Abstract
Background
DNA methylation is believed to be maintained in adult somatic cells. Recent findings, however, suggest that all methylation patterns are not stable. We demonstrate that stimulatory signals can change the DNA methylation status around transcription factor binding sites and a transcription start site and activate expression of the aldosterone synthase gene (CYP11B2).
Methods and Results
DNA methylation of CYP11B2 was analyzed in aldosterone‐producing adenomas, nonfunctioning adrenal adenomas, and adrenal glands and compared with the gene expression levels. CpG dinucleotides in the CYP11B2 promoter were found to be hypormethylated in tissues with high expression, but not in those with low expression, of CYP11B2. Methylation of the CYP11B2 promoter fused to a reporter gene decreased transcriptional activity. Methylation of recognition sequences of transcription factors, including CREB1, NGFIB (NR4A1), and NURR1 (NR4A2) diminished their DNA‐binding activity. A methylated‐CpG‐binding protein MECP2 interacted directly with the methylated CYP11B2 promoter. In rats, low salt intake led to upregulation of CYP11B2 expression and DNA hypomethylation in the adrenal gland. Treatment with angiotensin II type 1 receptor antagonist decreased CYP11B2 expression and led to DNA hypermethylation.
Conclusions
DNA demethylation may switch the phenotype of CYP11B2 expression from an inactive to an active state and regulate aldosterone biosynthesis.
Keywords: aldosterone, angiotensin II, CYP11B2, epigenetics, sodium
Subject Categories: Epigenetics
Clinical Perspective
What Is New?
CYP11B2 gene expression is epigenetically controlled.
Angiotensin II or low salt intake induces demethylation of CYP11B2 and increases gene expression and aldosterone synthesis.
Stimulatory signals such as angiotensin II or potassium can change the DNA methylation status around transcription factor binding sites and a transcription start site and activate expression of CYP11B2.
Type 1 angiotensin II receptor blockers reversibly influence the methylation status of CYP11B2 and decrease aldosterone synthesis.
What Are the Clinical Implications?
Circulating aldosterone depends on DNA methylation patterns of the CYP11B2 promoter in the adrenal gland.
The methylation patterns may determine genetic traits such as susceptibility to hypertension and target organ damage.
Aldosterone is produced in the zona glomerulosa of the adrenal gland largely in response to angiotensin II (Ang II) or high dietary potassium. The primary effect of aldosterone is to induce sodium and fluid retention, resulting in increases in intravascular volume. Aldosterone synthase catalyzes the final steps in the deoxycorticosterone‐to‐aldosterone reaction. A single gene (CYP11B2) encodes aldosterone synthase. A number of cis‐acting regulatory elements have been identified in the CYP11B2 promoter: Ad1 (cAMP response element) at −71/−64, Ad4 at −344/−336, Ad5 at −129/−114, and nerve growth factor‐induced clone B response element at −766/−759. Activating transcription factor 1 or cAMP responsive element binding protein 1 (CREB1) binds to Ad1/cAMP response element, leading to increased CYP11B2 transcription. Nuclear receptor subfamily 5, group A, member 1 (NR5A1) (steroidogenic factor 1 [SF1]) binds to the Ad4 element. The Ad5 element interacts with several nuclear proteins, including NR4A1, NR4A2 (NUR‐related factor 1 [NURR1]), NR2F1 (chicken ovalbumin upstream promoter‐transcription factor I [COUP‐TFI]), and NR5A1. NGFI‐B, NURR1, and COUP‐TFI activate,1, 2, 3 whereas SF1 represses, CYP11B2 transcription.4
DNA methylation at the 5′‐cytosine of CpG dinucleotides is a major epigenetic modification in eukaryotic genomes and is required for mammalian development. The patterns of DNA methylation are faithfully replicated at every cell division once they have been established. DNA methylation is associated with the formation of heterochromatin and gene silencing. The methyl‐CpG binding domain (MBD) proteins are capable of directly binding to methylated DNA.5, 6 Methyl‐CpG binding protein 2 (MECP2), MBD1, and MBD2 can repress methylation‐based transcription.7 A non‐CpG island promoter regulates CYP11B2 gene expression. However, Ad1 and Ad5 contain CpG dinucleotides, which are target sites for DNA methylation, leading to the hypothesis that CpG dinucleotide methylation may regulate CYP11B2 gene expression. Although many studies have been conducted on the mechanisms of CYP11B2 gene regulation, the contribution of DNA methylation to regulation of the CYP11B2 gene is unknown.
Here, we present evidence that CpG methylation is an epigenomic mechanism for regulating CYP11B2 gene expression and is associated with interorgan variation. DNA methylation is a critical determinant for the phenotype of CYP11B2 expression and aldosterone biosynthesis.
Materials and Methods
The data that support the findings of this study are available from the corresponding author on reasonable request.
Procurement of Human Tissue
Human tissue samples were obtained from Kanazawa University Hospital. Adrenal tumors were collected after removal by surgery. Patients with aldosterone‐producing adenoma (51±3 years old, n=8; plasma aldosterone concentration 200±57 pg/mL; plasma renin activity 0.3±0.1 ng/mL per hour; serum potassium 3.5±0.3 mEq/L) were diagnosed according to the guidelines for primary aldosteronism of the Japan Endocrine Society.8 Nonfunctioning adenomas (non‐APAs) (n=5) were found incidentally by computed tomography scan for unrelated reasons. Patients with a clinically nonfunctioning adenoma had no signs or symptoms of hormone excess, normal serum potassium levels, and plasma cortisol levels suppressible by 1 mg of dexamethasone. Human adrenal glands and renal arteries were obtained directly after total nephrectomy for renal cell carcinoma. All samples were frozen in liquid nitrogen and stored at −80°C. Both DNA and RNA were simultaneously isolated and used for analyses of CpG methylation status and mRNA expression, respectively.
The purpose of the study was explained, and written informed consent was obtained from all study participants. The use of these tissues was approved by the Institutional Review Boards of Kanazawa University Graduate School of Medical Science.
Cell Culture
Adrenocortical H295R cells were purchased from ATCC (Manassas, VA) and maintained in DMEM/F12 medium (Invitrogen Life Technologies Japan Ltd, Tokyo, Japan) supplemented with 2.5% Ultroser G (Life Sciences, Cergy, France), penicillin, streptomycin (Life Technologies), and 1% ITS premix (BD Biosciences, Bedford, MA).
Animal Experiments
Rat adrenal gland was obtained from Wistar rats (male, 6 weeks old) that were fed either low‐sodium (0.03%), normal‐sodium (0.45%), or high‐sodium (7%) chow for 1, 2, and 4 weeks (n=5, each group). Rats switched from a low‐salt diet for 4 weeks to a high‐salt diet for 4 weeks were also examined. Ang II type 1 receptor antagonist (ARB) candesartan (Takeda Pharmaceutical, Osaka, Japan) (1 mg/[kg·day]) or Ang II (Peptide Research Inc, Osaka, Japan) (200 ng/[kg·min]) was subcutaneously infused by osmotic minipumps (Alzet model; Alza Corp, Palo Alto, CA).9 After 4 weeks, blood was collected from the tail vein as previously reported.9 All animals were housed in a temperature‐controlled environment (25°C) with a 12‐hour light‐dark cycle. Urinary and plasma concentrations of aldosterone (PAC) were estimated by radioimmunoassay after extraction with a Sep‐pak C18 cartridge (Waters, Milford, MA).9 Plasma renin activity was measured using a commercial kit (Yamasa Corporation, Chohu, Japan).
All experiments were performed in accordance with the guidelines for the use of experimental animals of the Animal Research Committee of Kanazawa University (permit No. AP‐122576).
Real‐Time Reverse Transcriptase Polymerase Chain Reaction
Real‐time polymerase chain raction (PCR) for human CYP11B2 (Hs01597732_m1) and rat Cyp11b2 (Rn01767819_g1) was carried out using the TaqMan Gene Expression Assay (Applied Biosystems, Life Technologies Japan Ltd, Tsu, Japan). Real‐time PCR for internal controls was carried out using the SYBR Green method. The primer sequences are listed in Table 1. Triplicates of each cDNA sample and no template controls were added to each real‐time PCR run.
Table 1.
Oligonucleotide DNA Used in This Study
| Name | Forward Primer (5′‐3′) | Reverse Primer (5′‐3′) | Footnotes |
|---|---|---|---|
| Real‐time reverse transcriptase PCR | |||
| GAPDH | tcattgacctcaactacatggttt | ttgattttggagggatctcg | Human |
| ACTB | tggcacccagcacaatgaa | ctaagtcatagtccgcctagaagca | Human |
| Rn18s | acggaagggcaccaccagga | caccaccacccacggaatcg | Rat |
| Bisulfite sequencing | |||
| Human CYP11B2 BM (−290 to +11) | gaaaggagaggttaggttttattatttt | accctttacaaaaacaaccaaaa | CpG1 to CpG3 |
| Human CYP11B2 BM (−278 to +69) | gaaaggagaggttaggttttattatttt | accctttacaaaaacaaccaaaa | CpG‐2 to CpG3 |
| Rat CYP11B2BM | ctcagacaagtccagggacagtttc | ggcaacactggcccccaagacactg | CpG5 to CpG8 |
| Luciferase assay | |||
| CYP11B2 (−167 KpnI/+9 NheI) | atggggtaccggcacctgacttctccttca | Atcggctagcattccaatgctccctccac | |
| CYP11B2 (−475 KpnI/+9 NheI) | atggggtaccaaccagtgcgctccgt | Atcggctagcattccaatgctccctccac | |
| pGL4.10 BM | ttttagtgtaagtgtaggtg | aaccaaatcttaatatcct | |
| Name | Biotinylated Double‐Stranded Oligonucleotide (5′‐3′) | ||
|---|---|---|---|
| NoShift assay | |||
| Unmethylated Ad1 (CpG1) | agccagttctcccatgacgtgatatgtttccag‐biotin | The bold Cs indicate cytosine residues methylated in vitro. | |
| Methylated Ad1 (CpG1) | agccagttctcccatgaCgtgatatgtttccag‐biotin | ||
| Unmethylated Ad5 (CpG2) | ggctgggggcctccagccttgaccttcgctctgaga‐biotin | ||
| Methylated Ad5 (CpG2) | ggctgggggcctccagccttgaccttCgctctgaga‐biotin | ||
| Unmethylated CpG3 | gtccagaccccacgccttttct‐biotin | ||
| Methylated CpG3 | gtccagaccccaCgccttttct‐biotin | ||
| Chromatin immunoprecipitation‐quantitative PCR | |||
| R1 (Ad1/CpG1) | gagtctcaggcaggcaggtccaga | attccaatgctccctccac | 113 bp |
| R2 (Ad5/CpG2) | ggcacctgacttctccttca | gagaactggctctggacctg | 93 bp |
| R3 (CpG3) | gagaaaggagaggccaggtc | gtcctgctggtgtgaggatg | 94 bp |
| R4 | aaaaagaaaccaggggttgg | ttgcttgcatggtttctgag | 95 bp |
| GNAS CpG island region | ttctcgtccgaagatacgaaa | gccgaggtaggtgtcagaga | 76 bp |
| R1 (Luc‐Ad1) | gagtctcaggcaggtccaga | cttaatgtttttggcatcttcca | |
| R2 (Luc‐Ad5) | ctagcaaaataggctgtccc | gagaactggctcggacctg | |
| Name | Sequencing Primer | Footnotes |
|---|---|---|
| Pyrosequencing | ||
| Rat CYP11B2(1) | aagagtaggtgtagtg | CpG2 to 4CpG2 to 7CpG2 to 10 |
| Rat CYP11B2(2) | ggaagaaattgtttttgtg | |
| Rat CYP11B2(3) | tgttttgggggttag | |
| Name | Forward Primer (5′‐3′) | Reverse Primer (5′‐3′) (Biotinylated) | Footnotes |
|---|---|---|---|
| Pyrosequencing | |||
| Rat CYP11B2(1) | tttgagttttggttatggttggggtttaat | aaaaaataatttcttccccttctacct | CpG2 to 4 |
| Rat CYP11B2(2) | gtttttggagtattgtggtatgg | ttctaccccccaacaaactttac | CpG2 to 7 |
| Rat CYP11B2(3) | gttatttgtttttggagtattgtggta | actaacaattaaaccccaaccat | CpG2 to 10 |
ACTD indicates beta actin; GAPDH, glyceraldehyde‐3‐phosphate dehydrogenase; PCR, polymerase chain reaction; Rn18, 18s ribosomal RNA.
Bisulfite Sequencing
Genomic DNA was treated with bisulfite and amplified. The human CYP11B2 promoter regions spanning from −290 to +11 (CpG1 to CpG3) or from −278 to +69 (CpG‐2 to CpG3) were amplified using specific primers (Table 1). The CpG sites of rat CYP11B2 promoter regions are shown in Figure 1. Each site was amplified using specific primers (Table 1). Ten PCR product clones generated using DNA from each sample (CpG1‐3, 5, 6, 8) were picked to analyze DNA methylation status, as we previously described.10 Methylation analysis (CpG 4, 7, 9, 10) by pyrosequencing was performed with PyroMarkGold Q96 Reagents and the PyroMarkQ24 pyrosequencing system (Qiagen, Hilden, Germany).
Figure 1.
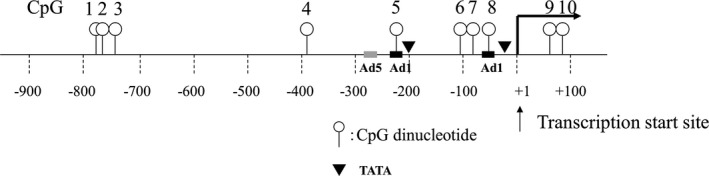
Schema of CpG dinucleotides within the rat CYP11B2 gene promoter.
CYP11B2 Promoter Constructs
Luciferase reporter plasmids containing 2 fragments of the flanking region of the CYP11B2 promoter were prepared from human genomic DNA using PCR (Figure 2). After digestion with KpnI and NheI, PCR‐amplified fragments were subcloned into the pGL4.10 luciferase reporter plasmid. Inserts were sequenced to ensure fidelity of the amplified sequences. The primers are listed in Table 1.
Figure 2.
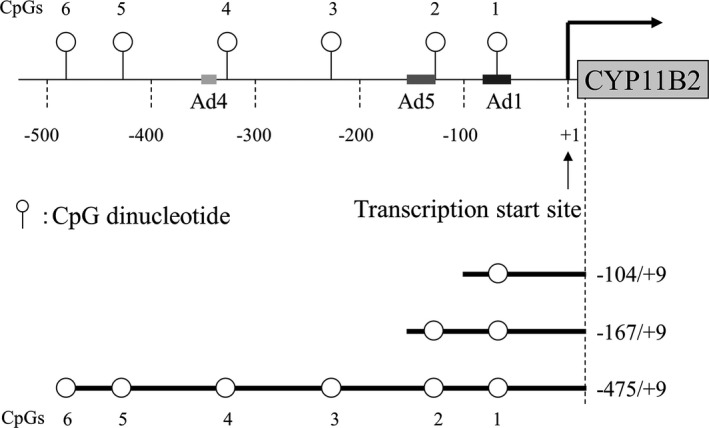
Schema of CpG dinucleotides with in the human CYP11B2 gene promoter and luciferase constructs.
Luciferase Assay
Sss I methylase (New England Biolabs Japan Inc, Tokyo, Japan) was used for in vitro methylation of CYP11B2 promoter luciferase constructs (pCYP11B2‐Luc) in the pGL4.10 vector. In each case, half of the DNA sample was methylated using Sss I, and the other half was incubated with Sss I methylase in the absence of S‐adenosylmethionine (mock methylation). The efficiency of methylation or mock methylation was determined using bisulfite sequencing and a primer pair (pGL4.10 BM) specific for a pGL4.10 vector. Adrenocortical H295R cells were transfected using FuGene HD (Promega, Madison, WI). Cells were incubated for 24 hours following transfection. After transfection, the cells were treated with 0.1, 1, 10, and 100 nmol/L Ang II, 16 mmol/L KCl, or 1 mmol/L cAMP for additional 24 hours according to the published paper.11 Firefly and Renilla luciferase assays were performed with 10 μL of cell lysates using a dual‐luciferase reporter assay system kit (Promega). Results are reported as the average of triplicate assays and are expressed as a ratio to the internal standard Renilla luciferase.
Enzyme‐Linked Immunosorbent Assay–Based Electrophoretic Mobility Shift Assay (NoShift Assay)
Nuclear extracts from H295R cells were prepared according to the manufacturer's protocol with the NE‐PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Yokohama, Japan). Protein concentration was determined using a BCA protein assay (Thermo Fisher Scientific) with BSA as a standard. The enzyme‐linked immunosorbent assay (ELISA)‐based electrophoretic mobility shift assay was carried out using NoShift transcription factor assay kits (Takara Bio Inc, Otsu, Japan) according to the manufacturer's protocol. Sequences of the biotinylated double‐stranded oligonucleotides are shown in Table 1. Antibodies used in this study are listed in Table 2.
Table 2.
Antibodies Used in This Study
| NoShift assay | |
| Anti‐CREB1 | sc‐58; Santa Cruz Biotechnology, Santa Cruz, CA |
| Anti‐NR4A1 (NGFI‐B) | CBX00289; Cosmo Biolabs Inc, Tokyo, Japan |
| Anti‐NR4A2 (NURR1) | sc‐991; Santa Cruz Biotechnology |
| Anti‐NR4A3 (NOR1) | PP‐H7833‐00; Perseus Proteomics Inc, Tokyo, Japan |
| Anti‐NR5A1 (SF1) | A generous gift of Dr Ken‐ichiro Morohashi |
| Anti‐MECP2 | pAb‐052‐050; Diagenode sa, Liège, Belgium |
| Chromatin immunoprecipitation quantitative PCR | |
| Anti‐MECP2 | pAb‐052‐050; Diagenode sa |
| Anti‐MBD1 | ab2846; Abcam, Cambridge, UK |
| Anti‐MBD2 | A‐1007; Epigentek Group Inc, Brooklyn, NY |
| Anti‐CREB1 | sc‐240; Santa Cruz Biotechnology |
| Anti‐phospho‐CREB1 (Ser133) | #17‐10131; Millipore, Billerica, MA |
| Anti‐NR4A1 (NGFI‐B) | ab13851; Abcam |
CREB1 indicates cAMP response element binding protein1; MBD, methyl‐CpG‐binding domain protein; MECP, methyl‐CpG binding protein; NGFI‐B, nerve growth factor‐induced clone B; NOR1, neuron‐derived orphan receptor 1; NURR1, NUR‐related factor 1; SF1, steroidogenic factor 1.
Chromatin Immunoprecipitation Quantitative PCR
The chromatin immunoprecipitation (ChIP) assays were performed with the ChIP‐IT High Sensitivity Kit (Active Motif, Carlsbad, CA) in accordance with the manufacturer's protocol. Antibodies used in this study are listed in Table 2. Quantification of ChIP DNA was accomplished by quantitative PCR with primers specific for the CYP11B2 promoter region (Table 1). The regions R1, R2, and R3 contain Ad1/CpG1, Ad5/CpG2, and CpG3, respectively. The distal promoter region R4 was used as a control and contained no CpG sites. Primers that amplify the 76‐bp guanine nucleotide‐binding protein Gs CpG island region (nt 27659851‐27659926, NT_011362.10) were used as a control for a condensed methylated region (Table 1). The 76‐bp region contains 11 methylated cytosine residues of CpG.
For detection of the −167/+9 CYP11B2 promoter that was inserted into the pGL4.10‐Luc plasmid, R1 (Luc‐Ad1) and R2 (Luc‐Ad5) were amplified using specific primers (Table 1). The results are presented as fold enrichment of IgG. Each sample was amplified in triplicate, and all experiments were repeated at least 3 times.
Human CYP11B2 ELISA
To quantify human CYP11B2 protein in cell lysate, a human CYP11B2 ELISA was performed with a Human CYP11B2 ELISA kit (EL006391HU; CUSABIO, Wuhan, China). Cytoplasmic extracts were isolated from H295R cells. Data were normalized to the total protein concentration of the cytoplasmic extracts.
Statistical Analysis
The data are expressed as mean±SEM. Data were compared by a Mann‐Whitney U test, 2‐way ANOVA or Friedman test, and Fisher protected least significance or Scheffe F test was performed which reach ANOVA‐indicated significance. Statistical significance was accepted for P<0.05.
Results
Inverse Correlation Between CpG Methylation and CYP11B2 mRNA Levels
We identified a number of CpG DNA methylation target sites within the CYP11B2 promoter. CpG1 (−68/−67) and CpG2 (−112/−111) were located within the Ad1 (−71/−64) and directly after Ad5 (−128/−113) cis‐acting elements, respectively. No transcription factors that bind to CpG3 (−219/−218) have been identified (Figure 3A). We analyzed both CpG methylation status and CYP11B2 mRNA levels using simultaneously isolated DNA and RNA from adrenal tumor, artery, and adrenal tissues. The CYP11B2 mRNA levels of APA were significantly high compared with those of nonfunctioning adenomas, adrenal glands, and arteries (Figure 3B). Methylation ratios of CpG1, CpG2, and CpG3 in APA (n=9) were approximately half of those in non‐APAs (n=5), normal adrenal glands (n=4), or normal arteries (n=3) (Figure 3C). Collectively, CpG methylation within the CYP11B2 promoter appeared to be inversely associated with CYP11B2 gene expression.
Figure 3.
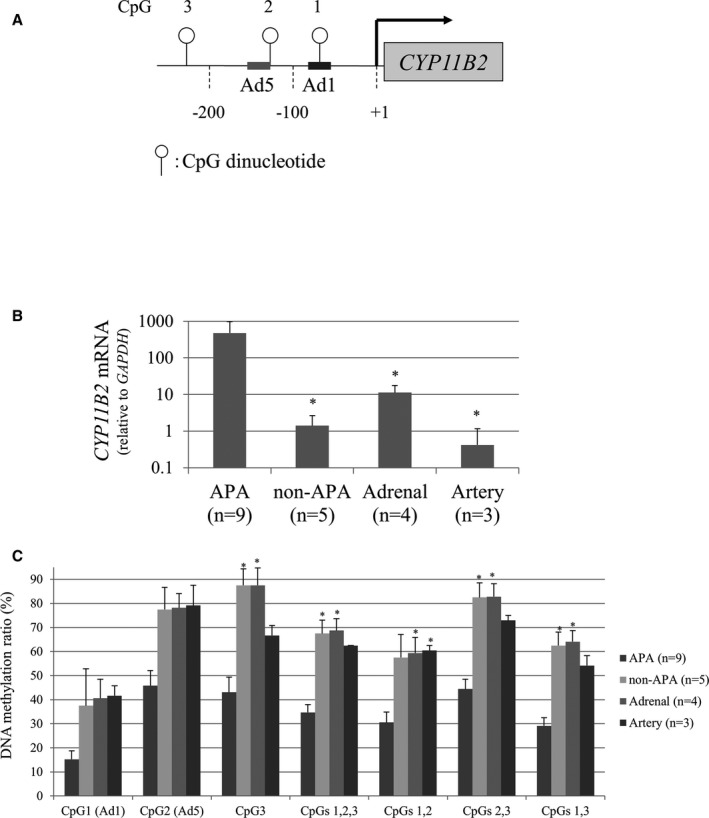
Inverse correlation between DNA methylation and CYP11B2 mRNA levels. A, Schema of 3 CpG dinucleotides within the CYP11B2 gene promoter. Ad1 and Ad5 represent cis‐acting elements previously reported. B, CYP11B2 mRNA level. *P<0.05 compared to aldosterone‐producing adenoma (APA). C, CpG methylation ratios of the CYP11B2 promoter. Data are presented as mean±SEM (n=9, APA; n=5, non‐APA; n=4, the adrenal gland; n=3, the artery). *P<0.05 compared to APA.
Repression of CYP11B2 Promoter Transcription by CpG Methylation
To assess the transcriptional ability of the CYP11B2 promoter with or without DNA methylation, luciferase reporter constructs containing 3 different lengths of the CYP11B2 promoter and its 5′‐flanking sequence were transfected into H295R cells (Figure 2). The CYP11B2 stimuli including Ang II, KCl, and cAMP activated transcriptional activities of the unmethylated reporter constructs. However, complete CpG methylation totally abolished luciferase expression levels of methylated constructs (Figure 4A). Incomplete CpG methylation reduced responsiveness to Ang II, KCl, and cAMP (Figure 4B). Ang II dose‐dependently increased transcriptional activities of the partially methylated reporter constructs (Figure 4C). These results indicated that the CYP11B2 promoter activity depended on CpG methylation.
Figure 4.
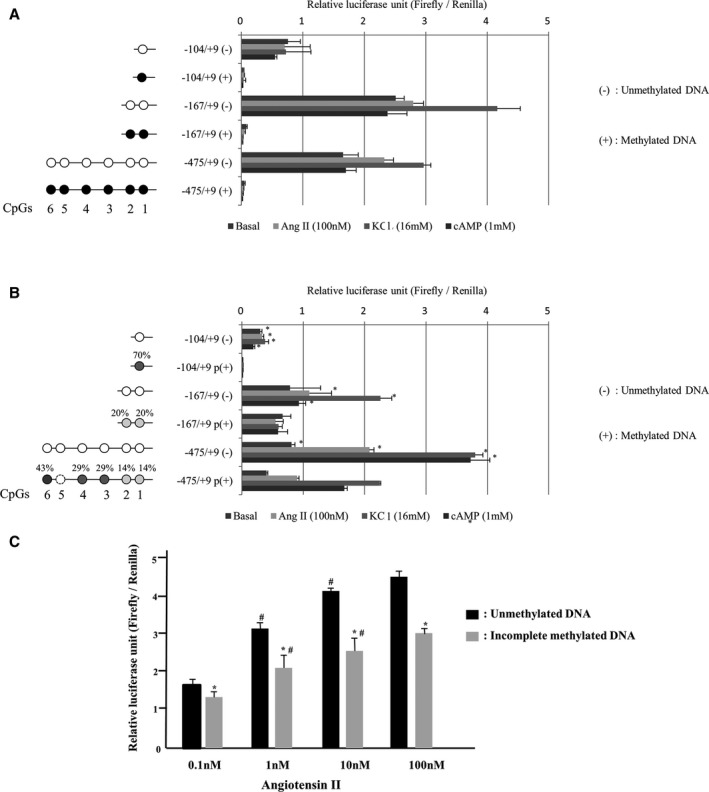
Effect of DNA methylation on the CYP11B2 promoter activity. A, Effect of complete CpG methylation. B, Effect of incomplete CpG methylation. C, Effect of angiotensin II. Bisulfite sequencing determined CpG methylation ratios. A methylation ratio at CpG5, dotted cycle was not able to be measured. Different luciferase constructs were methylated (filled circles) or mock‐methylated (open circles) in vitro and were transfected into H295R cells. Filled circles denote methylated cytosine residues; open circles, mock‐methylated. Results are given as luciferase activity normalized to cotransfected pRL‐TK Renilla luciferase activity. Results were mean±SEM of triplicate replicates. The numbering was based on the transcription start site of the CYP11B2 promoter. Complete CpG methylation totally abolished luciferase expression levels of methylated constructs (A). *P<0.01 vs incomplete methylation (B). *P<0.01 vs unmethylated DNA constructs (C). # P<0.01 vs compared 0.1 or 10 nmol/L of angiotensin II (Figure 2C).
Effect of DNA Methylation on Protein‐DNA Interaction in Vitro
A number of nuclear receptors families, including the CREB/ATF and NR4A (NGFIB, NURR1, NOR1) families as well as NR5A1 (SF1), have been reported to play a pivotal role in CYP11B2 gene transcription. CREB/ATF and NR4A families function as activators, whereas NR5A1 acts as a repressor.
NoShift transcriptional assays showed that DNA methylation at CpG1 reduced CREB1 binding to Ad1 (CpG1) by nearly half (Figure 5). A methylated cytosine residue at CpG2 inhibited interactions between NR4A1 (NGFIB) and NR4A2 (NURR1) in the presence of Ad5 by 30% and 50%, respectively (Figure 5). CpG methylation did not decrease NR4A3 (NOR1) and NR5A1 (SF1) binding activity in nuclear extracts.
Figure 5.
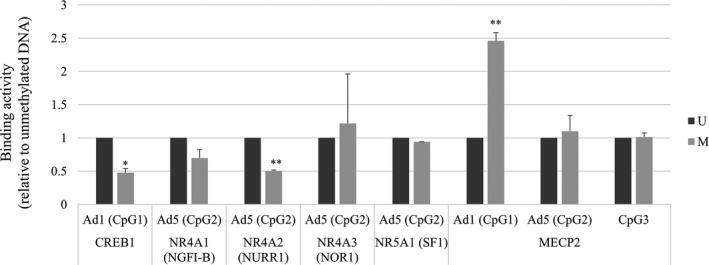
In vitro nuclear protein interactions with the CYP11B2 promoter (NoShift transcriptional assay). Binding activities to Ad1 or Ad5 with or without DNA methylation. Values are means±SD. M indicates methylated DNA; NGFI‐B, nerve growth factor‐induced clone B; NOR1, neuron‐derived orphan receptor 1; NURR1, NUR‐related factor 1; SF1, steroidogenic factor 1; U, unmethylated DNA. *P<0.05, **P<0.001.
We also measured MECP2 binding activities to CpG1 (Ad1), CpG2 (Ad5), and CpG3 under basal conditions, showing that MECP2 bound to methylated cytosine residues in the order CpG1, 2, and 3. DNA methylation increased MECP2 binding to CpG1 (Ad1) and CpG2 (Ad5) by 112% and 10%, respectively (Figure 5). No apparent difference in MECP2 binding activity to CpG 3 was observed between methylated and unmethylated states. It has been reported that an [A/T]≥4, a sequence adjacent to methyl‐CpG, is necessary for high‐affinity MECP2 binding.12 Four A/T bases were observed within the 3 bases of CpG1 and within 4 bases of CpG3, whereas CpG2 contained no A/T run. Despite A/T runs adjacent to both CpG1 and 3, we observed increased binding of MECP2 to methylated CpG1, but not to CpG3.
Increased Recruitment of MECP2 to the CYP11B2 Promoter in H295R Cells
ChIP PCR analysis was performed to determine whether endogenous MBDs interacted with the CYP11B2 promoter region, which contains 3 CpGs (Figure 6A). Methylation status of the 3 CpGs in H295R cell is shown in Figure 6B. Sheared chromatin from H295R cells was immunoprecipitated using antibodies against MECP2, MBD1, and MBD2, and enrichment of CYP11B2 promoter DNA in the immunoprecipitate was determined using real‐time PCR. The regions of R1, 2, and 3 contained CpG1, 2, and 3, respectively. As shown in Figure 6A, the ChIP PCR assay, using a MECP2‐specific antibody, revealed that there was significantly increased binding of MECP2 to the CYP11B2 promoter in the order of R1 (CpG1), R2 (CpG2), and R3 (CpG3) compared with R4. These results are consistent with those of NoShift assays. No enrichment was observed for the antibodies against MBD1 and MBD2 using primers that amplify the endogenous CYP11B2 promoter (Figure 6C).
Figure 6.
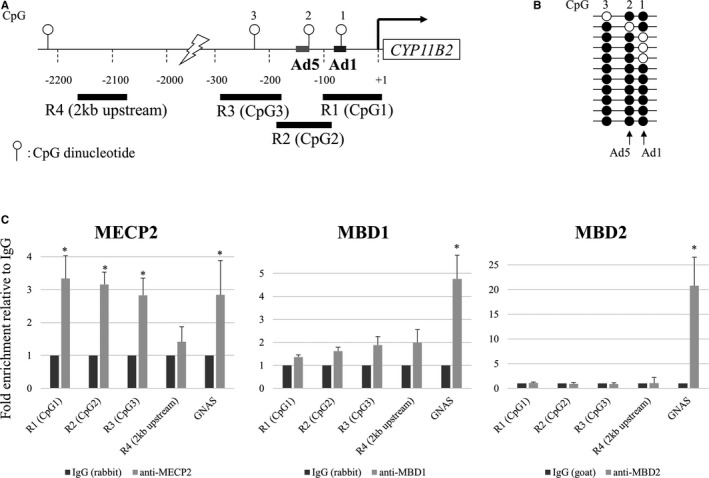
In vivo interactions of methyl CpG binding domain (MBD) proteins with the CYP11B2 promoter (chromatin immunoprecipitation assay). A, Schema of the CYP11B2 promoter. B, CpG methylation status of CpG1, CpG2, and CpG3 in H295R cells. C, Recruitment of MBD proteins. R1, R2, and R3 denote regions 1, 2, 3 amplified to measure MBDs interaction with CpG1 (Ad1), CpG2 (Ad5), and CpG3, respectively. R4 was used as a control that contained no CpG site. Primers for the 76‐bp GNAS CpG island region (nt 27659851‐27659926, NT_011362.10) were used as a control for a condensed methylated region. The 76‐bp region contains 11 methylated cytosine residues of CpG. Values are means±SD. *P<0.05.
To increase specificity, we also performed ChIP PCR in H295R cells transfected with the −167/+9 CYP11B2 Luc‐promoter plasmid, either with or without CpG methylation. We amplified the promoter regions of R1 (Luc‐Ad1) and R2 (Luc‐Ad5) using primers specific for the plasmid sequences (Figure 7A and Table 1). CpG methylation significantly increased the interaction of MECP2 with the exogenous promoter sequence containing CpG1 (Ad1) and CpG2 (Ad5) (Figure 7B). MECP2 binding to methylated CpG1 (Ad1) was high compared with methylated CpG2 (Ad5). No obvious enrichment was observed using antibodies against MBD1 and MBD2 between the unmethylated and methylated exogenous promoter sequence (Figure 7B). These results strongly suggested that CpG methylation increased the interaction between MECP2 and CpG1 (Ad1).
Figure 7.
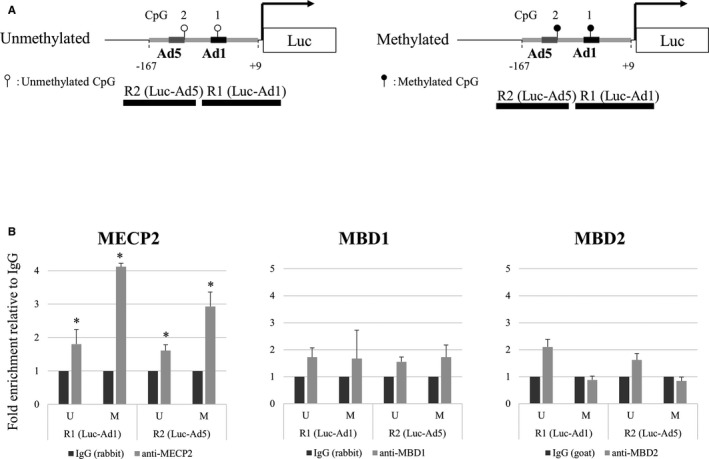
In vivo interactions of methyl CpG binding domain (MBD) proteins with the luc‐CYP11B2 promoter with or without DNA methylation (chromatin immunoprecipitation assay). A, Schema of the luc‐CYP11B2 promoter constructs with or without DNA methylation. B, Recruitment of MBD proteins. R1 and R2 denote regions 1 and 2 amplified to measure MBDs interaction with CpG1 (luc‐Ad1) and CpG2 (luc‐Ad5), respectively. Values are means±SD. *P<0.05.
Low Salt Intake Induces DNA Demethylation of the CYP11B2 Promoter and Increases CYP11B2 Expression in Rat Adrenal Glands
Urinary aldosterone excretion, PAC, CYP11B2 mRNA levels, and DNA methylation ratio are shown in Figure 8A through 8D. Urinary aldosterone excretion, PAC, and CYP11B2 mRNA levels were significantly increased in the rats fed with a low‐salt diet compared with rats fed a high‐salt diet over 4 weeks (P<0.05). Plasma renin activity was significantly increased in a low‐salt diet (6.8±0.6) compared with a high‐salt diet (0.81±0.2) or diet from low salt to high salt (0.7±0.1) (P<0.05). Treatment with candesartan significantly decreased urinary aldosterone excretion, PAC, and CYP11B2 mRNA levels (P<0.05). Switching from a low‐salt diet to a high‐salt diet decreased urinary aldosterone excretion, PAC, and CYP11B2 mRNA levels (P<0.05). We determined both DNA methylation status of the CYP11B2 promoter in the adrenal glands from Wistar rats fed either a low‐, normal‐, or high‐salt diet. The CpG1, 2, and 3 were almost methylated in each experimental group (data not shown). DNA methylation at Ad1 (CpG 5 or 8) was significantly lower in rats fed a low‐salt diet compared with those fed a high‐salt diet (P<0.05). Treatment with candesartan significantly increased the methylation ratio (P<0.05). Switching from a low‐salt diet to a high‐salt diet significantly increased the methylation ratio (P<0.05). The methylation ratios of CpG4, 6, 7, 9, and 10 were not significantly different in each group (data not shown). A low‐salt diet for 3 days did not influence aldosterone synthesis, whereas a low‐salt diet for 1 and 2 weeks or 4 weeks significantly increased aldosterone synthesis and induced hypomethylation (Figure 9). Ang II infusion for 3 days did not influence aldosterone synthesis, however, Ang II infusion for 1, 2, or 4 weeks significantly increased aldosterone synthesis and induced hypomethylation (Figure 9).
Figure 8.
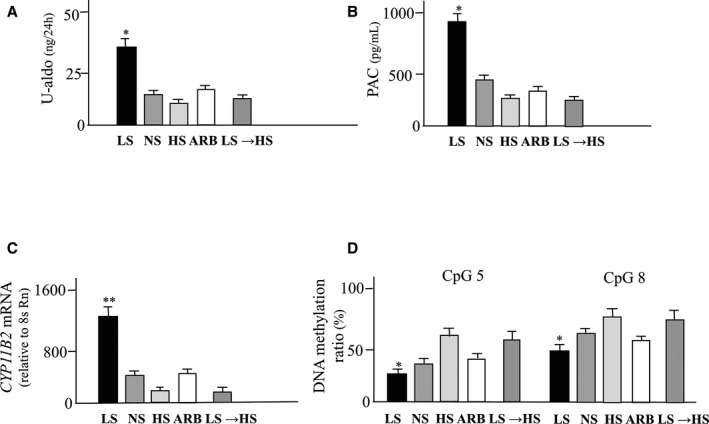
Effects of a low‐salt diet on aldosterone synthesis and methylation of CYP11B2. A, Urinary excretion of aldosterone (U‐aldo); (B) Plasma aldosterone concetration (PAC); (C) CYP11B2 mRNA levels; (D) DNA methylation ratios. Values are means±SEM. **P<0.01 vs NS, HS, ARB and LS →HS; *P<0.05 vs NS, HS, ARB and LS →HS. ARB indicates Angiotensin II type1 receptor antagonist; HS, high salt diet; LS, low salt diet; LS →HS, rats switched from LS to HS; NS, normal salt diet.
Figure 9.
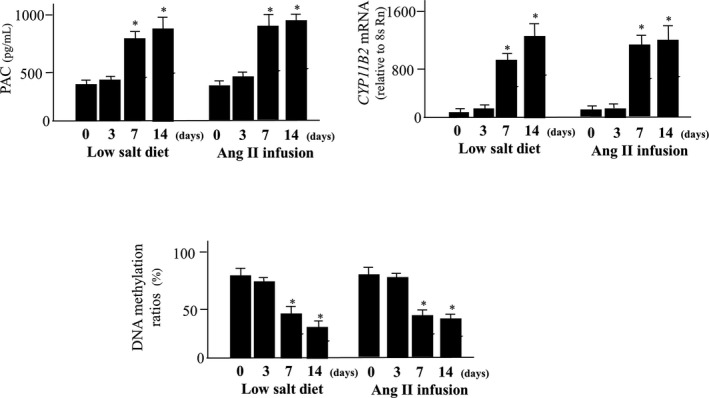
Effects of a low‐salt diet or angiotensin II on aldosterone synthesis and methylation of CYP11B2 during 14 days. Treatment with a low‐salt diet or angiotensin II for 7 and 14 days significantly increased plasma aldosterone concentration (PAC) and CYP11B2 mRNA levels and decreased methylation ratio of CYP11B2. *P<0.05 vs 0 or 3 days.
Discussion
The present study describes a previously unknown mechanism of CYP11B2 gene regulation associated with CpG methylation. A non‐CpG island promoter regulates CYP11B2 expression. However, 2 crucial cis‐acting elements, Ad1 and Ad5, are present in the CYP11B2 promoter and contain CpG dinucleotides, which are putative sites for DNA methylation. DNA methylation at CpG dinucleotides has been reported to alter gene expression by affecting transcription factor binding activity.13 This mechanism would be of particular importance in understanding regulation of non‐CpG island promoters. Our study revealed that differences in CYP11B2 promoter methylation within the Ad1 and Ad5 determined its expression levels.
CREB1/ATF family members and NR4A family members bind the CYP11B2 promoter at Ad1 and Ad5, respectively, leading to activation of transcription. We found methylation of CpG1 greatly decreased CREB1 binding to Ad1. DNA methylation at CpG2 reduced basal binding activities of NR4A1 (NGF1B) and NR4A2 (NURR1) with Ad5 by 30% and 50%, respectively. Previous work has shown that both NGF1B and NURR1 immunoreativities are high in the zona glomerulosa of the adrenal gland.2 The pattern of CpG methylation and expression levels of NGF1B and NURR1 appear to produce a synergistic effect on the CYP11B2 transcription in the adrenal cortex.
Hypomethylation of CYP11B2 was detected in APAs, and a negative correlation between CYP11B2 mRNA levels and methylation ratio was seen in our clinical samples. Hypomethylation of CYP11B2 may be an important regulatory mechanism of gene expression not only in the normal adrenal gland but also in aldosterone‐producing adenomas. Howard et al14 reported that CpG island in the promoter region of CYP11B2 was hypomethylated in APAs but not in blood DNA from the same patients. Yoshii et al15 reported the methylation rate in CpG (−116), CpG (−471), and CpG (−757) were lowered in APAs compared with nonfunctioning tumors, but no significant correlation between methylation rates and mRNA levels was seen. They also demonstrated somatic KCNJ5 mutation did not influence the methylation rate in APAs. Nishimoto et al16 reported an interesting case who showed nodule development from subcapsular aldosterone‐producing cell clusters caused hyperaldosteronism. The epigenome of CYP11B2 between APA and the aldosterone‐producing cell clusters should be clarified.
In the adrenal cortex, expression of the aldosterone synthase gene, CYP11B2, is mainly controlled by body sodium status via the renin‐angiotensin system. Sodium restriction or Ang II infusion significantly increased CYP11B2 mRNA levels in the rat adrenal gland.17 In our data, Ang II infusion in rats decreased the methylation ratio of CYP11B2 and increased gene expression in the adrenal gland. A low‐salt diet induced hypomethylation of rat CYP11B2 and increased CYP11B2 mRNA levels parallel with aldosterone synthesis. Plasma renin activity was also increased by a low‐salt diet. Treatment with angiotensin receptor blocker decreased aldosterone synthesis and induced hypermethylation of CYP11B2. These results clearly indicated the influence of Ang II to the epigenesis of CYP11B2 in the adrenal gland. Nishimoto et al18 reported that the rat zona glomerulosa transcriptome changed by dietary sodium intake and more than 280 differentially regulated genes. Gene ontology analysis identified 3 different gene groups: cell proliferation, response to a stimulus, and cholesterol/steroid metabolism. They suggested that the transcriptome changes were caused not only by activation of the renin‐angiotensin‐aldosterone system but also by the neurological responses.
DNA methylation is widely recognized for gene repression. Recent findings support the emerging consensus that DNA methylation patterns are not always stable in CpG island promoters.19 However, very little is known about where and how changes in DNA methylation take place in non‐CpG island promoters. We have recently shown that interleukin 6 stimulation can cause DNA demethylation of the angiotensinogen gene promoter in H295R cells.20 In our data, switching from a low‐salt diet for 4 weeks to a high‐salt diet for 4 weeks decreased aldosterone synthesis and induced hypermethylation of CYP11B2 in the adrenal glands. Stimulatory signals will orchestrate local recruitment of transcription factors and global reduction in DNA methylation activity. This recruitment helps open the chromatin configuration and promotes DNA demethylation in a locus‐specific manner. DNA methylation status can be used to trace the amount of chromatin that is opened by transcription factors at specific sites and functions as a memory to maintain responsiveness of gene expression to additional signals. DNA demethylation converts the gene expression phenotype within a tissue from an inactive to an active state.
Tissue renin‐angiotensin‐aldosterone system (RAAS) is activated in salt‐sensitive hypertensive rats.21 Pei et al22 reported hypomethylation of the promoter regions of the Ang II type 1a receptor gene in vascular tissues of hypertensive rats. Hypomethylation of the promoter regions of the Ang II type 1b receptor gene in the adrenal glands of the maternal low‐protein rat promoted hypertension through an exaggerated response to salt.23 The influences of salt intake on epigenomic alterations of CYP11B2 in the adrenal and cardiovascular tissues of salt‐sensitive hypertension should be clarified.
Sources of Funding
This study was supported by the Japan Society for the Promotion of Science (Grant‐in‐Aid for Young Scientists [B] #21790889 to Demura) and the Ministry of Health, Labor, and Welfare of Japan (Yoshiyu Takeda).
Disclosures
None.
Acknowledgments
We are grateful to Professor Ken‐ichiro Morohashi for providing anti‐SF1 antibody and to Cheng Wu for animal experimental assistance.
(J Am Heart Assoc. 2018;7:e008281 DOI: 10.1161/JAHA.117.008281.)29739797
Contributor Information
Yoshimichi Takeda, Email: takeday@med.kanazawa-u.ac.jp.
Yoshiyu Takeda, Email: takeday@med.kanazawa-u.ac.jp.
References
- 1. Bassett MH, Suzuki T, Sasano H, White PC, Rainey WE. The orphan nuclear receptors NURR1 and NGFIB regulate adrenal aldosterone production. Mol Endocrinol. 2004;18:279–290. [DOI] [PubMed] [Google Scholar]
- 2. Lu L, Suzuki T, Yoshikawa Y, Murakami O, Miki Y, Moriya T, Bassett MH, Rainey WE, Hayashi Y, Sasano H. Nur‐related factor 1 and nerve growth factor‐induced clone B in human adrenal cortex and its disorders. J Clin Endocrinol Metab. 2004;89:4113–4118. [DOI] [PubMed] [Google Scholar]
- 3. Shibata H, Kobayashi S, Kurihara I, Suda N, Yokota K, Murai A, Ikeda Y, Saito I, Rainey WE, Saruta T. COUP‐TF and transcriptional co‐regulators in adrenal steroidogenesis. Endocr Res. 2004;30:795–801. [DOI] [PubMed] [Google Scholar]
- 4. Ye P, Nakamura Y, Lalli E, Rainey WE. Differential effects of high and low steroidogenic factor‐1 expression on CYP11B2 expression and aldosterone production in adrenocortical cells. Endocrinology. 2009;150:1303–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl‐CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Laget S, Joulie M, Le Masson F, Sasai N, Christians E, Pradhan S, Roberts RJ, Defossez PA. The human proteins MBD5 and MBD6 associate with heterochromatin but they do not bind methylated DNA. PLoS One. 2010;5:e11982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim MS, Kondo T, Takada I, Youn MY, Yamamoto Y, Takahashi S, Matsumoto T, Fujiyama S, Shirode Y, Yamaoka I, Kitagawa H, Takeyama K, Shibuya H, Ohtake F, Kato S. DNA demethylation in hormone‐induced transcriptional derepression. Nature. 2009;461:1007–1012. [DOI] [PubMed] [Google Scholar]
- 8. Nishikawa T, Omura M, Satoh F, Shibata H, Takahashi K, Tamura N, Tanabe A. Guidelines for the diagnosis and treatment of primary aldosteronism—the Japan Endocrine Society 2009. Endocr J. 2011;58:711–721. [DOI] [PubMed] [Google Scholar]
- 9. Takeda Y, Yoneda T, Demura M, Miyamori I, Mabuchi H. Cardiac aldosterone production in genetically hypertensive rats. Hypertension. 2000;36:495–500. [DOI] [PubMed] [Google Scholar]
- 10. Demura M, Takeda Y, Yoneda T, Furukawa K, Tachi A, Mabuchi H. Completely skewed X‐inactivation in a mentally retarded young female with pseudohypoparathyroidism type IB and juvenile renin‐dependent hypertension. J Clin Endocrinol Metab. 2003;88:3043–3049. [DOI] [PubMed] [Google Scholar]
- 11. Clyne CD, Zhang Y, Slutsker L, Mathis JM, White PC, Rainey WE. Angiotensin II and potassium regulate human CYP11B2 transcription through common cis‐elements. Mol Endocrinol. 1997;11:638–649. [DOI] [PubMed] [Google Scholar]
- 12. Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl‐CpG. Mol Cell. 2005;19:667–678. [DOI] [PubMed] [Google Scholar]
- 13. Demura M, Bulun SE. CpG dinucleotide methylation of the CYP19 I.3/II promoter modulates cAMP‐stimulated aromatase activity. Mol Cell Endocrinol. 2008;283:127–132. [DOI] [PubMed] [Google Scholar]
- 14. Howard BH, Wang Y, Xekouki P, Faucz FR, Jain M, Zhang L, Meltzer PG, Stratakis CA, Kebebew E. Integrated analysis of genome‐wide methylation and gene expression shows epigenetic regulation of CYP11B2 in aldosteronomas. J Clin Endocrinol Metab. 2014;99:E536–E543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yoshii Y, Oki K, Gomez‐Sanchez CE, Ohno H, Itcho K, Kobuke K, Yoneda M. Hypomethylation of CYP11B2 in aldosterone‐producing adenoma. Hypertension. 2016;68:1432–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nishimoto K, Seki T, Kurihara I, Yokota K, Omura M, Nishikawa T, Shibata H, Kosaka T, Oya M, Suematsu M, Mukai K. Case report: nodule development from subcapsular aldosterone producing cell clusters causes hyperaldosteronism. J Clin Endocrinol Metab. 2016;101:6–9. [DOI] [PubMed] [Google Scholar]
- 17. Ping Y, Kenyon CJ, Mackenzie SM, Seckl JR, Fraser R, Connell JMC, Davies E. Regulation of aldosterone synthase gene expression in the rat adrenal gland and central nervous system by sodium and angiotensin II. Endocrinology. 2003;144:3321–3328. [DOI] [PubMed] [Google Scholar]
- 18. Nishimoto K, Harris RBS, Rainey WE, Tsugio Seki T. Sodium deficiency regulates rat adrenal zona glomerulosa gene expression. Endocrinology. 2014;155:1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thillainadesan G, Chitilian JM, Isovic M, Ablack JN, Mymryk JS, Tini M, Torchia J. TGF‐β‐dependent active demethylation and expression of the p15ink4b tumor suppressor are impaired by the ZNF217/CoREST complex. Mol Cell. 2012;46:636–649. [DOI] [PubMed] [Google Scholar]
- 20. Wang F, Demura M, Cheng Y, Zhu A, Karashima S, Yoneda T, Demura Y, Maeda Y, Namiki M, Ono K, Nakamura Y, Sasano H, Akagi T, Yamagishi M, Saijoh K, Takeda Y. Dynamic CCAAT/enhancer binding protein (CEBP)‐Associated changes of DNA methylation in the angiotensinogen gene. Hypertension. 2014;63:281–288. [DOI] [PubMed] [Google Scholar]
- 21. Takeda Y, Yoneda T, Demura M, Usukura M, Mabuchi H. Calcineurin inhibition attenuates mineralocorticoid‐induced cardiac hypertrophy. Circulation. 2002;105:677–679. [DOI] [PubMed] [Google Scholar]
- 22. Pei F, Wang X, Yue R, Chen C, Huang J, Huang J, Li X, Zeng C. Differential expression and DNA methylation of angiotensin type 1A receptors in vascular tissues during genetic hypertension development. Mol Cell Biochem. 2015;402:1–8. [DOI] [PubMed] [Google Scholar]
- 23. Bogdarina I, Welham S, King PJ, Burns SP, Clark AJ. Epigenetic modification of the renin‐angiotensin system in the fetal programming of hypertension. Circ Res. 2007;100:520–526. [DOI] [PMC free article] [PubMed] [Google Scholar]


