Abstract
DNA-encoded libraries have emerged as a widely used resource for discovery of bioactive small molecules and offer substantial advantages compared to conventional small-molecule libraries. Here we developed and streamlined multiple fundamental aspects of DNA-encoded and DNA-templated library synthesis methodology, including computational identification and experimental validation of a 20×20×20×80 set of orthogonal codons, chemical and computational tools for enhancing the structural diversity and drug-likeness of library members, a highly efficient polymerase-mediated template library assembly strategy, and library isolation and purification methods. We integrated these improved methods to produce a second-generation DNA-templated library of 256,000 small-molecule macrocycles with improved drug-like physical properties. In vitro selection of this library for insulin-degrading enzyme (IDE) affinity resulted in novel IDE inhibitors including one of unusual potency and novel macrocycle stereochemistry (IC50 = 40 nM). Collectively, these developments enable DNA-templated small-molecule libraries to serve as more powerful, accessible, streamlined, and cost-effective tools for bioactive small-molecule discovery.
Keywords: DNA-templated synthesis, DNA-encoded libraries, macrocycles, in vitro selections, insulin-degrading enzyme
Graphical Abstract
The discovery of new bioactive small-molecule ligands remains a central endeavor of the life-sciences research community. Common small-molecule discovery approaches rely on screening large collections (libraries) of chemical compounds1. In a typical screening campaign, library members are individually assayed in separate locations for a desired biological activity, and therefore the time, effort, and expense associated with screening is proportional to the library size. While chemical library screening has yielded many important successes2, the development, maintenance, and high-throughput screening of large chemical libraries require infrastructure, resources, and logistics that are unavailable to most research groups3. Moreover, the discrete nature of screening assays can require prohibitively large quantities of unstable biological materials that need to be scaled up to match the size of the screened library. In contrast, selection methods evaluate an entire library in a single experiment, typically requiring an amount of biological material less than that of a single plate of a microtiter assay. Moreover, selections do not require infrastructure to separate, assay, or manipulate individual library members, and consume resources in a manner that is largely independent of library size.
DNA-encoded chemical libraries (DELs), mixtures of synthetic molecules that are each encoded by a covalently attached DNA tag, were developed to bring the advantages of selections and DNA sequencing to bear on biomedical targets that are best suited to synthetic small-molecule ligands. DNA encoding of chemical libraries was first proposed as a theoretical solid-phase peptide synthesis encoding strategy in 19924. The use of DNA encoding for general solution-phase small-molecule libraries suitable for in vitro selection was conceived and developed over the next decade5, 6. Since then, the field of selectable DNA-encoded libraries has greatly expanded to include a wide variety of small-molecule and synthetic polymer structures, as well as a number of different strategies to ensure the correspondence between a library member’s structure and the attached DNA barcode sequence, including DNA-templated synthesis, DNA routing, DNA tagging (ligation of DNA barcodes after each synthesis step), and variants and combinations of these concepts7–16. Selections using DELs are typically conducted by incubating an immobilized or epitope-tagged target with the library, washing unbound library members away from library members with target affinity, and isolating the latter by eluting or denaturing the target, or by adding an excess of a known ligand or free target15–17. The DNA sequences encoding enriched library members are typically amplified by PCR and analyzed by high-throughput DNA sequencing to identify the inferred structures of the active library members. DELs therefore enable rapid and inexpensive simultaneous testing of an entire library in one solution for binding to a target of interest and require only small amounts of biological material (~5–50 μg of a typical target protein per selection).
Despite these advantages, the vast majority of DNA-encoded libraries remain confined to pharmaceutical companies, and much of the research progress surrounding their development and use in industry remains undisclosed8. A number of original strategies to synthesize DNA-encoded libraries have been reported18–22. In some cases, these approaches enable the construction of libraries with vast theoretical sizes exceeding millions or even billions of compounds, with the trade-off that as library size increases, the fraction of the library components that can be confirmed to undergo anticipated reaction pathways decreases. Importantly, the quality of a DNA-encoded library is determined by the proportion of properly synthesized molecules that are correctly encoded by their DNA tags, and model studies demonstrated that library quality directly affects the reliability of selections results23, 24. In most cases, purification of products after each chemical coupling step is not viable, which results in truncated byproducts linked to DNA tags that contaminate or even dominate the finished library9. This limitation can become especially problematic when challenging chemical transformations such as macrocyclizations or coupling reactions using inefficient building blocks are part of the library design. As a result, the use of conventional approaches to generate high-quality DNA-encoded libraries of macrocycles can be a particular challenge, unless the bulk of the scaffold is pre-formed and combinatorial variation is limited to the introduction of substituents, a strategy that substantially limits library structural diversity.
The development of approaches that yield highly diverse libraries of DNA-encoded macrocycles25 represents a challenging goal that can potentially provide access to underexplored chemical space. The potential of such libraries is further highlighted by the favorable biomedical properties of macrocyclic molecules26–30. Macrocycles are generally known to display better stability in vivo than their linear counterparts27, 30, and to offer a balance between flexibility and pre-organization that allows macrocycles to interact across extended protein binding sites with entropic penalties that are lower than corresponding linear molecules. The latter feature renders them promising in targeting surfaces or protein-protein interactions, which can be difficult to target with conventional small-molecules libraries31, 32. Indeed, approximately 70 macrocyclic drugs have already been approved for human use and more than 35 macrocycles are in various stages of clinical development33.
Beginning in 2001, we developed DNA-templated synthesis (DTS) as a strategy to bring the substantial strengths of reactivity programming, in vitro selection, PCR amplification, and DNA sequence analysis to the synthesis and evaluation of synthetic molecules5, 34, 35. DNA-templated synthesis is based on the principle that highly diluted DNA-tagged reactants experience a greatly increased effective molarity upon DNA hybridization36. This phenomenon allows many independently DNA-programmed reactions to take place simultaneously in the same solution in a highly selective fashion37, so that products are formed only between reactants linked to complimentary DNA sequences. A diverse set of reactions have been successfully validated in a DNA-templated manner36, 38 and a broader toolbox of DNA-compatible chemistry is steadily growing.8, 39, 40 Our group has applied this concept to create libraries of DNA-templated small molecules6, in which the DNA tags not only function as barcodes, but also as templates that orchestrate the synthesis of each library member.
The first discovery-oriented DNA-templated small-molecule library contained up to 13,824 macrocyclic molecules (Fig. 1)41. This DNA-templated macrocycle library was notable for the use of DNA hybridization to assist macrocyclization, the development of a final DNA-templated reaction step that simultaneously results in a one-pot purification of the library, thereby eliminating truncated and uncyclized byproducts, and compatibility with macrocycles of variable sizes and structures. Despite its modest size compared with subsequent industrial DNA-encoded libraries9, 11, 42, this initial library was of sufficient quality and diversity to serve as a source of potent and selective inhibitors of proteins including kinases and insulin-degrading enzyme (IDE) protease, ultimately leading to biological discoveries and the validation in vivo of new targets for therapeutic intervention.43–46
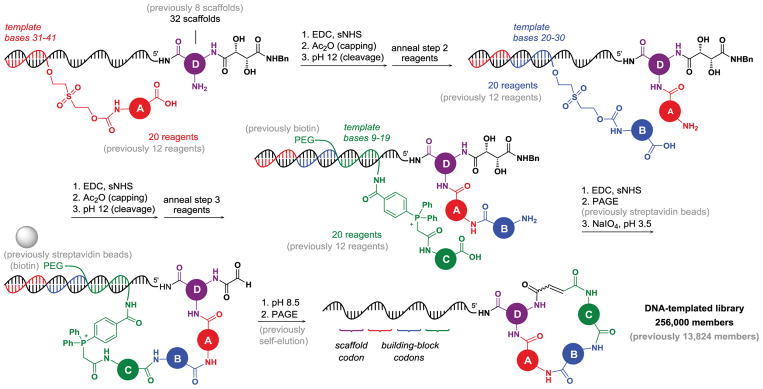
Figure 1. DNA-templated macrocycle library synthesis scheme.
Key aspects of the previously described first-generation (grey) and second-generation (black, color) library syntheses are shown. In the first step, scaffold building block D attached to 5′ end of the template undergoes coupling with building block A, which is initially attached to the corresponding “anticodon” DNA via a cleavable bis(2-(succinimidooxycarbonyloxy)ethyl) sulfone (BSOCOES) linker. Unreacted templates are capped with acetic anhydride. The linker is cleaved at high pH, liberating the amino group of building block A, which subsequently undergoes the step 2 coupling with building block B followed by capping and linker cleavage. After coupling to biotin- or PEG-labeled Wittig reagent building block C, pulldown with streptavidin-tagged beads (first-generation procedure) or gel purification (second-generation procedure) enables isolation of those templates that successfully reacted at all three steps. Periodate treatment cleaves the diol fragment of the tartaramide moiety to furnish a glyoxyloyl group, which undergoes Wittig cyclization under mildly basic conditions. Successfully cyclized products are eluted off the beads on cyclization (first-generation procedure) or are purified on a polyacrylamide gel (second-generation procedure).
In this study, we substantially improved and streamlined virtually every aspect of DNA-templated library technology and we feature the resulting advances to generate a larger, more diverse, and more drug-like 256,000-membered DNA-templated macrocycle library. As a test of the ability of this second-generation DNA-templated library to enable the discovery of bioactive macrocycles, in vitro selection of the library yielded potent and structurally unique macrocyclic inhibitors of insulin-degrading enzyme (IDE). These methodological advances collectively represent the state-of-the-art in DNA-templated library synthesis and provide improved access to a rich set of diverse, drug-like molecules.
RESULTS
General Design of the DNA-Templated Library Architecture
The DNA-templated library synthesis is summarized in Fig. 1, with changes compared to the first-generation library shown in grey41. The template architecture of the library is shown in Fig. 2a. The coding region is flanked with 10-mer primer-binding sites and consists of three building block codons and a scaffold codon interspaced with three constant regions. Codons 1, 2, and 3 determine the identity of three macrocycle building blocks introduced by DTS, while codon 4 identifies the bis-amino acid scaffold at the 5′ end of the template. After each templated coupling reaction, unreacted templates are capped by acetylation (Fig. 1). Capture with streptavidin-linked beads separates templates that successfully reacted at all three steps from those that failed to react at any step. During macrocyclization, the library is purified again by a capture-and-release strategy that causes successfully macrocyclized DNA-linked library members to self-elute from beads, whereas uncyclized material remains bound. This capping and macrocycle purification strategy furnishes material of sufficiently high purity to support DNA-encoded library selections and accurate post-selection decoding9.
Figure 2. Identification of an orthogonal codon set for second-generation DNA-templated libraries.
a, General architecture of second-generation template libraries. Consecutive Ns do not represent randomized sequences but indicate the location of individual codons. b, The coding system for the second-generation library. c, Proposed model of DNA templates used to calculate an orthogonal codon set. d, The ideal outcome of DNA-templated synthesis codon reactivity tables (1). Numbers represent apparent conversions of reactions between the corresponding DNA templates (horizontal) and DNA-linked reagents (vertical). Green and purple fields represent apparent conversions and annealing factors, respectively, that are acceptable because they correspond to mismatched reactivity below the 7% threshold. e, Deconvolution approach based on the model of additive annealing factors (7): experimentally obtained reactivity tables (3) are converted into anticipated affinity tables (4), which are refined with additional DTS reactions (5). Geometrical shapes represent various codons and anticodons; equations 2 and 5 denote apparent conversions of the corresponding DTS reactions (α, β, γ). See the Supplementary Information for details of the deconvolution process leading to the final codon set.
Identification of an Orthogonal Codon Set
One factor that limits the size of DNA-templated libraries is the requirement of codon orthogonality. A DTS reagent’s anticodon must efficiently anneal only with the corresponding complimentary codon of the template. Moreover, the template requires a certain degree of secondary structure in order for the hybridized reacting groups to experience optimal effective molarity47. To design the codon set for the second-generation DNA-templated macrocycle library, we began with a set of 30 × 30 × 30 putatively orthogonal codons that we previously derived41 computationally to impart template folding energies in the range we found to be optimal for DNA-templated synthesis47. All codons were designed to have identical 50% G/C content to minimize melting temperature differences. We used the Visual OMP platform (DNA Software, Inc.) to identify a set of 30 scaffold codon candidates out of 256 possible sequences of the form NNNN that avoided hairpin formation with the adjacent codons and minimized predicted off-target hybridization to reagent anticodons (Fig. 2a, b). The resulting building block and scaffold codons were arbitrarily assigned number and letter codes (Supplementary Table 1). Codons involved in DTS steps 1, 2, and 3 were designated codons 1, 2, and 3 respectively, while the scaffold codon was defined as codon 4 (Fig. 2a).
We synthesized and purified 90 DNA-linked phenylalanine model reagents, each containing one of the 90 different anticodon oligonucleotides (1a…1z, 1ww…1zz, 2a…2z, 2ww…2zz, and 3a…3z, 3ww…3zz), and 30 DNA templates (3a-2a-1a-4a … 3zz-2zz-1zz-4zz) that collectively contain codons for all 90 reagents in order to validate all possible codon-anticodon combinations for their ability to support efficient and sequence-specific DTS. We performed 2,700 individual DNA-templated amine acylation reactions between each of the 90 DNA-linked model reagents and each of the 30 test templates that collectively contain all 90 possible building block codons and all 30 possible scaffold codons. Based on our previous work41, we chose a threshold of 7% or greater conversion of non-complementary reagent and template as unacceptable. The DTS reactivity tables for codons 1 and 2 obtained at the previously used temperature regimes (25 °C for steps 1 and 2, and 37 °C for step 3)41 resulted in prohibitively high levels of mismatched cross-reactivity, with 31% of mismatched step 1 reagent-template combinations and 22% of mismatched step 2 combinations yielding apparent DTS conversions above the 7% threshold at 25 °C (Supplementary Table 3).
Therefore, we repeated the set of reactions at elevated temperatures (30 °C for steps 1 and 2), resulting in a substantial reduction in cross-reactive mismatched reagent-template combinations for DTS steps 1 and 2 (23% and 16%, respectively) (Supplementary Table 4). While elevating the temperature of step 3 to 43 °C dramatically reduced the frequency of unacceptable mismatched product formation from 5.3% to 0.1%, the yields of matched reactions also decreased substantially from 92% to 53% average apparent conversion (Supplementary Table 4). As a result, temperatures of 30 °C, 30 °C and 37 °C were chosen for DTS reactions 1, 2, and 3, respectively. Despite these sequence specificity improvements, the remaining number of templates not involved in any mismatched conversions provided an insufficient number of codons to support the DTS of 256,000 macrocycles (Supplementary Tables 3 and 4).
Mismatched product formation likely arises from a single problematic codon:anticodon combination, and thus some codons in excluded templates were likely innocent bystanders that did not contribute to mismatched product formation. We sought to identify the smallest possible set of problematic codons that, once removed from the codon pool, would enable all remaining reagent-template combinations to satisfy the above orthogonality criteria. To identify the problematic codons, we assumed a model in which each template behaves as a chain of four independent codons (Fig. 2c) and that contributions of DNA hybridization between a given anticodon and each of the four codons to reaction conversion were additive. These assumptions allowed us to convert experimental reactivity tables (Supplementary Tables 3 and 4) into an anticipated “annealing factor” table that assigns the expected contribution of each individual codon-anticodon hybridization to overall conversion (Supplementary Table 5). This process is summarized in Fig. 2e. For each case of a template-reagent combination that resulted in unacceptable mismatched product formation, we designed and synthesized new templates containing each of the four original codons in a different surrounding codon context and performed new DTS reactions with the original reagent. The resulting iterative deconvolution used 80 templates and 1,890 additional DTS reactions (Supplementary Tables 7–9), and resulted in the refinement of annealing factors for 1,372 codon-anticodon pairs initially identified as potentially problematic. The refined annealing factors confirmed that 813 of these codon-anticodon pairs do not cause ≥ 7% mismatched product formation, which substantially contributed to the identification of a maximum set of orthogonal codons (Supplementary Table 8).
We excluded the most promiscuous codons by inspection from further consideration, resulting in the removal of 7, 5, and 1 codons from reactions 1, 2, and 3, respectively. The least promiscuous codons (12, 15, and 6 codons from reactions 1, 2, and 3, respectively, and all 30 scaffold codons), showing no mismatched reactivity were directly included into the final orthogonal codon set. The remaining 44 “grey-area” codons (Supplementary Table 11) could not be excluded or included by inspection because their suitability was mutually dependent on the inclusion or exclusion of other grey-area codons and instead were further analyzed by a computational approach. We developed a mathematical model in which the presence (1) or absence (0) of each of the remaining 44 codons was represented by a binary digit in a 44-digit binary string, which was divided into three sub-strings corresponding to each codon group. Each of the possible binary sub-strings (211, 210, and 223 sub-strings for codon group 1, 2, and 3 respectively) representing a candidate set of viable codons was scored computationally using the annealing factor table to identify the number of incompatible sequence pairs contained within each codon set (see Supplementary Fig. 1 for details). The three pools that passed the filter were then combined and subjected to an identical refinement process. The codon set containing the minimum number of problematic reagent-template combinations (those predicted to result in ≥ 7% conversion) contained 27 of the 44 grey zone codons (Supplementary Table 12) and was added to the previously accepted subset of 12, 15, and 6 codons for reactions 1, 2, and 3, respectively. The resulting orthogonal codon set contained 20 × 20 × 20 × 30 codons for reactions 1, 2, 3, and the scaffold, respectively. After separate validation of additional two scaffold codons with 2 × 60 DTS reactions, we obtained a final orthogonal 20 × 20 × 20 × 32 codon set (Supplementary Tables 13–14) capable of encoding 256,000 unique DNA-templated reaction products. This final codon set was renamed as 1A…1T; 2A…2T; 3A…3T; and 4A…4Z, 4UU…4ZZ (Fig. 3, Supplementary Tables 13 and 14).
Figure 3. Building blocks for the second-generation DNA-templated macrocycle library.
a, Synthetic routes enabling incorporation of new scaffold structures into DNA templates, exemplified with scaffolds 4I and 4L. b, Scaffolds validated and used in the second-generation library of macrocycles. Red and green spheres represent connectivity with building blocks 1 and 3, respectively. Scaffolds 4A–4H (dashed boxes) were used in the first-generation library. c, Iteratively selected building blocks maximizing overlap of the library with Kihlberg’s parameter space for orally bioavailable molecules55, 57.
To validate the final orthogonal codon set, we re-analyzed the results of 4,068 DNA-templated reactions performed at the optimized temperatures collectively involving all of these codons and tested if the empirical conversion data matched the result predicted by the final annealing factor table. The predicted apparent conversions of only 178 of the 3,929 mismatched reactions (4.5%) were substantially (> 50%) different from the observed experimental values, out of which only 108 (2.7%) corresponded to selected codons, suggesting the validity of the codon set and the codon derivation methodology (Fig. 2c). Finally, we noticed that the scaffold codon is rarely problematic in our DTS architecture due to its distal location. Therefore, we repeated the in silico codon analysis described above including only the 20 + 20 + 20 final codons encoding reagents for steps 1, 2, and 3 resulted in the identification of an additional 48 scaffold codons predicted to not interfere with codon orthogonality (Supplementary Table 17). These additional scaffold codons expand the theoretical capacity of future DNA-templated libraries of this format to 640,000 members.
Expanding the Diversity of Macrocycle Scaffolds
We sought to expand the functional and stereochemical diversity of simple bis-amino acid scaffolds41, which were previously chosen based on the commercial availability of Fmoc- and trityl-protected derivatives suitable for on-bead DNA conjugation. Previously, we were unable to use Boc-protected bis-amino acids as scaffolds due to DNA-incompatible deprotection conditions. We found that a 1-minute exposure to 50% trifluoroacetic acid in dichloromethane deprotects Boc-functionalized scaffolds and yields sufficient amounts of intact DNA for subsequent DNA-templated library synthesis steps. (Fig. 3a and Supplementary Fig. 4). We confirmed that these conditions did not isomerize a variety of candidate new scaffolds (Supplementary Table 28), allowing us to add 12 aminomethyl phenylalanine scaffolds and four aminoprolines in addition to 8 stereoisomers of previously used scaffolds. These additions expanded the set of scaffolds from 8 used in our original library41 to 32 (Fig. 3b and Supplementary Table 18) and also substantially increased the structural diversity of the resulting library.
Selection of Building Blocks to Improve Cell Permeability
Lipinski and coworkers developed guidelines commonly known as “the rule of 5”, which postulates that a molecule is more likely to be orally active (and, by inference, cell-permeable) if molecular weight, octanol/water partition coefficient (LogP), numbers of hydrogen bond donors, and number of hydrogen bond acceptors lie within the ranges listed in Table 1.48 Additional limitations for the number of rotatable bonds and polar surface area were subsequently introduced49. Multiple examples of orally bioavailable molecules violating rule-of-5 principles, especially including macrocycles33, 50–54, have led researchers including Kihlberg and co-workers to develop alternative, expanded guidelines (MW ≤ 1,000 Da, # of H-bond donors below 6; # of H-bond acceptors below 15, cLogP from -2 to 10, # of rotatable bonds below 20, polar surface area below 250 Å2) that are especially relevant to macrocyclic molecules such as those in our DNA-templated libraries33, 53–57 (Table 1).
Table 1.
| Parameter | Lipinski | Kihlberg |
|---|---|---|
| molecular weight | < 500 Da | < 1000 Da |
| cLogP | 0 < × <5 | −2 < × < 10 |
| # hydrogen bond donors | < 5 | < 6 |
| # hydrogen bond acceptors | < 10 | < 15 |
| # rotatable bonds | < 10 | < 20 |
| polar surface area | < 140 Å2 | < 250 Å2 |
We chose building blocks for the second-generation library such that the resulting macrocycles are consistent with Kihlberg rules55 in order to increase the likelihood of compatibility with cell-based assays and to facilitate subsequent hit-to-lead optimization. We developed a method to calculate the influence of any building block candidate on the predicted Kihlberg conformity of the resulting library using widely available chemistry software (ChemBioOffice from CambridgeSoft). We designed code for the VBA platform (an integrated part of Microsoft Office) that generates SDF files, a widely used structure-data file format, containing the building block connectivities of all 256,000 macrocycles. We programmed ChemBioDraw to recognize the letter codes of a given selection of building block candidates and used this software to convert SDF files into drawn chemical structures. A VBA program then exported the resulting files into ChemFinder, which calculated the Kihlberg parameters. We iteratively optimized the set of building blocks to comply with Kihlberg’s guidelines through minimization of the number of highly polar functional groups and hydrogen bond donors, as well as liberal use of N-alkylated amino acids (Fig. 3c and Supplementary Table 19).
In order to access underexplored macrocycle chemical space, we also introduced building blocks with increased steric bulk or reduced conformational freedom compared to standard alpha amino acids, including alicyclic (1O, 1R, 1S, 2K, 2N, 2P, 3B, 3C, 3D, 3F, 3G, 3J, 3K, 3M), aromatic (1J, 1L, 1M, 1N, 1T, 3E, 3H, 3L, 3R), and spirogenic (1I, 2E, 2Q, 2S, 3N, 3O, 3Q) building blocks. Amino acids with less nucleophilic nitrogen centers were mostly used in reaction 3, since the corresponding amide bond is not formed through DNA-templated amine acylations requiring nucleophilic amines (Fig. 1). To maximize library diversity and take full advantage of DNA-templated macrocyclization, we chose building blocks that include α (29 building blocks), β (12), γ (8), δ (7) and ε+ (4) amino acids. Likewise, we incorporated a comparable number of building blocks from both L- and D-amino acid pools for each structural type (13 and 12 amino acids, respectively).
To maximize the quality of the resulting library, all candidate building blocks not previously tested were validated in model single-macrocycle DNA-templated syntheses and only those that provided at least 30% yield of coupling product (typically 50–80%) and at least 45% yield of cyclization (typically 80–90%) were considered further. The final sets of selected scaffolds and building blocks are shown in Fig. 3b and 3c.
The resulting final macrocyclic products were calculated to possess bioavailability-correlated parameters that are greatly improved compared to our first-generation DNA-templated macrocycle library (Fig. 4). The difference is particularly striking for cLogP, polar surface area, and the number of hydrogen bond donors. In addition, the methodology developed here enables rapid generation of large virtual libraries using widely available, economical software and thus could assist the broader small-molecule library research community (see Supporting Information for programming code and detailed protocols).
Figure 4. Distribution of physical parameters among library members from the second-generation macrocycle library (above the X-axis) and the first-generation library (below the X-axis).
Colors represents values that lie within (blue) or outside (red) desirable “beyond rule-of-five” (bRo5) parameter space described by Kihlberg and coworkers55, 57.
Novel DNA Template Assembly Methodology
The previously established strategy of assembling the library of DNA templates used split-pool oligonucleotide synthesis of phosphorylated 3′ fragments, followed by enzymatic splint-assisted ligation with chemically modified 5′ fragments41. Applying the same approach to the preparation of a 256,000-membered library would require many more oligonucleotide syntheses and split-pool events; for example, 1,280 vs. 192 oligonucleotide syntheses alone would be required for the preparation of the 3′ fragment (Fig. 5a). Splitting the template into three parts rather than two (Fig. 5b) could mitigate the problem, however, we sought to provide a more convenient template library assembly to popularize application of DNA-templated libraries. We sought to reduce the number of required manipulations, enable quality control before the final stages of the library assembly, avoid the use of splint ligations, which are inconvenient on preparative scale, and enable template library synthesis components to be reused wherever possible for subsequent library preparation efforts. Furthermore, we sought to eliminate the need to isolate and characterize complex mixtures of chemically modified oligonucleotides, which is problematic in the case of low-yielding reactions with multiple by-products (such as those involving some of the novel scaffolds). We therefore developed a novel approach to template library assembly based on polymerase-mediated extension of chemically modified primers.
Figure 5. Approaches to the assembly of DNA template libraries.
a, Assembly of the first-generation library of DNA templates. For each scaffold codon, a sub-library of templates was previously assembled via splint ligation of phosphorylated 33- or 34-mers (generated on a DNA synthesizer in a split-pool manner) and 21-mers chemically modified with the scaffold amino acid. b, Modified version of the splint ligation assembly for the second-generation DTS library. Increasing the number of ligated fragments from two to three greatly reduces the number of required oligonucleotide syntheses. c, Template library assembly strategy via preparative enzymatic primer extensions. An 8,000-membered library of templates with four deoxyinosines at the scaffold codon is prepared by split-pool oligonucleotide synthesis. Each primer extension with one of 32 poly-dA-tagged primers followed by strand separation via PAGE yields a heavy strand sub-library with an individual scaffold codon sequence. Another round of primer extensions with the corresponding chemically modified primers followed by strand separation results in 32 sub-libraries of templates, which are combined to obtain a 256,000-membered template library. A shortened method involves direct preparation of the heavy strands by split-pool oligonucleotide synthesis. Methods for template assembly are described in detail in Supplementary Fig. 8.
For a 32×20×20×20 library this route would involve separate primer extensions of thirty-two 8,000-membered libraries with different scaffold codons. To avoid synthesizing multiple initial libraries, we exploited the ability of deoxyinosine to pair in vitro with all four natural nucleobases.58 Indeed, a creative way of using deoxyinosine in DNA-templated synthesis has been recently reported.20 We reasoned that a 256,000-membered template library could be generated from a single universal 8,000-membered starting library (tetradeoxyinosine library or I4 library, Supplementary Fig. 5–7) by allowing the 32 scaffold codons to each hybridize to the I4 region of a DNA template containing codons 1, 2, and 3 in a primer extension reaction (Fig. 5c). For each of the 32 primer extensions, the identity of the scaffold on the 5′-scaffold-linked primer is encoded by the sequence information introduced by the other primer (Fig. 5c). After extensive experimentation, we found that the I4 template could be successfully converted to the desired library by consecutive primer extensions with Klenow(exo–) fragment of DNA polymerase I and Vent polymerase. We also found that appending a sufficiently long oligonucleotide tail (e.g. A30) on one primer allows separation of the two product strands (55-mer light strand and 55-mer + 30-mer tail heavy strand) in a library format using denaturing PAGE. These results together provide streamlined access to libraries of single-stranded DNA templates suitable for DTS (Fig. 5c and Supplementary Fig. 8).
Improved Synthesis and Recovery of DNA-Templated Libraries
We developed a solution-phase alternative to the on-bead macrocyclization of immobilized DTS intermediates (Fig. 1). Instead of using a biotin group to capture intermediates prior to macrocyclization, we equipped each reagent 3 oligonucleotide with 18 ethylene glycol units and developed an efficient PAGE purification protocol for intermediates that successfully reacted in all three DTS steps (Supplementary Fig. 11). The macrocyclization step occurs in solution, and macrocyclized products are separated from uncyclized intermediates by PAGE isolation. This strategy allowed more accurate control over library preparation and avoided uncertainties associated with solid-phase capture and heterogeneous on-bead reactions. Moreover, this solution-phase approach enables library syntheses on nmol to μmol scales, which would previously have required prohibitive quantities of expensive streptavidin-conjugated magnetic beads.
To isolate template-linked macrocycles or intermediates from dilute solutions with minimal losses we developed a simple chaotropic buffer (4 vol. saturated aqueous guanidine hydrochloride + 6 vol. isopropanol) that efficiently promotes the association of DNA-linked species with commercially available silica membranes such as Omega HiBind or Qiagen Qiaquick columns. For example, we were able to achieve 99% recovery and 50-fold concentration of 4.8 nmol of single-stranded 55-mer oligonucleotide from a dilute (120 nM) solution. This methodology has proven instrumental for DTS, which requires multiple isolations of dilute short oligonucleotide-linked products that were previously recovered by less reliable alcohol precipitation41. Importantly, this approach also enables efficient recycling of DNA-templated libraries from in vitro selections, as the vast majority of library members (both target binders and non-binders) end up in dilute flowthrough and wash solutions, from which they can be salvaged using the chaotropic buffer and silica membranes. For example, we were able to recover 867 pmol (51%) of the final DNA-templated macrocycle library described below from the combined flowthrough volumes of 98 selections (averaging 17 pmol library each). The purity of the recovered material as evaluated by PAGE was very similar to that of freshly made library, and selections for target protein binding described below yielded qualitatively similar selection results using freshly synthesized or recovered library (Supplementary Fig. 20). This recycling capability greatly reduces the resources expended in each DNA-templated library selection and should also facilitate the recycling of other DNA-encoded libraries.
DNA-templated synthesis of a library of 256,000 macrocycles
To confirm that the second-generation DNA-templated library synthesis methodology and materials generate compounds consistent with the target macrocycles, we subjected 20×1×1×1 and a 1×1×20×1 template subsets of the library to the DNA-templated library synthesis methodology, followed by removal of DNA templates with S1 nuclease to afford macrocycles made conjugates to a guanine nucleotide. MALDI mass spectrometry revealed product masses consistent with the presence of 32/40 expected macrocycles (Supplementary Fig. 14). These results confirmed the ability of the DNA-templated library synthesis methodology to generate expected macrocycles, as we have previously shown.41, 43, 45
The second-generation DTS library of macrocycles was prepared by integrating the above methodologies. The DNA template library was generated by two sequential series of 32 primer extensions/PAGE purifications (Fig. 5c) starting with 32×50 nmol of 8,000-membered universal library of I4 templates and yielding 250 nmol of the 5′-scaffold modified template library. The improved DNA-templated synthesis protocol with two sequential PAGE purifications allowed isolation of the final macrocycle library in a total yield of 1.5% relative to the DNA template library entering the process. Assuming two regeneration cycles per library member, this library synthesis (2 × 1.83 nmol) is sufficient to conduct > 300 selections using a validated quantity of 20 pmol library per selection (see below). Importantly, the developed methodology enables facile scale-up of the library synthesis, as well as swapping of building blocks or scaffolds in subsequent library syntheses.
High-throughput DNA sequencing of the final library revealed the presence of 255,954 (>99%) library member templates. We observed a distribution of DNA sequences in the final library that was consistent with the anticipated reactivities of individual building blocks and the expected efficiency of macrocyclization. For example, large and flexible scaffolds, which are expected to result in the most facile cyclizations (α-Lys, 4H, 4X) were more highly represented than structures expected to macrocyclize less efficiently (α-Dap, 4E, 4U; aminoprolines 4O, 4P, 4YY, 4ZZ). Likewise, N-alkylated amino acids and other building blocks predicted to be less reactive also were found in lower representation of the library (Supplementary Fig. 12 and 13).
In vitro selection and validation of the library of 256,000 macrocycles
We chose insulin-degrading enzyme (IDE) as a protein target for library selection and validation. From our first-generation DNA-encoded macrocycle library41, we previously identified macrocycles 6b and 5b containing D-4-benzoylphenylalanine and L-3-cyclohexylalanine as potent ligands and inhibitors of IDE45. We performed in vitro selections for IDE binding using the 256,000-membered macrocycle library. His-tagged IDE (10 μg) was immobilized on 25 μL of magnetic Dynabeads, treated with yeast total RNA to minimize non-specific binding to DNA templates, and incubated with 1 to 20 pmol macrocycle library in TBST buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 0.05% Tween-20) for 1 h. Three washes with TBST were followed by elution with 300 mM imidazole in TBST. The eluate was directly used in PCR reactions introducing adapter sequences and barcodes for high-throughput sequencing (Illumina MiSeq and NextSeq). Selections were highly reproducible using 20 pmol of library (Supplementary Fig. 20), which corresponds to an amount of each library member less than or similar to our previously reported selections using 5 pmol of the 13,824-membered DNA-templated macrocycle library43, 45.
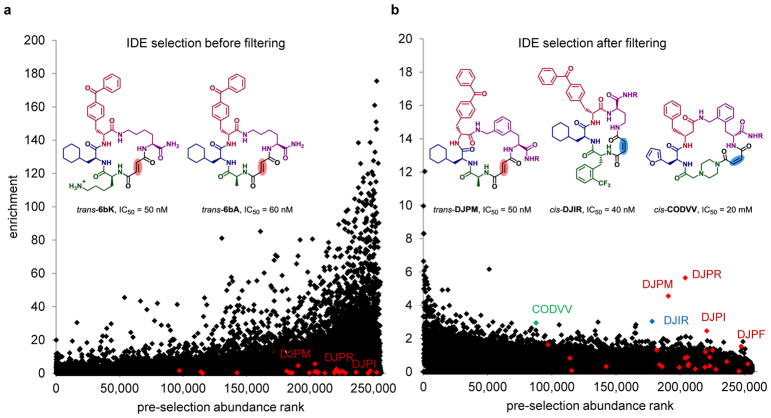
The initial raw IDE selection results revealed several building blocks (1J, 1L, 1M, 1N, 1T, 3E, 3H, 3L, 3R) that consistently demonstrated unusually high enrichments across all amounts of library tested (Supplementary Fig. 15 and 16). We hypothesized that these building blocks formed excessively hydrophobic macrocycles prone to IDE binding, possibly as promiscuous aggregators59. Indeed, analysis of in vitro selections of the 256,000-membered library on unrelated proteins revealed that those building blocks introducing substituted phenyl rings into the macrocycle backbone were unusually represented among non-specific hits. Plotting the selections results after computational filtering of the nine building blocks highlighted in Supplementary Table 37 (1J, 1L, 1M, 1N, 1T, 3E, 3H, 3L, 3R) greatly reduced background binding and restored the normal enrichment range and distribution (Fig. 6b). The most strongly enriched macrocycles after this filtering step shared the codon combination of the form DJP(*), which encode structures closely resembling a previously discovered family of IDE-inhibiting macrocycles including 6b and 5b.
Figure 6. In vitro selection of the 256,000-membered DNA-templated macrocycle library for binding to insulin-degrading enzyme (IDE).
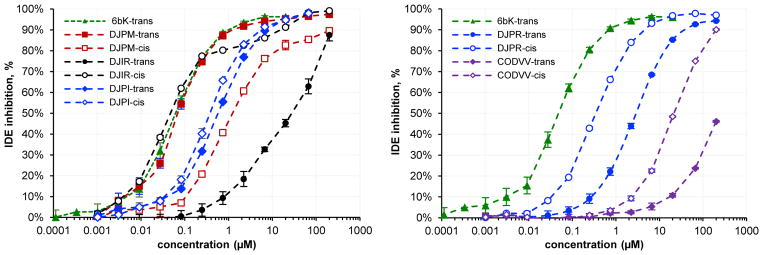
a, b, Results of the selection against IDE before (a) and after (b) computational filtering of nine structurally similar hydrophobic building blocks (1J, 1L, 1M, 1N, 1T, 3E, 3H, 3L, 3R) that were unusually represented among hits across multiple unrelated selections. Removal of the substantial non-specific noise revealed an enriched DJP* series of macrocycles. Compounds trans-DJPM and cis-DJIR were chemically synthesized in a DNA-free form and were found to be equipotent to the structurally similar trans-6bK and trans-6bA macrocycles developed from the first-generation DNA-templated library.45 The identified hits also included unrelated CODVV macrocycles of a new structural family. R = (CH2)2O(CH2)2NH2. c, Concentration-dependent IDE inhibition profiles of macrocyclic hits determined by fluorogenic decapeptide cleavage assay (see the Supplementary Information). Error bars reflect to standard error of the mean. The plots for a cis- and a trans-isomer of each hit are of the same color and marker shape, with filled markers for trans-isomers, and empty markers for cis-isomers). Whereas DJPM trans isomers were more potent than cis isomers), the opposite trend was observed for other tested hits.
To test if these new hits from the in vitro selection of the 256,000-membered library represent bona fide IDE inhibitors, we synthesized several of the corresponding cis- and trans-macrocycles (DJPR, DJPM, DJPI, DJIR, CODVV) in a DNA-free format and assayed their ability to inhibit IDE activity. All tested hits demonstrated inhibition of IDE over a range of concentrations using a fluorogenic decapeptide cleavage assay (Fig. 6b,c and Supplementary Fig. 19). Notably, the 21-membered trans-DJPM macrocycle (Fig. 6b) is comparable in potency to our previously optimized 20-membered inhibitor 6bK45 (IC50 = 50 nM, Fig. 6c) and is more potent than the original lead compound 6b45. We also observed enrichment of the related macrocycle DJPI, which features an unusual ortho-substituted backbone (cis/trans IDE IC50 = 400 nM/600 nM). Smaller, 18-membered DJPR macrocycles were also less potent (cis/trans IC50 = 400 nM/2 μM) than DJPM, consistent with our previous characterization of the crystal structure of IDE bound to related DNA-templated macrocycles45. Weak inhibition was observed for unrelated 24-membered CODVV macrocycles encoding a new structural family (cis/trans IC50 = 30 μM/>100 μM).
Interestingly, whereas trans-isomers of all previously screened IDE inhibitors45 were much more potent than their cis-analogs, CODVV and DJ*R families demonstrated the opposite stereochemistry-activity relationship. Cis-DJIR (IC50 = 40 nM) was found to be at least as potent as 6bK and thereby serves as the first example of a highly potent macrocyclic IDE inhibitor containing a backbone alkene with cis configuration. Together these results validated the new library and demonstrated the ability of the DTS library of macrocycles to identify new ligands for targets of biomedical interest, as well as to provide new structure-activity insights that facilitate medicinal chemistry efforts.
DISCUSSION
We developed and synthesized a second-generation DNA-templated and DNA-encoded library of 256,000 macrocycles suitable for in vitro selection and high-throughput DNA sequencing. During the course of this library’s synthesis, we developed and extensively optimized many fundamental aspects of DNA-encoded and DNA-templated library technology. These advances include: (1) We proposed and experimentally validated a new model for identifying orthogonal codons for DTS library syntheses, which resulted in a 20×20×20×80 codon set sufficient to support up to 640,000-membered libraries. (2) In addition to increasing the number of scaffold codons (from 8 to 32 or 80), we developed an efficient DNA-compatible on-bead Boc-deprotection method and validated a number of new building blocks that together substantially expand scaffold and building block diversity of DNA-templated macrocycles. (3) We developed computer scripts to help generate in silico databases of compound libraries and to select building blocks that enhance the predicted bioavailability of the resulting molecules. (4) We developed new isolation and purification methods for DNA-linked small molecules that allow more reliable, scalable, high-yielding, and cost-effective preparation of DTS libraries and also enable the recovery and recycling of libraries after selection. (5) We developed new polymerase-assisted methods to synthesize libraries of DNA templates with 5′ chemical modifications. These methods provide more precise control of the library quality, eliminate the necessity of conducting reactions with oligonucleotide mixtures, and minimize material losses through unreliable immobilization on streptavidin-linked beads and poor recovery from standard precipitation methods. (6) Finally, we validated the new library synthesis protocols by in vitro selection against insulin-degrading enzyme (IDE), resulting in the discovery of macrocycle trans-DJPM, which is equipotent to the previously optimized IDE inhibitor 6bK (IC50 = 50 nM), and the discovery of cis-DJIR (IC50 = 40 nM), an unexpectedly potent IDE inhibitor of cis macrocycle backbone configuration that represents a new class of macrocycles that bind IDE.
The successful application of DNA-encoded libraries and the development of macrocycles emerging from our first-generation library has already resulted in highly potent and selective macrocycles that modulate the activity of a variety of targets of biomedical interest, in some cases with activity in mammalian cells and in mice43–45. We anticipate that this second-generation macrocycle library will prove a fertile source of new bioactive small molecules. An extensive selection campaign against biomedically important targets is underway, and the results will be reported in due course as separate studies focused on the corresponding biological investigations. In addition, we hope that the comprehensively improved methodology of DNA-templated libraries reported in this work will stimulate the use of this unique, accessible, and convenient tool for molecular discovery.
METHODS
All computational and experimental procedures are described in detail in the Supporting Information.
Buffer UM for the isolation of oligonucleotides from dilute solutions with minimal losses
We developed a simple alternative to commercially available spin-column binding buffers (such as PN and PNI from Qiagen). Our alternative can be easily prepared from readily accessible components and facilitates manipulations of relatively short oligonucleotides. 1 volume of the DNA solution is combined with a mixture of 4 volumes of saturated aqueous guanidinium chloride solution (natural pH ~6.4) and 6 volumes of isopropanol. The buffer enables spin-column isolation of oligonucleotides from very dilute solutions with minimal losses. For instance, we were able to achieve 99% recovery of 4.8 nmol of single-stranded 55-mer oligonucleotide from 40 mL of 120 nM solution (400 mL of Buffer UM was used, Omega HiBind Midi column).
General procedure for DNA-templated library selections
For a His-tagged protein, 25 μL of Dynabeads (His-Tag Isolation and Pulldown, 10103D) was washed with 2×300 μL PBST (50 mM sodium phosphate pH 8.0, 300 mM NaCl, 0.01% Tween-20, ± 5 mM DTT depending on whether the target needs reductive media). 10–40 μg of the His-tagged protein was diluted into 300 μL PBST and incubated with the beads at 4 °C for 30 min. The beads were washed twice with 200 μL TBST (50 mM Tris-HCl pH 8, 150 mM NaCl, 0.05% Tween-20, ± 5 mM DTT) followed by a 15-min incubation with the blocking solution at 4 °C (100 μL TBST, 0.6 mg/mL yeast RNA). The required amount of the DNA-encoded library (20 pmol) was incubated with the beads in 50 μL TBST w/RNA for 60 min at 4 °C. The flowthrough solutions from this point on are saved for library recovery. The beads were washed three times with 200 μLTBST and eluted with 50 μL of TBST containing 300 mM imidazole (5 min). The eluate was directly used for qPCR with adaptor primers for HTS barcoding (Supplementary Table 35) in order to find the maximum number of cycles within the exponential amplification range. Preparative PCR was performed with the identified number of cycles without addition of SYBR Green. Preparation of the resulting material for high-throughput sequencing followed standard protocols (Illumina).
Code availability
Developed scripts for the generation of electronic databases of macrocycles and analysis of selection sequencing results are provided in the Supplementary Information.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information file. High-throughput sequencing raw data is available upon request.
Supplementary Material
Acknowledgments
This paper is dedicated to Professor Hisashi Yamamoto on the occasion of his 75th birthday. This work was supported by U.S. National Institutes of Health (NIH) R35 GM118062, DARPA HR0011-17-2-0049, the Howard Hughes Medical Institute, and the F-Prime Biomedical Research Initiative.
Footnotes
Author Contributions
D.L.U. and D.R.L. designed the research and wrote the manuscript. D.L.U. conducted all the experimental, analytical and computational work for the development and synthesis of the library. Selections and library regeneration were optimized and conducted by D.L.U. and A.I.C. Macrocyclic hits were synthesized and purified by A.I.C., IDE inhibition assays were conducted by J.P.M. All authors edited the manuscript.
Competing financial interests
The authors declare the following competing financial interest(s): the authors are inventors on patents and patent applications describing DNA-templated synthesis methods and applications.
Any supplementary information, chemical compound information and source data are available in the online version of the paper.
References
- 1.Hüser J, Mannhold R, Kubinyi H, Folkers G. High-throughput screening in drug discovery. Wiley; 2006. [Google Scholar]
- 2.Macarron R, et al. Impact of high-throughput screening in biomedical research. Nat Rev Drug Discovery. 2011;10:188–195. doi: 10.1038/nrd3368. [DOI] [PubMed] [Google Scholar]
- 3.Dandapani S, Marcaurelle LA. Grand Challenge commentary: accessing new chemical space for ‘undruggable’ targets. Nat Chem Biol. 2010;6:861–863. doi: 10.1038/nchembio.479. [DOI] [PubMed] [Google Scholar]
- 4.Brenner S, Lerner RA. Encoded combinatorial chemistry. Proc Natl Acad Sci USA. 1992;89:5381–5383. doi: 10.1073/pnas.89.12.5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J Am Chem Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gartner ZJ, et al. DNA-templated organic synthesis and selection of a library of macrocycles. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zimmermann G, Neri D. DNA-encoded chemical libraries: foundations and applications in lead discovery. Drug Discovery Today. 2016;21:1828–1834. doi: 10.1016/j.drudis.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goodnow RA. A handbook for DNA-encoded chemistry: theory and applications for exploring chemical space and drug discovery. Wiley; 2014. [Google Scholar]
- 9.Franzini RM, Neri D, Scheuermann J. DNA-encoded chemical libraries: advancing beyond conventional small-molecule libraries. Acc Chem Res. 2014;47:1247–1255. doi: 10.1021/ar400284t. [DOI] [PubMed] [Google Scholar]
- 10.Krall N, Scheuermann J, Neri D. Small targeted cytotoxics: current state and promises from DNA-encoded chemical libraries. Angew Chem, Int Ed. 2013;52:1384–1402. doi: 10.1002/anie.201204631. [DOI] [PubMed] [Google Scholar]
- 11.Mannocci L, Leimbacher M, Wichert M, Scheuermann J, Neri D. 20 Years of DNA-encoded chemical libraries. Chem Commun. 2011;47:12747–12753. doi: 10.1039/c1cc15634a. [DOI] [PubMed] [Google Scholar]
- 12.Kleiner RE, Dumelin CE, Liu DR. Small-molecule discovery from DNA-encoded chemical libraries. Chem Soc Rev. 2011;40:5707–5717. doi: 10.1039/c1cs15076f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheuermann J, Neri D. DNA-encoded chemical libraries: a tool for drug discovery and for chemical biology. ChemBioChem. 2010;11:931–937. doi: 10.1002/cbic.201000066. [DOI] [PubMed] [Google Scholar]
- 14.Clark MA. Selecting chemicals: the emerging utility of DNA-encoded libraries. Curr Opin Chem Biol. 2010;14:396–403. doi: 10.1016/j.cbpa.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Buller F, Mannocci L, Scheuermann J, Neri D. Drug discovery with DNA-encoded chemical libraries. Bioconjugate Chem. 2010;21:1571–1580. doi: 10.1021/bc1001483. [DOI] [PubMed] [Google Scholar]
- 16.Clark MA, et al. Design, synthesis and selection of DNA-encoded small-molecule libraries. Nat Chem Biol. 2009;5:647–654. doi: 10.1038/nchembio.211. [DOI] [PubMed] [Google Scholar]
- 17.Doyon JB, Snyder TM, Liu DR. Highly sensitive in vitro selections for dna-linked synthetic small molecules with protein binding affinity and specificity. J Am Chem Soc. 2003;125:12372–12373. doi: 10.1021/ja036065u. [DOI] [PubMed] [Google Scholar]
- 18.Scheuermann J, Neri D. Dual-pharmacophore DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:99–103. doi: 10.1016/j.cbpa.2015.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Wrenn SJ, Weisinger RM, Halpin DR, Harbury PB. Synthetic ligands discovered by in vitro selection. J Am Chem Soc. 2007;129:13137–13143. doi: 10.1021/ja073993a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zhao P, Zhang M, Zhao X, Li X. Multistep DNA-templated synthesis using a universal template. J Am Chem Soc. 2013;135:17727–17730. doi: 10.1021/ja409936r. [DOI] [PubMed] [Google Scholar]
- 21.Hansen MH, et al. A yoctoliter-scale DNA reactor for small-molecule evolution. J Am Chem Soc. 2009;131:1322–1327. doi: 10.1021/ja808558a. [DOI] [PubMed] [Google Scholar]
- 22.Chan AI, McGregor LM, Liu DR. Novel selection methods for DNA-encoded chemical libraries. Curr Opin Chem Biol. 2015;26:55–61. doi: 10.1016/j.cbpa.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Satz AL. DNA encoded library selections and insights provided by computational simulations. ACS Chem Biol. 2015;10:2237–2245. doi: 10.1021/acschembio.5b00378. [DOI] [PubMed] [Google Scholar]
- 24.Satz AL. Simulated screens of DNA encoded libraries: the potential influence of chemical synthesis fidelity on interpretation of structure–activity relationships. ACS Comb Sci. 2016;18:415–424. doi: 10.1021/acscombsci.6b00001. [DOI] [PubMed] [Google Scholar]
- 25.Connors WH, Hale SP, Terrett NK. DNA-encoded chemical libraries of macrocycles. Curr Opin Chem Biol. 2015;26:42–47. doi: 10.1016/j.cbpa.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 26.Levin JI. Macrocycles in drug discovery. Royal Society of Chemistry; 2014. [Google Scholar]
- 27.Driggers EM, Hale SP, Lee J, Terrett NK. The exploration of macrocycles for drug discovery - an underexploited structural class. Nat Rev Drug Discovery. 2008;7:608–624. doi: 10.1038/nrd2590. [DOI] [PubMed] [Google Scholar]
- 28.Marsault E, Peterson ML. Macrocycles are great cycles: applications, opportunities, and challenges of synthetic macrocycles in drug discovery. J Med Chem. 2011;54:1961–2004. doi: 10.1021/jm1012374. [DOI] [PubMed] [Google Scholar]
- 29.White CJ, Yudin AK. Contemporary strategies for peptide macrocyclization. Nat Chem. 2011;3:509–524. doi: 10.1038/nchem.1062. [DOI] [PubMed] [Google Scholar]
- 30.Yudin AK. Macrocycles: lessons from the distant past, recent developments, and future directions. Chem Sci. 2015;6:30–49. doi: 10.1039/c4sc03089c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Villar EA, et al. How proteins bind macrocycles. Nat Chem Biol. 2014;10:723–731. doi: 10.1038/nchembio.1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dougherty PG, Qian Z, Pei D. Macrocycles as protein–protein interaction inhibitors. Biochem J. 2017;474:1109–1125. doi: 10.1042/BCJ20160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giordanetto F, Kihlberg J. Macrocyclic drugs and clinical candidates: what can medicinal chemists learn from their properties? J Med Chem. 2014;57:278–295. doi: 10.1021/jm400887j. [DOI] [PubMed] [Google Scholar]
- 34.Gartner ZJ, Kanan MW, Liu DR. Expanding the reaction scope of DNA-templated synthesis. Angew Chem, Int Ed. 2002;41:1796–1800. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 35.Gartner ZJ, Kanan MW, Liu DR. Multistep small-molecule synthesis programmed by DNA templates. J Am Chem Soc. 2002;124:10304–10306. doi: 10.1021/ja027307d. [DOI] [PubMed] [Google Scholar]
- 36.Li X, Liu DR. DNA-templated organic synthesis: Nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew Chem, Int Ed. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 37.Calderone CT, Puckett JW, Gartner ZJ, Liu DR. Directing otherwise incompatible reactions in a single solution by using DNA-templated organic synthesis. Angew Chem, Int Ed. 2002;41:4104–4108. doi: 10.1002/1521-3773(20021104)41:21<4104::AID-ANIE4104>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 38.O’Reilly RK, Turberfield AJ, Wilks TR. The evolution of dna-templated synthesis as a tool for materials discovery. Acc Chem Res. 2017;50:2496–2509. doi: 10.1021/acs.accounts.7b00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malone ML, Paegel BM. What is a “DNA-compatible” reaction? ACS Comb Sci. 2016;18:182–187. doi: 10.1021/acscombsci.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Satz AL, et al. DNA compatible multistep synthesis and applications to DNA encoded libraries. Bioconjugate Chem. 2015;26:1623–1632. doi: 10.1021/acs.bioconjchem.5b00239. [DOI] [PubMed] [Google Scholar]
- 41.Tse BN, Snyder TM, Shen Y, Liu DR. Translation of DNA into a library of 13 000 synthetic small-molecule macrocycles suitable for in vitro selection. J Am Chem Soc. 2008;130:15611–15626. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mullard A. DNA tags help the hunt for drugs. Nature. 2016;530:367–369. doi: 10.1038/530367a. [DOI] [PubMed] [Google Scholar]
- 43.Kleiner RE, Dumelin CE, Tiu GC, Sakurai K, Liu DR. In vitro selection of a DNA-templated small-molecule library reveals a class of macrocyclic kinase inhibitors. J Am Chem Soc. 2010;132:11779–11791. doi: 10.1021/ja104903x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Georghiou G, Kleiner RE, Pulkoski-Gross M, Liu DR, Seeliger MA. Highly specific, bisubstrate-competitive Src inhibitors from DNA-templated macrocycles. Nat Chem Biol. 2012;8:366–374. doi: 10.1038/nchembio.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maianti JP, et al. Anti-diabetic activity of insulin-degrading enzyme inhibitors mediated by multiple hormones. Nature. 2014;511:94–98. doi: 10.1038/nature13297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aleem SU, et al. Structural and biochemical basis for intracellular kinase inhibition by Src-specific peptidic macrocycles. Cell Chem Biol. 2016;23:1103–1112. doi: 10.1016/j.chembiol.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Snyder TM, Tse BN, Liu DR. Effects of template sequence and secondary structure on DNA-templated reactivity. J Am Chem Soc. 2008;130:1392–1401. doi: 10.1021/ja076780u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 49.Veber DF, et al. Molecular properties that influence the oral bioavailability of drug candidates. J Med Chem. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- 50.Pye CR, et al. Nonclassical size dependence of permeation defines bounds for passive adsorption of large drug molecules. J Med Chem. 2017;60:1665–1672. doi: 10.1021/acs.jmedchem.6b01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bockus AT, et al. Probing the physicochemical boundaries of cell permeability and oral bioavailability in lipophilic macrocycles inspired by natural products. J Med Chem. 2015;58:4581–4589. doi: 10.1021/acs.jmedchem.5b00128. [DOI] [PubMed] [Google Scholar]
- 52.Hewitt WM, et al. Cell-permeable cyclic peptides from synthetic libraries inspired by natural products. J Am Chem Soc. 2015;137:715–721. doi: 10.1021/ja508766b. [DOI] [PubMed] [Google Scholar]
- 53.Matsson P, Kihlberg J. How big is too big for cell permeability? J Med Chem. 2017;60:1662–1664. doi: 10.1021/acs.jmedchem.7b00237. [DOI] [PubMed] [Google Scholar]
- 54.Over B, et al. Structural and conformational determinants of macrocycle cell permeability. Nat Chem Biol. 2016;12:1065–1074. doi: 10.1038/nchembio.2203. [DOI] [PubMed] [Google Scholar]
- 55.Doak BC, Over B, Giordanetto F, Kihlberg J. Oral druggable space beyond the rule of 5: insights from drugs and clinical candidates. Chem Biol. 2014;21:1115–1142. doi: 10.1016/j.chembiol.2014.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Doak BC, Zheng J, Dobritzsch D, Kihlberg J. How beyond rule of 5 drugs and clinical candidates bind to their targets. J Med Chem. 2016;59:2312–2327. doi: 10.1021/acs.jmedchem.5b01286. [DOI] [PubMed] [Google Scholar]
- 57.Matsson P, Doak BC, Over B, Kihlberg J. Cell permeability beyond the rule of 5. Adv Drug Delivery Rev. 2016;101:42–61. doi: 10.1016/j.addr.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Watkins NE, SantaLucia J. Nearest-neighbor thermodynamics of deoxyinosine pairs in DNA duplexes. Nucleic Acids Res. 2005;33:6258–6267. doi: 10.1093/nar/gki918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Irwin JJ, et al. An aggregation advisor for ligand discovery. J Med Chem. 2015;58:7076–7087. doi: 10.1021/acs.jmedchem.5b01105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information file. High-throughput sequencing raw data is available upon request.