Abstract
Autologous human cardiac stem/progenitor cell (hCPC) therapy is a promising treatment that has come into use in recent years for patients with cardiomyopathy. Though innovative in theory, a major hindrance to the practical application of this treatment is that the hCPCs of elderly patients, who are most susceptible to myocardial disease, are senescent and prone to cell death. Rejuvenating hCPCs from elderly patients may help overcome this obstacle, and can be accomplished by reversing entry into the cellular stage of senescence. p16INK4A, a cyclin dependent kinase inhibitor, is an important player in the regulation of cell senescence. In this study, we investigated whether knockdown of p16INK4A will rejuvenate aging hCPCs to a youthful phenotype. Our data indicated that upregulation of p16INK4A is associated with hCPC senescence. Both cell proliferation and survival capacity were significantly increased in hCPCs infected with lentivirus expressing p16INK4A shRNA when compared to control hCPCs. The knockdown of p16INK4A also induced antioxidant properties as indicated by a 50% decrease in ROS generation at basal cell metabolism, and a 25% decrease in ROS generation after exposure to oxidative stress. Genes associated with cell senescence (p21CIP1), anti-apoptosis (BCL2 and MCL1), anti-oxidant (CYGB, PRDX1 and SRXN1), and NFκB signal pathway (p65, IKBKB, HMOX1, etc.), were significantly upregulated after the p16INK4A knockdown. Knocking down the NFκB-p65 expression also significantly diminished the cytoprotective effect caused by the p16INK4A knockdown. Our results suggest that genetic knockdown of p16INK4A may play a significant role in inducing antioxidant effects and extending lifespan of aging hCPCs. This genetic modification may enhance the effectiveness of autologous hCPC therapy for repair of infarcted myocardium.
Keywords: p16INK4A, Human cardiac stem/progenitor cells, Heart failure, Myocardial infarction, cell rejuvenation, Lentiviral knockdown
Introduction
About 1 in every 4 deaths in the USA can be attributed to cardiovascular disease [1]. Though symptomatic treatment is available, there is no permanent cure for heart disease [2]. Autologous cardiac stem/progenitor cell (CPC) transplant is a promising treatment that involves extraction of the patient’s own cardiac progenitor cells, expansion ex vivo, and subsequent injected back into the cardiac infarct site, in a bid to promote healing and repair of the myocardial infarct site [3]. A major obstacle hindering the efficacy of stem/progenitor cell transplant therapy is the age of the patients suffering from myocardial infarction and cardiovascular disease. This patient population has a median age of 56 years in men and 65 years in women [4]. Because the patients are of advanced age, the progenitor cells harvested from the tissues of this patient population tend to be pre-senescent or already in senescence. Cells in this state are in growth arrest, and are likely to undergo necrotic and/or apoptotic processes, contributing to loss of cardiac function [5, 6]. Senescence can be induced by environmental and genetic factors and has been an evolutionary tool for the prevention of tumor formation and suppression of oncogenes [7]. It is comprised of molecular pathways that can be turned on by various signals and lead to semi-permanent growth arrest. One of the biomarkers for aging and senescent cells is p16INK4A [5, 8–13].
The gene p16INK4A, located on human chromosome 9, functions as a cyclin-dependent kinase (CDK) inhibitor and a significant player in regulation of the cell cycle [8]. Its expression is known to increase proportionally with age in most mammalian tissues [7, 14]. p16INK4A operates via a retinoblastoma (Rb)-mediated signal transduction cascade whose endpoint is cell cycle arrest and subsequent onset of cellular senescence [15]. One of p16INK4A’s major functions consists of acting as a cyclin dependent kinase inhibitor (CKI) by binding to and inhibiting cyclin-dependent kinases (CDKs) 4 and 6 [16, 17]. When p16INK4A is active, it inhibits the CDK complexes, causing Rb to remain bound to E2F and thereby inhibiting progression of the cell cycle [7, 16, 18]. Thus, Rb and p16INK4A are among the most important proteins involved in the checkpoints between stages G1 and S of the cell cycle and the onset of cellular senescence [6, 19, 20]. Over time, mass inhibition of the cell cycle causes cycle arrest and therefore senescence. Senescent cells that show elevated p16INK4A expression eventually lose ability to proliferate and cell population growth is halted [9]. In cells genetically modified to overexpress p16INK4A, growth arrest is similarly observed [21]. This inverse relationship between p16INK4A expression and proliferation ability has been observed to be age-dependent in certain types of stem cells [17]. Through its cell cycle inhibition effects, p16INK4A has also proven itself to be a major regulator of self-renewal of various stem cell types, and thus a target of control to inhibit cellular proliferation [18]. Its upregulation can be induced by stress stimuli such as DNA damage by UV light, oxygen radicals, or other factors, and is associated with both Ras protein activation and mitogen-activated protein (MAP) kinase activation [17, 18]. The promoter for the p16INK4A gene is also known to consist partly of a negative regulatory element for other transcription factors similarly involved in anti-apoptosis and cell cycle progression [18].
Since senescent cells from older patients are known to have decreased regenerative potential in stem cell therapy, it has been previously suggested that p16INK4A expression may play a role in the decreased efficacy of stem cells [17]. Studies have shown that p16INK4A silencing may significantly delay cellular entry into senescence [11, 22]. Silencing of p16INK4A, along with silencing of the Arf gene of the same locus, was even observed to increase hematopoietic stem cell proliferation and transplant efficacy by Stepanova et al [23]. Thus, we believe that silencing p16INK4A may have a similar positive effect on hCPC regenerative potential. Our lab has chosen knockdown of this gene as a method to attempt rejuvenation of aging hCPCs for myocardial repair. Our goal was to observe the effects of silencing p16INK4A in aged human cardiac progenitor cells with elevated endogenous levels of this gene. We hypothesized that knockdown of p16INK4A would rejuvenate hCPCs to a youthful phenotype by inducing antioxidant effects and increasing hCPC proliferation and survival.
Materials and Methods
Reagents
Ham’s F12 medium was from Invitrogen. Fetal bovine serum (FBS) was obtained from Hyclone. The primary antibodies were listed in the Supplemental Table S1. Quantitative PCR primers for target genes were obtained from Real Time Primers, LLC. Unless indicated otherwise, chemicals used in experiments were purchased from Sigma.
Harvesting of Human c-Kit+ CPCs
Human cardiac stem/progenitor cells (hCPCs), expressing c-Kit cell surface marker, were isolated and purified from atrial appendages of patients during open-heart surgery at Albany Medical Center. The procedures were followed exactly as described previously [24]. Our protocol has been approved by the Institutional Committee on Research Involving Human Subjects (IRB). All participants in the study provided written informed consent. Briefly, right atrial tissues (100 to 400 mg) were minced and enzymatically digested with collagenase II (30 U/ml) at 37°C in a shaking water bath. During the incubation, chopped tissues were mechanically disturbed by gently pipetting several times. After 1 hour of incubation, undigested clumps were separated by gravity on ice for 10 min, and the supernatant was carefully transferred into a 15-ml tube. Dissociated cells from digestion were collected by centrifuge at 1200 RPM for 5 minutes. Cell pellets were suspended and cultured in Ham’s F12 medium supplemented with 10% FBS, 10 ng/ml human basic FGF, 0.005 U/ml human erythropoietin, 0.2 mM L-glutathione, 100 U/ml penicillin and 100 μg/ml streptomycin. Cells were maintained in a humidified environment at 37°C and 5% CO2. The next day, the CPC growth medium was refreshed, and adherent cells were cultured with medium change every other day. Upon reaching approximately 80% confluence, cells were sorted using the c-kit MACS kit according to the manufacturer’s instructions (Miltenyi Biotec), expanded, and characterized by FACS analysis to obtain lineage-negative hCPCs. A total of 9 lines of hCPCs were used in this study, categorized into three groups based on the donor’s age as follows: the group with relative young age (40–50 years) (hCPCs-Y), the group with middle age (65–75 years) (hCPCs-M), and the group with advanced age (>80 years) (hCPCs-A). Please see the Supplemental Table S2 for basic demographic information for the cardiac tissue donors. Human CPCs at passage 6–10 were used to perform most of in vitro experiments in this study. CPCs at passage 15 were used only to assess the expression of p16INK4A during replicative senescence.
Cell Culture
c-Kit+ CPCs were isolated from tissue samples obtained from patients of various ages at Albany Medical Center. Cells were cultured in F12 media supplemented with 10% Fetal Bovine Serum (FBS), 10 ng/mL Fibroblast Growth Factor (FGF), 1% penicillin/streptomycin, 200 uM L-glutathione, and 0.005 units/mL human erythropoietin. Cells were cultured in T75 flasks at 37°C and 5% CO2. HEK293FT cells were grown in DMEM containing 10% FBS, 10mM Non-Essential Amino Acids (NEAA), and 1% penicillin/streptomycin.
CyQuant and Viability Assays
Cells were plated in quadruplicate (2000 cells/well) in a 96-well plate for the CyQuant assay with the CyQuant reagent (Thermo Fisher Scientific) as previously described [25]. Cell viability was assessed by trypan blue staining [25]. Population doubling times were calculated using a population doubling time online calculator (http://www.doubling-time.com/compute.php) based on the reading from CyQuant and viability assays.
Lentiviral Infection of hCPCs
pLenti X2 Blast/shp16 (w112-1) was a gift from Eric Campeau (Addgene plasmid # 22261). This plasmid containing the shRNA insert for p16INK4A knockdown was co-transfected with three packaging plasmids: 1.5ug each of PLP1, PLP2, and PLP/VSG using Gene Jammer Transfection Reagent (Agilent). The lentiviral supernatant from the transfected cells was harvested, sterile filtered, and concentrated using the Lenti-X Concentrator (Clontech). It was subsequently used for infection of hCPCs at a ratio of 1:100. 24 hours prior to infection, hCPCs were seeded at the density of 1 × 105 per well in a 6-well plate. At 50–60% confluence, cells were refreshed with 2 ml of culture medium followed by the direct addition of 0.5 μg/ml Polybrene and 20 μl of lentiviral particles with gentle orbital shaking. The next day, the culture medium was replaced with normal growth medium for recovery.
Senescence-associated Beta-galactosidase Assay
Senescence was qualitatively measured by using the SA-beta-gal staining kit (Cell Signaling Technology, Inc.). After lentiviral infection for 48 hours, hCPCs were washed 2 times with 1X PBS and fixed with the solution provided according to manufacturer’s instructions. Beta-galactosidase staining solution was added, and cells were incubated in the plate at 37°C. After 24 hours, the Beta-gal positive cells were identified by blue color under 200X magnification.
ROS Generation Assay
Cellular ROS generation was measured by staining of hCPCs using the Cellular ROS Detection Assay Kit (Abcam). Prior to staining, hCPCs were plated at a density of 2 × 105 per well in a 6-well plate and infected with lentiviral particles carrying the shRNA to knockdown p16INK4A. After 48-hour infection, hCPCs were challenged with or without 1mM H2O2 in 1% FBS F12 medium for 1 hour. Cells were then detached by TrypLE solution (Invitrogen), washed twice with PBS, and suspended by gentle mixing in 100 μl of ROS Assay Buffer supplemented with 1 μl 200x ROS Deep Red Stock solution. Cells were incubated in this suspension at 37°C and 5% CO2 for 30 minutes, 100 μl additional ROS Assay Buffer was added, and samples were immediately analyzed by flow cytometry.
RNA Isolation and Quantitative Analysis
Real-Time PCR was performed to determine the basal p16INK4A expression levels in cells of different ages. Primers used in this study were obtained from IDT (Integrated DNA Technologies). The total RNA of each sample was extracted and purified from whole cell lysates using the Aurum™ Total RNA Mini Kit (Bio-Rad). The quality and quantity of RNA was detected by a NanoDrop 2000C spectrophotometer (Thermo Scientific). The reverse transcription of 1 μg of RNA to cDNA was established using Bio-Rad iScript™ cDNA synthesis Kit, and samples for Real-time PCR were prepared according to the manufacturer’ instructions of the iQ SYBR Green Supermix kit (Bio-Rad). Real-time PCR was run in a 384-well plate with a Bio-Rad iQ5 optical module in the CFX96 Touch™ Real-Time PCR Detection System. Cycling conditions were: 95°C for 2 minutes as initial denaturation, 40 cycles of denaturation at 95°C for 15 seconds, and annealing/extension at 60°C for 40 seconds. Melting Curve analysis was set between 65°C and 95°C with 0.5°C increments at 5 seconds per step. The gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control for quantitative RT-PCR expression analysis.
Telomere Length Measurements
Telomere length was examined by real-time qPCR detection system using the modified monochrome multiplex quantitative PCR method as described previously [26]. In this PCR method, albumin is amplified with the telomere template at the same time. Briefly, 20ng of DNA, 1X syber green with albumin and telomere primer were combined in a reaction volume of 10ul and the samples were run in a set of 6 replicates at minimum. The primer sequences used were 5′-ACA CTA AGG TTT GGG TTT GGG TTT GGG TTT GGG TTA GTG T (forward) and 5′-TGT TAG GTA TCC CTA TCC CTA TCC CTA TCC CTA TCC CTA ACA (reverse) for telomere, and 5′-CGG CGG CGG GCG GCG CGG GCT GGG CGG AAA TGC TGC ACA GAA TCC TTG (forward) and 5′-GCC CGG CCC GCC GCG CCC GTC CCG CCG GAA AAG CAT GGT CGC CTG TT (reverse) for albumin. The PCR conditions included initial denaturing at 95°C for 15 minutes, followed by 2 cycles of 15 sec at 94°C, and 15 sec at 49°C, and then 26 cycles of 15 sec at 94°C, 10 sec at 62°C, and 15 sec at 73°C with signal acquisition for telomere, 10 sec at 84°C, and 15 sec at 87°C with signal acquisition for albumin. After PCR amplification, melting curves were analyzed to confirm the size of PCR products.
Immunoblotting
Human CPCs were grown to confluence in 6-well plates. Whole cell lysates were washed twice with ice-cold PBS, then collected using Radioimmunoprecipitation (RIPA) buffer (150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 20 mM Tris-HCl, PH 7.5) mixed with Proteinase Inhibitor Cocktail (PIC). NuPAGE® LDS Sample Buffer (4X) were added to samples and they were boiled for 5 minutes. 20 μg of protein from each sample was loaded onto a 10% SDS-PAGE gel and run at 120V for 1 hour, and then transferred to a nitrocellulose membrane at 200 mA for 2 hours at 4°C. The membrane was then blocked with 5% milk/TBST for 1 hour, placed into primary antibodies (Please see the list of antibodies in the Supplemental Table S1), and left shaking overnight at 4°C. The next day, secondary antibody was added at a dilution of 1:2000 for 2-hour shaking at room temperature. After extensive washing between each step, the signals were detected with the chemiluminescent Amersham ECL Detection Reagent (GE Healthcare Life Sciences). The loading control was α-tubulin detected in the same sample.
Statistics
Data were presented as means ± SD of results taken from at least 3 independent experiments. Statistical significance was assessed by analysis of variance using unpaired student t-test. A p value less than 0.05 was considered statistically significant.
Results
p16INK4A expression is associated with senescence
To attempt any cellular rejuvenation technique, it is necessary to first obtain senescent hCPCs. Cellular senescence in vitro can be qualitatively measured by examining of senescence-associated β-galactosidase (SAβG) activity at pH 6.0. SAβG is a lysosomal enzyme whose function is to hydrolyze β-galactoside molecules into monosaccharide sugars in senescent cells exclusively [27]. In the presence of a chromogenic substrate known as X-gal, this biochemical reaction produces a blue-color byproduct which can be easily visualized in vitro under phase-contrast microscopy [27]. In this study, a total of 9 lines of hCPCs were randomly selected and catagorized into three groups based on the donor’s age, including the relatively young aged group (40–50 years) (hCPCs-Y), the relatively middle aged group (65–75 years) (hCPCs-M), and the advanced age group (>80 years) (hCPCs-A). Basic demographic information of the cardiac tissue donor is listed in the Supplemental Table S2. Only age and sex information are available based on our initial IRB protocol. Human CPCs isolated from atrial appendages from the group of hCPCs-A were larger, flatter, more multinucleated, and displayed an increase in intensity of blue color stain originating from the presence of β-galactosidase (Fig. 1A–D). Each of these findings is correlated with advanced patient age, and known to be related to senescence in this assay [7]. These results confirmed that the hCPCs selected for this study were indeed senescent, as each cell line was obtained from male patients increasing in age from 44 years old (Fig. 1A), 71 years old (Fig. 1B), to 93 years old (Fig. 1C). Assessment of the population doubling time of the 9 lines of hCPCs showed a range from 19.1 to 31.5 hours. Interestingly, the growth kinetics were ~30% slower in the group of hCPCs-A when compared to the group of hCPCs-Y as measured by population doubling time (Fig. 1E). Proliferation rates were also observed to be decreased in the group of hCPCs-A compared to the group of hCPCs-Y (Fig. 1F).
Figure 1. P16INK4A expression is associated with the aging/senescence of cardiac progenitor cells.
Representative images showed the senescence-associated β-galactosidase staining in hCPCs from patients at the age of 44 years old (A), 71 years old (B), and 93 years old (C). D. Quantitative analysis of total β-galactosidase staining. E. Multiple lines of hCPCs showed variation in population doubling times as measured by CyQuant and viability assay cell counts from different groups of patients (n=3 independent experiments). See the Supplemental Table S2 for the basic demographic information for the cardiac tissue donors. F. Differences in proliferation rates were observed in three different hCPC groups (n=3 lines of hCPCs per group). G. Real time qPCR data showed the p16INK4A gene expression in hCPCs from different groups of patients (n=3 independent experiments). * indicates p<0.05 versus the group of hCPCs-Y for panel E, F, G. H. Quantitative analysis of Western blot showing the significant increase of p16INK4A expression in the group of hCPCs-A versus the group of hCPCs-Y at passage 6 (P6) (n=3 lines of hCPCs per group). I. Quantitative analysis of Western blot revealed an increase in p16INK4A protein after replicative passaging (P15 versus P6) of three lines of hCPCs in the hCPCs-Y group. J. Quantitative analysis of Western blot showed an increase in p16INK4A protein expression in three lines of hCPCs at passage 6 (P6) in the hCPCs-Y group following induction of stress-induced premature senescence through treatment in 0.5 μM doxorubicin for 18 h. * indicates p<0.05. All data were presented as means ± SD from either 3 independent experiments or 3 lines of hCPCs per group.
Quantitative PCR was subsequently run on these cell lines to examine the expression levels of p16INK4A and determine whether this gene was indeed correlated with age and cellular senescence. RT-PCR analysis confirmed that normalized p16INK4A mRNA expression is significantly higher in both the group of hCPCs-M and the group of hCPCs-A compared to the group of hCPCs-Y (Fig. 1G). This increase in expression was a step-wise change, very similar to the step-wise increase in SABG staining (Fig. 1D). The mRNA level differences coincide with changes in protein expression, with significant increases of p16INK4A (>3.5-fold; p<0.05) in the group of hCPCs-A relative to the group of hCPCs-Y (Fig. 1H). Once p16INK4A’s association with the advanced age was established, it was important to confirm whether this association held true with replicative senescence. Three lines of the hCPCs-Y group were cultured from passage 6 (P6) through passage 15 (P15), and Western blot confirmed that p16INK4A expression was significantly higher in the cells that had been cultured for longer time (Fig. 1I). Going by p16INK4A’s correlation with senescence, this finding suggests that senescence had been “induced” by replicative passaging. To examine whether stress induced senescence would similarly be associated with increased p16INK4A expression, three lines of the hCPCs-Y group, which have a low baseline level of p16INK4A expression at passage 6 (P6), were exposed to stress and then re-examined for p16INK4A levels. The “younger” hCPCs were treated with 0.5 μM doxorubicin for 18 hours, and Western blot was performed to check p16INK4A expression after the treatment. Doxorubicin is a cardiotoxic anti-neoplastic drug that can be used to induce stress in myocardial origin cells [28]. Quantitative analysis shows that p16INK4A expression was tripled with the stress-inducing doxorubicin treatment (Fig. 1J), showing that p16INK4A is associated with both natural progression of aging, as well as stress induced senescence.
p16INK4A knockdown reverses the senescent phenotype of hCPCs
Our data correlated higher p16INK4A expression with age and therefore senescence. This is consistent with the current literature [5, 8–12]. Based on this information, our lab chose to perform a knockdown of p16INK4A by lentiviral infection to attempt a reversal of cell cycle arrest. It was our hope that this would encourage the progenitor cells to enter cell division, increase cell proliferation, and lead to an overall increase in cell survival. The lentiviral infection was successful in the hCPCs (Fig. 2A–B) and showed significant decrease in p16INK4A expression in both RT-PCR as well as Western blot analysis. The experimental groups consisted of hCPCs from patients of advanced age greater than 80 years old (AMC20) infected with the lentivirus containing sh-RNA to knock down p16INK4A compared to the same line of hCPCs infected by a non-coding (scramble) sh-RNA. The infection was performed as described briefly in the Methods section and the Supplemental Method document. To evaluate the effect of p16 knockdown on the hCPC replicative senescence, we performed the qPCR for hCPCs at various passages after infected with lentivirus expressing shRNA against p16INK4A at passage 6. As shown in the Supplemental Figure S1, hCPCs stably expressing p16INK4A shRNA were able to be expanded to at least 14 passages, and the expression of p16INK4A remained similar throughout the passages. Telomere shortening has been associated with cell senescent phenotypes and is known to contribute to the loss of regenerative potential with age [29]. Therefore, the monochrome multiplex quantitative PCR method [26] was employed to assess the effect of p16INK4A knockdown on telomere length in CPCs with advanced age. As shown in the Supplemental Figure S2, the telomere length increased significantly in three advanced aging hCPC lines (AMC20, AMC54, AMC61) at passage 6 after knocking down p16INK4A, compared with the hCPCs of the same lines that were instead infected with lentiviral particles expressing scramble shRNA.
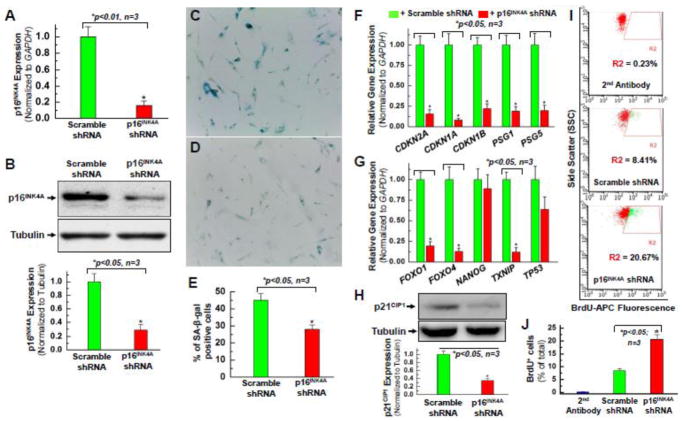
Figure 2. Knocking down the expression of p16INK4A reverses the senescent phenotype of hCPCs.
Real time qPCR data (A) and quantitative analysis of Western blot (B) confirmed that p16INK4A protein expression was knocked down in one line (AMC20) of the hCPCs-A group at passage 8 (P8) infected with lentivirus expressing shRNA against p16INK4A compared to the control which was infected with scramble shRNA. Senescence-associated β-galactosidase staining showed a decrease in the number of senescent cells in the group of hCPCs that were infected with lentivirus expressing shRNA against p16INK4A (D) compared to the group of hCPCs expressing the scramble shRNA (C). E. Quantitative analysis of β-galactosidase positive staining in panel C and D. In hCPCs with 48-hour knockdown of p16INK4A, senescence-associated genes (F) and quiescence-associated genes (G) exhibited decreased expression compared to hCPCs with 48-hour infection by a scramble control. Data labels shown are for the gene expression levels in p16INK4A shRNA-infected hCPCs, relative to those in scramble shRNA-infected hCPCs. H. Quantitative analysis of Western blot confirmed a decrease in p21CIP1 protein expression after knocking down p16INK4A in hCPCs, consistent with its decreased gene expression (CDKN1A) shown in panel F. I. Representative FACS analysis showed the increased proliferation ability of hCPCs after knocking down p16INK4A. J. Quantitative data analysis for panel I. Data were presented as means ± SD from 3 independent experiments (n=3); * indicates p<0.05 vs. scramble shRNA.
Interestingly, SAβG staining showed decreased numbers of senescent cells in the experimental hCPCs infected with lentivirus expressing shRNA against p16INK4A (Fig. 2D) when compared to the control hCPCs expressing scramble shRNA (Fig. 2C). Since the hCPCs expressing shRNA against p16INK4A showed decreased expression of p16INK4A as well as decreased SAβG positive staining (Fig. 2E), this further suggests that p16INK4A is involved with senescence. The next step was to perform genetic library screens for some of the various other genes associated either with the cell cycle (Fig. 2F) or with senescence (Fig. 2G). As expected, CDKN2A (an alias for p16INK4A) was significantly knocked down after the infection. Interestingly, CDKN1A was also significantly downregulated after the knockdown (Fig. 2F). CDKN1A’s alias, p21, showed greatly decreased post-infection expression on Western Blot (Fig. 2H). This is likely due to p21/CDKN1A’s role as a cyclin dependent kinase inhibitor (CKI). p21/CDKN1A is known to induce senescence by inhibiting progression of the cell cycle much in the same way that p16INK4A does [5, 10, 30, 31]. CDKN1B (p27) gene expression was also downregulated, likely by a similar mechanism as the one mentioned previously for p21. This is the most logical explanation given p27’s similar role as a CKI [30, 31]. Other known senescence associated genes such as FOXO1, FOXO4, and TxNIP were also downregulated (Fig. 2G), indicating that the knockdown of p16INK4A may help reverse hCPC senescence.
p16INK4A knockdown’s success in decreasing the senescence phenotype (Fig. 2C–E) likely lies in its ability to cause downstream downregulation of other cell cycle inhibitors, and to allow progression of the cell cycle. 5-Bromo-2-deoxyuridine (BrdU) incorporation was used as a marker for cell proliferation, as it contains a nucleic acid (thymine) that is incorporated in only cells that are actively replicating their DNA. CPCs at various conditions were incubated with 2mM BrdU solution for 12 hours, and then samples were harvested and fixed with a paraformaldehyde-based fixative. After p16INK4A knockdown, an increase of nearly 12% in BrdU staining was observed on FACS analysis (Fig. 2I–J). The percent of total cells stained with BrdU more than doubled in hCPCs in whom p16INK4A was silenced compared to those infected with the control lentivirus. This suggests that the knockdown can start cell cycle progression.
p16INK4A knockdown exhibits an anti-apoptotic effect in aging hCPCs
Once the knockdown was confirmed, infected hCPCs were incubated with 2mM H2O2 for 3 hours and then evaluated by lactate dehydrogenase (LDH) assay. A decrease in LDH release of approximately 13% was observed in the p16INK4A knockdown cells compared to the controls, suggesting that the knockdown may be cytoprotective for aging hCPCs (Fig. 3A). Similarly, the infected hCPCs were challenged with 1mM H2O2 for 1.5 hours, and then evaluated by Annexin/PI FACS analysis to compare the effect of the knockdown on apoptosis. An increase of nearly 15% was observed in the number of “Live Cells” when comparing the p16INK4-knockdown cells to the control cells (Fig. 3B). Because of this, the knockdown cells showed an overall decrease in all other categories of the FACS analysis: 8% decrease in “early apoptosis” cell detection, 4% decrease in “late apoptosis”, 3% decrease in “necrotic cells”. These results corroborate the suggestion from the LDH release assay: that p16INK4A knockdown leads to pro-survival effects in hCPCs.
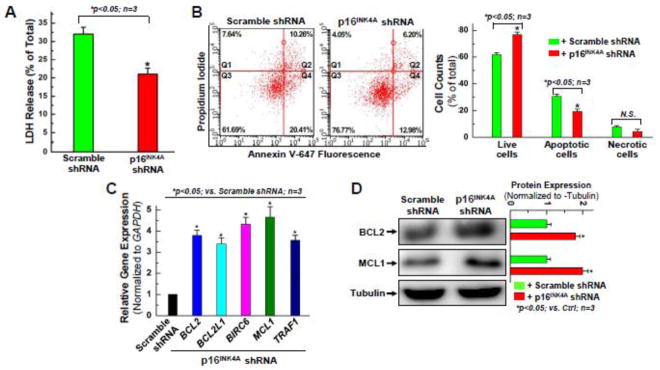
Figure 3. Knocking down the expression of p16INK4A exhibits an anti-apoptotic effect in aging hCPCs.
A. Aging hCPCs (AMC20) were infected with lentivirus expressing shRNA against p16INK4A, or scramble shRNA for 48 hours, challenged with 2 mM H2O2 for 3 hours, then evaluated by LDH assay. Data presented in this figure were mean values on a ratio of H2O2-induced LDH release to total LDH in cells with standard deviation (means ± SD). B. Human CPCs were infected with p16INK4A, or scramble shRNA for 48 hours, and challenged with 1 mM H2O2 for 1.5 hours, then evaluated by Annexin/PI FACS assay with quantitative analysis. C. Real time qPCR was performed to examine the expression of anti-apoptotic genes at the mRNA level after p16INK4A was knocked down in hCPCs. D. Representative images and quantitative data of Western blot showing the expression level of anti-apoptotic proteins, including BCL2 and MCL1 after p16INK4A knockdown in hCPCs. Data were presented as means ± SD from 3 independent experiments (n=3); * indicates p<0.05 vs. scramble shRNA.
To confirm whether these pro-survival, anti-apoptotic effects truly exist, and if so, what their mechanisms are, Real-Time qPCR and Western Blot were performed for anti-apoptotic genes. Quantitative PCR analysis showed a significant (3~5-fold) increase in the expression of BCL2, BCL2L1 (BCL-XL), BIRC6, MCL1 and TRAF1 in response to p16INK4A knockdown (Fig. 3C). This expression pattern of mRNA transcripts was confirmed at the protein level for BCL2 and MCL1 (Fig. 3D), which are known anti-apoptotic proteins [32–35]. The upregulation of MCL-1 is likely a major factor in cytoprotective effects of the p16INK4A knockdown, as loss of MCL-1 has been implicated in myocardial failure [36]. Prior evidence has suggested that the release of paracrine factors from CPCs plays an important role in mediating cardiac repair [32, 37]. To test the effect of p16INK4A knockdown on the expression of growth factors, qPCR for two human cytokine primer libraries was performed for the advanced age hCPC lines after knocking down p16INK4A. As shown in the Supplemental Figure S3, results from qPCR library array showed a significant increase in the gene expression of many cytoprotective cytokines in one advanced age hCPC line after knocking down p16INK4A, compared with those infected with lentiviral particles expressing scramble shRNA.
p16INK4A knockdown Displays Anti-Oxidant Effects in Aging hCPCs
It is possible that the same cytoprotective effect that led to pro-survival, anti-apoptotic gene upregulation may also lead to anti-oxidant gene upregulation. Reactive oxygen species (ROS) have been widely implicated in DNA damage and cellular senescence, and are a source of intracellular oxidative stress [38]. ROS production has also been previously linked to increased expression of CDK inhibitors, p16INK4A in particular [39]. We hypothesized that the p16INK4A knockdown would decrease intracellular ROS generation levels, induce a protective effect on the hCPCs and prevent excess cell damage and cell death. Both basal and oxidative stress-induced cellular ROS generation levels were examined by flow cytometric ROS Detection Assay Kit (Deep Red Fluorescence), and merged histograms of the ROS levels in the control hCPCs vs sh-p16INK4A -infected hCPCs were subsequently generated (Fig. 4A–B). The mean fluorescent intensity (MFI) is representative of ROS levels in each experiment. As seen in Fig. 4A–B, the MFI for the control hCPCs is significantly higher than the MFI for the shRNA-p16INK4A knockdown hCPCs under normal conditions (−H2O2) as well as under oxidative stress (+H2O2). It is notable that at basal levels before oxidative stress induction, the p16INK4A knockdown cells showed approximately 50% less ROS generation compared to the control (Fig. 4A). When both groups of hCPCs were challenged by oxidative stress, ROS levels rose in both groups, but ROS in the sh-p16INK4A infected cells was still 25% less than that of the control hCPCs (Fig. 4B). This data suggests that the p16INK4A knockdown may induce cytoprotection by lowering ROS production in aging hCPCs, which further highlights the significance of p16INK4A knockdown’s role in rejuvenation of senescent hCPCs.
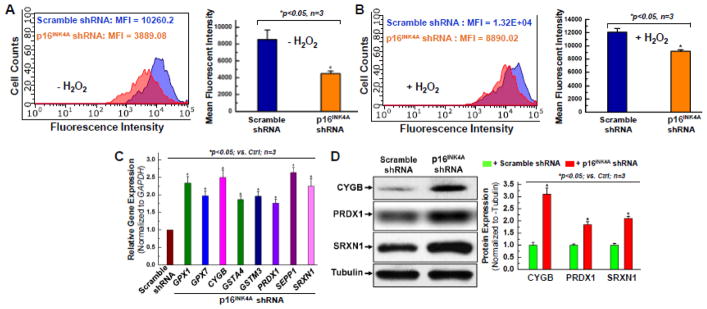
Figure 4. Knocking down the expression of p16INK4A displays the anti-oxidant effect in aging hCPCs.
Representative merged histogram and quantitative analysis for ROS measurement under the condition without oxidative stress (A) or with oxidative stress induced by 1mM H2O2 (B) for aging hCPCs (AMC20) expressing shRNA against p16INK4A vs. scramble shRNA by the FACS analysis with the ROS Detection Assay Kit (Deep Red Fluorescence). C. Real time qPCR was performed to examine the expression of anti-oxidant genes at the mRNA level after p16INK4A was knocked down in hCPCs. D. Representative images and quantitative data of Western blot showing the expression level of anti-oxidant proteins, including CYGB, PRDX1, and SRXN1 after p16INK4A knockdown in aging hCPCs. Data were presented as means ± SD from 3 independent experiments (n=3); * indicates p<0.05 vs. scramble shRNA.
To expand on these results, we screened for the expression of genes classically associated with anti- and pro-oxidant functions using an oxidative stress targeted PCR primer library (Supplemental Table S3). Changes in gene expression between the two groups were analyzed. In total, 38 genes related to oxidative stress were upregulated, and 11 genes related to oxidative stress were downregulated in the p16INK4A knockdown hCPCs as compared to the hCPCs that had been infected with a control virus. The most significant upregulated genes were glutathione peroxidase 1 and 7 (GPX1, GPX7), CYGB, GSTA4, GSTM3, PRDX1, SEPP1, and SRXN1 (Fig. 4C). The function of glutathione peroxidase (GPX1, GPX7) is to detoxify hydrogen peroxide [40]. GPX family genes are among the most significant anti-oxidant genes in humans, and so this upregulation is strongly predictive of p16INK4A knockdown’s protective effect on hCPCs. Cytoglobin (CYGB) is a hemoprotein that responds to oxidative stress by modulating nitric oxide metabolism and apoptosis pathways [41, 42]. A previous study by our lab found that CYGB is expressed in hCPCs, and that it encompasses an anti-oxidant role particularly in the face of oxidative stress [43]. In Fig. 3C–D, MCL1 and BCL2 were significantly upregulated after the p16INK4A knockdown, and CYGB expression was almost tripled after the same knockdown (Fig. 4D). This data suggests that p16INK4A may be inhibitory in this pro-survival pathway. Previous studies showed that recombinant human CYGB upregulated superoxide dismutase and glutathione peroxidase activity, and was associated with decreased NO and ROS levels [42]. PRDX1 has been implicated in protecting telomeres from damage due to ROS, contributing to longer cell life and increased cell cycles [44]. Along with other peroxiredoxin proteins, PRDX1 can reduce peroxides and protect cells from oxidative damage [45–47]. Since PRDX1 expression was doubled upon p16INK4A knockdown (Fig. 4D), it likely contributed to the decreased ROS generation at basal levels as well as after the induction of oxidative stress. PRDX1’s known function suggests that it may play a role in pro-survival and pro-proliferation pathways. Sulfiredoxin 1 (SRXN1) is an anti-oxidant enzyme participant in signaling pathways, and is regulated by the Srx-Prx axis [48]. It balances ROS production and elimination and is known to have anti-apoptotic properties when exposed to oxidative stress [49]. SRXN1 expression more than doubled after the p16INK4A knockdown (Fig. 4D), supporting a possible antioxidant role for p16INK4A knockdown.
Upregulation of NFκB signal pathway is associated with the rejuvenating effect of knocking down p16INK4A in aging hCPCs
Both pro-survival and anti-oxidant pathways have been linked with the NFκB signaling pathway [50–52]. Since p16INK4A knockdown has shown considerable antiapoptotic and antioxidant effects, it is of utmost importance to elucidate whether p16INK4A plays any role in the interactions of these pathways. RT-PCR and Western Blot with cytokines and NFκB related genes was performed (Fig. 5A–B) and showed upregulation in NFKB1, TBK1, RELA, IKBKB, and TICAM2. NFKB2 and HMOX1 showed 3~4-fold increased mRNA expression, while TLR3 showed almost 5.5-fold increased mRNA expression in the sh-p16INK4A infected aging hCPCs compared to the control hCPCs.
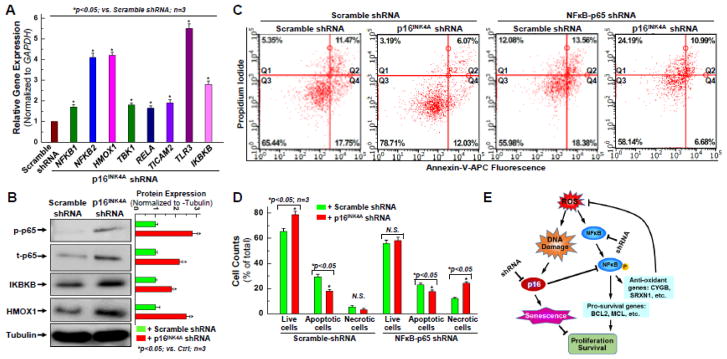
Figure 5. Up-regulation of NFκB signal pathway is associated with the rejuvenating effect of knocking down p16INK4A in aging hCPCs.
A. Real time qPCR was performed to examine the expression of NFκB signal pathway associated genes at the mRNA level after p16INK4A was knocked down in aging hCPCs. B. Representative images and quantitative data of Western blot showing the expression level of NFκB-associated proteins, including total and phosphorylated p65, IKBKB, and HMOX1 after p16INK4A was knocked down in aging hCPCs. C. Representative FACS analysis with annexin-V/PI staining showing H2O2-induced apoptosis in hCPCs stably expressing scrambled or NFκB-p65 shRNA following with or without knocking down p16INK4A. D. Quantitative data analysis for panel C. E. Descriptive diagram of proposed molecular mechanism underlying the rejuvenating effect of knocking down p16INK4A in aging hCPCs. Data were presented as means ± SD from 3 independent experiments (n=3); * indicates p<0.05 vs. scramble shRNA.
Inhibitor of Nuclear Factor Kappa B (IKBKB), also known as IKKbeta and IKK2, functions as a kinase for IKB which is an essential modulator of the NFκB pathway that regulates the immune response and cellular differentiation [53–55]. IKBKB was doubled in the cells with the p16INK4A knockdown (Fig 5A–B), suggesting that p16INK4A may also have a role in this pathway. Expression of HMOX1, or heme oxygenase-1 (HO-1), was also almost tripled after the knockdown (Fig 5A–B). This gene is known to have cytoprotective effects by cleaving the rings of free heme, which are known to make cells more susceptible to apoptosis [56, 57]. HO-1 expression exhibits significant anti-inflammatory and pro-survival properties, providing a secondary pathway for the aging hCPCs to be protected from apoptosis. Its significant upregulation in the p16INK4A knockdown hCPCs compared to the control hCPCs is certainly a favorable effect of this knockdown. Expression of phospho-p65 and total p65 were also increased significantly after the knockdown, with a ~3-fold increase compared to the control (Fig 5A–B). p65 is known to activate NFκB and is the starting point for various effector molecules that eventually give rise to downstream antiapoptotic and antioxidant effects of NFκB [58, 59]. Therefore, this data provides additional evidence for the involvement of p16INK4A molecule in the NFκB pathway.
To confirm these results, we used a previously established hCPC line with stable knockdown of NFκB (denoted NFkB-p65 shRNA in Fig. 5C) and compared it to its control (scramble-shRNA) using Annexin V/PI staining and flow cytometry analysis [60]. One group of hCPCs was infected with an shRNA to silence the form of NFκB which specifically is activated by p65 (NFκB-p65). That group was compared to a control which was infected only with a scrambled shRNA. Each of those groups were then infected with the shRNA to silence p16INK4A, or another scramble shRNA. Essentially, the effects of silencing p16INK4A with activated NFκB-p65 were compared to the effects of silencing p16INK4A without activation of NFκB-p65. As seen in Fig. 5C–D, the group with activated NFκB-p65 and silenced p16INK4A had 12% more live cells, 11% less apoptotic cells, and 2% less necrotic cells than the group with both NFκB-p65 activation and unsilenced p16INK4A. When NFκB-p65 was silenced, there was no significant difference in live cells between the cells that had p16INK4A expression and the cells that had p16INK4A silencing. There was, however, a 14% decrease in apoptotic cells after the p16INK4A silencing, but that was counterbalanced by a 12% increase in necrotic cells as compared to the control. From this data, it appears that the activation of NFκB-p65 is essential for the antiapoptotic and antioxidant effects observed in Fig. 2–4. With NFκB first externally silenced, there was virtually no difference in live cell count in the hCPCs with p16INK4A expression (scramble shRNA) and without p16INK4A expression (p16INK4A shRNA). However, there was an increase in necrotic cells when NFκB was silenced first. The control for this experiment consisted of comparing live, apoptotic, and necrotic cell count in each group of hCPCs without silencing NFκB. In this control, there was a statistically significant increase in live cell count, and decrease in apoptotic and necrotic cell count in the p16INK4A knockdown hCPCs when compared to the scramble shRNA hCPCs. This is strong evidence that NFκB is essential to any pro-survival and cytoprotective effects that p16INK4A knockdown might have, and thus we propose the model shown in Fig. 5E. We propose that, in addition to its known CKI properties, p16INK4A may have an additional physiological role in inhibiting NFκB activation. It is previously known and can also be seen from our data that aging, DNA damage, and ROS induction increases p16INK4A expression (Fig 1). If p16INK4A truly does inhibit NFκB, this may be a major mechanism that can be used to explain the decreased cell survival and ROS generation seen in hCPCs that have unsilenced p16INK4A expression. This would also explain the variety of pro-survival and anti-oxidant effects that were observed after p16INK4A was knocked down by shRNA lentiviral infection.
Discussion
The effects of p16INK4A downregulation via lentiviral shRNA mediated knockdown were analyzed thoroughly to obtain insight into possible models of cellular rejuvenation, and to elucidate downstream pathway changes that occur after silencing this CDK inhibitor. Phenotypes observed from this genetic modification should contribute a better understanding of how to rejuvenate the cellular pathways involved in senescence. Thus far, effects of p16INK4a silencing have shown to improve cell survival and proliferation abilities in hematopoietic and other stem cells [8], but it is yet unclear whether this mechanism will retain the same effect in hCPCs. There is a consensus that p16INK4A expression is increased with age and is correlated with cellular senescence [8–11, 31, 61]. Our studies have confirmed this correlation through RT-PCR and Western blotting. Using p16INK4A’s role as a cyclin dependent kinase inhibitor to cause G1 mitotic arrest, and the knowledge that p16INK4A is a reliable biomarker for senescence, logic follows in wondering if downregulating p16INK4A may slow down the aging process, and lead to rejuvenation of senescent hCPCs.
Most senescence associated genes, including CDKN1A, CDKN1B, PSG1/5, FOXO1, FOXO4, and TxNIP, were downregulated after the p16INK4A knockdown. Among them, FOXO (Forkhead) family genes are transcription factors known to blunt cardiac growth and inhibit the cell cycle, and are usually expressed concurrently with p21, p27, and other CKIs [62, 63]. However, NANOG was not significantly downregulated after the p16INK4A knockdown, perhaps because NANOG is a pluripotent-associated transcription factor associated with induced pluripotent stem cell (iPSC) generation from senescent fibroblasts [64]. NANOG has also been associated with reversal of “aging effects” in mesenchymal stem cells, and with enhanced cell proliferation [65, 66]. These specific changes are consistent among species – Castaldi et al observed increased expression of both SAβG as well as p16INK4A mRNA in c-kit+ mouse cardiac progenitor cells (mCPC) [67]. The c-kit+ mCPCs were compared after isolation from young (3 months old) and aged (24 months old) mice, and the mCPCs from the aged mice were found to have significantly decreased rates of proliferation, decreased expression of cardiac lineage marker genes and mitochondrial proteins, decreased paracrine factor expression, and decreased angiogenic effect [67]. These findings stand as evidence to further the idea that age and cellular senescence is a major hindrance to the efficacy of CPCs.
ROS generation is known to contribute to DNA mutation and cause oxidative damage-related aging [68–70]. Our knockdown of p16INK4a showed antioxidant properties in decreasing the basal presence of ROS by approximately 50% as discussed above. Because of ROS’s role in acceleration of cellular senescence, this is significant in showing that knockdown of the p16INK4A gene may exert beneficial effects in extending lifespan and inducing cellular rejuvenation of the aging hCPCs. Expression of the genes glutathione peroxidase reductase, cytoglobin, and peroxiredoxin were upregulated in our data. Each of these genes is known to harbor significant protective effects in the face of oxidative stress [41, 71–76]. The knockdown of p16INK4A may confer resistance to oxidative stress via upregulation of cytoprotective, anti-oxidant genes. This is evidenced by the decrease in ROS production compared to control. p16INK4A knockdown may also be able to induce cellular growth, which may prove beneficial in helping hCPCs to survive within harsh environments such as infarcted myocardium.
Purified c-Kit+/Lin− CPC populations have become increasingly favorable for in vitro manipulations to enhance cardiac repair, and CPC transplantation assays in vivo have shown vast improvement in cardiac function [77]. This is the first conclusive study to elucidate the role of p16INK4A silencing in aging c-kit+ hCPCs by examining cellular proliferation and survival in vitro. However, CPC commitment to cardiomyocyte differentiation is still debated. Multiple lineage-tracing studies indicated that the proportion of cardiomyocytes derived from c-kit+ cells is limited [78, 79]. These interesting results push us to reconsider the physiological role of c-kit+ CPCs in the heart. However, there is no question in the literature that c-kit+ CPCs are beneficial in improving myocardial repair. Several clinical trials have employed c-kit+ CPCs and have shown enhancements in cardiac function [77, 80]. In addition, significant elevation in cytokine expression have been observed in preconditioned CPCs [32] as well as in rejuvenated hCPCs by knocking down p16INK4A, suggesting that the therapeutic effect of CPCs may be mainly due to cytokine effects. The focus of our current research is to enhance the regenerative potential of aging hCPCs by knocking down p16INK4A, which may result in increased cell survival capability, faster cell proliferation, and more cytokine release after cell transplantation for future in vivo therapeutic application. However, elucidating the potential mechanisms for the therapeutic effect of cardiac progenitor cell therapy will require a considerable effort, which is beyond the scope of the present study.
In conclusion, our results demonstrate that knockdown of p16INK4A in hCPCs reverses the senescent phenotype and has an antioxidant effect on aging hCPCs. As described above, this knockdown leads to the activation of both antiapoptotic and antioxidant pathways via NF-kB signaling that hopefully will stimulate faster recovery after transplantation into infarcted myocardium in the future in vivo studies.
Supplementary Material
Acknowledgments
This study was supported by the NIH Grant R01HL114951 (to C.C.). We appreciate all our colleagues from the Divisions of Pediatric and Adult Cardiothoracic Surgery at the Albany Medical Center, including Kyla Philbrook and Karen Larsen, for their assistance in obtaining consent from patients for the cardiac tissue collection.
Footnotes
Disclosure statement: The authors report no potential conflicts of interest.
Compliance with Ethical Standards
Disclosure statement: The authors report no potential conflicts of interest.
Research involving Human Participants: The Institutional Review Board (#3728) of the Albany Medical Center approved this study and all human samples were obtained with informed consent.
References
- 1.CDC. and NCHS., Underlying Cause of Death 1999–2013 on CDC WONDER Online Database, released 2015. 2015, Data are from the Multiple Cause of Death Files, 1999–2013, as compiled from data provided by the 57 vital statistics jurisdictions through the Vital Statistics Cooperative Program.
- 2.Friede A, Reid JA, Ory HW. CDC WONDER: a comprehensive on-line public health information system of the Centers for Disease Control and Prevention. Am J Public Health. 1993;83(9):1289–94. doi: 10.2105/ajph.83.9.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Segers VF, Lee RT. Stem-cell therapy for cardiac disease. Nature. 2008;451(7181):937–42. doi: 10.1038/nature06800. [DOI] [PubMed] [Google Scholar]
- 4.Anand SS, et al. Risk factors for myocardial infarction in women and men: insights from the INTERHEART study. Eur Heart J. 2008;29(7):932–40. doi: 10.1093/eurheartj/ehn018. [DOI] [PubMed] [Google Scholar]
- 5.Cesselli D, et al. Effects of age and heart failure on human cardiac stem cell function. Am J Pathol. 2011;179(1):349–66. doi: 10.1016/j.ajpath.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayess H, Wang MB, Srivatsan ES. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130(8):1715–25. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuilman T, et al. The essence of senescence. Genes Dev. 2010;24(22):2463–79. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Janzen V, et al. Stem-cell ageing modified by the cyclin-dependent kinase inhibitor p16INK4a. Nature. 2006;443(7110):421–6. doi: 10.1038/nature05159. [DOI] [PubMed] [Google Scholar]
- 9.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530(7589):184–9. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alcorta DA, et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4a) in replicative senescence of normal human fibroblasts. Proc Natl Acad Sci U S A. 1996;93(24):13742–7. doi: 10.1073/pnas.93.24.13742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479(7372):232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brookes S, et al. Contribution of p16(INK4a) to replicative senescence of human fibroblasts. Exp Cell Res. 2004;298(2):549–59. doi: 10.1016/j.yexcr.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 13.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–7. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 14.Bayoglu B, et al. The severity of internal carotid artery stenosis is associated with the cyclin-dependent kinase inhibitor 2A gene expression. J Atheroscler Thromb. 2014;21(7):659–71. doi: 10.5551/jat.21774. [DOI] [PubMed] [Google Scholar]
- 15.Lowe SW, Cepero E, Evan G. Intrinsic tumour suppression. Nature. 2004;432(7015):307–15. doi: 10.1038/nature03098. [DOI] [PubMed] [Google Scholar]
- 16.Hara E, et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16(3):859–67. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim WY, Sharpless NE. The Regulation of <em>INK4</em>/<em>ARF</em> in Cancer and Aging. Cell. 2006;127(2):265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 18.D’Arcangelo D, Tinaburri L, Dellambra E. The Role of p16(INK4a) Pathway in Human Epidermal Stem Cell Self-Renewal, Aging and Cancer. Int J Mol Sci. 2017;18(7) doi: 10.3390/ijms18071591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rastogi S, et al. Prohibitin Facilitates Cellular Senescence by Recruiting Specific Corepressors To Inhibit E2F Target Genes. Mol Cell Biol. 2006;26(11):4161–71. doi: 10.1128/MCB.02142-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X, et al. Upregulation of lactate-inducible snail protein suppresses oncogene-mediated senescence through p16(INK4a) inactivation. J Exp Clin Cancer Res. 2018;37(1):39. doi: 10.1186/s13046-018-0701-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani N, et al. The p16INK4a-RB pathway: molecular link between cellular senescence and tumor suppression. The Journal of Medical Investigation. 2004;51(3,4):146–153. doi: 10.2152/jmi.51.146. [DOI] [PubMed] [Google Scholar]
- 22.Ben-Porath I, Weinberg RA. The signals and pathways activating cellular senescence. The International Journal of Biochemistry & Cell Biology. 2005;37(5):961–976. doi: 10.1016/j.biocel.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 23.Stepanova L, Sorrentino BP. A limited role for p16Ink4a and p19Arf in the loss of hematopoietic stem cells during proliferative stress. Blood. 2005;106(3):827–32. doi: 10.1182/blood-2004-06-2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bearzi C, et al. Human cardiac stem cells. Proc Natl Acad Sci U S A. 2007;104(35):14068–73. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohsin S, et al. Rejuvenation of human cardiac progenitor cells with Pim-1 kinase. Circ Res. 2013;113(10):1169–79. doi: 10.1161/CIRCRESAHA.113.302302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cawthon RM. Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res. 2009;37(3):e21. doi: 10.1093/nar/gkn1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dimri GP, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci U S A. 1995;92(20):9363–7. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Renu K, et al. Molecular mechanism of doxorubicin-induced cardiomyopathy - An update. Eur J Pharmacol. 2017;818:241–253. doi: 10.1016/j.ejphar.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 29.Moslehi J, DePinho RA, Sahin E. Telomeres and mitochondria in the aging heart. Circ Res. 2012;110(9):1226–37. doi: 10.1161/CIRCRESAHA.111.246868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Di Stefano V, et al. Knockdown of cyclin-dependent kinase inhibitors induces cardiomyocyte re-entry in the cell cycle. J Biol Chem. 2011;286(10):8644–54. doi: 10.1074/jbc.M110.184549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Park SH, Lim JS, Jang KL. All-trans retinoic acid induces cellular senescence via upregulation of p16, p21, and p27. Cancer Lett. 2011;310(2):232–9. doi: 10.1016/j.canlet.2011.07.009. [DOI] [PubMed] [Google Scholar]
- 32.Cai C, et al. The heme oxygenase 1 inducer (CoPP) protects human cardiac stem cells against apoptosis through activation of the extracellular signal-regulated kinase (ERK)/NRF2 signaling pathway and cytokine release. J Biol Chem. 2012;287(40):33720–32. doi: 10.1074/jbc.M112.385542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basu A, Sridharan S. Regulation of anti-apoptotic Bcl-2 family protein Mcl-1 by S6 kinase 2. PLoS One. 2017;12(3):e0173854. doi: 10.1371/journal.pone.0173854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meyerovich K, et al. MCL-1 Is a Key Antiapoptotic Protein in Human and Rodent Pancreatic beta-Cells. Diabetes. 2017;66(9):2446–2458. doi: 10.2337/db16-1252. [DOI] [PubMed] [Google Scholar]
- 35.Cerella C, et al. Early downregulation of Mcl-1 regulates apoptosis triggered by cardiac glycoside UNBS1450. Cell Death Dis. 2015;6:e1782. doi: 10.1038/cddis.2015.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thomas RL, et al. Loss of MCL-1 leads to impaired autophagy and rapid development of heart failure. Genes Dev. 2013;27(12):1365–77. doi: 10.1101/gad.215871.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mirotsou M, et al. Paracrine mechanisms of stem cell reparative and regenerative actions in the heart. J Mol Cell Cardiol. 2011;50(2):280–9. doi: 10.1016/j.yjmcc.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beckman KB, Ames BN. The free radical theory of aging matures. Physiol Rev. 1998;78(2):547–81. doi: 10.1152/physrev.1998.78.2.547. [DOI] [PubMed] [Google Scholar]
- 39.Sasaki M, et al. Reactive oxygen species promotes cellular senescence in normal human epidermal keratinocytes through epigenetic regulation of p16(INK4a.) Biochem Biophys Res Commun. 2014;452(3):622–8. doi: 10.1016/j.bbrc.2014.08.123. [DOI] [PubMed] [Google Scholar]
- 40.Brigelius-Flohe R, Maiorino M. Glutathione peroxidases. Biochim Biophys Acta. 2013;1830(5):3289–303. doi: 10.1016/j.bbagen.2012.11.020. [DOI] [PubMed] [Google Scholar]
- 41.Singh S, et al. Calcineurin activates cytoglobin transcription in hypoxic myocytes. J Biol Chem. 2009;284(16):10409–21. doi: 10.1074/jbc.M809572200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ou L, et al. Recombinant Human Cytoglobin Prevents Atherosclerosis by Regulating Lipid Metabolism and Oxidative Stress. J Cardiovasc Pharmacol Ther. 2017:1074248417724870. doi: 10.1177/1074248417724870. [DOI] [PubMed] [Google Scholar]
- 43.Zhang S, et al. Cytoglobin Promotes Cardiac Progenitor Cell Survival against Oxidative Stress via the Upregulation of the NFkappaB/iNOS Signal Pathway and Nitric Oxide Production. Sci Rep. 2017;7(1):10754. doi: 10.1038/s41598-017-11342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aeby E, et al. Peroxiredoxin 1 Protects Telomeres from Oxidative Damage and Preserves Telomeric DNA for Extension by Telomerase. Cell Rep. 2016;17(12):3107–3114. doi: 10.1016/j.celrep.2016.11.071. [DOI] [PubMed] [Google Scholar]
- 45.Neumann CA, Cao J, Manevich Y. Peroxiredoxin 1 and its role in cell signaling. Cell Cycle. 2009;8(24):4072–8. doi: 10.4161/cc.8.24.10242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mullen L, et al. Cysteine Oxidation Targets Peroxiredoxins 1 and 2 for Exosomal Release through a Novel Mechanism of Redox-Dependent Secretion. Mol Med. 2015;21:98–108. doi: 10.2119/molmed.2015.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carvalho LAC, et al. Urate hydroperoxide oxidizes human peroxiredoxin 1 and peroxiredoxin 2. J Biol Chem. 2017;292(21):8705–8715. doi: 10.1074/jbc.M116.767657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Findlay VJ, et al. A novel role for human sulfiredoxin in the reversal of glutathionylation. Cancer Res. 2006;66(13):6800–6. doi: 10.1158/0008-5472.CAN-06-0484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baek JY, et al. Sulfiredoxin protein is critical for redox balance and survival of cells exposed to low steady-state levels of H2O2. J Biol Chem. 2012;287(1):81–9. doi: 10.1074/jbc.M111.316711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Morgan MJ, Liu ZG. Crosstalk of reactive oxygen species and NF-kappaB signaling. Cell Res. 2011;21(1):103–15. doi: 10.1038/cr.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Birben E, et al. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5(1):9–19. doi: 10.1097/WOX.0b013e3182439613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakano H, et al. Reactive oxygen species mediate crosstalk between NF-kappaB and JNK. Cell Death Differ. 2006;13(5):730–7. doi: 10.1038/sj.cdd.4401830. [DOI] [PubMed] [Google Scholar]
- 53.Schmid JA, Birbach A. IkappaB kinase beta (IKKbeta/IKK2/IKBKB)--a key molecule in signaling to the transcription factor NF-kappaB. Cytokine Growth Factor Rev. 2008;19(2):157–65. doi: 10.1016/j.cytogfr.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 54.Tsuchiya Y, et al. Nuclear IKKbeta is an adaptor protein for IkappaBalpha ubiquitination and degradation in UV-induced NF-kappaB activation. Mol Cell. 2010;39(4):570–82. doi: 10.1016/j.molcel.2010.07.030. [DOI] [PubMed] [Google Scholar]
- 55.Clark K, et al. Novel cross-talk within the IKK family controls innate immunity. Biochem J. 2011;434(1):93–104. doi: 10.1042/BJ20101701. [DOI] [PubMed] [Google Scholar]
- 56.Yoshida T, et al. Human heme oxygenase cDNA and induction of its mRNA by hemin. Eur J Biochem. 1988;171(3):457–61. doi: 10.1111/j.1432-1033.1988.tb13811.x. [DOI] [PubMed] [Google Scholar]
- 57.Gozzelino R, Jeney V, Soares MP. Mechanisms of cell protection by heme oxygenase-1. Annu Rev Pharmacol Toxicol. 2010;50:323–54. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 58.Scott ML, et al. The p65 subunit of NF-kappa B regulates I kappa B by two distinct mechanisms. Genes Dev. 1993;7(7A):1266–76. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 59.Christian F, Smith EL, Carmody RJ. The Regulation of NF-kappaB Subunits by Phosphorylation. Cells. 2016;5(1) doi: 10.3390/cells5010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teng L, Bennett E, Cai C. Preconditioning c-Kit-positive Human Cardiac Stem Cells with a Nitric Oxide Donor Enhances Cell Survival through Activation of Survival Signaling Pathways. J Biol Chem. 2016;291(18):9733–47. doi: 10.1074/jbc.M115.687806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Narita M, et al. Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell. 2003;113(6):703–16. doi: 10.1016/s0092-8674(03)00401-x. [DOI] [PubMed] [Google Scholar]
- 62.Ni YG, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114(11):1159–68. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Evans-Anderson HJ, Alfieri CM, Yutzey KE. Regulation of cardiomyocyte proliferation and myocardial growth during development by FOXO transcription factors. Circ Res. 2008;102(6):686–94. doi: 10.1161/CIRCRESAHA.107.163428. [DOI] [PubMed] [Google Scholar]
- 64.Lapasset L, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25(21):2248–53. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mistriotis P, et al. NANOG Reverses the Myogenic Differentiation Potential of Senescent Stem Cells by Restoring ACTIN Filamentous Organization and SRF-Dependent Gene Expression. Stem Cells. 2017;35(1):207–221. doi: 10.1002/stem.2452. [DOI] [PubMed] [Google Scholar]
- 66.Han J, et al. Nanog reverses the effects of organismal aging on mesenchymal stem cell proliferation and myogenic differentiation potential. Stem Cells. 2012;30(12):2746–59. doi: 10.1002/stem.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castaldi A, et al. Decline in cellular function of aged mouse c-kit(+) cardiac progenitor cells. J Physiol. 2017;595(19):6249–6262. doi: 10.1113/JP274775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cadet J, Wagner JR. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a012559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen JH, Hales CN, Ozanne SE. DNA damage, cellular senescence and organismal ageing: causal or correlative? Nucleic Acids Res. 2007;35(22):7417–28. doi: 10.1093/nar/gkm681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cooke MS, et al. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17(10):1195–214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 71.Li C, et al. Heme oxygenase 1 induction protects myocardiac cells against hypoxia/reoxygenation-induced apoptosis: The role of JNK/c-Jun/Caspase-3 inhibition and Akt signaling enhancement. Herz. 2016;41(8):715–724. doi: 10.1007/s00059-016-4424-6. [DOI] [PubMed] [Google Scholar]
- 72.Issan Y, et al. Heme oxygenase-1 induction improves cardiac function following myocardial ischemia by reducing oxidative stress. PLoS One. 2014;9(3):e92246. doi: 10.1371/journal.pone.0092246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paradis M, et al. The effects of nitric oxide-oxidase and putative glutathione-peroxidase activities of ceruloplasmin on the viability of cardiomyocytes exposed to hydrogen peroxide. Free Radic Biol Med. 2010;49(12):2019–27. doi: 10.1016/j.freeradbiomed.2010.09.030. [DOI] [PubMed] [Google Scholar]
- 74.Singh S, et al. Cytoglobin modulates myogenic progenitor cell viability and muscle regeneration. Proc Natl Acad Sci U S A. 2014;111(1):E129–38. doi: 10.1073/pnas.1314962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Iskusnykh I, Popova TN, Musharova OS. Intensity of cardiac free-radicals processes and expression of glutathione peroxidase and glutathione reductase genes in rats with adrenaline. Biomed Khim. 2012;58(5):530–8. doi: 10.18097/pbmc20125805530. [DOI] [PubMed] [Google Scholar]
- 76.Yan YF, et al. DJ-1 upregulates anti-oxidant enzymes and attenuates hypoxia/re-oxygenation-induced oxidative stress by activation of the nuclear factor erythroid 2-like 2 signaling pathway. Mol Med Rep. 2015;12(3):4734–42. doi: 10.3892/mmr.2015.3947. [DOI] [PubMed] [Google Scholar]
- 77.Bolli R, et al. Cardiac stem cells in patients with ischaemic cardiomyopathy (SCIPIO): initial results of a randomised phase 1 trial. Lancet. 2011;378(9806):1847–57. doi: 10.1016/S0140-6736(11)61590-0. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 78.van Berlo JH, et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature. 2014;509(7500):337–41. doi: 10.1038/nature13309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sultana N, et al. Resident c-kit(+) cells in the heart are not cardiac stem cells. Nat Commun. 2015;6:8701. doi: 10.1038/ncomms9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Makkar RR, et al. Intracoronary cardiosphere-derived cells for heart regeneration after myocardial infarction (CADUCEUS): a prospective, randomised phase 1 trial. Lancet. 2012;379(9819):895–904. doi: 10.1016/S0140-6736(12)60195-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.