Abstract
Rationale
Cardiac fibroblasts do not form a syncytium but reside in the interstitium between myocytes. This topological relationship between fibroblasts and myocytes is maintained throughout post-natal life until acute myocardial injury occurs, when fibroblasts are recruited to, proliferate and aggregate in the region of myocyte necrosis. The accumulation or aggregation of fibroblasts in the area of injury thus represents a unique event in the life cycle of the fibroblast but little is known about how changes in the topological arrangement of fibroblasts following cardiac injury affect fibroblast function.
Objective
The objective of the study was to investigate how changes in topological states of cardiac fibroblasts (such as following cardiac injury) affect cellular phenotype.
Methods and Results
Using two and three-dimensional (2D vs 3D) culture conditions, we show that simple aggregation of cardiac fibroblasts is sufficient by itself to induce genome wide changes in gene expression and chromatin remodeling. Remarkably, gene expression changes are reversible following the transition from a 3D back to 2D state demonstrating a topological regulation of cellular plasticity. Genes induced by fibroblast aggregation are strongly associated and predictive of adverse cardiac outcomes and remodeling in mouse models of cardiac hypertrophy and failure. Using solvent based tissue clearing techniques to create optically transparent cardiac scar tissue, we show that fibroblasts in the region of dense scar tissue express markers that are induced by fibroblasts in the 3D conformation. Finally, using live cell interferometry, a quantitative phase microscopy technique to detect absolute changes in single cell biomass, we demonstrate that conditioned medium collected from fibroblasts in 3D conformation compared to that from a 2D state significantly increases cardiomyocyte cell hypertrophy.
Conclusions
Taken together, these findings demonstrate that simple topological changes in cardiac fibroblast organization are sufficient to induce chromatin remodeling and global changes in gene expression with potential functional consequences for the healing heart.
Keywords: Remodeling, fibrosis, gene expression and regulation, cell biology, hypertrophy, fibroblasts, stem cell plasticity
Subject Terms: Heart Failure, Myocardial Infarction, Remodeling
INTRODUCTION
Cardiac fibroblasts develop from epithelial-mesenchymal-transition (EMT) of epicardial cells during cardiac development[1]. Following adoption of the mesenchymal phenotype, they migrate into the developing myocardium and as the myocardium compacts, they get trapped between the myocyte interstitium to become resident cardiac fibroblasts. This topological arrangement of fibroblasts and myocytes persists throughout post-natal life. However, this spatial relationship is disrupted following acute myocardial necrosis, when fibroblasts are recruited to, proliferate and aggregate in the region of injury, resulting in a much higher density of fibroblasts in the region of necrosis[2]. Aggregating fibroblasts in the region of injury are known to express gap junctions that facilitate intercellular communication between physically apposed fibroblasts[3]. Tumor cells and cancer cell lines, when cultured in 3D conditions to promote aggregation exhibit altered phenotypic features such as migration, proliferation and chemo resistance associated with changes in gene expression profiles[4]. However, little is known about how spatial rearrangement of fibroblasts such as that occurs after acute myocardial injury affects the cellular and genetic outputs of the fibroblast and the cardiac wound healing response.
METHODS
All data and supporting materials are within the article and online supplementary files. In addition, RNA-seq and ATAC-seq data for the study are available in NCBI’s Gene Expression Omnibus and have been made publicly available through GEO series accession number GSE113277 at https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE113277.
Cardiac fibroblasts were isolated from adult wild type mice (both male and female) as well as Col1a2CreERT:R26Rtdtomato and TCF21MerCreMer:R26Rtdtomato as described[5]. Isolated cardiac fibroblasts (< 3 passages) were grown on standard polystyrene coated tissue culture plates (2D) (plates not coated with collagen or other matrix proteins) or seeded onto ultra-low attachment plates (not coated with any extracellular matrix protein), whereby they formed spheres within 24 hours of seeding (3D). Subsequently, the cardiac fibroblasts were again transferred back to regular tissue culture plates, on which the spheres attached and fibroblasts migrated out of the spheres to form monolayers within 4–5 days (3D-2D). Reseeding of the fibroblasts onto ultra-low attachment plates again resulted in formation of spheres within 24 hours (3D-2D-3D). Fibroblasts in 2D or 3D maintained for 5 days served as temporally adjusted controls for 3D-2D states. 3D-2D fibroblasts trypsinized and reseeded onto 2D states served as additional controls for 3D-2D-3D states. RNA-seq and ATAC-seq was performed at each topological state of the cardiac fibroblast and on temporally adjusted controls for each time point. Transcripts upregulated in 3D states were correlated to clinical traits across a mouse population (HMDP) following isoproterenol infusion[6]. Cardiac fibroblasts were also seeded onto tissue culture plates of varying stiffness (0.5kPa, 8kPa and 64kPa elastic moduli) to determine whether 2D-3D gene expression changes were recapitulated by modulating substrate stiffness. Optical transparency of the heart was performed with solvent based tissue clearing[7] and imaging performed with a Nikon C2+ confocal microscope. Immunofluorescent staining was performed using standard methods[5]. Conditioned medium was collected from 2D or 3D cardiac fibroblasts exactly 24 hours after initial seeding. LCI was performed to track changes in cell biomass of single neonatal rat ventricular cardiomyocytes with 2D or 3D conditioned medium.
RESULTS
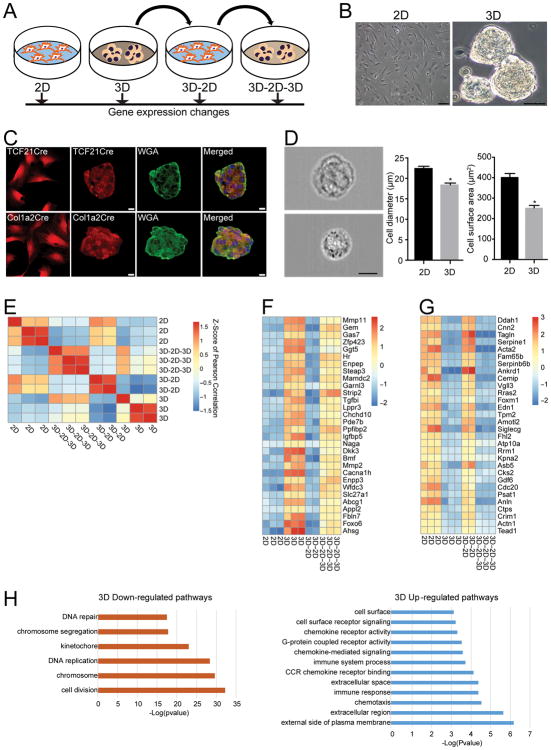
To determine whether aggregation of cardiac fibroblasts affects the cellular phenotype, we first created a scaffold-free 3D system using ultra-low attachment tissue culture dishes where a covalently bonded hydrogel layer on the surface of the dish prevents cell attachment[8]. Cardiac fibroblasts were isolated from adult mice, and cells that had not undergone more than 3 passages were used for experiments. Seeding of primary adult mouse cardiac fibroblasts onto ultra-low attachment dishes resulted in fibroblasts aggregating together within 24 hours to form 3D spherical clusters (Fig 1A, B). To confirm that cardiac fibroblasts alone were capable of forming these spherical clusters, we next isolated cardiac fibroblasts from uninjured hearts of TCF21MerCreMer:R26RtdTomato and Col1a2CreERT:R26RtdTomato mice[5, 9, 10]. We and others have shown that the inducible Cre drivers are specific for genetic labeling of cardiac fibroblasts following tamoxifen administration. Similar to cardiac fibroblasts from wild type animals, genetically labeled cardiac fibroblasts within 24 hours of seeding onto ultra-low attachment plates also formed spherical clusters confirming the ability of cardiac fibroblasts to form 3D spherical aggregates under defined conditions (Fig 1C). Imaging Flow cytometry[11] demonstrated that aggregation into a 3D state resulted in significantly smaller small cell size (cell diameter: 22.45±0.30um in 2D versus 18.41±0.26; mean±S.E.M; p<0.001) and surface area (449.85±3.05um2 in 3D versus 297.97±8 um2; mean±S.E.M; p<0.001) (Fig 1D) suggestive of cellular remodeling as fibroblasts adopt the 3D state. To determine whether a switch from a 2D to a 3D state changes fibroblast phenotype, we first compared global gene expression changes by RNA-seq between cardiac fibroblasts cultured under standard 2D conditions on regular tissue culture dishes and 3D conditions as mentioned above (Fig 1A). For this purpose, cardiac fibroblasts were seeded onto standard tissue culture plates or ultra-low attachment plates with similar seeding density and identical cell culture medium and cells were harvested 24 hours later for gene expression analysis. To ask whether observed changes were reversible, we transferred 3D cardiac fibroblasts to regular tissue culture plates to put them back in 2D conditions (group termed 3D-2D) (Fig 1A). Spherical clusters of 3D fibroblasts attached to regular tissue culture plates and the fibroblasts migrated from spherical cluster to a monolayer within 4–5 days. We again determined gene expression of 3D-2D fibroblasts (following transition from 3D to a monolayer) to determine whether the gene expression pattern reverted to that of the 2D state (Fig 1A). Finally, cardiac fibroblasts which had been grown under 3D conditions and then transferred to 2D conditions (3D-2D) were put back under 3D conditions (group termed 3D-2D-3D). Sphere formation occurred within 24 hours of reseeding on ultra-low attachment plates and RNA-seq was performed to determine whether re-adoption of the 3D state was associated with gene expression signatures flipping back to the 3D state (Fig 1A). These experiments would thus determine whether changes in topological states or spatial arrangement of cardiac fibroblasts are associated with reversible and dynamic changes in gene expression. RNA-seq was performed for all the different topological states of the cardiac fibroblast and clustering of sample correlations demonstrated a grouping of all 2D fibroblast states, and a separate grouping of 3D fibroblast states (Fig 1E). The gene expression profile of 2D cardiac fibroblasts was like that of the 3D-2D fibroblast group while the gene expression profile of 3D fibroblasts was like that of the 3D-2D-3D group (Fig 1E). We observed a remarkable dynamic and reversible plasticity between the 2D and 3D states (Online Table I, Fig 1F, G). Out of 997 genes that were upregulated in 3D fibroblasts, expression of 996 genes reverted back when the 3D fibroblasts were transitioned back to the 2D state (3D-2D group) and re-induced following transition to 3D (3D-2D-3D group) (Fig 1F). Similarly, genes downregulated in 3D state exhibited increased expression following transition to the 2D state (3D-2D group) and silencing upon transitioning back to 3D (3D-2D-3D group) (Fig 1G). To adjust for potential temporal changes in gene expression, the gene expression pattern of the 3D-2D group was also compared to that of 2D and 3D fibroblasts cultured for 5 days. A cluster analysis demonstrated distinct clustering of 2D and 3D states (Online Fig IA). In addition, for the 3D-2D-3D group, temporally adjusted controls of 3D-2D cells lifted and reseeded back onto 2D instead of 3D conditions was also used (Online Fig IB). Again, cluster analysis demonstrated distinct 2D and 3D states making it unlikely that differential gene expression was simply secondary to temporal dependent changes in gene expression of cardiac fibroblasts in culture.
Figure 1. Cardiac fibroblasts exhibit dynamic changes in gene expression in different topological states.
(A) Schematic of how fibroblasts were transitioned from a 2D to 3D state and then back to 2D and 3D respectively. For each topological state, fibroblasts were harvested for RNA-seq. (B) Bright phase image of cardiac fibroblasts in 2D and 3D (Scale bar: 50μm) (C) Pure population of genetically labeled (tdTomato) fibroblasts isolated by flow cytometry from hearts of TCF21MerCreMer:R26Rtdtomato or Col1a2CreERT:R26Rtdtomato mice were subjected to sphere formation (3D) and spheres stained with wheat germ agglutinin (WGA), that stains cell membranes (Scale bar: 20μm). (D) Cardiac fibroblasts in 2D or 3D states dissociated and subjected to image flow cytometry showing representative image of fibroblast from 2D or 3D state (3000 cells imaged in each group, scale bar:10μm) and corresponding mean diameter and surface area of fibroblasts in 2D or 3D states (*p<0.001, mean ± S.E.M., n=3) (E) Heat map demonstrating clustering of sample correlations of fibroblasts (shown by Z scores) in different topological states (F,G) Heat map comparing (F) expression of the most upregulated 3D genes in different topological states and (G) 3D downregulated genes in different topological states (H) GO analysis showing cellular pathways most affected by genes upregulated or downregulated in 2D/3D states.
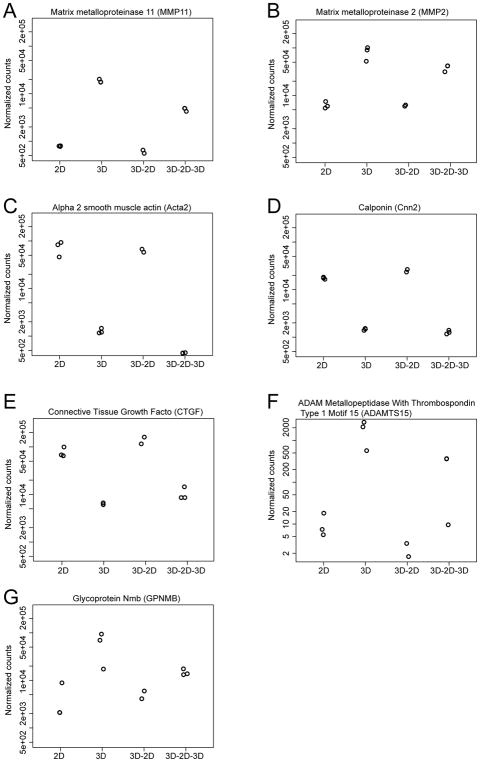
We next examined whether dynamic changes in gene expression in different topological states can be simply explained by sudden changes in substrate stiffness as the fibroblasts transition from a 2D adherent state to a 3D spherical non-adherent state. To answer this question, we seeded cardiac fibroblasts onto tissue culture plates coated with biocompatible silicone controlled elastic moduli recapitulating environments similar to tissue[12]. We seeded cardiac fibroblasts on tissue culture plates with stiffness of 0.8kpA, 8kPa and 64kPa (Online Fig IIA) and following 24 hours of seeding, harvested the cells to compare changes in gene expression to that of 2D and 3D topological states. Analysis of global gene expression demonstrated a clustering of 2D states with that of cells seeded at different substrate stiffness (0.5, 8, 64 kpA) and were distinct from gene expression signature of cardiac fibroblasts in 3D states (Online Fig IIB). We specifically examined the set of genes that displayed the highest degree of differential expression between 2D and 3D states and observed that the expression pattern of such genes was similar between cells seeded at 0.8,8 and 64kPA and 2Dstates and distinct from that seen in 3D states (Online Fig IIC, D). Taken together, these observations suggest that topological changes in cardiac fibroblasts drive gene expression patterns and changes in substrate stiffness are unlikely to underlie differences in gene expression between 2D and 3D states. We next examined the pool of genes that were the most upregulated (Fig 1F) or downregulated (Fig 1G) in 3D versus 2D fibroblast states. Gene ontology (GO) analysis demonstrated that genes downregulated in the 3D state mainly comprised cell cycle processes such as DNA replication, chromosomal condensation/segregation and cytokinesis (Fig 1H). Transcripts differentially upregulated in the 3D state involved pathways regulating extracellular matrix metabolism/proteolysis, surface proteins, chemotaxis and immune response. (Fig 1H, Online Table II). We next specifically examined several genes which were highly differentially expressed between 3D versus 2D fibroblasts and that are also known to regulate extracellular matrix such as metalloproteinases/metallopeptidases, [Metalloproteinase (MMP11, MMP2), ADAMTS15 (metallopeptidase with thrombospondin motif 15)], connective tissue growth factor (CTGF) and fibroblast contractility, alpha smooth muscle actin 2 (Acta2), Calponin (Cnn2) and modulators of inflammatory response (Glycoprotein non metastatic b, GPNMB). Based on RNA-seq patterns, expression of these genes was reversible and highly dependent upon the topological state of the fibroblast (Fig 2). For instance, MMP11 and MMP2 were highly induced following aggregation and sphere formation of fibroblasts but expressions declined to 2D levels when the 3D fibroblasts were allowed to attach and grow out as a monolayer for a few days (Fig 2A, B). However, reseeding the cells back to a 3D conformation led to rapid re-induction of MMP2/MMP11 expression illustrating the dynamic plasticity of the system (Fig 2A, B). Gene expression of Acta2, Cnn2, ADAMTS15, GPNMB and CTGF, which are thought to play a role in fibroblast contractility and regulation of inflammation and extracellular matrix, demonstrated similar patterns of changes of gene expression dependent upon the topological state (Fig 2C–G).
Figure 2. Dynamic changes in expression of myofibroblast and extracellular matrix genes between 2D and 3D cardiac fibroblast states.
(A, B) normalized gene counts on RNA-seq demonstrating rapid changes in gene expression of (A) MMP11 (B) MMP2, (C) Acta2 (D) Calponin (E) Connective tissue growth factor (CTGF) (F) ADAMTS15 and (G) GPNMB in cardiac fibroblasts in different topological states.
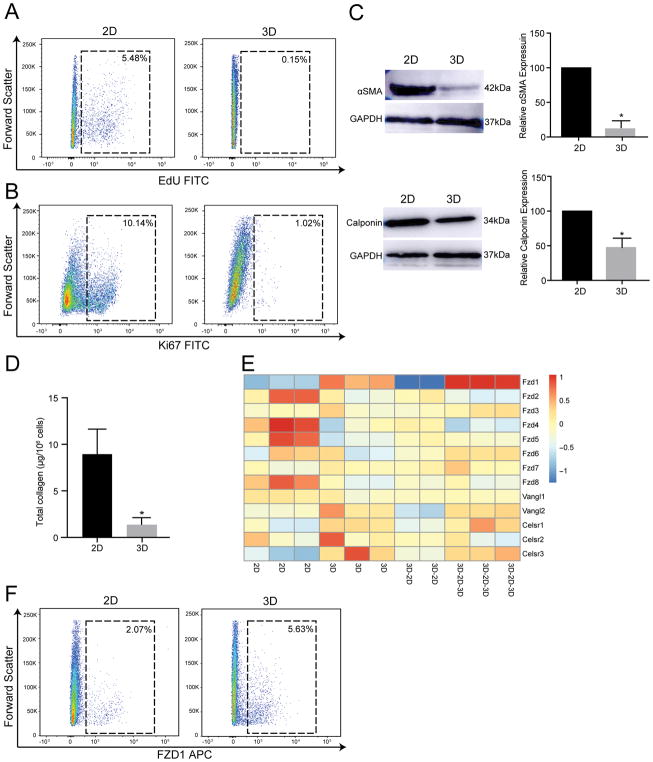
To determine whether such gene expression changes are associated with changes in phenotype, we first determined changes in cardiac fibroblast proliferation in the 3D versus 2D state. For this purpose, cardiac fibroblasts either in the 2D or 3D state were treated with EdU for 4 hours followed by determination of EdU uptake by flow cytometry. Consistent with decreased expression of cell cycle genes in the 3D state, we observed that 5.47±1.4% of cardiac fibroblasts in the 2D state were cycling (EdU uptake) versus 0.15±0.05% in the 3D state (p<0.05) (Fig 3A). Similarly, the fraction of cells expressing Ki67 (marker of proliferation) significantly decreased from 10.94±3.0% of cardiac fibroblasts in the 2D state to 1.0±0.08% in the 3D state (p<0.05) (Fig 3B). Western blotting with quantitative densitometry demonstrated that fibroblasts in the 3D state exhibit decreased expression of contractile proteins alpha smooth muscle actin (88±6% decrease in 3D versus 2D, p<0.001) and calponin (54±6% decrease in 3D versus 2D; p<0.001) (Fig 3C), consistent with gene expression data demonstrating decreased expression of myofibroblast proteins. Differentially expressed genes between the 2D and 3D states included genes affecting extracellular matrix catabolism. Collagen is the most common abundant extracellular matrix protein secreted by cardiac fibroblasts and we next determined how adoption of the 3D state affects collagen production. We measured total collagen using the Sircoll assay in 2D and 3D fibroblasts and observed that the total cellular collagen content significantly decreased from 8.40±2.8ug/106 cells in 3D states to 1.32±0.71/106 cells in 2D states (p<0.05) (Fig 3D). Cardiac fibroblasts secrete extracellular matrix proteins but are also known to express matrix degrading enzymes and can undergo de-differentiation as well[13]. These data suggest that a transition from a 2D to a 3D state leads to a switch of cardiac fibroblast phenotype from a matrix synthetic to a non-synthetic de-differentiated state. Recent evidence suggests that aggregation of cardiac fibroblasts in the area of myocardial injury is associated with fibroblasts exhibiting evidence of polarization[14]. Polarization or alignment of cardiac fibroblasts is thought to play a critical role in appropriate cardiac wound healing[15]. We thus examined whether 3D cardiac fibroblasts exhibited any evidence of polarization compared to 2D fibroblasts. To address this question, we examined expression of genes that are members of the Frizzled (Fzd), Van Gogh (Vangl in vertebrates) and Flamingo (Celsr in vertebrates) (Fig 3E). These families of genes initially identified in drosophila are now known to play critical role in planar cell polarity and cellular orientation in epithelial and mesenchymal cells of vertebrates as well[16]. Within this subset of genes known to regulate cellular polarity, we observed that Frizzled 1 (Fzd1) expression was significantly higher in 3D compared to 2D states (Fig 3E). Fzd1 is a cell surface receptor and we performed flow cytometry to demonstrate that Fzd 1 expression was significantly up-regulated in 3D fibroblasts (Fig 3F) consistent with gene expression changes. Members of the frizzled family are known to be expressed in fibroblasts in the area of injury following myocardial injury and thought to contribute to cardiac remodeling and have been considered as therapeutic targets for augmenting cardiac repair[14, 17, 18]. In this regard, cardiac fibroblasts in 3D states recapitulate to a certain extent the expression of polarity genes known to be important for wound healing in vivo. Taken together, these observations demonstrate that aggregation and changes in spatial arrangement of cardiac fibroblasts can drive rapid, dynamic and reversible expression of genes affecting a panoply of processes regulating wound healing such as fibroblast proliferation, activation, collagen content and cell polarity.
Figure 3. Changes in fibroblast phenotype in 3D versus 2D topological state.
(A, B) Flow cytometry to determine fraction of proliferating fibroblasts in 2D and 3D states by (A) EdU uptake (5.48±1.4% in 2D versus 0.15±0.05% in 3D, mean ± S.E.M, p<0.05, n=3) or (B) Ki67 expression (10.14±3.0% in 2D versus 1.02±0.01% in 3D, mean ± S.E.M, p<0.05, n=3). (C) Western blotting and quantitative densitometry of expression of Alpha smooth muscle actin and calponin expression by cardiac fibroblasts in 2D or 3D states (mean ± S.E.M, *p<0.001, n=3). (D) Estimation of total collagen content of cardiac fibroblasts in 2D or 3D state (8.40±2.8μg/106 cells in 3D versus 1.32±0.71/106 cells in 2D, mean ± S.E.M, *p<0.05, n=3). (E) Heat map demonstrating expression of members of the Frizzled, Vangl and Celsr family in different topological states of cardiac fibroblasts. (F) Flow cytometry demonstrating Fzd1 expression in 3D versus 2D cardiac fibroblasts (2.07±0.33% in 2D versus 5.63±0.24% in 3D, mean ± S.E.M, p<0.05, n=3).
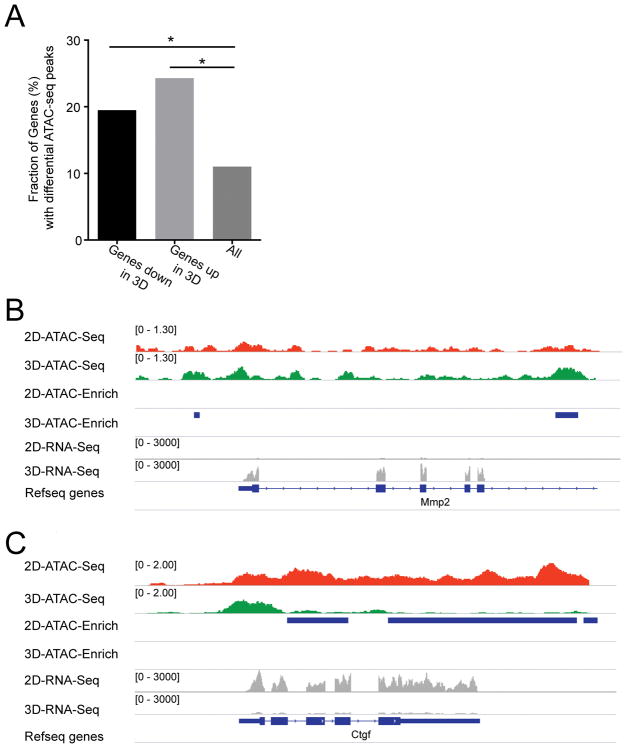
We next investigated the mechanistic basis of such rapid and reversible changes in gene expression. We hypothesized that dynamic changes in chromatin structure may contribute at least in part to the rapid changes in gene expression seen following the transition of fibroblasts from a 2D to 3D state. Therefore, changes in chromatin organization and DNA accessibility (open and closed chromatin) were examined between cardiac fibroblasts in 2D versus 3D states by performing an assay for transposase accessible chromatin (ATAC-seq)[19]. ATAC-seq enables identification of open and closed regions of chromatin across the genome and provides insights about regions of the genome that are more (open) or less accessible (closed) to transcription factors[19]. We observed that there were significant changes in global chromatin organization (Fig 4A). Approximately 23% of the genes differentially upregulated in fibroblast 3D states and 18% of the genes downregulated in fibroblast 3D state (i.e. upregulated in 2D states) underwent significant changes in chromatin accessibility (Fig 4A) with remarkable concordance with their RNA-seq profiles. Both these values were significantly enriched over background levels as we observed that only 10% of all genes had differential ATAC-seq peaks upon transition from a 2D to 3D state (Fig 4A). We next examined differential ATAC-seq peaks for specific genes such as MMP2 and CTGF that demonstrated significant induction and silencing of gene expression respectively in the 3D state and observed significant differences in ATAC-seq peaks in their respective genomic loci, correlating with changes in gene expression (Fig 4B, C). These observations suggest that fibroblast aggregation and changes in spatial arrangement of cardiac fibroblasts are sufficient to induce changes in chromatin structure or organization that contributes to the global changes in genes expression between the 2D and 3D state.
Figure 4. Chromatin changes underlie altered gene expression of fibroblasts in 3D versus 2D states.
(A) ATAC seq performed to demonstrate fraction of genes demonstrating differential ATAC-seq peaks in either 2D or 3D cardiac fibroblast states (B, C) ATAC-seq peaks and RNA-seq showing expression of (B) MMP2 and (C) CTGF in 2D and 3D states demonstrating differential ATAC-seq peaks in loci of MMP2 and CTGF genes (numbers listed refer to scales of enrichment).
Having demonstrated that changes in fibroblast aggregation and spatial arrangement are associated with concordant changes in the epigenome and gene expression, we next investigated the functional connotations of such global changes in gene expression for cardiac wound healing. Like humans, genetically diverse strains of mice differ in the degree of fibrosis or cardiac remodeling following pathological cardiac stressors and offer the advantage of tissue availability and experimental manipulation. The Hybrid Mouse Diversity Panel (HMDP) is a collection of genetically diverse mouse strains and allows sufficient power for genome wide association analysis to determine how genetic architecture impacts phenotypic traits [20–23]. A single pathologic stressor can be thus applied across all strains within the HMDP to perform genome wide association studies and determine how genetic and environmental interactions contribute to global gene expression and clinical phenotypes[24]. In these studies, 96 strains of mice were administered a three-week continuous infusion of isoproterenol via osmotic pump[6]. Throughout the study, various physiological characteristics including cardiac functional indices (e.g. ejection fraction, left ventricular internal dimensions in end systole and diastole), metabolic parameters and tissue weights (61 traits in all) were measured (Online Table III) and the left ventricle of each mouse strain was subjected to global expression arrays[6, 24]. In this study, the mice responded dramatically to isoproterenol, as nearly every individual showed increased left ventricular mass following treatment. This dataset enabled us to assess whether differentially expressed genes between 3D and 2D states of cardiac fibroblasts could inform phenotypic traits known to predict outcomes or disease severity in isoproterenol induced cardiac hypertrophy and failure.
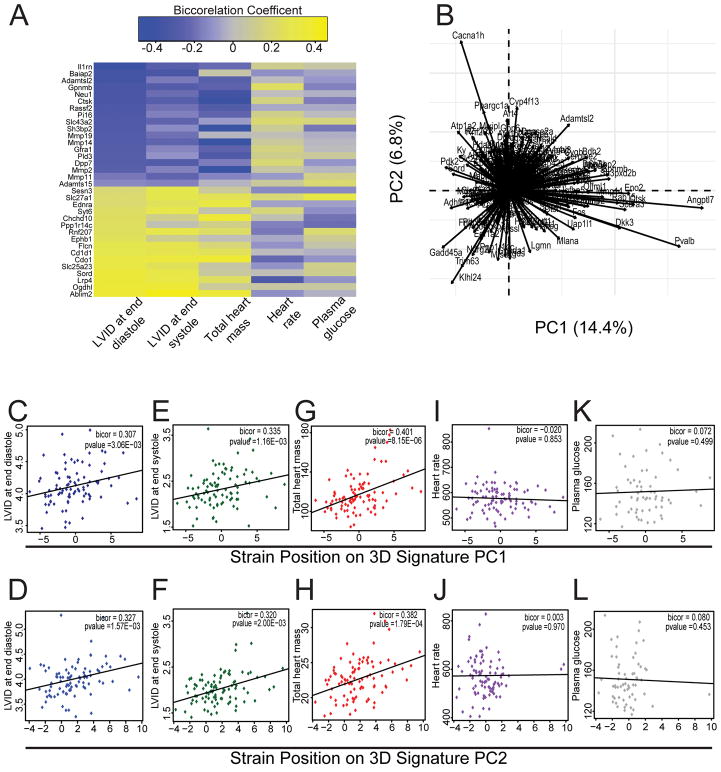
Initially we asked whether significantly upregulated transcripts in all 3D fibroblast states (compared to 2D) were correlated with heart failure traits in the HMDP. By simply correlating individual 3D up-regulated genes from our RNA-sequencing experiment across clinical traits in the mouse population (Online Fig III), we observed striking patterns of significance (Fig 5A). Since these patterns are difficult to interpret on a gene by-gene basis, we used a data reduction method to establish vectors which represent 3D specific gene signatures. Principle component (PC) approaches provide a means of data-reduction whereby variation across any number of dimensions can be aggregated into single or multiple vectors. Similar approaches utilizing a principle component to represent large gene sets are commonly utilized in population-based studies [6, 25]. These produce a series of vectors which represent a given pattern of variation, referred to as eigenvectors. Here, we applied this approach to gene expression, where the genes identified from the 2D vs 3D analysis were analyzed across a mouse population. We generated principle component (PC) eigengenes which captured 14.4% (PC1) and 6.8% (PC2) of the variation of all 3D-upregulated transcripts within the HMDP expression arrays (Fig 5B). It is worth mentioning that these values are fairly typical when performing principle component analysis on population-wide data (here, we use ~600 genes within ~100 strains of mice), especially given the significant variation observed in gene expression profiles. Using these eigengenes (PC1 & PC2) as signatures of 3D fibroblast genes, we plotted the position of each strain against various cardiac and non-cardiac clinical traits. Cardiac fibroblasts are known to affect cardiac hypertrophy and play a major role in adverse cardiac remodeling and dilatation of the cardiac chambers, clinically determined by the left ventricular dimensions in end systole and diastole. Consistent with this notion, we observed highly significant positive correlations between 3D fibroblast derived gene signatures and left ventricular dimensions in both end diastole (Fig 5C, D) and end systole (Fig 5E, F) as well as cardiac mass (Fig 5G, H). Notably these 3D fibroblast eigengene signatures did not correlate with either heart rate (Fig 5I, J) or non-cardiac traits such as plasma glucose (Fig 5K, L) demonstrating specificity of these eigengene signatures to cardiac remodeling traits. Collectively, these data show that 3D fibroblast enriched transcripts show striking patterns of correlation with adverse cardiac indices such as cardiac hypertrophy and chamber dilatation across the murine population following isoproterenol infusion.
Figure 5. Genes enriched in 3D fibroblast states show significant correlation with indices of adverse ventricular modeling in HMDP studies following isoproterenol infusion.
(A) Correlation heat map (yellow: positive and blue: negative correlation) of top 15 differentially upregulated genes in 3D/2D states versus clinical traits of left ventricular dimensions, heart mass, plasma glucose and heart rate following infusion of isoproterenol (B) Individual gene contribution to eigengene signatures PC1 and PC2 using transcripts enriched in 3D states. (C–H) Correlation of both eigengene signatures against cardiac and non-cardiac traits with significant correlation between both eigengenes and (C, D) LVID at end diastole (E, F) LVID at end systole and (G, H) total heart mass with no significant correlation between either eigengene and (I–J) heart rate and (K, L) plasma glucose (LVID: left ventricular internal diameter; bicor: bicorrelation coefficient).
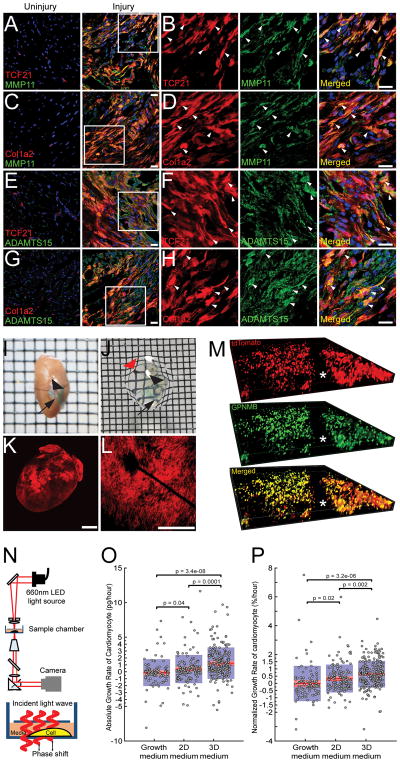
So far, our data demonstrates that cardiac fibroblasts exhibit a high degree of dynamic plasticity with induction and silencing of genes following transition from a 2D to 3D state. Genes induced in the 3D state significantly correlated with clinical indices of adverse ventricular remodeling. Therefore, we next determined whether genes differentially expressed in the 3D state were also upregulated in regions of fibroblast aggregation in vivo at the time of wound healing. For this purpose, we performed cryo-injury on hearts of Col1a2CreERT:R26Rtdtomato and TCF21MerCreMer:R26Rtdtomato mice following tamoxifen mediated labeling of cardiac fibroblasts. Tamoxifen was administered for 10 days to label the cardiac fibroblasts and stopped 5 days prior to cryo-injury. We chose cryo-injury as cryo-injury unlike ischemic myocardial injury creates a highly well-defined compact transmural scar on the left ventricle and the tdTomato labeling of cardiac fibroblasts can easily identify regions of compact scarring. Hearts were harvested at 7 days following cryo-injury and immunofluorescent staining performed to determine whether genes highly upregulated in 3D fibroblasts in vitro were expressed by labeled cardiac fibroblasts or expressed in abundance in the region of fibroblast aggregation. We observed abundant expression of MMP11 by tdTomato labeled cardiac fibroblasts but minimal MMP11 expression in uninjured regions (Fig 6A–D). ADAMTS15, a secreted protein that regulates extracellular matrix, is expressed in the developing heart and highly induced in the 3D fibroblast state (Supplementary Table 1), was also found to be abundantly present in the injured region and expressed by tdTomato labeled fibroblasts (Fig 6E–H). To study the expression of 3D enriched transcripts in aggregating fibroblasts in regions of injury in greater detail, we subjected the harvested heart to solvent based tissue clearing techniques to make the heart optically transparent[7]. This allows the entire 3D structure of the scar to be visualized in detail without having to extrapolate and reconstruct a 3D structure from conventional analysis of histological sections. We again performed cryo-injury on Col1a2CreERT:R26Rtdtomato mice following fibroblast labeling. We harvested the heart 7 days after cryo-injury and made them optically transparent and a non-absorbable suture (placed at the time of injury) was used to identify area of cryo injury in the heart after tissue clearing (Fig 6I–K). The region of injury could be identified easily as an area with accumulation of tdTomato labeled cardiac fibroblasts (Fig 6L). On the optically cleared heart, we performed immunostaining for another marker GPNMB, a gene upregulated in the 3D state, involved with immune response pathways and that strongly correlated with adverse cardiac remodeling indices in our murine model of isoproterenol induced heart failure. Analysis of Z stacked confocal images taken sequentially through the whole depth of the scar demonstrated expression of GPNMB by tdTomato labeled fibroblasts throughout the depth of the scar (Fig 6M). These observations demonstrate that genes expressed by aggregating fibroblasts in the region of injury at least partially recapitulate the gene expression signatures of 3D fibroblasts.
Figure 6. Genes enriched in 3D fibroblasts are expressed in vivo in regions of fibroblast aggregation after heart injury and affect cardiomyocyte hypertrophy.
(A–D) Immunofluorescent staining for MMP11 on uninjured and cryo-injured hearts of (A, B) TCF21MerCreMer:R26Rtdtomato and (C, D) Col1a2CreERT:R26Rtdtomato mice (B, D) area of injury shown in higher magnification demonstrating tdTomato labeled fibroblasts expressing MMP11 (arrows). (E–H) Immunofluorescent staining for ADAMTS15 on uninjured and cryo-injured hearts of (E, F) TCF21MerCreMer:R26Rtdtomato and (G, H) area of injury shown in higher magnification demonstrating tdTomato labeled fibroblasts expressing ADAMTS15 (arrows) (Scale bars: 20μm). (I, J) Cryo-injured heart of Col1a2CreERT:R26Rtdtomato mouse (I) prior to and (J) following optical clearing (arrowhead points to suture for identifying injured region, arrow points to green dye to identify area adjacent to injury; note the wire mesh on which the heart lies is now visible through the transparent heart; red arrow) (K) tdTomato fluorescence observed on cryo-injured Col1a2CreERT:R26Rtdtomato heart and (L) confocal image through area of injury showing intense tdTomato fluorescence (Scale bar: 500μm) (M) Immunofluorescent staining for GPNMB on optically cleared Col1a2CreERT:R26Rtdtomato heart after injury. The entire depth of the scar was imaged with a confocal microscope and sequential Z stack images demonstrating distribution of tdTomato (red), GPNMB (green) and merged (yellow) image demonstrating distribution of fluorophores across the depth of the scar (asterisk corresponds to position of suture). (N) Set up of live cell interferometry with phase shift of light being a read out for changes in cell biomass (O) Absolute and (P) normalized single cell cardiomyocyte (NRVM) biomass accumulation rates determined by serial measurements with interference microscopy over 48 hours following treatment of NRVM with growth medium (non-conditioned) or conditioned medium from fibroblasts in 2D or 3D state (each circle represents a single cardiomyocyte; Number of single cardiomyocytes tracked: 103 for growth medium, 142 for 2D medium and 231 for 3D medium).
Fibroblasts are known to affect cardiac hypertrophy[26] and the gene expression signatures of 3D fibroblasts strongly correlated with clinical indices of heart mass and remodeling across mouse strains. We next investigated whether fibroblasts in 3D exert pro-hypertrophic effects on cardiomyocytes compared to fibroblasts cultured in 2D. For this purpose, we collected conditioned medium from fibroblasts grown in 3D or 2D conditions for 24 hours. We treated neonatal rat cardiomyocytes with 3D or 2D conditioned medium to determine effects on cardiomyocyte hypertrophy over the next 48 hours. Live cell interferometry (LCI), a validated version of quantitative phase microscopy [27, 28] is an extremely sensitive tool for determining changes in total cellular biomass. LCI is based on the principle that light slows as it interacts with matter. As light traverses through a cell that has greater biomass (i.e. hypertrophied), the light slows and its waveform shifts in phase compared to light not passing through the cell [29](Fig 6N). The change in phase shift over time is directly related to the change in biomass of the cell over time and this quantitative phase shift has been used to precisely and reproducibly determine the dry biomass of cells including T cells, stem cells, cancer cells and fibroblasts [27, 28]. Neonatal rat ventricular cardiomyocytes (NRVM) were treated with conditioned medium as above and each cardiomyocyte was subjected to repeated measurements by LCI to obtain a growth rate. We observed that 2D conditioned medium significantly increased the rate of cardiomyocyte biomass accumulation compared to non-conditioned medium (0.4 picogram/hr for 2D compared to −0.12 picogram/hour for growth medium, p=0.04) (Fig 6O). However, treatment with 3D conditioned medium tripled the rate of growth versus treatment with 2D conditioned medium (1.25 pg/hr for 3D versus 0.4 pg/hr for 2D, p=0.0001) (Fig 6O). As cardiomyocytes after isolation can exhibit significant difference in cell size, we normalized the growth rate of each cardiomyocyte to initial cell biomass. Again, we observed a significant 34% increase in cell biomass of NRVM following treatment with 2D conditioned medium compared to ‘non-conditioned’ growth medium (0.34% for 2D vs −0.01% for growth medium, p=0.02) (Fig 6P). However, 3D fibroblast conditioned medium significantly increased the cell biomass accumulation rate of NRVM by a further 88% compared to NRVM treated with 2D conditioned medium (0.64% for 3D versus 0.34% for 2D, p=0.002) (Fig 6P). These observations demonstrate that the secretome of fibroblasts in 3D is sufficient to induce cardiomyocyte hypertrophy and are broadly consistent with the genome-wide association data shown earlier demonstrating high correlation between genes induced in the 3D state and indices of cardiac hypertrophy and remodeling after isoproterenol infusion. Our observations also suggest that the gene expression signatures adopted by aggregating fibroblasts may have a direct causal effect on a hypertrophic response after cardiac injury.
We next analyzed our RNA-seq data to obtain insight into transcription factors or transcriptional regulators that could be contributing to changes in gene expression between the 2D and 3D states and affecting myocyte hypertrophy. Genes differentially upregulated in the 3D versus 2D state were assayed for enrichment of upstream transcriptional factors or regulators using TRRUSTv2[30]. This analysis queries hundreds of published Chip-Seq and/or open chromatin data to infer regulatory elements from gene expression patterns. The 3D upregulated genes were used to identify enrichment of regulation by specific transcription factors or DNA binding elements known to regulate expression. We observed significant representation of several transcription factors predicted to regulate 3D-specific genes (Online Figure IV) and some of these are also known to regulate or be associated with the cardiac hypertrophic response such as Microphthalmia associated transcription factor (MITF), Beta catenin (CTNNB1) and serum response factor (SRF)[31–33]. Next, to obtain insight into secreted factors present in 3D conditioned medium that induced or contributed to myocyte hypertrophy, we filtered the differentially upregulated genes in the 3D state for secreted factors and observed expression of proteins known to affect the myocyte hypertrophic response such as angiotensinogen, pyrophosphatases affecting purinergic signaling (ENPP3) and members of the Wnt signaling family (Dkk3) (Online Table IV)[34–36].
DISCUSSION
Cardiac fibroblasts are known to be highly plastic and our study suggests that simple aggregation of fibroblasts may be sufficient to induce genome wide changes in chromatin reorganization and gene expression. We show that gene expression signatures adopted by aggregating cardiac fibroblasts at least in part recapitulates changes in gene expression in the injured region in vivo and that such altered genetic outputs may have functional consequences for cardiac wound healing and remodeling. Cardiac fibroblasts are the principal contributors towards deposition of extracellular matrix but are also known to secrete metalloproteinases and extracellular proteases that leads to degradation of extracellular matrix[37]. Acute myocardial injury is associated with significant upregulation in metalloproteinase activity[38] and MMP expression significantly increased in 3D cardiac fibroblasts mirroring such in vivo changes. A balance between the synthetic and proteolytic phenotype of the fibroblasts determines extracellular matrix content or burden of scar tissue in pathologic states. Augmented matrix synthetic and matrix degrading properties of cardiac fibroblasts can lead to high turnover of extracellular matrix, as seen in heart failure. Such fibroblast phenotypes with opposing effects on matrix synthesis and degradation, as seen in our 2D and 3D model could determine the burden of scar after acute and chronic injury and serve as a model for obtaining further mechanistic insight [39]. Although little is known about signaling mechanisms that regulate resolution of fibrosis, de-differentiation of contractile elements of fibroblasts with decreased expression of alpha smooth muscle actin is thought to represent a key event for fibrosis resolution[40]. In this regard, our model of fibroblast aggregation with decreased expression of smooth muscle actin and induction of various matrix degrading enzymes demonstrates phenotypic features consistent with myofibroblast de-differentiation and a proteolytic rather than a synthetic phenotype. The expression of alpha smooth muscle actin and other contractile proteins in fibroblasts in the injury region allows for wound contraction in vivo, a mechanism that enables reduction in the area of injury. Conversely, impaired expression of fibroblast contractile proteins or defects in fibroblast polarization in vivo can cause impaired wound contraction, dysregulated wound healing and lead to expansion of the infarcted region, a dreaded complication after myocardial infarction. Our model that demonstrates a rapid and reversible expression of contractile proteins in fibroblasts could serve as a platform for investigating the molecular events that abruptly can switch a cardiac fibroblast from a synthetic and contractile phenotype to a proteolytic and de-differentiated phenotype. Hypertrophy of surviving cardiac myocytes at the edges of the injured region occurs after myocardial infarction and the 3D fibroblasts can serve as a platform for interrogating the paracrine effects of cardiac fibroblasts on myocyte hypertrophy. Given the global changes in gene expression and substantial changes in the 3D cardiac fibroblast secretome, it is likely that rather than a single driver, activity of multiple transcription factors and secreted proteins synergistically affect gene expression changes and the myocyte hypertrophic response.
Study of cells in spheroids have been performed for cancer cells and cells with progenitor potential. Our study suggests that studying fibroblasts in a 3D state in contrast to conventional analysis of 2D fibroblasts may be more informative of cellular changes in the injury region in vivo. Potentially, our model could also be used as a tool or a primary screening system to determine how drugs or small molecules affect changes in expression of specific genes that are upregulated in the 3D state or affect phenotypic transitions between matrix synthetic (2D) and matrix degrading (3D) states of a cardiac fibroblast.
Supplementary Material
Novelty and Significance.
What Is Known?
Unlike cardiac myocytes, cardiac fibroblasts do not form a syncytium but reside in the interstitium among myocytes.
This topological relationship is altered after heart injury when fibroblasts are recruited to and aggregate at the area of injury.
Aggregation of fibroblasts after injury thus represents a unique event in the life cycle of the cardiac fibroblast but whether such topological rearrangement affects fibroblast function is not clear
What New Information Does This Article Contribute?
Aggregation of cardiac fibroblasts leads to global changes in gene expression and chromatin reorganization.
Changes in the transcriptome are reversible upon aggregation, disaggregation and reaggregation of cardiac fibroblasts.
Genes induced by fibroblast aggregation are expressed in the injured heart and correlate with poor cardiac outcomes in mouse models of hypertrophy and heart failure.
The secretome of aggregated cardiac fibroblasts can induce hypertrophy of cardiac myocytes.
Cardiac fibroblasts reside in the interstitium of the heart and do not form a syncytium. Following injury, they however are recruited to aggregate in the area of injury, but the physiological significance of fibroblast aggregation remains unknown. Here, we demonstrate that simple aggregation of cardiac fibroblasts induces wide spread changes in gene expression and chromatic reorganization. Such transcriptional changes are reversible when cardiac fibroblasts are disaggregated or subsequently reaggregated. Genes upregulated in the aggregated state are expressed in the region of injury and correlate with indices of adverse cardiac remodeling in murine models of cardiac hypertrophy and failure. Finally, we demonstrate that the secretome of aggregated cardiac fibroblasts induces hypertrophy of cardiac myocytes. Taken together these observations demonstrate that topological changes in the spatial organization of cardiac fibroblasts drives chromatin reorganization, gene expression patterns and has functional consequences for cardiac wound healing.
Acknowledgments
We thank the UCLA Heart lab cell core for providing freshly isolated cardiomyocytes and the UCLA Clinical Microarray Core for RNA-sequencing. Imaging flow cytometry was performed in the UCLA Jonsson Comprehensive Cancer Center and Center for AIDS Research Flow Cytometry Core Facility. We thank Dr. Eric Olson, University of Texas Southwestern Medical Center and Dr. Andrew Leask, University of Western Ontario Canada for sharing the TCF21MerCreMer and Col1a2CreERT mice.
SOURCES OF FUNDING
The project was supported by grants from the NIH (HL129178, HL137241 to AD, CA185189, GM073981, GM114188 to MT, HL30568 and HL123295 to AJL, Department of Defense (PR152219, PR161247 to AD), Air Force Office of Scientific Research (FA9550-15-1-0406 to MT), California Institute of Regenerative Medicine (DISC1-08790 to AD), research award from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research & Rose Hills Foundation to AD and planning award from the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research and California Nanosystems Institute at UCLA to AD and DC. The project also received support from NIH/NCATS UCLA CTSI (ULTR00024). Imaging flow cytometry at the UCLA Jonsson Comprehensive Cancer Center is supported by NIH awards P30 CA016042 and 5P30 AI028697.
Nonstandard Abbreviations and Acronyms
- Acta2
alpha smooth muscle actin 2
- ADAMTS15
metallopeptidase with thrombospondin motif 15
- ATAC
assay for transposase accessible chromatin
- Cnn2
Calponin 2
- CTGF
connective tissue growth factor
- EMT
epithelial-mesenchymal-transition
- GO
Gene ontology
- GPNMB
Glycoprotein non metastatic b
- HMDP
Hybrid Mouse Diversity Panel
- LCI
Live cell interferometry
- MMP
Matrix Metalloproteinase
- NRVM
Neonatal rat ventricular cardiomyocytes
- PC
principle component
- WGA
wheat germ agglutinin
Footnotes
DISCLOSURES
None.
References
- 1.Manner J, Perez-Pomares JM, Macias D, Munoz-Chapuli R. The origin, formation and developmental significance of the epicardium: a review. Cells Tissues Organs. 2001;169:89–103. doi: 10.1159/000047867. [DOI] [PubMed] [Google Scholar]
- 2.Christia P, Bujak M, Gonzalez-Quesada C, Chen W, Dobaczewski M, Reddy A, Frangogiannis NG. Systematic characterization of myocardial inflammation, repair, and remodeling in a mouse model of reperfused myocardial infarction. J Histochem Cytochem. 2013;61:555–70. doi: 10.1369/0022155413493912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camelliti P, Borg TK, Kohl P. Structural and functional characterisation of cardiac fibroblasts. Cardiovasc Res. 2005;65:40–51. doi: 10.1016/j.cardiores.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 4.Shoval H, Karsch-Bluman A, Brill-Karniely Y, Stern T, Zamir G, Hubert A, Benny O. Tumor cells and their crosstalk with endothelial cells in 3D spheroids. Scientific Reports. 2017;7:10428. doi: 10.1038/s41598-017-10699-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pillai IC, Li S, Romay M, Lam L, Lu Y, Huang J, Dillard N, Zemanova M, Rubbi L, Wang Y, Lee J, Xia M, Liang O, Xie YH, Pellegrini M, Lusis AJ, Deb A. Cardiac Fibroblasts Adopt Osteogenic Fates and Can Be Targeted to Attenuate Pathological Heart Calcification. Cell stem cell. 2017;20:218–232e5. doi: 10.1016/j.stem.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rau CD, Romay MC, Tuteryan M, Wang JJ, Santolini M, Ren S, Karma A, Weiss JN, Wang Y, Lusis AJ. Systems Genetics Approach Identifies Gene Pathways and Adamts2 as Drivers of Isoproterenol-Induced Cardiac Hypertrophy and Cardiomyopathy in Mice. Cell Syst. 2017;4:121–128e4. doi: 10.1016/j.cels.2016.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung K, Ding Y, Ma J, Chen H, Huang V, Cheng M, Yang CF, Kim JT, Eguchi D, Di Carlo D, Hsiai TK, Nakano A, Kulkarni RP. Simplified three-dimensional tissue clearing and incorporation of colorimetric phenotyping. Sci Rep. 2016;6:30736. doi: 10.1038/srep30736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang YJ, Bailey JM, Rovira M, Leach SD. Sphere-forming assays for assessment of benign and malignant pancreatic stem cells. Methods Mol Biol. 2013;980:281–90. doi: 10.1007/978-1-62703-287-2_15. [DOI] [PubMed] [Google Scholar]
- 9.Ubil E, Duan J, Pillai IC, Rosa-Garrido M, Wu Y, Bargiacchi F, Lu Y, Stanbouly S, Huang J, Rojas M, Vondriska TM, Stefani E, Deb A. Mesenchymal-endothelial transition contributes to cardiac neovascularization. Nature. 2014;514:585–90. doi: 10.1038/nature13839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kanisicak O, Khalil H, Ivey MJ, Karch J, Maliken BD, Correll RN, Brody MJ, SCJL, Aronow BJ, Tallquist MD, Molkentin JD. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat Commun. 2016;7:12260. doi: 10.1038/ncomms12260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Basiji D, O’Gorman MR. Imaging flow cytometry. J Immunol Methods. 2015;423:1–2. doi: 10.1016/j.jim.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Gutierrez E, Groisman A. Measurements of elastic moduli of silicone gel substrates with a microfluidic device. PLoS One. 2011;6:e25534. doi: 10.1371/journal.pone.0025534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baudino TA, Carver W, Giles W, Borg TK. Cardiac fibroblasts: friend or foe? Am J Physiol Heart Circ Physiol. 2006;291:H1015–26. doi: 10.1152/ajpheart.00023.2006. [DOI] [PubMed] [Google Scholar]
- 14.Blankesteijn WM, Essers-Janssen YP, Verluyten MJ, Daemen MJ, Smits JF. A homologue of Drosophila tissue polarity gene frizzled is expressed in migrating myofibroblasts in the infarcted rat heart. Nat Med. 1997;3:541–4. doi: 10.1038/nm0597-541. [DOI] [PubMed] [Google Scholar]
- 15.Kong P, Shinde AV, Su Y, Russo I, Chen B, Saxena A, Conway SJ, Graff JM, Frangogiannis NG. Opposing Actions of Fibroblast and Cardiomyocyte Smad3 Signaling in the Infarcted Myocardium. Circulation. 2018;137:707–724. doi: 10.1161/CIRCULATIONAHA.117.029622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Mlodzik M. Wnt-Frizzled/planar cell polarity signaling: cellular orientation by facing the wind (Wnt) Annu Rev Cell Dev Biol. 2015;31:623–46. doi: 10.1146/annurev-cellbio-100814-125315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laeremans H, Hackeng TM, van Zandvoort MA, Thijssen VL, Janssen BJ, Ottenheijm HC, Smits JF, Blankesteijn WM. Blocking of frizzled signaling with a homologous peptide fragment of wnt3a/wnt5a reduces infarct expansion and prevents the development of heart failure after myocardial infarction. Circulation. 2011;124:1626–35. doi: 10.1161/CIRCULATIONAHA.110.976969. [DOI] [PubMed] [Google Scholar]
- 18.Daskalopoulos EP, Hermans KC, Janssen BJ, Matthijs Blankesteijn W. Targeting the Wnt/frizzled signaling pathway after myocardial infarction: a new tool in the therapeutic toolbox? Trends Cardiovasc Med. 2013;23:121–7. doi: 10.1016/j.tcm.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 19.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nature methods. 2013;10:1213–8. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ghazalpour A, Rau CD, Farber CR, Bennett BJ, Orozco LD, van Nas A, Pan C, Allayee H, Beaven SW, Civelek M, Davis RC, Drake TA, Friedman RA, Furlotte N, Hui ST, Jentsch JD, Kostem E, Kang HM, Kang EY, Joo JW, Korshunov VA, Laughlin RE, Martin LJ, Ohmen JD, Parks BW, Pellegrini M, Reue K, Smith DJ, Tetradis S, Wang J, Wang Y, Weiss JN, Kirchgessner T, Gargalovic PS, Eskin E, Lusis AJ, LeBoeuf RC. Hybrid mouse diversity panel: a panel of inbred mouse strains suitable for analysis of complex genetic traits. Mamm Genome. 2012;23:680–92. doi: 10.1007/s00335-012-9411-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lusis AJ, Seldin MM, Allayee H, Bennett BJ, Civelek M, Davis RC, Eskin E, Farber CR, Hui S, Mehrabian M, Norheim F, Pan C, Parks B, Rau CD, Smith DJ, Vallim T, Wang Y, Wang J. The Hybrid Mouse Diversity Panel: a resource for systems genetics analyses of metabolic and cardiovascular traits. J Lipid Res. 2016;57:925–42. doi: 10.1194/jlr.R066944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome research. 2010;20:281–90. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patterson M, Barske L, Van Handel B, Rau CD, Gan P, Sharma A, Parikh S, Denholtz M, Huang Y, Yamaguchi Y, Shen H, Allayee H, Crump JG, Force TI, Lien CL, Makita T, Lusis AJ, Kumar SR, Sucov HM. Frequency of mononuclear diploid cardiomyocytes underlies natural variation in heart regeneration. Nature genetics. 2017;49:1346–1353. doi: 10.1038/ng.3929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang JJ, Rau C, Avetisyan R, Ren S, Romay MC, Stolin G, Gong KW, Wang Y, Lusis AJ. Genetic Dissection of Cardiac Remodeling in an Isoproterenol-Induced Heart Failure Mouse Model. PLoS genetics. 2016;12:e1006038. doi: 10.1371/journal.pgen.1006038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol. 2007;1:54. doi: 10.1186/1752-0509-1-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiu K, Nagai R. Fibroblast-mediated pathways in cardiac hypertrophy. J Mol Cell Cardiol. 2014;70:64–73. doi: 10.1016/j.yjmcc.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 27.Zangle TA, Burnes D, Mathis C, Witte ON, Teitell MA. Quantifying biomass changes of single CD8+ T cells during antigen specific cytotoxicity. PLoS One. 2013;8:e68916. doi: 10.1371/journal.pone.0068916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zangle TA, Chun J, Zhang J, Reed J, Teitell MA. Quantification of biomass and cell motion in human pluripotent stem cell colonies. Biophys J. 2013;105:593–601. doi: 10.1016/j.bpj.2013.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bon P, Maucort G, Wattellier B, Monneret S. Quadriwave lateral shearing interferometry for quantitative phase microscopy of living cells. Opt Express. 2009;17:13080–94. doi: 10.1364/oe.17.013080. [DOI] [PubMed] [Google Scholar]
- 30.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic acids research. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tshori S, Gilon D, Beeri R, Nechushtan H, Kaluzhny D, Pikarsky E, Razin E. Transcription factor MITF regulates cardiac growth and hypertrophy. J Clin Invest. 2006;116:2673–2681. doi: 10.1172/JCI27643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bergmann MW. WNT signaling in adult cardiac hypertrophy and remodeling: lessons learned from cardiac development. Circ Res. 2010;107:1198–208. doi: 10.1161/CIRCRESAHA.110.223768. [DOI] [PubMed] [Google Scholar]
- 33.Madonna R, Geng YJ, Bolli R, Rokosh G, Ferdinandy P, Patterson C, De Caterina R. Co-activation of nuclear factor-kappaB and myocardin/serum response factor conveys the hypertrophy signal of high insulin levels in cardiac myoblasts. J Biol Chem. 2014;289:19585–98. doi: 10.1074/jbc.M113.540559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burnstock G. Purinergic Signaling in the Cardiovascular System. Circ Res. 2017;120:207–228. doi: 10.1161/CIRCRESAHA.116.309726. [DOI] [PubMed] [Google Scholar]
- 35.Wang AY, Chan JC, Wang M, Poon E, Lui SF, Li PK, Sanderson J. Cardiac hypertrophy and remodeling in relation to ACE and angiotensinogen genes genotypes in Chinese dialysis patients. Kidney international. 2003;63:1899–907. doi: 10.1046/j.1523-1755.2003.00933.x. [DOI] [PubMed] [Google Scholar]
- 36.Foulquier S, Daskalopoulos EP, Lluri G, Hermans KCM, Deb A, Blankesteijn WM. WNT Signaling in Cardiac and Vascular Disease. Pharmacol Rev. 2018;70:68–141. doi: 10.1124/pr.117.013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Travers JG, Kamal FA, Robbins J, Yutzey KE, Blaxall BC. Cardiac Fibrosis: The Fibroblast Awakens. Circ Res. 2016;118:1021–40. doi: 10.1161/CIRCRESAHA.115.306565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DeLeon-Pennell KY, Meschiari CA, Jung M, Lindsey ML. Matrix Metalloproteinases in Myocardial Infarction and Heart Failure. Prog Mol Biol Transl Sci. 2017;147:75–100. doi: 10.1016/bs.pmbts.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan D, Takawale A, Lee J, Kassiri Z. Cardiac fibroblasts, fibrosis and extracellular matrix remodeling in heart disease. Fibrogenesis Tissue Repair. 2012;5:15. doi: 10.1186/1755-1536-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jun JI, Lau LF. Resolution of organ fibrosis. J Clin Invest. 2018;128:97–107. doi: 10.1172/JCI93563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.