Abstract
Eukaryotic cells respond to DNA damage by arresting the cell cycle and modulating gene expression to ensure efficient DNA repair. The human ATR kinase and its homolog in yeast, MEC1, play central roles in transducing the damage signal. To characterize the role of the Mec1 pathway in modulating the cellular response to DNA damage, we used DNA microarrays to observe genomic expression in Saccharomyces cerevisiae responding to two different DNA-damaging agents. We compared the genome-wide expression patterns of wild-type cells and mutants defective in Mec1 signaling, including mec1, dun1, and crt1 mutants, under normal growth conditions and in response to the methylating-agent methylmethane sulfonate (MMS) and ionizing radiation. Here, we present a comparative analysis of wild-type and mutant cells responding to these DNA-damaging agents, and identify specific features of the gene expression responses that are dependent on the Mec1 pathway. Among the hundreds of genes whose expression was affected by Mec1p, one set of genes appears to represent an MEC1-dependent expression signature of DNA damage. Other aspects of the genomic responses were independent of Mec1p, and likely independent of DNA damage, suggesting the pleiotropic effects of MMS and ionizing radiation. The complete data set as well as supplemental materials is available at http://www-genome.stanford.edu/mec1.
INTRODUCTION
The integrity of genomic information is critical to the survival and propagation of all cellular organisms. DNA damage that compromises genomic stability can result from environmental stresses and from cellular processes that occur during normal growth. Thus, cells have evolved complex surveillance mechanisms that monitor genomic integrity during normal cell-cycle progression and in response to DNA damage, and they orchestrate a multifaceted response to DNA damage to ensure accurate transmission of genetic information (Hartwell and Weinert, 1989; Hartwell et al., 1994; reviewed in Elledge, 1996).
The multiple facets of the DNA damage response include cell-cycle arrest, alterations in gene expression, DNA damage repair, and cell death. These responses are mediated by a kinase cascade that appears to have been conserved through eukaryotic evolution. At the top of this cascade is a family of phospho-inositol kinase-related proteins, which includes the ATR and ATM kinases in mammals and their homologs in yeast, Mec1p and Tel1p (Kato and Ogawa, 1994; Weinert et al., 1994; Savitsky et al., 1995; Bentley et al., 1996; Cimprich et al., 1996). Downstream of the phospho-inositol kinase-related kinases are two classes of checkpoint kinases, including CHK1 and CHK2 in mammals and Chk1p and Rad53p in yeast (Allen et al., 1994; Sanchez et al., 1996, 1997, 1999; Matsuoka et al., 1998). An additional kinase in yeast, named Dun1p, acts downstream of Rad53p and is involved in both cell-cycle arrest and transcriptional regulation in the DNA damage response (Zhou and Elledge, 1993; Pati et al., 1997). Mutations in components of the ATR/Mec1 pathways result in hypersensitivity to DNA-damaging agents and, in higher organisms, predisposition to cancer (Cliby et al., 1998; Smith et al., 1998; Wright et al., 1998). Yeast cells harboring mutations in components of this pathway are defective in both cell-cycle arrest and gene expression responses, and these mutants display severe sensitivity to DNA-damaging agents (Kato and Ogawa, 1994; Desany et al., 1998; Bashkirov et al., 2000).
In yeast, the DNA-damage and DNA-replication-stress pathway activates checkpoints at four points in the cell cycle: at the G1/S transition (the G1 checkpoint), during S phase to prevent DNA replication (the S-phase progression checkpoint) and mitosis (the S/M checkpoint), and at the G2/M boundary (the G2/M checkpoint) (reviewed in Elledge, 1996; Longhese et al., 1998; Weinert, 1998). In addition, cells responding to DNA damage or blocks in replication induce the expression of a set of genes thought to facilitate DNA synthesis and repair. Which checkpoint becomes activated may be linked to the recognition of the type of DNA lesion as well as its consequences. For example, double-strand breaks resulting from ionizing radiation trigger G2/M arrest before mitotic entry, preventing loss of chromosome fragments during division (Weinert and Hartwell, 1988; Weinert and Hartwell, 1989), whereas base modifications that inhibit DNA replication activate the S-phase-progression checkpoint (Paulovich and Hartwell, 1995). Although different sets of proteins seem to be involved in sensing DNA damage at different phases of the cell cycle, the transduction of all resulting signals is thought to require the kinase cascade consisting of Mec1p, Rad53p, Chk1p, and Dun1p (reviewed in Elledge, 1996).
The Mec1 pathway also affects gene expression. Among the best characterized gene targets of this pathway are the RNR genes, which encode subunits of ribonucleotide reductase, the enzyme that controls the rate-limiting step of deoxyribonucleotide synthesis (reviewed in Stubbe, 1990; Stubbe and van der Donk, 1995). In yeast, three of the four RNR genes are repressed by the Crt1 repressor under normal conditions, but they become derepressed after the Mec1p-dependent hyperphosphorylation and inactivation of Crt1p in response to DNA damage (Huang et al., 1998). Besides these gene targets, little is known about the regulation of gene expression governed by the Mec1-Rad53-Dun1 pathway in response to DNA damage.
We used DNA microarrays to characterize the genomic expression programs in wild-type and mec1 mutant cells responding to two different DNA-damaging agents: the methylating agent methylmethane sulfonate (MMS) and ionizing radiation. MMS and ionizing radiation inflict different types of DNA damage by distinct mechanisms; therefore, we identified gene expression responses that were dependent on Mec1p in response to both conditions. We also characterized the involvement of downstream regulators dependent on Mec1p by observing genomic expression patterns in dun1 mutant cells responding to MMS and in cells lacking the Crt1 repressor. By comparing these expression programs to genomic responses induced by other experimental conditions, we have identified expression responses that are specific to DNA damage and dependent on the Mec1 pathway, as well as responses that are independent of Mec1p and likely independent of DNA damage. The complete data set and supplemental materials are available at http://www-genome.stanford.edu/mec1.
MATERIALS AND METHODS
Strains
Strains are listed in Table 1.
Table 1.
Strains used in this study
| Name | Genotype | Source |
|---|---|---|
| Y580 | MATa trp1::GAP-RNR1-TRP1, can1-100, ade2-1, his3-11,15, leu2-3,12, trp1-1, ura3-1 | Desany et al., 1998 |
| Y581 | MATa mec1 Δ::HIS3 trp1::GAP-RNR1-TRP1 can1-100, ade2-1, his3-11,15, leu2-3,12, trp1-1, ura3-1 | Desany et al., 1998 |
| Y578 | MATa dun1Δ::HIS3, can1-100, ade2-1, his3-11,15, leu2-3,12, trp1-1, ura3-1 | Desany et al., 1998 |
| Y300 | MATa can1-100, ade2-1, his3-11,15, leu2-3,12, trp1-1, ura3-1 | Zhou and Elledge, 1993 |
| Y577 | MATa can1-100, ade2-1, his3-11,15, leu2-3,12, trp1-1, ura3-1, crt1 Δ::LEU2 | Huang et al., 1998 |
| DBY9439 | MATa ura3-52 GAL2 pRS416 | Gasch et al., 2000 |
| DBY9518 | MATa ura3-52 GAL2 pTS3 | Tae Bum Shin |
Sample Collection, RNA Isolation, and Microarray Analysis
Culture sample collection, cell lysis, and mRNA isolation were performed as previously described (Gasch et al., 2000). Probes for microarray analysis were prepared as described (DeRisi et al., 1997), with the use of Cy5-conjugated or Cy3-conjugated dUTP (Amersham Pharmacia Biotech, Piscataway, NJ) with Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). Genomic DNA probes were prepared by labeling 2 μg of DNA in 50-μl of reactions, with 25 μM dATP, dCTP, dGTP, 10 μM dTTP, and Cy-conjugated dUTP with the use of Klenow DNA polymerase (New England Biolabs, Beverly, MA) similar to a previously published method (Pollack et al., 1999). Microarrays, constructed as previously described (Shalon et al., 1996), contained polymerase chain reaction-amplified DNA fragments representing ∼6200 predicted yeast open reading frames identified at the time of our analysis. Microarray hybridizations were performed as described previously (DeRisi et al., 1997), and data were collected with the use of a scanning laser microscope from Axon Instruments (Foster City, CA) and the program Scanalyze (available at http://rana.stanford.edu/).
Fluorescence-activated Cell Sorting (FACS) Analysis
A 250-μl aliquot from each wild-type and mec1 culture sample (∼1.5 × 106–4 × 106 cells) was added directly to 0.75 ml of ethanol and allowed to stand 1 h for fixation. Cells were rehydrated in 1× phosphate-buffered saline (PBS) buffer for at least 1 h and washed once with FACS buffer (0.2 M Tris pH 7.5, 20 mM EDTA). In a volume of 100 μl of FACS buffer, cells were treated with 1 mg/ml RNase A at 37°C for 4 h. Cells were then washed in 1× PBS, treated with 5 μg/ml propidium iodide in a final volume of 1 ml of PBS, and analyzed for fluorescence content with the use of a Coulter model Epics XL-MCL. The DNA content of ∼30,000 cells was determined for each sample.
Strain Comparisons Using Microarrays
To verify that the growth conditions and microarray analyses used in this study resulted in reproducible genomic expression programs in cells, the wild-type cells were grown on separate days in different batches of YPD medium to an optical density at 600 nm of 0.35–0.45 (∼8 × 106 cells/ml). Poly-adenylated RNA isolated from each culture was labeled with Cy3-dUTP or Cy5-dUTP, and the two samples were combined and analyzed by comparative hybridization to the yeast genome microarrays. Fewer than 25 transcripts differed in abundance more than twofold between the two samples, and no transcripts differed in abundance greater than threefold between the two samples (see Web supplement Figure i), revealing that the growth conditions and microarray analysis methods used in this study resulted in highly reproducible gene expression measurements.
Genomic expression patterns in untreated wild-type and mec1 cells were compared in duplicate experiments by analyzing mRNA isolated from the untreated cells that were used as the microarray reference samples in the MMS and ionizing radiation time courses (see below). Poly-adenylated RNA isolated from the mec1 cells was used to prepare a cDNA probe labeled with Cy5-dUTP, and poly-adenylated RNA isolated from the wild-type was used to prepare probe labeled with Cy3-dUTP. The two differentially labeled probes were mixed and analyzed by comparative hybridization to yeast genome microarrays.
To compare the strains' genomic DNA content, genomic DNA was isolated from wild-type, mec1, and dun1 cells grown at 30°C in YPD medium with the use of Qiagen genomic DNA preparative columns (QIAGEN, Valencia, CA). DNA from the wild-type cells was labeled with Cy3-dUTP, and DNA from the mec1 and dun1 mutants was labeled with Cy5-dUTP.
Comparison of the wild-type and crt1 strains was done in duplicate by isolating mRNA from cells grown at 30°C in YPD medium to mid-log phase. Poly-adenylated RNA collected from the crt1 cells was used to prepare a cDNA probe labeled with Cy5-dUTP, and poly-adenylated RNA isolated from the wild-type was used to prepare a cDNA probe labeled with Cy3-dUTP. The differentially labeled probes were combined and hybridized to the yeast genomic microarrays.
MMS Time Courses
YPD was inoculated with overnight cultures of either wild-type or mec1 cells, and grown at 30°C to an optical density at 600 nm of ∼0.5. An aliquot of each culture was collected for FACS analysis, and a separate aliquot was frozen to serve as the untreated microarray reference sample for each respective time course. To the remainder of each culture, 0.02% MMS (Sigma, St. Louis, MO) was added and the culture growth was resumed. Cells were collected for FACS and microarray analysis at 5, 15, 30, 45, 60, 90, and 120 min. A similar time course was performed for dun1 cells, except that samples were collected at 30, 90, and 120 min for microarray analysis. mRNA isolated from each time point sample was used to generate a cDNA probe labeled with Cy5-dUTP, and mRNA from the untreated reference sample was used to prepare probe labeled with Cy3-dUTP. The two differentially labeled probes were mixed and analyzed by comparative hybridization to yeast genome microarrays.
Ionizing Radiation Time Courses
All the ionizing radiation experiments were done at room temperature to minimize temperature fluctuation during gamma irradiation. YPD was inoculated with overnight cultures of either wild-type or mec1 cells, and grown to an optical density at 600 nm of ∼0.5. Cells were collected by centrifugation and resuspended in YPD at 1/10 the original culture volume. Cells were irradiated with 170 Gray and subsequently resuspended in fresh YPD at the original culture volume. The process of irradiation was completed within 20 min, and irradiated cells were returned to growth conditions. Samples from the wild-type and mec1 cultures were collected for FACS and microarray analysis at 5, 10, 20, 30, 45, 60, 90, and 120 min after completion of the irradiation process. Mock-irradiation time courses of wild-type and mec1 cells were conducted without irradiation but otherwise identically to the irradiated samples; microarray analyses were performed on the 5-, 30-, 60-, and 90-min samples after mock treatment in the wild type, and on the 5-, 30-, and 60-min samples after mock irradiation in the mec1 cells. mRNA from each time point was used to generate a cDNA probe labeled with Cy5-dUTP, whereas mRNA from asynchronous, untreated cells was used to prepare a probe labeled with Cy3-dUTP. The differentially labeled probes were combined and hybridized to the yeast genomic microarrays.
37°C Heat Transfer
Wild-type, mec1, and dun1 cells were grown to early log phase at 30°C in YPD medium, and an aliquot was collected to serve as the 30°C sample. Cells were rapidly collected by centrifugation, resuspended in 37°C YPD medium, and returned to growth at 37°C for 20 min. Cy5-labeled cDNA was prepared from total RNA isolated from the 37°C samples, and Cy3-labeled cDNA was generated from total RNA isolated from the 30°C samples. The corresponding, differentially labeled probes were combined and hybridized to the yeast genomic microarrays.
ROX1 Overexpression
Cells harboring the plasmid pTS-3, containing ROX1 under control of the Gal promoter, and cells harboring the empty vector pRS416 were grown at 30°C in minimal medium supplemented with 2% glucose to early log phase. Cells collected by centrifugation were washed three times in minimal medium supplemented with 2% galactose, resuspended in minimal medium containing 2% galactose, and returned to 30°C growth for 4 h, at which time the samples were collected. A cDNA probe generated from total RNA isolated from the strain carrying pTS-3 was labeled with Cy5-dUTP, and a cDNA probe made from total RNA isolated from the strain harboring pRS416 was labeled with Cy3-dUTP.
Hierarchical Clustering
Hierarchical clustering of the microarray data was performed as previously described (Eisen et al., 1998) with the use of the program Cluster (available at http://rana.stanford.edu). Unless otherwise noted in the figure legends, data for ∼6200 genes recovered from 40 microarrays, including the time course experiments following the responses of wild-type and mec1 cells to MMS, ionizing radiation, and mock irradiation and the dun1 response to MMS, were analyzed. In all clustering analyses, average-linkage clustering was used to organize the genes, with the use of microarray weights generated by Cluster with a correlation cutoff of 0.8 and an exponent of 1.0 (see Cluster manual for details). The resulting clusters were visualized with the use of the program TreeView (available at http://rana.stanford.edu/software/). For clarity, some of the experiments used in the clustering analyses have been omitted from the figures. The complete clustered data sets can be found on the supplemental Web site.
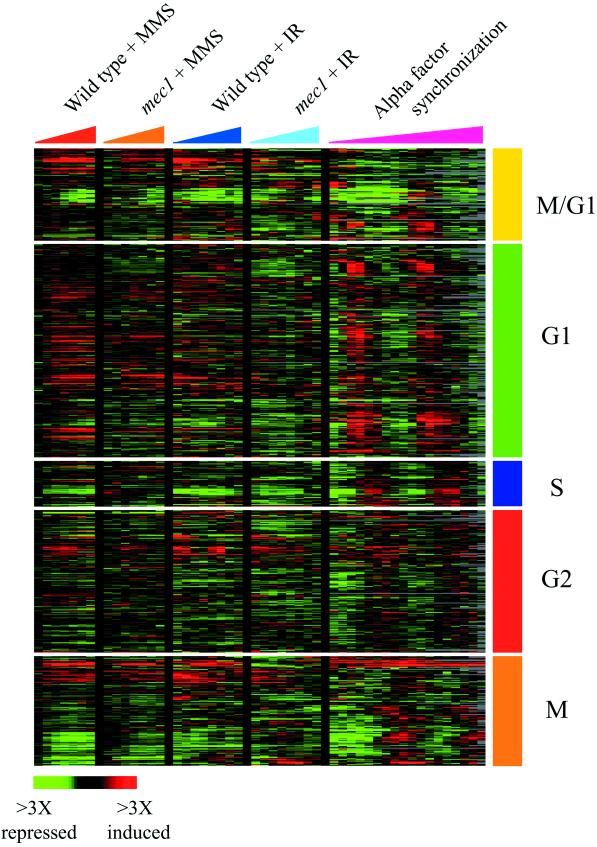
To characterize the expression responses of cell-cycle regulated genes, hierarchical clustering analyses were also performed on a smaller set of 700 genes whose expression had previously been shown to vary periodically during the cell cycle (Spellman et al., 1998). For the data shown in Figure 3, the genes were divided into five classes defined by Spellman et al. (1998) (M/G1,G1, S, G2, and M), reflecting the cell cycle phase during which those transcript levels peak in cycling cells. Each class of genes was clustered separately with the use of data recovered from a total of 86 microarrays, including 30 microarrays representing the wild-type and mec1 responses to MMS and ionizing radiation and 56 microarray analyses performed by Spellman et al. (1998) that followed genomic expression in cycling cells synchronized by alpha factor block and release, cdc15 block and release, and elutriation. The diagram shown in Figure 4 was derived from a separate hierarchical clustering analysis, performed by clustering all of the 700 cell cycle genes together with the use of data recovered from the 86 arrays described.
Figure 3.
Expression of cell cycle-regulated genes. The expression of ∼700 cell cycle-regulated genes (Spellman et al., 1998) was analyzed by hierarchical clustering, as described in MATERIALS AND METHODS. The genes were divided into five classes defined by Spellman et al. (1998) (M/G1, G1, S, G2, and M), reflecting the cell-cycle phase during which those transcript levels peak in cycling cells. Each class of genes was clustered separately with the use of data recovered from a total of 86 microarrays. For this display, data are shown for wild-type and mec1 cells responding to MMS treatment and ionizing radiation (IR) (this study) and cells progressing through the cell cycle after alpha factor synchronization (Spellman et al., 1998). The time points represented for the DNA damage responses are 5, 15, 30, 45, 60, 90, and 120 min after the MMS treatment and at 5, 10, 20, 30, 45, 60, 90, and 120 min after ionizing radiation. Changes in transcript levels are represented with the use of the color scale indicated at the bottom of the figure.
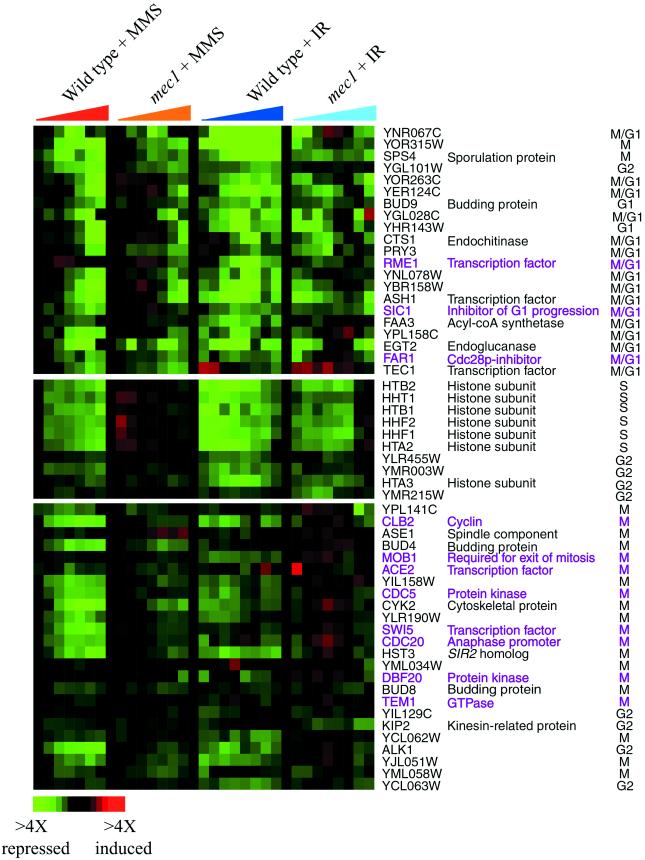
Figure 4.
Expression of cell-cycle genes that correlated with cell-cycle arrest. Hierarchical clustering analysis of all 700 previously identified cell-cycle genes revealed three clusters of genes whose transcript levels seemed to correlate with the cell-cycle arrest point induced by MMS and ionizing radiation (IR). The time points represented for the DNA-damage responses are the same described in Figure 3. Genes that are known to regulate cell-cycle progression and cell-cycle–dependent gene expression are labeled in purple. The color scale used to represent variations in transcript abundance is shown in the key at the bottom of the figure.
A comparison of genomic expression responses to DNA damage with the response to diverse environmental stresses was made by hierarchical cluster analysis of 95 arrays, including the 40 DNA damage micorarray experiments described above, and microarrays following the response of cells to heat shock, oxidative stress, reductive stress, osmotic shock, and amino acid starvation (Gasch et al., 2000).
RESULTS
To characterize the Mec1p-dependent response to DNA damage, the response of wild-type and mec1 mutant cells to 0.02% MMS or 170 Gray units of ionizing radiation was observed over the course of 2 h. The dosage of each treatment was calibrated to result in >45% cell viability in wild-type cells; the mec1 mutant was exposed to identical doses, but only 1 and 3% of the mutant cells survived MMS treatment and ionizing radiation, respectively. The process of irradiation involved extensive cell handling, and therefore a control, mock-irradiation time course was also examined to identify the gene expression responses that were due to cell handling and independent of irradiation. Each sample collected in the time series was analyzed by DNA microarray hybridization to observe gene expression, and FACS analysis to characterize cell-cycle progression.
We also analyzed the genomic expression pattern in mec1 mutant cells growing in the absence of exogenous DNA damage. Analysis of the data revealed that most of the genes on chromosome IV were expressed at levels approximately twofold higher in the mutant strain than the nominally isogenic MEC1 strain. Microarray analysis comparing genomic DNA in the mutant and MEC1 cells revealed that chromosome IV was duplicated in the mec1 strain (see Web supplement Figure ii). Recent evidence from Hughes et al. (2000) suggests that chromosomal duplication occurs frequently in mutant strains, apparently to ameliorate growth or survival disadvantages imposed by the mutations. In the mec1 mutant strain used in these experiments, a cassette carrying RNR1 under control of the GAP1 promoter, which suppresses mec1 lethality (Desany et al., 1998), is integrated at the trp1-1 locus on chromosome IV. Duplication of chromosome IV may enhance the fitness of the mutant cells by duplication of the RNR1 cassette, but it is also possible that other genes on chromosome IV may offer a selective advantage to the mutant when present at higher copy number.
Investigation of other mec1 null mutant strains in our lab revealed that all of these strains contained a duplication of chromosome IV (Huang, Elledge, unpublished data), and attempts to isolate a mec1 strain without the chromosomal duplication failed, highlighting the fact that deletion of the essential MEC1 gene imposes strong selection for additional suppressor mutations in cells. To distinguish the genomic expression responses to DNA damage that were dependent on the Mec1 pathway but independent of chromosome IV duplication, we characterized the genomic expression response to MMS in a dun1 null mutant strain, which does not have chromosomal duplications (see Web supplement Figure ii). The genomic expression response to MMS was very similar in the mec1 and dun1 cells (see Web supplement Figure iii). As discussed below, almost all of the Mec1p-dependent genes that responded to MMS treatment and ionizing radiation were similarly affected in the dun1 mutant, including genes localized to chromosome IV. Thus, the majority of the observed effects in the mec1 deletion strain are clearly independent of chromosome IV duplication.
Overview of Genomic Expression Responses to MMS and Ionizing Radiation
The expression patterns of the ∼6200 predicted yeast genes in response to MMS treatment and ionizing radiation, as measured in a total of 40 microarray hybridizations, were analyzed by hierarchical clustering (Eisen et al., 1998). A hierarchical clustering method was used to organize genes according to their similarity in expression profiles across all of the microarray experiments, such that genes with similar expression patterns are “clustered” together. The data are graphically displayed in tabular format in which each row of colored boxes represents the variation in transcript abundance for each gene, and each column represents the variation in transcript levels of every gene in a given mRNA sample as detected on one array. The variations in transcript abundance for each gene are represented by a color scale, in which shades of red represent increases and shades of green represent decreases in mRNA levels, relative to the untreated reference culture. The saturation of the color is proportionate to the magnitude of the variation in transcript levels. Black indicates no detectable change in transcript level, whereas gray represents missing data. In addition, the clustering algorithm generates a dendrogram that indicates the relationships between the expression patterns of genes; the branch lengths of the tree indicate the degree of similarity between the genes' expression profiles. Genes with similar patterns of expression over multiple experiments are thus grouped together on a common branch of the dendrogram and can also be recognized by an obvious pattern of contiguous patches of color in the cluster diagram.
The results of hierarchical clustering revealed that both MMS and ionizing radiation triggered rapid and extensive changes in the genomic expression program in wild-type cells (Figure 1), as has been previously observed (Jelinsky and Samson, 1999; Jelinsky et al., 2000). Transcripts of >750 genes changed at least twofold in abundance after MMS treatment. These alterations in gene expression began within 15 min of MMS exposure and persisted over the course of the experiment, reflecting the constant presence and effects of MMS in the culture medium. Ionizing radiation also provoked significant changes in the gene expression program in wild-type cells, with the relative abundance of >1300 transcripts changing by twofold or more. The changes in the genomic expression program after irradiation were transient; transcripts returned to near pre-irradiation levels with the passage of time after the transient exposure of cells to ionizing radiation. Large changes in gene expression were also observed in the mock-irradiated control; transcripts of >800 genes changed by more than twofold in relative abundance. Differences between the irradiated- and mock-treated samples, including differences in the magnitude and choreography of gene expression changes, revealed responses that were specifically dependent on ionizing radiation.
Figure 1.
Genomic responses to MMS and ionizing radiation. The expression patterns of ∼2000 genes whose transcript levels changed more than twofold in response to either MMS or ionizing radiation (IR) are shown. Hierarchical clustering was performed as described in MATERIALS AND METHODS, analyzing genes whose transcript levels changed at least twofold from the pretreatment level on at least one microarray experiment. For clarity, the mock-irradiated time courses that were used in the clustering analysis have been omitted from this display. The results of five time courses are shown, as indicated by the legend at the top of the figure. In each time course, the columns in the figure represent samples taken at 5, 15, 30, 45, 60, 90, and 120 min after the MMS treatment in the wild-type and mec1 cells; at 30, 90, and 120 min after MMS treatment in dun1 cells; and at 5, 10, 20, 30, 45, 60, 90, and 120 min after ionizing radiation exposure in wild-type and mec1 cells. The fold changes in transcript abundance relative to the pretreament levels are represented by a color scale, as indicated by the key at the bottom of the figure.
Deletion of MEC1 affected the expression of >1000 genes in response to both MMS and ionizing radiation (Figure 1; see below). All but a handful of these genes were equally affected by a dun1 mutation in cells responding to MMS, indicating that these effects were independent of chromosome IV duplication in the mec1 strain. The Mec1p dependence was seen for induced as well as repressed genes, revealing that the Mec1 pathway can direct both increases and decreases in gene expression. Because the microarrays measure changes in transcript levels, which are determined both by synthesis and degradation of mRNAs, the Mec1p-dependent effects on gene expression could be controlled either at the level of transcription or RNA turnover. As discussed in detail below, genes dependent on Mec1p for expression in response to these conditions were involved in a variety of processes, including cell-cycle progression, DNA damage repair, stress responses, and others. Although many of these responses are likely to be directly regulated by Mec1p, some may be affected by secondary consequences of the loss of Mec1p function.
Expression of Cell Cycle-regulated Genes
Both MMS and ionizing radiation induced complete cell-cycle arrest in wild-type cells, as indicated by FACS analysis (Figure 2). After exposure to MMS, wild-type cells accumulated with a DNA content between 1N and 2N, indicative of S-phase arrest (Figure 2A). The cells started to accumulate in S phase within 30 min of exposure to MMS and remained in S phase at 120 min. In response to ionizing radiation, the wild-type cells showed delayed progression through S phase 45 min after irradiation, and had completely arrested with a 2N DNA content by 90 min, indicative of arrest at the G2/M boundary (Figure 2B). This G2/M arrest was dependent on irradiation, because cells exposed to mock treatment did not arrest their cell cycle. Cell-cycle arrest in response to both MMS and ionizing radiation was dependent on MEC1, because mec1 null mutants failed to exhibit any observable cell-cycle arrest after identical treatments, in agreement with previous studies (Allen et al., 1994; Weinert et al., 1994; Paulovich and Hartwell, 1995).
Figure 2.
Cell-cycle progression in response to MMS and ionizing radiation. FACS analysis was performed on cells collected at each time point indicated. FACS profiles for wild-type cells (left) and mec1 cells (right) are shown after MMS treatment (A) and ionizing radiation (B). The FACS profile of the asynchronous cells used as the reference sample in each time course is shown in gray. Arrows indicate peaks in the profiles that represent cells with 1N or 2N DNA content, corresponding to cells in G1 phase or G2/M phase, respectively.
In wild-type cells, the expression of many genes that are regulated according to cell-cycle progression was affected by both MMS treatment and ionizing radiation. However, the genomic transcript profile in cells arrested by these agents in S phase or G2/M phase did not mimic the transcript profile in cells cycling through those phases. Figure 3 shows a comparison of the expression patterns of ∼700 cell-cycle–regulated genes identified by Spellman et al. (1998) in synchronously dividing cells (Spellman et al., 1998) and in asynchronous cells responding to MMS treatment and ionizing radiation (this study). In response to the DNA-damaging agents tested here, the expression of most of the cell-cycle–regulated genes did not appear to correlate in a simple way with the cell-cycle arrest point. Of the cell-cycle–regulated genes that were induced in response to these agents, most genes were similarly induced in cells arrested in S phase after MMS treatment and cells arrested in G2/M after irradiation, revealing that the expression changes seen for these genes were not specific to the cell-cycle arrest point. A small set of genes normally expressed during G1 phase was slightly induced specifically in response to MMS treatment, whereas a small cluster of genes normally expressed during G2 was specifically induced in irradiated cells; however the expression changes of these genes were largely unaffected by MEC1 deletion, suggesting that they were not related to the Mec1p-dependent cell-cycle arrest (see Web supplement Figure iv). Only a small set of repressed genes appeared to relate in a simple way to the cell-cycle stage at which arrest occurred (see below). The discordant features probably reflect the fact that genes apparently coregulated during cell-cycle progression in exponentially growing cells may actually be controlled by distinct mechanisms (Iyer et al., 2001). Further studies will be required to differentiate between gene expression changes that were related to cell-cycle arrest and changes that were triggered by other cellular consequences of MMS exposure or ionizing radiation.
A small number of repressed genes, including known regulators of the cell cycle, showed temporal patterns of expression that seemed to correlate with the cell-cycle arrest point. Hierarchical clustering of the complete set of 700 cell-cycle regulated genes in cells responding to MMS and ionizing radiation revealed three clusters of genes, many of which are normally expressed during M/G1, S, or G2/M phases of the cell cycle (Figure 4). In the wild-type strain, transcripts of these genes decreased over time in response to both MMS and ionizing radiation, but with a different temporal profile for each cluster of genes and each DNA damaging agent. For example, in wild-type cells responding to MMS exposure, histone transcripts were the first to decrease (at 15 min), followed by a decrease in some transcripts normally expressed in M phase (at 15–45 min), and followed in turn by a decrease in transcripts normally expressed in M/G1 phase (at 45–90 min). In response to ionizing radiation, histone transcripts decreased immediately, followed by a decrease in some M/G1 transcripts (at 20–45 min); transcripts of many genes that are characteristically expressed in M phase were immediately reduced in abundance in response to ionizing radiation, and their transcript levels remained low throughout the experiment. Differences in the temporal patterns of expression observed for the different cell-cycle classes may simply reflect differences in the proportions of the cell population in each cell-cycle stage as the asynchronous cell population proceeds through the cell cycle to the arrest point. Alternatively, yeast cells may actively repress these cell-cycle genes in response to the DNA-damaging agents.
The mec1 mutant cells failed to arrest progression through the cell cycle in response to MMS or ionizing radiation, and many of the effects of DNA damage on cell-cycle gene expression seen in the wild-type were muted in the mec1 mutant (Figures 3 and Figure 4). For example, the reduced expression of genes encoding histone subunits that was seen in wild-type cells responding to MMS was completely lost in the mec1 mutant responding to the drug, whereas the reduction of these transcripts after irradiation was only slightly muted in the mec1 strain relative to the wild type. Similarly, the decrease in M-phase transcripts (including genes required for the exit of mitosis; McCollum and Gould 2001) after irradiation of the wild-type cells was absent in the irradiated mec1 mutant. The cell-cycle–dependent expression of most of these M-phase–specific genes was recently shown to be governed by the transcription factors Fkh1p and Fkh2p (Zhu et al., 2000), and the anomalous expression of these genes in the irradiated mec1 cells may result from defects in Fkh-dependent signaling under these conditions. The defects in the expression of cell-cycle–regulated genes in the mec1 mutant is consistent with the failure of these cells to arrest the cell cycle in S and G2/M phases after MMS treatment and irradiation. Interestingly, residual changes in expression of most of the cell-cycle genes were still observed in the mec1 cells, with temporal profiles similar to those seen in the wild type.
DNA Damage Responses
We were surprised to find that there were few observable effects of MMS and ionizing radiation on transcripts of genes known to be involved in DNA damage repair. Transcripts of most genes in this category were negligibly increased in response to either MMS or radiation (less than threefold), consistent with previous observations (reviewed in Bachant and Elledge, 1999). Among the exceptions were genes previously implicated in the expression response to DNA damage, such as MAG1, PHR1, DDR2, DDR48, RAD51, RAD52, and RAD54. The induction of many of these genes is not specific to DNA damage, however, but rather a feature of the expression response to diverse stressful conditions (Gasch et al., 2000).
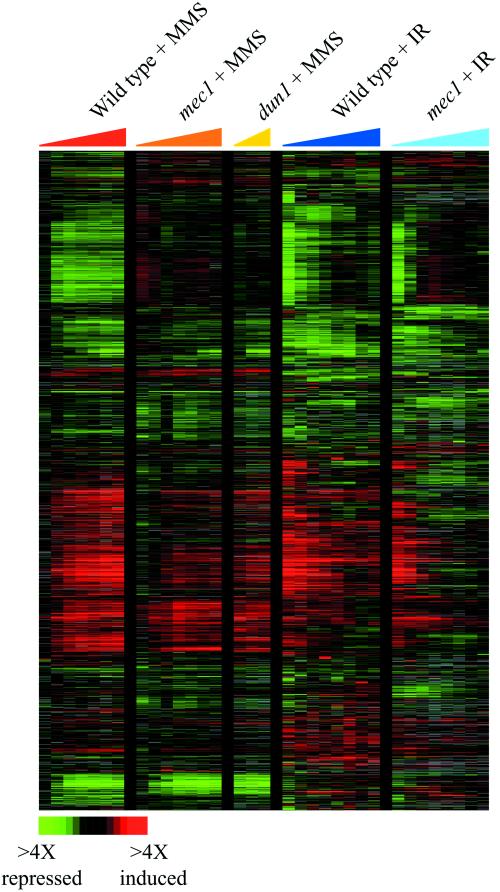
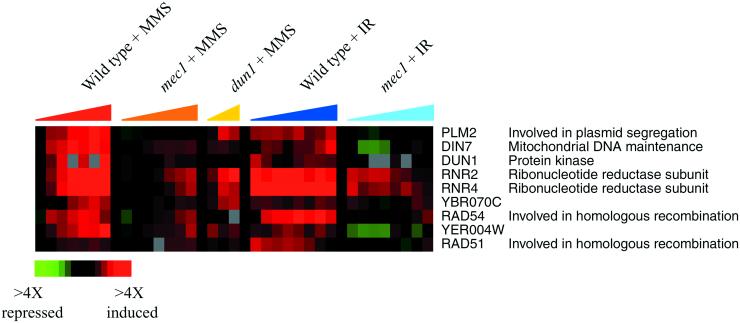
To identify genes that were induced specifically in response to DNA damage, we compared the expression programs observed in this study with the responses evoked by a variety of environmental perturbations, including heat shock, oxidative stress, reductive stress, osmotic shock, and amino acid starvation (Gasch et al., 2000). In total, results from 95 microarray hybridizations were analyzed by hierarchical clustering (see Web supplement Figure v). The results revealed a single cluster of genes whose induction was largely specific to MMS and ionizing radiation (Figure 5). This cluster included the DNA damage repair genes RAD51 and RAD54, the DNA damage inducible ribonucleotide reductase subunits RNR2 and RNR4, the DNA damage-activated kinase DUN1, and the uncharacterized genes YER004W and YBR070C. The induction of these genes in response to either MMS or ionizing radiation was muted in the mec1 cells, and their induction in response to MMS was also muted in the dun1 mutant cells, indicating that these genes are targets of the Mec1 pathway. These data corroborate the known Mec1p-dependence of RNR2 and RNR4 induction (Huang and Elledge, 1997), and also identify additional DNA damage-specific targets of the Mec1 pathway. Interestingly, induction of these genes was only partially dependent on Mec1p, pointing to the existence of additional mechanisms involved in controlling their response to DNA damage.
Figure 5.
Genes in the DNA Damage Signature. The expression patterns of the genes that were substantially induced in response to both MMS and ionizing radiation (IR), but whose expression was largely unaffected in response to other environmental stresses, are shown as described in Figure 1. The color scale used to represent changes in transcript levels is shown in the key at the bottom of the figure.
Environmental Stress Response
The environmental stress response (ESR) involves >900 genes whose expression is stereotypically altered in response to diverse environmental stresses (Gasch et al., 2000). Many of the genes repressed in this program are involved in protein synthesis and metabolism, and their repression in response to stressful environments probably conserves energy in the cell, whereas genes induced in the ESR may protect critical features of the internal homeostasis. As expected, the ESR was rapidly initiated in wild-type cells responding to MMS and ionizing radiation, and it was sustained for at least 2 h after MMS exposure and at least 45–60 min after ionizing radiation (Figure 6).
Figure 6.
Mec1p controls the ESR in response to DNA damage. Representative genes induced in the ESR (A) or repressed in the ESR (B) are shown for the time points described in Figure 1, and 20 min after heat transfer. The color scale used to indicate changes in transcript abundance is shown at the bottom of each figure.
Activation of the ESR in response to these DNA-damaging agents was dependent on the Mec1 pathway. Initiation of the ESR was greatly attenuated in both the mec1 and dun1 strains responding to MMS, and the prolonged alterations in ESR gene expression in the irradiated wild-type cells were strongly muted in the mec1 cells. Among the genes whose induction was attenuated in the mutant was the gene encoding the transcription factor Msn4p, known to regulate the induction of many ESR genes (Gasch et al., 2000). Many of the genes normally repressed in the ESR were slightly induced (less than twofold) in the mec1 mutant responding to these agents, but not the dun1 strain, for reasons that are not understood.
In contrast to the response to MMS and ionizing radiation, the DNA damage–independent initiation of the ESR in response to cell handling was not affected by deletion of the Mec1 kinase (see Web supplement to Figure 6). That initiation of the ESR by mock irradiation was independent of Mec1p suggested that Mec1p governs this program only in response to DNA damage. We therefore compared the expression response of the wild-type, mec1, and dun1 strains to a to 37°C temperature shift, a viable heat transfer known to trigger a substantial genomic expression response in wild-type cells (Gasch et al., 2000). As shown in Figure 6, the gene expression program in the mec1 cells responding to the temperature shift was nearly identical to that seen in wild-type cells, except for the slightly greater amplitude of the response in the mec1 strain. Thus, the Mec1 pathway is not required for proper ESR expression after heat transfer, supporting the hypothesis that the Mec1 pathway governs the ESR specifically in response to DNA damage.
Regulators Downstream of Mec1p
As discussed above, the DNA damage response of cells defective in Dun1p activity is largely the same as the response seen in the mec1 mutant, suggesting that most of the Mec1p-dependent effects on genomic expression observed here are mediated by the downstream Dun1 kinase. In contrast to deletion of DUN1, deletion of the Dun1p-dependent repressor CRT1 resulted in the reproducible constitutive induction of only a few of the Mec1p-dependent genes (see Web supplement Figure vi). The genes most strongly affected were the known Crt1p-targets RNR2 and RNR4. Most of the other genes in the DNA Damage Signature were unaffected by deletion of CRT1, despite the presence of the putative Crt1p-binding sequences in some of their promoters (Huang et al., 1998). It is possible that we have missed legitimate targets of the repressor because simple deletion of CRT1 in the absence of DNA damage may affect only a subset of Crt1p targets.
Mec1p-independent Responses to MMS and Ionizing Radiation
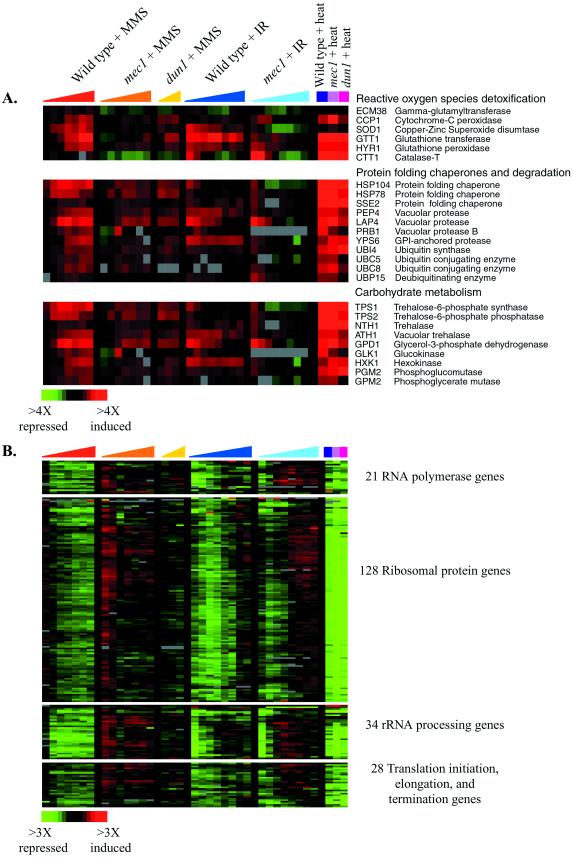
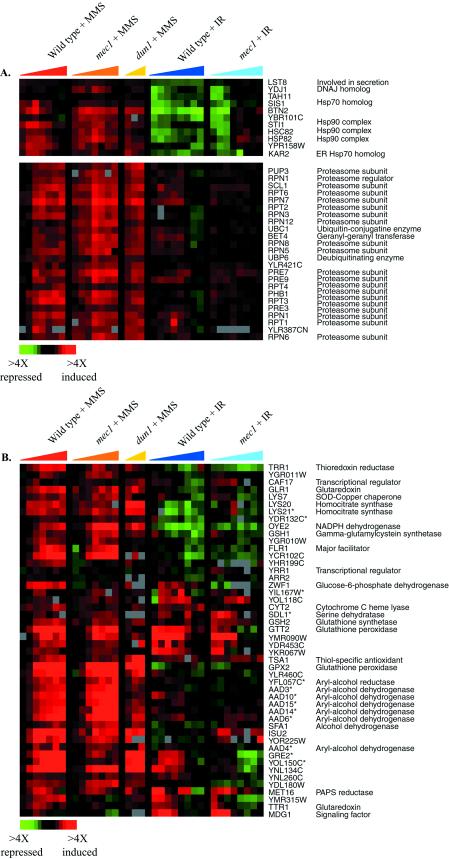
Many features of the gene expression programs evoked by treatment with MMS and ionizing radiation appeared to be unrelated to DNA damage. Although usually thought of as a DNA-damaging agent, MMS can also methylate other cellular targets. The induction of protein-folding chaperones localized to the cytoplasm, mitochondria, and endoplasmic reticulum suggests that MMS-induced methylation may affect protein structures, resulting in protein unfolding or misfolding (Figure 7A). Chaperones in multiple families, including the Hsp90 and Hsp70 families, were induced in both the wild-type and mec1 strains. Like other stressful conditions, MMS treatment also induced genes encoding proteasome subunits, as previously observed (Jelinsky et al. 2000). The induction of the protein chaperone and proteasome genes was independent of the Mec1 pathway, however, suggesting that their induction was not a specific response to DNA damage.
Figure 7.
Genes induced specifically by MMS treatment. The expression pattern of genes encoding protein-folding chaperones and proteasome components (A), and targets of the transcription factor Yap1p (B), after MMS treatment and ionizing radiation (IR) are shown, organized according to the hierarchical cluster analysis of the complete data set. This figure omits a subset of the Yap1p targets that are induced in the ESR by other regulators (Gasch et al., 2000). The time points represented for the DNA damage responses are the same as described in Figure 1, and the color scale that represents the changes in transcript levels is shown in the key at the bottom left of each figure. Adjacent genes on this display that share high sequence similarity and may therefore be subject to cross-hybridization in the microarray analysis are indicated by an asterisk.
MMS also induced targets of the transcription factor Yap1p, which is characteristically activated in response to agents that alter the cellular redox potential (Figure 7B) (Stephen et al., 1995; Kuge et al., 1997; Coleman et al., 1999). Most of the known targets of Yap1p (Gasch et al., 2000) were induced by MMS, as was the YAP1 gene itself, raising the possibility that Yap1p is activated in response to MMS, leading to the induction of its transcriptional targets. Previous studies in mammalian systems suggest that MMS reduces cellular glutathione pools, leading to perturbation of the cellular redox potential (Mizumoto et al., 1993; Wilhelm et al., 1997). A similar situation may occur in yeast: Yap1p targets include genes involved in glutathione synthesis and conjugation, genes encoding putative transporters required for resistance to various drugs, and genes involved in thiol oxidation and reduction. It is unclear whether Yap1p is induced in response to an imbalance of the cellular redox potential or oxidation of thiol groups after glutathione conjugation to MMS, directly by MMS, or both.
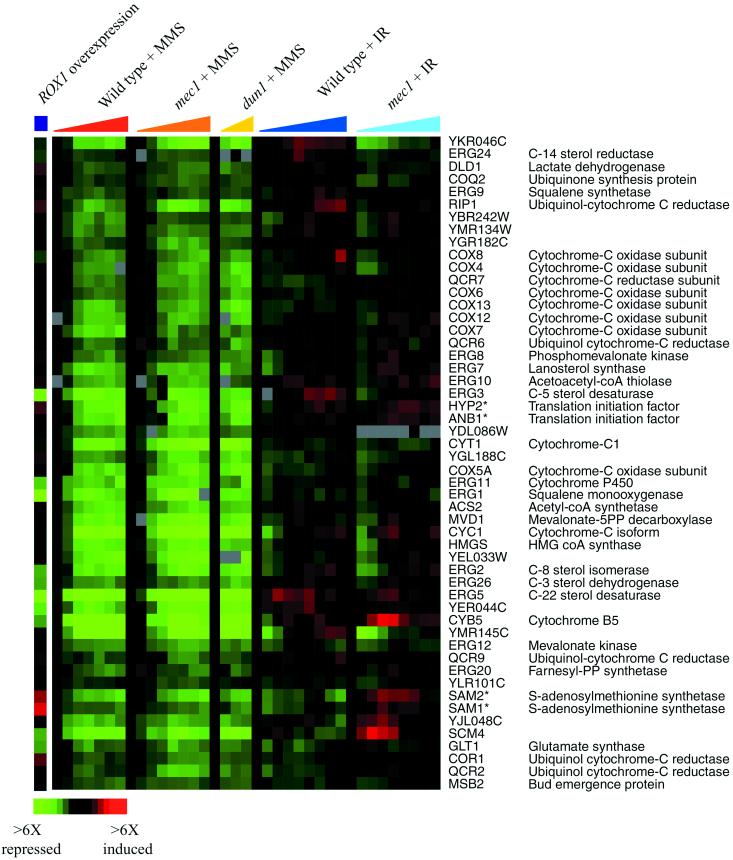
Hierarchical clustering of the complete DNA damage data set revealed that one large cluster of genes was strongly and uniquely repressed by MMS. This cluster included genes encoding ergosterol biosynthetic enzymes, components of the mitochondrial electron transport system, the transcriptional repressor ROX1, and others (Figure 8). Many of these genes are involved in oxygen- and heme-dependent processes, and are known to be regulated by the transcriptional activator Hap1p and the Hap1p-dependent repressor Rox1p (reviewed in Zitomer and Lowry, 1992). A small fraction of these genes was repressed by overexpression of the Rox1 repressor (Figure 8), whereas coordinate expression of most of these genes correlates with the absence of HAP1 (Uhlik and Brown, unpublished data). Indeed, most of these genes contain the known binding site for Hap1p within their promoter regions (see Web supplement to Figure 8). One explanation for the repression of these genes is that MMS artificially affects Hap1p-dependent signaling by directly methylating Hap1p or Rox1p; alternatively, MMS could methylate, or directly or indirectly alter the oxidation state of other cellular targets, such as lipids or heme, to affect the activity of this signaling system.
Figure 8.
Genes repressed specifically by MMS treatment. The expression patterns of genes repressed by MMS treatment are shown after MMS exposure and ionizing radiation (IR). The genes are organized as seen in the hierarchical cluster of the entire DNA damage data set. The responses of these genes to ROX1 overexpression are also shown. Time points are the same as described in Figure 1, and the color scale key indicating the changes in transcript levels is shown at the bottom of the figure. Adjacent genes on this display that share high sequence similarity and may therefore be subject to cross-hybridization in the microarray analysis are indicated by an asterisk.
Few features of the genomic response to ionizing radiation were specific to the level of irradiation used in this study. Notably absent from the response observed here were the genes involved in responding to oxidative stress (Gasch et al., 2000). It has been proposed that DNA damage resulting from ionizing radiation may be mediated by hydroxyl radicals, formed during irradiation (Ward, 1985, 1988; Wallace, 1998). Because hydroxyl radicals are also formed after exposure to hydrogen peroxide (H2O2) (Hauptmann and Cadenas, 1997), we expected that there would be corresponding similarities between the genomic responses to ionizing radiation and H2O2, perhaps reflecting protein damage and oxidative stress in addition to DNA damage. Instead, there were few similarities between the responses to ionizing radiation and to H2O2. For example, neither genes implicated in the detoxification of reactive oxygen species nor many of the Yap1p targets, involved in the response to oxidative stress, were strongly induced (Figure 7B). In contrast to the response to H2O2, genes encoding protein-folding chaperones were slightly repressed in response to ionizing radiation (Figure 7A), suggesting that the effects of ionizing radiation on protein stability were not enough to induce these genes. Our results provided no evidence for widespread hydroxyl-radical formation throughout the cell, and support the view that, at the level of ionizing radiation used in this study, DNA is specifically susceptible to the effects of ionizing radiation.
DISCUSSION
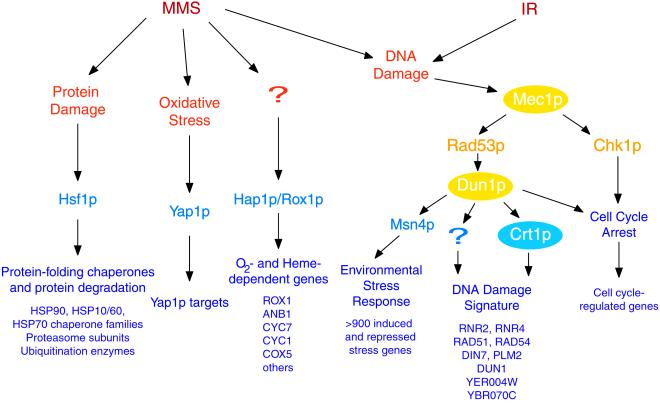
The results of this study provide a survey of the genomic expression programs elicited by MMS and ionizing radiation (Figure 9). Many cellular targets are subject to methylation by MMS. Its effects on DNA activate the DNA damage response pathway, leading to S-phase arrest of wild-type cells. In contrast, ionizing radiation induces double-strand breaks (Weinert et al., 1994), delaying S phase and leading to complete arrest in G2/M phase to prevent missegregation of broken chromosomes. Although the cellular responses to MMS and ionizing radiation were distinct, the genomic expression response to both of these agents was largely dependent upon Mec1p, highlighting the importance of this pathway in the DNA damage response. Thus, cells can sense the specific consequences of MMS and ionizing radiation to activate related but distinct genomic expression responses and to arrest the cell cycle at specific checkpoints.
Figure 9.
Summary of genomic responses to MMS and ionizing radiation. This diagram summarizes the functional features of the genomic expression responses observed in this study (purple), transcription factors (blue) and protein kinases (yellow) that have been implicated in those genomic responses, and the hypothetical cellular signals that trigger the responses (orange). Regulatory factors that were investigated in this study are indicated as colored ellipses.
The DNA Damage Signature Cluster
We identified one cluster of induced genes that appears to provide a specific signature of DNA damage. In addition to the conditions reported in this study, we have found that this cluster of genes is also induced by the UV-mimetic drug 4-nitroquinone and the RNR inhibitor hydroxyurea (Gasch, Huang, Elledge, Brown, unpublished results). A similar set of genes was also identified as being induced by HO endonuclease–induced double-strand breaks (Lee et al., 2000). Thus, these genes are apparently induced by DNA damage regardless of the type of DNA insult and irrespective of the resulting cell-cycle arrest point.
The genes in the DNA Damage Signature cluster encode proteins with a variety of functions, some of which have been related to DNA repair. Rad51p and Rad54p are involved in homologous recombination and the repair of double-strand DNA breaks, and both are required for repair of DNA damage inflicted by multiple agents (Budd and Mortimer 1982; Rattray and Symington 1995; Jiang et al. 1996; Petukhova et al. 1999, Simon et al., 2000). Ribonucleotide reductase catalyzes the rate-limiting step of deoxyribonucleotide biosynthesis (reviewed in Stubbe, 1990; Stubbe and van der Donk, 1995), and induction of RNR subunits may support increased or altered synthesis of nucleotide pools for DNA replication. Other genes in the DNA Damage Signature cluster include DIN7, encoding a protein with homology to nucleases that is proposed to localize to and function in mitochondria, and PLM2, which has homology to the forkhead-associated domain found in a number of transcription factors and kinases, and which has been implicated in the maintenance of the 2μ plasmid in yeast (Hofmann and Bucher, 1995; Mieczkowski et al., 1997; Fikus et al., 2000; Cashmore, unpublished data). Interestingly, the gene YER004W has homology to the human protein Tip30, a tumor suppressor that mediates apoptosis (Shtivelman, 1997; Xiao et al., 1998, 2000), but its function in yeast remains to be discovered. Clearly, the role of these genes in the response to DNA damage warrants careful investigation. Finally, the presence of DUN1 in this cluster reveals that this gene is itself induced by DNA damage and suggests that it is autoregulated by Dun1p and its upstream regulator, Mec1p. Increased synthesis of Dun1p after an increase in its transcript levels probably promotes signaling through the Mec1 pathway, further enabling a rapid and efficient cellular response to DNA damage.
Role of Mec1p in Regulating Responses to DNA Damage
The role of the Mec1-Rad53 pathway in response to DNA damage has previously been recognized through its effects on RNR gene expression and cell-cycle arrest (Elledge et al., 1993; Weinert et al., 1994; Paulovich and Hartwell, 1995; Sanchez et al., 1996; Huang and Elledge, 1997). In our study, the mec1 mutant cells failed to arrest their cell cycle in response to either DNA-damaging agent. Furthermore, the response of cell cycle-regulated genes to DNA damage was muted, although still detectible in most cases, in the mec1 cells. One possible explanation is that, after DNA damage, the Mec1 pathway actively represses these cell-cycle genes to promote cell-cycle arrest (Sidorova and Breeden, 1997). In this model, the muted repression of cell-cycle genes in the mutant cells is not enough to induce the appropriate checkpoint arrest. An alternative (but not mutually exclusive) model is that the decrease in cell-cycle transcripts is simply a reflection of the decreased fraction of the cells in the culture populating specific cell-cycle phases. In this model, the fact that the mec1 mutant retains some features of the wild-type decrease in cell-cycle transcripts suggests that either a small fraction of the culture is arresting at the appropriate checkpoint or that these cells expire at a specific point in the cell cycle. The severe sensitivity of the mec1 cells to DNA damage may confound accurate FACS analysis, obscuring any shifts in mec1 FACS profiles due to either checkpoint arrest or cell death.
In addition to governing cell-cycle arrest, Mec1p regulates the expression of genes induced specifically by DNA-damaging agents, as well as a large set of genes responsive to diverse environmental stresses. Genes in the DNA Damage Signature cluster were partially dependent on Mec1p and Dun1p in response to MMS and ionizing radiation, and these genes are probably induced to promote DNA repair and to enhance signaling through the Mec1 pathway. In addition to its role in mediating these specific responses to DNA damage, the Mec1 pathway is also involved in initiating the ESR after DNA damage. This general stress response has been proposed to protect internal homeostasis in the cell under diverse stressful conditions (Gasch et al., 2000). Indeed, most of the genomic expression response to DNA damage is accounted for by activation of the ESR, rather than DNA damage-specific features. The ESR is activated through different signaling pathways in response to different stressful conditions (Gasch et al., 2000). Expression of the ESR in response to heat stress is independent of the Mec1 pathway, suggesting that the Mec1 regulatory system governs ESR regulation specifically after DNA damage. Activated Mec1p therefore plays a multifaceted role in the response to DNA damage, simultaneously initiating cell-cycle arrest and activating genomic expression responses, including a few genes specific to the response to DNA damage, and a much larger set of genes that comprise the ESR, which protects many physiological systems during this stress.
The effects of Mec1p activity on gene expression reveal the complexity of Mec1p-dependent responses and suggest the involvement of other regulators. Aspects of Mec1-controlled cell-cycle arrest and gene expression responses were condition specific, suggesting that activation of Mec1p can have variable consequences in the cell. One possibility is that in response to different upstream signals, Mec1p activates different downstream regulators, resulting in differential effects on gene expression and cell-cycle arrest. Alternatively, Mec1p may routinely activate the same downstream regulators, whose subsequent activity is affected by other factors through regulatory systems that converge at downstream steps. Interestingly, almost all of the Mec1p-affected genes showed residual induction in the absence of Mec1p, pointing to other regulators that can partially substitute for Mec1p activity. One candidate for such a regulator is the Mec1p paralog Tel1p (Greenwell et al., 1995; Morrow et al., 1995; Sanchez et al., 1996). Future studies characterizing the involvement of Tel1p in response to DNA-damaging agents, both in the presence and absence of Mec1p, will be needed to clarify its role in governing these responses.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tae Bum Shin for supplying the overexpression constructs; Barbara Dunn, Caroline Uhlik, and Gavin Sherlock for experimental help; and Sarah Martinez, who participated in a related project as an undergraduate summer researcher. This work was supported by grants from the National Institutes of Health (HG-00983, HG-00450, GM-44664, and GM-46406), and by the Howard Hughes Medical Institute. M.H. was supported by a Damon Runyon-Walter Winchell posdoctoral fellowship. S.J.E. is an investigator, and P.O.B. is an associate investigator of the Howard Hughes Medical Institute.
Abbreviations used:
- ESR
environmental stress response
- MMS
methylmethane sulfonate
- RNR
ribonucleotide reductase
REFERENCES
- Allen JB, Zhou Z, Siede W, Friedberg EC, Elledge SJ. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2401–2415. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- Bachant JB, Elledge SJ. Mitotic treasures in the nucleolus. Nature. 1999;398:757–758. doi: 10.1038/19641. [DOI] [PubMed] [Google Scholar]
- Bashkirov VI, King JS, Bashkirova EV, Schmuckli-Maurer J, Heyer WD. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley NJ, Holtzman DA, Flaggs G, Keegan KS, DeMaggio A, Ford JC, Hoekstra M, Carr AM. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- Budd M, Mortimer RK. Repair of double-strand breaks in a temperature conditional radiation-sensitive mutant of Saccharomyces cerevisiae. Mutat Res. 1982;103:19–24. doi: 10.1016/0165-7992(82)90080-x. [DOI] [PubMed] [Google Scholar]
- Cimprich KA, Shin TB, Keith CT, Schreiber SL. cDNA cloning and gene mapping of a candidate human cell cycle checkpoint protein. Proc Natl Acad Sci USA. 1996;93:2850–2855. doi: 10.1073/pnas.93.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cliby WA, Roberts CJ, Cimprich KA, Stringer CM, Lamb JR, Schreiber SL, Friend SH. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman ST, Epping EA, Steggerda SM, Moye-Rowley WS. Yap1p activates gene transcription in an oxidant-specific fashion. Mol Cell Biol. 1999;19:8302–8313. doi: 10.1128/mcb.19.12.8302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Desany BA, Alcasabas AA, Bachant JB, Elledge SJ. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elledge SJ. Cell cycle checkpoints: preventing an identity crisis. Science. 1996;274:1664–1672. doi: 10.1126/science.274.5293.1664. [DOI] [PubMed] [Google Scholar]
- Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays. 1993;15:333–339. doi: 10.1002/bies.950150507. [DOI] [PubMed] [Google Scholar]
- Fikus MU, Mieczkowski PA, Koprowski P, Rytka J, Sledziewska-Gójska E, Ciela Z. The product of the DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, specifically functions in mitochondria. Genetics. 2000;154:73–81. doi: 10.1093/genetics/154.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell. 2000;11:4241–4257. doi: 10.1091/mbc.11.12.4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwell PW, Kronmal SL, Porter SE, Gassenhuber J, Obermaier B, Petes TD. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- Hartwell LH, Weinert TA. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989;246:629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Hartwell L, Weinert T, Kadyk L, Garvik B. Cell cycle checkpoints, genomic integrity, and cancer. Cold Spring Harb Symp Quant Biol. 1994;59:259–263. doi: 10.1101/sqb.1994.059.01.030. [DOI] [PubMed] [Google Scholar]
- Hauptmann N, Cadenas E. The oxygen paradox: biochemistry of active oxygen. In: Scandalios JG, editor. Oxidative Stress and the Molecular Biology of Antioxidant Defenses. Raleigh, NC: Cold Spring Harbor Laboratory Press; 1997. [Google Scholar]
- Hofmann K, Bucher P. The FHA domain: a putative nuclear signaling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- Huang M, Elledge SJ. Identification of RNR4, encoding a second essential small subunit of ribonucleotide reductase in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6105–6113. doi: 10.1128/mcb.17.10.6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Hughes TR, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet. 2000;25:333–337. doi: 10.1038/77116. [DOI] [PubMed] [Google Scholar]
- Iyer VR, Horak CE, Scafe CS, Botstein D, Snyder M, Brown PO. Genomic binding sites of the yeast cell-cycle transcription factors SBF and MBF. Nat Genet. 2001;409:533–538. doi: 10.1038/35054095. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Estep P, Church GM, Samson LD. Regulatory networks revealed by transcriptional profiling of damaged Saccharomyces cerevisiae cells: Rpn4 links base excision repair with proteasomes. Mol Cell Biol. 2000;20:8157–8167. doi: 10.1128/mcb.20.21.8157-8167.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Samson LD. Global response of Saccharomyces cerevisiae to an alkylating agent. Proc Natl Acad Sci USA. 1999;96:1486–1491. doi: 10.1073/pnas.96.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem. 1996;271:33181–33186. doi: 10.1074/jbc.271.52.33181. [DOI] [PubMed] [Google Scholar]
- Kato R, Ogawa H. An essential gene, ESR1, is required for mitotic cell growth, DNA repair and meiotic recombination in Saccharomyces cerevisiae. Nucleic Acids Res. 1994;22:3104–3112. doi: 10.1093/nar/22.15.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuge S, Jones N, Nomoto A. Regulation of yAP-1 nuclear localization in response to oxidative stress. EMBO J. 1997;16:1710–1720. doi: 10.1093/emboj/16.7.1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S.E., et al. (2000). Arrest, Adaptation, and Recovery following a chromosome double-strand break in Saccharomyces cerevisiae. Cold Spring Harb. Symp. Quant. Biol. (in press). [DOI] [PubMed]
- Longhese MP, Foiani M, Muzi-Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka S, Huang M, Elledge SJ. Linkage of ATM to cell cycle regulation by the Chk2 protein kinase. Science. 1998;282:1893–1897. doi: 10.1126/science.282.5395.1893. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- Mieczkowski PA, Fikus MU, Ciesla Z. Characterization of a novel DNA damage-inducible gene of Saccharomyces cerevisiae, DIN7, which is a structural homolog of the RAD2 and RAD27 DNA repair genes. Mol Gen Genet. 1997;253:655–665. doi: 10.1007/s004380050369. [DOI] [PubMed] [Google Scholar]
- Mizumoto K, Glascott PA, Jr, Farber JL. Roles for oxidative stress and poly(ADP-ribosyl)ation in the killing of cultured hepatocytes by methyl methanesulfonate. Biochem Pharmacol. 1993;46:1811–1818. doi: 10.1016/0006-2952(93)90587-m. [DOI] [PubMed] [Google Scholar]
- Morrow DM, Tagle DA, Shiloh Y, Collins FS, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- Pati D, Keller C, Groudine M, Plon SE. Reconstitution of a MEC1-independent checkpoint in yeast by expression of a novel human fork head cDNA. Mol Cell Biol. 1997;17:3037–3046. doi: 10.1128/mcb.17.6.3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulovich AG, Hartwell LH. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Van Komen S, Vergano S, Klein H, Sung P. Yeast Rad54 promotes Rad51-dependent homologous DNA pairing via ATP hydrolysis-driven change in DNA double helix conformation. J Biol Chem. 1999;274:29453–29462. doi: 10.1074/jbc.274.41.29453. [DOI] [PubMed] [Google Scholar]
- Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- Rattray AJ, Symington LS. Multiple pathways for homologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:45–56. doi: 10.1093/genetics/139.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y, Bachant J, Wang H, Hu F, Liu D, Tetzlaff M, Elledge SJ. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Desany BA, Jones WJ, Liu Q, Wang B, Elledge SJ. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Wong C, Thoma RS, Richman R, Wu Z, Piwnica-Worms H, Elledge SJ. Conservation of the Chk1 checkpoint pathway in mammals: linkage of DNA damage to Cdk regulation through Cdc25 [see comments] Science. 1997;277:1497–1501. doi: 10.1126/science.277.5331.1497. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Sfez S, Tagle DA, Ziv Y, Sartiel A, Collins FS, Shiloh Y, Rotman G. The complete sequence of the coding region of the ATM gene reveals similarity to cell cycle regulators in different species. Hum Mol Genet. 1995;4:2025–2032. doi: 10.1093/hmg/4.11.2025. [DOI] [PubMed] [Google Scholar]
- Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- Shtivelman E. A link between metastasis and resistance to apoptosis of variant small cell lung carcinoma. Oncogene. 1997;14:2167–2173. doi: 10.1038/sj.onc.1201059. [DOI] [PubMed] [Google Scholar]
- Sidorova JM, Breeden LL. Rad53-dependent phosphorylation of Swi6 and down-regulation of CLN1 and CLN2 transcription occur in response to DNA damage in Saccharomyces cerevisiae. Genes Dev. 1997;11:3032–3045. doi: 10.1101/gad.11.22.3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Szankasi P, Nguyen DK, Ludlow C, Dunstan HM, Roberts CJ, Jensen EL, Hartwell LH, Friend SH. Differential toxicities of anticancer agents among DNA repair and checkpoint mutants of Saccharomyces cerevisiae. Cancer Res. 2000;60:328–333. [PubMed] [Google Scholar]
- Smith L, et al. Duplication of ATR inhibits MyoD, induces aneuploidy and eliminates radiation-induced G1 arrest. Nat Genet. 1998;19:39–46. doi: 10.1038/ng0598-39. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephen DW, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- Stubbe J. Ribonucleotide reductases: amazing and confusing. J Biol Chem. 1990;265:5329–5332. [PubMed] [Google Scholar]
- Stubbe J, van der Donk WA. Ribonucleotide reductases: radical enzymes with suicidal tendencies. Chem Biol. 1995;2:793–801. doi: 10.1016/1074-5521(95)90084-5. [DOI] [PubMed] [Google Scholar]
- Wallace SS. Enzymatic processing of radiation-induced free radical damage in DNA. Radiat Res. 1998;150:S60–S79. [PubMed] [Google Scholar]
- Ward JF. Biochemistry of DNA lesions. Radiat Res Suppl. 1985;8:S103–S111. [PubMed] [Google Scholar]
- Ward JF. DNA damage produced by ionizing radiation in mammalian cells: identities, mechanisms of formation, and reparability. Prog Nucleic Acid Res Mol Biol. 1988;35:95–125. doi: 10.1016/s0079-6603(08)60611-x. [DOI] [PubMed] [Google Scholar]
- Weinert T. DNA damage checkpoints update: getting molecular. Curr Opin Genet Dev. 1998;8:185–193. doi: 10.1016/s0959-437x(98)80140-8. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Hartwell LH. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- Weinert T, Hartwell L. Control of G2 delay by the rad9 gene of Saccharomyces cerevisiae. J Cell Sci Suppl. 1989;12:145–148. doi: 10.1242/jcs.1989.supplement_12.12. [DOI] [PubMed] [Google Scholar]
- Weinert TA, Kiser GL, Hartwell LH. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Bender K, Knebel A, Angel P. The level of intracellular glutathione is a key regulator for the induction of stress-activated signal transduction pathways including Jun N-terminal protein kinases and p38 kinase by alkylating agents. Mol Cell Biol. 1997;17:4792–4800. doi: 10.1128/mcb.17.8.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JA, Keegan KS, Herendeen DR, Bentley NJ, Carr AM, Hoekstra MF, Concannon P. Protein kinase mutants of human ATR increase sensitivity to UV and ionizing radiation and abrogate cell cycle checkpoint control. Proc Natl Acad Sci USA. 1998;95:7445–7450. doi: 10.1073/pnas.95.13.7445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Palhan V, Yang Y, Roeder RG. TIP30 has an intrinsic kinase activity required for up-regulation of a subset of apoptotic genes. EMBO J. 2000;19:956–963. doi: 10.1093/emboj/19.5.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Tao Y, Greenblatt J, Roeder RG. A cofactor, TIP30, specifically enhances HIV-1 Tat-activated transcription. Proc Natl Acad Sci USA. 1998;95:2146–2151. doi: 10.1073/pnas.95.5.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. DUN1 encodes a protein kinase that controls the DNA damage response in yeast. Cell. 1993;75:1119–1127. doi: 10.1016/0092-8674(93)90321-g. [DOI] [PubMed] [Google Scholar]
- Zitomer RS, Lowry CV. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol Rev. 1992;56:1–11. doi: 10.1128/mr.56.1.1-11.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G, Spellman PT, Volpe T, Brown PO, Botstein D, Davis TN, Futcher B. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406:90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.