SUMMARY
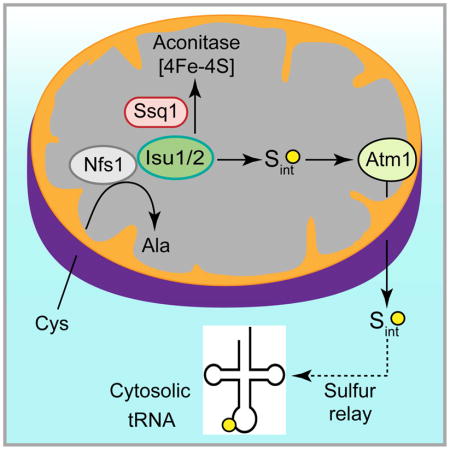
In eukaryotes, mitochondria have been hypothesized to generate sulfur species required for tRNA thiolation in the cytosol, although no direct evidence thus far exists. Here we have detected these sulfur species, making use of our observation that isolated yeast cytosol alone is unable to thiolate tRNAs but can do so upon addition of mitochondria. Mitochondria were found to utilize the cysteine desulfurase Nfs1 to produce sulfur-containing species with masses from 700 – 1100 Da. Mitochondria export these species via the Atm1 transporter in the inner membrane. Once exported to the cytosol, these sulfur species can promote cytosolic tRNA thiolation with no further requirement of mitochondria. Furthermore, we found that the Isu1/2 scaffolds but not the Ssq1 chaperone of the mitochondrial iron-sulfur cluster machinery were required for cytosolic tRNA thiolation, and thus the sulfur utilization pathway bifurcates at the Isu1/2 site for intra-organellar use in mitochondria or export to the cytosol.
In Brief
Critical signals and metabolites move between mitochondria and cytosol, and such communications are essential for cellular homeostasis. Pandey et al. demonstrate that mitochondria generate activated sulfur intermediates (Sint). The intermediates are exported to the cytosol in an ATP-dependent fashion, and there they are utilized for thio-modification of tRNAs.
INTRODUCTION
Correct and efficient protein translation is essential to cell survival, and a variety of post-transcriptional modifications of tRNAs are vital for their folding, stability, and decoding functions (El Yacoubi et al., 2012). In the cytosol of eukaryotes, the wobble uridine (U34) of three tRNAs specific for lysine (tRNALysUUU), glutamate (tRNAGluUUC), and glutamine (tRNAGlnUUG) contain a methoxycarbonylmethyl (mcm5) functional group and a thio-modification (s2) at positions 5 and 2, respectively (Nakai et al., 2017). The thiolation of these cytosolic tRNAs promotes their binding to the ribosomal A-site (Rezgui et al., 2013), prevents frameshifting (Urbonavicius et al., 2001), ensures accurate translation at an optimal rate (Nedialkova and Leidel, 2015), and senses cellular sulfur availability (Laxman et al., 2013). In yeast, impaired s2U34 modification leads to pleiotropic phenotypes including genome instability (Dewez et al., 2008). In humans, impaired thiolation of cytosolic tRNAs has been linked to familial dysautonomia (Karlsborn et al., 2014). Additional thio-modifications have been identified in mitochondrial tRNAs such as 5-carboxymethylaminomethyl-2-thiouridine (cmnm5s2U34) in yeast, and 5-taurinomethyl-2-thiouridine (τm5s2U34) in mammals (Nakai et al., 2017). Non-thiolated mitochondrial tRNAs in humans are associated with myoclonic epilepsy with ragged-red fibers (Yasukawa et al., 2001). Thus, thiolation of tRNAs is critical to maintaining overall cellular homeostasis. Here we focus on cytosolic tRNA thiolation in yeast, particularly the contribution of mitochondria to this process.
The amino acid cysteine is the source of sulfur for tRNA thiolation, and cysteine desulfurase is the only enzyme capable of abstracting sulfur from cysteine. It generates an enzyme-bound persulfide from cysteine and donates the activated sulfur to designated recipients (Nakai et al., 2004; Pandey et al., 2012). In yeast, the Nfs1 cysteine desulfurase is present primarily in mitochondria (Li et al., 1999). Trace amounts are probably also present in nucleus and cytosol, as shown by sensitive genetic methods (Naamati et al., 2009). In vivo depletion of the mitochondrial form of Nfs1 leads to deficiencies of both mitochondrial and cytosolic tRNA thio-modifications (Nakai et al., 2007; Nakai et al., 2004). The implication is that mitochondrial Nfs1 may generate some species, perhaps a sulfur intermediate that is utilized directly or indirectly for cytosolic tRNA thiolation. However, none of the steps regarding the hypothesized sulfur intermediate, such as its generation, export or utilization, has been demonstrated. Progress has been hampered because available assays mostly rely on measuring steady state levels of thiolated tRNAs in vivo (Leidel et al., 2009; Nakai et al., 2007; Noma et al., 2009), making it difficult to identify the steps involved in mitochondrial formation of sulfur intermediates and export to the cytosol.
The idea that mitochondria might export some sulfur species originated from studies of iron-sulfur (Fe-S) cluster biogenesis (Lill et al., 2015). Fe-S clusters are essential cofactors of proteins involved in vital processes including respiration, DNA repair, protein translation, and iron sensing. As with tRNA modifications, the biogenesis of Fe-S clusters in eukaryotes is also compartmentalized (Lill et al., 2015). Fe-S cluster assembly is mediated by a multi-subunit machinery inside mitochondria, termed the ISC (Iron Sulfur Cluster), and outside of mitochondria in the cytosol, termed the CIA (Cytosolic Iron-Sulfur Protein Assembly). Importantly, mitochondrial Nfs1 is required for mitochondrial as well as cytosolic FeS cluster synthesis (Kispal et al., 1999), implying that a substrate or signal derived from Nfs1 activity is exported from mitochondria and utilized in the cytosol, similar to the proposed mechanism of tRNA thiolation. The species exported from mitochondria is unknown, but it likely involves sulfur in view of its dependence on the Nfs1 cysteine desulfurase (Lill et al., 2015). The best candidate for the exporter is Atm1, an ABC transporter situated in the mitochondrial inner membrane and oriented with its substrate-binding site and ATP-binding site facing the matrix (Leighton and Schatz, 1995). The phenotype of Atm1-depleted cells is characterized by deficiency of cytosolic Fe-S proteins. (Lill et al., 2015). The crystal structure of yeast Atm1 (Srinivasan et al., 2014) has been solved but its export substrate remains elusive. Candidate substrates such as small thiol compounds were evaluated for their ability to stimulate the Atm1 ATPase activity or to be transported into Atm1-containing vesicles (Kuhnke et al., 2006; Li and Cowan, 2015; Schaedler et al., 2014). However, none qualifies as a truly exported and functional sulfur-containing molecule (Lill et al., 2015). Interestingly, the sulfur export hypothesis has recently been extended to cytosolic tRNA thiolation as well (Nakai et al., 2017), although no supporting evidence thus far exists. Here we have developed a strategy to track mitochondria-generated sulfur intermediates required for cytosolic tRNA thiolation. The experimental approach involves detecting de novo cytosolic tRNA thiolation using mitochondria and cytosol isolated from yeast cells, and [35S]cysteine as the sulfur source.
RESULTS
Radiolabeling of an endogenous species in the cytosol occurs only in the presence of mitochondria
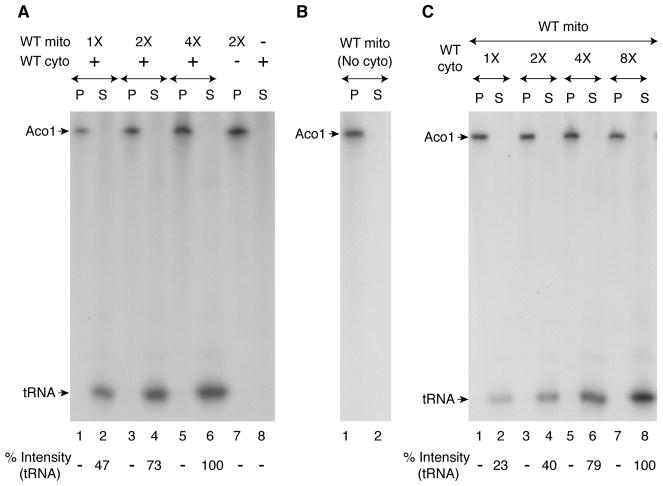
Aconitase (Aco1) is a mitochondrial matrix protein that requires a [4Fe-4S] cluster for enzymatic activity in the Krebs cycle. Mitochondria were isolated from wild-type yeast cells (WT mito) and incubated with [35S]cysteine, nucleotides and iron. Samples were analyzed by native PAGE, followed by autoradiography. Fe-35S clusters were made and inserted into a population of apoaconitase within the organelle (Figure 1A, lane 7) (Amutha et al., 2008). When wild-type cytosol (WT cyto) was likewise incubated, there was no signal (Figure 1A, lane 8). Increasing concentrations of WT mitochondria were then added to a fixed concentration of WT cytosol and incubated with [35S]cysteine under identical conditions. The reaction mixtures were centrifuged to re-separate mitochondria (pellet “P”; Figure 1A, lanes 1, 3 and 5) and cytosol (supernatant “S”; Figure 1A, lanes 2, 4 and 6, respectively). As expected, radiolabeled aconitase was found in the mitochondrial pellets. Surprisingly, however, a new radiolabeled band (determined to be thiolated tRNAs; see below) was detected in the supernatant/cytosol fractions, and the radioactive signal was enhanced with increasing concentrations of added mitochondria. No such band was detected in the supernatant after centrifugation of a reaction mixture containing mitochondria alone incubated with [35S]cysteine (Figure 1B, lane 2), showing that the radioactive band was not a simple mitochondrial export product; rather it depended on a mitochondrial export product and was a resident of the cytosol. Furthermore, the signal of the radiolabeled band in the cytosol was greatly enhanced with increasing concentrations of cytosol added to a fixed concentration of mitochondria, consistent with its cytosolic dependence (Figure 1C).
Figure 1. Radiolabeling of a cytosolic component requires mitochondrial contribution of sulfur species.
(A) Reaction mixtures containing wild-type mitochondria (WT mito) alone, wild-type cytosol (WT cyto) alone, or both were supplemented with [35S]cysteine (10 μCi), nucleotides (4 mM ATP, 1 mM GTP, 2 mM NADH and 1 mM NADPH), and ferrous ascorbate (10 μM). After incubation at 30°C for 30 min, samples were centrifuged, and the pellet (“P”; mitochondria) and supernatant (“S”; cytosol) fractions were analyzed by native PAGE and autoradiography. WT mito 1X = 50 μg of proteins; WT cyto = 300 μg of proteins.
(B) WT mitochondria (200 μg) alone were incubated with [35S]cysteine, nucleotides and iron. After centrifugation, the pellet and supernatant fractions were analyzed as in (A) above.
(C) Reaction mixtures containing different amounts of cytosol (1X = 50 μg) were supplemented with a fixed amount of mitochondria (200 μg), and incubated with [35S]cysteine, nucleotides and iron. Following centrifugation, samples were analyzed as in (A) above.
The radiolabeled endogenous species in the cytosol are tRNAs
The radiolabeling of the cytosolic species could be due either to insertion of newly made Fe-35S clusters into a protein or to the thio-modification of tRNAs. Fe-S clusters are non-covalently attached to proteins via cysteine residues in most cases and are destroyed by denaturants such as SDS. Thus, Fe-35S labeled proteins can only be analyzed by native gels (Amutha et al., 2008). By contrast, the 35S label is covalently attached to tRNAs and therefore thiolated tRNAs can be analyzed by either native PAGE or SDS-PAGE. To determine the nature of 35S label associated with the cytosolic species, samples of the following experiment were analyzed by SDS-PAGE. Isolated WT cytosol alone was incubated with [35S]cysteine, nucleotides and iron, but no radiolabeled band was detected (Figure 2A, lane 1). WT mitochondria were then added to WT cytosol and the mixture was incubated under identical conditions, with or without an added substrate for tRNA thiolation, namely tRNALys. After centrifugation, the cytosolic supernatants were analyzed. When mitochondria were included, an endogenous species was radiolabeled and the intensity of the band was enhanced with the addition of tRNALys (Figure 2A, compare lanes 2 and 3). In a separate experiment, nuclease pretreatment of the cytosol abolished radiolabeling of the endogenous band (Figure 2A, lanes 4 and 5) and tRNALys added after nuclease inhibition was thiolated (Figure 2A, lanes 6 and 7). Thus, the radiolabeled endogenous species in the cytosol most likely represent thiolated tRNAs.
Figure 2. The radiolabeled signal in the cytosol is due to thiolation of tRNAs.
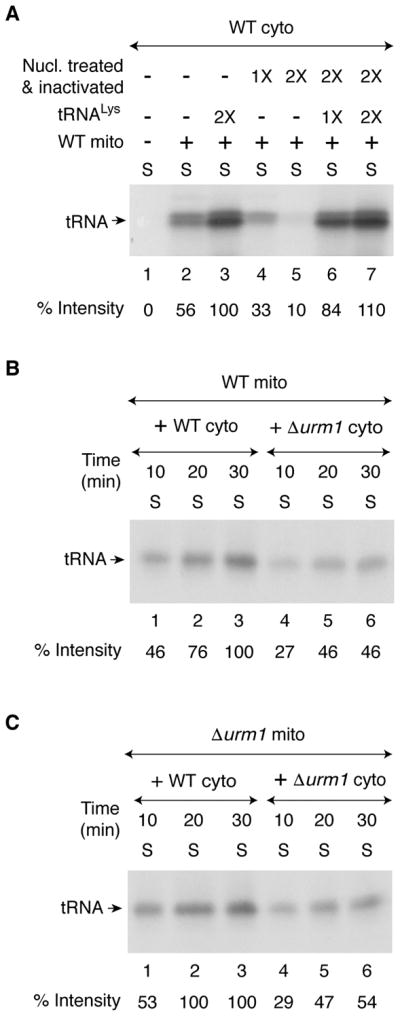
(A) As indicated, WT cytosol (200 μg) was treated with S7 micrococcal nuclease (1X = 450 units/ml) in the presence of 0.5 mM CaCl2 for 10 min at 30°C, and the nuclease was inactivated by the addition of EGTA (2 mM). Nuclease-untreated (lanes 1–3), or nuclease-treated and subsequently inactivated (lanes 4–7) cytosolic samples were supplemented with [35S]cysteine, nucleotides and iron as in Figure 1A legend. Reaction mixtures were incubated at 30°C for 30 min, with or without added mitochondria (200 μg) and purified yeast tRNALys (1X = 0.25 A260 unit). After centrifugation, the supernatant/cytosol fractions were analyzed by SDS-PAGE and autoradiography.
(B) WT mitochondria (200 μg) were added to WT or Δurm1 cytosol (200 μg), and reaction mixtures were incubated with [35S]cysteine, nucleotides and iron at 30°C for different time periods as indicated. After centrifugation, the resulting supernatant (cytosol) fractions were analyzed as in Figure 1A legend.
(C) Δurm1 mitochondria (200 μg) were added to WT or Δurm1 cytosol (200 μg) and reactions were carried out as in (B) above.
The in vivo thio-modification (s2U34) of cytosolic tRNAs is mediated by a sulfur transfer system that consists of several cytosolic proteins including a ubiquitin-related modifier Urm1 (Leidel et al., 2009; Nakai et al., 2017; Noma et al., 2009). To further confirm that tRNA thiolation was taking place in our in vitro assays, both mitochondria and cytosol were isolated from cells lacking Urm1 (Δurm1; Table S1). WT mitochondria strongly promoted tRNA thiolation in WT cytosol but slowly and inefficiently in Δurm1 cytosol (Figure 2B). Likewise, Δurm1 mitochondria efficiently promoted tRNA thiolation in WT cytosol but at a significantly reduced level in Δurm1 mutant cytosol (Figure 2C). Thus, regardless of mitochondria (WT or Δurm1) used, tRNA thiolation in cytosol lacking Urm1 occurred inefficiently. A key advantage of our in vitro assays is that they allow mixing of mitochondria and cytosol from different sources, thereby specifically identifying compartmental contributions or defects.
Effects of impaired mitochondrial Fe-S cluster assembly on cytosolic tRNA thiolation
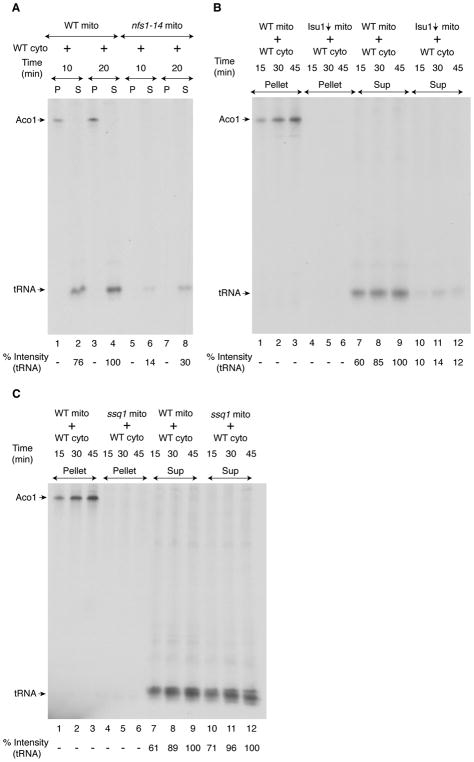
In mitochondria, the core ISC machinery, composed of cysteine desulfurase Nfs1 in a complex with Isd11/Acp1 (Van Vranken et al., 2016), generates a persulfide intermediate (Pandey et al., 2012) that is subsequently transferred to Isu scaffold proteins (Isu1 or Isu2 isoforms in yeast) where it combines with iron to form an Fe-S cluster intermediate. Frataxin (Yfh1) likely plays a role at this stage. Ferredoxin reductase (Arh1) and ferredoxin (Yah1) work together to supply reducing equivalents that are needed for assembly of the Fe-S cluster scaffold intermediate. The cluster intermediate is then transferred to apoproteins (e.g. apoaconitase) in a step involving chaperones (Ssq1, Jac1) and glutaredoxins (Grx5) (Lill et al., 2015). To determine the role of the ISC machinery in cytosolic tRNA thiolation, we focused on Nfs1, Isu1/2, and Ssq1. In S. cerevisiae, the NFS1 gene is essential for cell survival (Li et al., 1999). We therefore used a hypomorphic nfs1 mutant (nfs1-14) with a missense NFS1 allele (I191S). This mutant exhibited Fe-S cluster deficiencies and other iron-related phenotypes because of compromised cysteine desulfurase activity resulting from the point mutation (Li et al., 1999). To determine the role of mitochondrial Nfs1 in cytosolic tRNA thiolation, mitochondria isolated from the nfs1-14 strain were added to WT cytosol and incubated with [35S]cysteine, nucleotides and iron. No radiolabeled aconitase was detected in nfs1-14 mitochondria. Compared with WT mitochondria, nfs1-14 mitochondria also failed to efficiently promote 35S-labeling of cytosolic tRNAs, and thus tRNA thiolation occurred at a greatly reduced level (Figure 3A). We conclude that mitochondrial Nfs1 cysteine desulfurase activity is essential for generating the sulfur species utilized for cytosolic tRNA thiolation.
Figure 3. Nfs1 and Isu1, but not Ssq1, are required for cytosolic tRNA thiolation.
(A) WT cytosol (200 μg) was supplemented with WT or nfs1-14 mitochondria (100 μg). After addition of [35S]cysteine, nucleotides and iron, samples were incubated at 30°C and centrifuged. The resulting pellet (“P”, mitochondria) and supernatant (“S”, cytosol) fractions were analyzed as in Figure 1A legend.
(B) Isu1-depleted (Isu1↓; Isu2 absent) or WT mitochondria (250 μg) were added to WT cytosol (200 μg), and reactions mixtures were incubated with [35S]cysteine, nucleotides and iron at 30°C. After centrifugation, the pellet (mitochondria) and supernatant (cytosol) fractions were analyzed as in Figure 1A legend.
(C) WT or ssq1 mutant mitochondria (250 μg) were added to WT cytosol (200 μg), and assays were performed as in (B) above. See also Figure S1.
To determine if Isu1/2 proteins participate in cytosolic tRNA thiolation, mitochondria were isolated from a GAL1-ISU1/Δisu2 strain after depleting Isu1 (Yoon et al., 2015). As expected, no aconitase activity was detected (Figure S1A). These Isu1-depleted (Isu1↓) mitochondria were then incubated with WT cytosol in the presence of [35S]cysteine, nucleotides and iron. No radiolabeled Aco1 was detected (Figure 3B, lanes 4–6), and little cytosolic tRNAs was radiolabeled (Figure 3B, lanes 10–12). Thus, mitochondrial Isu1 is required for de novo cytosolic tRNA thiolation consistent with an earlier finding that depletion of Isu proteins was associated with greatly reduced levels of in vivo thiolated tRNALys (Nakai et al., 2007). Together these results suggest that Isu1/2 participate in delivering sulfur species for mitochondrial aconitase and cytosolic tRNA thiolation.
Surprisingly, similar experiments with ssq1 mutant mitochondria (Knight et al., 1998) uncovered a clear-cut two-sided phenotype. On the one hand, ssq1 mitochondria behaved just like Isu1-depleted (Isu1↓) mitochondria and did not exhibit aconitase activity (Figure S1B) or radiolabeling of aconitase (Figure 3C, lanes 4–6). On the other hand, ssq1 mitochondria conferred strong cytosolic tRNA labeling (Figure 3C, compare lanes 7–9 with lanes 10–12, respectively), thus behaving like WT and not like Isu1↓ mitochondria. Altogether these results suggest the existence of a branch point in the sulfur delivery pathway. Nfs1 and Isu1/2 are required for generating sulfur species for mitochondrial Fe-S clusters and for cytosolic uses. However, Ssq1 is required for mitochondrial Fe-S cluster assembly but not for cytosolic tRNA thiolation.
Effects of disrupted cytosolic Fe-S cluster assembly on cytosolic tRNA thiolation
The cytosol carries a dedicated CIA machinery for Fe-S cluster assembly. This machinery is composed of essential reductase (Dre2/Tah18), scaffold (Cfd1/Nbp35), and other components (Lill et al., 2015). To determine the role of this machinery in cytosolic tRNA thiolation, Dre2 expression in a GAL1-DRE2 strain was repressed (Zhang et al., 2008), and Dre2-depleted (Dre2↓) cytosol was isolated. WT mitochondria were added to WT cytosol or Dre2↓ cytosol, and incubated with [35S]cysteine, nucleotides and iron. After centrifugation, supernatant/cytosol fractions were analyzed for 35S-labeling of tRNAs. Compared with WT cytosol, little tRNA was thiolated by Dre2↓ cytosol (Figure 4, compare lanes 1–4 with lanes 5–8, respectively). Poor thiolation was not due to lack of endogenous tRNA substrates in Dre2↓ cytosol, because addition of tRNALys did not enhance the thiolation signal (Figure 4, lanes 9–12). These results can be explained either by a direct effect of Dre2 on sulfur utilization for cytosolic tRNA thiolation or an indirect effect on assembly of downstream components involved in sulfur utilization and containing Fe-S cluster cofactors e.g. Ncs6 [3Fe-4S] (Liu et al., 2016), Elp3 [4Fe-4S] (Paraskevopoulou et al., 2006) and/or other proteins. These possibilities are consistent with reduced steady state levels of thiolated tRNAs in cells lacking other CIA components (Nakai et al., 2007).
Figure 4. Dre2 is required for cytosolic tRNA thiolation.
WT mitochondria (200 μg) were added to WT cytosol or Dre2-depleted (Dre2↓) cytosol (200 μg). As indicated, Dre2↓ cytosol was supplemented with purified yeast tRNALys (0.15 A260 unit). Assay conditions were as in Figure 1A legend. After centrifugation, the supernatant/cytosol fractions were analyzed.
Detection of active sulfur species accumulated within mitochondria
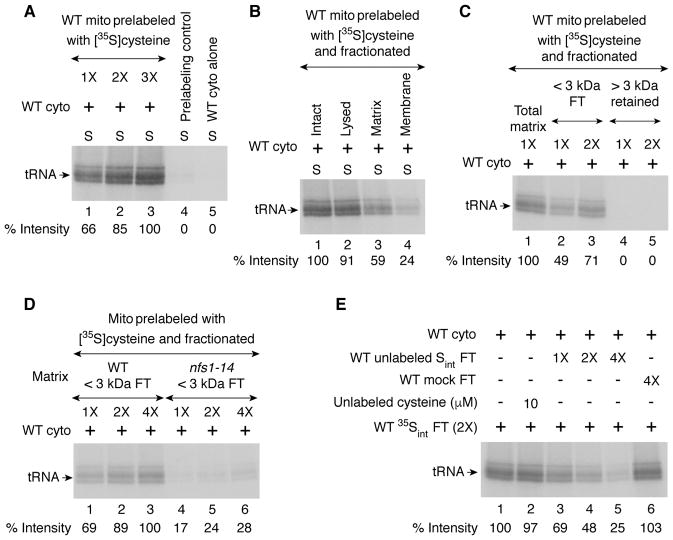
The persulfide-forming activity of Nfs1 does not require nucleotides whereas subsequent use of the persulfide sulfur for Fe-S cluster assembly of aconitase depends on the availability of nucleotides (Pandey et al., 2012). To track formation of sulfur intermediates, mitochondria alone (no cytosol) were incubated with [35S]cysteine in the absence of any added nucleotides. Mitochondria were recovered and washed to remove excess [35S]cysteine. These prelabeled mitochondria were then incubated with cytosol in the presence of added nucleotides. Increasing amounts of cytosolic tRNAs were radiolabeled with increasing concentrations of prelabeled mitochondria (Figure 5A, lanes 1–3). Thus, mitochondria were able to generate and accumulate active sulfur species (called Sint), and release them to the cytosol when allowed to do so. As a control, mitochondria were incubated with cytosol in the presence of nucleotides but no [35S]cysteine. After removal of mitochondria, the cytosolic fraction was supplemented with [35S]cysteine and incubated. No radiolabeled tRNA was detected, further substantiating that the 35Sint species is coming from mitochondria (Figure 5A, lane 4). Note that the samples were analyzed by SDS gels. Under these conditions, radiolabeled tRNAs often appear as doublets/triplets, likely due to separation of newly thiolated tRNAs for Lys, Glu, and Gln. Such a separation does not usually occur on native gels.
Figure 5. Detection of active sulfur species in mitochondrial lysate.
(A) WT mitochondria (1X = 200 μg; lanes 1–3) were incubated with [35S]cysteine at 30°C for 15 min with no added nucleotides. Mitochondria were recovered and washed. These prelabeled mitochondria were then incubated with WT cytosol (200 μg) and nucleotides at 30°C for 30 min. After centrifugation, the supernatant/cytosol fractions were analyzed by SDS-PAGE and autoradiography. As a “Prelabeling control” (lane 4), WT mitochondria (2X) were incubated with WT cytosol (200 μg) and nucleotides at 30°C for 30 min. After centrifugation to remove mitochondria, the supernatant/cytosol was supplemented with [35S]cysteine and incubated again at 30°C for 30 min, and analyzed. As another control, WT cytosol alone (lane 5) was incubated with [35S]cysteine and nucleotides, and analyzed.
(B) WT mitochondria (200 μg) were prelabeled with [35S]cysteine as in (A) above. Mitochondria were lysed (lane 2), and soluble matrix (lane 3) and membranes (lane 4) were isolated. These fractions were separately added to WT cytosol (200 μg) and incubated with nucleotides at 30°C for 30 min. After centrifugation, the supernatant fractions were analyzed as in (A) above.
(C) The matrix fraction isolated from 35S-prelabeled WT mitochondria was centrifuged through a 3 kDa cutoff filter. The flow through (“FT”; lanes 2 and 3) and retained (lanes 4 and 5) fractions were separately incubated with WT cytosol (200 μg) and nucleotides, and analyzed as in (A) above. 1X = 160 μg of starting mitochondrial proteins.
(D) The <3 kDa flow through (“FT”) fractions isolated from the matrix of 35S-prelabeled WT (lanes 1–3) or nfs1-14 (lanes 4–6) mitochondria were incubated with WT cytosol (200 μg) and nucleotides, and analyzed as in (A) above. 1X = 160 μg of starting mitochondrial proteins.
(E) The FT fraction obtained from 35S-prelabeled WT mitochondria as in (D) above is referred to as “WT 35Sint FT”. WT mitochondria were preloaded with unlabeled cysteine (10 μM), and the unlabeled FT fraction was isolated (“WT unlabeled Sint FT”). A mixture of WT 35Sint FT and nucleotides was added to WT cytosol (200 μg) and incubated without (lane 1) or with increasing amounts of WT unlabeled Sint FT (lanes 3–5) at 30°C for 30 min. Unlabeled FT fraction was also isolated from WT mitochondria without prior loading with unlabeled cysteine (“WT mock FT”) and tested as above (lane 6). Furthermore, a reaction mixture containing WT cytosol, WT 35Sint FT, nucleotides, and directly added unlabeled cysteine (10 μM) was included (lane 2). Samples were analyzed as in (A) above. 1X = 80 μg of starting mitochondrial proteins. See also Figures 7 and S3, and Table S2.
The accumulated intermediate Sint was present predominantly in the matrix fraction of the mitochondria (Figure 5B). Sint was of low molecular mass, as evidenced by its ability to pass through a 3 kDa cutoff filter (Figure 5C). As predicted by the Nfs1 requirement for the entire process, mitochondrial matrix derived from the nfs1-14 mutant did not efficiently form or accumulate Sint (Figure 5D). Importantly, unlabeled Sint generated by incubating mitochondria with unlabeled cysteine could compete with labeled intermediate (35Sint) generated with [35S]cysteine when added to WT cytosol in the thiolation assay (Figure 5E, lanes 3–5). The inhibition of 35S-labeling of tRNAs was not due to contaminating unlabeled cysteine that might have been carried over during isolation of unlabeled Sint species, since a large excess of unlabeled cysteine that was directly added to cytosol failed to inhibit 35Sint-mediated tRNA thiolation (Figure 5E, compare lanes 1 and 2). Likewise, no inhibition was observed when mitochondrial preloading with unlabeled cysteine was omitted (Figure 5E, lane 6; “WT mock FT”).
Mitochondria lacking Atm1 cannot promote efficient cytosolic tRNA thiolation
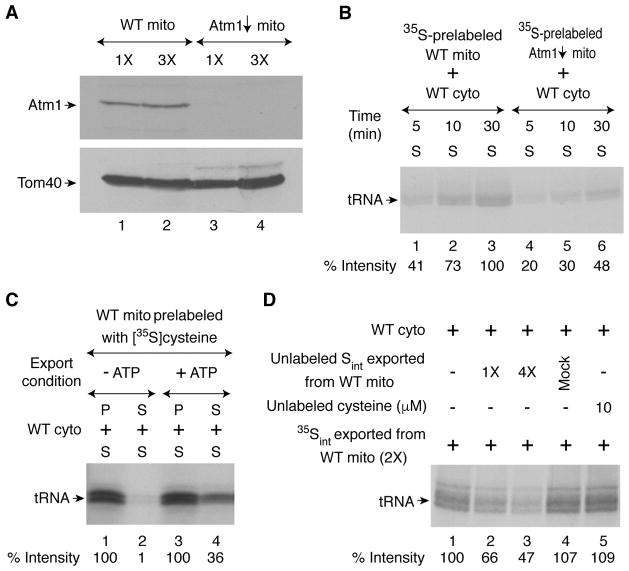
We sought to determine whether Atm1 is involved in exporting Sint from mitochondria to cytosol for tRNA thiolation. A GAL1-ATM1 strain was grown under repressing conditions for Atm1 expression, and mitochondria were isolated. No Atm1 protein was detected by immunoblotting (Figure 6A). These Atm1-depleted (Atm1↓) mitochondria were prelabeled with [35S]cysteine in the absence of added nucleotides. After removal of excess [35S]cysteine, prelabeled mitochondria were incubated with WT cytosol in the presence of nucleotides. Compared with WT mitochondria, the Atm1↓ mitochondria-mediated radiolabeling of cytosolic tRNAs was less efficient and occurred slowly (Figure 6B). In the absence of adequate levels of Atm1, mitochondria were unable to efficiently export active 35Sint species required for cytosolic tRNA thiolation.
Figure 6. Active sulfur species exported from intact mitochondria: Atm1 and ATP dependence.
(A) WT and Atm1-depleted (Atm1↓) mitochondria were analyzed by immunoblotting using anti-Atm1 and anti-Tom40 antibodies. 1X = 75 μg of proteins.
(B) WT or Atm1↓ mitochondria (200 μg) were incubated with [35S]cysteine (10 μCi) in the presence of ATPγS (2 mM) at 30°C for 15 min. Mitochondria were recovered, washed, and then incubated with WT cytosol (200 μg) and nucleotides at 30°C for 30 min. After centrifugation, the supernatant/cytosol fractions were analyzed by SDS-PAGE and autoradiography.
(C) WT mitochondria (2 mg) were incubated with [35S]cysteine (20 μCi) and ATPγS (2 mM) to generate 35Sint inside the organelle (1st step). Mitochondria were recovered, washed, and then incubated without (lanes 1 and 2) or with (lanes 3 and 4) ATP (4 mM) at 30°C for 15 min, and centrifuged (2nd step; Sint export). The pellet (mitochondria with residual 35Sint; lanes 1 and 3) or supernatant (exported 35Sint; lanes 2 and 4) fractions were separately mixed with WT cytosol (300 μg). Nucleotides were maintained at a final concentration of 4 mM ATP, 1 mM GTP, 2 mM NADH and 1 mM NADPH. After incubation at 30°C for 30 min (3rd step), samples were centrifuged, and the supernatant/cytosol fractions were analyzed as in (B).
(D) Export reactions were performed with 4 mM ATP. Unlabeled Sint exported from WT mitochondria was obtained as in (C) above except mitochondria were preloaded with 10 μM unlabeled cysteine instead of [35S]cysteine during the 1st step. A mixture of exported 35Sint and nucleotides was added to WT cytosol (200 μg) and incubated without (lane 1) or with (lanes 2 and 3) unlabeled Sint at 30°C for 30 min. Unlabeled exported material was also isolated from WT mitochondria without preloading with unlabeled cysteine during the 1st step (“Mock”; 4X) and tested for competing activity with 35Sint (lane 4). Likewise, a reaction mixture containing cytosol, 35Sint, nucleotides, and directly added 10 μM unlabeled cysteine (lane 5) served as another control. Samples were analyzed by SDS-PAGE and autoradiography. 1X = 200 μg of starting mitochondrial proteins. See also Figure S2.
Detection of Sint exported from intact mitochondria in active form
To detect the sulfur species exported from mitochondria, we developed a three-step assay that allowed us to dissect the mitochondria-cytosol interaction. Intact WT mitochondria were incubated with [35S]cysteine to generate 35Sint inside the organelle. ATPγS was included to prevent intra-organellar use of 35Sint thus generated and to prevent its export at this stage (1st step). Mitochondria were recovered and washed to remove free [35S]cysteine and ATPγS. Samples were then incubated with or without ATP (2nd step), and centrifuged. The mitochondrial pellets or the supernatant fractions thus obtained were separately incubated with WT cytosol and nucleotides but no [35S]cysteine was added (3rd step). After centrifugation, the supernatant fractions were analyzed for cytosolic tRNA thiolation. 35Sint was generated within WT mitochondria and exported from mitochondria in a manner completely dependent on added ATP (Figure 6C, compare supernatants in lane 2 with no ATP and lane 4 with ATP). These results are consistent with Atm1 being involved in the export process because Atm1 is an ATP binding cassette transporter expected to require ATP hydrolysis for its export activity. A key result is that once exported, 35Sint by itself promoted tRNA thiolation in cytosol with no further requirement of mitochondria (Figure 6C, lane 4). When unlabeled cysteine was utilized, unlabeled intermediate (Sint) was generated and exported, and this competed with labeled intermediate (35Sint) for promoting cytosolic tRNA thiolation (Figure 6D, lanes 1–3). As a control, unlabeled cysteine was ineffective when added directly to the cytosol (Figure 6D, lane 5). Mitochondria incubated with no added cysteine during the 1st step (“Mock”) served as another negative control (Figure 6D, lane 4).
Characterization of Sint
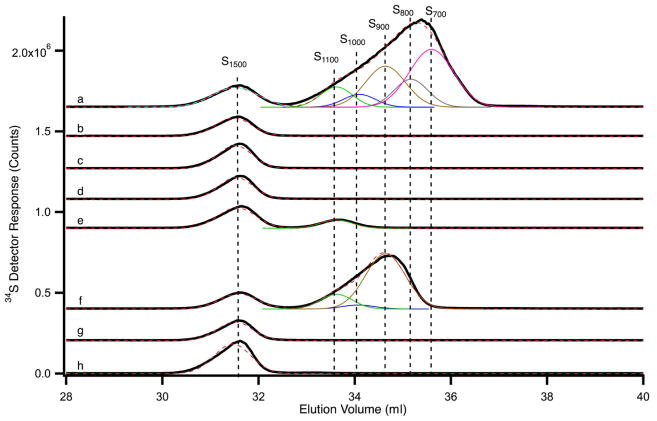
Unlabeled materials exported from various mitochondria under different conditions were analyzed for competing activity with 35Sint (Figure S2), and in parallel by SEC-ICP-MS chromatography (Figure 7) (McCormick et al., 2015). The material exported from WT mitochondria (preloaded with unlabeled cysteine) exhibited competing activity (Figure S2, lanes 2 and 3), and this sample contained six sulfur-containing species with approximate molecular masses of 1500, 1100, 1000, 900, 800, and 700 Da (Figure 7, trace a). These species are referred to as S1500, S1100, S1000, S900, S800, and S700, respectively. Some of these species may represent degradation or oxidatively damaged products. Uncertainties in these masses are significant (± 25%), and so the numbers should be viewed mainly as a means of distinguishing one species from another. S1500 was present in all traces, including a trace of the Hepes/KOH buffer alone (Figure 7, trace h), and so it likely originated from Hepes, which contains a sulfur molecule. However, the mass of Hepes (238 Da) is significantly less than 1500 Da, suggesting that it formed a multimeric species, as has been reported (Saraiva et al., 2010). A peak corresponding to the authentic mass of Hepes (238 Da) was not detected, likely due to unusual migration effects of very low molecular mass sulfur species down the LC column (McCormick et al., 2015).
Figure 7. SEC-ICP-MS chromatogram of sulfur species.
Solid black lines are data, and dashed red lines are composite fits. Simulations of individual peaks are color-coded as follows: S1100, green; S1000, blue; S900, brown; S800, grey; S700, pink. a) Exported material from WT mitochondria (cysteine-preloaded); b) exported material from nfs1-14 mitochondria (cysteine-preloaded); c) exported material from WT mitochondria with no cysteine preloading; d) exported material from WT mitochondria (cysteine-preloaded) in which ATPγS rather than ATP was used during the export process; e) exported material from Atm1↓ mitochondria (cysteine-preloaded); f) material isolated from WT mitochondrial matrix after preloading with cysteine; g) material isolated from WT mitochondrial matrix with no cysteine preloading; h) sample buffer only (20 mM Hepes/KOH, pH 7.5). See also Figures 5, S2 and S3, and Table S2.
As a control, no sulfur species besides S1500 was detected in the material exported from nfs1-14 mutant mitochondria (preloaded with unlabeled cysteine) or from WT mitochondria with no preloaded cysteine (Figure 7, traces b and c, respectively), consistent with lack of competing activity of these samples (Figure S2, lanes 6 and 7, and lanes 10 and 11, respectively). Likewise, when WT mitochondria were preloaded with unlabeled cysteine but the export process was blocked with ATPγS, no sulfur species other than S1500 was detected (Figure 7, trace d), nor was there any competing activity (Figure S2, lanes 8 and 9). The material exported from Atm1↓ mitochondria (preloaded with unlabeled cysteine) did not exhibit any detectable competing activity (Figure S2, lanes 4 and 5). It completely lacked 1000, 900, 800, and 700 Da species and contained a small amount of S1100 in addition to the buffer peak (Figure 7, trace e). We speculate that S1100 might have been exported by a different ABC transporter in the mitochondrial inner membrane such as Mdl1 (Young et al., 2001). In fact, S1100 was detected in exported material from WT mitochondria only when ATP, and not ATPγS, was included during the export process (Figure 7, compare traces a and d). In mitochondria-cytosol mixing assays using [35S]cysteine, cytosolic tRNA thiolation occurred poorly but a small signal was still detectable with both Atm1↓ (Figure 6B) and nfs1-14 (Figure 3A) mitochondria because of high sensitivity. However, no relevant unlabeled sulfur species able to compete with 35Sint was detected in the materials exported from these mitochondria (Figure 7 and Figure S2). This could be due to loss of limited material during the multi-step isolation procedure. Regardless, all of the species exported from WT mitochondria as described above were consistently detected in another set of chromatograms from a separate experiment (Figure S3), even though an additional species (S600) was present (Figure S3, trace a versus Figure 7, trace a). Unlabeled and active material isolated from WT mitochondrial matrix with a molecular mass of <3 kDa (see Figure 5E, lanes 3–5) also contained similar sulfur species, although the subspecies composition appeared slightly altered (Figure 7, trace f; Figure S3, trace f). The sulfur species that accumulated in the matrix might differ slightly from the exported forms. Alternatively, these subtle differences could be due to sample preparation – exported material from intact mitochondria versus fractionation of mitochondrial lysate/matrix. The relative concentrations of various sulfur species are presented in Table S2.
DISCUSSION
Thio-modification of the wobble uridines of a subset of cytosolic tRNAs is vital for their roles in protein synthesis. Here we demonstrate that mitochondria play a direct and essential role in cytosolic tRNA thiolation. Specifically, mitochondria generate low molecular mass sulfur-containing intermediate species (Sint) and export them to the cytosol. We detected the exported Sint species in active form that are utilized by the cytosol to thiolate tRNAs. In prior work, other laboratories defined various mitochondrial and cytosolic proteins involved in thiolation of cytosolic tRNAs (Leidel et al., 2009; Nakai et al., 2007; Nakai et al., 2004; Noma et al., 2009). They measured steady state levels of thiolated tRNAs in vivo but did not assess the individual contributions of mitochondria and cytosol to cytosolic tRNA thiolation and how they are functionally connected. We resolved these issues here by developing assays for de novo cytosolic tRNA thiolation, using metabolically active mitochondria and cytosol isolated from yeast cells. In these assays, [35S]cysteine was used as the source of sulfur to simultaneously monitor radiolabeling of aconitase (Fe-S cluster assembly) in mitochondria and radiolabeling of tRNAs (s2U thiolation) in cytosol.
In yeast, the Nfs1 cysteine desulfurase is found primarily in mitochondria (Li et al., 1999) with a small amount located in the cytosol (Naamati et al., 2009). An experiment was performed in which mitochondrial Nfs1 was depleted while maintaining expression of cytosolic Nfs1 (Nakai et al., 2007). This mitochondrial Nfs1 depleted strain lacked cytosolic tRNA thiolation, and based on these results, it was concluded that mitochondrial Nfs1 is essential for cytosolic tRNA thiolation (Nakai et al., 2007). However, depletion of mitochondrial Nfs1 (cytosolic Nfs1 present) also leads to deficiency in cytosolic Fe-S cluster assembly (Kispal et al., 1999). Furthermore, in vivo depletion of some CIA components is associated with greatly reduced levels of thiolated tRNAs in the cytosol (Nakai et al., 2007). Therefore, the in vivo studies could not rule out the possibility that depletion of mitochondrial Nfs1 might indirectly block cytosolic tRNA thiolation via inhibitory effects on cytosolic Fe-S cluster assembly. This is an important issue because the [4Fe-4S] cluster-containing Elp3 (Paraskevopoulou et al., 2006) and the [3Fe-4S] cluster-containing Ncs6 (Liu et al., 2016) are involved in mcm5s2U34 modification of cytosolic tRNAs (Nakai et al., 2017). In the absence of cytosolic Fe-S cluster assembly, these proteins are unlikely to be functional. Data presented here settle the issue and conclusively show that mitochondrial Nfs1 is directly required for cytosolic tRNA thiolation independent of its effects on cytosolic Fe-S cluster assembly. For example, no radiolabeled tRNA was detected when isolated wild-type cytosol alone was incubated with [35S]cysteine (Figures 1A and 2). Thus, cytosolic Fe-S proteins, together with any trace amounts of cytosolic Nfs1, were unable to thiolate tRNAs. Upon addition of wild-type mitochondria, cytosol acquired the thiolation activity because Nfs1 in mitochondria generated the critical sulfur species required for cytosolic tRNA thiolation. This conclusion was further substantiated by the observation that nfs1-14 mutant mitochondria with compromised cysteine desulfurase activity failed to efficiently promote cytosolic tRNA thiolation (Figures 3A and 5D).
In mitochondria, Nfs1 binds its substrate cysteine through the cofactor pyridoxal phosphate. The cysteine sulfur is removed and used to form a covalent persulfide on an active site cysteine (Nfs1-S-SH) (Pandey et al., 2012). The persulfide sulfur is then transferred to the scaffold Isu1/2, where it combines with iron. The iron-sulfur cluster intermediates thus formed are donated to apoproteins (e.g. apoaconitase), a process that requires chaperones (Lill et al., 2015). Our data show that Isu1-depleted mitochondria (Isu2 absent) failed to support Fe-S cluster biogenesis within the organelle or tRNA thiolation in the cytosol (Figure 3B). Thus, in addition to mitochondrial Nfs1, mitochondrial Isu1/2 are also required for generating sulfur intermediates (Sint) necessary for thio-modification of cytosolic tRNAs. The ssq1 mutant mitochondria did not support Fe-S cluster biogenesis of aconitase but surprisingly were able to efficiently promote cytosolic tRNA thiolation to WT levels (Figure 3C). Thus, unlike Nfs1 and Isu1/2, Ssq1 is not involved in generating Sint. These results argue for a hitherto unidentified branch point in the sulfur delivery pathway at the Isu1/2 site for intra-organellar use in mitochondria or export to the cytosol.
The sulfur species generated within mitochondria involving Nfs1 and Isu1/2 must be exported to the cytosol for tRNA thiolation. The Atm1 ATPase in the mitochondrial inner membrane has been hypothesized to export a sulfur-containing molecule that is utilized for both tRNA thiolation and Fe-S cluster assembly in the cytosol (Lill et al., 2015; Nakai et al., 2017). However, none of the candidate Atm1 substrates thus far reported is supported by convincing biochemical data (Lill et al., 2015). The Sint species that we detected here were exported from intact WT mitochondria in an ATP-dependent manner (Figure 6C). The molecular masses of exported Sint ranged from 700 Da to 1100 Da (Figure 7). Mitochondria lacking Atm1 failed to efficiently support cytosolic tRNA thiolation because they were unable to export active Sint species (Figure 6B). Importantly, once exported by WT mitochondria, Sint species were able to promote cytosolic tRNA thiolation by themselves, with no further dependence on mitochondria (Figures 6C and 6D). Thus, the primary contribution of mitochondria in this process is to generate and export of Sint species to cytosol.
Importantly, the cytosolic use of the exported species depended on Dre2, an essential component of the CIA machinery (Figure 4). Dre2 may participate in utilization of exported Sint either directly or indirectly via effects on Fe-S cluster-containing CIA components such as Nbp35 and/or other proteins such as Elp3 and Ncs6 (Liu et al., 2016; Nakai et al., 2017; Paraskevopoulou et al., 2006). Although the principal function of the cytosol in tRNA thiolation is to utilize exported Sint, it may also enhance Sint export from mitochondria. In the absence of cytosol, some of the Sint species were exported in an ATP-dependent manner but a significant portion of Sint still remained trapped within mitochondria in active form (Figure 6C). It is therefore reasonable to speculate that cytosol is required for efficient Sint export, mimicking the in vivo situation. Cytosolic proteins or small molecules might “grab” exiting Sint, making the export process unidirectional and more efficient under physiological conditions within cells. How mitochondria sense the cytosolic need for exported sulfur species remains to be determined.
Most of the S. cerevisiae proteins involved in cytosolic tRNA thiolation or Fe-S cluster assembly are conserved and have human orthologs (Leimkuhler et al., 2017; Nakai et al., 2017; Netz et al., 2014). As in yeast, human NFS1 is also thought to be present in mitochondria with a small amount in cytosol/nucleus (Land and Rouault, 1998). Furthermore, yeast Nfs1/human NFS1 forms an active enzyme complex with accessory proteins Isd11/ISD11 and Acp1/ACP1 (Cory et al., 2017; Van Vranken et al., 2016). More importantly, depletion of the mitochondrial Nfs1/NFS1 leads to Fe-S cluster deficiency in yeast or HeLa cells, and the plasmid-borne expression of the respective proteins without their mitochondrial targeting sequence cannot rescue the defect (Biederbick et al., 2006; Kispal et al., 1999). Thus, the underlying mechanism for cytosolic tRNA thiolation in humans appears to be very similar to that in yeast, involving mitochondrial NFS1. That similarity can now be tested using mitochondria and cytosol isolated from human cells.
SIGNIFICANCE
Critical signals and metabolites move between mitochondria and cytosol, and such communications are essential for cellular homeostasis (Chandel, 2015). For example, mitochondrial biogenesis depends on importing precursor proteins from cytosol to mitochondria. ATP flows from mitochondria to cytosol, and cytochrome c release from mitochondria induces apoptosis. Here we have elucidated the mitochondrial contribution of sulfur-containing intermediates (Sint) to cytosolic tRNA thiolation. The Sint species that we have detected: 1) is generated within mitochondria in a manner that depends on organellar Nfs1 activity, 2) accumulates inside mitochondria under conditions that minimize use within the organelle, 3) is inefficiently exported from mitochondria lacking Atm1, 4) requires CIA components for utilization, and 5) is utilized in the cytosol for tRNA thiolation. Thus, Sint satisfies stringent criteria for a truly exported molecule and qualifies as a bona fide Atm1 export substrate. These findings could be relevant to human diseases. For example, impaired thiolation of cytosolic tRNAs has been associated with familial dysautonomia, a neurodegenerative disease affecting sympathetic nerves (Karlsborn et al., 2014), and details of the Sint export process may shed light on the pathophysiology. ABCB7 is the human ortholog of yeast Atm1, and mutations in ABCB7 are the cause of X-linked sideroblastic anemia and ataxia syndrome (Bekri et al., 2000). Isolation of the ABCB7 export substrate will very likely provide mechanistic insights into this disease.
STAR*METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Debkumar Pain (painde@njms.rutgers.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Yeast strains, cell growth, and isolation of mitochondria and cytosol
Yeast strains are described in Table S1. For Dre2 depletion, a strain was generated as follows. Promoter swap was performed (Longtine et al., 1998), thereby placing DRE2 under control of the GAL1 promoter using the kanMX6 marker in the BY4741 genetic background, and the correctness of the genomic insertion site was checked by PCR (strain #105-59).
All cells were grown at 30°C. YP raffinose, and synthetic complete (SC) medium containing raffinose, with or without dextrose or galactose, were made as described (Sherman, 2002). Matched WT and mutant S. cerevisiae strains were examined (BY4741 and Δurm1 or GAL1-DRE2; YPH499 and nfs1-14, GAL1-ISU1/Δisu2 or GAL1-ATM1; 61 and 33C (ssq1 mutant), see Table S1). BY4741, YPH499, Δurm1, nfs1-14, 61, and 33C strains were grown in SC medium containing 2% raffinose and 0.5% dextrose. Protein depletions were performed as described below. In all of these cases, matched WT strains were similarly grown.
a) Isu1 depletion in GAL1-ISU1/Δisu2 strain (strain #115-10): Cells were initially grown in SC/2% raffinose/0.5% galactose (200 ml) for 26 h. Cells were harvested, washed, switched to SC/2% raffinose/0.5% dextrose medium (2 liter), and grown for 11 h. b) Dre2 depletion in GAL1-DRE2 strain (strain #105-59): Cells were initially grown in SC/2% raffinose/0.5% galactose (15 ml) for 24 h. Cells were harvested, washed, switched to SC/2% raffinose (1 liter) and grown for 22 h. c) Atm1 depletion in GAL1-ATM1 strain (strain #83-25): Cells were grown in YP/2 % raffinose medium for 80 h.
Intact mitochondria were isolated from various strains (Amutha et al., 2009; Amutha et al., 2008) and stored in isotonic HS buffer (20 mM Hepes/KOH, pH 7.5 containing 0.6 M sorbitol) with 10% DMSO at −80°C. The post-mitochondrial supernatant was centrifuged at 153,000 × g for 45 min, and the supernatant thus obtained (cytosol) was also stored at −80°C.
METHOD DETAILS
Mitochondria and cytosol mixing assays
A typical one step mixing assay mixture (100 μl) contained isolated and intact mitochondria (50–200 μg of proteins) added to isolated cytosol (50–400 μg of proteins) in HS buffer (20 mM Hepes/KOH, pH 7.5, 0.6 M sorbitol) containing 10 mM Mg(OAc)2, 40 mM KOAc, nucleotides (4 mM ATP, 1 mM GTP, 2 mM NADH and 1 mM NADPH), and 1 mM TCEP (Tris(2-carboxyethyl)phosphine hydrochloride). Ferrous ascorbate was added to a final concentration of 10 μM from a stock mixture of ammonium ferrous sulfate (0.5 mM) and sodium ascorbate (5 mM). Following addition of [35S]cysteine (10 μCi), samples were incubated at 30°C for different time periods (10–45 min). Control assays were performed with mitochondria alone or cytosol alone. The reaction mixtures were centrifuged at 15,000 × g for 10 min at 4°C, and the resulting pellet (“P”, mitochondria) and supernatant (“S”, cytosol) fractions were processed as follows.
The mitochondrial pellets were washed once with ice-cold HS buffer and then resuspended in 35 μl of 50 mM Tris/HCl, pH 8.0 containing 0.5 mM PMSF. Mitochondrial membranes were ruptured by freezing the samples on dry ice followed by thawing and mild bath sonication for 20–30 sec at 4°C. After repeating this procedure for two more times, samples were centrifuged at 15,000 × g for 30 min at 4°C, and supernatant fractions were used for gel analysis (Amutha et al., 2009; Amutha et al., 2008). The cytosolic fractions were subjected to precipitation with ammonium sulfate (final concentration 67%) for 2 h on ice. Samples were centrifuged at 15,000 × g for 45 min, and the pellets thus obtained were dissolved in 35 μl of 50 mM Tris/HCl, pH 8.0. Mitochondrial and cytosolic samples were analyzed by 12% native polyacrylamide gel electrophoresis (native PAGE). The gel was fixed with 20% methanol in 50 mM Tris/HCl, pH 8.0 for 1 h at 4°C, dried, and exposed to Carestream BIOMAX MR film for autoradiography (Amutha et al., 2009; Amutha et al., 2008). In some cases, cytosolic samples, obtained after removal of mitochondria from the reaction mixtures, were precipitated with ice-cold 10% trichloroacetic acid (TCA), and these samples were analyzed by either 14% or 15% SDS-PAGE. Numerous variations of these assays are indicated in figure legends.
Isolation of unlabeled materials exported from mitochondria for SEC-ICP-MS chromatography
A schematic outline for isolating materials exported from intact mitochondria is presented in Figure S2, top panel. Briefly, reactions were performed in six batches. Each batch contained intact WT mitochondria (5 mg of proteins) in a final volume of 0.5 ml HS buffer containing 40 mM KOAc, 10 mM Mg(OAc)2, 2 mM ATPγS and 10 μM unlabeled cysteine. Samples were incubated at 30°C for 30 min to allow generation of unlabeled Sint and to have it “trapped” within mitochondria. The reaction mixtures were diluted with ice-cold HS buffer and centrifuged at 15,000 × g for 5 min at 4°C. The mitochondrial pellets thus obtained were washed twice with HS buffer to remove any residual ATPγS and unlabeled cysteine. Mitochondria were then resuspended in 100 μl of HS buffer containing 4 mM ATP and incubated at 30°C for 30 min to allow the export process. Samples were centrifuged at 15,000 × g for 10 min at 4°C. The supernatant fractions containing exported Sint were pooled from six identical reaction mixtures (total volume ~0.6 ml) and extracted with an equal volume of water-saturated chloroform. After centrifugation at 15,000 × g for 2 min at 4°C, the organic layer containing Sint was dried down in SpeedVac. The dried material was dissolved with 200 μl of 20 mM Hepes/KOH, pH 7.5 and stored at −80°C until further use.
The nfs1-14 and Atm1-depleted (Atm1↓) mitochondria were similarly processed to obtain corresponding exported materials. Likewise, exported material from WT mitochondria with no preloaded cysteine served as a negative control. As another control, the export reaction with WT mitochondria (preloaded with unlabeled cysteine) was performed in the presence of ATPγS (2 mM) rather than ATP. All of these exported materials were tested in a competition assay for their ability to inhibit 35Sint-mediated radiolabeling of cytosolic tRNAs (see Figure S2, bottom panel), and further characterized by SEC-ICP-MS chromatography as described below.
Aconitase activity measurement
Aconitase activity was measured by an in-gel assay essentially as described (Amutha et al., 2008; Tong and Rouault, 2006). Isolated mitochondria were lysed by incubating with 50 mM Tris/HCl, pH 8.0 containing 50 mM NaCl, 1% Triton X-100, 10% glycerol, 2 mM sodium citrate, 200 U/ml catalase, and 1 mM PMSF in a total volume of 50 μl for 30 min on ice. Samples were centrifuged at 15,000 × g for 10 min at 4°C. Supernatant fractions with added bromophenol blue (0.025%) were subjected to electrophoresis on a native gel. The separating gel contained 6% acrylamide, 132 mM Tris base, 132 mM boric acid and 3.6 mM sodium citrate, and the stacking gel contained 4% acrylamide, 67 mM Tris base, 67 mM boric acid and 3.6 mM sodium citrate. The running buffer contained 25 mM Tris/192 mM glycine, pH 8.3 and 3.6 mM sodium citrate. Electrophoresis was carried out at 90 V for 12 h at 4°C. After electrophoresis, the gel was incubated with 100 mM Tris/HCl, pH 8.0, 1 mM NADP, 2.5 mM cis-aconitic acid, 5 mM MgCl2, 1.2 mM thiazolyl blue tetrazolium bromide (MTT), 0.3 mM phenazine methosulfate and 5 U/ml isocitrate dehydrogenase in the dark at 37°C with gentle shaking for 5–30 min. Bands displaying aconitase activity appeared purple/blue. The gel was washed with distilled water, and scanned.
Immunoblotting
Proteins were separated on a 12% SDS gel and then transferred to nitrocellulose (GE Healthcare Health Sciences). The blot was stained with 0.002% amido black (Sigma), washed with water, and blocked with 5% non-fat dry milk in 100 mM Tris/HCl, pH 7.4 containing 150 mM NaCl and 0.1% Tween-20 (Sigma). The blot was horizontally cut around 55 kDa region into two pieces. The top half of the blot was probed with rabbit anti-Atm1 antibodies (1:500 dilution), and the bottom half was probed with rabbit anti-Tom40 antibodies (1:2000 dilution). After washing, the blots were incubated with Horseradish peroxidase (HRP)-conjugated donkey anti-rabbit IgG (1:5000 dilution), and the proteins were detected using chemiluminescence. Tom40 is a mitochondrial outer membrane protein (Schulke et al., 1999) and served as an internal control.
Chromatography and elemental spectroscopy
Instrumentation and parameters: SEC-ICP-MS chromatography was performed on an Agilent 1260 Bio-inert HPLC quaternary pump system that was installed in an anaerobic and refrigerated glovebox (MBRAUN LabMaster, NH, USA). The glovebox was maintained with a nitrogen atmosphere at <1 ppm O2 (Teledyne Analytical Instruments, CA, USA) and at 6°C. The HPLC system was equipped with a bio-inert multivalve injector and a 200 μl PEEK injection loop. All column tubing and fittings were made of PEEK. Two Superdex Peptide 10/300 GL columns (GE Healthcare Life Science, PA, USA) were connected in series via a male-to-male fitting. Ammonium bicarbonate (20 mM), degassed via a Schlenk line, was used as the mobile phase and run at a flow rate of 0.350 ml min−1. The eluent from the column was fed into the Diode Array and then into a splitter. A major portion (70%) of the eluent flowed to the fraction collector and the rest 30% was exported from the glovebox and into an Agilent 7700x ICP-MS in an on-line configuration. This allowed for simultaneous separation, fraction collection, and elemental detection. Elements detected included 31P, 34S, 48Ti, 55Mn, 56/57Fe, 59Co, 60Ni, 63/65Cu, 66/68Zn, and 95Mo. The ICP-MS was fitted with a micromist nebulizer, Scott-type spray chamber, and nickel sampling/skimmer cones. ICP-MS operational parameters are as follows: RF Power 1550 W; Ar flow rate, 15 l min−1; collision cell He flow rate, 4.1 ml min−1; carrier gas flow rate, 1.05 l min−1; dilution mode, OFF; spray chamber temperature, 2°C; analysis time, 9600 sec.
All analytical solutions used high purity water, made from distilled, triple filtered, deionized water. The sample loop was prepared by passing 3 loop volumes of a 10% nitric acid solution and incubating for a minimum of 2 h prior to analysis, after which the loop was rinsed with 3 loop volumes of high purity water. Samples were stored at −80°C and thawed inside the glove box for analysis. The columns were regenerated after each day of use by running 10 column volumes of a chelator cocktail composed of 10 mM ascorbic acid and 10 μM each of EDTA, EGTA, 2,2′-bipyridine, 1,10 phenanthroline, and bathocuproinedisulfonic acid.
Species molecular mass and peak fitting: Molecular masses were determined by fitting the peak elution volume to a calibration curve, in which standards of known molecular masses were passed through the SEC-ICP-MS system. The calibration curve was generated by plotting the log of the molecular mass vs the elution volume divided by the void volume of the standards. The elution volume of blue dextran determined the void volume. The standards used included: AMP, ADP, ATP, cyanocobalamin, insulin, and cytochrome C.
QUANTIFICATION AND DATA ANALYSIS
Radiolabeled tRNA bands were quantified by densitometric analysis of autoradiographs using the NIH ImageJ software, and the variation between repeat experiments was in the range of 5–10%. In most cases, data as presented are internally controlled. The reproducibility of various assays was confirmed with biological replicates from different batches of isolated mitochondria and/or cytosol. The in-gel aconitase activity was also quantified by densitometric analysis of scanned gels using the ImageJ software. For the SEC-ICP-MS chromatography, the chromatogram peak fitting was performed with the Fityk software (Wojdyr, 2010). The Levenberg-Marquardt method was used and Gaussian line shapes were assumed. The minimum number of peaks required to fit the entire data set were used.
Supplementary Material
Highlights.
Mitochondria generate activated sulfur species (Sint) with masses from 700–1100 Da
Sint synthesis requires Nfs1 and Isu1/2, but not Ssq1, of the Fe-S cluster machinery
Mitochondria export Sint to the cytosol via the ABC transporter, Atm1
Cytosol uses exported Sint for thiolation of tRNAs specific for Lys, Glu and Gln
Acknowledgments
This work was supported by National Institutes of Health Grants RO1 GM107542 (to D.P. and A.D.) and R35 GM127021 (to P.A.L.). We thank M. M. Balamurali, Arnab K. Ghosh, Simon A.B. Knight, and Anjaneyulu Murari for participation at different stages of this work. This paper is dedicated in loving memory of Günter Blobel.
Footnotes
AUTHOR CONTRIBUTIONS
D.P. conceived the study and designed the experiments. A.P., J.P., N.D., and A.K.P. performed the experiments. A.D. generated some of the yeast strains. D.P. wrote the paper with A.D. and P.A.L.
DECLARATION OF INTERESTS: The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amutha B, Gordon DM, Dancis A, Pain D. Chapter 14 Nucleotide-dependent iron-sulfur cluster biogenesis of endogenous and imported apoproteins in isolated intact mitochondria. Methods Enzymol. 2009;456:247–266. doi: 10.1016/S0076-6879(08)04414-5. [DOI] [PubMed] [Google Scholar]
- Amutha B, Gordon DM, Gu Y, Lyver ER, Dancis A, Pain D. GTP is required for iron-sulfur cluster biogenesis in mitochondria. J Biol Chem. 2008;283:1362–1371. doi: 10.1074/jbc.M706808200. [DOI] [PubMed] [Google Scholar]
- Bekri S, Kispal G, Lange H, Fitzsimons E, Tolmie J, Lill R, Bishop DF. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood. 2000;96:3256–3264. [PubMed] [Google Scholar]
- Biederbick A, Stehling O, Rosser R, Niggemeyer B, Nakai Y, Elsasser HP, Lill R. Role of human mitochondrial Nfs1 in cytosolic iron-sulfur protein biogenesis and iron regulation. Mol Cell Biol. 2006;26:5675–5687. doi: 10.1128/MCB.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS. Evolution of Mitochondria as Signaling Organelles. Cell Metab. 2015;22:204–206. doi: 10.1016/j.cmet.2015.05.013. [DOI] [PubMed] [Google Scholar]
- Cory SA, Van Vranken JG, Brignole EJ, Patra S, Winge DR, Drennan CL, Rutter J, Barondeau DP. Structure of human Fe-S assembly subcomplex reveals unexpected cysteine desulfurase architecture and acyl-ACP-ISD11 interactions. Proc Natl Acad Sci U S A. 2017;114:E5325–E5334. doi: 10.1073/pnas.1702849114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewez M, Bauer F, Dieu M, Raes M, Vandenhaute J, Hermand D. The conserved Wobble uridine tRNA thiolase Ctu1-Ctu2 is required to maintain genome integrity. Proc Natl Acad Sci U S A. 2008;105:5459–5464. doi: 10.1073/pnas.0709404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Yacoubi B, Bailly M, de Crécy-Lagard V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu Rev Genet. 2012;46:69–95. doi: 10.1146/annurev-genet-110711-155641. [DOI] [PubMed] [Google Scholar]
- Karlsborn T, Tukenmez H, Chen C, Bystrom AS. Familial dysautonomia (FD) patients have reduced levels of the modified wobble nucleoside mcm5s2U in tRNA. Biochem Biophys Res Commun. 2014;454:441–445. doi: 10.1016/j.bbrc.2014.10.116. [DOI] [PubMed] [Google Scholar]
- Kispal G, Csere P, Prohl C, Lill R. The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 1999;18:3981–3989. doi: 10.1093/emboj/18.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SA, Sepuri NB, Pain D, Dancis A. Mt-Hsp70 homolog, Ssc2p, required for maturation of yeast frataxin and mitochondrial iron homeostasis. J Biol Chem. 1998;273:18389–18393. doi: 10.1074/jbc.273.29.18389. [DOI] [PubMed] [Google Scholar]
- Kuhnke G, Neumann K, Muhlenhoff U, Lill R. Stimulation of the ATPase activity of the yeast mitochondrial ABC transporter Atm1p by thiol compounds. Mol Membr Biol. 2006;23:173–184. doi: 10.1080/09687860500473630. [DOI] [PubMed] [Google Scholar]
- Land T, Rouault TA. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol Cell. 1998;2:807–815. doi: 10.1016/s1097-2765(00)80295-6. [DOI] [PubMed] [Google Scholar]
- Laxman S, Sutter BM, Wu X, Kumar S, Guo X, Trudgian DC, Mirzaei H, Tu BP. Sulfur amino acids regulate translational capacity and metabolic homeostasis through modulation of tRNA thiolation. Cell. 2013;154:416–429. doi: 10.1016/j.cell.2013.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S, Pedrioli PG, Bucher T, Brost R, Costanzo M, Schmidt A, Aebersold R, Boone C, Hofmann K, Peter M. Ubiquitin-related modifier Urm1 acts as a sulphur carrier in thiolation of eukaryotic transfer RNA. Nature. 2009;458:228–232. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- Leighton J, Schatz G. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 1995;14:188–195. doi: 10.1002/j.1460-2075.1995.tb06989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leimkuhler S, Buhning M, Beilschmidt L. Shared Sulfur Mobilization Routes for tRNA Thiolation and Molybdenum Cofactor Biosynthesis in Prokaryotes and Eukaryotes. Biomolecules. 2017;7 doi: 10.3390/biom7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Cowan JA. Glutathione-coordinated [2Fe-2S] cluster: a viable physiological substrate for mitochondrial ABCB7 transport. Chem Commun (Camb) 2015;51:2253–2255. doi: 10.1039/c4cc09175b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Kogan M, Knight SA, Pain D, Dancis A. Yeast mitochondrial protein, Nfs1p, coordinately regulates iron-sulfur cluster proteins, cellular iron uptake, and iron distribution. J Biol Chem. 1999;274:33025–33034. doi: 10.1074/jbc.274.46.33025. [DOI] [PubMed] [Google Scholar]
- Lill R, Dutkiewicz R, Freibert SA, Heidenreich T, Mascarenhas J, Netz DJ, Paul VD, Pierik AJ, Richter N, Stumpfig M, et al. The role of mitochondria and the CIA machinery in the maturation of cytosolic and nuclear iron-sulfur proteins. Eur J Cell Biol. 2015;94:280–291. doi: 10.1016/j.ejcb.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Liu Y, Vinyard DJ, Reesbeck ME, Suzuki T, Manakongtreecheep K, Holland PL, Brudvig GW, Soll D. A [3Fe-4S] cluster is required for tRNA thiolation in archaea and eukaryotes. Proc Natl Acad Sci U S A. 2016;113:12703–12708. doi: 10.1073/pnas.1615732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- McCormick SP, Moore MJ, Lindahl PA. Detection of Labile Low-Molecular-Mass Transition Metal Complexes in Mitochondria. Biochemistry. 2015;54:3442–3453. doi: 10.1021/bi5015437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naamati A, Regev-Rudzki N, Galperin S, Lill R, Pines O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J Biol Chem. 2009;284:30200–30208. doi: 10.1074/jbc.M109.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Lill R, Suzuki T, Hayashi H. Thio modification of yeast cytosolic tRNA is an iron-sulfur protein-dependent pathway. Mol Cell Biol. 2007;27:2841–2847. doi: 10.1128/MCB.01321-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Nakai M, Yano T. Sulfur Modifications of the Wobble U34 in tRNAs and their Intracellular Localization in Eukaryotic Cells. Biomolecules. 2017;7 doi: 10.3390/biom7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai Y, Umeda N, Suzuki T, Nakai M, Hayashi H, Watanabe K, Kagamiyama H. Yeast Nfs1p is involved in thio-modification of both mitochondrial and cytoplasmic tRNAs. J Biol Chem. 2004;279:12363–12368. doi: 10.1074/jbc.M312448200. [DOI] [PubMed] [Google Scholar]
- Nedialkova DD, Leidel SA. Optimization of Codon Translation Rates via tRNA Modifications Maintains Proteome Integrity. Cell. 2015;161:1606–1618. doi: 10.1016/j.cell.2015.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz DJ, Mascarenhas J, Stehling O, Pierik AJ, Lill R. Maturation of cytosolic and nuclear iron-sulfur proteins. Trends Cell Biol. 2014;24:303–312. doi: 10.1016/j.tcb.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Noma A, Sakaguchi Y, Suzuki T. Mechanistic characterization of the sulfur-relay system for eukaryotic 2-thiouridine biogenesis at tRNA wobble positions. Nucleic Acids Res. 2009;37:1335–1352. doi: 10.1093/nar/gkn1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Yoon H, Lyver ER, Dancis A, Pain D. Identification of a Nfs1p-bound persulfide intermediate in Fe-S cluster synthesis by intact mitochondria. Mitochondrion. 2012;12:539–549. doi: 10.1016/j.mito.2012.07.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paraskevopoulou C, Fairhurst SA, Lowe DJ, Brick P, Onesti S. The Elongator subunit Elp3 contains a Fe4S4 cluster and binds S-adenosylmethionine. Mol Microbiol. 2006;59:795–806. doi: 10.1111/j.1365-2958.2005.04989.x. [DOI] [PubMed] [Google Scholar]
- Rezgui VA, Tyagi K, Ranjan N, Konevega AL, Mittelstaet J, Rodnina MV, Peter M, Pedrioli PG. tRNA tKUUU, tQUUG, and tEUUC wobble position modifications fine-tune protein translation by promoting ribosome A-site binding. Proc Natl Acad Sci U S A. 2013;110:12289–12294. doi: 10.1073/pnas.1300781110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M, Borges CM, Florencio MH. Behaviour of 4-(-2-hydroxyethyl)-1-piperazineethanesulfonic acid under electrospray ionization mass spectrometry conditions. Eur J Mass Spectrom (Chichester) 2010;16:199–213. doi: 10.1255/ejms.1061. [DOI] [PubMed] [Google Scholar]
- Schaedler TA, Thornton JD, Kruse I, Schwarzlander M, Meyer AJ, van Veen HW, Balk J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J Biol Chem. 2014;289:23264–23274. doi: 10.1074/jbc.M114.553438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulke N, Sepuri NB, Gordon DM, Saxena S, Dancis A, Pain D. A multisubunit complex of outer and inner mitochondrial membrane protein translocases stabilized in vivo by translocation intermediates. J Biol Chem. 1999;274:22847–22854. doi: 10.1074/jbc.274.32.22847. [DOI] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 2002;350:3–41. doi: 10.1016/s0076-6879(02)50954-x. [DOI] [PubMed] [Google Scholar]
- Srinivasan V, Pierik AJ, Lill R. Crystal structures of nucleotide-free and glutathione-bound mitochondrial ABC transporter Atm1. Science. 2014;343:1137–1140. doi: 10.1126/science.1246729. [DOI] [PubMed] [Google Scholar]
- Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Urbonavicius J, Qian Q, Durand JM, Hagervall TG, Bjork GR. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vranken JG, Jeong MY, Wei P, Chen YC, Gygi SP, Winge DR, Rutter J. The mitochondrial acyl carrier protein (ACP) coordinates mitochondrial fatty acid synthesis with iron sulfur cluster biogenesis. Elife. 2016;5:e17828. doi: 10.7554/eLife.17828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojdyr M. Fityk: a general-purpose peak fitting program. J Appl Cryst. 2010;43:1126–1128. [Google Scholar]
- Yasukawa T, Suzuki T, Ishii N, Ohta S, Watanabe K. Wobble modification defect in tRNA disturbs codon-anticodon interaction in a mitochondrial disease. EMBO J. 2001;20:4794–4802. doi: 10.1093/emboj/20.17.4794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon H, Knight SA, Pandey A, Pain J, Turkarslan S, Pain D, Dancis A. Turning Saccharomyces cerevisiae into a Frataxin-Independent Organism. PLoS Genet. 2015;11:e1005135. doi: 10.1371/journal.pgen.1005135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Lyver ER, Nakamaru-Ogiso E, Yoon H, Amutha B, Lee DW, Bi E, Ohnishi T, Daldal F, Pain D, et al. Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol Cell Biol. 2008;28:5569–5582. doi: 10.1128/MCB.00642-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.