 | |||
| cg19944656 | 17 | 76876040 | TIMP2 | Body | 0.18 | 0.171 | 0.17 | 0.18 | |||
| cg11707084a | 19 | 48345263 | CRX | 3'UTR | 0.22 | 0.04 | 0.23 | 0.22 | |||
| cg03566291 | 21 | 44831040 | - | - | 0.23 | 0.201 | 0.21 | 0.29 | |||
Note: Ch., chromosome; CpG, cytosine-phosphate-guanine methylation site; FDR, false discovery rate adjusted p-value; NHBCS, New Hampshire Birth Cohort; RICHS, Rhode Island Child Health Study; TSS200, within 200 bp of the transcriptions start site; TSS1500, within 1500 bp of the transcription start site; UTR, untranslated region.
CpG Sites that yielded meta-analysis in models before and after heterogeneity adjustments.
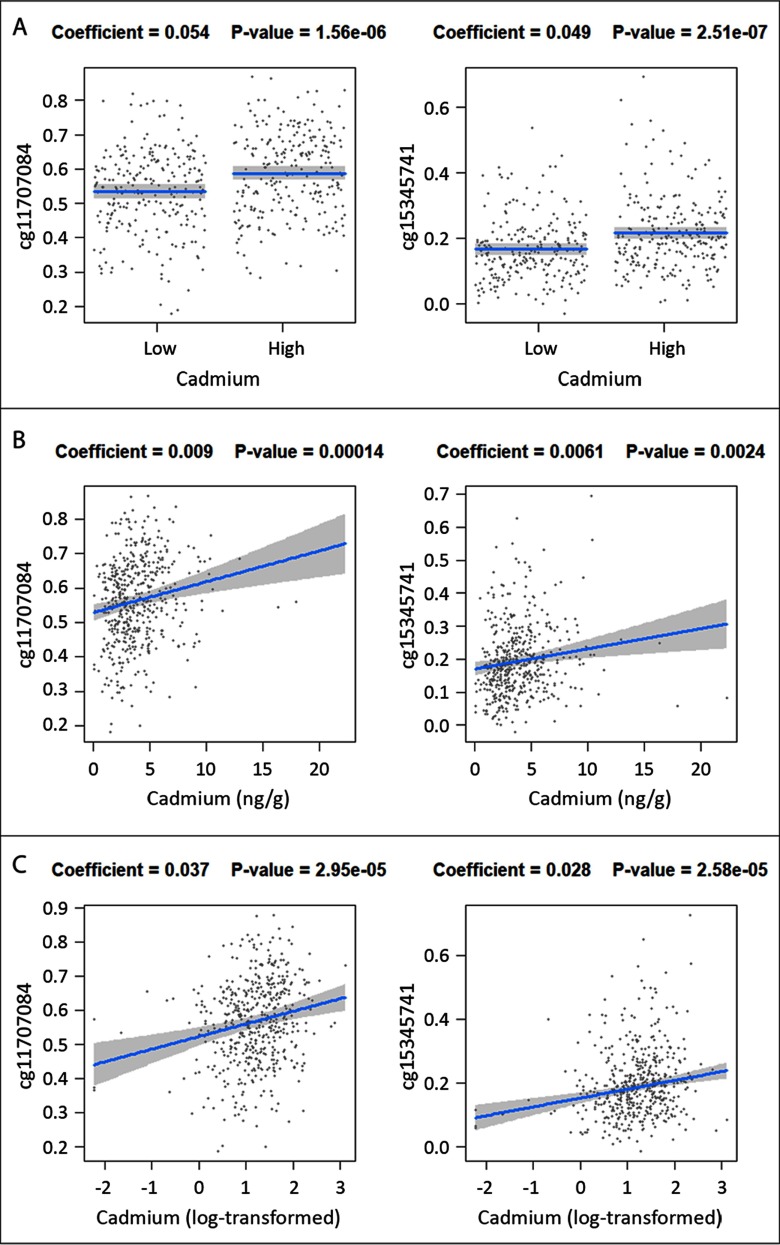
Figure 2.
Differences in average DMA methylation (DNAM) by cadmium (Cd) as a dichotomous, continuous, and log-transformed variable (panels A, B, and C, respectively), at the two false discovery rate (FDR)-significant CpGs (left, cg11701084; right, cg15345741). Robust linear regression models were adjusted for fetal sex, maternal smoking during pregnancy, mother’s highest achieved education level, putative placental cell types, and an indicator for study [New Hampshire Birth Cohort Study (NHBCS) and Rhode Island Child Health Study (RICHS)]. For models of dichotomous Cd, values were classified as low or high according to the median value for each cohort (3.13 and for NHBCS and RICHS, respectively.).
Figure 3.
Manhattan plot of Cd meta-analysis results surrounding the GAS7 gene. Annotated with Ensembl gene names and RefSeq IDs for GAS7 variants, Spearman correlations between GAS7 CpG sites, and CpG-expression correlations. Cadmium (Cd) associations at CpGs outside of the GAS7 coding region () have triangles, whereas CpGs within GAS7 have squares; hits with meta-analysis are indicated with an asterisk (*). Directions of association between DNA methylation (DNAM) and GAS7 expression are indicated with and signs for those relationships whose Spearman correlation p-values were nominally significant ().
We also investigated the underlying biology that may be affected by Cd-associated variations in the placental epigenome. Because we had too few FDR-significant findings to perform an enrichment analysis on only those hits, we tested for GO term and KEGG pathway enrichment among the top 250 CpGs as-sociated with placental Cd after PCT adjustment. The top GO terms included dimethylarginase activity, eye and heart valve development, and heparan sulfate proteoglycan and polysaccharide biosynthetic processing (see Excel Table S5), although none of these enrichments was significant at an FDR threshold of 5%. In contrast, our gene set was enriched with genes from numerous KEGG pathways (see Excel Table S6), 12 of which yielded p-values within an FDR threshold of 5% (Table 4); these included cell adhesion molecules (CAMs) and tight junctions, multiple cancer pathways, multiple G-protein and nitric oxide signaling pathways, and cytotoxic processes.
Table 4.
Significantly enriched KEGG pathways (FDR 5%) associated with for genes annotated to the top 250 CpG sites from the PCT-adjusted Meta-analysis.
| KEGG pathway description | n | DE | FDR |
|---|---|---|---|
| RAS signaling pathway | 217 | 6 | 0.0058 |
| Tight junction | 163 | 5 | 0.0058 |
| Platelet activation | 122 | 4 | 0.019 |
| Cell adhesion molecules (CAMs) | 133 | 4 | 0.019 |
| Taste transduction | 79 | 3 | 0.019 |
| Apelin signaling pathway | 136 | 4 | 0.019 |
| Pancreatic cancer | 61 | 3 | 0.026 |
| Oxytocin signaling pathway | 150 | 4 | 0.026 |
| cGMP-PKG signaling pathway | 157 | 4 | 0.026 |
| Viral carcinogenesis | 191 | 4 | 0.030 |
| Metabolic pathways | 1189 | 8 | 0.034 |
| Natural killer cell mediated cytotoxicity | 107 | 3 | 0.042 |
Note: N represents the total genes in the KEGG path, DE represents the number of genes within our top 250 sites. CpG, cytosine-phosphate-guanine methylation site; DE, number of genes within our top 250 sites; FDR, false discovery rate; KEGG, Kyoto Encyclopedia of Genes and Genomes; n, total genes in KEGG path.
To explore whether gene expression levels might be associated with variations in DNAM levels at Cd-associated CpGs with meta-analysis , we performed an expression quantitative trait methylation (eQTM) (Teschendorff and Relton 2017) analysis. We regressed the expression levels of any gene within of a candidate CpG site on the DNAM levels of that CpG. Three of these CpGs (cg24000528, cg03917020, and cg23193177) were not within of any genes that yielded detectable RNA from our placental samples. Thus, 14 CpGs and 32 genes were included in this analysis; only GAS7 was cis to multiple CpGs within our set of candidates. We produced 36 linear models ranging between 1 and 7 genes tested per CpG (Table 5). Of the 36 eQTM models, 10 yielded FDR-significant linear associations between DNAM and expression of six unique genes (see Figure S5). Of particular note were the associations between our top hit from the meta-analysis and the expression levels of EXOC3L4 (, ) and TNFAIP2 (, ). Additionally, all five of the Cd-associated GAS7 CpGs (within or nearby the first exons of variants a, b, and d) were associated with increased GAS7 expression (Table 5). In contrast, DNAM surrounding the first exon of GAS7 variant c tended to be inversely correlated with mRNA levels (Figure 3).
Table 5.
Expression quantitative trait methylation (eQTM) results for CpGs with PCT-adjusted meta-analysis .
| CpG site | Gene | p-Value | FDR | |
|---|---|---|---|---|
| cg15345741 | EXOC3L4 | 2.28 | 0.019 | |
| cg15345741 | TNFAIP2 | 2.56 | 0.019 | |
| cg11707084 | EHD2 | 0.11 | ||
| cg11707084 | GLTSCR2 | 0.68 | 0.66 | |
| cg11707084 | SEPW1 | 0.89 | 0.66 | |
| cg16768966 | GAS7 | 4.18 | ||
| cg06100161 | RORA | 4.18 | ||
| cg19944656 | CYTH1 | 0.77 | 0.66 | |
| cg19944656 | USP36 | 0.99 | 0.64 | |
| cg19944656 | TIMP2 | 0.97 | ||
| cg19944656 | LGALS3BP | 0.99 | ||
| cg16034168 | RPL22 | 0.97 | ||
| cg16034168 | RNF207 | 0.94 | 0.37 | |
| cg16034168 | ICMT | 0.66 | ||
| cg16034168 | GPR153 | 0.97 | ||
| cg16034168 | ACOT7 | 2.20 | 0.0096 | |
| cg03762242 | GAS7 | 4.24 | ||
| cg11851174 | RAI1 | 0.14 | 0.97 | |
| cg11851174 | SREBF1 | 4.19 | 0.0099 | |
| cg11851174 | TOM1L2 | 2.35 | 0.25 | |
| cg24696183 | KCNQ1 | 0.25 | 0.97 | |
| cg24696183 | CDKN1C | 0.20 | ||
| cg24696183 | SLC22A18 | 0.28 | ||
| cg24696183 | PHLDA2 | 0.065 | ||
| cg24696183 | NAP1L4 | 0.97 | ||
| cg02600679 | RNF166 | 0.17 | 0.97 | |
| cg02600679 | CTU2 | 0.42 | 0.78 | |
| cg02600679 | PIEZO1 | 1.77 | 0.068 | |
| cg02600679 | CDT1 | 0.66 | ||
| cg02600679 | APRT | 0.97 | ||
| cg02600679 | GALNS | 1.17 | 0.38 | |
| cg02600679 | TRAPPC2L | 0.15 | 0.97 | |
| cg10618704 | GAS7 | 3.84 | ||
| cg10517535 | GAS7 | 3.16 | ||
| cg04267691 | GAS7 | 9.17 | ||
| cg03566291 | SIK1 | 0.17 | 0.97 |
Note: Analysis conducted with Rhode Island Child Health Study (RICHS) placental samples for which RNAseq and Illumina Infinium 450K were performed (). CpG, cytosine-phosphate-guanine methylation site; FDR, false discovery rate; PCT, putative constituent cell type.
We then investigated whether the expression levels of the FDR-significant genes from the eQTM analysis were associated with fetal growth metrics standardized by sex and gestational age (Fenton and Kim 2013) (Table 6). We found that higher placental expression levels of TNFAIP2 and acyl-CoA thioesterase 7 (ACOT7) were modestly associated with decreasing birth weight z-scores, and higher expression of retinoic acid receptor-related orphan receptor alpha (RORA) was modestly associated with increased birth weight z-scores (). Higher expression of ACOT7 was also associated with decreasing birth length and head circumference z-scores (). None of the other gene expression levels was associated with birth metrics.
Table 6.
Associations between expression levels with growth metrics at birth in RICHS ().
| Gene | BW | BL | HC |
|---|---|---|---|
| Tau (p-Value) | Tau (p-Value) | Tau (p-Value) | |
| TNFAIP2 | (0.039) | (0.24) | (0.061) |
| EXOC3L4 | (0.62) | 0.005 (0.91) | (0.54) |
| GAS7 | 0.012 (0.80) | 0.032 (0.51) | 0.032 (0.51) |
| RORA | 0.106 (0.026) | 0.068 (0.16) | 0.077 (0.12) |
| ACOT7 | (0.0048) | (0.029) | (0.0032) |
| SREBF1 | 0.032 (0.50) | 0.003 (0.95) | (0.30) |
Note: BW, BL, and HC were sex-specific z-scores that have been adjusted for gestational age. Tau represents the Kendall correlation coefficient. BL, birth length; BW, birth weight; HC, head circumference; RICHS, Rhode Island Child Health Study.
Finally, we explored whether top hits from a previous small-sample study () of placental Cd and DNAM levels (Mohanty et al. 2015) demonstrated associations in our study. In our meta-analysis, one of the top six loci from that study, cg04528060 on chromosome 4 within the ADP-ribosylation factor-like 9 (ARL9) gene, demonstrated a nominally significant association with Cd (, ), with the same direction of effect. However, we did not observe any evidence of sex-specific effects at this or the other 5 CpG sites. We also explored whether any CpGs from our meta-analysis with yielded statistically significant interactions between Cd and fetal sex (see Excel Table S5). Of the 17 models, only cg02600679 within the gene body of piezo type mechanosensitive ion channel component 1 (PIEZO1) produced a significant interaction (). Stratifying by fetal sex revealed that Cd was associated with increased DNAM at cg02600679 among males (, ), whereas there was no association among females ().
Sensitivity Analyses
We performed sensitivity analyses to a) evaluate the linear relationships between -Cd and DNAM, and b) evaluate the potential confounding effects of maternal smoking during pregnancy. To evaluate the linear relationships, we tested the associations between DNAM and log-Cd at the 17 CpG sites from Table 3 while adjusting for sex, maternal education, maternal smoking, and PCTs within each cohort; then, we meta-analyzed the results (see Table S1). All 17 models produced nominally significant () linear associations with log-Cd with the same direction of effect, although p-values for these associations (ranging between 0.014 and 0.000019) did not produce suggestive significance (). To evaluate whether our observed associations between dichotomized Cd and DNAM were driven by the effects of maternal smoking during pregnancy, we reproduced the fully adjusted study-specific models and meta-analyses after excluding all mothers who reported smoking during pregnancy. The associations we observed between Cd and DNAM did vary somewhat after excluding smokers, but there was no strong attenuation of towards the null (see Figures S6 and S7). Among our top hits, the Cd-DNAM associations were only modestly attenuated (see Table S2): coefficients changed by for only 3 CpGs and by for an additional 5 CpGs; however, only 3 CpGs retained meta-analysis . In addition, maternal smoking was not significantly associated with estimated placental heterogeneity in either cohort (see Table S3; minimum T-test ) or with the expression of our top genes (see Table S4; minimum T-test ). Our top two hits from the original analysis retained strong associations with Cd when utilizing log-Cd as the independent variable (cg15345741 , ; cg11707084 , ) and when excluding mothers who reported smoking during pregnancy (cg15345741 , ; cg11707084 , ).
Discussion
We conducted a large EWAS study across two cohorts to examine associations between Cd concentrations and DNAM from human placentae while considering the potential confounding effects of placental tissue heterogeneity and maternal smoking. We further assessed eQTM and associations with birth metrics. We used identical measurement techniques for both DNAM and trace metal measurement in the two cohorts, and the placental concentrations of Cd were similar to the previously reported in the full NHBCS cohort (Punshon et al. 2016), and towards the lower end but consistent with other cohorts that have reported concentrations ranging from as low as to as high as (Esteban-Vasallo et al. 2012). We identified Cd-associated variations at two CpGs within a 5% FDR threshold and an additional 15 CpGs with meta-analysis . At all but one of these loci, higher Cd was associated with increased DNAM levels, and each of these sites produced linear associations with log-Cd and in the same direction of effect at a nominal significance level () We also found that the DNAM levels at many of these CpGs were associated with the expression patterns of six genes, and the expression of three genes (TNFAIP2, ACOT7, and RORA) was associated with birth metrics.
The observed preference for Cd-associated hypermethylation, as opposed to hypomethylation, is consistent with the findings of previous studies that observed higher Cd to be associated with a higher proportion of hypermethylation at gene promoters from fetal and maternal blood samples (Sanders et al. 2014) and with increased DNAM at regulatory regions for imprinted genes from cord blood samples (Vidal et al. 2015). In contrast, others have observed Cd-associated hypomethylation in placental DNA; however, this finding was from a relatively small study that focused on sex-specific associations (Mohanty et al. 2015). Cd has been shown to enhance DNA methyltransferase activity and thus to increase the preponderance of hypermethylated loci, although these effects on DNAM may vary by timing and duration of exposure (Takiguchi et al. 2003).
The top hit from our study, cg15345741 is on chromosome 14, approximately and downstream of the TNFAIP2 and EXOC3L4 genes, respectively, which encode tumor necrosis factor alpha-induced protein 2 and the exocyst complex component 3 like 4 protein. Higher DNAM at cg15345741 was associated with higher expression of both TNFAIP2 and EXOC3L4 and thus may be involved in the regulation of these genes or as a possible marker of their expression, although our findings are not consistent with the promoter methylation paradigm. Other investigators have observed that arsenic-associated DNAM variations in the first exon, particularly within CpG islands, have been associated with increased gene expression in cord blood leukocytes (Rojas et al. 2015). Furthermore, environmental toxicant–associated DNAM variations in nonpromoter regions have been suggested to represent transcription factor binding activity (Martin and Fry 2016). Interestingly, the location of our top CpG does overlap with a DNase hypersensitivity region and with multiple putative transcription factor binding sites, which may explain the apparent relationship between DNAM and the expression of TNFAIP2 and EXOC3L4. We also identified the Cd-associated locus cg11707084 on chromosome 19, which is cis to six genes, although the expression levels of most of these genes were not detectable in placental tissue, and those that were did not yield associations with DNAM at our candidate loci. Thus, the potential functional role of DNAM at cg11707084 is unclear.
Both TNFAIP2 and EXOC3L4 also serve other biological roles that may be important to Cd-associated toxicity. For example, the expression of TNFAIP2 can be up-regulated by , retinoic acid, , and other pro-inflammatory and cytotoxic signals (Rusiniak et al. 2000; Sarma et al. 1992). SNPs within EXOC3L4 have been implicated in affecting transferase (GGT) activity (Middelberg et al. 2012), and elevated levels of GGT are recognized markers of oxidative stress (Fentiman 2012), which plays a central role in Cd toxicity. Disrupted epigenetic regulation of these genes and their involvement in placental growth, development, inflammation, and/or oxidative stress responses present compelling potential mechanisms through which Cd may elicit some of its reproductive toxicity.
Our top 250 sites included genes that were significantly enriched for nitric oxide (NO) and G-protein signaling pathways (RAS, apelin, oxytocin, and cGMP-PKG), cancerous processes, cellular adhesion, cellular metabolism, and cytotoxicity. These pathways are consistent with currently recognized mechanisms of Cd-associated toxicity that have primarily been studied in animal or in vitro models: altered signal transduction, impaired cellular adhesion, disruptions to the cell cycle, increased oxidative stress, and cytotoxic/apoptotic signaling (Thompson and Bannigan 2008). We provide evidence that these biologic processes in the human placenta may be perturbed by Cd-associated differential DNAM, despite overall Cd exposures being low and our study samples primarily consisting of healthy pregnancies.
Expression of the GAS7 gene, which encodes the growth arrest specific 7 protein, was strongly associated with DNAM at all five candidate CpGs that were associated with Cd (). Interestingly, the strongest Cd associations were localized near the first exons of transcript variants a, b, and d for GAS7, and thus these variations may play roles in Cd-associated alternative splicing of this gene. However, the read-depth of our RNA-seq data was not sufficient to accurately test for variant-specific expression levels; this should be investigated further in future studies given the potential for variant-specific functions that is described below. The GAS7 gene, which is highly expressed in embryonic cells (Moorthy et al. 2005), mature brain cells, and Purkinje neurons, plays an essential role in neurite outgrowth (You and Lin-Chao 2010) and in inducing neuronal cell death (Hung and Chao 2010); specific functions may be isoform dependent. In the mouse embryo, levels of GAS7 decrease as the embryo grows, except in neuronal-specific epidermal cells, in which expression remains high (Moorthy et al. 2005). GAS7 expression also plays an essential role in other nonneuronal growth functions, such as osteoblast differentiation and bone development (Chao et al. 2013). Furthermore, neural outgrowth and arborization are shared functions between the exocyst and GAS7 (Martin-Urdiroz et al. 2016; You and Lin-Chao 2010), whereas TNFAIP2 expression has been associated with some neurodegenerative conditions (Brohawn et al. 2016; Colangelo et al. 2002). Relatively few studies have investigated relationships between perinatal Cd exposure and neurodevelopment, although some studies suggest an association with impaired cognition (Kippler et al. 2012a; Sanders et al. 2015), which raises some questions about the possible roles of the above-mentioned genes in Cd-associated cognitive and neurobehavioral deficits. However, the functional roles of these genes in human placental tissue are not well understood. Thus, it is unclear how, or whether, Cd-associated differential placental DNAM and expression may affect early-life cognitive function; this should be the focus of additional research.
Genes known to contribute to neurite outgrowth are sometimes also involved in regulating placental angiogenesis, as has been shown for vascular endothelial growth factor (VEGF) and neurotrophins (Sahay et al. 2015). Interestingly, sterol regulatory element binding proteins (SREBPs) such as SREBF1, whose expression was strongly correlated with one of our top hits, play key roles in VEGF-induced angiogenesis (Zhou et al. 2004). Additionally, inappropriate placental vascularization can result in fetal growth restriction or other pregnancy complications (Sahay et al. 2015). In our study, the expression of GAS7, EXOC3L4, and SREBF1 was not associated with standardized percentiles for birth weight, head circumference, or birth length and thus may not be directly related to fetal growth. In contrast, expression of TNFAIP2, ACOT7, and RORA was associated with fetal growth. ACOT7, which encodes acyl-CoA thioesterase 7, is involved in lipid metabolism in neuronal cells and may prevent neurotoxicity (Ellis et al. 2013); it also promotes pro-inflammatory activities by up-regulating arachidonic acid production (Brocker et al. 2010). RORA, which encodes the retinoic acid receptor-related orphan receptor alpha, is involved in numerous developmental processes including innate immune system development, regulation of inflammatory responses, and neuronal survival and growth, as well as in lipid and glucose metabolism (Cook et al. 2015). One common biological thread that differentiates TNFAIP2, ACOT7, and RORA from GAS7, EXOC3L4, and SREBF1 is their role in mediating inflammatory and immune responses. Thus, although we observed numerous Cd-associated variations in DNAM among genes involved in neurodevelopment and cellular growth processes, variations involved in inflammatory and immune signaling disruption may affect fetal growth. Additionally, these genes are involved in glucose and insulin signaling, which is consistent with the endocrine-disruptive effects of cadmium, and may have implications for metabolic health. GAS7 influences adiposity, fat metabolism, and insulin signaling (Yang et al. 2009). SREBF1 mediates insulin action, and polymorphisms and DNA methylation variations within SREBF1 increase predisposition to type II diabetes (Grarup et al. 2008; Muftah et al. 2016). ACOT7 is a key regulator of glucose response and is more highly expressed in pancreatic of those with type II diabetes (Martinez-Sanchez et al. 2016). RORA is involved in lipid metabolism and insulin sensitivity, and it is associated with obesity and type II diabetes (Jetten et al. 2013).
Other epidemiologic studies of maternal Cd and birth/pregnancy outcomes have found that higher Cd may affect essential metals transport (Kippler et al. 2010); reduce expression of a protocadherin involved in neuronal development (Everson et al. 2016); and affect the DNAM of genes involved in cell death, lipid metabolism, and cancer pathways (Sanders et al. 2014). Similarly, electronic waste (e-waste) exposure, which is associated with higher placental concentrations of toxic metals including Cd, may influence differential expression of proteins involved in immune and metabolic processes (Xu et al. 2016). The top hits from a small-sample EWAS of placental Cd and DNAM indicated differential methylation of genes involved in cellular metabolism, growth, and cell damage response (Mohanty et al. 2015). We were able to reproduce Cd-associated DNAM variations for one top hit from that study, cg04528060 in the 5′ untranslated region (UTR) of the ARL9 gene (meta-analysis ), with the same direction of effect. Notably, the ARL9 gene encodes a GTP-binding protein that is similar to members of the RAS superfamily (Louro et al. 2004), and RAS signaling was the most highly enriched KEGG pathway from our study.
Several studies have investigated relationships between maternal Cd biomarkers and DNAM in cord blood. One study found that maternal blood Cd was associated with higher cord blood DNAM at the paternally expressed gene 3 (PEG3) differentially methylated region (DMR) and observed indiciations of sex-specific effects and possible antagonism of effects by Zn (Vidal et al. 2015). In our study, the four CpGs within the PEG3 DMR (Chr. 19: 57,351,945–57,352,096; UCSC Genome Browser, GRCH37/hg19) (Nye et al. 2013) produced . A genome-wide study found that genes involved in regulating expression, cell cycle, cell death, and nervous system development were differentially methylated by Cd exposure (Sanders et al. 2014); many of the same processes were identified in our enrichment analysis. Another genome-wide study observed higher DNAM at nine of their top loci associated with maternal Cd, consistent with our observation of hypermethylation among top hits (Kippler et al. 2013). The CpG with the second-strongest effect in that study, cg16667631, showed a strong effect in the same direction of effect in our meta-analysis.
The preponderance of similar biological processes associated with Cd across multiple studies with different technologies, populations, and tissues, is striking and suggests that common mechanisms may be affected by Cd exposure during pregnancy. The findings from our study should be interpreted within the context of this study’s limitations. This was an observational study in which placental Cd concentrations and DNAM levels were both measured in placenta at term. Thus, temporality between the Cd concentrations and variations in DNAM could not be established, and the observed associations at term may not be representative of Cd and DNAM associations throughout development. Although we did estimate and adjust for cellular heterogeneity, we used a reference-free approach that was data-driven rather than biology-driven. Future studies of placental DNAM would benefit greatly from the production of an appropriate cell-type-specific methylome for placental tissues. Although we adjusted for likely confounders in our study, we also cannot rule out the possibility that unmeasured or residual confounding may have affected our observations; one of the most likely confounders of these associations was maternal smoking, which is a common source of high Cd exposure. However, we adjusted for maternal smoking in our primary analysis and further evaluated potential bias with a sensitivity analysis in which we excluded mothers who reported smoking during pregnancy. These analyses indicated that the observed associations between Cd and DNAM were largely independent of maternal smoking during pregnancy. We utilized a 5% FDR cutoff to identify our top statistically significant loci; however, given the small number of significant hits, we performed follow-up DNAM expression analyses on all loci with raw . Thus, there are likely some false positive findings among these hits. Power is a common problem in epigenetic epidemiology given the high dimensionality of the data and the small effect sizes (Bakulski and Fallin 2014); therefore, we recommend further replication in other independent study samples. Further, both samples utilized in this study were from the New England region of the United States and consisted predominantly of healthy white mothers. Thus, the generalizability of our findings to populations in different regions of the world and to those with different racial and ethnic backgrounds may be limited. However, to our knowledge, this was the largest study yet to examine the relationships between placental Cd and DNAM and the first to account for and evaluate associations with estimated cellular heterogeneity.
The loci that we did identify are involved in biological processes known to be affected by Cd toxicity and thus are strong candidates for future studies. We observed these associations in two healthy populations with relatively low exposure levels. In the United States, exposure to Cd has declined since the late 1980s, which is primarily attributed to declining smoking rates (Tellez-Plaza et al. 2012). However, Cd is still detectable in numerous commonly consumed foods (Awata et al. 2016), and exposure prevalence remains quite high (Satarug et al. 2017). Additionally, e-waste recycling and disposal are emerging sources of environmental Cd (and other toxic metals) contamination, particularly in developing countries (Sthiannopkao and Wong 2013). These sources of exposure raise some concerns about the potential threshold for Cd-associated reproductive toxicity and whether current dietary intake guidelines and efforts to reduce tobacco smoke exposure during pregnancy adequately protect against adverse birth and pregnancy outcomes. We encourage further molecular epidemiologic studies of the associations between Cd and DNAM in placental tissue, particularly in populations with higher exposure levels and different ethnic backgrounds; we also encourage studies of the associations between dietary intake– and smoking-related Cd exposure with the placental accumulation of Cd. Understanding these relationships can help us determine whether changes to current guidelines and recommendations could reduce Cd-associated adverse reproductive outcomes. Finally, the genes whose expression levels were associated with DNAM at our top hits are multifunctional and are involved in neurodevelopmental and/or neurodegenerative processes, cellular growth processes, and inflammatory/immune signaling. We also found that the expression levels of three of these genes were associated with infant birth weight. Thus, additional future studies should investigate whether differential placental epigenetic regulation and expression of GAS7, EXOC3L4, SREBF1, RORA, ACOT7, and TNFAIP2 are associated with cognitive/neurobehavioral, growth, and immune outcomes in children.
Conclusion
Cadmium is a toxic environmental pollutant that can impair fetal development, although mechanisms underlying this developmental toxicity are unclear. Because cadmium can accumulate in the placenta during pregnancy, we hypothesized that some of this toxicity may be due to disrupted placental functions. We showed that placental cadmium was associated with differential placental DNAM at multiple loci that are involved in cellular growth processes, metabolism, and inflammatory signaling. We also observed corresponding DNAM-expression associations at many of the genes in close genomic proximity to these loci. The expression of the genes involved in inflammatory and immune signaling (RORA, ACOT7 and TNFAIP2) was also associated with birth size. The present study provides supporting evidence that disrupted placental epigenetic regulation of cellular growth and immune/inflammatory signaling could play a role in Cd-associated reproductive toxicity in human pregnancies.
Supplemental Material
Supplemental Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH-NIMH R01MH094609, NIH-NIEHS R01ES022223, NIH-NIGMS P20 GM104416, and NIH-NIEHS P01 ES022832) and by the U.S. Environmental Protection Agency (EPA) (U.S. EPA grant RD83544201). Its contents are solely the responsibility of the grantee and do not necessarily represent the official views of the U.S. EPA. Further, the U.S. EPA does not endorse the purchase of any commercial products or services mentioned in the presentation.
References
- Al-Saleh I, Al-Rouqi R, Obsum CA, Shinwari N, Mashhour A, Billedo G. 2015. Interaction between cadmium (Cd), selenium (Se) and oxidative stress biomarkers in healthy mothers and its impact on birth anthropometric measures. Int J Hyg Environ Health 218(1):66–90, PMID: 25239374, 10.1016/j.ijheh.2014.08.001. [DOI] [PubMed] [Google Scholar]
- Al-Saleh I, Shinwari N, Mashhour A, Rabah A. 2014. Birth outcome measures and maternal exposure to heavy metals (lead, cadmium and mercury) in Saudi Arabian population. Int J Hyg Environ Health 217(2–3):205–218, PMID: 23735463, 10.1016/j.ijheh.2013.04.009. [DOI] [PubMed] [Google Scholar]
- ATSDR. 2012. Toxicological profile for cadmium. https://www.atsdr.cdc.gov/toxprofiles/tp5.pdf [accessed 17 February 2016].
- Aulchenko YS, De Koning DJ, Haley C. 2007. Genomewide rapid association using mixed model and regression: A fast and simple method for genomewide pedigree-based quantitative trait loci association analysis. Genetics 177(1):577–585, PMID: 17660554, 10.1534/genetics.107.075614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awata H, Linder S, Mitchell LE, Delclos GL. 2016. Association of dietary intake and biomarker levels of arsenic, cadmium, lead, and mercury among Asian populations in the U.S.: NHANES 2011–2012. Environ Health Perspect 125(3):314–323, PMID: 27586241, 10.1289/EHP28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakulski KM, Fallin MD. 2014. Epigenetic epidemiology: promises for public health research. Environ Mol Mutagen 55(3):171–183, PMID: 24449392, 10.1002/em.21850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Gormley NA, Baybayan PA, Balasubramanian S, Swerdlow HP, Benoit VA, et al. 2008. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456(7218):53–59, PMID: 18987734, 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocker C, Carpenter C, Nebert DW, Vasiliou V. 2010. Evolutionary divergence and functions of the human acyl-CoA thioesterase gene (ACOT) family. Hum Genomics 4(6):411–420, PMID: 20846931, 10.1186/1479-7364-4-6-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn DG, O’Brien LC, Bennett JP Jr. 2016. RNAseq analyses identify tumor necrosis factor-mediated inflammation as a major abnormality in ALS spinal cord. PLoS One 11(8):1–25, PMID: 27487029, 10.1371/journal.pone.0160520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao CCK, Hung FC, Chao JJ. 2013. Gas7 is required for mesenchymal stem cell-derived bone development. Stem. Stem Cells Int 2013:137010, PMID: 23840221, 10.1155/2013/137010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YA, Lemire M, Choufani S, Butcher DT, Grafodatskaya D, Zanke BW, et al. 2013. Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 8(2):203–209, PMID: 23314698, 10.4161/epi.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, Lukiw WJ. 2002. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res 70(3):462–473, PMID: 12391607, 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- Cook DN, Kang HS, Jetten AM. 2015. Retinoic acid-related orphan receptors (RORs): Regulatory functions in immunity, development, circadian rhythm, and metabolism. Nucl Recept Res 2:101185, PMID: 26878025, 10.1038/nbt.3121.ChIP-nexus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du P, Zhang X, Huang C-C, Jafari N, Kibbe WA, Hou L, et al. 2010. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics 11:587, PMID: 21118553, 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis JM, Wong GW, Wolfgang MJ. 2013. Acyl coenzyme A thioesterase 7 regulates neuronal fatty acid metabolism to prevent neurotoxicity. Mol Cell Biol 33(9):1869–1882, PMID: 23459938, 10.1128/MCB.01548-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erboga M, Kanter M. 2016. Effect of cadmium on trophoblast cell proliferation and apoptosis in different gestation periods of rat placenta. Biol Trace Elem Res 169(2):285–293, PMID: 26170172, 10.1007/s12011-015-0439-8. [DOI] [PubMed] [Google Scholar]
- Esteban-Vasallo MD, Aragonés N, Pollan M, López-Abente G, Perez-Gomez B. 2012. Mercury, cadmium, and lead levels in human placenta: a systematic review. Environ Health Perspect 120(10):1369–1377, PMID: 22591711, 10.1289/ehp.1204952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everson TM, Armstrong DA, Jackson BP, Green BB, Karagas MR, Marsit CJ. 2016. Maternal cadmium, placental PCDHAC1, and fetal development. Reprod Toxicol 65:263–271, PMID: 27544570, 10.1016/j.reprotox.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fentiman IS. 2012. Gamma-glutamyl transferase: risk and prognosis of cancer. Br J Cancer 106(9):1467–1468, PMID: 22531718, 10.1038/bjc.2012.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton TR, Kim JH. 2013. A systematic review and meta-analysis to revise the Fenton growth chart for preterm infants. BMC Pediatr 13:59, PMID: 23601190, 10.1186/1471-2431-13-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin J-P, Labbe A, Lemire M, Zanke BW, Hudson TJ, Fertig EJ, et al. 2014. Functional normalization of 450k methylation array data improves replication in large cancer studies. Genome Biol 15(12):503, PMID: 25599564, 10.1186/s13059-014-0503-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grarup N, Stender-Petersen KL, Andersson EA, Jørgensen T, Borch-Johnsen K, Sandbaek A, et al. 2008. Association of variants in the sterol regulatory element-binding factor 1 (SREBF1) Gene with Type 2 diabetes, glycemia, and insulin resistance: a study of 15,734 Danish subjects. Diabetes 57(4):1136–1142, PMID: 18192539, 10.2337/db07-1534.N.G. [DOI] [PubMed] [Google Scholar]
- Gundacker C, Hengstschläger M. 2012. The role of the placenta in fetal exposure to heavy metals. Wien Med Wochenschr 162(9–10):201–206, PMID: 22717874, 10.1007/s10354-012-0074-3. [DOI] [PubMed] [Google Scholar]
- Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, et al. 2012. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86, PMID: 22568884, 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseman EA, Kile ML, Christiani DC, Ince TA, Kelsey KT, Marsit CJ. 2016. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics 17:259, PMID: 27358049, 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R, Jaritz M, Guenzl P, Vlatkovic I, Sommer A, Tamir IM, et al. 2011. An RNA-Seq strategy to detect the complete coding and non-coding transcriptome including full-length imprinted macro ncRNAs. PLoS One 6(11):e27288, PMID: 22102886, 10.1371/journal.pone.0027288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung FC, Chao CCK. 2010. Knockdown of growth-arrest-specific gene 7b (gas7b) using short-hairpin RNA desensitizes neuroblastoma cells to cisplatin: implications for preventing apoptosis of neurons. J Neurosci Res 88(16):3578–3587, PMID: 20890993, 10.1002/jnr.22504. [DOI] [PubMed] [Google Scholar]
- Järup L, Åkesson A. 2009. Current status of cadmium as an environmental health problem. Toxicol Appl Pharmacol 238(3):201–208, PMID: 19409405, 10.1016/j.taap.2009.04.020. [DOI] [PubMed] [Google Scholar]
- Jetten AM, Kang HS, Takeda Y. 2013. Retinoic acid-related orphan receptors α and γ: key regulators of lipid/glucose metabolism, inflammation, and insulin sensitivity. Front Endocrinol (Lausanne) 4:1, PMID: 23355833, 10.3389/fendo.2013.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiménez-Ortega V, Cano Barquilla P, Fernández-Mateos P, Cardinali DP, Esquifino AI. 2012. Cadmium as an endocrine disruptor: correlation with anterior pituitary redox and circadian clock mechanisms and prevention by melatonin. Free Radic Biol Med 53(12):2287–2297, PMID: 23085516, 10.1016/j.freeradbiomed.2012.10.533. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. 2007. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 8(1):118–127, PMID: 16632515, 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kippler M, Engström K, Mlakar SJ, Bottai M, Ahmed S, Hossain MB, et al. 2013. Sex-specific effects of early life cadmium exposure on DNA methylation and implications for birth weight. Epigenetics 8(5):494–503, PMID: 23644563, 10.4161/epi.24401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Hoque AMW, Raqib R, Öhrvik H, Ekström E, Vahter M. 2010. Accumulation of cadmium in human placenta interacts with the transport of micronutrients to the fetus. Toxicol Lett 192(2):162–168, PMID: 19854248, 10.1016/j.toxlet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Kippler M, Tofail F, Hamadani JD, Gardner RM, Grantham-McGregor SM, Bottai M, et al. 2012a. Early-life cadmium exposure and child development in 5-year-old girls and boys: a cohort study in rural Bangladesh. Environ Health Perspect 120(10):1462–1468, PMID: 22759600, 10.1289/ehp.1104431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippler M, Wagatsuma Y, Rahman A, Nermell B, Persson L, åke Raqib R, et al. 2012b. Environmental exposure to arsenic and cadmium during pregnancy and fetal size: a longitudinal study in rural Bangladesh. Reprod Toxicol 34(4):504–511, PMID: 22985739, 10.1016/j.reprotox.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Laine JE, Ray P, Bodnar W, Cable PH, Boggess K, Offenbacher S, et al. 2015. Placental cadmium levels are associated with increased preeclampsia risk. PLoS One 10(9):1–9, PMID: 26422011, 10.1371/journal.pone.0139341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louro R, Nakaya HI, Paquola ACM, Martins EAL, Da Silva AM, Verjovski-Almeida S, et al. 2004. RASL11A, member of a novel small monomeric GTPase gene family, is down-regulated in prostate tumors. Biochem Biophys Res Commun 316(3):618–627, PMID: 15033445, 10.1016/j.bbrc.2004.02.091. [DOI] [PubMed] [Google Scholar]
- Martin EM, Fry RC. 2016. A cross-study analysis of prenatal exposures to environmental contaminants and the epigenome: support for stress-responsive transcription factor occupancy as a mediator of gene-specific CpG methylation patterning. Environ Epigenet 2(1):dvv011, PMID: 27066266, 10.1093/eep/dvv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin TC, Yet I, Tsai P-C, Bell JT. 2015. coMET: visualisation of regional epigenome-wide association scan results and DNA co-methylation patterns. BMC Bioinformatics 16:131, PMID: 25928765, 10.1186/s12859-015-0568-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Urdiroz M, Deeks MJ, Horton CG, Dawe HR, Jourdain I. 2016. The Exocyst Complex in Health and Disease. Front Cell Dev Biol 4:24, PMID: 27148529, 10.3389/fcell.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Sanchez A, Pullen TJ, Chabosseau P, Zhang Q, Haythorne E, Cane MC, et al. 2016. Disallowance of Acot7 in β-cells is required for normal glucose tolerance and insulin secretion. Diabetes 65(5):1268–1282, PMID: 26861785, 10.2337/db15-1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menai M, Heude B, Slama R, Forhan A, Sahuquillo J, Charles M-A, et al. 2012. Association between maternal blood cadmium during pregnancy and birth weight and the risk of fetal growth restriction: the EDEN mother–child cohort study. Reprod Toxicol 34(4):622–627, PMID: 23017269, 10.1016/j.reprotox.2012.09.002. [DOI] [PubMed] [Google Scholar]
- Middelberg RP, Benyamin B, de Moor MHM, Warrington NM, Gordon S, Henders AK, et al. 2012. Loci affecting gamma-glutamyl transferase in adults and adolescents show age × SNP interaction and cardiometabolic disease associations. Hum Mol Genet 21(2):446–455, PMID: 22010049, 10.1093/hmg/ddr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty AF, Farin FM, Bammler TK, MacDonald JW, Afsharinejad Z, Burbacher TM, et al. 2015. Infant sex-specific placental cadmium and DNA methylation associations. Environ Res 138:74–81, PMID: 25701811, 10.1016/j.envres.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorthy PP, Kumar AA, Devaraj H. 2005. Expression of the Gas7 gene and Oct4 in embryonic stem cells of mice. Stem Cells Dev 14(6):664–670, PMID: 16433621, 10.1089/scd.2005.14.664. [DOI] [PubMed] [Google Scholar]
- Muftah WA, Al-Shafai M, Zaghlool SB, Visconti A, Tsai P-C, Kumar P, et al. 2016. Epigenetic associations of type 2 diabetes and BMI in an Arab population. Clin Epigenetics 8:13, PMID: 26823690, 10.1186/s13148-016-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nye MD, Hoyo C, Huang Z, Vidal AC, Wang F, Overcash F, et al. 2013. Associations between methylation of paternally expressed gene 3 (PEG3), cervical intraepithelial neoplasia and invasive cervical cancer. PLoS One 8(2):e56325, PMID: 23418553, 10.1371/journal.pone.0056325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phipson B, Maksimovic J, Oshlack A. 2015. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics 32(2):286–288, PMID: 26424855, 10.1093/bioinformatics/btv560. [DOI] [PubMed] [Google Scholar]
- Punshon T, Li Z, Marsit CJ, Jackson BP, Baker ER, Karagas MR. 2016. Placental metal concentrations in relation to maternal and infant toenails in a U.S. cohort. Environ Sci Technol 50(3):1587–1594, PMID: 26727403, 10.1021/acs.est.5b05316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D, Schwartz K, Sherlock G, Dudoit S. 2011. GC-Content Normalization for RNA-Seq Data. BMC Bioinformatics 12:480, PMID: 22177264, 10.1186/1471-2105-12-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140, PMID: 19910308, 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas D, Rager JE, Smeester L, Bailey KA, Drobná Z, Rubio-andrade M, et al. 2015. Prenatal Arsenic Exposure and the Epigenome: Identifying Sites of 5-methylcytosine Alterations that Predict Functional Changes in Gene Expression in Newborn Cord Blood and Subsequent Birth Outcomes. Toxicol Sci 143(1):97–106, PMID: 25304211, 10.1093/toxsci/kfu210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiniak ME, Yu M, Ross DT, Tolhurst EC, Slack JL. 2000. Identification of B94 (TNFAIP2) as a potential retinoic acid target gene acute promyelocytic leukemia. Cancer Res 60(7):1824–1829, PMID: 10766166. [PubMed] [Google Scholar]
- Sahay AS, Sundrani DP, Joshi SR. 2015. Regional changes of placental vascularization in preeclampsia: a review. IUBMB Life 67(8):619–625, PMID: 26269153, 10.1002/iub.1407. [DOI] [PubMed] [Google Scholar]
- Sanders AP, Claus Henn B, Wright RO. 2015. Perinatal and childhood exposure to cadmium, manganese, and metal mixtures and effects on cognition and behavior: a review of recent literature. Curr Envir Health Rpt 2(3)284–294, PMID: 26231505, 10.1007/s40572-015-0058-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AP, Smeester L, Rojas D, DeBussycher T, Wu MC, Wright FA, et al. 2014. Cadmium exposure and the epigenome: exposure-associated patterns of DNA methylation in leukocytes from mother–baby pairs. Epigenetics 9(2):212–221, PMID: 24169490, 10.4161/epi.26798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma V, Wolf FW, Marks RM, Shows TB, Dixit VM. 1992. Cloning of a novel tumor necrosis factor-alpha-inducible primary response gene that is differentially expressed in development and capillary tube-like formation in vitro. J Immunol 148(10):3302–3312, PMID: 1374453. [PubMed] [Google Scholar]
- Satarug S, Vesey DA, Gobe GC. 2017. Health risk assessment of dietary cadmium intake: do current guidelines indicate how much is safe? Environ Health Perspect 125(3):284–288, PMID: 28248635, 10.1289/EHP108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sthiannopkao S, Wong MH. 2013. Handling e-waste in developed and developing countries: initiatives, practices, and consequences. Sci. Total Environ 463–464:1147–1153, PMID: 22858354, 10.1016/j.scitotenv.2012.06.088. [DOI] [PubMed] [Google Scholar]
- Takiguchi M, Achanzar WE, Qu W, Li G, Waalkes MP. 2003. Effects of cadmium on DNA-(Cytosine-5) methyltransferase activity and DNA methylation status during cadmium-induced cellular transformation. Exp Cell Res 286(2):355–365, PMID: 12749863. [DOI] [PubMed] [Google Scholar]
- Tellez-Plaza M, Navas-Acien A, Caldwell KL, Menke A, Muntner P, Guallar E. 2012. Reduction in cadmium exposure in the United States population, 1988–2008: the contribution of declining smoking rates. Environ Health Perspect 120(2):204–209, PMID: 22062584, 10.1289/ehp.1104020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, et al. 2013. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450 k DNA methylation data. Bioinformatics 29(2):189–196, PMID: 23175756, 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff AE, Relton CL. 2017. Statistical and integrative system-level analysis of DNA methylation data. Nat Rev Genet, PMID: 29129922, 10.1038/nrg.2017.86. [DOI] [PubMed] [Google Scholar]
- Thompson J, Bannigan J. 2008. Cadmium: toxic effects on the reproductive system and the embryo. Reprod Toxicol 25(3):304–315, PMID: 18367374, 10.1016/j.reprotox.2008.02.001. [DOI] [PubMed] [Google Scholar]
- Valko M, Jomova K, Rhodes CJ, Kuča K, Musílek K. 2016. Redox- and non-redox-metal-induced formation of free radicals and their role in human disease. Arch Toxicol 90(1):1–37, PMID: 26343967, 10.1007/s00204-015-1579-5. [DOI] [PubMed] [Google Scholar]
- Vidal AC, Semenova V, Darrah T, Vengosh A, Huang Z, King K, et al. 2015. Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol Toxicol 16:20, PMID: 26173596, 10.1186/s40360-015-0020-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilahur N, Vahter M, Broberg K. 2015. The Epigenetic Effects of Prenatal Cadmium Exposure. Curr Environ Health Rpt 2(2):195–203, PMID: 25960943, 10.1007/s40572-015-0049-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virani S, Rentschler KM, Nishijo M, Ruangyuttikarn W, Swaddiwudhipong W, Basu N, et al. 2016. DNA methylation is differentially associated with environmental cadmium exposure based on sex and smoking status. Chemosphere 145:284–290, PMID: 26688266, 10.1016/j.chemosphere.2015.10.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang Q, Zhang X, Luo S, Ye D, Guo Y, et al. 2014. Preeclampsia induced by cadmium in rats is related to abnormal local glucocorticoid synthesis in placenta. Reprod Biol Endocrinol 12:77, PMID: 25108313, 10.1186/1477-7827-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Liu L, Hu YF, Hao JH, Chen YH, Su PY, et al. 2016a. Maternal serum cadmium level during pregnancy and its association with small for gestational age infants: a population-based birth cohort study. Sci Rep 6: 22631, PMID: 26934860, 10.1038/srep22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Wang Y, Bo Q-L, Ji Y-L, Liu L, Hu Y-F, et al. 2016b. Maternal cadmium exposure reduces placental zinc transport and induces fetal growth restriction in mice. Reprod Toxicol 63:174–182, PMID: 27319394, 10.1016/j.reprotox.2016.06.010. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang H, Xu ZM, Ji Y-L, Chen Y-H, Zhang Z-H, et al. 2012. Cadmium-induced teratogenicity: Association with ROS-mediated endoplasmic reticulum stress in placenta. Toxicol Appl Pharmacol 259(2):236–247, PMID: 22252055, 10.1016/j.taap.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Li Y, Abecasis GR. 2010. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26(17):2190–2191, PMID: 20616382, 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Ge J, Huo X, Zhang Y, Lau ATY, Xu X. 2016. Differential proteomic expression of human placenta and fetal development following e-waste lead and cadmium exposure in utero. Sci Total Environ 550:1163–1170, PMID: 26895036, 10.1016/j.scitotenv.2015.11.084. [DOI] [PubMed] [Google Scholar]
- Yang X, Deignan JL, Qi H, Zhu J, Qian S, Zhong J, et al. 2009. Validation of candidate causal genes for obesity that affect shared metabolic pathways and networks. Nat Genet 41(4):415–423, PMID: 19270708, 10.1038/ng.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You JJ, Lin-Chao S. 2010. Gas7 functions with N-WASP to regulate the neurite outgrowth of hippocampal neurons. J Biol Chem 285(15):11652–11666, PMID: 20150425, 10.1074/jbc.M109.051094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R-H, Yao M, Lee T-S, Zhu Y, Martins-Green M, Shyy JY-J. 2004. Vascular endothelial growth factor activation of sterol regulatory element binding protein: a potential role in angiogenesis. Circ Res 95(5):471–478, PMID: 15271857, 10.1161/01.RES.0000139956.42923.4A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.