Abstract
A subset of chromosomal translocations related to B cell malignancy in human patients arise due to DNA breaks occurring within defined 20 to 600 basepair zones. Several factors influence the breakage rate at these sites including transcription, DNA sequence, and topological tension. These factors favor non-B DNA structures that permit formation of transient single-stranded DNA (ssDNA), making the DNA more vulnerable to agents such as the enzyme activation-induced cytidine deaminase (AID) and reactive oxygen species (ROS). Certain DNA lesions created during the ssDNA state persist after the DNA resumes its normal duplex structure. We propose that factors favoring both formation of transient ssDNA and persistent DNA lesions are key in determining the DNA breakage mechanism.
MULTIPLE FACTORS FAVOR BREAKAGE AT HIGHLY LOCALIZED FRAGILE ZONES
Chromosomal translocations involving the BCL2 or CCND1 loci occur in nearly all cases of follicular lymphoma and mantle cell lymphoma, respectively [1]. These translocations occur when a DNA double-strand break (DSB) generated near these oncogenes joins with an induced DSB created at the immunoglobulin heavy chain (IgH) locus during V(D)J recombination. While the V(D)J DSBs are initiated by the recombination activating gene (RAG) complex, the cause and mechanism of DSB formation at the non-IgH loci is unknown. Translocations involving RAG cutting at the non-IgH sites are not common (relative to mechanisms discussed below) and are mostly confined to a subset of T-cell lymphomas [1].
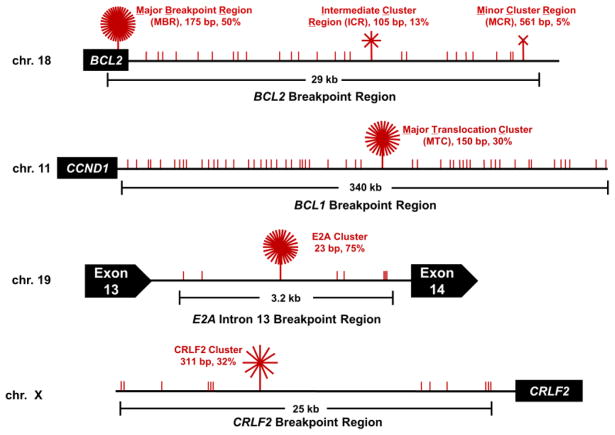
Sequencing data from over 2000 human patient translocations has revealed that DSBs resulting in neoplastic translocations can arise at some of these non-IgH loci over spans of tens to hundreds of kilobases, yet are clustered up to 1000-fold more frequently in regions of only 20 to 600 bp (Figure 1) [2]. We refer to these focal regions of DSB formation as fragile zones. The major breakpoint region (MBR) of BCL2 and the major translocation cluster (MTC) of CCND1 are two such fragile zones, but several other fragile zones relevant to chromosomal translocations in other human B cell malignancies have also been described (Figure 1) [1, 2].
Figure 1. Fragile Zones in Human Lymphoid Chromosomal Translocations.
Schematics of the BCL2 break cluster regions on chromosome 18, the BCL1 breakpoint region (which is downstream of the cyclin D1 (CCND1) gene) on chromosome 11, Exon 13 of E2A (also known as TCF3) on chromosome 19, and the CRLF2 breakpoint region upstream of the CRLF2 gene on the X chromosome illustrate the accumulation of breakpoints within the various regions. Breakpoint junctions sequenced from human patients demonstrate that DSBs anywhere within these regions can result in chromosomal translocations relevant for various B cell malignancies (red vertical lines represent a mapped breakpoint), within these regions, however, are regions where breakpoints appear to cluster in a non-random fashion (red starbursts). In the BCL2 region (top), relative proportions of breakpoints at the BCL2 major breakpoint region (MBR), intermediate cluster region (ICR), and minor cluster region (MCR) are shown. The MBR is located in the third exon of the BCL2 gene within the 3′ untranslated region (UTR), while the ICR and MCR are further downstream from the translated region. The 175 bp MBR, 105 bp ICR, and 561 bp MCR account for about 50%, 13%, and 5% of the BCL2 translocation breakpoints. Within the BCL1 region, the major translocation cluster (MTC) is located about 110 kb from the CCND1 gene. The 150 bp MTC contains about 30% of breakpoints, whereas the remaining 70% of events are distributed widely over the surrounding 340 kb as recently mapped and sequenced [33]. In the E2A cluster, which occurs in intron 13 of the E2A gene [34], 75% of breakpoints occur in the 23 bp E2A cluster, while the surrounding 3 kb only account for 25%. The CRLF2 cluster lies upstream of the CRLF2 gene with 32% of mapped break in the 25 kb region occurring within a 311 bp cluster. These sites of breakpoint accumulation that range from 23 to 561 bp are termed ‘fragile zones’ and every CG sequence motif in each of these fragile zones is a hotspot for human translocation [2, 33].
The fragile zones we describe are naturally occurring, meaning they are fragile (DSB-prone) in wild-type cells and not induced by replication stress. This distinguishes them from regions known as common fragile sites (FRA sites) that are not generally involved in chromosomal translocations. In other words, these are breaks that occur in healthy human patients and are not induced by underlying drug treatment or genetic defects. Therefore, understanding the mechanism that makes relatively small and defined regions of DNA prone to DSBs has the potential to reveal the disease etiology at the crossroads of naturally occurring biological processes and environmental exposure.
We have identified several factors that increase the rate of DSBs occurring within these fragile zones. Four of the factors affecting fragility are considered here. First, fragile zones display a transient, non-B conformation containing single-stranded DNA (ssDNA)[3, 4]. Second, the transient ssDNA structures are susceptible to damage from agents such as reactive oxygen species (ROS) or activation-induced cytidine deaminase (AID)[5]. Third, this non-B conformation can be stabilized by changes in torsional stress that favor unwinding of the duplex, thereby increasing the lifetime of the transient structure [6]. Fourth, the rate of lesion repair and the proximity of two or more lesions on each of the two anti-parallel DNA strands determines the frequency of DSB formation [5].
FORMATION OF SINGLE-STRANDED, NON-B DNA STRUCTURES
We have yet to definitively identify a specific non-B DNA structure because they are likely to be very short-lived (on the order of milliseconds or less) [6]. We note, however, that many of the fragile zone sequences have runs of cytosines (C’s) in several locations on the same strand (C-strings) [1–4]. One consequence of these C-strings on the DNA duplex are structural and electrostatic changes between the stacked C’s and guanines (G’s) that leads to a change from the typical B-form helix to a conformation that shifts between B- and A-form (B/A intermediate) [7]. This state causes increased single-strandedness, evidenced by the fact that C’s in these regions are more susceptible to reaction with sodium bisulfite under native conditions (increased accessibility of C’s to bisulfite under non-denaturing conditions indicates an increased tendency for the C’s not being protected within the duplex and, thus, not base paired) [4].
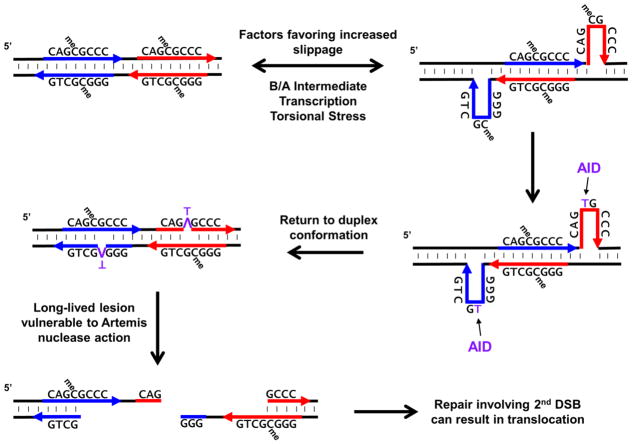
The B/A intermediate and any mismatches will favor breathing, potentiating the formation of higher-order single-stranded structures [4]. One such structure is slipped-strand DNA where, following strand separation, directly repeated sequences can re-anneal out of register such that the top strand of an upstream repeat anneals with the bottom strand of a downstream repeat [6] (Figure 2). Since they are less thermodynamically stable than a duplex structure, these slipped structures are expected to be transient and short-lived, quickly snapping back into register. While formed, however, they create regions of ssDNA on both strands. The C-strings within fragile zones not only affect the structure of the duplex, but also act as direct repeats. Furthermore, other closely-spaced direct repeat sequences of 6 to 8 bp are also present at the MBR and MTC fragile zones. Thus, the C-strings and the direct repeats are two elements that distinguish fragile zones from other regions in the genome, though there may be others (see Outstanding Questions). Such elements increase the likelihood of forming vulnerable non-B, slipped-stand DNA structures.
Figure 2. Sequence-Level View of ssDNA Formation via a Slipped-Strand Structure Leading to an AID-Induced Persistent Lesion.
Transcription or increased torsional stress in C-string sequences that display a B/A intermediate conformation can lead to transient strand slippage at direct repeats and generate ssDNA on each strand of DNA. While these transient slippage events may last only milliseconds or less before returning to a duplex conformation, AID can act to deaminate either C’s or meC’s within preferred single-stranded target sequences (WRC). The T’s generated from deamination of meC’s are removed more slowly than U’s and are thus more long-lived lesions. After the DNA resumes its duplex conformation, the resulting T:G mismatch is vulnerable to DNA repair enzymes, such as activated Artemis, that may convert these to DSBs. For chromosomal translocations that occur in human B cells, Artemis is activated during DSB repair at chromosome 14 (e.g., during V(D)J recombination or Ig class switch recombination). Failure to efficiently repair the chromosome 14 break coupled with simultaneous generation of DSB at a fragile zone can lead to events that favor chromosomal translocations.
Outstanding Questions.
What additional genetic and epigenetic elements distinguish fragile zones from other CpG containing sites in the genome?
Are the non-B DNA structures at different fragile zones similar to one another or do they include a range of various non-B DNA structures?
By altering the sequence of fragile zones, can we alter the chemical probing profile or susceptibility to DSB formation?
In non-lymphoid cells, what non-B DNA structures and enzymes are important for DSBs and translocations?
What are the various factors that generate the localized torsional strain that favors non-B DNA conformations?
Do certain heritable or acquired genetic defects predispose an individual to cancers involving breaks at fragile zones?
Other non-B structures are possible, such as triplex or G quadruplex [8, 9], but these structures require the disruption and unwinding of much greater lengths of duplex DNA, markedly reducing the likelihood of their formation. Another structure that can form is an R-loop, which forms when a newly transcribed RNA strand reanneals with the template DNA to form an RNA:DNA hybrid that results in a single-stranded non-template DNA strand [10, 11]. R-loops may be relevant to long fragile zones, such as the 2 kb region in MYC [12, 13]. However, the non-B structure at the 20 to 600 bp zones survives RNase H treatment, ruling out an R-loop at these zones [3, 4]. Thus, a slipped-strand structure is a simpler and more thermodynamically favored non-B structure.
Importantly, we have shown that transcription through any DNA sequence increases the propensity for breakage by >10-fold in a sensitive yeast genetic assay [5]. Transcription separates two DNA strands. If this occurs in a sequence with C-strings, the B/A intermediate conformation and repetitive C’s would favor slipped-strand formation. Many of the human fragile zones occur in known regions of transcription, including the MBR at the BCL2 locus and the intronic E2A at the TCF3 locus. However, this is not true for all breakage sites as the BCL2 intermediate and minor breakpoint cluster regions (icr and mcr) are not known to be transcribed in humans and neither is the BCL1 MTC (Figure 1). Therefore, for a subset of DSBs, other factors must be as important as transcription in the human genome, or, alternatively, low levels of spurious transcription from variable start sites or non-coding transcripts may be responsible.
SOURCES OF DAMAGE THAT TARGET ssDNA
Bases in duplex DNA are more protected from some types of damage than bases in ssDNA because of the stacking intrinsic in a double helical structure. Indeed, the enzyme activation-induced cytidine deaminase (AID) requires a ssDNA substrate because of its binding site [14], and it preferentially targets the C in WRC (W = adenine or thymine, R = purine) sequences for deamination to uracil (U) [15, 16]. AID is thought to be a major source of DSB induction at the MBR and MTC fragile zones since analysis of translocation breakpoint junctions from over 2,000 human patients suffering a variety of hematopoietic malignancies revealed that a majority of DSBs occur near C’s vulnerable to AID when DNA is single-stranded [2].
While AID expression is highest in mature B cells undergoing class switch recombination (CSR) and somatic hypermutation (SHM), sufficient levels of AID are present in pro-B and pre-B cells to lead to DSBs at the fragile zones [1, 17]. Why translocations involving the MBR and MTC fragile zones occur in pre-B cells, where they join with J segments of unrepaired V(D)J events, and not in mature B cells, where they could potentially join with unrepaired DSBs initiated by AID in the IgH switch regions, is not entirely clear. Zones that do form translocations with the IgH switch regions, such as MYC and BCL6, are larger (>2 kb) than the MBR and MTC zones, and the breaks are more often at WGCW sequences rather than CpG [18]. This suggests a slightly different breakage mechanism [1]. Additionally, selection of the translocation product may be a factor, as joins involving J segments place the IgH μ enhancer near the oncogenes associated with the MBR and MTC zones while joins with switch regions would put this enhancer on the other reciprocal chromosome [2].
AID is not a nuclease, so how does it cause a DSB? During CSR, the U’s generated by deamination of C’s are efficiently removed by uracil DNA glycosylase (UDG) to produce an abasic site that is targeted by AP-endonuclease, which nicks the phosphate backbone. If this occurs in close proximity on both DNA strands, as is likely for symmetrical CpG sites, the two nicks become a DSB [11, 19].
In our yeast genetic system, we found that any human sequence that was transcribed also was targeted by ectopically expressed human AID in a manner that led to a DSB [5]. Furthermore, DSB formation was completely dependent on the presence of functional UDG. Thus, transcription is a source of ssDNA, which may account for the increased instability of transcription measured in our genetic assay.
In addition to AID, we also found reactive oxygen species (ROS) were another source of damage to ssDNA. By deleting the gene encoding the major peroxiredoxin in yeast (TSA1) we were able to measure how increased levels of ROS affected the breakage rate [5]. Similar to AID, we found that transcribed regions were significantly more prone to DSBs due to increased ROS. ROS could directly nick each DNA strand or lead to oxidized bases that are excised by glycosylases with DSBs created by subsequent AP-endonuclease or AP-lyase activity.
TOPOLOGICAL TENSION INCREASES THE LONGEVITY OF TRANSIENT ssDNA STRUCTURES
Topological tension is another important factor that influences overall breakage propensity. Bisulfite reactivity is more pronounced at fragile zones on a supercoiled plasmid versus linearized DNA, indicating that increased torsional stress can lead to a further increase in single-strandedness at fragile zones [3, 4]. Under conditions of increased negative supercoiling, the relaxed DNA duplex may be more prone to forming slipped DNA structures because the negative supercoiling favors unwinding of the duplex. Therefore, torsional stress may trap transient ssDNA structures, increasing their lifespan and leaving them more vulnerable to agents that target ssDNA. Transcription and promoter orientation relative to other promoters can lead to changes in supercoiling, and this can likely be further exacerbated by defects in topoisomerase activity, either due to mutants or drugs that specifically target the enzymes [20, 21].
In our yeast system, we tested the effect of increased torsional stress by deleting the gene encoding topoisomerase 1 (TOP1) [5]. Interestingly, none of the individual factors described thus far (transcription, AID, ROS) were able to specifically increase the breakage rate in the inserted human fragile zones relative to the “no insert” control, nor did deletion of TOP1 alone. It was not until we combined these factors with the top1Δ mutation that we observed significant effects. Expression of AID in a top1Δ mutant significantly increased the rate of DSB formation at the MTC fragile zone while combining the top1Δ mutant with increased ROS led to a significant increase in DSBs at the MBR fragile zone. In both cases, the increases were dependent on transcription opening-up the region of DNA [5]. The increased torsional stress due to a combination of transcription and loss of TOP1 likely increases the frequency and the lifespan of slipped-strand DNA structures, allowing a greater opportunity for AID or ROS to damage the exposed ssDNA.
LONGEVITY OF LESIONS THAT INITIATE TRANSLOCATIONS
Lastly, we can ask if certain types of damage to ssDNA are more likely to lead to a DSB due to a slower repair rate, making some lesions more persistent. As stated above, AID deaminates C to U, and the U is efficiently processed by UDG, which can act on both single- and double-stranded DNA, creating an abasic site for AP-endonuclease cleavage. During CSR, AID has access to abundant ssDNA due to the formation of kilobase-long R-loops at switch regions, making it more likely that AP-endonuclease nicking occurs on each strand of the duplex, leading to a DSB. ssDNA formed at fragile zones, however, will be much more localized and transient, thus a different mechanism of DSB formation may be occurring.
It has been demonstrated that AID can also act within CpG sequences and, importantly, on a 5-methylcytosine (meC) in CpG sequences, where it deaminates the base to thymine [15, 22]. This leads to a T:G mismatch that can be repaired by thymine DNA glycosylase (TDG) or methyl-CpG binding protein domain 4 (MBD4), but approximately 2500-fold less efficiently than UDG repair of a U:G mismatch [23, 24]. Thus, the T:G mismatch caused by AID acting at meC is an example of a persistent or long-lived lesion. As translocation breakpoints in human patients frequently map at or near CpG sites, which are often sites of meC, persistent T:G mismatches due to AID expression are likely common in both pre- and mature B cells.
These persistent lesions or mismatches are a potential source of instability as activated Artemis nuclease can recognize these structures [25], albeit at reduced efficiency compared to overhang structures [26], and create a DSB. Artemis is a structure-specific nuclease that cleaves at the boundaries of single- and double-stranded DNA [26, 27]. In pre-B and pro-B cells, when DSBs at the fragile zones occur, Artemis is activated by DNA-PKcs during the failed attempt to repair the DSB created during V(D)J recombination via non-homologous end joining (NHEJ) [28]. This activated Artemis is present to cleave at persistent lesions within fragile zones to generate the second DSB required for translocation formation (Figure 3, Key Figure).
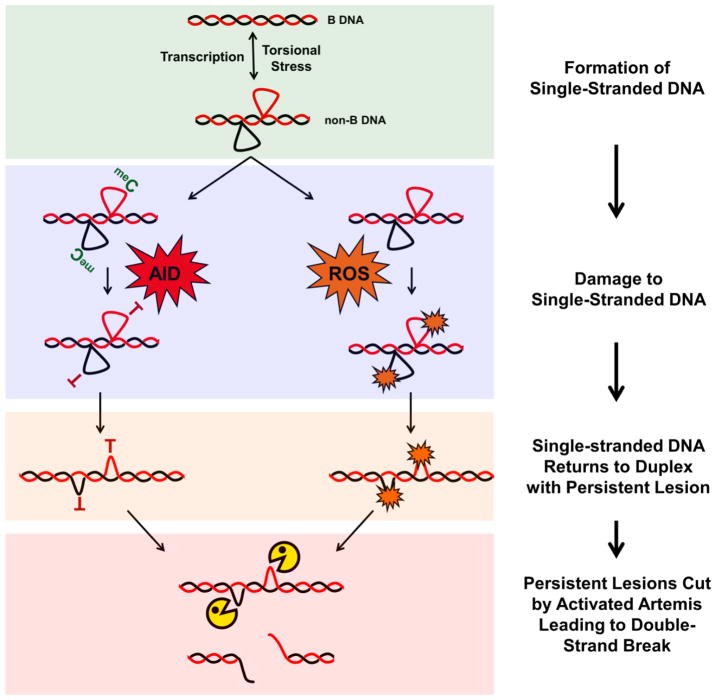
Figure 3, Key Figure. Model of DSB Formation at Human Fragile Zones.
Fragile zones are prone to forming transient, non-B DNA structures based on biochemical [3, 4], enzymatic [2, 5, 25, 33], and genetic [5] data. Formation of ssDNA in the non-B state is enhanced by processes that separate the DNA strands (i.e., transcription) and increased torsional stress. The non-B structures (such as the slipped-strand structure depicted here) are short-lived, but, while formed, the ssDNA is vulnerable to damage. AID can attack ssDNA in human B cells and reactive oxygen species (ROS) can more easily oxidize DNA not protected within a DNA duplex. Oxidized bases generated by ROS and T:G mismatches generated by AID deamination of 5-methylcytosine represent long-lived, persistent lesions (relative to the efficient processing of uracil by uracil glycosylase) that, upon collapse of the non-B structure back to duplex DNA, create distortions recognized by activated Artemis nuclease and are cleaved. When the persistent lesions are adjacent on opposite strands of the DNA duplex, this process can result in a DSB.
Persistent DNA lesions generated by AID deamination of meC are not amenable to study in our yeast genetic assay as S. cerevisiae lacks DNA methylation, but our data suggests that base damage due to increased ROS is another source of persistent DNA lesions [5] (Figure 3, Key Figure). This is based on our results expressing a constitutively active form of Artemis, Artemis-413 (a 413 amino acid C-terminal truncation mutant also called ARM37) that does not require DNA-PKcs for endonuclease activity [29–31]. While Artemis-413 expression alone did not lead to an increase in the breakage rate, expressing Artemis-413 in a tsa1Δ mutant (i.e., increased ROS) led to a synergistic increase in the rate of DSBs. DSBs occurred at a significantly higher rate in the MBR and MTC fragile zones, indicating these sequences are vulnerable to formation of these lesions. Since oxidized bases are repaired more slowly than an incorporated U [32], lesions resulting from increased ROS create a persistent lesion that Artemis can recognize.
Lesion persistence or longevity is key to Artemis-413 having time to recognize the lesion. The result of co-expression of AID and Artemis-413 in the yeast system is no different than expression of AID alone since the U’s are not persistent due to a much higher rate of lesion processing [5]. Interestingly, by deleting UDG (encoded by UNG1 in yeast), U’s will persist longer in the DNA, and in this context, we see an increase in DSB formation at the MBR and MTC regions. Furthermore, co-expression of AID and Artemis-413 in an ung1Δ mutant significantly increases the DSB rate at some sequences that were not elevated in the absence of Artemis-413 expression.
By slowing the repair rate, lesions become more vulnerable to DSBs, demonstrating that lesion longevity is a factor in sequence fragility. Mismatches themselves may be cut by Artemis or other nucleases, but we reason that mismatches would also increase the fraction of time that a region of DNA is prone to forming a non-B structure. This may be because a small distortion (mismatch) favors transitions between a normal and a more extensive non-B DNA state. Another possibility is that during the repair of an ROS lesion, there may be obstruction of transcription or presence of proteins that favor the non-B DNA state [5].
CONCLUDING REMARKS
Transient non-B DNA structures may form during transcription or other strand separation processes, such as DNA repair. We favor slipped-strand DNA as the simplest such structure. While these structures may last only milliseconds, this is sufficiently long for AID to generate T:G mismatches at methyl C’s. When the duplex reforms, the T:G mismatches persist longer than most mismatches due to slow T:G mismatch repair. Mismatches may be converted to DSBs if they encounter activated Artemis (Figure 3, Key Figure), which may arise at another chromosomal location during V(D)J recombination or Ig class switch recombination. This would represent the two partner chromosomes in a reciprocal translocation.
In S. cerevisiae, there are no methyl C’s. But ROS generate lesions in transient ssDNA that are long-lived in the reformed duplex. These can become DSBs either due to attempts to repair the oxidized lesion or by a structure-specific nuclease recognizing the ssDNA-containing distortion due to the lesion. Thus, increased ROS recapitulates key aspects of the process described above in B cells and provides a way to further study DSB mechanisms involving persistent lesions. ROS may be involved in some DSBs at fragile zones in human cells but are likely occurring much less frequently than the mechanism involving AID.
In summary, short-lived DNA structures are vulnerable to damage, and when the DNA duplex reforms, long-lived lesions are the site of DSBs. Future work will be required to fully test all aspects of this model across a range of biological circumstances (see Outstanding Questions). Our yeast system provides the opportunity to manipulate known fragile sequences and test the potential of various sequence elements (C-strings, direct repeats) to cause DSBs. Furthermore, in-depth meta-analyses may uncover genetic defects that predispose patients to breaks at lymphoma-related fragile zones, which can then be tested in our genetic system.
Highlights.
Topological factors and transcriptional-dependence provide in vivo support for non-B DNA conformations in human neoplastic fragile zones.
Slowly repaired DNA lesions arise because transient non-B DNA structures are vulnerable to enzymes and agents that create such lesions.
The slowly-repaired DNA lesions likely are converted to DSBs after DNA resumes normal duplex structure.
Exposure to reactive oxygen species may preferentially affect vulnerable DNA regions and may be important for disease etiology.
Acknowledgments
We thank Dr. Raymond D. Mosteller for comments on the manuscript. The work was supported by grants to MRL from NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lieber MR. Mechanisms of human lymphoid chromosomal translocations. Nat Rev Cancer. 2016;16(6):387–98. doi: 10.1038/nrc.2016.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsai AG, et al. Human chromosomal translocations at CpG sites and a theoretical basis for their lineage and stage specificity. Cell. 2008;135(6):1130–42. doi: 10.1016/j.cell.2008.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raghavan SC, et al. A non-B-DNA structure at the bcl-2 major break point region is cleaved by the RAG complex. Nature. 2004a;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 4.Tsai AG, et al. Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites. J Biol Chem. 2009;284(11):7157–64. doi: 10.1074/jbc.M806866200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pannunzio NR, Lieber MR. AID and Reactive Oxygen Species Can Induce DNA Breaks within Human Chromosomal Translocation Fragile Zones. Mol Cell. 2017;68(5):901–912e3. doi: 10.1016/j.molcel.2017.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sinden RR. DNA Structure and Function. Academic Press; 1994. [Google Scholar]
- 7.Calladine CR, et al. Understanding DNA: The Molecule and How it Works. 3. Elsevier Academic Press; 2004. [Google Scholar]
- 8.Nambiar M, et al. Formation of a G-quadruplex at the BCL2 major breakpoint region of the t(14;18) translocation in follicular lymphoma. Nucleic Acids Res. 2011;39(3):936–48. doi: 10.1093/nar/gkq824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams JD, et al. Sites of instability in the human TCF3 (E2A) gene adopt G-quadruplex DNA structures in vitro. Front Genet. 2015;6:177. doi: 10.3389/fgene.2015.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu K, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nature Immunol. 2003;4:442–451. doi: 10.1038/ni919. [DOI] [PubMed] [Google Scholar]
- 11.Yu K, Lieber MR. Nucleic acid structures and enzymes in the immunoglobulin class switch recombination mechanism. DNA Repair. 2003;2:1163–1174. doi: 10.1016/j.dnarep.2003.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Duquette ML, et al. AID binds to transcription-induced structures in c-MYC that map to regions associated with translocation and hypermutation. Oncogene. 2005;24:5791–5798. doi: 10.1038/sj.onc.1208746. [DOI] [PubMed] [Google Scholar]
- 13.Yang Y, et al. Arginine Methylation Facilitates the Recruitment of TOP3B to Chromatin to Prevent R Loop Accumulation. Mol Cell. 2014;53(3):484–97. doi: 10.1016/j.molcel.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiao Q, et al. AID Recognizes Structured DNA for Class Switch Recombination. Mol Cell. 2017;67(3):361–373e4. doi: 10.1016/j.molcel.2017.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pham P, et al. Processive AID-catalyzed cytosine deamination on single-stranded DNA stimulates somatic hypermutation. Nature. 2003;424:103–107. doi: 10.1038/nature01760. [DOI] [PubMed] [Google Scholar]
- 16.Yu K, et al. DNA substrate length and surrounding sequence affect the activation induced deaminase activity at cytidine. J Biol Chem. 2004;279(8):6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 17.Swaminathan S, et al. Mechanisms of clonal evolution in childhood acute lymphoblastic leukemia. Nat Immunol. 2015;16(7):766–74. doi: 10.1038/ni.3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Z, et al. BCL6 breaks occur at different AID sequence motifs in Ig-BCL6 and non-Ig-BCL6 rearrangements. Blood. 2013;121:4551–4. doi: 10.1182/blood-2012-10-464958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masani S, et al. Apurinic/apyrimidinic endonuclease 1 is the essential nuclease during immunoglobulin class switch recombination. Mol Cell Biol. 2013;33(7):1468–73. doi: 10.1128/MCB.00026-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pannunzio NR, Lieber MR. RNA Polymerase Collision versus DNA Structural Distortion: Twists and Turns Can Cause Break Failure. Mol Cell. 2016;62(3):327–334. doi: 10.1016/j.molcel.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pannunzio NR, Lieber MR. Dissecting the Roles of Divergent and Convergent Transcription in Chromosome Instability. Cell Rep. 2016;14(5):1025–31. doi: 10.1016/j.celrep.2015.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bransteitter R, et al. Activation-induced cytidine deaminase deaminates deoxycytidine on single-stranded DNA but requires the action of RNase. Proc Natl Acad Sci. 2003;100(7):4102–4107. doi: 10.1073/pnas.0730835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmutte C, et al. Base excision repair of U:G mismatches at a mutational hotspot in the p53 gene is more efficient than base excision repair of T:G mismatches in extracts of human colon tumors. Cancer Res. 1995;55:3742–3746. [PubMed] [Google Scholar]
- 24.Walsh CP, Xu GL. Cytosine methylation and DNA repair. Curr Top Microbiol Immunol. 2006;301:283–315. doi: 10.1007/3-540-31390-7_11. [DOI] [PubMed] [Google Scholar]
- 25.Cui X, et al. Both CpG Methylation and AID are Required for the Fragility of the Human Bcl-2 Major Breakpoint Region: Implications for the Timing of the Breaks in the t(14;18) Mol Cell Biol. 2013;33(5):947–957. doi: 10.1128/MCB.01436-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chang HH, Lieber MR. Structure-Specific nuclease activities of Artemis and the Artemis: DNA-PKcs complex. Nucleic Acids Res. 2016;44(11):4991–7. doi: 10.1093/nar/gkw456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang HH, et al. Unifying the DNA End-processing Roles of the Artemis Nuclease: Ku-Dependent Artemis Resection at Blunt DNA Ends. J Biol Chem. 2015;290(40):24036–50. doi: 10.1074/jbc.M115.680900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pannunzio NR, et al. Nonhomologous DNA End Joining for Repair of DNA Double-Strand Breaks. J Biol Chem. 2017 doi: 10.1074/jbc.TM117.000374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niewolik D, et al. DNA-PKcs dependence of artemis endonucleolytic activity: differences between hairpins and 5′ or 3′ overhangs. J Biol Chem. 2006;281:33900–33909. doi: 10.1074/jbc.M606023200. [DOI] [PubMed] [Google Scholar]
- 30.Niewolik D, et al. Autoinhibition of the Nuclease ARTEMIS Is Mediated by a Physical Interaction between Its Catalytic and C-terminal Domains. J Biol Chem. 2017;292(8):3351–3365. doi: 10.1074/jbc.M116.770461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma Y, et al. The DNA-PKcs phosphorylation sites of human artemis. J Biol Chem. 2005;280:33839–46. doi: 10.1074/jbc.M507113200. [DOI] [PubMed] [Google Scholar]
- 32.Cravens SL, Stivers JT. Comparative Effects of Ions, Molecular Crowding, and Bulk DNA on the Damage Search Mechanisms of hOGG1 and hUNG. Biochemistry. 2016;55(37):5230–42. doi: 10.1021/acs.biochem.6b00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Greisman HA, et al. IgH Partner Breakpoint Sequences Provide Evidence that AID Initiates t(11;14) and t(8;14) Chromosomal Breaks in Mantle Cell and Burkitt Lymphomas. Blood. 2012;120(14):2864–2867. doi: 10.1182/blood-2012-02-412791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiemels JL, et al. Site-specific translocation and evidence of postnatal origin of the t(1;19) E2A-PBX1 fusion in childhood acute lymphoblastic leukemia. Proc Natl Acad Sci U S A. 2002;99(23):15101–6. doi: 10.1073/pnas.222481199. [DOI] [PMC free article] [PubMed] [Google Scholar]