Abstract
Identification of transcription factors that directly convert pluripotent stem cells (PSCs) into endothelial and blood cells and advances in the chemical modifications of messenger RNA (mRNA) offer alternative nucleic acid-based transgene-free approach for scalable production of these cells for drug screening and therapeutic purposes. Here we evaluated the effect of 5′ and 3′ RNA untranslated regions (UTRs) on translational efficiency of chemically-modified synthetic mRNA (modRNA) in human PSCs and showed that an addition of 5′UTR indeed enhanced protein expression. With the optimized modRNAs expressing ETV2 or ETV2 and GATA2, we are able to produce VE-cadherin+ endothelial cells and CD34+CD43+ hematopoietic progenitors, respectively, from human PSCs as well as non-human primate (NHP) PSCs. Overall, our findings provide valuable information on the design of in vitro transcription templates being used in PSCs and its broad applicability for basic research, disease modeling, and regenerative medicine.
Keywords: Hematopoiesis, untranslated region, stem cells, modified RNA, UTR, half-life, endothelial, primate, ETV2, GATA2
Introduction
Pluripotent stem cell (PSC) technologies have opened possibilities for generation of patient-specific cells and tissues for the study of diseases and therapeutic applications. Typically, PSC differentiation protocols employ morphogens and growth factors to induce differentiation. However, recent advances in identification of key transcriptional regulators involved in cell fate commitment make it possible to directly convert somatic cells (1–3) and PSCs (4) to desired cell lineage simply by forced expression of transcription factors. Nevertheless, the widely used viral gene delivery systems are associated with genome integration and reactivation of the viral proteins, making it unsafe for therapeutic purposes. In recent years, advances in chemical synthesis have improved the stability of synthetic RNA to allow its use in nucleic acid transfer (5). Use of modRNA is currently an attractive alternative and has become more prevalent in many applications (5–9). However, further optimization of mRNA stability and translational efficiency would be essential to advancing modRNA-based technologies for in vitro PSC differentiation.
Because mRNA untranslated regions (UTRs) are known to regulate gene expression and affect translation in eukaryotic cells, different UTR modifications have been proposed to increase translational efficiency of modRNA. Guanine (G)-rich nucleic acid sequences are prevalent in the human genomes (10). Located within the 5′-UTR of the RNA, these sequences form stable four-stranded secondary structures called G-quadruplexes and inhibits NRAS protein expression in vitro (11) suggesting that 5′UTR might be unfavorable for the modRNA. Recent demonstration that effects of UTRs on modRNA translation depends on cell type (6), suggests that evaluation of the UTR impact should be performed using the appropriate target cells. Although several research using different types of modRNA has been published in somatic cells, its optimization has not yet determined in the PSCs. In present study, we evaluated the translational efficiency of modRNA synthesized from different in vitro transcription (IVT) templates encoding enhanced green fluorescence protein (eGFP) and demonstrated that modRNA containing 5′UTR and single 3′UTR is most efficiently translated into protein in human PSCs. Additionally, neither of the modifications affected eGFP half-life.
Recently, we revealed that human PSCs can be converted to endothelial cells by overexpression of ETV2, while overexpression of ETV2 and GATA2 using lentiviral or modRNA gene delivery lead to formation of multipotent CD34+CD45+ hematopoietic progenitors (4). We adopted the optimized modRNA expressing ETV2 or ETV2 and GATA2 to generate endothelial and hematopoietic cells, respectively, from human PSCs under defined xeno-free conditions. In addition, we showed that similar approach can successfully produce endothelial and hematopoietic cells from non-human primate (NHP) induced PSC (iPSC).
Materials and Methods
Cell culture
Human embryonic stem cell (WA01), bone marrow-derived iPSC (IISH2i-BM9), and fibroblast-derived iPSC (DF19-9-7T) were all procured from WiCell (Madison, WI). Nonhuman primate iPSC (MnCy0669 iPSC) was previously derived by our group (12). Human bone marrow and cord blood mononuclear cells from healthy donors were purchased commercially from Cell Processing Core (Cincinnati, OH). All PSCs were cultured on vitronectin-coated tissue culture plates in E8 medium (Stem Cell Technologies) (13) and an addition of 2.5 μM of IWR1 (ApexBio Technology) for MnCy0669 iPSC. Cells were subcultured every 4–5 days using 0.5 mM EDTA in PBS. For modRNA transfection, single cell suspension was prepared using either HyQTase (Hyclone, UT) or Accutase (Innovative Cell Technologies, San Diego, CA). Endothelial cells were cultured on collagen coated-plate with endothelium medium (Cell Science).
Construction of in vitro transcription (IVT) templates
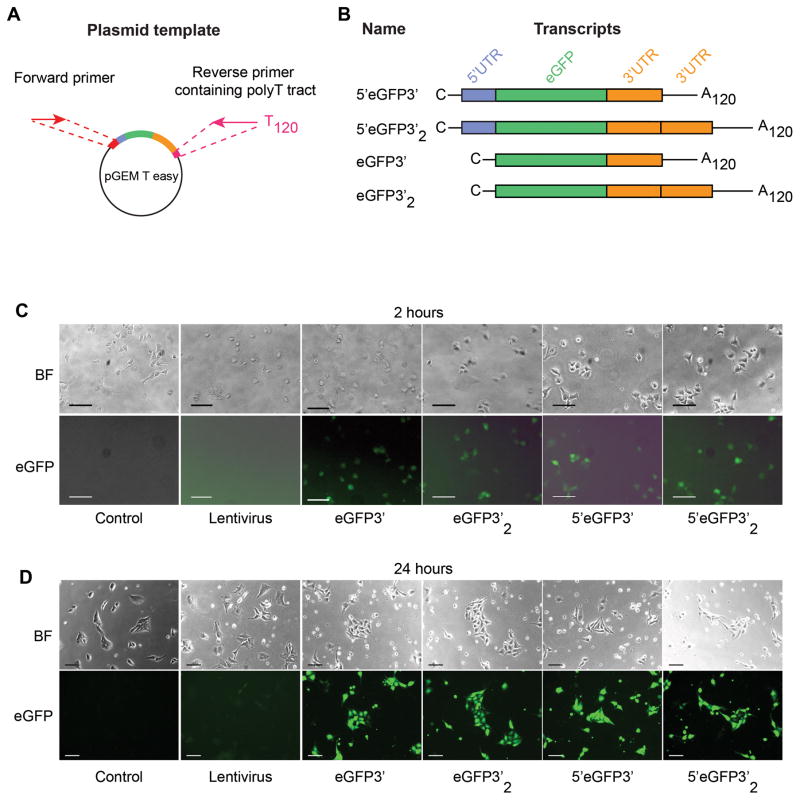
All vectors and their RNA transcripts are shown in Figure 1A and 1B. A series of vectors for the IVT was modified from pGEM-T Easy (Promega, Madison, WI). In brief, a cassette of 5′UTR of β-globin and multiple cloning sites (MCS) was synthesized by GeneArt service (Thermo Fisher Scientific) and cloned into the AatII/SpeI restriction sites of pGEM-T Easy. A single or double fragment of β-globin 3′UTR was inserted downstream of the MCS to generate 5′-MCS-1β and 5′-MCS-2β constructs respectively. AatII/BamHI restriction followed by blunt-end ligation was performed to generate MCS-1β and MCS-2β. eGFP flanked by BamHI restriction sites was cloned into the MCS of each constructs. Human ETV2 transcript variant 1(NM_014209.3) and GATA2 isoform 1 (NM_001145661.1) were cloned into 5′-MCS-1β construct to generate IVT template for ETV2. To generate IVT templates with 120-A tract, a reverse primer containing 120 T base pairs and forward primer were used in a PCR reaction (primers are listed in Supplemental Table 1). All the PCR reactions were carried out using Phusion (Thermo Fisher Scientific), 30 cycles of 95°C for 10 seconds; 52°C for ETV2; 58°C for GATA2 for 10 seconds; 72°C for 50 seconds. PCR products were run on the agarose gel and extracted using QIAEX II gel extraction kit (Qiagen, Valencia, CA) before further processing.
Figure 1. Plasmids for IVT templates and RNA transcripts.
A) A pGEM T easy vector served as a backbone to generate all IVT templates using PCR. A single pair of forward and reverse primers binding to specific sequences at the 5′ upstream of the T7 promoter and 3′ downstream of the 3′UTR was used to amplified all IVT templates.
B) Diagram showing a composition of each modRNA and their names used throughout the study. All templates were transcribed in vitro in the presence of ARCA cap analog and pseudouridine. C; capped analog, UTR; untranslated region, A120; 120-base pair polyadenylation.
C) Bright field and immunofluorescence images of WA01 cell line at 2 hours and D) 24 hours after transfection. Scale bars represent 50 μm. BF; bright field.
Synthesis of modRNA
modRNA was synthesized using the MEGAscript T7 kit (Ambion, Austin, TX), using a custom ribonucleoside cocktail comprising 3′-0-Me-m7G(5′)ppp(5′)G ARCA cap analog, pseudouridine triphosphate (TriLink Biotechnologies, San Diego, CA), adenosine triphosphate, guanosine triphosphate and cytidine triphosphate. The synthesis reactions were setup according to the manufacturer’s instructions. Reactions were incubated 3 hours at 37°C and DNAse-treated. RNA was purified using PureLink RNA Micro kit (Thermo Fisher Scientific) and adjusted with RNase-free water to 100 ng/μl working concentration before freezing at −80°C.
eGFP virus production
PCR product of eGFP was subcloned into pSIN/EF1α lentiviral expression vector. Virus production was carried out by calcium phosphate transfection of Lenti-X 293T cells (Clontech, Mountain View, CA). Packaged lentiviral units were concentrated using Amicon Ultra-15 Centrifugal Filter Units (Millipore, Billerica, MA). Viral titer was assessed by transduction of Lenti-X 293T cells.
modRNA transfection
For eGFP optimization, a total of 5×104 WA01 single cell suspension in basal E8 medium was transfected with 200 ng of RNA using TransIT-mRNA (Mirus Bio, Madison, WI) according to the manufacturer’s instructions. Cell/RNA mixture was plated into one well of vitronectin-coated 24-well plate. Complete E8 medium was added to bring total volume to 550 μl/well. ROCK inhibitor (StemCell Technologies, Vancouver, Canada) was added at 10 μM to improve cell viability as described in (4). eGFP virus with a titer of 7×107 transducing unit/ml was used at 1 μl per well, and complete medium was changed 24 hours after transduction. For direct induction of endothelial cells, 2×105 of indicated cells were transfected with 200 ng of ETV2 modRNA and cultured in vitronectin-coated 6-well plate. For generation of hematopoietic cells, 2×105 of indicated cells were transfected with 400 ng of each ETV2 and GATA2 modRNA and cultured under the same conditions. After 24 hours, cells were harvested with Accutase and transferred to collagen coated 6-well plate in IF9S medium (14) supplemented with hematopoietic cytokines including 10 ng/ml IL3, 20 ng/ml FGF2, 50 ng/ml VEGF, Il6, SCF, and TPO. Second transfection was performed on the same day with 400 ng of each modRNA.
Flow Cytometry
Cells were harvested using Accutase and stained with Ghost Dye™ (Tonbo Biosciences, Madison, WI), CD31, CD34, and CD144 (BD Bioscience) for 30 minutes at 4°C. Cells were washed before analyses using MACSQuant Analyzer 10 (Miltenyi Biotec Inc, San Diego, CA). Non-transfected cell was included to establish a threshold for positive gating. Single live cells were gated and analyzed by FlowJo software (Tree Star, Ashland, OR). eGFP positive cells were gated for the quantitative measurement of mean fluorescence intensity (MFI).
Live staining of endothelial cells and AcLDL uptake assay
ETV2-transfected cells were transferred to a collagen-coated plate and cultured in endothelial medium (PromoCell). Ten μg/ml of Alexa-594-conjugated AcLDL (Thermo Fisher Scientific) was added to culture at 24 hours after transfection and incubated at 37 °C for 4 hours as previously described (4). VE-cadherin was then added to the culture and incubated on the rocker at room temperature for 15 minutes. Cells were washed twice with PBS prior to fluorescence imaging.
Colony-forming cell assay
CFC assays were performed in 35-mm low-adherent culture dishes (StemCell Technologies) using MethoCult 4435 (StemCell Technologies). Colonies were scored after 14 days of incubation. Wright staining was used to evaluate the morphology of cells within colonies on cytospins.
Statistical analysis
Data obtained from multiple experiments were reported as mean ± SD. Significant levels were determined by one-way ANOVA followed by Tukey post hoc test as appropriate. p < 0.05 was considered significant. All the graphs and statistics were performed using GraphPad Prism software (GraphPad, San Diego, CA).
Results
Optimization of in vitro transcription (IVT) template for RNA synthesis
The translational efficiency of mRNA is tightly regulated to ensure a proper amount of protein is being produced at the right time and terminated when the cells no longer need it. 5′ and 3′UTR have been shown to play important roles in the regulatory mechanics during the translational process (i.e., initiation, elongation, and termination) and post-translational modifications. Initially, the ribosomal complex is formed at the 5′UTR by a direction of the cap-binding protein eIF4F complex, which is a rate-limiting step of cap-dependent initiation (reviewed in (15)). Kozak demonstrated that the α- and β-globin 5′UTR can impact translational efficiency (16), and alternative splice variants of the 5′UTR can affect translation of the bovine growth hormone receptor over an 80-fold range (17). 5′UTR together with 3′UTR, which contains AU-rich elements, polyadenylation signals, cis- and trans regulatory elements, offer an unlimited range of control possibilities within a single mRNA that influences polyadenylation, translation efficiency, localization, and stability of the mRNA (reviewed in (18)).
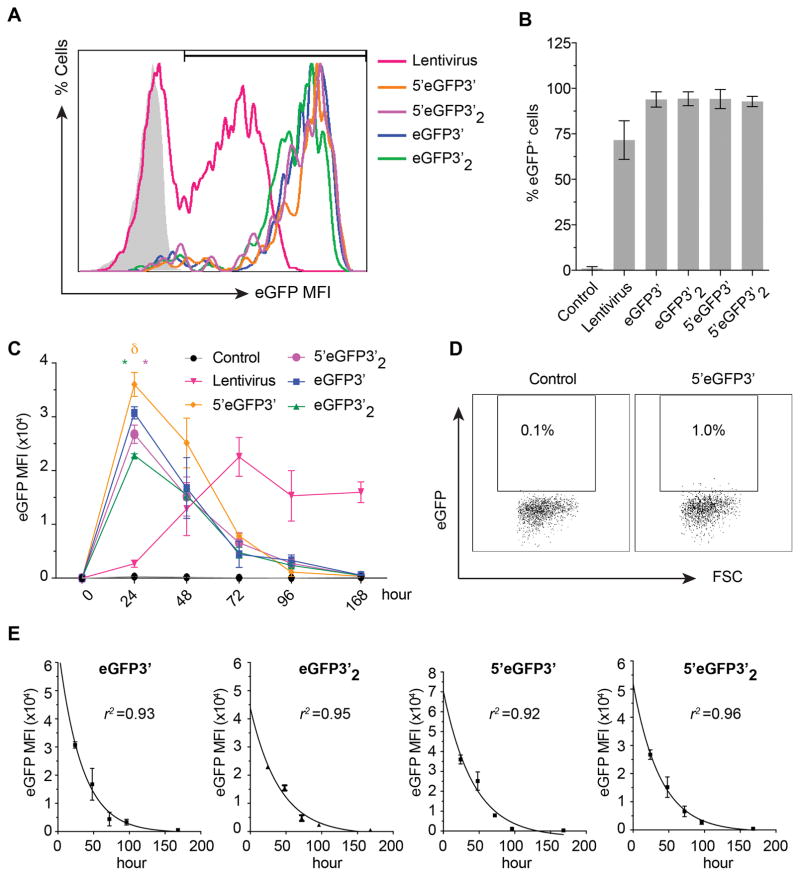
Despite current understanding of translational control regulation, precise roles of the UTRs on translational efficiency in human PSCs have not been systematically demonstrated. To evaluate the effect of UTRs on translational efficiency of modRNA, we generated 4 different eGFP-encoding in vitro transcription (IVT) templates with and without 5′UTR. We also incorporate 2 repeats of artificially added β-globin 3′UTR into the templates (Figure 1A and 1B). To assess how different mRNA modifications affect the onset, duration and the level of gene expression, we transfected WA01 human PSC line with modRNA synthesized from all IVT templates and analyzed eGFP expression by immunofluorescense and flow cytometry. We observed that the onset of eGFP expression was approximately 90 minutes after transfection regardless of the modifications, but the fluorescence intensity became decent for imaging after 2 hours (Figure 1C). In contrast, the signal intensity from lentivirus remained low at 24 hours after transduction (Figure 1D). We found that IVT modifications did not affect transfection efficiency (>90% for all modifications) (Figure 2A and 2B). However, in contrast to previous findings in dendritic cells (9), we found the presence of 5′UTR of human β-globin gene significantly improved eGFP protein levels at 24 hours after transfection, whereas artificially added repeat of β-globin 3′UTR significantly diminished maximum eGFP intensity (Figure 2C). To precisely evaluate the impact of these modifications on the level of protein expression, we performed time kinetic study of eGFP expression at a single cell level using flow cytometry. We found that eGFP intensity reached maximum level at 24 hours after transfection and started declining after since prior to fading out by day 7 (Figure 2C and 2D). Neither of the modification showed significant advantages over the others in term of duration of eGFP expression suggesting the modifications did not significantly affect RNA stability in WA01. We observed that the first 72 hours is a golden period for modRNA gene delivery showing high protein expression compared to lentivirus. In contrast, the signal from lentiviral delivery was increasing and reached steady expression at 96 hours (Figure 2C).
Figure 2. Mean fluorescence intensity, transfection efficiency, and time kinetics of eGFP expression.
A) Flow cytometric histograms show eGFP expression pattern and gating threshold used for subsequent analyses of eGFP. Shaded histogram represents non-transfection control.
B) Percentage of eGFP-positive cells at 24 hours after transfection. Mean±SD from 3 experiments is shown.
C) Time kinetic study of eGFP levels. eGFP mean fluorescence intensity (MFI) from all samples were plotted as 0 at time 0. Green and pink * indicate significant different in eGFP expression compared to their single 3′UTR mimics. δ indicates significance compared to all others. Results are mean ±SD from 3 experiments
D) Representative flow cytometric dot plot showing eGFP expression in WA01 cell line at 1 week after transfection with 5′eGFP3′. E) Non-linear curve fit was performed using the first-order kinetic model (A(t)=A0e−kt). eGFP MFI were plotted on a log scale vs. time. Data represents mean±SD from 3 experiments.
In principle, the time kinetic curve of eGFP represents the summation of several cellular processes such as mRNA transfer rate, distribution, and protein degradation. While the early phase of the plot involves protein production and degradation, the late phase generally represents the degradation of the protein (19). We further analyzed time kinetic data to evaluate whether modifications of IVT transcripts affect a protein half-life. Based on degradation pattern analyses of the eGFP fluorescence intensity from all modifications, we found that eGFP was a stable protein of which its degradation followed first-order kinetics (Figure 2E, R2 >0.95); A(t) = A0e−kt where A(t) is a mean fluorescence intensity (MFI) at time t, A0 is MFI at time 0, k is a constant of decay. To accurately estimate eGFP half-life, we used data from the late phase of the kinetic curve (24–96 hours) where protein decay is the major process. Although eGFP products were encoded from different IVT templates, they are theoretically the same protein. In agreement with Corish P et al (20), our estimated eGFP half-life was ~26 hours (average from all IVT templates) (Table 1).
Table 1.
Estimated parameters of protein physical properties
| Transcripts | k (hour−1) | Half-life (hour) | |
|---|---|---|---|
| Mean | Mean | 95% CI | |
| eGFP3′ | 0.03094 | 22.41 | 15.71–39.04 |
| eGFP3′2 | 0.02439 | 28.42 | 20.81–44.77 |
| 5′eGFP3′ | 0.02383 | 29.08 | 19.80–54.79 |
| 5′eGFP3′2 | 0.02647 | 26.19 | 19.80–38.65 |
Next, we assessed whether similar modifications of RNA have similar effect in NHP. Using the same set of modRNAs, we found a significant increase in expression of eGFP when 5′UTR is present. However, in contrast to human PSCs, we did not observe decline in eGFP signal following addition of the extra repeat of β-globin 3′UTR (Supplemental Figure 1). Overall these experiments revealed that 5′UTR and single 3′UTR allow superior protein expression in human and NHP iPSCs. Thus, we selected this modification for our endothelial and blood cell induction experiments.
Generation of endothelial cells and hematopoietic progenitors from PSCs
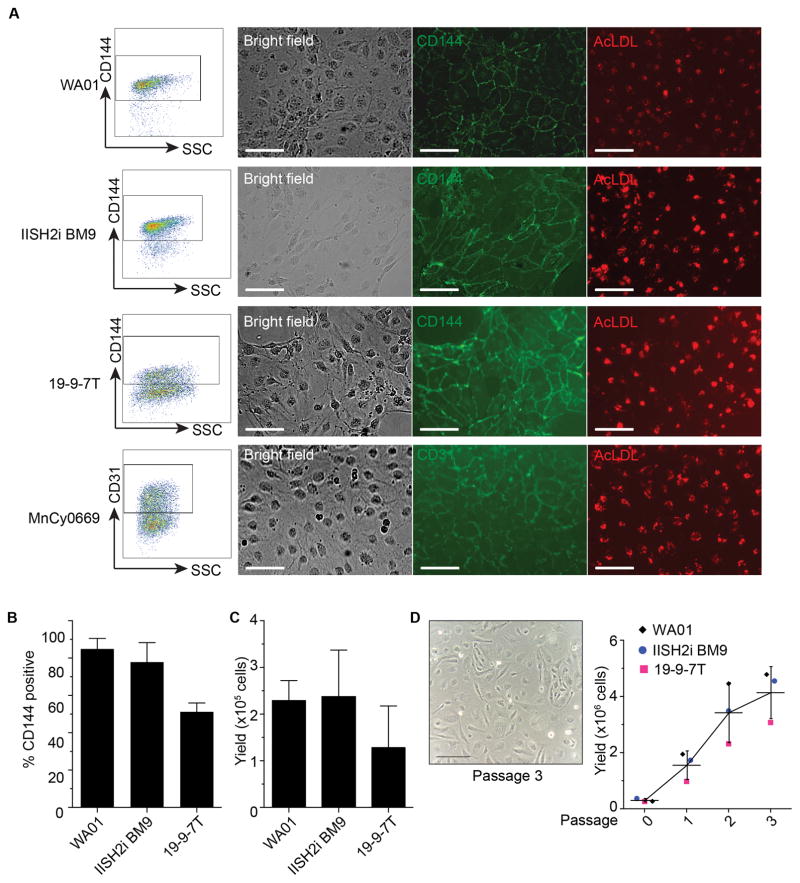
ETS variant 2 (ETV2) is well-known as a single transcription factor essential for endothelial program (4, 21, 22). Our group previously demonstrated that using lentiviral vector to force express a single transcription factor ETV2 in pluripotent stem cells requires 48 hours after transduction to bypass mesodermal stage and become endothelial cells. Since modRNA can directly translate into protein, we evaluate whether this process could be reproduced more rapidly. We synthesized ETV2 modRNA from the optimized IVT template containing 5′UTR and single 3′UTR and performed a single transfection using a cationic polymer/lipid reagent. Twenty-four hours after transfection with the ETV2 modRNA, morphology of pluripotent stem cells from 3 irrelevant human PSCs (WA01, IISH2i-BM9, and DF19-9-7T) and cynomolgus macaque MnCy0669 NHP-iPSC transformed into typical endothelial phenotype, acquired endothelial markers (CD31 and CD144 for human PSCs; CD31 for NHP iPSCs), and endocytosed AcLDL (Figure 3A). Interestingly, WA01 and IISH2i-BM9 achieved >80% conversion efficiency while DF19-9-7T achieved >40 % yielding up to 3×105 cells on passage 0 (Figure 3B and 3C). These endothelial cells are able to expand under endothelial condition for up to 3 passages and retain their typical endothelial cell morphology (Figure 3D).
Figure 3. Generation of Endothelial cells with ETV2 modRNA.
A) Representative flow cytometric dot plots and immunofluorescence images showing endothelial markers and AcLDL uptake on ETV2-transfected PSCs at 24 hours after transfection. Cells were transferred to collagen-coated plate and cultured in endothelial medium for live staining. AcLDL = acetylated-low density lipoprotein. Scale bar = 50 μm.
B) Percentage of CD144+ cells shown in the gate from A). Results are mean±SD from 3 experiments.
C) Yield of total live CD144+ cells from ETV2 transfected cultures. Mean±SD from 3 experiments is shown.
D) Expansion of endothelial cells generated in ETV2-transduced cultures. Representative picture shows endothelial cells at passage 3 after transfection. Scatter dot plot showing accumulative number of endothelial cells after expansion. Scale bar = 100 μm.
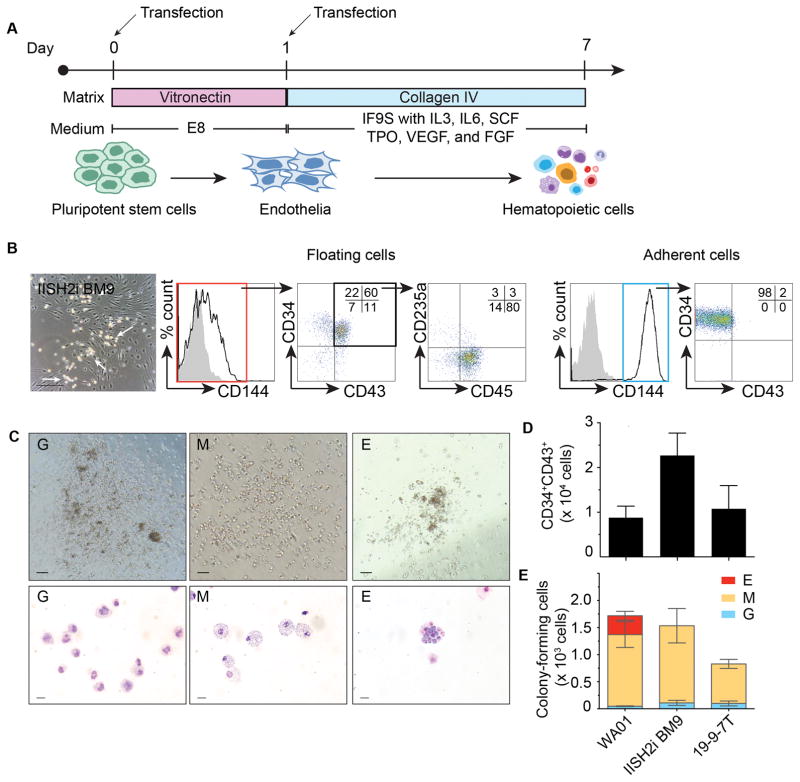
As we previously showed, overexpression of ETV2 and GATA2 in human PSCs induces formation of CD34+CD45+ multipotent hematopoietic progenitors through hemogenic endothelium intermediates (4). To further improve direct blood induction protocol and avoid the use of xenogenic components such as Matrigel, we evaluated the efficacy of blood production from different human PSCs lines in our new system which is based on vitronectin and collagen under serum-free and xeno-free conditions (Figure 4A). Following human PSC transfection with ETV2 and GATA2 modRNA, we observed the first hematoendothelial cluster as early as 4 days of culture. Flow cytometric analyses on day 7 after transfection showed that most of the early hematopoietic progenitors (CD34+43+45+235a−) have detached from their parental endothelial cells and expand in suspended culture (Figure 4B). These cells are able to form typical granulocyte colony-forming cell (CFC, CFC-G), macrophage (CFC-M), and erythroid (CFC-E) colonies in clonogenic cultures (Figure 4C). Comparison of gene expression between endothelial/hematopoietic cells generated from our protocol and primary human cells (i.e., HUVEC, cord blood (CB), and bone marrow (BM) mononuclear cell) demonstrated similarity in molecular signature of endothelial (TIE2, FLI1, and SCL) and hematopoietic lineages (C-Kit, LMO2, and MYB) (Supplemental Figure 2)
Figure 4. Generation of hematopoietic cells with ETV2/GATA2 modRNA.
A) Schematic diagram showing in vitro hematopoietic differentiation protocol. Pluripotent stem cells were transfected twice with ETV2/GATA2 combination (arrow). Transfected cells were transferred to collagen coated plate on day 1 in IF9S medium supplemented with hematopoietic cytokines.
B) Representative picture of day 7 hematoendothelial clusters (arrow) from IISH2i-BM9 iPSCs. Floating and adherent cells were analyzed separately with CD34, CD43, CD45, CD144, and CD253a. Dot plot showing live cells gated from the left histogram.
C) Types of hematopoietic colonies formed by floating cells from day 7 post-transfection with ETV2/GATA2 and corresponding Wright-stained cytospin. Granulocytic colonies (G), macrophage colonies (M), and erythroid colonies (E). Scale bar for CFC assay = 200 μm; cytospin = 20 μm.
D) Yields of CD34+43+ cells from human PSCs and E) Yields of colony-forming cells obtained from 2×105 undifferentiated pluripotent stem cells and their CFC potential. Mean±SD from 3 experiments is shown.
We estimated to obtain 1–2×104 CD34+CD43+ cells or 0.8–1.8×103 colony-forming cells from 2×105 initial undifferentiated human PSCs (Figure 4D and 4E). Similar combination of ETV2 and GATA2 modRNA induced formation of floating CD34+ hematopoietic cells from MnCy0669 iPSC. However, direct blood induction with modRNA from NHP iPSC growing under defined xeno-free condition was less efficient than from human iPSCs, probably due to lower efficiency of human transcription factors in NHP cells (Supplemental Figure 3).
Discussion
In these studies, we evaluated translational efficiency of different mRNA modifications in order to improve direct induction of endothelium and blood from PSCs. Using eGFP modRNA, we found that IVT template containing 5′UTR and single 3′UTR from β globin gene provided maximum protein levels in human PSCs while IVT template containing artificial two repeats of 3′UTRs yielded the least. The results indicated that the presence of β globin 5′UTR enhanced the translational efficiency of the transgenes, whereas double repeats of β globin 3′UTR is notably disadvantageous in human, but not in NHP iPSCs. Analysis of the β globin 5′UTR sequence reveals a low G-C content (44%) suggesting the potential of secondary structure formation that can suppress protein translation is unlikely. Point mutations within the 5′UTR of the β globin gene reduced the rate of transcription resulting in thalassemia diseases, but did not interfere with mRNA transport from the nucleus to the cytoplasm, 3′ end processing, and mRNA stability (23). These findings indicate the translational enhancement mechanism interacts on the specific sequence within 5′UTR during the transcription process. We found neither of the modifications demonstrated any significant benefit on the duration of functional eGFP protein expression. Study in dendritic cells showed that β globin UTRs on modRNA contribute to the translational efficiency and RNA stability (6); however, they affect differently between immature and mature dendritic cells indicating the impact of UTRs is likely cell-type specific. The differences in modRNA stability were also observed in our studies, which revealed variable effect of double repeats of β globin 3′UTR in human and NHP iPSCs. Therefore, the impact of UTRs on translation should be determined on a cell-to-cell basis.
Currently available methods to generate hematoendothelial cells from PSCs are a) embryoid body formation which is labor-intensive and time consuming (24, 25), b) monolayer differentiation (14, 26–28) which requires either hypoxic condition, stromal cells, or cytokine cocktail, and c) direct programming using transcription factors (4, 21). Since differentiation process is stochastic and depends on several factors, the generation efficiency become inconsistent between experiments and cell lines (29). Here we showed that productions of endothelial and blood cells from PSCs can be achieved in defined conditions by using ETV2 or ETV2 and GATA2 modRNA, respectively. Compared to other protocols for endothelial and hematopoietic cell generation, our modRNA-based approach does not require morphogens and provides experimental consistency and reliability regardless of types of PSCs and species. Because we use defined conditions, our method allows a production of clinical-grade cells for therapeutics. Estimated cost of reagents including synthesis kit, modified nucleotide, cap analog, and transfection chemicals (not including medium and culture vessels) to generate approximately 1–2×105 endothelial cells from PSCs is much lower than typical differentiation protocol using combination of cytokines (14). Additionally, these endothelial cells can be further expanded to obtain cells in large numbers.
Assessment on efficacy, safety, immunogenicity and toxicity of iPSC-based therapies in preclinical models would be essential to advance these technologies to the clinic. Demonstration in our studies that modRNA protocol can be adopted to generate endothelial and blood cells from NHP iPSCs, open opportunity to evaluate modRNA-induced blood and endothelial products in NHP preclinical model. In current studies, we used iPSCs from Mauritian cynomolgus monkeys. Mauritian cynomolgus macaques are descendent from a small founder population and have very limited MHC diversity consisting of only seven common haplotypes (M1-M7) (30–32). This provides a unique opportunity to rapidly select MHC and blood group-identical animals, or animals with well-defined MHC/blood group mismatches by genetic screening to test immunogenicity of PSC-derived products.
Overall, our study will assist stem cell researchers in designing experiments to modulate gene expressions using modRNA and provides a scalable protocol to generate hematoendothelial cells from human and NHP iPSCs for a range of applications such as developmental study, disease modeling, high-throughput screening and eventually cellular therapies where the availability of large cell numbers is a key prerequisite.
Supplementary Material
Acknowledgments
We thank the Cell Processing and Manipulation Core in the Translational Cores, and Physicians and Nurses at University of Wisconsin Carbone Cancer Center and Cincinnati Children’s Hospital Medical Center for obtaining and processing bone marrow samples and Translational Research Trials Office for providing the regulatory and administrative support for this endeavor. This work was supported by funds from the National Institute of Health (1R01HL132891, 4R01HL116221, and P51OD011106).
Footnotes
Author Contributions
K.S. designed and conducted experiments and wrote the manuscript, L.T. conducted eGFP experiments, V.B.V. conducted hematopoietic developmental assays, S.S.D. conducted NHP-related experiments, A.K. conducted endothelial developmental assays, I.S. conceptualized and supervised all aspects of the studies and wrote the manuscript.
Disclosure Statement
Authors declare no conflict of interest.
Contributor Information
Kran Suknuntha, Department of Pharmacology, Faculty of Science, Mahidol University, Bangkok, Thailand, 10400. Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715.
Lihong Tao, Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715.
Vera Brok-Volchanskaya, Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715.
Saritha S. D’Souza, Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715
Akhilesh Kumar, Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715.
Igor Slukvin, Department of Pathology and Laboratory Medicine, University of Wisconsin, Madison, WI 53792. Wisconsin National Primate Research Center, University of Wisconsin, Madison, WI 53715.
References
- 1.Szabo E, Rampalli S, Risueno RM, et al. Direct conversion of human fibroblasts to multilineage blood progenitors. Nature. 2010;468(7323):521–6. doi: 10.1038/nature09591. [DOI] [PubMed] [Google Scholar]
- 2.Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature. 2010;463(7284):1035–41. doi: 10.1038/nature08797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lis R, Karrasch CC, Poulos MG, et al. Conversion of adult endothelium to immunocompetent haematopoietic stem cells. Nature. 2017;545(7655):439–45. doi: 10.1038/nature22326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Elcheva I, Brok-Volchanskaya V, Kumar A, et al. Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun. 2014;5:4372. doi: 10.1038/ncomms5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oh S, Kessler JA. Design, Assembly, Production, and Transfection of Synthetic Modified mRNA. Methods. 2017 doi: 10.1016/j.ymeth.2017.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holtkamp S, Kreiter S, Selmi A, et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood. 2006;108(13):4009–17. doi: 10.1182/blood-2006-04-015024. [DOI] [PubMed] [Google Scholar]
- 7.Kranz LM, Diken M, Haas H, et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature. 2016;534(7607):396–401. doi: 10.1038/nature18300. [DOI] [PubMed] [Google Scholar]
- 8.Warren L, Manos PD, Ahfeldt T, et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 2010;7(5):618–30. doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kormann MS, Hasenpusch G, Aneja MK, et al. Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol. 2011;29(2):154–7. doi: 10.1038/nbt.1733. [DOI] [PubMed] [Google Scholar]
- 10.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–16. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumari S, Bugaut A, Huppert JL, et al. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3(4):218–21. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Souza SS, Maufort J, Kumar A, et al. GSK3beta Inhibition Promotes Efficient Myeloid and Lymphoid Hematopoiesis from Non-human Primate-Induced Pluripotent Stem Cells. Stem Cell Reports. 2016;6(2):243–56. doi: 10.1016/j.stemcr.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen G, Gulbranson DR, Hou Z, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods. 2011;8(5):424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uenishi G, Theisen D, Lee JH, et al. Tenascin C promotes hematoendothelial development and T lymphoid commitment from human pluripotent stem cells in chemically defined conditions. Stem Cell Reports. 2014;3(6):1073–84. doi: 10.1016/j.stemcr.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5(10):827–35. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozak M. Features in the 5′ non-coding sequences of rabbit α and β-globin mRNAs that affect translational efficiency. Journal of Molecular Biology. 1994;235(1):95–110. doi: 10.1016/s0022-2836(05)80019-1. [DOI] [PubMed] [Google Scholar]
- 17.Jiang H, Lucy MC. Variants of the 5′-untranslated region of the bovine growth hormone receptor mRNA: isolation, expression and effects on translational efficiency. Gene. 2001;265(1–2):45–53. doi: 10.1016/s0378-1119(01)00356-0. [DOI] [PubMed] [Google Scholar]
- 18.Conne B, Stutz A, Vassalli JD. The 3′ untranslated region of messenger RNA: A molecular ‘hotspot’ for pathology? Nat Med. 2000;6(6):637–41. doi: 10.1038/76211. [DOI] [PubMed] [Google Scholar]
- 19.Leonhardt C, Schwake G, Stogbauer TR, et al. Single-cell mRNA transfection studies: delivery, kinetics and statistics by numbers. Nanomedicine. 2014;10(4):679–88. doi: 10.1016/j.nano.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng. 1999;12(12):1035–40. doi: 10.1093/protein/12.12.1035. [DOI] [PubMed] [Google Scholar]
- 21.Morita R, Suzuki M, Kasahara H, et al. ETS transcription factor ETV2 directly converts human fibroblasts into functional endothelial cells. Proc Natl Acad Sci U S A. 2015;112(1):160–5. doi: 10.1073/pnas.1413234112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamamizu K, Matsunaga T, Katayama S, et al. PKA/CREB signaling triggers initiation of endothelial and hematopoietic cell differentiation via Etv2 induction. Stem Cells. 2012;30(4):687–96. doi: 10.1002/stem.1041. [DOI] [PubMed] [Google Scholar]
- 23.Sgourou A, Routledge S, Antoniou M, et al. Thalassaemia mutations within the 5′UTR of the human beta-globin gene disrupt transcription. Br J Haematol. 2004;124(6):828–35. doi: 10.1111/j.1365-2141.2004.04835.x. [DOI] [PubMed] [Google Scholar]
- 24.Levenberg S, Zoldan J, Basevitch Y, et al. Endothelial potential of human embryonic stem cells. Blood. 2007;110(3):806–14. doi: 10.1182/blood-2006-08-019190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ng ES, Azzola L, Bruveris FF, et al. Differentiation of human embryonic stem cells to HOXA+ hemogenic vasculature that resembles the aorta-gonad-mesonephros. Nat Biotechnol. 2016;34(11):1168–79. doi: 10.1038/nbt.3702. [DOI] [PubMed] [Google Scholar]
- 26.Vodyanik MA, Bork JA, Thomson JA, et al. Human embryonic stem cell-derived CD34+ cells: efficient production in the coculture with OP9 stromal cells and analysis of lymphohematopoietic potential. Blood. 2005;105(2):617–26. doi: 10.1182/blood-2004-04-1649. [DOI] [PubMed] [Google Scholar]
- 27.Wang ZZ, Au P, Chen T, et al. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat Biotechnol. 2007;25(3):317–8. doi: 10.1038/nbt1287. [DOI] [PubMed] [Google Scholar]
- 28.Patsch C, Challet-Meylan L, Thoma EC, et al. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat Cell Biol. 2015;17(8):994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orlova VV, Drabsch Y, Freund C, et al. Functionality of endothelial cells and pericytes from human pluripotent stem cells demonstrated in cultured vascular plexus and zebrafish xenografts. Arterioscler Thromb Vasc Biol. 2014;34(1):177–86. doi: 10.1161/ATVBAHA.113.302598. [DOI] [PubMed] [Google Scholar]
- 30.Budde ML, Wiseman RW, Karl JA, et al. Characterization of Mauritian cynomolgus macaque major histocompatibility complex class I haplotypes by high-resolution pyrosequencing. Immunogenetics. 2010;62(11–12):773–80. doi: 10.1007/s00251-010-0481-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiseman RW, Karl JA, Bohn PS, et al. Haplessly hoping: macaque major histocompatibility complex made easy. ILAR J. 2013;54(2):196–210. doi: 10.1093/ilar/ilt036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiseman RW, Wojcechowskyj JA, Greene JM, et al. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81(1):349–61. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.