Abstract
Purpose
INHBA, which encodes a member of the TGF-beta superfamily of proteins, has been identified to play a critical role in different types of cancer. However, its clinical significance in esophageal squamous cell carcinoma (ESCC) has never been reported.
Patients and methods
In this study, we collected 239 ESCC paraffin-embedded specimens and measured the expression of INHBA with immunohistochemistry (IHC). The clinical and prognostic significance of INHBA expression was statistically analyzed. What is more, we conducted a meta-analysis to study the prognostic value of INHBA expression in multiple types of solid tumors.
Results
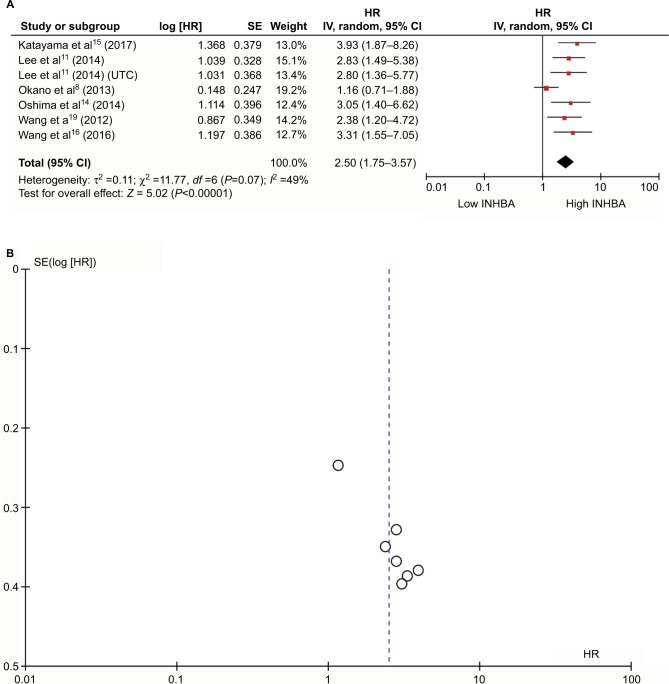
The results showed that INHBA expression was observed predominantly in the cytoplasm of cells in the ESCC specimens. INHBA expression was closely correlated with N categories (P=0.026). Kaplan–Meier analysis showed that ESCC patients in the low INHBA expression subgroup had significantly better prognosis than those with high INHBA level. Subgroup analysis revealed that INHBA distinguished the disease-free survival (DFS) and overall survival (OS) when patients were stratified by TNM stage status and N status. Multivariate analysis results suggested that INHBA expression was an independent factor that affected OS (HR =1.679, P=0.022) and DFS (HR =1.715, P=0.017). In the meta-analysis, six papers with 1321 patients were included and patients with high INHBA level had worse prognosis than patients with low INHBA level (HR 2.50, 95% CI 1.75–3.57, P<0.0001).
Conclusion
High INHBA level predicts poor prognosis in ESCC and other solid tumors. More studies are required to elucidate the role of INHBA and its clinical application in cancer settings.
Keywords: INHBA, ESCC, meta-analysis, prognosis
Introduction
Esophageal cancer (EC) is one of the most lethal types of malignancy, leading to the death of 400.000 people worldwide in 2012.1 It could be primarily divided into squamous cell carcinoma and adenocarcinoma. Esophageal squamous cell carcinoma (ESCC) is the primary histological type of EC in most EC high-incidence areas, including the People’s Republic of China, Middle East, and southern Africa. The molecular mechanism of EC tumorigenesis remains unclear, and the primary risk factors for ESCC are smoking and alcohol consumption.2 Most EC patients are diagnosed at advanced stages because of its silent onset and aggressive nature. Current treatments for EC patients are surgery and chemoradiotherapy. Different from other types of cancer, in which molecular markers have been extensively studied and applied in clinics to assist diagnosis and prognosis prediction, very few advances have been made in ESCC. A strong need for the discovery of new biomarkers in ESCC is emerging.
INHBA is a gene located at 7p14.1, encoding a member of the TGF-beta superfamily of proteins.3 The ligand could be further homo-dimerized to form activin A or hetero-dimerized to form inhibin with inhibin beta B. The involvement of activin A has been observed in multiple biological activities, such as stem cell differentiation,4 glucose metabolism,5 immune response,6 and tumor development.7 Recently, the overexpression of INHBA has been reported in multiple types of cancers, including breast cancer,7 colorectal cancer,8 gastric cancer,9 lung cancer,10 and bladder cancer.11 However, the prognostic effects of INHBA in ESCC remain unclear.
In this study, we collected 239 ESCC patient samples and measured the expression of INHBA with IHC to study the clinical significance of INHBA expression in ESCC. To further explore the prognostic meaning of INHBA expression, we gathered six already published manuscripts and conducted a meta-analysis of INHBA expression in multiple types of solid tumors. The aim of this study is to have a better understanding of the clinical significance of INHBA expression.
Material and methods
Meta-analysis – literature search
A search was performed for eligible literature in PubMed and Web of Science from 2000 to November 2017. All literature had to be published in English. The keywords used in the search included “INHBA”, “Inhibin beta A”, “cancer”, and “prognosis”. The last date of search was November 18, 2017. We retrieved eligible manuscripts for data extraction and analysis.
Meta-analysis – selection criteria
Literature was collected according to the following criteria: 1) published in English; 2) primary cancer which had been previously histopathologically diagnosed; 3) level of INHBA had been detected before surgery; 4) HRs and their 95% CIs were calculated in all studies; and 5) only the most recent data were included if the same group published multiple studies.
Meta-analysis – data extraction and quality assessment
Two independent researchers (Lyu and Jiang) assessed the quality of the selected studies, using Revman5.3, as recommended by Cochrane Collaboration. Different factors were extracted for quality assessments, including clear description of purpose/objectives, clear description of patients’ eligibility criteria (including/excluding), clear description of INHBA detection, predefinition of predictors (disease-free survival [DFS]/cancer-specific survival/overall survival [OS]) and outcome measurements, long enough follow-up period, and limitations considered, modified from Perisanidis et al.12 We ranked the included papers according to the quality items used in each study (score range 0–6, Table S1). Quality assessment was not used as exclusion criterion for the eligible studies. We (Lyu and Xu) extracted data from each study for analysis, including author, country, year of publication, study type and study period, cancer type and stage of cancers, number of patients and male patient ratio, and INHBA HR with its 95% CI. Disagreements were resolved by discussion. Publication bias was assessed by Begg’s funnel plot in Revman5.3 and Egger’s test in R programming language. “Trim and fill” method was used to adjust HRs if publication bias was found.
Tissue and patient sources
The study was approved by the medical ethics committee of Sun Yat-sen University Cancer Center. In total, 239 specimens from ESCC patients who underwent surgery in the hospital were sectioned and confirmed by pathological review of IHC. All the ESCC patients were seen from 2000 to 2007 at the hospital and were clinically and histologically diagnosed. All patients were treated with radical surgery without neoadjuvant/adjuvant treatments. We used the 7th edition of the TNM classification of the International Union Against Cancer (2009) to record the histologic grade and the clinical stage of the tumors. The selection criteria are as follows: 1) histologically confirmed primary ESCC without any previous treatment; 2) no familial malignancies or other synchronous malignancies history; 3) underwent resection plus lymphadenectomy (limited or extended); and 4) clinical information and follow-up data were documented. Clinical data were obtained from the hospital records, and all the patients were contacted in 2012 to determine their vital status. Patients whose tissues and medical records were used for this study had provided written informed consent.
Tissue microarray construction
The tissue microarray was constructed according to methods that were described previously.13 Tissues from all the patients in this study were collected, fixed in ethanol, and embedded in paraffin. A senior pathologist reviewed and defined representative tumor regions through hematoxylin and eosin staining. Two targeted core samples of each specimen were obtained using a tissue array instrument (MiniCore instruments; Alphelys, Plaisir, France). Tissue cylinders with a diameter of 10 mm were punched and arrayed on a recipient paraffin block. Sections of the tissue array were cut and placed onto glass slides.
Immunohistochemical staining and assessment
The tissue slides were rehydrated by a graded alcohol series. Then the slides were incubated in 0.3% hydrogen peroxide for 15 minutes to quench the endogenous peroxidase activity. Tris (hydroxymethyl) aminomethane-EDTA buffer (pH 8.0) was used in antigen retrieval. We used 10% normal goat serum for 10 minutes to block nonspecific binding. The slides were incubated with polyclonal antibody against INHBA (1:1000, ab56057, Abcam, USA) for 12 hours at 4°C in a moist chamber. We used blocking solutions without primary antibody as a control. The slides were then incubated with horseradish peroxidase for 30 minutes and 3,30-diaminobenzidine solution for visualization. Mayer’s hematoxylin was applied as a counterstain. Internal positive and negative controls, including normal squamous mucosa of the esophagus from cancer-free patients, were also used to further support the staining patterns. Two independent pathologists (Lyu and Jiang), who were blinded to the clinicopathologic data, generated the immunoreactivity score (IRS) for INHBA expression. They scored the results based on the following criteria: 1) percentage of positive tumor cells in the tumor tissue: zero (0%), 1 (1%–10%), 2 (11%–50%), 3 (51%–70%), and 4 (71%–100%); and 2) signal intensity: zero (no signal), 1 (weak), 2 (median), and 3 (strong). The IRS was calculated by multiplying the score for the percentage of positive cells by the intensity score (range, 0–12). The specimens were rescored if the difference between the scores by the two pathologists was >3.
Statistical analysis
We performed the meta analysis with Revman5.3 (The Nordic Cochrane Centre, the Cochrane Collaboration, Copenhagen, Denmark). The details have been previously described.14 Statistical heterogeneity between studies was determined by Cochran’s Q test and Higgins I2. Sensitivity analysis was performed by omitting each study or specific studies to find potential outliers in R, using “metainf ” and “forest” function in “meta” package. P<0.1 or I2>50% was considered as heterogeneity. A random effects model was used to combine data if statistically significant heterogeneity was found. We did not do subgroup analysis as the research papers are few. Publication bias was assessed by funnel plot as well as Begg’s and Egger’s test in R software.
Survival analysis was performed with SPSS software (standard version 16.0, SPSS Inc., Chicago, IL, USA). The ROC method was used to define the cutoff value for the INHBA IRS. The ROC curve was generated by the MedCalc statistical software package 11.0.1 (MedCalc Software bvba, Mariakerke, Belgium). Pearson’s χ2 test was used to define the correlation between INHBA expression and clinicopathological features of the patients. A binary logistic regression model was used to analyze the variables that correlated with INHBA expression. DFS survival was defined as the time from surgery to regional relapse or the development of distant metastasis. OS was defined as the time from surgery to death. DFS and OS were assessed with the Kaplan–Meier method and were compared by the log-rank test. A multivariate survival analysis was performed for all of the variables that were significant in the univariate analysis using the Cox regression model. For all analyses, a two-sided P-value <0.05 was considered statistically significant.
Results
Literature search
After the initial literature search, 15 were included for further analysis after reviewing the abstracts. Eight full text articles were retrieved for data extraction. Finally, six studies were included in this meta-analysis. The flow chart in Figure 1 shows the process of identifying studies. A total of 1321 patients were included and the characteristics of the studies were summarized in Table 1.8,9,11,15–17
Figure 1.
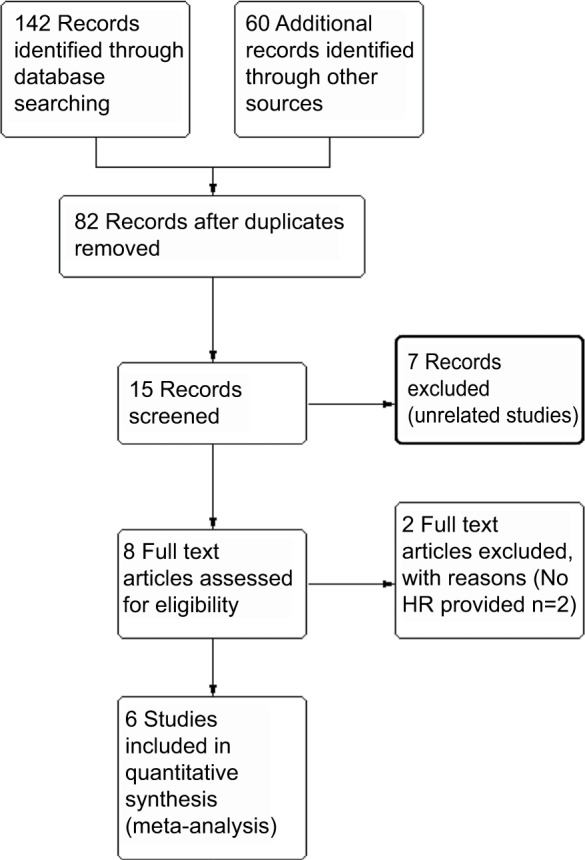
Flow chart of meta-analysis literature search.
Table 1.
The characteristics of the included studies
| Study | Year | Country | Study type (R or P) | Study Period | Cancer site | Stage | N | Sex (male) | U/M | HRs | Outcome 1 | In (HR) | SE |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lee et al11 | 2014 | Taiwan | R | 1996–2004 | Urinary bladder urothelial carcinoma | 1–4 | 340 | 0.465 | U | 2.825 | DSS | 1.039 | 0.328 |
| Lee et al11 | 2014 | Taiwan | R | 1996–2004 | Upper urinary tract carcinoma | 1–4 | 295 | 0.732 | U | 2.803 | DSS | 1.031 | 0.368 |
| Wang et al16 | 2016 | People’s Republic of China | R | 2001–2012 | Breast cancer | 1–3 | 126 | U | 3.311 | OS | 1.197 | 0.386 | |
| Okano et al8 | 2013 | Japan | R | 1990–2002 | Colorectal cancer | 1–4 | 126 | 0.595 | M | 1.160 | OS | 0.148 | 0.247 |
| Oshima et al14 | 2014 | Japan | R | 2002–2007 | Gastric cancer | 1–3 | 168 | 0.673 | U | 3.048 | OS | 1.114 | 0.396 |
| Wang et al9 | 2012 | People’s Republic of China | R | 2004–2006 | Gastric cancer | 132 | 0.621 | U | 2.380 | OS | 0.867 | 0.349 | |
| Katayama et al15 | 2017 | Japan | R | 2002–2010 | Gastric cancer | 2–3 | 134 | 0.687 | U | 3.926 | OS | 1.368 | 0.379 |
Abbreviations: OS, overall survival; DSS, disease-specific survival; R, research; P, prospective cohort study; U, univariate analysis; M, multivariate analysis; HR, hazard ration.
Meta-analysis results
The average quality assessment was fair (4.2/6.0), with a median score of 4 (Table S1). To evaluate the prognostic effect of INHBA in ESCC, we used the multivariate HRs and their 95% CIs in these studies to calculate a combined HR. The estimated proportion of heterogeneity (I2) between six INHBA studies was 49% (P=0.07) for the survival rates. Therefore, significant heterogeneity existed, and a random model was applied. We did the sensitivity analysis (Figure S1A) and we established that Okano et al’s8 study was very sensitive in this analysis. All other studies had an HR >2 and 95% CI >1. However, Okano et al’s study had an HR 1.16 and 95% CI from 0.71–1.88. Apart from this study, all other studies were very consistent.
Patients with a higher INHBA level had lower survival rates than patients with a lower INHBA level. High INHBA was a rick factor for poor prognosis (HR 2.50, 95% CI 1.75–3.57, P<0.0001, Figure 2A). Begg’s funnel test was performed to estimate the existing publication bias of the literature in this meta-analysis. As shown in Figure 2B, the shape of the Funnel plots showed some evidence of asymmetry, which was confirmed by Egger’s regression test (P=0.0010) and Begg’s test, (P=0.0004). Small studies normally show larger effect sizes. Some small studies with insignificant findings are harder to publish, and therefore they were not included in our meta-analysis. After we used the “trim and fill” method to adjust for publication bias under the random effects model, the corrected pooled HR for survival was 2.1409 (95% CI =1.5271–3.0014, P<0.0001, Figure S1B).
Figure 2.
Meta-analysis plots.
Notes: Forest plot of HR for the association of high plasma INHBA level and survival (A). Begg’s funnel plots of publication bias (B).
Abbreviation: UTC, upper tract urothelial carcinoma.
Expression of INHBA in ESCC tissue
The INHBA protein expression in ESCC was examined by IHC in 239 ESCC patients. The average age of the patients was 57.53 years old (range, 32–80). The clinicopathological characteristics of the population were listed in Table 2 and the representative figures of INHBA expression in ESCC and the adjacent normal mucosal tissues were listed in Figure 3. INHBA expression in the adjacent normal tissues was nonexistent (Figure 3A) while weak, median, and strong INHBA expression was observed predominantly in the cytoplasm of cells in the ESCC tissues (Figure 3C-H).
Table 2.
INHBA expression and other important clinicopathological parameters in esophageal squamous cell carcinoma
| Variables | Cases (n=239) | INHBA expression
|
P-valuea | |
|---|---|---|---|---|
| Low (n=66, %) | High (n=173, %) | |||
| Age, yearsb | 0.504 | |||
| ≤58 | 127 | 39 (30.7) | 88 (69.3) | |
| >58 | 112 | 30 (26.8) | 82 (73.2) | |
| Sex | 0.634 | |||
| Female | 64 | 17 (26.6) | 47 (73.4) | |
| Male | 175 | 52 (28.0) | 123 (72.0) | |
| Tumor location | 0.171 | |||
| Upper | 12 | 4 (33.3) | 8 (66.7) | |
| Middle | 163 | 41 (25.2) | 122 (74.8) | |
| Lower | 64 | 24 (37.5) | 40 (62.5) | |
| Histological gradec | 0.1 | |||
| Grade 1 | 58 | 23 (39.7) | 37 (60.3) | |
| Grade 2 | 154 | 38 (24.7) | 116 (75.3) | |
| Grade 3 | 27 | 8 (29.6) | 19 (70.4) | |
| pT statusc | 0.458 | |||
| pT 1 | 6 | 3 (50.0) | 3 (50.0) | |
| pT 2 | 55 | 15 (27.3) | 40 (72.7) | |
| pT 3 | 175 | 51 (29.1) | 124 (70.9) | |
| pT 4 | 3 | 0 (0.00) | 2 (100.0) | |
| N categories | 0.026 | |||
| Negative | 129 | 45 (34.9) | 84 (65.1) | |
| Positive | 110 | 24 (21.8) | 86 (78.2) | |
| TNM stage | 0.172 | |||
| Stage 1 | 9 | 3 (33.3) | 6 (66.7) | |
| Stage 2 | 138 | 46 (33.3) | 92 (66.7) | |
| Stage 3 | 91 | 20 (22.0) | 71 (78.0) | |
Notes:
Probability value of <0.05 indicates statistical significance shown in bold. Probability values were calculated by Pearson’s χ2 test.
Age was divided according to the median age of 58 years.
The grading and histopathology staging of ESCC specimens was based on the World Health Organization classification published in 2009.25
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Figure 3.
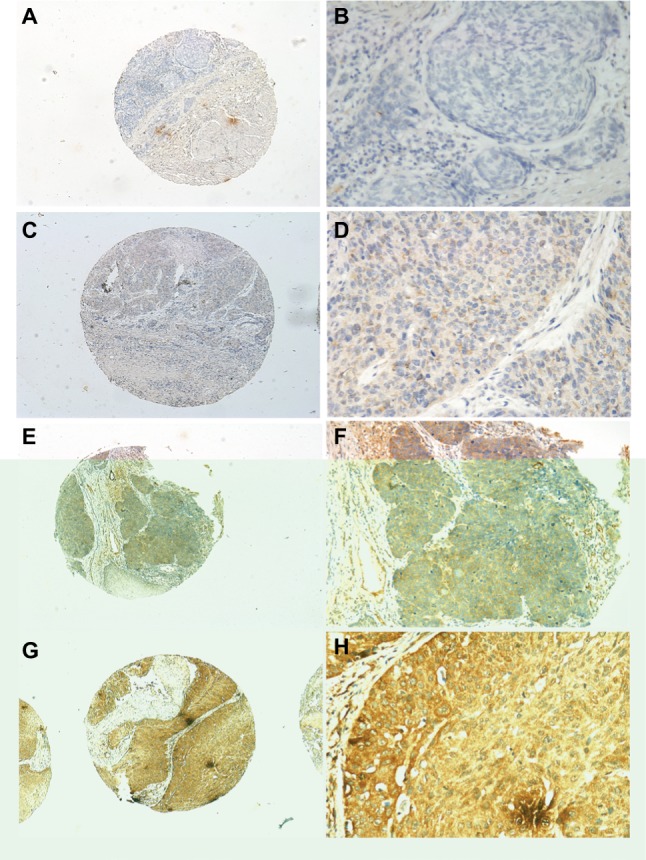
INHBA expression determined by IHC.
Notes: Normal esophageal tissue demonstrated no expression of INHBA protein in the cytoplasm of esophageal squamous cells (magnification: A, ×40; B, ×200). Low expression level of INHBA in ESCC tissues (magnification: C, ×40; D, ×200). Median expression of INHBA in ESCC tissues (magnification: E, ×40; F, ×200). High expression levels of INHBA were detected in ESCC tissues (magnification: G, ×40; H, ×200).
Abbreviations: ESCC, esophageal squamous cell carcinoma; IHC, immunohistochemistry.
Selection of the cutoff value
We used ROC curve to define the cutoff value for the INHBA IRS. The point with both maximum sensitivity and specificity was selected as the cutoff value. In this study, the IRS cutoff value was 8. The sensitivity was 0.800 and specificity was 0.632. The area under the ROC curve was 0.593. Thus, high expression of CEP55 was defined when the IRS was ≥8 (P<0.05); otherwise, the expression was defined as low.
INHBA expression and clinicopathological variables
The relationships between clinicopathological features of patients with ESCC and INHBA expression were summarized in Table 2. The INHBA expression closely correlated with N categories (P=0.026). No statistical correlations were found between INHBA expression and age, sex, tumor location, histological grade, T status, and TNM stage (P>0.05).
INHBA expression and patients’ survival
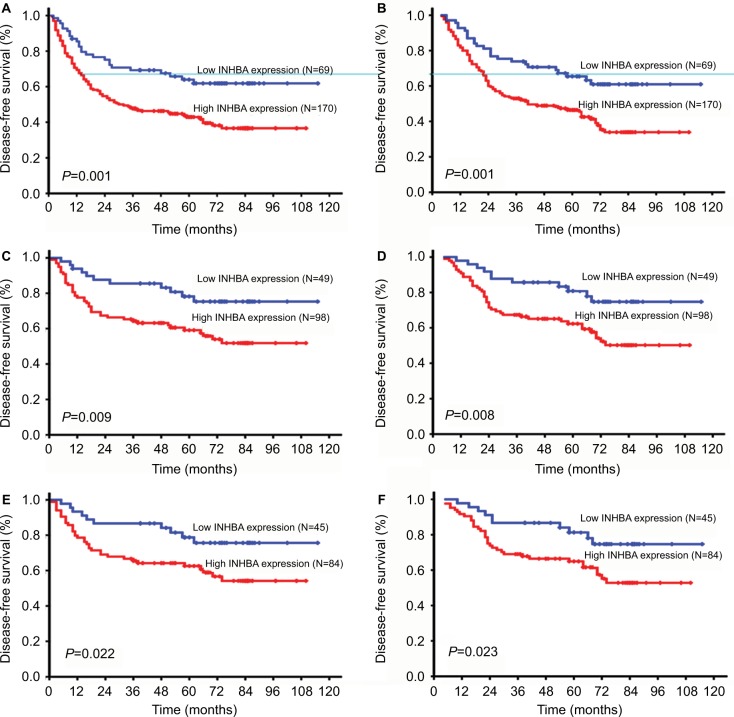
No patients were lost to follow-up among the 239 patients. The median observation period was 40 months (range 4–115 months). A total of 125 patients were deceased at the end of the follow-up period while 114 were alive. The 5-year DFS and OS of the entire cohort were 31% and 36%, with median survival times of 38 months and 40 months for patients with high and low expression.
According to the Kaplan–Meier analysis, INHBA expression in ESCC was closely related to DFS and OS. The median DFS was 58 months for patients with low INHBA expression, whereas it was 31 months for patients with high INHBA expression (P=0.001; Figure 4A). The median OS was 58 months for patients with low INHBA expression, whereas it was 37 months for patients with high INHBA expression (P=0.001; Figure 4B). In the INHBA subgroup analysis, INHBA distinguished DFS and OS when patients were stratified by TNM stage status and N status. For stage 1 and stage 2 patients, the average DFS and OS were 94 and 95 months for patients with low INHBA expression, whereas it was 68 and 71 months for patients with high INHBA expression (P=0.009 and P=0.008, Figure 4C, D). For N0 patients, the average DFS and OS were 94 and 95 months, respectively, for patients with low INHBA expression, whereas they were 71 and 73 months, respectively, for patients with high INHBA expression (P=0.022 and P=0.023, Figure 4E, F).
Figure 4.
Disease-free survival (DFS) and overall survival (OS) curves according to patients’ INHBA expression.
Notes: DFS curve in all patients with different levels of INHBA expression (A, P=0.001). OS curve in all patients with different levels of INHBA expression (B, P=0.001). DFS and OS curves in patients in TNM stage 1 and 2 with low and high INHBA expression (C, D, P=0.009 and P=0.008, respectively). DFS and OS curves in patients in N0 stage with low and high INHBA expression (E, F, P=0.022 and P=0.023, respectively).
A univariate analysis using the Cox proportional hazards model was applied to assess the importance of multiple factors in the survival times of ESCC patients. The result showed that the following parameters correlated significantly with DFS and OS: T category, N category, TNM stage, and INHBA expression (Table 3, P<0.001, P<0.001, P<0.01, and P=0.001, respectively). When the aforementioned parameters were included in a multivariate analysis, the results suggested that T category, N category, and INHBA expression were independent factors that affected OS and PFS (Table 4).
Table 3.
Univariate Cox regression analysis in ESCC patients for disease-free survival and overall survival
| Variables | Disease-free survival
|
Overall survival
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| Age, yearsa | 0.704 | 0.691 | ||
| ≤58 | Reference | Reference | ||
| >58 | 0.935 (0.658–1.328) | 0.932 (0.656–1.324) | ||
| Sex | 0.293 | 0.218 | ||
| Male | Reference | Reference | ||
| Female | 1.241 (0.827–1.863) | 1.288 (0.858–1.933) | ||
| Tumor location | 0.323 | 0.288 | ||
| Upper | 1.586 (0.758–3.317) | 1.551 (0.742–3.243) | ||
| Middle | 0.945 (0.631–1.415) | 0.905 (0.604–1.356) | ||
| Lower | Reference | Reference | ||
| Histological gradeb | 0.131 | 0.181 | ||
| Grade 1 | Reference | Reference | ||
| Grade 2 | 1.028 (0.674–1.568) | 1.016 (0.666–1.550) | ||
| Grade 2 | 1.688 (0.940–3.031) | 1.609 (0.896–2.891) | ||
| pT statusb | <0.001 | <0.001 | ||
| pT 1 | Reference | Reference | ||
| pT 2 | 1.496 (0.354–6.318) | 1.660 (0.393–7.013) | ||
| pT 3 | 1.960 (0.483–7.958) | 2.251 (0.554–9.146) | ||
| pT 4 | 35.323 (5.438–229.431) | 63.931 (9.762–418.677) | ||
| pT1-2 | 0.005 | 0.005 | ||
| Low expression | Reference | Reference | ||
| High expression | 4.709 (1.413–15.690) | 4.745 (1.425–15.804) | ||
| pT3-4 | 0.034 | 0.030 | ||
| Low expression | Reference | Reference | ||
| High expression | 1.657 (1.030–2.666) | 1.673 (1.040–2.692) | ||
| pN statusb | <0.001 | <0.001 | ||
| 0 | Reference | Reference | ||
| 1–3 | 3.257 (2.243–4.730) | 3.410 (2.347–4.954) | ||
| pN0 | 0.022 | 0.023 | ||
| Low expression | Reference | Reference | ||
| High expression | 2.216 (1.097–4.479) | 2.200 (1.089–4.445) | ||
| pN1-3 | 0.230 | 0.134 | ||
| Low expression | Reference | Reference | ||
| High expression | 1.403 (0.798–2.466) | 1.527 (0.867–2.689) | ||
| pStageb | <0.001 | <0.001 | ||
| Stage 1 | Reference | Reference | ||
| Stage 2 | 1.095 (0.342–3.509) | 1.073 (0.335–3.440) | ||
| Stage 3 | 3.852 (1.209–12.275) | 4.070 (1.276–12.986) | ||
| pStage 1/2 | 0.009 | 0.008 | ||
| Low expression | Reference | Reference | ||
| High expression | 2.352 (1.212–4.565) | 2.365 (1.219–4.590) | ||
| pStage 3 | 0.277 | 0.223 | ||
| Low expression | Reference | Reference | ||
| High expression | 1.379 (0.763–2.492) | 1.436 (0.792–2.603) | ||
| CEP55 expression | 0.001 | 0.001 | ||
| Low | Reference | Reference | ||
| High | 2.023 (1.304–3.139) | 2.053 (1.323–3.185) | ||
Notes:
Age is divided according to the median age of the study.
The grading and histopathology stage of ESCC specimens are based on the World Health Organization classification published in 2009.25
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Table 4.
Multivariate Cox regression analysis for disease-free survival and overall survival in patients with esophageal squamous cell carcinoma
| Variables | Disease-free survival
|
Overall survival
|
||
|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | |
| pT statusa | <0.001 | <0.001 | ||
| pT 1 | 1.000 | 1.000 | ||
| pT 2 | 1.395 (0.330–5.903) | 1.665 (0.393–7.057) | ||
| pT 3 | 1.734 (0.426–7.058) | 2.137 (0.524–8.718) | ||
| pT 4 | 32845 (5.021–214.879) | 69.057 (10.246–465.429) | ||
| pN categories | <0.001 | <0.001 | ||
| N0 | 1.000 | 1.000 | ||
| N1–3 | 3.094 (2.123–4.510) | 3.294 (2.258–4.804) | ||
| CEP55 expression | 0.022 | 0.017 | ||
| Low | 1.000 | 1.000 | ||
| High | 1.679 (1.077–2.619) | 1.715 (1.100–2.675) | ||
Note:
The grading and histopathology stage of ESCC specimens are based on the World Health Organization classification published in 2009.25
Abbreviation: ESCC, esophageal squamous cell carcinoma.
Discussion
In this study, we studied the relations of relative expression of the INHBA gene to clinicopathological factors and outcomes in patients with ESCC after surgery. In addition, we conducted a meta-analysis to further elucidate the prognostic role of INHBA in solid tumors.
Previous studies have shown that the expression of INHBA is associated with prognosis of different types of cancer patients. Okano et al8 reported that high expression of INHBA gene was associated with significantly poorer 5-year OS in colorectal cancer. Oshima et al15 found that gastric cancer patients with high INHBA expression showed significantly worse OS when compared to those with low INHBA expression. Two other independent groups reported similar results, showing that INHBA expression could be a useful tool to predict outcomes in gastric cancer.9,16 Consistently, we found that INHBA expression is higher in ESCC samples when compared with corresponding normal tissues and correlated with lymph node metastasis. One earlier study mentioned elevated levels of INHBA in ESCC patients, but we are the first to identify IHNBA as an independent prognostic factor in ESCC patients.18 What is more, the meta-analysis comprised nine studies of INHBA expression in six different types of cancer, showing that INHBA expression is a promising predictor of patient survival in solid tumors. Our results confirmed and extended the fact that high INHBA expression predicts poor prognosis in solid tumors to ESCC.
However, the molecular mechanism and tumor promoting functions of INHBA remain elusive. Most of the hypotheses are focused on tumor metastasis. Wamsley et al reported that activin is required to sustain cancer stem-like cells phenotypes and contributes to metastasis in non-small cell lung cancer.19 Chang et al also found that activin A promotes tumor invasion and metastasis in head and neck squamous cell carcinoma.20 Yoshinaga et al reported that INHBA promoted tumor aggressiveness by associating with N-cadherin.21 Our findings confirmed that INHBA level correlated with lymph node metastasis in ESCC and might contribute to a worse prognosis. On the other hand, Seder et al found that INHBA was overexpressed in esophageal adenocarcinoma and promoted cell proliferation by promoter demethylation and histone acetylation.22 Our own study and meta-analysis supported the oncogenic effects of INHBA in cancers. On the contrary, roles played by INHBA vary in different contexts of cancer. Panopoulou et al found that activin A suppresses neuroblastoma xenograft tumor growth via ALK4/SMAD2-SMAD3 pathways.23 Kaneda et al demonstrated that activin A inhibits vascular endothelial cell growth and suppresses tumor angiogenesis in gastric cancer.24 The molecular mechanism and pleiotropic functions of INHBA in cancers warrant further studies.
This study shed light on the clinical role of INHBA in ESCC, providing new evidence of its oncogenic functions. However, it also has several limitations. We only included ESCC samples in the study since it was the primary subtype in our patients. All patients were recruited from one single institute. Further multicenter studies are needed to verify our observations. In addition, the number of papers included in the meta-analysis is relatively low. Finally, IHC method is useful to evaluate clinical significance of INHBA, but it failed to answer mechanistic questions.
Conclusion
Our study showed that INHBA gene expression levels were higher in cancer tissue of patients with ESCC than in normal mucosa. High expression of this gene was significantly related to lymph node metastasis and poor prognosis. Combined with the meta-analysis, our findings suggest that the overexpression of INHBA is a useful predictor of prognosis in patients with ESCC.
Supplementary materials
Sensitivity analysis.
Notes: Sensitivity test of the meta-analysis (A). “Trim and fill” method to adjust for publication bias in this meta study (B).
Table S1.
Quality assessment of the selected papers
| Study | Year | Country | Clear description of purpose/objectives | Clear description of patient selection criteria | Clear description of INHBA detection | Predefinition of predictors and outcome measurements | Whether or not multivariate analysis and/or univariate analysis was used | Long enough follow-up period | Limitations considered | Quality (0–6) |
|---|---|---|---|---|---|---|---|---|---|---|
| Lee et al3 | 2014 | Taiwan | Yes | No | Yes | No | Yes | Yes | Yes | 5 |
| Wang et al6 | 2016 | People’s Republic of China | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Okano et al1 | 2013 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Oshima et al4 | 2014 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Wang et al2 | 2012 | People’s Republic of China | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Katayama et al5 | 2017 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |
References
- 1.Okano M, Yamamoto H, Ohkuma H, et al. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013;30(6):2903–2908. doi: 10.3892/or.2013.2761. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 3.Lee HY, Li CC, Huang CN, et al. INHBA overexpression indicates poor prognosis in urothelial carcinoma of urinary bladder and upper tract. J Surg Oncol. 2015;111(4):414–422. doi: 10.1002/jso.23836. [DOI] [PubMed] [Google Scholar]
- 4.Oshima T, Yoshihara K, Aoyama T, et al. Relation of INHBA gene expression to outcomes in gastric cancer after curative surgery. Anticancer Res. 2014;34(5):2303–2309. [PubMed] [Google Scholar]
- 5.Katayama Y, Oshima T, Sakamaki K, et al. Clinical significance of INHBA gene expression in patients with gastric cancer who receive curative resection followed by adjuvant S-1 chemotherapy. In Vivo. 2017;31(4):565–571. doi: 10.21873/invivo.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Huang Z, Liu R, et al. The expression status of INHBA as a prognostic marker for human breast cancer. [Accessed June 01, 2018];Int J Clin Exp Pathol. 2016 9(11):9. Available from: http://www.ijcep.com/files/ijcep0036680.pdf. [Google Scholar]
Acknowledgments
We are grateful to Affiliated Cancer Hospital & Institute of Guangzhou Medical University and our funding source: Guangzhou Medical University’s scientific research project 2016C40.
Footnotes
Availability of data and materials
The key raw data have been deposited into the Research Data Deposit (http://www.researchdata.org.cn), with the Approval Number as RDDB2018000307.
Author contributions
Shanshan Lyu and Chao Jiang are co-first authors. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Domper Arnal MJ, Ferrandez Arenas A, Lanas Arbeloa A. Esophageal cancer: risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–7943. doi: 10.3748/wjg.v21.i26.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaddy-Kurten D, Tsuchida K, Vale W. Activins and the receptor serine kinase superfamily. Recent Prog Horm Res. 1995;50:109–129. doi: 10.1016/b978-0-12-571150-0.50010-x. [DOI] [PubMed] [Google Scholar]
- 4.Beattie GM, Lopez AD, Bucay N, et al. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells. 2005;23(4):489–495. doi: 10.1634/stemcells.2004-0279. [DOI] [PubMed] [Google Scholar]
- 5.Totsuka Y, Tabuchi M, Kojima I, Shibai H, Ogata E. A novel action of activin A: stimulation of insulin secretion in rat pancreatic islets. Biochem Biophys Res Commun. 1988;156(1):335–339. doi: 10.1016/s0006-291x(88)80845-3. [DOI] [PubMed] [Google Scholar]
- 6.Robson NC, Phillips DJ, McAlpine T, et al. Activin-A: a novel dendritic cell-derived cytokine that potently attenuates CD40 ligand-specific cytokine and chemokine production. Blood. 2008;111(5):2733–2743. doi: 10.1182/blood-2007-03-080994. [DOI] [PubMed] [Google Scholar]
- 7.Bashir M, Damineni S, Mukherjee G, Kondaiah P. Activin-A signaling promotes epithelial-mesenchymal transition, invasion, and metastatic growth of breast cancer. NPJ Breast Cancer. 2015;(1):15007. doi: 10.1038/npjbcancer.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Okano M, Yamamoto H, Ohkuma H, et al. Significance of INHBA expression in human colorectal cancer. Oncol Rep. 2013;30(6):2903–2908. doi: 10.3892/or.2013.2761. [DOI] [PubMed] [Google Scholar]
- 9.Wang Q, Wen YG, Li DP, et al. Upregulated INHBA expression is associated with poor survival in gastric cancer. Med Oncol. 2012;29(1):77–83. doi: 10.1007/s12032-010-9766-y. [DOI] [PubMed] [Google Scholar]
- 10.Seder CW, Hartojo W, Lin L, et al. Upregulated INHBA expression may promote cell proliferation and is associated with poor survival in lung adenocarcinoma. Neoplasia. 2009;11(4):388–396. doi: 10.1593/neo.81582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HY, Li CC, Huang CN, et al. INHBA overexpression indicates poor prognosis in urothelial carcinoma of urinary bladder and upper tract. J Surg Oncol. 2015;111(4):414–422. doi: 10.1002/jso.23836. [DOI] [PubMed] [Google Scholar]
- 12.Perisanidis C, Psyrri A, Cohen EE, et al. Prognostic role of pretreatment plasma fibrinogen in patients with solid tumors: a systematic review and meta-analysis. Cancer Treat Rev. 2015 Dec;41(10):960–970. doi: 10.1016/j.ctrv.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Yan SM, Wu HN, He F, et al. High expression of zinc-binding protein-89 predicts decreased survival in esophageal squamous cell cancer. Ann Thorac Surg. 2014;97(6):1966–1973. doi: 10.1016/j.athoracsur.2014.01.058. [DOI] [PubMed] [Google Scholar]
- 14.Xu L, He F, Wang H, et al. A high plasma D-dimer level predicts poor prognosis in gynecological tumors in East Asia area: a systematic review and meta-analysis. Oncotarget. 2017;8(31):51551–51558. doi: 10.18632/oncotarget.17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshima T, Yoshihara K, Aoyama T, et al. Relation of INHBA gene expression to outcomes in gastric cancer after curative surgery. Anticancer Res. 2014;34(5):2303–2309. [PubMed] [Google Scholar]
- 16.Katayama Y, Oshima T, Sakamaki K, et al. Clinical significance of INHBA gene expression in patients with gastric cancer who receive curative resection followed by adjuvant S-1 chemotherapy. In Vivo. 2017;31(4):565–571. doi: 10.21873/invivo.11095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang J, Huang Z, Liu R, et al. The expression status of INHBA as a prognostic marker for human breast cancer. [Accessed June 01, 2018];Int J Clin Exp Pathol. 2016 9(11):9. Available from: http://www.ijcep.com/files/ijcep0036680.pdf. [Google Scholar]
- 18.Kashyap MK, Pawar HA, Keerthikumar S, et al. Evaluation of protein expression pattern of stanniocalcin 2, insulin-like growth factor-binding protein 7, inhibin beta A and four and a half LIM domains 1 in esophageal squamous cell carcinoma. Cancer Biomark. 2012;12(1):1–9. doi: 10.3233/CBM-120289. [DOI] [PubMed] [Google Scholar]
- 19.Wamsley JJ, Kumar M, Allison DF, et al. Activin upregulation by NF-kappaB is required to maintain mesenchymal features of cancer stem-like cells in non-small cell lung cancer. Cancer Res. 2015;75(2):426–435. doi: 10.1158/0008-5472.CAN-13-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang WM, Lin YF, Su CY, et al. Dysregulation of RUNX2/activin-A axis upon miR-376c downregulation promotes lymph node metastasis in head and neck squamous cell carcinoma. Cancer Res. 2016;76(24):7140–7150. doi: 10.1158/0008-5472.CAN-16-1188. [DOI] [PubMed] [Google Scholar]
- 21.Yoshinaga K, Inoue H, Utsunomiya T, et al. N-cadherin is regulated by activin A and associated with tumor aggressiveness in esophageal carcinoma. Clin Cancer Res. 2004;10(17):5702–5707. doi: 10.1158/1078-0432.CCR-03-0262. [DOI] [PubMed] [Google Scholar]
- 22.Seder CW, Hartojo W, Lin L, et al. INHBA overexpression promotes cell proliferation and may be epigenetically regulated in esophageal adenocarcinoma. J Thorac Oncol. 2009;4(4):455–462. doi: 10.1097/JTO.0b013e31819c791a. [DOI] [PubMed] [Google Scholar]
- 23.Panopoulou E, Murphy C, Rasmussen H, et al. Activin A suppresses neuroblastoma xenograft tumor growth via antimitotic and antiangiogenic mechanisms. Cancer Res. 2005;65(5):1877–1886. doi: 10.1158/0008-5472.CAN-04-2828. [DOI] [PubMed] [Google Scholar]
- 24.Kaneda H, Arao T, Matsumoto K, et al. Activin A inhibits vascular endothelial cell growth and suppresses tumour angiogenesis in gastric cancer. Br J Cancer. 2011;105(8):1210–1217. doi: 10.1038/bjc.2011.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bosman FT, Carneiro F, Hruban RH, et al. WHO classification of tumours of the digestive system. IARC Press; Lyon, France: 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sensitivity analysis.
Notes: Sensitivity test of the meta-analysis (A). “Trim and fill” method to adjust for publication bias in this meta study (B).
Table S1.
Quality assessment of the selected papers
| Study | Year | Country | Clear description of purpose/objectives | Clear description of patient selection criteria | Clear description of INHBA detection | Predefinition of predictors and outcome measurements | Whether or not multivariate analysis and/or univariate analysis was used | Long enough follow-up period | Limitations considered | Quality (0–6) |
|---|---|---|---|---|---|---|---|---|---|---|
| Lee et al3 | 2014 | Taiwan | Yes | No | Yes | No | Yes | Yes | Yes | 5 |
| Wang et al6 | 2016 | People’s Republic of China | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Okano et al1 | 2013 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Oshima et al4 | 2014 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Wang et al2 | 2012 | People’s Republic of China | Yes | No | Yes | No | Yes | Yes | No | 4 |
| Katayama et al5 | 2017 | Japan | Yes | No | Yes | No | Yes | Yes | No | 4 |