Abstract
In budding yeast, the release of the protein phosphatase Cdc14 from its inhibitor Cfi1/Net1 in the nucleolus during anaphase triggers the inactivation of Clb CDKs that leads to exit from mitosis. The mitotic exit pathway controls the association between Cdc14 and Cfi1/Net1. It is comprised of the RAS-like GTP binding protein Tem1, the exchange factor Lte1, the GTPase activating protein complex Bub2-Bfa1/Byr4, and several protein kinases including Cdc15 and Dbf2. Here we investigate the regulation of the protein kinases Dbf2 and Cdc15. We find that Cdc15 is recruited to both spindle pole bodies (SPBs) during anaphase. This recruitment depends on TEM1 but not DBF2 or CDC14 and is inhibited by BUB2. Dbf2 also localizes to SPBs during anaphase, which coincides with activation of Dbf2 kinase activity. Both events depend on the mitotic exit pathway components TEM1 and CDC15. In cells lacking BUB2, Dbf2 localized to SPBs in cell cycle stages other than anaphase and telophase and Dbf2 kinase was prematurely active during metaphase. Our results suggest an order of function of mitotic exit pathway components with respect to SPB localization of Cdc15 and Dbf2 and activation of Dbf2 kinase. BUB2 negatively regulates all 3 events. Loading of Cdc15 on SPBs depends on TEM1, whereas loading of Dbf2 on SPBs and activation of Dbf2 kinase depend on TEM1 and CDC15.
INTRODUCTION
The final stage of the cell cycle is exit from mitosis (reviewed in Morgan, 1999; Zachariae, 1999). During this time, the mitotic spindle disassembles, chromosomes decondense, and cytokinesis occurs. In the budding yeast Saccharomyces cerevisiae, these events are brought about by inactivation of Clb cyclin-dependent kinases (Clb-CDKs), which, earlier in the cell cycle, trigger entry into mitosis (reviewed in Zachariae and Nasmyth, 1999). Inactivation of Clb-CDKs is brought about by 2 redundant mechanisms. Ubiquitin-dependent degradation of Clb cyclins by the anaphase promoting complex (APC) or cyclosome (C) complexed with the APC activator Cdh1 (APC/CCdh1) and binding of the CDK inhibitor Sic1 to the Clb-CDK complex (Schwab et al., 1997; Visintin et al., 1997; Fang et al., 1998; Shirayama et al., 1998; Kotani et al.1999; Kramer et al., 2000; Yudkovsky et al., 2000). The protein phosphatase Cdc14 plays an important role in both stimulating APC-dependent degradation of mitotic Clb cyclins and accumulation of Sic1 (Visintin et al., 1998; Jaspersen et al., 1999). Cdc14 dephosphorylates the APC-specificity factor Cdh1, the CDK inhibitor Sic1 and its transcription factor Swi5, allowing for activation of Cdh1 and transcription and stabilization of Sic1, respectively.
Cdc14 is regulated by Cfi1/Net1 (Shou et al., 1999; Straight et al., 1999; Visintin et al., 1999; Traverso et al., 2001). Cfi1/Net1 inhibits Cdc14 in the nucleolus during G1, S phase, and early mitosis. During nuclear division, Cdc14 is released from Cfi1/Net1, allowing Cdc14 to reach its targets. The mitotic exit pathway is required for the release of Cdc14 from Cfi1/Net1 (Shou et al., 1999; Visintin et al., 1999). This pathway includes the Ras-like GTP binding protein Tem1, a putative exchange factor, Lte1; the 2 component GTPase-activating protein complex (GAP) Bub2-Bfa1/Byr4; the protein kinases Cdc5, Cdc15, Dbf2 and Dbf20; and the Dbf2-associated protein Mob1 (Kitada et al., 1993; Donovan et al., 1994; Toyn and Johnston, 1994; Shirayama et al., 1994a, b; Charles et al., 1998; Komarnitsky et al., 1998; Luca and Winey, 1998; Alexandru et al., 1999; Fesquet et al., 1999; Fraschini et al., 1999; Li, 1999). Pds1 and Esp1, both regulators of sister-chromatid separation are also required for release of Cdc14 from Cfi1/Net1 and inactivation of mitotic kinases (Cohen-Fix and Koshland, 1999; Shirayama et al., 1999; Tinker-Kulberg and Morgan, 1999). In addition to release of Cdc14 from the nucleolus, Clb5-CDK1 (Cdc28) kinase, a potent antagonist of Cdc14, needs to be inactivated to allow for exit from mitosis to occur. This is achieved by APC/CCdc20 which degrades Clb5 during anaphase (Shirayama et al., 1999).
One of the functions of the mitotic exit pathway is to ensure that exit from mitosis does not occur before partitioning of the genetic material between mother and daughter cell. The GTPase Tem1 and Bub2-Bfa1/Byr4 localize onto spindle pole bodies (SPBs), predominantly the 1, which migrates into the daughter cell. Lte1 localizes to the bud (Fraschini et al., 1999; Li 1999; Bardin et al., 2000; Bloecher et al., 2000; Daum et al., 2000; Pereira et al., 2000; Wang et al., 2000). The changes of Tem1 protein levels during the cell cycle and the localization patterns of Tem1 and Lte1 result in the 2 proteins being present in the same cellular compartment only after the nucleus enters the bud during nuclear division. This mode of localization ensures that mitotic exit does not occur before chromosome partitioning. Bub2-Bfa1/Byr4 also play a critical role in preventing exit from mitosis before partitioning of the genetic material between mother and daughter cell. However, whether Bub2-Bfa1/Byr4 activity is regulated remains to be determined.
The protein kinases Cdc15, Dbf2, and Dbf20, and the Dbf2-associated protein Mob1 are thought to relay the signal generated at the cytoplasmic face of the SPB to bring about the release of Cdc14 from Cfi1/Net. Cdc15 shows a high degree of homology to the family of MAP kinase kinase kinases (Hunter and Plowman, 1997). The protein interacts with Tem1 as judged by coimmunoprecipitation (Bardin et al., 2000; Asakawa et al., 2001), but there is no evidence that its kinase activity is regulated by Tem1. Cdc15 protein levels and associated kinase activity are constant throughout the cell cycle (Jaspersen et al., 1998); however, Cdc15 localization and its phosphorylation status change during the cell cycle. Cdc15 associates with the cytoplasmic face of SPBs during anaphase and is dephosphorylated during exit from mitosis (Cenamor et al., 1999; Gruneberg et al., 2000; Jaspersen and Morgan, 2000; Xu et al., 2000; Menssen et al., 2001). The protein levels of the related protein kinases Dbf2 and Dbf20 are also constant throughout the cell cycle (Toyn and Johnston, 1994), but Dbf2 kinase activity fluctuates during the cell cycle, being low during G1, S phase, and early mitosis, and high during anaphase and exit from mitosis (Toyn and Johnston, 1994). Dbf2, like Cdc15 localizes to SPBs and to the bud neck during cytokinesis (Frenz et al., 2000). Dbf2 kinase activity is thought to be controlled at least in part by Mob1, which physically interacts with Dbf2 (Luca and Winey, 1998; Komarnitsky et al., 1998).
Although the regulation of individual components of the mitotic exit pathway is the focus of intense investigation, the order in which the components of the mitotic exit pathway function is not known. To address this question, we investigated how the localization and activity of the protein kinases Cdc15 and Dbf2 is regulated during the cell cycle and how mutations impairing the mitotic exit pathway affect the 2 protein kinases. We find that Cdc15 and Dbf2 are recruited to both SPBs during anaphase. Localization of Cdc15 on SPBs depends on TEM1, but not on DBF2 and CDC14. Loading of Dbf2 onto SPBs and activation of Dbf2 kinase activity depends on TEM1 and CDC15, but not CDC14. These findings suggest an order of function in the recruitment of mitotic exit pathway components to the SPB and activation of Dbf2 kinase activity: TEM1 functions upstream of CDC15 and CDC15 upstream of DBF2. Furthermore, we find that during a normal cell cycle localization of Dbf2 to SPBs and Dbf2 kinase activity correlate, suggesting that association of Dbf2 with Tem1 and Cdc15 on SPBs is a prerequisite for Dbf2 activation. In cells lacking BUB2, Dbf2 is localized to SPBs throughout the cell cycle, but Dbf2 kinase activity was prematurely activated only during metaphase. This finding indicates that SPB localization is not sufficient for Dbf2 activation and that other factors are necessary for Dbf2 to be active as a protein kinase.
MATERIALS AND METHODS
All strains were derivatives of strain W303 (K699) and are listed with relevant genotypes in Table 1. MYC tags were introduced at the N-terminus of endogenous DBF2 or DBF20 according to Schneider et al. (1995). The conserved lysine (amino acid 206) and the after isoleucine (amino acid 207) residues within the Dbf2 kinase were mutated by PCR to arginine and threonine, respectively, thereby creating a BsiWI site. Strains carrying a CDC15-HA fusion were as described in Jaspersen et al. (1998). Conditions for growth and release of synchronous cultures from arrest by α-factor were as described by Surana et al. (1993). For synchronous release of cells from a hydroxyurea (HU) arrest cells were arrested with 10 mg/ml HU. When arrest was complete, cells were washed and releases into medium lacking the drug. Cells were arrested with hydroxyurea and nocodazole by adding to the cultures 10 mg/ml hydroxyurea or 15 μg/ml nocodazole, respectively. Immunoblot analysis of the total amount of Clb2, Kar2 and Dbf2-Myc was performed as described in Cohen-Fix et al. (1996). Antibody dilutions were used as in Visintin et al. (1997). Anti-Myc antibodies were used at 1:50 dilution to immunoprecipitate Dbf2 either for phosphatase treatment or to measure Dbf2 kinase activity. Dbf2 kinase activity was assayed as Clb2 kinase activity. The method is described in Surana et al. (1993). Indirect in situ immunofluorescence methods and antibody concentrations were as described in Visintin et al. (1999).
Table 1.
Strains and relevant genotypes
| Strain | Relevant genotype |
|---|---|
| K699 | MATa, ade2-1, leu2-3, ura3, trp1-1, his3-11,15, can1-100, GAL |
| A852 | MATalpha, dbf2-2 |
| A1787 | MATa CDC15-3HA |
| A1931 | MATa, DBF2-3MYC |
| A2044 | MATa, nud1-44∷TRP1, nud1∷HIS5, CDC15-HA |
| A2096 | MATa, cdc15-2, DBF2-3MYC |
| A2107 | MATa, cdc5-1, DBF2-3MYC |
| A2110 | MATa, cdc28-4, DBF2-3MYC |
| A2111 | MATalpha, dbf2-2, CDC15-3HA |
| A2125 | MATa, tem1-3, CDC15-3HA |
| A2135 | MATa, tem1-3, DBF2-3MYC |
| A2143 | MATa, cdc20-3, DBF2-3MYC |
| A2152 | MATa, cdc5-1, CDC15-3HA |
| A2156 | MATa, cfi1∷URA3, DBF2-3MYC |
| A2179 | MATa, cdc14-3, DBF2-3MYC |
| A2181 | MATalpha, cdc13-1, DBF2-3MYC |
| A2184 | MATa, cdc23-1, DBF2-3MYC |
| A2184 | MATa, cdc23-1, DBF2-3MYC |
| A2212 | MATalpha, cdc14-3, CDC15-3HA |
| A2339 | MATa, nud1-44∷TRP1, nud1∷HIS5, DBF2-3MYC |
| A2340 | MATalpha, nud1-44∷TRP1, nud1∷HIS5, DBF2-3MYC |
| A2472 | MATa, bub2∷HIS3, DBF2-3MYC |
| A2840 | MATa, bub2∷HIS3, CDC15-3HA |
| A3044 | MATa, YCplac22/dbf2KI→RT-MYC |
| A3045 | MATa, dbf2-2, YCplac22/dbf2KI→RT-MYC |
| A3046 | MATa, dbf2-2, YCplac22/DBF2-3MYC |
RESULTS
Cdc15 Localizes to Both Spindle Pole Bodies during Anaphase
Several reports had shown that Cdc15 localizes to SPBs (Cenamor et al., 1999; Xu et al., 2000; Gruneberg et al., 2000; Menssen et al., 2001). However, slight differences in the dynamics of Cdc15 localization had been observed, depending on the epitope tag used. The localization pattern we observed was identical to that previously observed by Xu et al. (2000). Cdc15, whose chromosomal copy was tagged with 3 HA epitopes (Jaspersen et al., 1998), was not detectable in G1 cells, in cells that had not yet formed a mitotic spindle and cells with metaphase spindles. Cdc15 was present as a dot at both ends of mitotic spindles in cells with anaphase and telophase spindles (Figure 1B). Similar results were obtained when Cdc15 was analyzed in synchronous cultures. Cells were arrested in G1 with the pheromone α-factor followed by release into the cell cycle. Cdc15 was found on SPBs in cells containing anaphase and telophase spindles but not in cells in other cell cycle stages (Figure 1C).
Figure 1.
The subcellular localization of Cdc15-Ha during the cell cycle. Exponentially growing cells either lacking (A, no tag; K699) or carrying a CDC15-HA fusion (B, A1787) were fixed and subjected to indirect in situ immunofluorescence. Cdc15-Ha was visualized with the use of α-HA antibodies (BABCO). DAPI (4′6-diamidino-2-phenylindole) was used to stain DNA. (C) Cdc15-Ha localization in cells undergoing a synchronous cell cycle after an α-factor–induced G1 arrest. The percentage of cells with Cdc15-Ha on the SPB (spindle pole body) and the percentage of cells with metaphase and telophase spindles are shown. (D, E) nud1–44 (A2044) or tem1–3 (A2125) cells carrying a CDC15-HA fusion were arrested in G1 with α-factor (5 μg/ml) followed by release into medium lacking pheromone at the restrictive temperature (37°C). The percentage of cells with metaphase (closed circles) and anaphase spindles (closed squares) as well as the percentage of cells with Cdc15 on SPBs (open squares) or in the cytoplasm (open circles) was determined. (F) Wild-type (A1787) and bub2:: HIS3 cells (A2840) were arrested in early S phase with the use of hydroxyurea (10 mg/ml) followed by release into medium lacking the drug. The percentage of cells with metaphase (closed circles) and anaphase spindles (closed squares) as well as the percentage of cells with Cdc15 on SPBs (open squares) was determined at the indicated times. (G) Colocalization of Cdc15-Ha with the spindle pole body component Tub4 in bub2:: HIS3 cells. In all experiments, 200 cells were counted per time point (n = 200).
Previous studies have shown that Cdc15 localizes to the outer plaque of the SPB, as Cdc15 localization is impaired in cnm67 mutants in which the outer plaque of the SPB is disrupted (Cenamor et al., 1999). However, Cdc15 localization was not affected in nud1–2 mutants (Gruneberg et al., 2000). As localization of Tem1 is impaired in temperature sensitive nud1–44 mutants (Bardin et al., 2000) and Cdc15 localization to SPBs depends on TEM1 (Figure 1E, Table 2) we wished to examine the localization of Cdc15 in nud1–44 mutants. In nud1–44 mutants the outer plaque of the SPB dissociates from the rest of the organelle (Adams and Kilmartin, 1999). nud1–44 mutants were arrested in G1 with the use of α-factor pheromone and released into the cell cycle at 37°C. Cdc15 was detected on SPBs in only 15% of anaphase and telophase cells (Figure 1D). In 30 percent of cells, Cdc15 localized to a dot in the cytoplasm, indicating that Cdc15 dissociated from SPBs along with the rest of the outer plaque of the SPB (Figure 1D). These results indicate that Tem1 and Cdc15 localize to the outer plaque of the SPB in a NUD1-dependent manner.
Table 2.
Cdc15 localization in cdc mutants
| Genotype | SPB localization |
|---|---|
| WT | + |
| nud1-44 | − |
| tem1-3 | − |
| cdc5-1 | +/− |
| dbf2-2 | + |
| cdc14-3 | + |
| bub2∷HIS3 | prematurely* |
Prematurely indicates that Cdc15 is present on SPBs in stages of the cell cycle when it is not found on SPBs in wild-type cells.
Cdc15 Localization to SPBs Depends on TEM1 and Is Inhibited by BUB2
Next we analyzed whether SPB localization of Cdc15 depended on components of the mitotic exit pathway. Cdc15 was not even transiently detected on SPBs in tem1–3 cells as they progressed through the cell cycle (Figure 1E), but was present on SPBs in dbf2–2 and cdc14–3 mutants and on 50 percent of cdc5–1 mutants (Table 2). In exponentially growing bub2Δ cells, Cdc15 was present on SPBs throughout the cell cycle (data not shown). To examine the effects of BUB2 on Cdc15 localization in more detail, we first analyzed Cdc15 localization in cells released from an α-factor block. We noticed, however, that prolonged incubation of cells in G1 led to a dramatic decline of Cdc15 signal in the α-factor–arrested cells, whereas Cdc15 could be detected on SPBs in the next G1 phase of the cell cycle (data not shown). Similar results were obtained when cells were released from a hydroxyurea block. Cdc15 was not detected in cells that had not yet formed a mitotic spindle, whereas it was detected in cells entering the next G1 phase (Figure 1F). These findings indicate that the association of Cdc15 with SPBs during interphase is not stable in bub2Δ cells and disrupted by prolonged cell cycle arrest. However, the association of Cdc15 with SPBs organizing mitotic spindles was more stable. During the hydroxyurea arrest, Cdc15 localized to at least 1 SPB in 75% of cells that had formed a mitotic spindle. During mitosis Cdc15 localized to SPBs in the majority of cells and remained localized at SPBs during the after G1 stage (Figure 1F). We confirmed that the Cdc15 indeed localized to SPBs in bub2Δ cells, as the Cdc15 staining overlapped with the staining pattern observed with the SPB component Tub4 (Figure 1G). Our data suggest that localization of Cdc15 to SPBs is inhibited by BUB2 and depends on TEM1 but not on DBF2 and CDC14.
Dbf2 Localizes to SPBs during Anaphase
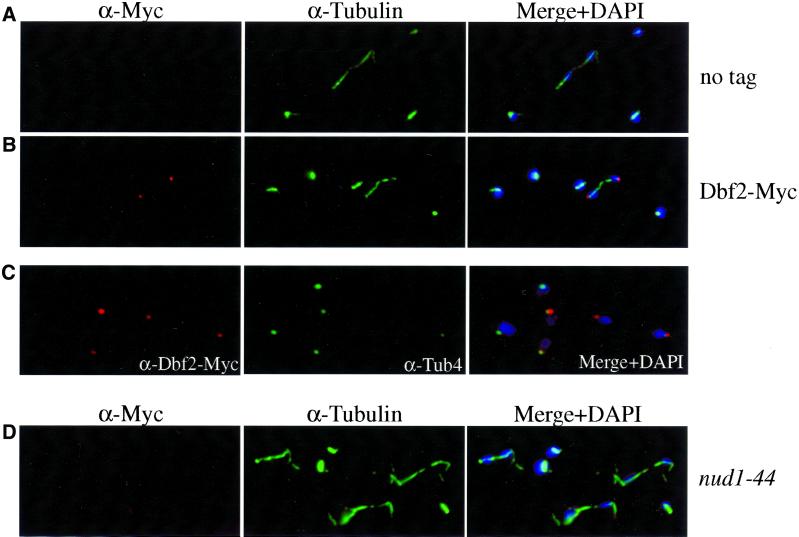
The chromosomal copies of DBF2 and DBF20 were tagged with 3 Myc epitopes We could not detect Dbf20 by indirect in situ immunofluorescence (data not shown); however, the subcellular localization of Dbf2 was similar to that of Cdc15 (Figure 2B). The protein was undetectable in G1, S phase, metaphase, and early anaphase cells. In most, although not all, late anaphase and telophase cells, Dbf2 was present as a dot at 1 or both ends of mitotic spindles (Figures 2B and 7A). Dbf2 staining overlapped with Tub4, indicating that Dbf2 also localizes to SPBs or a structure associated with it (Figure 2C). Dbf2 localization on SPBs also depended on NUD1, as Dbf2 was undetectable by indirect immunofluorescence in nud1–44 mutants (Figures 2D and 7F). These results suggest that Dbf2 localizes to the outer plaque of the SPB or a structure associated with it.
Figure 2.
The subcellular localization of Dbf2-Myc during the cell cycle. (A) Exponential growing cells lacking a Dbf2-Myc fusion (no tag; K699). (B) Dbf2-Myc and microtubules of strain A1931 were visualized with the use of anti-Myc antibodies (BABCO) and antitubulin antibodies (α-tubulin), respectively. DAPI was used to stain DNA (DAPI). (C) Colocalization of Dbf2-Myc with the spindle pole body component Tub4. (D) Dbf2-Myc localization in nud1–44 mutant (A2340) 120 min after shift to the restrictive temperature (37°C).
Figure 7.
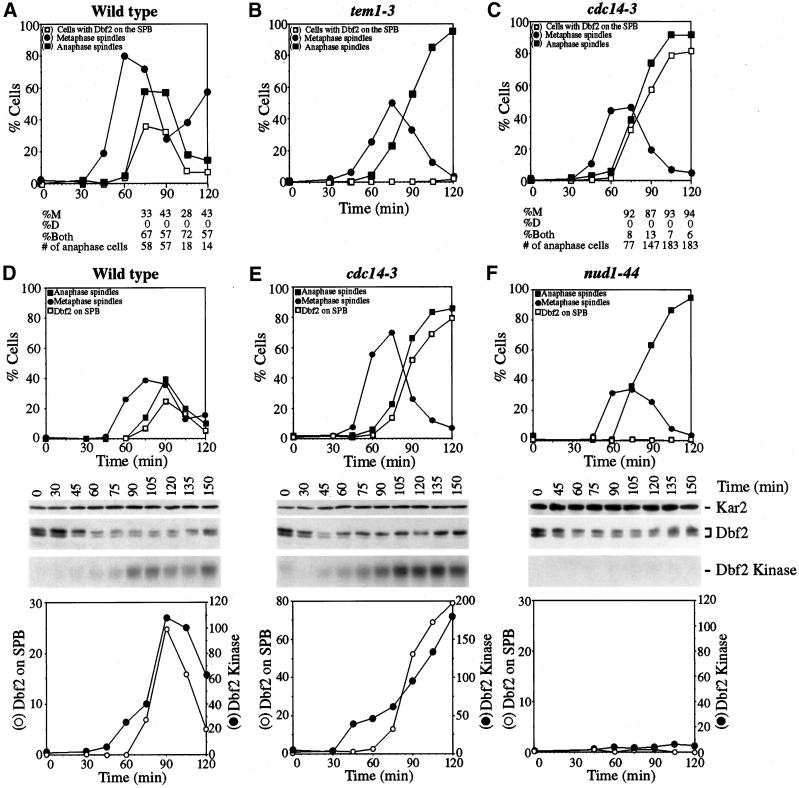
Dbf2 localization and kinase activity in tem1–3 and cdc14–3 mutants. (A-C) DBF2-MYC (A1931), tem1–3 DBF2-MYC (A2135) and cdc14–3 DBF2-MYC (A2179) cells were arrested in G1 with the use of 10 μg/ml (A-C) or 5 μg/ml (D-F) α-factor at 23°C in YEP medium containing glucose (YEPD). When the arrest was complete (after 2 h (D-F)) and a mating projection could be seen (after 3 h (A-C)), cells were released at the restrictive temperature (37°C) in YEPD. (A) Percentage of cells in metaphase (closed circles), anaphase (closed squares) and with Dbf2-Myc protein localized either on 1 or on both SPBs (open squares). The numbers underneath the graph represent the percentage of anaphase/telophase cells with the Dbf2-Myc protein on the SPB in the mother cell (%M), in the daughter cell (%D) or on both SPBs (%Both). “# of anaphase cells” indicates the number of anaphase/telophase cells analyzed. (B) Percentage of tem1–3 DBF2-MYC cells in metaphase (closed circles), anaphase (closed squares), and Dbf2-Myc protein localized either on 1 or both SPBs (open squares). (C) Percentage of cdc14–3 DBF2-MYC cells in metaphase (closed circles), anaphase (closed squares) and with Dbf2-Myc protein localized either on 1 or both SPBs (open squares). The numbers representing various types of Dbf2-Myc localization underneath the graph are as in (A). (D - F) Wild-type (A1931, D), cdc14–3 (A2179, E), and nud1–44 mutants (A2339, F), all carrying a DBF2-MYC fusion, were arrested in G1 with α-factor (5 μg/ml) followed by release into medium lacking pheromone. The graph at the top of each panel shows the percentage of cells with metaphase spindles (closed circles), with anaphase spindles (closed squares) and Dbf2 on SPBs (open squares). Dbf2-Myc protein levels and Dbf2 kinase activity are shown in the middle of each panel. The graphs at the bottom of each panel show a comparison between SPB localization of Dbf2 (open squares) and Dbf2 kinase/Dbf2 protein (closed squares).
Next we determined the fraction of cells containing Dbf2 on both SPBs versus 1 SPB and investigated whether Dbf2 localized to mother or daughter SPB in those cells containing Dbf2 on only 1 SPB. Cells were treated with α-factor pheromone until they formed a mating projection (shmoo). When cells were released into the cell cycle, the newly formed bud was spherical whereas the mother cell remained shmoo-shaped. In 60% to 70% of cells, Dbf2 was detected on both SPBs; in 30% to 40% of cells, Dbf2 was found on the SPB present in the mother cell (Figure 7A). Our results indicate that Dbf2 associates with the outer plaque of both SPBs during anaphase in the majority cells.
SPB Localization of Dbf2 Coincides with Activation of Dbf2 Kinase
Dbf2 kinase activity is low during early stages of the cell cycle but high during anaphase and exit from mitosis (Toyn and Johnston, 1994). In synchronous cultures, Dbf2 kinase activity correlated well with localization of Dbf2 on SPBs of late anaphase spindles and changes in Dbf2 mobility (Toyn and Johnston, 1994; Figure 3). This change in Dbf2 mobility was due to dephosphorylation of the protein (Toyn and Johnston, 1994; Figure 3C).
Figure 3.
Localization of Dbf2 to SPBs and activation of Dbf2 kinase activity occur concomitantly. Cells carrying a DBF2-MYC tag (A1931) were arrested in G1 with α-factor (5 μg/ml) followed by release into medium lacking pheromone. The amount of Dbf2-Myc protein (A), Dbf2 kinase activity (B), the percentage of cells carrying Dbf2 on SPBs (n = 200; D, E) and cells containing anaphase or telophase spindles (n = 200; D) were analyzed. Clb2 protein levels (A) were examined to assess cell cycle synchrony; Kar2 was used as an internal loading control in Western blots. (C) Dbf2-myc was immunoprecipitated with the use of anti-Myc antibodies and incubated with calf intestinal alkaline phosphatase (CIP) buffer alone (IP+buffer) or with 80 U of CIP (IP+CIP). An immunoprecipitation from cells without a DBF2-MYC fusion is shown in the lane labeled “no tag”.
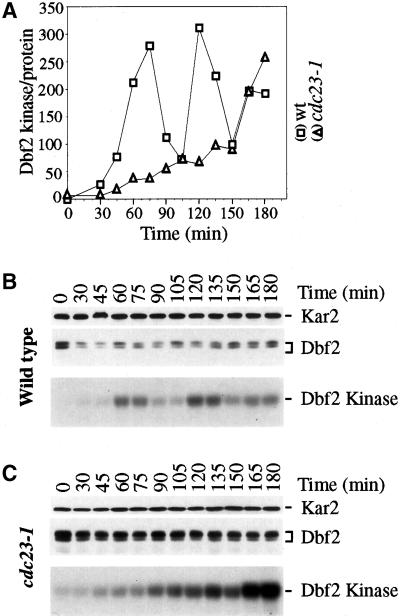
To further investigate whether a correlation between the presence of Dbf2 on SPBs and active Dbf2 kinase exists, we analyzed Dbf2 in various cell cycle arrests. Dbf2 was not present on SPBs and Dbf2 kinase activity was low in G1-arrested cells (α-factor, cdc28–4; Table 3), in S-phase-arrested cells (hydroxyurea), and G2/M phase arrested cells (nocodazole, cdc13–1; Table 3). In cdc23–1 and cdc20–1 mutants (both mutations inactivate the APC/CCdc20) Dbf2 kinase activity appeared high, yet Dbf2 was detected on SPBs in only a small fraction of cells (20% to 30%) and was present on only 1 SPB (data not shown). This finding suggested that Dbf2 kinase activity was high despite Dbf2 being largely absent from SPBs. To investigate this apparent noncorrelation between the presence of Dbf2 on SPBs and active Dbf2 kinase in these mutants in more detail, we analyzed Dbf2 in cdc23–1 mutants undergoing a synchronous cell cycle. cdc23–1 mutants were arrested in G1 at the permissive temperature and released into the cell cycle at the restrictive temperature. Dbf2 kinase activity was found to be unusually high but so were Dbf2 protein levels (Figure 4). Similar results were found in cdc20–1 mutants (data not shown). A subsequent analysis of the amount of Dbf2 kinase activity per Dbf2 protein revealed that Dbf2 kinase remained low in cdc23–1 mutants (Figure 4). Only after long periods of incubation at 37°C did Dbf2 kinase activity increase (Figure 4) indicating that Dbf2 kinase is eventually activated. We noticed that in these later time points, Dbf2 associated with 1 SPB in ∼30% of the cells (data not shown). These results show that Dbf2 protein is not active as a protein kinase in APC/CCdc20 mutants and that APC/CCdc20 is required to keep Dbf2 protein levels low. The high levels of Dbf2 protein observed in APC/CCdc20 mutants may arise from Dbf2 itself being targeted for degradation by APC/CCdc20. As Dbf2 protein levels do not fluctuate during the cell cycle (as is characteristic for APC/CCdc20 substrates), it is more likely that proteins required for Dbf2 synthesis are more abundant in APC/CCdc20 mutants. In summary, our results show that in synchronous cultures as well as cell cycle arrests, Dbf2 localization to SPBs correlates with activation of Dbf2 kinase suggesting that these 2 processes are tightly linked.
Table 3.
Dbf2 localization, phosphorylation, and kinase activity in cdc mutants
| Genotype | SPB localization | Kinase activity | Dbf2 phosphorylation° |
|---|---|---|---|
| WT | + | + | + |
| nud1-44 | − | − | + |
| α-factor | − | − | + |
| cdc28-4 | − | − | − |
| hydroxyurea | − | − | ++ |
| cdc13-1 | − | − | ++ |
| nocodazole | − | − | ++ |
| cdc23-1 | −/+** | −/+** | ++ |
| cdc20-3 | −/+** | −/+** | ++ |
| cfi1∷URA3 | + | + | − |
| cdc5-1 | +/− | +/− | + |
| tem1-3 | − | − | + |
| cdc15-2 | − | − | + |
| cdc14-3 | + | + | ++ |
| bub2∷HIS3 | prematurely* | ++ | + |
Indicates that Dbf2 is present on SPBs in stages of the cell cycle when it is not found on SPBs in wild-type cells.
In approximately 30% of cells Dbf2 is present on one SPB after prolonged incubation at 37°C.
Dbf2 kinase activity is low in these mutants but after prolonged incubation at the restrictive temperature becomes active.
° + indicates that overall phosphorylation of Dbf2 is as in exponentially growing wild-type cells; ++ indicates that Dbf2 is primarily found in the hyperphosphorylated form; − indicates that Dbf2 is primarily found in the hypophosphorylated form.
Figure 4.
APC/C Cdc20 is required to maintain low levels of Dbf2 protein. DBF2-MYC (A1931) and cdc23–1 DBF2-MYC (A2184) cells were arrested in G1 with the use of 5 μg/ml α-factor and released at the restrictive temperature (37°C) in medium lacking pheromone. Samples were taken at the indicated times to determine Dbf2 protein levels and Dbf2 kinase activity. The graph shows the amount of Dbf2 kinase activity per Dbf2 protein. Kar2 (top panel), Dbf2 protein (middle panel) and Dbf2 kinase activity (lower panel).
Dbf2 Lacking Kinase Activity Associates with SPBs during Anaphase
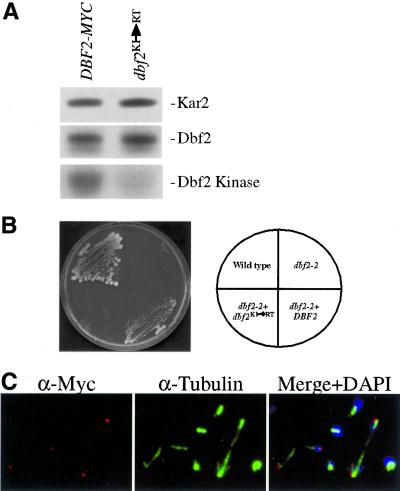
Owing to the correlation between activation of Dbf2 kinase activity and localization of Dbf2 to SPBs, we were interested in determining whether Dbf2 kinase activity was required for its association with SPBs or whether Dbf2 can associate with SPBs in the absence of kinase activity. We replaced the conserved lysine residue at position 206, which is critical for kinase activity by arginine (Dbf2KI ->RT, Materials and Methods) and analyzed the consequences on Dbf2 localization. The Dbf2KI ->RT mutation was inactive as a kinase in vitro (Figure 5A) and failed to complement the temperature sensitive lethality of a dbf2–2 mutant in vivo (Figure 5B), indicating that this mutation indeed inactivates Dbf2 kinase activity in vitro and in vivo. Immunolocalization of this allele in wild-type cells and dbf2–2 mutants showed that the Dbf2KI ->RT mutation did not affect Dbf2 localization (Figure 5C, data not shown). This result indicates that Dbf2 kinase activity is not required for localization of Dbf2 on SPBs.
Figure 5.
A kinase-dead Dbf2 localizes to SPBs. (B) Wild-type (K699), dbf2–2 (A852), dbf2–2 YcP DBF2-MYC (A3046), and dbf2–2 YcP Dbf2KI ->RT -MYC (A3045) cells grown at 37°C. (A) dbf2–2 cells carrying a DBF2-MYC fusion or a DBF2-MYC fusion with the conserved K206 mutated to R (Dbf2KI ->RT) were grown to exponential phase followed by shift to 37°C for 2 h and Dbf2 kinase activity was determined. (C) Localization of Dbf2KI ->RT -Myc in exponentially growing cells (DBF2, YcP Dbf2KI ->RT -MYC; A3044).
SPB Localization and Activation of Dbf2 Require TEM1, CDC15, and NUD1
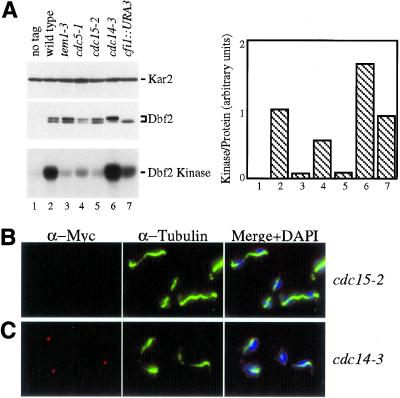
To determine whether SPB localization and kinase activity of Dbf2 were controlled by components of the mitotic exit pathway, we analyzed the localization of Dbf2 and its kinase activity in cells impaired for TEM1, NUD1, CDC5, CDC15, CFI1 and CDC14 (summarized in Table 3). In cells lacking CFI1, Dbf2 localization and Dbf2 kinase activity per Dbf2 protein was similar to that of wild-type cells but Dbf2 protein was predominantly in the dephosphorylated form (Figure 6A, Table 3). In cells defective for the polo-like kinase CDC5, Dbf2 was absent from SPBs in 50% of cells and found to be localized to 1 SPB in the other 50% of cells. Dbf2-associated kinase activity was 50% of the amount found in exponential growing wild-type cells (Figure 6A, Table 3). In tem1–3 and cdc15–2 mutants, Dbf2 was either very weakly or not at all present on SPBs. Dbf2 kinase activity was low in both mutants (Figure 6, A and B; Table 3). A detailed analysis of SPB localization in tem1–3 mutants undergoing a synchronous cell cycle showed that the protein was never observed on SPBs (Figure 7B). Similarly, in nud1–44 mutants progressing synchronously through the cell cycle, Dbf2 failed to localize to SPBs and Dbf2 kinase activity was low (Figure 7F).
Figure 6.
Dbf2 localization and kinase activity in mutants defective for the mitotic exit pathway. Cells lacking a DBF2-MYC fusion (K699, no tag; lane 1), DBF2-MYC cells (A1931, wild-type, lane 2), tem1–3 DBF2-MYC (A 2135, lane 3), cdc5–1 DBF2-MYC (A 2107, lane 4), cdc15–2 DBF2-MYC (A 2096, lane 5), cdc14–3 DBF2-MYC (A 2179, lane 6) and cfi1:: URA3 DBF2-MYC (A 2156, lane 7) cells were arrested at the restrictive temperature (37°C) for 2 h. Dbf2-Myc proteins levels and Dbf2 kinase activity was determined (A). The graph in (A) shows the amount of Dbf2 kinase activity/Dbf2 protein with the numbers in the graph corresponding to lane numbers. Dbf2-Myc localization was analyzed in cdc15–2 DBF2-MYC (B) and cdc14–3 DBF2-MYC (C) mutants.
In cdc14–3 mutants, Dbf2 localized to a single SPB in the majority of cells (Figures 6C and 7C) and Dbf2-associated kinase activity was high (Figures 6A and 7E), suggesting that CDC14 was dispensable for efficient activation of Dbf2 kinase. In synchronous cultures, Dbf2 associated with SPBs with identical kinetics as wild-type cells and activation of Dbf2 kinase correlated with localization to SPBs (Figure 7E). To determine whether Dbf2 localized to mother or daughter, SPB cells were treated with α-factor pheromone until they formed a mating projection (shmoo). When cells were released into the cell cycle, the newly formed bud was spherical whereas the mother cell remained shmoo-shaped. This analysis revealed that Dbf2 was found on the SPB that resides in the mother cell (Figure 7C). Thus, CDC14 is required either for recruiting or maintaining Dbf2 at the daughter SPB. Alternatively, other proteins that assemble onto the SPB may mask Dbf2. Our results suggest that CDC15 and TEM1 are required for Dbf2 to load onto SPBs and to become active as a kinase. CDC14 is dispensable for activation of Dbf2 kinase activity. However, CDC14 is required for Dbf2′s presence on the daughter SPB.
We also analyzed the phosphorylation status of Dbf2 in the various mitotic exit pathway mutants (summarized in Table 3). In synchronous cultures, dephosphorylation of Dbf2, the presence of Dbf2 on SPBs and Dbf2-associated kinase activity correlate suggesting that dephosphorylation of Dbf2 may play a role in Dbf2 localization and Dbf2 kinase activation (Toyn and Johnston, 1994; Figure 3). However, this correlation was not found in cdc14–3 mutants and nocodazole-arrested bub2Δ cells. In these mutants Dbf2 was hyperphosphorylated but was present at SPBs and active as a kinase (Figures 6A and 7E, data not shown). Thus, at least complete dephosphorylation of Dbf2 is not required for the protein to localize to SPBs and to be active as a protein kinase. These data, however, do not rule out the possibility that dephosphorylation at specific sites is important for Dbf2 activation.
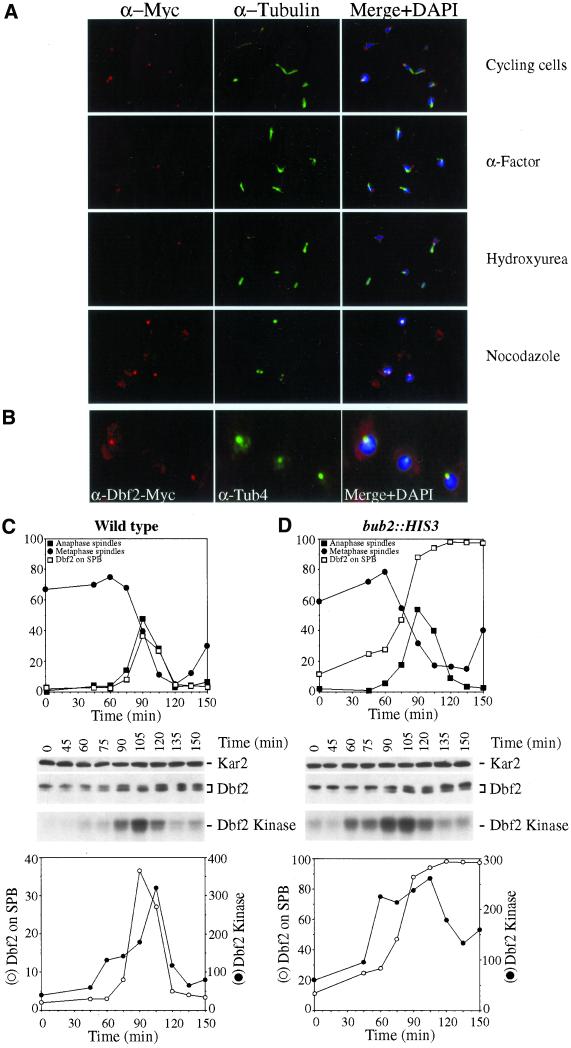
SPB Localization and Activation of Dbf2 Are Inhibited by BUB2
Our results indicated that Cdc15 localization to SPBs is inhibited by BUB2. We therefore wished to determine whether BUB2 had a similar effect on Dbf2 localization and Dbf2 kinase activity. Dbf2 was not only present on SPBs during anaphase and telophase but also during other cell cycle stages, and Dbf2 kinase was increased (Figure 8A and data not shown). Dbf2 colocalized with Tub4 (Figure 8B), indicating that the Dbf2 signal observed in cell cycle stages other than anaphase and telophase indeed overlapped with SPBs. To examine the effects of BUB2 on Dbf2 in more detail, we first analyzed Dbf2 localization and activity in cells released from an α-factor block. However, as observed for Cdc15, prolonged incubation of cells in G1 led to a dramatic decline of Dbf2 signal in the G1-arrested cells, whereas Dbf2 could be easily detected on SPBs in the next G1 phase of the cell cycle (data not shown). Similarly, in cells arrested with hydroxyurea, Dbf2 was not detected on SPBs that nucleated interphase microtubules, whereas it was detected in cells entering the next G1 phase (Figure 8D). Thus, as found for Cdc15, the association of Dbf2 with interphase SPBs appears unstable and sensitive to prolonged cell cycle arrest.
Figure 8.
Dbf2 loading onto SPBs and activation of Dbf2 kinase occur prematurely in cells lacking BUB2. (A) bub2:: HIS3 DBF2-MYC (A 2472) cells were either grown to exponential phase (Cyc), or arrested with α-factor (α-F), hydroxyurea (HU), or nocodazole (Noc) and Dbf2 localization was determined. (B) Colocalization of Dbf2-Myc with the SPB component Tub4 in bub2:: HIS3 cells. (C - D) Wild-type (A1931, C) and bub2:: HIS3 (A2472, D) cells all carrying a DBF2-MYC fusion were arrested in early S phase with hydroxyurea followed by release into medium lacking pheromone. The graph at the top of each panel shows the percentage of cells with metaphase spindles (closed circles), with anaphase spindles (closed squares) and Dbf2 on SPBs (open squares). Dbf2-Myc protein levels and Dbf2 kinase activity are shown in the middle of each panel. The graphs at the bottom of each panel show a comparison between SPB localization of Dbf2 (open squares) and Dbf2 kinase/Dbf2 protein (closed squares).
In 25% of bub2Δ cells that had formed a mitotic spindle during the hydroxyurea arrest, Dbf2 was found to be localized to at least 1 SPB. During mitosis, Dbf2 localized to SPBs in the majority of cells and remained localized at SPBs during the after G1 stage (Figure 8D). On release from the hydroxyurea block, Dbf2 kinase activity correlated well with the presence of Dbf2 on SPBs and appeared prematurely active during early stages of mitosis (Figure 8D). However, as cells exited, mitosis Dbf2 kinase activity decreased despite Dbf2 remaining on SPBs. Our results indicate that BUB2 is required to inhibit localization of Dbf2 on SPBs and activation of Dbf2 kinase activity during metaphase in an unperturbed cell cycle. Our findings further suggest that SPB localization is not sufficient for Dbf2 activation and that other factors are necessary for Dbf2 to be active as a protein kinase.
DISCUSSION
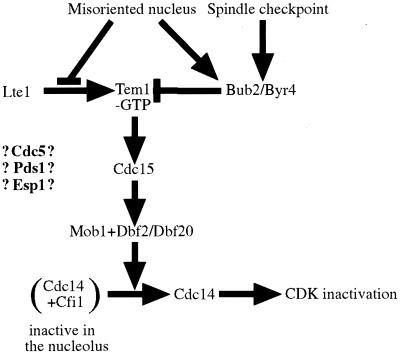
Our analysis of Cdc15 localization in various mitotic exit pathway mutants showed that Cdc15 localization to SPBs was independent of DBF2 and CDC14, but dependent on TEM1. A similar analysis of Dbf2 showed that Dbf2 localization to SPBs and Dbf2 kinase activity required TEM1 and CDC15 but not CDC14 function. In cells lacking BUB2, Cdc15 and Dbf2 were loaded onto SPBs prematurely and Dbf2 kinase activity was elevated. Previous studies have shown that Tem1 localization to SPBs is independent of CDC15, CDC5, DBF2, and CDC14 (Bardin et al., 2000). Together these findings suggest an order of function as to how Cdc15 is loaded onto SPBs and how Dbf2 localization to SPBs and Dbf2 kinase are controlled. BUB2 negatively regulates all 3 events. Furthermore, Cdc15 localization depends on TEM1 but not other mitotic exit pathway components, whereas activation of Dbf2 kinase activity and SPB localization depend on TEM1 and CDC15 but not CDC14. It is tempting to speculate that the dependencies observed for the loading of Cdc15 and Dbf2 onto SPBs and activation of Dbf2 kinase reflect the order of function in which these proteins function in promoting release of Cdc14 from Cfi1/Net1. In Figure 9, we propose a model where BUB2 negatively regulates the activity of the mitotic exit pathway and where TEM1 acts upstream of CDC15 and CDC15 upstream of DBF2. Similar dependencies have been found for the homologous pathway in S. pombe (Sparks et al., 1999). This pathway is termed septation initiation network and controls cytokinesis (reviewed in Balasubramanian et al., 2000). It is important to note that our findings do not imply direct activation of Cdc15 by Tem1-GTP or Dbf2 by Cdc15. Additional components of the mitotic exit pathway may await identification. It is also important to note that while our results clearly demonstrate an order of function in which Cdc15 and Dbf2 assemble onto SPBs and in which Dbf2 kinase is activated, we do not yet know whether association of these protein kinases with SPBs is critical for promoting release of Cdc14 from Cfi1/Net1. The analysis of mutants in Cdc15 and Dbf2 that can no longer associate with SPBs will be necessary to address this question.
Figure 9.
Order of function of the mitotic exit pathway - a model. Conversion of Tem1-GDP to Tem1-GTP causes activation of the mitotic exit pathway. The status of nucleotide binding of Tem1 is controlled by the nucleotide exchange factor (GEF) Lte1 and the 2 component GTPase activating enzyme (GAP) Bub2-Bfa1/Byr4. Mitotic spindle mis-orientation and mitotic spindle defects prevent Lte1 from activating Tem1 and protract Bub2/Byr4 activity causing Tem1 to be in the GDP-bound form. After nuclear division led to partitioning of the nucleus between mother and daughter cell, activation of Tem1 leads to recruitment of Cdc15 to SPBs which in turn causes recruitment of Dbf2 to SPBs and activation of Dbf2 kinase. Dbf2 kinase activity also requires Clb-CDK activity. How Clb-CDKs control Dbf2 activity is at present unclear. How CDC5 and the regulators of sister-chromatid separation PDS1 and ESP1 participate in this signal transduction pathway is also not known. Activation of the mitotic exit pathway eventually leads to release of Cdc14 from the nucleolus leading to inactivation of mitotic CDKs.
Our attempts to place CDC5 with respect to Cdc15 and Dbf2 localization onto SPBs were inconclusive, as Cdc15 and Dbf2 were localized to SPBs in 50% of cdc5–1 cells, but not in the other 50%. Dbf2 kinase activity was low in cdc5–1 mutants, however not as low as in tem1–3 mutants. These findings raise the possibility that CDC5 function is required at multiple steps during mitotic exit.
The Localization of Cdc15
Despite Cdc15 protein being present at constant levels throughout the cell cycle (Jaspersen et al., 1998), we and others found that Cdc15 only localized to SPBs during anaphase and telophase (Cenamor et al., 1999; Gruneberg et al., 2000; Xu et al., 2000; Menssen et al., 2001). However, there are slight variations in the localization pattern observed for Cdc15, which appears to depend on the epitope tag used in the different studies. Our localization data on endogenous Cdc15 are in agreement with previously published reports showing that Cdc15 localizes to both SPBs during anaphase and telophase (Xu et al., 2000). We also find that localization of Cdc15 to SPBs depends on the outer plaque component NUD1. In nud1–44 mutants, Cdc15 is largely delocalized from SPBs, as are Tem1 and Dbf2 (Bardin et al., 2000; Gruneberg et al., 2000; this study). Cdc15 localization was not affected in nud1–2 mutants (Gruneberg et al., 2000), indicating that different NUD1 alleles affect Cdc15 localization differently.
Regulation of Dbf2 Localization and Activity
Contrary to our observations, Frenz et al. (2000) showed that Dbf2 localizes to SPBs throughout the cell cycle. In this study, mutations in components of the mitotic exit pathway did not affect Dbf2 localization. We can envision several reasons or a combination thereof for these differences. (1) While Frenz et al. (2000) used strains containing 3 copies of a Dbf2-GFP fusion, we analyzed strains carrying a single copy of Dbf2 with 3 Myc epitopes. The employment of strains containing higher amounts of Dbf2 may cause constitutive association of Dbf2 with SPBs. This idea is consistent with the finding that in strains carrying a single copy of the DBF2-GFP fusion, the protein is found on SPBs only during anaphase and telophase (Frenz et al., 2000). (2) It is also possible that the signal generated by the Dbf2-Myc fusion is too weak or the Myc epitope is masked and, thus, escapes detection in cell cycle stages other than anaphase and telophase. However, we should note that we do detect Dbf2 on SPBs in cell cycle stages other than anaphase and telophase in cells lacking BUB2. Dbf2-GFP was also shown to localize to the bud neck during cytokinesis (Frenz et al., 2000). Why we fail to detect Dbf2 at the bud neck is at present unclear. However, it is worth noting that several proteins, such as Cap2, shown to be localized to the bud neck with the use of GFP fusion protein, cannot be detected with the use of conventional immunofluorescence protocols (Waddle et al., 1996; Lippincott and Li, 1998).
Although Dbf2 protein levels do not fluctuate during the cell cycle Dbf2 protein is only present at 1 or both SPBs during anaphase and telophase. The SPB localization of Dbf2 correlates with activation of Dbf2 kinase in synchronous cultures and various cell cycle arrests and mutant backgrounds. We also find that loading of Dbf2 onto SPBs does not require Dbf2 kinase activity. While these data suggest that localization of Dbf2 to SPBs, where Tem1 and Cdc15 localize, is important for Dbf2 kinase activation, it is also clear that it is not sufficient for Dbf2 kinase to be active. In cells lacking BUB2, Dbf2 protein is present on SPBs in G1 but Dbf2 is not active as a protein kinase. Only during metaphase is Dbf2 prematurely active in bub2Δ cells. The identity of these other factor(s) controlling Dbf2 activity is unclear. However, our results lead us to believe that 2 steps are necessary for Dbf2 to be active as a protein kinase. When the SPB enters the bud, the mitotic exit pathway components TEM1 and CDC15 not only recruit Dbf2 to SPBs but are also required for Dbf2 kinase to be active. In addition, a yet to be identified factor is required for Dbf2 to be active as a protein kinase.
The Spindle Pole Body—a Signaling Beacon
The GTPase Tem1 predominantly localizes to the SPB that migrates into the bud (Bardin et al., 2000; Pereira et al., 2000). It was, therefore, surprising that Cdc15 and Dbf2 localized to both SPBs and that localization to both SPBs depended on TEM1. It was also surprising that Dbf2 was present on only the mother cell SPB in some wild-type cells and almost exclusively present on the mother cell SPB in cdc14–3 mutants. How can we explain these observations? It is possible that Tem1 protein is present on the mother cell SPB but undetectable by indirect in situ immunofluorescence or by with the use of a Tem1-GFP fusion. It is also possible that Cdc15 and Dbf2 are somehow modified in a TEM1-dependent manner on the daughter cell SPB, which allows them to associate with the mother cell SPB. We also need to explain the finding that in some wild-type cells and cdc14–3 mutants, Dbf2 is found only on the mother cell SPB. One or more of the following ideas can explain this observation. (1) Dbf2 at the daughter cell SPB is more sensitive to fixation procedures or masked by other proteins. (2) In cdc14–3 mutants, the absence of Dbf2 from the daughter cell SPB may indicate that Cdc14 is required for efficient maintenance of Dbf2 localization on the daughter SPB. (3) We favor the idea that once Dbf2 is activated at the daughter SPB, it dissociates from this organelle and associates with its targets in other regions of the cell. This idea is consistent with the localization pattern of Sid2, the Dbf2 homolog in S. pombe, which is critical for the cytokinesis (Sparks et al., 1999). Sid2 first localizes to SPBs during mitosis and then localizes to the site of septation during septum formation.
Control of the Mitotic Exit Pathway
The activity of the mitotic exit pathway is sensitive to the status of a variety of different cellular events, such as nuclear position, integrity of the mitotic spindle, and onset of sister-chromatid separation (Figure 9; Alexandru et al., 1999; Cohen-Fix and Koshland, 1999; Fesquet et al., 1999; Fraschini et al., 1999; Li, 1999; Shirayama et al., 1999; Tinker-Kulberg and Morgan, 1999; Bardin et al., 2000; Bloecher et al., 2000; Daum et al., 2000; Pereira et al., 2000; Wang et al., 2000). Most of these signals are thought to control the nucleotide binding status of Tem1. Our analysis of Dbf2 kinase activity in cells lacking BUB2 suggests that Dbf2 activity is not only controlled by Tem1 and Cdc15 but by an as yet unknown factor. Furthermore, our and previous analyses of Dbf2 activity and localization in cells lacking BUB2 showed that there is at least 1 other factor controlling Cdc14 release from Cfi1/Net1, which functions downstream or in parallel to Dbf2. In bub2Δ cells, Dbf2 is present on SPBs already during metaphase and Dbf2 kinase activity is unusually high (Fesquet et al., 1999). However, only a small percentage of bub2Δ cells release Cdc14 prematurely during metaphase (Bardin et al., 2000). This result indicates that active Dbf2 kinase is not sufficient to trigger release of Cdc14 from Cfi1/Net1 and raises the interesting possibility that Cdc14 release for Cfi1/Net1 is regulated by factors other than mitotic exit pathway components. Determining the identity of these factors and the nature of signals they sense to regulate release of Cdc14 from Cfi1/Net1 will be critically important to understand how exit from mitosis is integrated with other cell cycle events.
ACKNOWLEDGMENTS
We thank John Kilmartin for generous gifts of strains and antibodies and review 1 for helpful comments. This work was conducted utilizing the W.M. Keck Foundation Biological Imaging Facility at the Whitehead Institute. We thank Frank Solomon, Dannel McCollum, and members of the Amon Lab for their critical reading of the manuscript. This research was supported by a National Institutes of Health grant GM-56800 to A.A.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandru G, Zachariae W, Schleiffer A, Nasmyth K. Sister chromatid separation and chromosome re-duplication are regulated by different mechanisms in response to spindle damage. EMBO J. 1999;18:2707–2721. doi: 10.1093/emboj/18.10.2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asakawa K, Yoshida S, Otake F, Toh-E A. A novel functional domain of Cdc15 kinase is required for its interaction with the Tem1 GTPase in Saccharomyces cerevisiae. Genetics. 2001;157:1437–1450. doi: 10.1093/genetics/157.4.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Surana U. Tying the knot: linking cytokinesis to the nuclear cycle. J Cell Sci. 2000;113:1503–1513. doi: 10.1242/jcs.113.9.1503. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Bloecher A, Venturi GM, Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- Cenamor R, Jimenez J, Cid VJ, Nombela C, Sanchez M. The budding yeast Cdc15 localizes to the spindle pole body in a cell-cycle-dependent manner. Mol Cell Biol Res Com. 1999;2:178–184. doi: 10.1006/mcbr.1999.0173. [DOI] [PubMed] [Google Scholar]
- Charles JF, Jaspersen SL, Tinker-Kulberg RL, Hwang L, Szidon A, Morgan DO. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr Biol. 1998;8:497–507. doi: 10.1016/s0960-9822(98)70201-5. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters J-M, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Koshland D. Pds1p of budding yeast has dual roles: inhibition of anaphase initiation and regulation of mitotic exit. Genes Dev. 1999;13:1950–1959. doi: 10.1101/gad.13.15.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daum JR, Gomez-Ospina N, Winey M, Burke DJ. The spindle checkpoint of Saccharomyces cerevisiae responds to separable microtubule-dependent events. Curr Biol. 2000;10:1375–1378. doi: 10.1016/s0960-9822(00)00780-6. [DOI] [PubMed] [Google Scholar]
- Donovan JD, Toyn JH, Johnson AL, Johnston LH. P40SDB25, a putative CDK inhibitor, has a role in the M/G1 transition in Saccharomyces cerevisiae. Genes Dev. 1994;8:1640–1653. doi: 10.1101/gad.8.14.1640. [DOI] [PubMed] [Google Scholar]
- Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- Fesquet D, Fitzpatrick PJ, Johnson AL, Kramer KM, Toyn JH, Johnston LH. A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J. 1999;18:2424–2434. doi: 10.1093/emboj/18.9.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R, Formenti E, Lucchini G, Piatti S. Budding yeast Bub2 is localized at spindle pole bodies and activates the mitotic checkpoint via a different pathway from Mad2. J Cell Biol. 1999;145:979–991. doi: 10.1083/jcb.145.5.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenz LM, Lee SE, Fesquet D, Johnston LH. The budding yeast dbf2 protein kinase localizes to the centrosome and moves to the bud neck in late mitosis. J Cell Sci. 2000;113:3399–3408. doi: 10.1242/jcs.113.19.3399. [DOI] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter T, Plowman GD. The protein kinases of budding yeast: six score and more. Trends Biochem Sci. 1997;22:18–22. doi: 10.1016/s0968-0004(96)10068-2. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Tinker-Kulberg RL, Morgan DO. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol Biol Cell, 1998;9:2803–2817. doi: 10.1091/mbc.9.10.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Jaspersen SL, Morgan DO. Cdc14 activates cdc15 to promote mitotic exit in budding yeast. Curr Biol. 2000;10:615–618. doi: 10.1016/s0960-9822(00)00491-7. [DOI] [PubMed] [Google Scholar]
- Kitada K, Johnson LA, Johnston LH, Sugino A. A multicopy suppressor gene of the Saccharomyces cerevisiae G1 cell cycle mutant dbf4 encodes a protein kinase and is identified as. CDC5. Mol Cell Biol. 1993;13:4445–4457. doi: 10.1128/mcb.13.7.4445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komarnitsky SI, Chiang Y-C, Luca FC, Chen J, Toyn JH, Winey M, Johnston LH, Denis CL. DBF2 protein kinase binds to and acts through the cell cycle-regulated MOB1 protein. Mol Cell Biol. 1998;18:2100–2107. doi: 10.1128/mcb.18.4.2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotani S, Tanaka H, Yasuda H, Todokoro K. Regulation of A.P.C. activity by phosphorylation and regulatory factors. J Cell Biol. 1999;146:791–800. doi: 10.1083/jcb.146.4.791. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Kramer ER, Scheuringer N, Podtelejnikov AV, Mann M, Peters J-M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol Biol Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott J, Li R. Sequential assembly of myosin II, an IQGAP-like protein, and filamentous actin to a ring structure involved in budding yeast cytokinesis. J Cell Biol. 1998;140:355–366. doi: 10.1083/jcb.140.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Winey M. MOB1, an essential yeast gene required for completion of mitosis and maintenance of ploidy. Mol Biol Cell. 1998;9:29–46. doi: 10.1091/mbc.9.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R, Neutzner A, Seufert W. Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr Biol. 2001;11:345–350. doi: 10.1016/s0960-9822(01)00095-1. [DOI] [PubMed] [Google Scholar]
- Morgan DO. Regulation of the APC and the exit from mitosis. Nat Cell Biol. 1999;1:E47–E53. doi: 10.1038/10039. [DOI] [PubMed] [Google Scholar]
- Pereira G, Hofken T, Grindlay G, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;13:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh-e A. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol Cell Biol. 1994a;14:7476–7482. doi: 10.1128/mcb.14.11.7476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Matsui Y, Toh-e A. Isolation of a Cdc25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 1994b;10:451–461. doi: 10.1002/yea.320100404. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Zachariae W, Ciosk R, Nasmyth K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998;17:1336–1349. doi: 10.1093/emboj/17.5.1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APCCdc20 promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Surana U, Amon A, Dowzer C, Mcgrew J, Byers B, Nasmyth K. Destruction of the CDC28/CLB kinase is not required for metaphase/anaphase transition in yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker-Kulberg RL, Morgan DO. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyn JH, Johnston LH. The Dbf2 and Dbf20 protein kinases of budding yeast are activated after the metaphase to anaphase cell cycle transition. EMBO J. 1994;13:1103–1113. doi: 10.1002/j.1460-2075.1994.tb06359.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traverso EE, Baskerville C, Liu Y, Shou W, James P, Deshaies RJ, Charbonneau H. Characterization of the Net1 cell cycle-dependent regulator of the Cdc14 phosphatase from budding yeast. J Biol Chem. 2001;276:21924–21931. doi: 10.1074/jbc.M011689200. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of CDK-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Visintin R, Hwang ES, Amon A. Cfi1 prevents premature exit from mitosis by anchoring Cdc14 phosphatase in the nucleolus. Nature. 1999;398:818–823. doi: 10.1038/19775. [DOI] [PubMed] [Google Scholar]
- Waddle JA, Karpova TS, Waterston RH, Cooper JA. Movement of cortical actin patches in yeast. J Cell Biol. 1996;132:861–870. doi: 10.1083/jcb.132.5.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu F, Elledge SJ. The Bfa1/Bub2 GAP complex comprises a universal checkpoint required to prevent mitotic exit. Curr Biol. 2000;10:1379–1382. doi: 10.1016/s0960-9822(00)00779-x. [DOI] [PubMed] [Google Scholar]
- Xu S, Huang HK, Kaiser P, Latterich M, Hunter T. Phosphorylation and spindle pole body localization of the Cdc15p mitotic regulatory protein kinase in budding yeast. Curr Biol. 2000;10:329–332. doi: 10.1016/s0960-9822(00)00382-1. [DOI] [PubMed] [Google Scholar]
- Yudkovsky Y, Shteinberg M, Listovsky J, Brandeis M, Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem Biophys Res Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]
- Zachariae W. Progression into and out of mitosis. Curr Opin Cell Biol. 1999;11:708–716. doi: 10.1016/s0955-0674(99)00041-1. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Nasmyth K. Whose end is destruction: cell division and the anaphase-promoting complex. Genes Dev. 1999;13:2039–2058. doi: 10.1101/gad.13.16.2039. [DOI] [PubMed] [Google Scholar]