Summary
In the nematodes Caenorhabditis elegans and Pristionchus pacificus, a modular library of small molecules control behavior, lifespan, and development. However, little is known about the final steps of their biosynthesis, in which diverse building blocks from primary metabolism are attached to glycosides of the dideoxysugar ascarylose, the ascarosides. We combine metabolomic analysis of natural isolates of P. pacificus with genome-wide association mapping to identify a putative carboxylesterase, Ppa-uar-1, that is required for attachment of a pyrimidine-derived moiety in the biosynthesis of ubas#1, a major dauer pheromone component. Comparative metabolomic analysis of wildtype and Ppa-uar-1 mutants showed that Ppa-uar-1 is required specifically for the biosynthesis of ubas#1 and related metabolites. Heterologous expression of Ppa-UAR-1 in C. elegans yielded a non-endogenous ascaroside, whose structure confirmed that Ppa-uar-1 is involved in modification of a specific position in ascarosides. Our study demonstrates the utility of natural variation-based approaches for uncovering biosynthetic pathways.
eTOC Blurb
A small molecule library, the ascarosides, regulates the life history of Caenorhabditis elegans and Pristionchus pacificus. GWAS combined with metabolomics of P. pacificus natural isolates revealed a putative carboxylesterase, Ppa-uar-1, involved in attaching a pyrimidine-derived moiety in the biosynthesis of a major dauer pheromone component.
Introduction
The size of metabolome, the collection of small molecules produced by an organism, easily rivals that of the genome and proteome (Schrimpe-Rutledge et al., 2016; Viant et al., 2017; Zampieri et al., 2017). Even for well-studied model organisms such as Caenorhabditis elegans, Drosophila, Arabidopsis, or mouse, the structures and biosynthetic pathways of most endogenously produced small molecules remain to be elucidated. As the functions of roughly half of the genes in model organisms remain unknown (Niehaus et al., 2015), it seems reasonable to assume that many of these are involved in the biosynthesis of known as well as yet unidentified small molecules. Natural genetic variation provides an important resource that can be leveraged to identify diverse genetic traits using genome-wide association (GWAS) mappings (Bush and Moore, 2012). The utility of GWAS mappings for identifying genetic loci involved in a variety of phenotypes in C. elegans and other model systems is well established (Andersen et al., 2012; Ghosh et al., 2012; McGrath et al., 2009; Rhee et al., 2013), and GWAS has contributed to the characterization of biosynthetic pathways in plants (Richter et al., 2016; Wen et al., 2014; Wu et al., 2016) and birds (Cooke et al., 2017). However, the potential of GWAS to uncover the biosyntheses of the vast numbers of new metabolites currently being identified in animal model systems has not been explored.
In this work, we aimed to investigate the biosynthesis of a structurally diverse class of metabolites produced by C. elegans, Pristionchus pacificus, and other nematode species, which are derived from modular assembly of building blocks from conserved primary metabolism (von Reuss and Schroeder, 2015). These nematode-derived modular metabolites (NDMMs) are based on glycosides of the dideoxysugars ascarylose and paratose, which serve as scaffolds for the attachment of additional building blocks from all major primary metabolic pathways, including lipid, amino acid, carbohydrate, neurotransmitter, and nucleoside metabolism (Fig. 1a). NDMMs play a central role in nematode life history by regulating social behaviors, lifespan, and development, including the formation of long-lived, arrested dauer larvae in both C. elegans and P. pacificus (Ludewig and Schroeder, 2013; von Reuss and Schroeder, 2015). The C. elegans dauer stage has served as a highly tractable model for the study of aging and development, and four decades of extensive analyses revealed that conserved signaling cascades, including insulin, TGF-ß and steroid hormone signaling, transmit environmental information downstream of NDMM perception to trigger entry and exit from the dauer stage (Fielenbach and Antebi, 2008; Hu, 2007). In P. pacificus, NDMMs further regulate adult phenotypic plasticity (Bose et al., 2014; Bose et al., 2012), which results in alternative feeding habits by predatory vs. bacteriovorous feeding (Serobyan et al., 2016; Sommer et al., 2017).
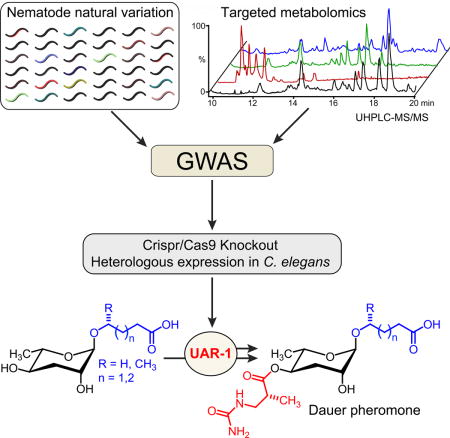
Figure 1. P. pacificus as a model system for linking genome and metabolome.
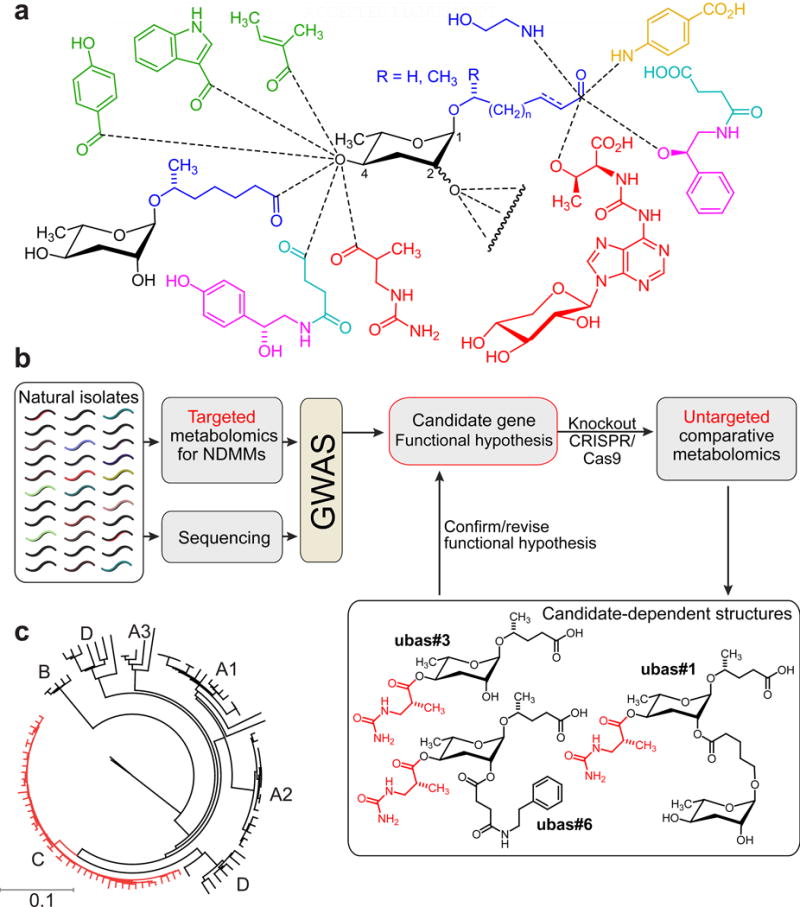
a) NDMMs form a modular library of signaling molecules, integrating building blocks derived from carbohydrate (black), fatty acid (blue), amino acid (green), neurotransmitter (purple), folate (orange), TCA cycle (cyan), and nucleoside metabolism (red). See www.smid-db.org for chemical structures of all NDMMs mentioned in the text. b) Strategy for using natural variation to link genes and metabolites. GWAS using relative abundances of NDMMs in natural isolates reveals candidate genes. Subsequent untargeted metabolomic comparison of candidate-gene knockout and wildtype strains reveals additional candidate-dependent metabolites that facilitate functional characterization. c) Neighbor joining dendrogram based on average number of substitutions within 1 million variable sites that were genotyped in all P. pacificus strains (modified from (Rodelsperger et al., 2014)). P. pacificus lineage C (red) is mainly associated with O. borbonicus on La Réunion.
Given the significance of NDMMs for the study of small molecule signaling, their biosynthesis and regulation are of great interest. Previous studies established the role of conserved primary metabolic pathways for the production of the building blocks required for NDMM assembly. For example, the fatty acid like side chains of ascaroside and paratoside glycosides have been shown to be derived from peroxisomal β-oxidation of long chain precursors (Markov et al., 2016; von Reuss et al., 2012; Zhang et al., 2015). The indole and octopamine moieties attached to the 4ʹ-position of the sugars in e.g. icas#3 and osas#9 have been shown to be derived from tryptophan (von Reuss et al., 2012) and tyrosine (Artyukhin et al., 2013), respectively. However, little is known about the origin of modularity and the enzymes that are involved in the highly specific assembly of primary-metabolism-derived building blocks via ester and amide bonds in the NDMMs. Since different combinations of building blocks encode different chemical messages, identification of the enzymes and regulatory factors that control modular assembly is of particular importance for understanding NDMM signaling.
Here, we describe a natural variation-based approach to study the biosynthetic origin of the modularity of NDMMs (Fig. 1b). Measuring intra-specific variation in the production of the major dauer pheromone component, ubas#1, in 264 wild isolates of P. pacificus enabled genome-wide association mapping (GWAS) of a locus encoding a putative esterase, Ppa-uar-1 required for the biosynthesis of dauer pheromone component ubas#1 (Fig. 1b). CRISPR-Cas9-induced mutation of Ppa-uar-1 converted a ubas#1-positive to a ubas#1-negative strain, and untargeted metabolomics of this Ppa-uar-1 mutant strain revealed the absence of several additional ubas#1-related NDMMs, clarifying the biosynthetic role of Ppa-uar-1. Lastly, heterologous expression of Ppa-uar-1 in C. elegans yielded a non-endogenous ascaroside, confirming that Ppa-uar-1 is involved in attachment of modules to the 4′-position of the ascarylose sugar moiety. Taken together, this work highlights the power of natural variation for linking genome and metabolome to uncover components of non-canonical biosynthetic pathways in animal model systems. Further, we provide a first example for the heterologous expression of ascaroside biosynthetic genes in C. elegans.
Results
Variation of NDMM Production Among Closely Related P. pacificus Strains
P. pacificus is a nearly cosmopolitan nematode species and is often found in association with scarab beetles (Herrmann et al., 2007). Genome sequencing and re-sequencing studies of 104 natural isolates from around the world revealed the existence of multiple clades of P. pacificus with substantial population structure (Fig. 1c) (McGaughran et al., 2016; Rodelsperger et al., 2014). As strong population structure complicates the identification of significant associations between phenotype and genotype (McGaughran et al., 2016), we concentrated on strains of one clade of P. pacificus, clade C, isolated from the volcanic island of La Réunion in the Indian Ocean. Extensive sampling of P. pacificus at La Réunion has resulted in the isolation of several hundred strains, many of which show natural variation of developmental characteristics including dauer induction, pheromone composition, and mouth-form plasticity, although they are genetically closely related to each other (Bose et al., 2014; Morgan et al., 2012; Ragsdale et al., 2013; Rodelsperger et al., 2014). Here, we only used strains collected from the long-term sampling site Trois Bassins and neighboring locations. All of these strains were isolated from the endemic rhinoceros beetle Oryctes borbonicus, a beetle species that is known to show an infestation rate of up to 90% with P. pacificus at Trois Bassins (Meyer et al., 2016; Morgan et al., 2012).
In total, we analyzed exo-metabolomes (the entirety of excreted small molecules) of 264 P. pacificus clade C strains using targeted high-pressure liquid chromatography coupled with mass spectrometry (HPLC-MS). We have previously demonstrated high reproducibility of relative abundances of NDMMs between biological replicates of mixed stage P. pacificus cultures (Bose et al., 2014), which we therefore chose for this comparison. HPLC-MS data for the metabolite extracts of the 264 strains revealed significant differences of NDMM abundances, whereby different compounds showed strikingly different patterns of variation (Fig. 2, Fig. S1). Most NDMMs were detected in all strains and their abundances followed a bell-shaped-like distribution (Fig. 2a–h). In contrast, distributions for ubas#1, ubas#2, and dasc#1, peaked at zero or very low abundances (Fig. 2i–k). Notably, the abundances of the structurally closely related NDMMs ubas#1 and ubas#2 were strongly correlated, whereas abundances of dasc#1 and ubas#1/ubas#2 were not correlated (Fig. 2m, n, Fig. S1a). The different shapes of distributions for ubas#1, ubas#2, and dasc#1 were not due to overall low abundance of these molecules (Fig. 2l). Furthermore, abolished or low production of ubas#1, ubas#2, or dasc#1 did not correlate with changes in the total abundance of all ascarosides in this set of strains (Fig. 2o). Given that most of the analyzed strains are phylogenetically extremely closely related (McGaughran et al., 2016; Rodelsperger et al., 2014), the almost complete absence of ubas#1, ubas#2, and dasc#1 in a large subset of strains suggested involvement of one or a small number of specific genetic loci.
Figure 2. Variation of NDMM abundances across P. pacificus natural isolates.
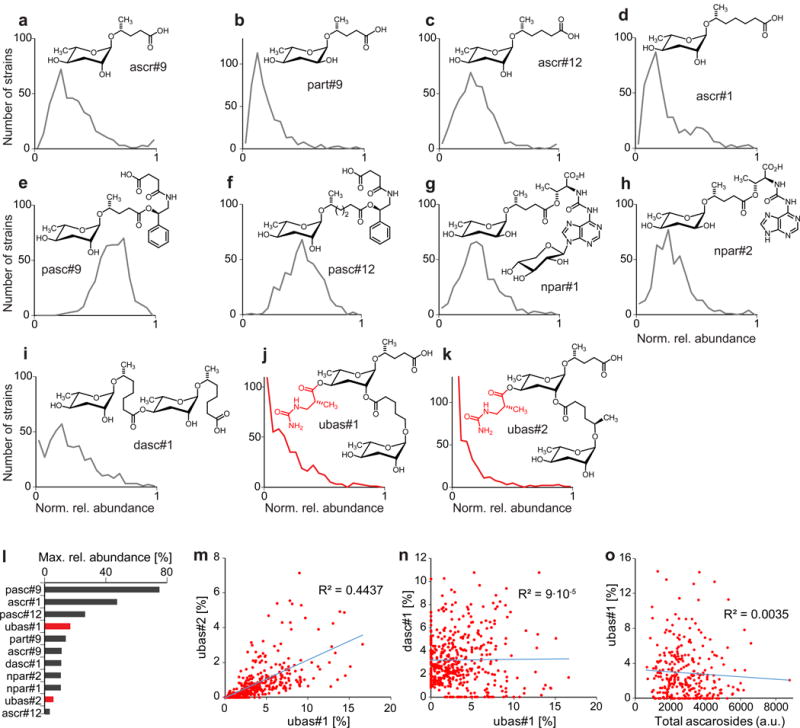
a) to k) Distributions of normalized relative abundances of individual NDMMs across natural isolates. For each NDMM, its percentage of all monitored NDMMs (based on MS ion current) for a particular strain was normalized to this compound’s maximum relative abundance across all strains, which is plotted in l). Number of bins for each NDMM distribution equals 20. m) Correlation between abundances of ubas#1 and ubas#2 across all strains. n) Correlation between abundances of dasc#1 and ubas#1 across all strains. o) Correlation between abundance of ubas#1 and total amount of all detected NDMMs across all strains. See also Figure S2.
Two easyGWAS Hits Associated with Variation of ubas#1 and ubas#2
To search for genetic loci involved in NDMM biosynthesis, we used easyGWAS, an integrated interspecies platform (Grimm et al., 2017). We began by genotyping all 264 P. pacificus strains for which the exo-metabolome had been analyzed using restriction site associated DNA (RAD) marker sequencing. RAD sequencing confirmed that almost all 264 strains belong to the clade C lineage, as suggested by their association with O. borbonicus (Fig. S1).
To identify genes involved in NDMM production we tested whether the variance in their abundance could be linked to genotypic variance. Indeed, using easyGWAS we identified two neighboring single nucleotide polymorphism (SNP) markers on chromosome I that were significantly correlated with variation of the abundances of ubas#1 and ubas#2 (Fig. 3a, Fig. S1d). Both SNP markers are located on Contig39 in close proximity to each other. We then used previously calculated pair-wise LD-values (McGaughran et al., 2016) to further narrow down the genomic region to a 30 kb-interval on Contig39. This region was found to contain a cluster of five genes that, based on sequence similarity, were predicted to encode esterase-like enzymes (Marchler-Bauer et al., 2015). Less significant GWAS mappings were obtained for variation of the abundance of dasc#1 (Fig. S1e) and other NDMMs. A higher marker density would likely enable additional mappings. For this study, we decided to focus on the SNPs associated with variation of ubas#1 and ubas#2 production, two important components of the P. pacificus dauer pheromone that are involved in intraspecific competition (Bose et al., 2014).
Figure 3. Identification of a gene involved in the biosynthesis of ubas#1 and ubas#2.
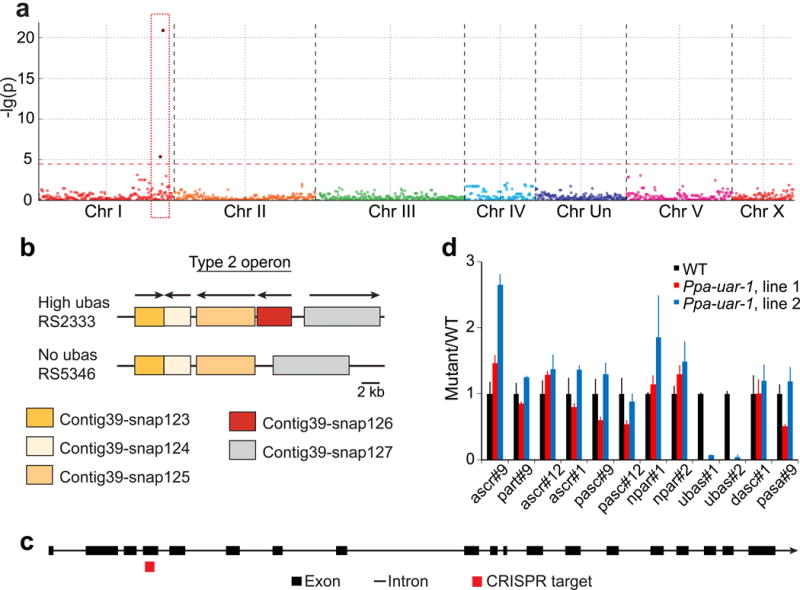
a) GWAS using genomic data created with RAD marker sequencing and metabolite abundance (here ubas#1) as phenotypic dataset. The Manhattan plot shows two significant SNP markers for the ubas#1 phenotype (red rectangle). Each dot represents the p-value for one SNP marker. b) Genomic structure of the candidate region in the wildtype strain RS2333, in the de novo assembled “high ubas” strain RSA016, and the “no ubas” strain RS5346. The gene Contig39-snap126 (Ppa-uar-1) is largely deleted in naturally occurring “no ubas” strains. c) Intron and exon structure of Ppa-uar-1 determined by RACE-PCR and CRISPR target side in exon 4. d) NDMM abundances of Ppa-uar-1 mutant strains, normalized to average levels in wild type (RS2333). Data are from three independent experiments. Error bars represent standard deviation. See also Figures S1-S5 and Tables S1-S5.
Identification and Inactivation of the Gene Involved in ubas Biosynthesis
The P. pacificus reference strain RS2333 (Dieterich et al., 2008) and the clade C strains studied here are known to be genetically divergent and might contain major genomic rearrangements (Rodelsperger et al., 2014). Therefore, we performed nanopore amplicon sequencing for one “no–ubas” strain (RS5346) and one “high ubas” strain (RSA016). We used the long nanopore reads in conjunction with the shorter Illumina reads to create a de-novo assembly of the target region of strains RS5346 and RSA016 (Fig. 3b), which resulted in detection of an approximately 2.6 kb-deletion that is specific to all “no ubas” strains. This deletion spans most of the coding region of the gene Contig39-snap126 and is part of an operon of two genes (Sinha et al., 2014), both of which encode putative esterases (Marchler-Bauer et al., 2015). Contig39-snap126 encodes a putative type-B carboxylesterase with homology to human liver carboxylesterases, e.g. cocaine esterase CES2 (Fig. S4, Table S1), which participates in the detoxification of xenobiotics through hydrolysis of ester bonds (Pindel et al., 1997). To investigate the potential function of this candidate gene in the biosynthesis of ubas#1 and ubas#2, we employed the CRISPR/Cas9 system (Witte et al., 2015) to create a Contig39-snap126 deletion mutant in the “high ubas” reference strain RS2333. The guide RNA was designed based on the exon and intron structure of Contig39-snap126 determined by RACE-PCR experiments targeting exon 4 (Fig. 3c). We obtained the allele tu611, which contains a 7-bp deletion resulting in a frame shift and premature termination of Contig39-snap126. We renamed Contig39-snap126 as Ppa-uar-1 (ubas-ascaroside-required).
Ppa-uar-1 Mutation Abolishes ubas#1 and ubas#2 Biosynthesis
Comparing the exo-metabolome of P. pacificus wild type (RS2333) with that of the Ppa-uar-1 null mutants by UPLC-HRMS (Panda et al., 2017), we found that the amounts of ubas#1 and ubas#2 in the Ppa-uar-1 exo-metabolome were reduced by a factor of at least 100 (Fig. 3d). We observed a similar reduction in the levels of ubas#1 and ubas#2 in worm pellet extracts (endo-metabolome) of the Ppa-uar-1 mutant (Fig. S1f). In contrast, amounts of ubas#1 and ubas#2 were unchanged from wild type amounts in a CRISPR-generated mutant for one of the neighboring esterase-encoding genes, Contig39-snap124 (Fig. S1g). Notably, the amounts of all other known ascarosides varied only slightly between wild type and Ppa-uar-1 mutant strains (Fig. 3d). Next we asked whether mutation of Ppa-uar-1 affects production of ureidoisobutyric acid, a likely precursor required for the biosynthesis of ubas#1 and ubas#2. However, analysis of the exo-metabolomes of Ppa-uar-1 mutants and the “high-ubas” reference strain, RS2333, did not reveal any differences in ureidoisobutyric acid production (Fig. 4a, Fig. S2a). Taken together, these results indicate that mutation of Ppa-uar-1 abolishes biosynthesis of ubas#1 and ubas#2, rather than their excretion or the availability of starting materials.
Figure 4. Untargeted metabolomics of Ppa-uar-1 reveals previously unrecognized ubas derivatives and heterologous expression of Ppa-uar-1 in C. elegans.
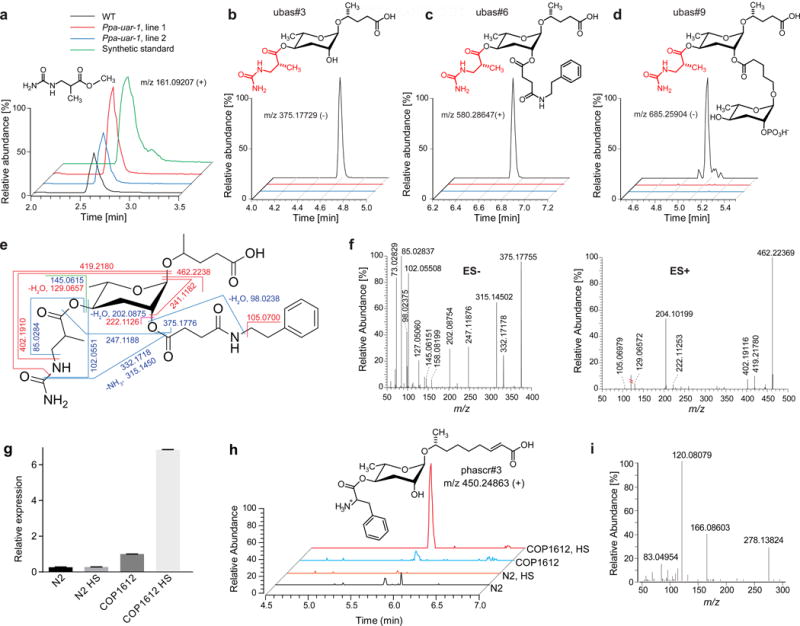
a) Extracted ion chromatograms (EICs) for methyl ester of ureidoisobutyric acid in WT (RS2333) and Ppa-uar-1 extracts. b)-d) Proposed structures and EICs of discovered metabolites ubas#3, ubas#6, and ubas#9. EICs in a) – d) were generated using a 5 ppm m/z window. e) Proposed structure and MS/MS fragmentation of ubas#6. Blue lines – fragmentation in negative mode, red lines – in positive mode, green – in both modes. f) MS/MS spectrum of ubas#6 in negative and positive ion modes. g) RT-qPCR for Ppa-uar-1 in hsp16.41::Ppa-uar-1 C. elegans (COP1612). h) LC-HRMS extracted ion chromatograms for phascr#3 in C. elegans wildtype and hsp16.41::Ppa-uar-1 (COP1612), under normal growth conditions and after heat shock (HS). i) ESI+ MS/MS spectrum of phascr#3. See also Figures S2 and S3 and Tables S2 and S5.
Untargeted Metabolomics Reveals Additional ubas Derivatives
To further characterize the effect of the Ppa-uar-1 mutation on metabolism, we performed an untargeted comparison of the RS2333 and Ppa-uar-1 exo-metabolomes by analyzing UPLC-HRMS data using the XCMS software package, which facilitates detection of compounds that are strongly up- or downregulated among samples (Tautenhahn et al., 2012). This comparative metabolomic analysis revealed seven compounds, in addition to ubas#1 and ubas#2, whose production was abolished in Ppa-uar-1 mutants (Fig. 4b–d and Fig. S2). Based on their MS/MS fragmentation spectra (Fig. 4e–g, Fig. S2), we proposed structures of these previously unrecognized metabolites, named ubas#3 through ubas#9, all of which represent ascaroside derivatives that feature an ureidoisobutyryl moiety attached to ascarylose (Fig. 4b–d, Fig. S2). Comparison of the MS/MS fragmentation patterns of ubas#6-9 with fragmentation patterns of ubas#1-3 and other NDMMs indicated that these metabolites represent derivatives of ubas#3 in which a variety of different building blocks are attached at the 2ʹ-position of the ascarylose (see Fig. 4c,d, and Fig. S2 for proposed structures). Among these newly detected Ppa-uar-1 dependent metabolites we selected ubas#6 for purification via preparative HPLC and more detailed structural characterization via 2D NMR spectroscopy. The dqfCOSY NMR spectrum of this sample is fully consistent with the proposed structure for ubas#6 (Fig. 4e, Table S2). Given that all additional Ppa-uar-1-dependent metabolites represent ascaroside derivatives that feature an ureidoisobutyryl side chain, whereas production of other metabolites is not affected, inactivation of Ppa-uar-1 appears to abolish specifically attachment of the ureidoisobutyryl side chain, suggesting that Ppa-uar-1 encodes an enzyme that directly participates in this biosynthetic step.
Heterologous Expression of Ppa-uar-1 C. elegans Furnishes new Ascarosides
Our attempts at heterologous expression of Ppa-uar-1 in bacteria and yeast as well as genetic complementation in P. pacificus failed. Similarly, we were unable to obtain C. elegans strains expressing Ppa-uar-1 under the C. elegans rpl-28 constitutive promoter. Therefore, we constructed four transgenic lines of C. elegans expressing Ppa-uar-1 under the heat shock-inducible hsp16 promoter (see Methods). Under standard growth conditions, transgenic hsp16.41::Ppa-uar-1 C. elegans reproduced normally, RT-qPCR indicated very low levels of Ppa-uar-1 expression (Fig. 4g), and the metabolome of hsp16.41::Ppa-uar-1 worms was found to be identical to that of C. elegans wildtype. However, following heat shock, RT-qPCR revealed starkly increased expression of Ppa-uar-1 in hsp16.41::Ppa-uar-1 worms, and untargeted comparative metabolomics of hsp16.41::Ppa-uar-1 and wildtype worms via UPLC-HRMS uncovered several new peaks in all four hsp16.41::Ppa-uar-1 strains that were absent in wildtype (Fig. 4g,h). Their MS/MS spectra indicated that these compounds represent ascarosides of varying chain length that, like ubas#1, are modified in the 4′-position of the ascarylose sugar, but bear a phenylalanyl instead of a ureidoisobutyryl group (Fig. 4i). We confirmed these structural assignments by synthesizing a representative member of the heat shock induced ascaroside family, phascr#3 (Fig. S3). The ascaroside phascr#3 and related phenylalanine ascarosides were the only heat shock-induced compounds, and formation of ubas#1 or other ureidoisobutyryl ascarosides was not detected, even when supplying β-ureidoisobutyic acid or its putative precursor, dihydrothymine, in the growth media. Taken together, these results indicate that Ppa-UAR-1 is directly involved in the attachment of acyl moieties to the 4′-position of ascarylose, whereby this enzyme appears to be somewhat promiscuous, catalyzing attachment of both ureidoisobutyryl and phenylalanyl moieties. Notably, the products of Ppa-UAR-1, ubas#1 and derivatives in P. pacificus and phascr#3 and derivatives in hsp16.41::Ppa-uar-1 C. elegans, are the only known ascarosides that bear a free amino group, suggesting that this structural feature is required for Ppa-UAR-1 activity.
Discussion
The recent identification of NDMMs provided a striking example of combinatorial generation of structural diversity, via modular assembly of primary metabolic building blocks (von Reuss and Schroeder, 2015). Although production of the primary metabolism-derived building blocks of the NDMMs is widely conserved across the animal kingdom, each nematode species appears to have evolved biosynthetic pathways for specific subsets of compounds. In this study, we identified Ppa-uar-1 as the first gene involved in the modular assembly of ubas#1 and ubas#2, NDMMs that function as part of the P. pacificus dauer pheromone (Bose et al., 2014; Bose et al., 2012). To that end, we have combined inputs from natural variation data for NDMM production in P. pacificus, GWAS, genetic engineering, and LC-MS analysis of resulting mutant metabolomes. The identified gene, Ppa-uar-1, is annotated to encode a type-B carboxylesterase (Marchler-Bauer et al., 2015), i.e. an enzyme catalyzing the hydrolysis of an ester of a carboxylic acid (Fig. S4, Table S1). The closest mammalian homologs of Ppa-UAR-1 are liver carboxylesterases, e.g. cocaine esterase CES2 (~30% amino acid sequence similarity), which is implicated in detoxification of xenobiotics through hydrolysis of ester bonds (Pindel et al., 1997). Homologous features of CES2 and Ppa-UAR-1 include the conserved active site residues S251, E378, and H456 as well as four cysteine residues required for two disulfide bridges (Fig. S4) (Larsen et al., 2002; Pindel et al., 1997). The high degree of similarity between Ppa-UAR-1 and other carboxylesterases, including the serine hydrolase consensus sequence motif GXSXG, supports its functional annotation as an esterase (Larsen et al., 2002; Pindel et al., 1997). However, our data suggest that Ppa-UAR-1 functions as an acyl-CoA transferase, catalyzing formation of 3-ureidoisobutyryl esters of ascaroside precursors (Fig. 5). Notably, several homeostatic esterases have been shown to be promiscuous and switch between acyl transferase and esterase functions (Saerens et al., 2006).
Figure 5. Proposed biosynthetic pathway of NDMMs incorporating a ureidoisobutyric acid moiety.
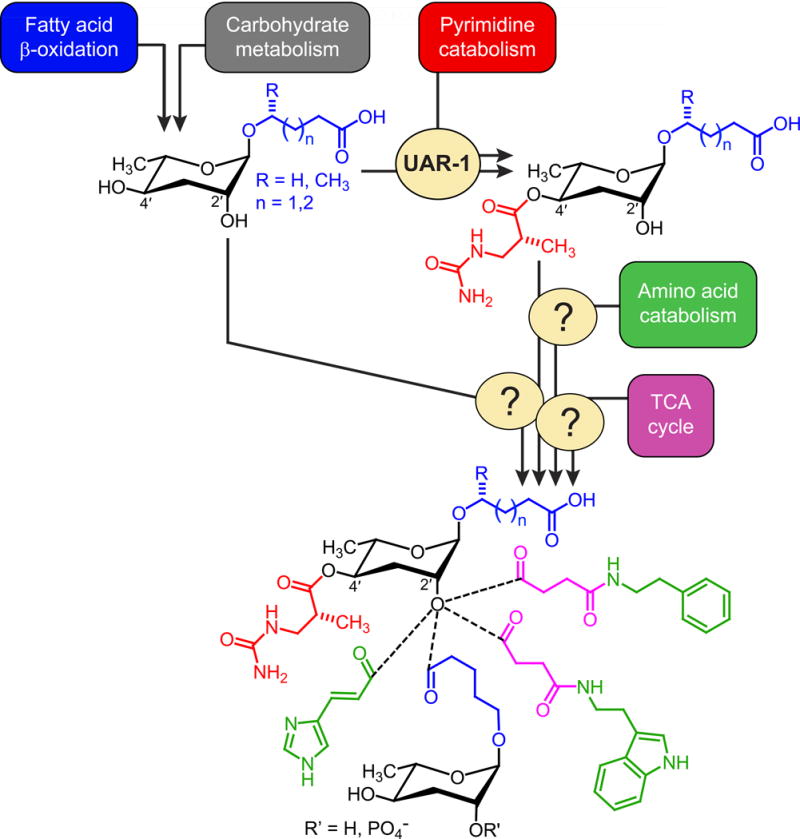
Unmodified ascarosides originating from carbohydrate and fatty acid metabolism undergo Ppa-UAR-1-dependent modification at the 4′-position. The resulting ureidoisobutyryl ascarosides are modified further via attachment of additional building blocks.
Our untargeted comparison of the metabolomes of WT and Ppa-uar-1 mutants revealed several previously undescribed Ppa-uar-1-dependent NDMMs, all of which feature a 3-ureidoisobutyryl moiety at the ascarylose 4ʹ-OH position. We further found that abolishment of all nine ‘ubas’ compounds in Ppa-uar-1 mutants is not associated with accumulation of only 2ʹ-OH functionalized ascarosides or free 3-ureidoisobutyrate. These observations suggest that attachment of the 3-ureidoisobutyryl moiety, which is likely derived from pyrimidine metabolism (Bose et al., 2012), to a simple ascaroside, e.g. ascr#9, constitutes the first step towards the biosynthesis of ‘ubas’ NDMMs (Fig. 5). Subsequent attachment of moieties from several other primary metabolic pathways then produces the ‘ubas’ family of NDMMs (Fig. 5). Detailed biological study of the newly identified ‘ubas’ derivatives will have to await their chemical synthesis; however, parallel work on small molecule signaling in C. elegans indicates that NDMMs with different combinations of modules often have distinct activities (von Reuss and Schroeder, 2015).
It is interesting to compare the biosynthetic roles of Ppa-UAR-1 and recently characterized Cel-ACS-7, an acyl-CoA synthetase required for the biosynthesis of icas#9 and osas#9 in C. elegans (Panda et al., 2017) (for structures, see www.smid-db.org). Cel-ACS-7 was found to be promiscuous with respect to the moiety it attaches to the 4ʹ-position (e.g. indole-3-carboxylic acid, N-succinyloctopamine, and p-hydroxybenzoic acid) but also specific in that it is only required for the production of 4ʹ-modified NDMMs featuring a 5-carbon fatty acid side chain, whereas biosynthesis of 4ʹ-modified NDMMs with other side chain lengths does not require ACS-7. In contrast, Ppa-UAR-1 is involved in the biosynthesis of NDMMs with a range of different chain lengths (e.g. ubas#3, ubas#4, and ubas#5) but specifically contributes to 4ʹ-attachment of a ureidoisobutyryl moiety in P. pacificus. The finding that Ppa-UAR-1 when expressed in C. elegans contributes to 4ʹ-attachment of a different moiety (phenylalanyl) suggests involvement of additional proteins in ureidoisobutyryl attachment, which also appears to be the case for the involvement of Cel-ACS-7 in 4ʹ-attachment of N-succinyloctopamine.
A phylogenetic analysis of putative type-B carboxylesterases from 14 nematode species including C. elegans and P. pacificus placed Ppa-uar-1 in a Pristionchus-specific subclade of esterases with no 1:1 orthologs in C. elegans (Fig. S5). This subclade has undergone a lineage specific expansion in Pristionchus, a pattern common for the evolution of enzyme-encoding genes in nematodes (Markov et al., 2015). We thus identified a lineage-specific enzyme of high apparent biochemical specificity indicating that the diversity in the chemical space is at least in part due to genes originating from gene duplication and the following functional diversification. It is not known whether related type-B carboxylesterases in C. elegans or higher animals participate in the biosynthesis of signaling molecules. Some esterases are involved in pheromone metabolism in felids (Miyazaki et al., 2006) and insects (Chertemps et al., 2012). Our identification of Ppa-uar-1 provides a strong incentive to investigate potential metabolic functions of the five closest Ppa-uar-1 homologs in C. elegans. More generally, untargeted comparative metabolomic analysis employed here to identify previously unrecognized metabolites derived from Ppa-uar-1 could be used to uncover potential roles of liver carboxylesterases and related enzymes in mammalian metabolism.
Prior studies of the biosynthesis of ascaroside-based NDMMs led to the identification of primary metabolic genes involved in the production of some of the building blocks, e.g. demonstrating the role of peroxisomal β-oxidation in the biosynthesis of the fatty acid-like side chains (Markov et al., 2016; von Reuss et al., 2012; Zhang et al., 2015). For the identification of genes involved in NDMM assembly, traditional mutant screens of candidate genes, selected based on functional predictions, have been of limited utility, which is not surprising given that NDMMs represent a recently discovered and structurally distinct class of metabolites. In the current study, GWAS identified several SNPs that correlate with the abundances of additional NDMMs, e.g, dasc#1, a dimeric ascaroside that is involved in adult phenotypic plasticity. Ongoing whole-genome sequencing of natural isolates will provide a higher marker density, which will help to further improve scope and accuracy of metabolite mappings. In addition, life stage-specific analyses of metabolomes from developmentally synchronized worm cultures could help reduce noise of the metabolomic data used for mapping.
Our study highlights the potential of natural variation-based approaches for the investigation of biosynthetic pathways without direct precedent. Considering the wealth of natural variants isolated for P. pacificus, C. elegans (Andersen et al., 2012; Cook et al., 2017), Drosophila (Lack et al., 2016), and other animal models, our HPLC-MS/MS, GWAS, and CRISPR-Cas9-based workflow could serve as a blueprint for elucidating the biosyntheses of the vast numbers of yet uncharacterized metabolites in these species. Lastly, we present a first example for the heterologous production of nematode-derived specialized metabolites, the phenylalanine ascarosides, via genetic engineering in C. elegans.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Frank Schroeder (fs31@cornell.edu).
Experimental Model and Subject Details
Nematode Strains and Cultures
Over the course of five years, 264 Pristionchus pacificus strains were isolated as described earlier (Morgan et al., 2012). The strains used in this study were all collected at the sample side Trois Bassins and surrounding sample sides on La Réunion Island (see Key Resources Table). Worms were grown on NGM agar plates seeded with 300 μl OP50, and liquid cultures of worms were prepared as described previously with the following alterations (Ogawa et al., 2009; Sommer et al., 1996). In brief, worms were washed off three fully grown ad-lib fed 6 cm NGM agar plates, incubated in a final volume of 10 ml S-medium with cholesterol (5 μg/ml), streptomycin (20 μg/ml), nystatin (20 μg/ml), and 250 μl 20% w/v E. coli OP50 for seven days at 22°C and 180 rpm. These conditions induce dauer formation in the different nematode strains under liquid culture conditions. The culture supernatant was separated from the worm pellet by centrifugation and used for the preparation of the metabolome extracts.
Method Details
Preparation of Metabolome Extracts
Based on previous studies demonstrating the high reproducibility of metabolome production (Bose et al., 2014), duplicates of 10 ml liquid cultures of each P. pacificus wild isolate were prepared. These cultures were harvested, centrifuged, and the resultant supernatant media and worm pellets were frozen over dry ice-acetone slush and lyophilized separately. The lyophilized materials from the supernatant were extracted with 50 ml of 95% ethanol at room temperature for 16 h. The resulting suspensions were filtered, and the filtrate was evaporated in vacuo at room temperature, producing supernatant (“exo-metabolome”) extracts. For analysis of Ppa-uar-1 metabolomes, two mutant lines resulting from CRISPR genome editing were selected. Mixed stage worms from a populated 10 cm NGM agar plate seeded with E. coli OP50 were washed into 25 ml of S-complete medium and fed with OP50 on days 1, 4, and 6 for a 9-day culture period, while shaking at 22 °C, 220 rpm. The cultures were then centrifuged, worm pellets and supernatant frozen separately and lyophilized. Worm pellets were then crushed to a fine powder in a mortar and pestle and extracted with 10 mL of methanol at room temperature for 12 h. The extracts were dried in vacuo, resuspended in 100 μL methanol and analyzed by HPLC-MS.
HPLC-MS Analysis
HPLC-MS quantification of known NDMMs was performed using an Agilent 1100 Series HPLC system equipped with an Agilent Eclipse XDB-C18 column (4.6 × 250 mm, 5 μm particle diameter) connected to a Quattro II spectrometer (Micromass/Waters). A 0.1% acetic acid-acetonitrile solvent gradient was used at a flow rate of 1 ml/min, starting with an acetonitrile content of 5% for 5 min which was increased to 100% over a period of 40 min. Exo-metabolome extracts were resuspended in 250 μL of methanol, filtered, and used directly for HPLC-MS injection. The metabolome extracts were analyzed by HPLC-ESI-MS in the negative ion mode as reported previously (Bose et al., 2012). Briefly, a capillary voltage of 3.5 kV and a cone voltage of −35 V were used, and the resulting data were analyzed using Waters MassLynx™ 4.1 software. Quantifications were based on integration of HPLC-MS signals from the corresponding ion-traces. The following ESI-ion traces were used m/z = 247 (ascr#9 and part#9), 261 (ascr#12), 275 (ascr#1), 466 (pasc#9), 480 (pasc#12), 494 (pasc#1), 509 (npar#2), 533 (dasc#1), 605 (ubas#1), 619 (ubas#2), and 641 (npar#1). Absolute concentrations were calculated using response factors determined for synthetic standards. Concentrations for minor compounds, for which synthetic standards were not available, were based on extrapolation of available standards of closely related structures.
High-resolution UPLC-MS for untargeted metabolomics was performed on a Dionex 3000 UPLC coupled to a Thermo Q Exactive high resolution mass spectrometer, using water-acetonitrile gradient on Agilent Zorbax Eclipse XDB-C18 column, 150 × 2.1 mm, particle size 1.8 μm, maintained at 40 °C. Solvent A: 0.1% formic acid in water, solvent B: 0.1% formic acid in acetonitrile. A/B gradient started at 5% B at 1.5 min after injection and increased linearly to 100% B at 12.5 min. Samples were analyzed in both positive and negative ionization modes. Molecular formulae were assigned based on their high resolution masses (< 2 ppm, also see Table S5), MS/MS fragmentation patterns, and comparison of retention times with those of authentic synthetic standards (Bose et al., 2012; von Reuss et al., 2012).
Methylation and Detection of UIBA
To facilitate detection and identification of free ureidoisobutyric acid (UIBA) in metabolome extracts it was converted to methyl ester with trimethylsilyl diazomethane (TMS-DM). To a 4-ml glass vial 100 μl of methanol, 2μl of a 68 mM solution of UIBA in methanol (for synthetic standard) or 50 μl of P. pacificus worm medium extract (for natural samples), 100 μl of toluene, and 50 μl of 2M TMS-DM in hexane were added. After vortexing the sample for ~30 s, the reaction was left at room temperature for half an hour. Subsequently, the solvent was removed in vacuum (down to 15 Torr), and the residue was redissolved in 100 μl of methanol for HPLC-MS analysis.
Isolation and Characterization of ubas#6
Medium from RS2333 mixed-stage culture (total volume 3 liters) was lyophilized and extracted with methanol. Dried methanol extract was loaded on Celite and fractionated on medium pressure reverse phase chromatography (86g C18 Combiflash RediSep column). A 0.1% aqueous acetic acid-acetonitrile solvent gradient was used at a flow rate of 60 ml/min, starting with acetonitrile content of 0% for 10 min which was increased to 100% over a period of 82 min. After assaying fractions for the presence of ubas#6 by UPLC-MS, the relevant fractions were combined, solvent removed, and the residue was fractionated further by preparative reverse-phase HPLC using an Agilent Zorbax Eclipse XDB-C8 column (9.4 × 250 mm, 5 μm particle diameter) and 0.1% aqueous acetic acid-acetonitrile solvent gradient at a flow rate of 3.6 ml/min. Isolated ubas#6 was redissolved in CD3OD for NMR spectroscopic analysis, using a Bruker AVANCE III HD (800 MHz) spectrometer. dqfCOSY spectra were acquired using an acquisition time of 0.6 s, sweep width of 8.5 ppm and 500 complex increments. Spectra were zero-filled to 10 16384×4096 data points and processed using cosine window functions in both dimensions. For NMR spectroscopic data see Table S2. Based on the comparison with 13C satellites of the solvent peak, we estimate the amount of isolated ubas#6 to be on the order of 1 μg, which was insufficient to obtain carbon chemical shift data.
DNA Extraction
Nematodes were washed off five fully grown unstarved 6 cm NGM agar plates using 0.9% NaCl. In a first wash step 0.9% NaCl solution containing 50 μg/ml ampicillin was added to the worms to a final volume of 40 ml. After incubating the worms for > 6 h the supernatant was discarded and the worms were washed the second time for > 6 h in 0.9% NaCl solution containing 50 μg/ml ampicillin and 25 μg/ml chloramphenicol. After the incubation, the supernatant was discarded and DNA was extracted from the worm pellet using the epicenter DNA extraction kit (Epicentre, Madison, USA) following the manufacturer’s instructions.
RNA Extraction and Sequencing
Nematodes were separated by centrifugation from the culture supernatant, washed once with S-medium and immediately transferred into Tri Reagent® (Sigma-Aldrich, Munich, Germany). After three rounds of freezing in liquid nitrogen and thawing at 37 °C to break open the cells RNA was extracted with the Direct-zol™ RNA MiniPrep (Zymo, Freiburg, Germany) following the manufacturer’s protocol. RNA quality was determined with a NanoDrop ND 1000 spectrometer (PeqLab, Erlangen, Germany) and RNA integrity was checked with the Agilent RNA Nano Chip Assay (Agilent, Santa Clara, USA). Only RNA samples of high quality (OD 260/280 > 2 and OD 260/230 > 1.8) and high integrity were used for the RACE PCR experiment. Further, the high quality RNA was used to prepare two libraries following Illumina’s truseq RNA-sequencing protocol (Illumina, Eindhoven, Netherlands). Both libraries were sequenced on one 101 bp pair-end flow cell lane of an Illumina HiSeq 2000 Sequencer.
RACE PCR and RAD Sequencing
A RACE-PCR experiment to determine the exact intro-exon structure and to identify potential isoforms was performed using the SMARTer® RACE 5′/3′kit (Clontech, Mountain View, USA) following the manufacturer’s protocol. RADseq was carried out using a modified version of the protocol employed by Poland et al. (Poland et al., 2012), as described in detail in Witte et al., (Witte et al., 2015). After library preparation and clean up with the QIAquick PCR Purification Kit (Qiagen, Hamburg, Germany) fragments 250-500 bp long were selected using bluepippin (Sage Science, Massachusetts, USA) following the manufacturer’s protocol. The library was validated by DNA quantification with a Qubit®2.0 fluorometer (Life Technologies GmbH, Darmstadt, Germany) and fragment length confirmation using Agilent Bioanalyzer (Agilent Technologies GmbH, Waldbronn, Germany). Finally, each library (48 plex) was normalized and single-end sequenced from the Pst1 ends on one lane of a Hiseq2000 sequencing platform (Illumina Inc., California, United States). The data analysis and SNP calling of the RAD-seq data from 264 strains was performed as described previously in Witte et al. (Witte et al., 2015).
DNA Whole Genome Re-sequencing
For each of 13 strains used in the high vs. low ubas comparison one paired-end library was created with an average insert size between 450 to 500 bp following Illumina’s paired-end protocol (Illumina, Eindhoven, Netherlands). A starting amount of 100 ng of DNA was sheared with Covaris S2 ultrasonicator (Covaris Inc. Woburn, San Diego) and the paired-end libraries were run on two 150 bp paired-end flow cell lanes of an Illumina HiSeq 3000 Sequencer (Illumina, Eindhoven, Netherlands) yielding average re-sequencing coverage of 40 to 70×.
Amplicon Long-Range PCR
Amplicons for the nanopore sequencing were generated using the Herculase II fusion DNA Polymerase kit (Agilent, Waldbronn, Germany) following the manufacturer’s protocol. The whole genomic region was divided into 10-15 kb fragments, which were amplified in both strains (RS5346 “no ubas” and RSA016 “high ubas”) using specific primers (see Key Resources Table). Primer design was carried out using Geneious software (version 8.0.1). In brief, the conditions for PCR experiment were the following: initial denaturation for 2 minutes at 95 °C, ten cycles of denaturation at 95 °C for 20 seconds, annealing at 60°C for 20 seconds, and extension at 68 °C for 8 min, followed by 20 cycles of denaturation at 95°C for 20 seconds, annealing at 60 °C for 20 seconds, and extension at 68 °C 8 min, plus 20 seconds longer per cycle and a final extension 68°C for 8 minutes.
Nanopore Amplicon Sequencing and Local Assembly
The DNA libraries for the ONT MinION were prepared according to the Nanopore Sequencing Kit protocol SQK-MAP005 (Version MN005_1115_revE_26Nov2014) for Genomic DNA with subsequent protocol modifications for improved yield and library performance. Five different amplicons were pooled for each strain RS5346 and RSA016 in equimolar amounts to obtain 1.5 μg of total input DNA. DNA fragmentation and DNA damage repair steps were skipped. Following the protocol, the DNA was end-repaired using the NEBNext® End Repair Module (NEB, catalog no. E6050) and dA-tailed with the NEBNext® dA-Tailing Module (NEB, catalog no. E6053). Between these steps, the DNA was purified with 1× AMPure XP beads (Beckman Coulter, catalog no. A63880) and the final bead incubation was performed each time with EB Buffer (QIAGEN, catalog no. 19086) at 37 °C. A mixture of sequencing adapters (ONT, SQK-MAP005.1) was ligated to the template using the Blunt/TA Ligase Master Mix (NEB, catalog no. M0367), followed by enrichment of successfully ligated molecules with Dynabeads® His-Tag Isolation and Pulldown (Thermo Fisher Scientific, catalog no. 10103D). The flowcell priming and library loading on the ONT MinION was performed according to the Nanopore Sequencing Kit protocol SQK-MAP005 with the R7.3 flowcell chemistry (ONT, FLO-MAP003). Finally, the libraries were sequenced using a 48 hour sequencing run and library reloading at 5, 15, 21, 27 and 41 hours into the sequencing run. The sequencing raw data was uploaded to the ONT’s proprietary cloud base calling service, which used the Metrichor Platform with the R7.X 2D workflow (rev 1.34) for base calling SQK-MAP005. After the base calling the 2D reads were classified as either “pass” or “fail” based on a preset quality filter to create a high quality sequencing dataset. We combined the Nanopore and Illumina data for one ubas#1 positive (RSA016) and one ubas#1 negative (RS5346) strain and generated local assemblies for both strains with the help of the PBcR (version 8.3rc2) software (Berlin et al., 2015). This revealed the deletion of large parts of the gene Contig39-snap.126 in RS5346.
GWAS and Linkage Disequilibrium (LD)
GWAS analysis was performed using the cloud-based platform easyGWAS (Grimm et al., 2017) with the same parameters as described in (McGaughran et al., 2016). For the GWAS analysis a subset of 153 strains was used to increase the marker density in all strains analyzed. LD estimates were obtained as described (McGaughran et al., 2016). In brief, direct measures of fine-scale population linkage disequilibrium (R2) were obtained using plink ver. 1.07 (Purcell et al., 2007). LD estimates were averaged over non-outlier 10-kb windows throughout the genome to obtain genome-wide baseline estimates.
CRISPR/Cas9 Deletion Mutants
CRISPR/Cas9-induced inactivation was performed as previously described (Witte et al., 2015). For contig39-snap126, 386 F1 animals were screened for deletions by Sanger sequencing after micro-injection. We obtained one mutant line Contig39-snap126 (tu611) with a 7 bp deletion. The mutant line was backcrossed twice.
Sequence Data Sampling and Phylogenetic Analyses
Phylogenetic analyses were performed as described previously (Markov et al., 2015). In short, protein sequences for C. elegans, C. briggsae, C. remanei, H. contortus, B. malayi, L. loa, A. suum and T. spiralis, were collected from BLAST searches in GenBank. Sequences for B. xylophilus, M. hapla, P. redivivus and S. ratti were collected from BLAST searches in Wormbase version WS240 (Yook et al., 2012). Sequences of P. pacificus and P. exspectatus are from http://pristionchus.org. Accession numbers for all used sequences can be found in Table S3. Collected sequences were aligned with Muscle (Edgar, 2004) and alignments were checked by eye and edited with Seaview (Gouy et al., 2010). Some sequence predictions were manually refined and are provided in Table S5. Phylogenetic trees were made using PHYML (Guindon and Gascuel, 2003), a fast and accurate maximum likelihood heuristic method, using the best estimated substitution models, that turned out to be the LG+G model (Le et al., 2008). Reliability of nodes was assessed by likelihood-ratio test (Anisimova and Gascuel, 2006).
Ppa-uar-1 Genetic Complementation Attempts
We conducted genetic complementation or “rescue” experiments using DNA-mediated transformation as described in Schlager et al., 2009 (Schlager et al., 2009). In Brief, we created an extra-chromosomal array using various concentrations between 2 ng/μl and 0.1 ng/μl of a genetic construct of the RS5346 and RSA016 variants of the uar-1 gene (Ppa-uar-1 Contig39-snap126), a Ppa-egl-20::rfp (red fluorescent protein) reporter (10 ng/μl), and genomic carrier DNA (60 ng/μl) from the recipient line. The two different constructs used (primers listed in Key Resources Table) consisted of a ~12-kb fragment and a ~14-kb fragment encompassing the predicted Contig39-snap126 coding region plus the genomic sequence upstream and downstream from the coding sequence between the neighbouring genes, or in the case of the longer construct, the upstream region covering parts of the neighbouring gene (Contig39-snap.127). The arrays were microinjected into the germlines of adult hermaphrodites of RS5346 and RSA016. In total, we injected four rounds of 40 Individual for each strain and construct. Individual worms containing a signal for the Ppa-egl-20::rfp co-injection marker were observed several times for both strains (RS5346 and RSA016) and both constructs; however, none of these animals were able to produce viable offspring. As a control, we also injected worms only with the Ppa-egl-20::rfp reporter and created lines in RS5346 and RSA016 resulting in wildtype-like animals that reproduced normally.
Ppa-uar-1 Heterologous Expression Attempts in Bacteria and Yeast
The protein coding sequence for Ppa-uar-1 was codon optimized for Escherichia coli (see Table S1) and chemically synthesized, (Biomatik, Wilmington, DE)) cloned into pET-28a(+) and pET-21a(+) utilizing ligase-independent PCR cloning, which provide N- and C-terminal hexa-histidine tags, respectively. A truncation was also designed to remove the cysteine-rich c-terminal providing Ppa-uar-1 from residues 1 to 570. Plasmids containing either full-length or truncated Ppa-uar-1 were sequenced and transformed into BL21 (DE3) (New England Biolabs) and Tuner (DE3) (EMD Millipore) chemically competent Escherichia coli. Single colonies were inoculated into 5 mL LB media, selected with the appropriate antibiotic, and cultured overnight at 37 °C. Overnight cultures were then diluted 1:100 v/v into 1 L of Terrific Broth (TB) supplemented with 10 mM MgCl2 and cultured at 37 °C in a 4 L flask shaking at 200 RPM. Once an OD of 0.5 was reached, the temperature is reduced to 18 °C. At approximately OD 1.0, various final concentrations (50 μM to 1 mM) of IPTG were added to induce protein production. Upon IPTG addition, culture growth was either halted or very strongly inhibited due to protein toxicity. Incubations were continued for 18 hours before harvesting at 5000 G 4 °C for 10 min. Pellets were stored at −80 °C until further processing. Western blot analysis probing with HRP conjugated anti-6-His antibody (QED Bioscience) showed very minor expression. Purifications were attempted with 50 mM Tris or Phosphates with pHs ranging from 6 to 8, NaCl ranging from 0 to 500 mM, BME ranging from 0 to 15 mM, glycerol ranging from 0 to 20%, and with or without 1 mM PMSF or cOmplete Mini EDTA-free Protease Inhibitor Cocktail (Roche), which failed to yield soluble protein. Additionally, co-transformation screening of pET-21a(+) Ppa-uar-1 was performed with the TaKaRa Chaperone Plasmid Set (Clontech, Mountain View, CA) including, pG-KJE8, pGro7, pKJE7, pG-Tf2, and pTf16. Each chaperone plasmid was individually transformed with pET-21a(+) Ppa-uar-1 into BL21(DE3) Escherichia coli, and chaperone were utilized per manufactures protocol, however expression was not improved.
Kluyveromyces lactis (yeast) heterologous expression was carried out with the K. lactis Protein Expression Kit (New England Biolabs) per protocol. Briefly, the protein coding sequence (codon optimized) of Ppa-uar-1 was cloned into pKLAC2 utilizing ligase-independent cloning, using PCR primers to include a decahistadine C-terminal tag (see Key Resources Table). Additionally, primers were designed to, both include, or exclude the N-terminal α-mating factor secretion signal to either promote or exclude protein secretion, respectively. Single colonies from YCB plates containing 5 mM acetamide were inoculated into 5 mL of YPD at 30 °C and cultured for 2 days. Small cultures were diluted into protein inducing media at 1:100 v/v into 2 L YPGalactose (1% Yeast Extract, 2% Bacto Peptone, 2% galactose) in a 4 L flask, and incubated at 30 °C at 200 RPM for 3 to 7 days. Reduced growth rates were noted as culture saturation times were roughly twice their typical length. The culture was harvested at 5000 G 10 min at 4 °C, with the yeast pellet and the culture supernatant stored separately at −80 °C until further processing. Western blot analysis probing with HRP conjugated anti-6-His antibody showed no detectable band in the yeast pellet or culture supernatant.
Heterologous Expression of Ppa-uar-1 in C. elegans
C. elegans transgenic worms were created by a custom transgenic service (Knudra Transgenics, Salt Lake City, UT). Briefly, MosSCI service initiated with the construction of hsp16.41::Ppa-uar-1::tbb2u, where the hsp16.41 promoter and tbb-2 3′-UTR were taken from the N2 genome, and Ppa-uar-1 was synthesized with codon and intron optimization for C. elegans. This construct was cloned into the pNU936 backbone for delivery to the ttTi5605 locus on chromosome II. The pNU936 backbone containing hsp16.41::Ppa-uar-1::tbb2u, which includes an unc-119 rescue cassette, was injected into the COP93 [unc-119(ed3); ttTi5605] strain. The MosSCI plasmid was injected at 15 ng/μL and eft-3p::mosase along with the following co-markers: myo-2p::mCherry at 1.5 ng/μL, myo-3p::mCherry at 10 ng/μL, and rab-3p::mCherry at 10 ng/μL. Injected animals are maintained at 15 °C and screened for unc-119 rescue. Candidates absent of array markers were homozygosed. The homozygosed lines were confirmed by PCR for amplicons specific to targeted insertion and identified as COP1610-1613: knuSi763 [ pnu1497 (hsp16.41::Ppa-uar-1::tbb-2utr, unc-119(+)) ] II ; unc-119(ed3) III.
Ppa-uar-1 Heat Shock Induction in C. elegans
N2, COP1610, COP1611, COP1612, and COP1613 worms were grown in 25-mL synchronized liquid cultures fed with E. coli HB101. Two cultures for each strain were grown, and each culture was initiated with 70,000–80,000 arrested L1 larvae (14–20 h at room temperature after isolation of eggs). Cultures were grown in a shaker at 22 °C, 220 rpm. 35 h after starting the cultures, one flask for each strain was transferred to a 34 °C shaker for one hour. The heat shock treatment (1 h at 34 °C) was repeated once again 15 h later (50 h after starting the cultures). Worms from all cultures were spun down at 59 h and worm pellets and growth medium were collected separately. Worm pellets and media were frozen, lyophilized, extracted with methanol, and analyzed by LC/MS as described.
Ppa-uar-1 Heat Shock RNA Analysis in C. elegans
Flash frozen worm pellets from the heat shock induction procedure were thawed and suspended in M9 buffer to a total volume of 2 mL. 400 μL of the resuspended worms were moved a separate tube and 600 μL of TRIzol Reagent (Invitrogen) was added. 5 rapid freeze-thaw cycles were carried out between liquid nitrogen and a 37 °C water bath, and quickly vortexed before freezing. 120 μL of chloroform was added and the tubes were inverted several times before leaving the sample to rest at RT for 3 minutes. The samples were then spun at 20000 G for 20 minutes at 4 °C. 330 μL of the RNA containing layer was carefully pipetted away and then further purified with the RNeasy Mini Kit (Qiagen) per manufactures protocol. 5 μg total RNA was moved to the SuperScript III Reverse Transcriptase Kit (Invitrogen) for conversion to cDNA per manufactures protocol. Real time quantitative PCR was performed with Power SYBR Green Master Mix (Life Technologies) per manufactures protocol on a Light Cycler 480 (Roche). All samples were run in triplicate. Gene expression was normalized to actin.
Quantification and Statistical Analysis
RT-qPCR data was quantified using the Roche LightCycler 480 Software, Version 1.5 utilizing the Abs Quant/2nd Derivative Max feature and duplicate values were averaged and analyzed in Microsoft Excel.
Chemical Syntheses
(R,E)-8-(((2R,3R,5R,6S)-5-(((tert-butoxycarbonyl)-L-phenylalanyl)oxy)-3-hydroxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)non-2-enoic acid
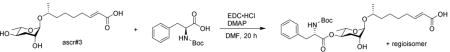
To a stirred solution of ascr#3 (10 mg, 0.033 mmol) in dry dichloromethane (1 mL), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC•HCl) (12.7 mg, 0.066 mmol), 4-dimethylaminopyridine (8.1 mg, 0.066 mmol) and Boc-L-phenylalanine (17.5 mg, 0.066 mmol) were added at room temperature. After 20 hours, the reaction was concentrated in vacuo. HPLC provided a pure product (5.1 mg, 28%).
1H NMR (600 MHz, methanol-d4): δ (ppm) 7.31−7.18 (m, 5H), 6.96 (dt, J = 15.6, 7.1 Hz, 1H), 5.83 (d, J = 15.6 Hz, 1H), 4.83 (ddd, J =11.3, 9.7, 4.7 Hz, 1H), 4.65 (br s, 1H), 4.32 (dd, J = 8.8, 6.4 Hz, 1H), 3.82−3.75 (m, 2H), 3.67−3.65 (m, 1H), 3.05 (dd, J = 13.9, 6.5 Hz, 1H), 2.94 (dd, J = 13.9, 8.7 Hz, 1H), 2.28−2.23 (m, 2H), 1.86 (dt, J = 13.1, 4.0 Hz, 1H), 1.66 (ddd, J = 13.1, 11.4, 3.0 Hz, 1H), 1.60−1.45 (m, 6H), 1.4 (s, 9H), 1.12 (d, J = 6.2 Hz, 6H).
(R,E)-8-(((2R,3R,5R,6S)-5-((L-phenylalanyl)oxy)-3-hydroxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)non-2-enoic acid

To a stirred solution of the BOC-protected compound (5.1 mg, 9.3 μmol) in dry dichloromethane (1 mL), trifluoroacetic acid (2.2 μL, 0.028 mmol) was added at room temperature. After 30 min, the reaction was concentrated in vacuo. Flash chromatograph on silica using a gradient of 0–20% methanol in dichloromethane afforded pure phascr#3 (4.0 mg, 96%).
1H NMR (600 MHz, methanol-d4): δ (ppm) 7.40−7.26 (m, 5H), 6.94 (dt, J = 15.6, 7.1 Hz, 1H), 5.81 (d, J = 15.6 Hz, 1H), 4.90 (m, 1H), 4.65 (br s, 1H), 4.35 (t, J = 7.2 Hz, 1H), 3.79−3.72 (m, 2H), 3.69−3.66 (m, 1H), 3.21 (dd, J = 14.2, 7.2 Hz, 1H), 3.17 (dd, J = 14.2, 7.5 Hz, 1H), 2.29−2.21 (m, 2H), 1.90 (dt, J = 12.9, 4.0 Hz, 1H), 1.66 (ddd, J = 12.9, 11.3, 3.1 Hz, 1H), 1.59−1.34 (m, 6H), 1.12 (d, J = 6.1 Hz, 3H), 1.10 (d, J =6.3 Hz, 3H). 13C NMR (600 MHz, chloroform-d): δ (ppm) 170.5, 169.7, 135.4, 130.6, 130.1, 128.9, 123.2, 97.6, 73.9, 72.9, 69.3, 67.8, 55.1, 38.0, 37.8, 33.1, 32.6, 29.1, 26.5, 19.4, 18.0.
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Goat Anti 6 - His Polyclonal Antibody, HRP Conjugate | QED Bioscience | Cat# 18812P |
| Bacterial and Virus Strains | ||
| E. coli OP50 | Lab of Leon Avery | N/A |
| E. coli HB101 | Lab of Leon Avery | N/A |
| NEB® 5 - alpha Competent E. coli (High Efficiency) | New England Biolabs | Cat# C2987H |
| BL21(DE3) Competent E. coli | New England Biolabs | Cat# C2527H |
| Tuner™(DE3) Competent E. coli | Novagen | Cat# 70623 - 3 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SMARTer® RACE 5′/3′ Kit | Clontech, Mountain View, USA | Cat#634859 |
| Master™ PureDNA extraction kit | Epicentre | Cat#MCD85201 |
| Direct-zolTM RNA MiniPrep | Zymo Research Germany | Cat# R2070 |
| QIAquick PCR Purification Kit | Qiagen, Germany | Cat# 28104 |
| Illumina’s paired-end protocol | Illumina, Netherlands | Cat#PE-102-1001 |
| Herculase II fusion DNA Polymerase kit | Agilent, Germany | Cat#600675 |
| Nanopore Sequencing Kit protocol SQK-MAP005 (Version MN005_1115_revE_26Nov2014) | Oxford Nanopore Technologies | Cat# SQK-LWP001 |
| NEBNext® End Repair Module | NEB | Cat# E6050 |
| Blunt/TA Ligase Master Mix | NEB | Cat# M0367 |
| Dynabeads® His-Tag Isolation and Pulldown | Thermo Fisher Scientific | Cat#10103D |
| TRIzol™ Reagent | Invitrogen | Cat# 15596018 |
| Critical Commercial Assays | ||
| K. lactis Protein Expression Kit | New England Biolabs | Cat# E1000S |
| Experimental Models: Organisms/Strains | ||
| C. elegans: COP93 [unc-119(ed3); ttTi5605] | Knudra Transgenics | N/A |
| C. elegans: N2 | Lab of Leon Avery | N/A |
| C. elegans: COP1610-1613: knuSi763 [ pnu1497 (hsp16.41::Ppa-uar-1::tbb-2utr, unc-119(+)) ] II ; unc-119(ed3) III. | This paper | N/A |
| P. pacificus: Ppa-uar-1 (tu611) | This paper | Contig39-snap126 |
| CostaR7 | P. pa. | Wild isolate |
| CostaRica3 | P. pa. | Wild isolate |
| JU482B | P. pa. | Wild isolate |
| PS1843 | P. pa. | Wild isolate |
| RS106B | P. pa. | Wild isolate |
| RS2333 | P. pa. | Wild isolate |
| RS5134B | P. pa. | Wild isolate |
| RS5188B | P. pa. | Wild isolate |
| RS5205 | P. pa. | Wild isolate |
| RS5208 | P. pa. | Wild isolate |
| RS5212B | P. pa. | Wild isolate |
| RS5215B | P. pa. | Wild isolate |
| RS5217B | P. pa. | Wild isolate |
| RS5221 | P. pa. | Wild isolate |
| RS5264 | P. pa. | Wild isolate |
| RS5265 | P. pa. | Wild isolate |
| RS5266 | P. pa. | Wild isolate |
| RS5275B | P. pa. | Wild isolate |
| RS5278 | P. pa. | Wild isolate |
| RS5279 | P. pa. | Wild isolate |
| RS5297 | P. pa. | Wild isolate |
| RS5333 | P. pa. | Wild isolate |
| RS5334 | P. pa. | Wild isolate |
| RS5336 | P. pa. | Wild isolate |
| RS5337 | P. pa. | Wild isolate |
| RS5338 | P. pa. | Wild isolate |
| RS5339 | P. pa. | Wild isolate |
| RS5340 | P. pa. | Wild isolate |
| RS5344 | P. pa. | Wild isolate |
| RS5346 | P. pa. | Wild isolate |
| RS5347 | P. pa. | Wild isolate |
| RS5348 | P. pa. | Wild isolate |
| RS5351 | P. pa. | Wild isolate |
| RS5353 | P. pa. | Wild isolate |
| RS5355 | P. pa. | Wild isolate |
| RS5377 | P. pa. | Wild isolate |
| RS5378 | P. pa. | Wild isolate |
| RS5379 | P. pa. | Wild isolate |
| RS5380 | P. pa. | Wild isolate |
| RS5382 | P. pa. | Wild isolate |
| RS5383 | P. pa. | Wild isolate |
| RS5384 | P. pa. | Wild isolate |
| RS5385 | P. pa. | Wild isolate |
| RS5386 | P. pa. | Wild isolate |
| RS5387 | P. pa. | Wild isolate |
| RS5388 | P. pa. | Wild isolate |
| RS5389 | P. pa. | Wild isolate |
| RS5390 | P. pa. | Wild isolate |
| RS5391 | P. pa. | Wild isolate |
| RS5392 | P. pa. | Wild isolate |
| RS5393 | P. pa. | Wild isolate |
| RS5394 | P. pa. | Wild isolate |
| RS5395 | P. pa. | Wild isolate |
| RS5396 | P. pa. | Wild isolate |
| RS5397 | P. pa. | Wild isolate |
| RS5398 | P. pa. | Wild isolate |
| RS5399 | P. pa. | Wild isolate |
| RS5400 | P. pa. | Wild isolate |
| RS5401 | P. pa. | Wild isolate |
| RS5402 | P. pa. | Wild isolate |
| RS5409 | P. pa. | Wild isolate |
| RS5410 | P. pa. | Wild isolate |
| RS5410 | P. pa. | Wild isolate |
| RS5412 | P. pa. | Wild isolate |
| RS5413 | P. pa. | Wild isolate |
| RS5415 | P. pa. | Wild isolate |
| RS5416 | P. pa. | Wild isolate |
| RS5417 | P. pa. | Wild isolate |
| RS5418 | P. pa. | Wild isolate |
| RS5419 | P. pa. | Wild isolate |
| RS5421 | P. pa. | Wild isolate |
| RS5423 | P. pa. | Wild isolate |
| RS5427 | P. pa. | Wild isolate |
| RS5428 | P. pa. | Wild isolate |
| RSA002 | P. pa. | Wild isolate |
| RSA003 | P. pa. | Wild isolate |
| RSA004 | P. pa. | Wild isolate |
| RSA005 | P. pa. | Wild isolate |
| RSA006 | P. pa. | Wild isolate |
| RSA007 | P. pa. | Wild isolate |
| RSA008 | P. pa. | Wild isolate |
| RSA009 | P. pa. | Wild isolate |
| RSA010 | P. pa. | Wild isolate |
| RSA011 | P. pa. | Wild isolate |
| RSA012 | P. pa. | Wild isolate |
| RSA014 | P. pa. | Wild isolate |
| RSA016 | P. pa. | Wild isolate |
| RSA017 | P. pa. | Wild isolate |
| RSA018 | P. pa. | Wild isolate |
| RSA018 | P. pa. | Wild isolate |
| RSA019 | P. pa. | Wild isolate |
| RSA020 | P. pa. | Wild isolate |
| RSA021 | P. pa. | Wild isolate |
| RSA022 | P. pa. | Wild isolate |
| RSA023 | P. pa. | Wild isolate |
| RSA027 | P. pa. | Wild isolate |
| RSA028 | P. pa. | Wild isolate |
| RSA029 | P. pa. | Wild isolate |
| RSA030 | P. pa. | Wild isolate |
| RSA031 | P. pa. | Wild isolate |
| RSA045 | P. pa. | Wild isolate |
| RSA045 | P. pa. | Wild isolate |
| RSA046 | P. pa. | Wild isolate |
| RSA047 | P. pa. | Wild isolate |
| RSA048 | P. pa. | Wild isolate |
| RSA056 | P. pa. | Wild isolate |
| RSA057 | P. pa. | Wild isolate |
| RSA069 | P. pa. | Wild isolate |
| RSA071 | P. pa. | Wild isolate |
| RSA073 | P. pa. | Wild isolate |
| RSA075 | P. pa. | Wild isolate |
| RSA076 | P. pa. | Wild isolate |
| RSA077 | P. pa. | Wild isolate |
| RSA079 | P. pa. | Wild isolate |
| RSA089 | P. pa. | Wild isolate |
| RSA091 | P. pa. | Wild isolate |
| RSA093 | P. pa. | Wild isolate |
| RSA094 | P. pa. | Wild isolate |
| RSA095 | P. pa. | Wild isolate |
| RSA096 | P. pa. | Wild isolate |
| RSA097 | P. pa. | Wild isolate |
| RSA098 | P. pa. | Wild isolate |
| RSA099 | P. pa. | Wild isolate |
| RSA100 | P. pa. | Wild isolate |
| RSA101 | P. pa. | Wild isolate |
| RSA102 | P. pa. | Wild isolate |
| RSA103 | P. pa. | Wild isolate |
| RSA104 | P. pa. | Wild isolate |
| RSA105 | P. pa. | Wild isolate |
| RSA108 | P. pa. | Wild isolate |
| RSA109 | P. pa. | Wild isolate |
| RSA110 | P. pa. | Wild isolate |
| RSA111 | P. pa. | Wild isolate |
| RSA113 | P. pa. | Wild isolate |
| RSA114 | P. pa. | Wild isolate |
| RSA115 | P. pa. | Wild isolate |
| RSA116 | P. pa. | Wild isolate |
| RSA117 | P. pa. | Wild isolate |
| RSA624 | P. pa. | Wild isolate |
| RSA626 | P. pa. | Wild isolate |
| RSA639 | P. pa. | Wild isolate |
| RSB001 | P. pa. | Wild isolate |
| RSB008 | P. pa. | Wild isolate |
| RSB013 | P. pa. | Wild isolate |
| RSB018 | P. pa. | Wild isolate |
| RSB020 | P. pa. | Wild isolate |
| RSB025 | P. pa. | Wild isolate |
| RSB026 | P. pa. | Wild isolate |
| RSB031 | P. pa. | Wild isolate |
| RSB034 | P. pa. | Wild isolate |
| RSB038 | P. pa. | Wild isolate |
| RSB040 | P. pa. | Wild isolate |
| RSB060 | P. pa. | Wild isolate |
| RSB070 | P. pa. | Wild isolate |
| RSB071 | P. pa. | Wild isolate |
| RSB072 | P. pa. | Wild isolate |
| RSB073 | P. pa. | Wild isolate |
| RSB079 | P. pa. | Wild isolate |
| RSB080 | P. pa. | Wild isolate |
| RSB091 | P. pa. | Wild isolate |
| RSB092 | P. pa. | Wild isolate |
| RSB094 | P. pa. | Wild isolate |
| RSB096 | P. pa. | Wild isolate |
| RSB097 | P. pa. | Wild isolate |
| RSB098 | P. pa. | Wild isolate |
| RSB099 | P. pa. | Wild isolate |
| RSB100 | P. pa. | Wild isolate |
| RSB101 | P. pa. | Wild isolate |
| RSB102 | P. pa. | Wild isolate |
| RSB103 | P. pa. | Wild isolate |
| RSB104 | P. pa. | Wild isolate |
| RSB105 | P. pa. | Wild isolate |
| RSB106 | P. pa. | Wild isolate |
| RSB107 | P. pa. | Wild isolate |
| RSB108 | P. pa. | Wild isolate |
| RSB109 | P. pa. | Wild isolate |
| RSB110 | P. pa. | Wild isolate |
| RSB111 | P. pa. | Wild isolate |
| RSB112 | P. pa. | Wild isolate |
| RSB113 | P. pa. | Wild isolate |
| RSB114 | P. pa. | Wild isolate |
| RSB115 | P. pa. | Wild isolate |
| RSB116 | P. pa. | Wild isolate |
| RSB117 | P. pa. | Wild isolate |
| RSB118 | P. pa. | Wild isolate |
| RSB119 | P. pa. | Wild isolate |
| RSB120 | P. pa. | Wild isolate |
| RSB120 | P. pa. | Wild isolate |
| RSB121 | P. pa. | Wild isolate |
| RSB122 | P. pa. | Wild isolate |
| RSB123 | P. pa. | Wild isolate |
| RSB124 | P. pa. | Wild isolate |
| RSB125 | P. pa. | Wild isolate |
| RSB126 | P. pa. | Wild isolate |
| RSC015 | P. pa. | Wild isolate |
| RSC046 | P. pa. | Wild isolate |
| RSC047 | P. pa. | Wild isolate |
| RSC048 | P. pa. | Wild isolate |
| RSC049 | P. pa. | Wild isolate |
| RSC050 | P. pa. | Wild isolate |
| RSC130 | P. pa. | Wild isolate |
| RSC141 | P. pa. | Wild isolate |
| RSC142 | P. pa. | Wild isolate |
| RSC143 | P. pa. | Wild isolate |
| RSC144 | P. pa. | Wild isolate |
| RSC145 | P. pa. | Wild isolate |
| RSC146 | P. pa. | Wild isolate |
| RSC147 | P. pa. | Wild isolate |
| RSC148 | P. pa. | Wild isolate |
| RSC149 | P. pa. | Wild isolate |
| RSC150 | P. pa. | Wild isolate |
| RSC151 | P. pa. | Wild isolate |
| RSC152 | P. pa. | Wild isolate |
| RSC154 | P. pa. | Wild isolate |
| RSC155 | P. pa. | Wild isolate |
| RSC156 | P. pa. | Wild isolate |
| RSC157 | P. pa. | Wild isolate |
| RSC158 | P. pa. | Wild isolate |
| RSC159 | P. pa. | Wild isolate |
| RSC160 | P. pa. | Wild isolate |
| RSC161 | P. pa. | Wild isolate |
| RSC162 | P. pa. | Wild isolate |
| RSD078 | P. pa. | Wild isolate |
| RSD104 | P. pa. | Wild isolate |
| RSD106 | P. pa. | Wild isolate |
| RSD115 | P. pa. | Wild isolate |
| RSD116 | P. pa. | Wild isolate |
| RSD122 | P. pa. | Wild isolate |
| RSD123 | P. pa. | Wild isolate |
| RSD124 | P. pa. | Wild isolate |
| RSD125 | P. pa. | Wild isolate |
| RSD139 | P. pa. | Wild isolate |
| RSD141 | P. pa. | Wild isolate |
| RSD144 | P. pa. | Wild isolate |
| RSD146 | P. pa. | Wild isolate |
| RSD147 | P. pa. | Wild isolate |
| RSD156 | P. pa. | Wild isolate |
| RSD157 | P. pa. | Wild isolate |
| RSD159 | P. pa. | Wild isolate |
| RSD165 | P. pa. | Wild isolate |
| RSD166 | P. pa. | Wild isolate |
| RSD171 | P. pa. | Wild isolate |
| RSD175 | P. pa. | Wild isolate |
| RSD178 | P. pa. | Wild isolate |
| RSD182 | P. pa. | Wild isolate |
| RSD182- | P. pa. | Wild isolate |
| RSD184 | P. pa. | Wild isolate |
| RSD185 | P. pa. | Wild isolate |
| RSD187 | P. pa. | Wild isolate |
| RSD188 | P. pa. | Wild isolate |
| RSD190 | P. pa. | Wild isolate |
| RSD192 | P. pa. | Wild isolate |
| RSD196 | P. pa. | Wild isolate |
| RSD197 | P. pa. | Wild isolate |
| RSD200 | P. pa. | Wild isolate |
| RSD201 | P. pa. | Wild isolate |
| RSD204 | P. pa. | Wild isolate |
| RSD205 | P. pa. | Wild isolate |
| RSD209 | P. pa. | Wild isolate |
| RSD213 | P. pa. | Wild isolate |
| RSD217 | P. pa. | Wild isolate |
| RSD218 | P. pa. | Wild isolate |
| RSD220 | P. pa. | Wild isolate |
| RSD221 | P. pa. | Wild isolate |
| RSD224 | P. pa. | Wild isolate |
| RSD225 | P. pa. | Wild isolate |
| RSD228 | P. pa. | Wild isolate |
| RSD229 | P. pa. | Wild isolate |
| RSD230 | P. pa. | Wild isolate |
| RSD233 | P. pa. | Wild isolate |
| RSD234 | P. pa. | Wild isolate |
| RSD235 | P. pa. | Wild isolate |
| RSD236 | P. pa. | Wild isolate |
| RSD237 | P. pa. | Wild isolate |
| RSD239 | P. pa. | Wild isolate |
| RSD240 | P. pa. | Wild isolate |
| RSD241 | P. pa. | Wild isolate |
| RSD242 | P. pa. | Wild isolate |
| RSD243 | P. pa. | Wild isolate |
| RSD246 | P. pa. | Wild isolate |
| RSD249 | P. pa. | Wild isolate |
| RSD250 | P. pa. | Wild isolate |
| RSD252 | P. pa. | Wild isolate |
| RSD257 | P. pa. | Wild isolate |
| RSD260 | P. pa. | Wild isolate |
| RSD261 | P. pa. | Wild isolate |
| RSD263 | P. pa. | Wild isolate |
| RSD264 | P. pa. | Wild isolate |
| RSD268 | P. pa. | Wild isolate |
| RSD272 | P. pa. | Wild isolate |
| RSD275 | P. pa. | Wild isolate |
| RSD276 | P. pa. | Wild isolate |
| RSD277 | P. pa. | Wild isolate |
| RSD278 | P. pa. | Wild isolate |
| RSD281 | P. pa | Wild isolate |
| RSD283 | P. pa | Wild isolate |
| RSD284 | P. pa | Wild isolate |
| SB5880B | P. pa | Wild isolate |
| Oligonucleotides: Please see Supplemental Table S6 for oligonucleotide/primer sequences | ||
| Recombinant DNA | ||
| Plasmid: pNU936 hsp16.41::Ppa-uar-1::tbb2u | Knudra Transgenics | N/A |
| Plasmid: pNU936 rpl-28::Ppa-uar-1::tbb2u | Knudra Transgenics | N/A |
| Plasmid: pET-28a(+) | Novagen | Cat# 69864-3 |
| Plasmid: pET-21a(+) | Novagen | Cat# 69770-3 |
| TaKaRa Chaperone Plasmid Set: pG-KJE8, pGro7, pKJE7, pG-Tf2, and pTf16. | Clontech | Cat# 3340 |
| Software and Algorithms | ||
| Geneious® | Biomatters Limited, version 8.0.4 | https://www.geneious.com/download/ |
| easyGWAS | https://easygwas.ethz.ch/ | |
| BWA | Version 0.6.1-r104 | https://github.com/lh3/bwa |
| samtools | Version 0.1.18 | http://www.htslib.org/ |
| PHYML | Version 3.0 | http://www.atgc-montpellier.fr/phyml/ |
| Muscle | Version 3.8.31 | https://www.drive5.com/muscle/ |
| Seaview | Version 1:4.4.2-1 | http://doua.prabi.fr/software/seaview |
| PBcR | Version 8.3rc2 | http://wgs-assembler.sourceforge.net/wiki/index.php/PBcR |
| XCMS R code | Bioconductor (Tautenhahn et al., 2012) | http://bioconductor.org/packages/release/bioc/html/xcms.html |
| Thermo XCalibur | ThermoFisher | https://www.thermofisher.com/order/catalog/product/OPTON-30487 |
| MestReNova | Mestrelab Research, version 11.0 | http://mestrelab.com/download/mnova/ |
Supplementary Material
Table S3. Accession numbers for all used sequences. Related to Figure 3.
Table S4. Manually improved protein sequence models. Related to Figure 3.
Table S6. Oligonucleotide sequences. Related to Figure 3.
Significance.
Natural genetic variation provides an important resource that can be leveraged to identify diverse genetic traits using genome-wide association (GWAS) mappings. However, the potential of GWAS to uncover the biosyntheses of the vast numbers of new metabolites currently being identified in animal model organisms has not been explored. In our study, we employ targeted metabolomics to measure natural variation of important nematode signaling molecules as input for GWAS mapping to identify biosynthetic loci. The signaling molecules we selected for this study, the nematode-derived modular metabolites (NDMMs), are a recently discovered class of metabolites that serve as central regulators of development and aging in the model organisms C. elegans and P. pacificus. Little is known about the pathways involved in the modular assembly of simple primary metabolic building blocks in the NDMMs, a feature that distinguishes NDMMs from other natural product classes. Importantly, the identification of enzymes involved in NDMM assembly will help clarify whether higher animals employ analogous strategies for the biosynthesis of modular signaling molecules.
Our approach led to the identification of uar-1, a gene that mediates a highly specific acylation reaction required for the biogenesis of NDMMs featuring a 3-ureidoisobutyrate (pyrimidine-metabolism-derived) moiety. Further, comparative metabolomic analysis of wildtype and uar-1 mutants, generated via CRISPR-Cas9-induced genetic engineering, revealed several previously unrecognized uar-1-dependent metabolites demonstrating that the biosynthesis of signaling molecules in nematodes proceeds via an interconnected network of primary metabolic pathways. Taken together, our study demonstrates the potential of natural variation-based approaches for the elucidation of the biosynthesis of important signaling molecules in animal model systems.
Highlights.
-
-
GWAS identifies a gene required for modular ascaroside biosynthesis.
-
-
Ppa-UAR-1 is required for attachment of a nucleobase-derived moiety to ascarylose.
-
-
Heterologous expression of Ppa-UAR-1 in C. elegans produced a non-natural ascaroside.
-
-
Natural variation-based approaches can uncover unanticipated biosynthetic pathways.
Acknowledgments
This work was supported in part by the NIH (GM113692, GM088290 to FCS, T32 GM008500 to JAB) and HHMI. We thank Maro J. Kariya for providing natural fractions containing ubas#6. We are grateful to David Kiemle for help with NMR spectroscopy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
J.M.F., N.B., R.J.S., and F.C.S. designed experiments. J.M.F., N.B., A.B.A., D.J., M.C., H.H.L., and O.P. conducted biological experiments. J.M.F., D.G., G.V.M., and C.R. performed bioinformatic analyses. N.B., A.B.A., J.J.Y., O.P., and J.A.B. performed HPLC-MS analyses. Y.K.Z. synthesized phascr#3. J.M.F., N.B., A.B.A., R.J.S. and F.C.S. wrote the manuscript. All authors read and commented on the manuscript.
DECLARATION OF INTERESTS
The authors declare no competing interests.
Supplemental Information
Document S1. Supplemental Experimental Procedures, Figures S1–S5, and Tables S1, S2, and S5.
References
- Andersen EC, Gerke JP, Shapiro JA, Crissman JR, Ghosh R, Bloom JS, Felix MA, Kruglyak L. Chromosome-scale selective sweeps shape Caenorhabditis elegans genomic diversity. Nat Genet. 2012;44:285–290. doi: 10.1038/ng.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- Artyukhin AB, Yim JJ, Srinivasan J, Izrayelit Y, Bose N, von Reuss SH, Jo Y, Jordan JM, Baugh LR, Cheong M, et al. Succinylated octopamine ascarosides and a new pathway of biogenic amine metabolism in Caenorhabditis elegans. J Biol Chem. 2013;288:18778–18783. doi: 10.1074/jbc.C113.477000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol. 2015;33:623–630. doi: 10.1038/nbt.3238. [DOI] [PubMed] [Google Scholar]
- Bose N, Meyer JM, Yim JJ, Mayer MG, Markov GV, Ogawa A, Schroeder FC, Sommer RJ. Natural variation in dauer pheromone production and sensing supports intraspecific competition in nematodes. Curr Biol. 2014;24:1536–1541. doi: 10.1016/j.cub.2014.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bose N, Ogawa A, von Reuss SH, Yim JJ, Ragsdale EJ, Sommer RJ, Schroeder FC. Complex small-molecule architectures regulate phenotypic plasticity in a nematode. Angew Chem Int Ed Engl. 2012;51:12438–12443. doi: 10.1002/anie.201206797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush WS, Moore JH. Chapter 11: Genome-wide association studies. PLoS computational biology. 2012;8:e1002822. doi: 10.1371/journal.pcbi.1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertemps T, Francois A, Durand N, Rosell G, Dekker T, Lucas P, Maibeche-Coisne M. A carboxylesterase, Esterase-6, modulates sensory physiological and behavioral response dynamics to pheromone in Drosophila. BMC Biol. 2012;10:56. doi: 10.1186/1741-7007-10-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DE, Zdraljevic S, Roberts JP, Andersen EC. CeNDR, the Caenorhabditis elegans natural diversity resource. Nucleic Acids Res. 2017;45:D650–D657. doi: 10.1093/nar/gkw893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke TF, Fischer CR, Wu P, Jiang TX, Xie KT, Kuo J, Doctorov E, Zehnder A, Khosla C, Chuong CM, et al. Genetic Mapping and Biochemical Basis of Yellow Feather Pigmentation in Budgerigars. Cell. 2017;171:427–439. doi: 10.1016/j.cell.2017.08.016. e421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich C, Clifton SW, Schuster LN, Chinwalla A, Delehaunty K, Dinkelacker I, Fulton L, Fulton R, Godfrey J, Minx P, et al. The Pristionchus pacificus genome provides a unique perspective on nematode lifestyle and parasitism. Nat Genet. 2008;40:1193–1198. doi: 10.1038/ng.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fielenbach N, Antebi A. C. elegans dauer formation and the molecular basis of plasticity. Genes Dev. 2008;22:2149–2165. doi: 10.1101/gad.1701508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh R, Andersen EC, Shapiro JA, Gerke JP, Kruglyak L. Natural variation in a chloride channel subunit confers avermectin resistance in C. elegans. Science. 2012;335:574–578. doi: 10.1126/science.1214318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- Grimm DG, Roqueiro D, Salome PA, Kleeberger S, Greshake B, Zhu W, Liu C, Lippert C, Stegle O, Scholkopf B, et al. easyGWAS: A Cloud-Based Platform for Comparing the Results of Genome-Wide Association Studies. Plant Cell. 2017;29:5–19. doi: 10.1105/tpc.16.00551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Herrmann M, Mayer WE, Hong RL, Kienle S, Minasaki R, Sommer RJ. The nematode Pristionchus pacificus (Nematoda: Diplogastridae) is associated with the oriental beetle Exomala orientalis (Coleoptera: Scarabaeidae) in Japan. Zoolog Sci. 2007;24:883–889. doi: 10.2108/zsj.24.883. [DOI] [PubMed] [Google Scholar]
- Hu PJ. Dauer. WormBook. 2007:1–19. doi: 10.1895/wormbook.1.144.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JB, Lange JD, Tang AD, Corbett-Detig RB, Pool JE. A Thousand Fly Genomes: An Expanded Drosophila Genome Nexus. Mol Biol Evol. 2016;33:3308–3313. doi: 10.1093/molbev/msw195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen NA, Turner JM, Stevens J, Rosser SJ, Basran A, Lerner RA, Bruce NC, Wilson IA. Crystal structure of a bacterial cocaine esterase. Nat Struct Biol. 2002;9:17–21. doi: 10.1038/nsb742. [DOI] [PubMed] [Google Scholar]
- Le SQ, Lartillot N, Gascuel O. Phylogenetic mixture models for proteins. Philos Trans R Soc Lond B Biol Sci. 2008;363:3965–3976. doi: 10.1098/rstb.2008.0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludewig AH, Schroeder FC. Ascaroside signaling in C. elegans. WormBook. 2013:1–22. doi: 10.1895/wormbook.1.155.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, Geer LY, Geer RC, He J, Gwadz M, Hurwitz DI, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Research. 2015;43:D222–D226. doi: 10.1093/nar/gku1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markov GV, Baskaran P, Sommer RJ. The same or not the same: lineage-specific gene expansions and homology relationships in multigene families in nematodes. J Mol Evol. 2015;80:18–36. doi: 10.1007/s00239-014-9651-y. [DOI] [PubMed] [Google Scholar]
- Markov GV, Meyer JM, Panda O, Artyukhin AB, Claassen M, Witte H, Schroeder FC, Sommer RJ. Functional Conservation and Divergence of daf-22 Paralogs in Pristionchus pacificus Dauer Development. Mol Biol Evol. 2016;33:2506–2514. doi: 10.1093/molbev/msw090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughran A, Rodelsperger C, Grimm DG, Meyer JM, Moreno E, Morgan K, Leaver M, Serobyan V, Rakitsch B, Borgwardt KM, et al. Genomic Profiles of Diversification and Genotype-Phenotype Association in Island Nematode Lineages. Mol Biol Evol. 2016;33:2257–2272. doi: 10.1093/molbev/msw093. [DOI] [PubMed] [Google Scholar]
- McGrath PT, Rockman MV, Zimmer M, Jang H, Macosko EZ, Kruglyak L, Bargmann CI. Quantitative mapping of a digenic behavioral trait implicates globin variation in C. elegans sensory behaviors. Neuron. 2009;61:692–699. doi: 10.1016/j.neuron.2009.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JM, Markov GV, Baskaran P, Herrmann M, Sommer RJ, Rodelsperger C. Draft Genome of the Scarab Beetle Oryctes borbonicus on La Reunion Island. Genome biology and evolution. 2016;8:2093–2105. doi: 10.1093/gbe/evw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazaki M, Yamashita T, Suzuki Y, Saito Y, Soeta S, Taira H, Suzuki A. A major urinary protein of the domestic cat regulates the production of felinine, a putative pheromone precursor. Chem Biol. 2006;13:1071–1079. doi: 10.1016/j.chembiol.2006.08.013. [DOI] [PubMed] [Google Scholar]
- Morgan K, McGaughran A, Villate L, Herrmann M, Witte H, Bartelmes G, Rochat J, Sommer RJ. Multi locus analysis of Pristionchus pacificus on La Reunion Island reveals an evolutionary history shaped by multiple introductions, constrained dispersal events and rare out-crossing. Mol Ecol. 2012;21:250–266. doi: 10.1111/j.1365-294X.2011.05382.x. [DOI] [PubMed] [Google Scholar]
- Niehaus TD, Thamm AM, de Crecy-Lagard V, Hanson AD. Proteins of Unknown Biochemical Function: A Persistent Problem and a Roadmap to Help Overcome It. Plant Physiol. 2015;169:1436–1442. doi: 10.1104/pp.15.00959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa A, Streit A, Antebi A, Sommer RJ. A conserved endocrine mechanism controls the formation of dauer and infective larvae in nematodes. Curr Biol. 2009;19:67–71. doi: 10.1016/j.cub.2008.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda O, Akagi AE, Artyukhin AB, Judkins JC, Le HH, Mahanti P, Cohen SM, Sternberg PW, Schroeder FC. Biosynthesis of Modular Ascarosides in C. elegans. Angew Chem Int Ed Engl. 2017;56:4729–4733. doi: 10.1002/anie.201700103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindel EV, Kedishvili NY, Abraham TL, Brzezinski MR, Zhang J, Dean RA, Bosron WF. Purification and cloning of a broad substrate specificity human liver carboxylesterase that catalyzes the hydrolysis of cocaine and heroin. J Biol Chem. 1997;272:14769–14775. doi: 10.1074/jbc.272.23.14769. [DOI] [PubMed] [Google Scholar]
- Poland JA, Brown PJ, Sorrells ME, Jannink JL. Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One. 2012;7:e32253. doi: 10.1371/journal.pone.0032253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragsdale Erik J, Müller Manuela R, Rödelsperger C, Sommer Ralf J. A Developmental Switch Coupled to the Evolution of Plasticity Acts through a Sulfatase. Cell. 2013;155:922–933. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- Rhee EP, Ho JE, Chen MH, Shen D, Cheng S, Larson MG, Ghorbani A, Shi X, Helenius IT, O’Donnell CJ, et al. A genome-wide association study of the human metabolome in a community-based cohort. Cell Metab. 2013;18:130–143. doi: 10.1016/j.cmet.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A, Schaff C, Zhang Z, Lipka AE, Tian F, Kollner TG, Schnee C, Preiss S, Irmisch S, Jander G, et al. Characterization of Biosynthetic Pathways for the Production of the Volatile Homoterpenes DMNT and TMTT in Zea mays. Plant Cell. 2016;28:2651–2665. doi: 10.1105/tpc.15.00919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodelsperger C, Neher RA, Weller AM, Eberhardt G, Witte H, Mayer WE, Dieterich C, Sommer RJ. Characterization of genetic diversity in the nematode Pristionchus pacificus from population-scale resequencing data. Genetics. 2014;196:1153–1165. doi: 10.1534/genetics.113.159855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saerens SM, Verstrepen KJ, Van Laere SD, Voet AR, Van Dijck P, Delvaux FR, Thevelein JM. The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J Biol Chem. 2006;281:4446–4456. doi: 10.1074/jbc.M512028200. [DOI] [PubMed] [Google Scholar]
- Schlager B, Wang X, Braach G, Sommer RJ. Molecular cloning of a dominant roller mutant and establishment of DNA-mediated transformation in the nematode Pristionchus pacificus. Genesis. 2009;47:300–304. doi: 10.1002/dvg.20499. [DOI] [PubMed] [Google Scholar]
- Schrimpe-Rutledge AC, Codreanu SG, Sherrod SD, McLean JA. Untargeted Metabolomics Strategies-Challenges and Emerging Directions. J Am Soc Mass Spectrom. 2016;27:1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serobyan V, Xiao H, Namdeo S, Rodelsperger C, Sieriebriennikov B, Witte H, Roseler W, Sommer RJ. Chromatin remodelling and antisense-mediated up-regulation of the developmental switch gene eud-1 control predatory feeding plasticity. Nat Commun. 2016;7:12337. doi: 10.1038/ncomms12337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha A, Langnick C, Sommer RJ, Dieterich C. Genome-wide analysis of trans-splicing in the nematode Pristionchus pacificus unravels conserved gene functions for germline and dauer development in divergent operons. RNA. 2014;20:1386–1397. doi: 10.1261/rna.041954.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer RJ, Carta LK, Kim SY, Sternberg PW. Morphological, genetic and molecular description of Pristionchus pacificus sp n (Nematoda: Neodiplogastridae) Fundamental and Applied Nematology. 1996;19:511–521. [Google Scholar]
- Sommer RJ, Dardiry M, Lenuzzi M, Namdeo S, Renahan T, Sieriebriennikov B, Werner MS. The genetics of phenotypic plasticity in nematode feeding structures. Open biology. 2017;7 doi: 10.1098/rsob.160332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautenhahn R, Patti GJ, Rinehart D, Siuzdak G. XCMS Online: A Web-Based Platform to Process Untargeted Metabolomic Data. Analytical Chemistry. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viant MR, Kurland IJ, Jones MR, Dunn WB. How close are we to complete annotation of metabolomes? Curr Opin Chem Biol. 2017;36:64–69. doi: 10.1016/j.cbpa.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reuss SH, Bose N, Srinivasan J, Yim JJ, Judkins JC, Sternberg PW, Schroeder FC. Comparative metabolomics reveals biogenesis of ascarosides, a modular library of small molecule signals in C. elegans. J Am Chem Soc. 2012;134:1817–1824. doi: 10.1021/ja210202y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Reuss SH, Schroeder FC. Combinatorial chemistry in nematodes: modular assembly of primary metabolism-derived building blocks. Nat Prod Rep. 2015;32:994–1006. doi: 10.1039/c5np00042d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun. 2014;5:3438. doi: 10.1038/ncomms4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte H, Moreno E, Rodelsperger C, Kim J, Kim JS, Streit A, Sommer RJ. Gene inactivation using the CRISPR/Cas9 system in the nematode Pristionchus pacificus. Dev Genes Evol. 2015;225:55–62. doi: 10.1007/s00427-014-0486-8. [DOI] [PubMed] [Google Scholar]
- Wu S, Alseekh S, Cuadros-Inostroza A, Fusari CM, Mutwil M, Kooke R, Keurentjes JB, Fernie AR, Willmitzer L, Brotman Y. Combined Use of Genome-Wide Association Data and Correlation Networks Unravels Key Regulators of Primary Metabolism in Arabidopsis thaliana. PLoS Genet. 2016;12:e1006363. doi: 10.1371/journal.pgen.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yook K, Harris TW, Bieri T, Cabunoc A, Chan J, Chen WJ, Davis P, de la Cruz N, Duong A, Fang R, et al. WormBase 2012: more genomes, more data, new website. Nucleic Acids Res. 2012;40:D735–741. doi: 10.1093/nar/gkr954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zampieri M, Sekar K, Zamboni N, Sauer U. Frontiers of high-throughput metabolomics. Curr Opin Chem Biol. 2017;36:15–23. doi: 10.1016/j.cbpa.2016.12.006. [DOI] [PubMed] [Google Scholar]
- Zhang X, Feng L, Chinta S, Singh P, Wang Y, Nunnery JK, Butcher RA. Acyl-CoA oxidase complexes control the chemical message produced by Caenorhabditis elegans. Proceedings of the National Academy of Sciences. 2015;112:3955–3960. doi: 10.1073/pnas.1423951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S3. Accession numbers for all used sequences. Related to Figure 3.
Table S4. Manually improved protein sequence models. Related to Figure 3.
Table S6. Oligonucleotide sequences. Related to Figure 3.


