Semaphorins are a family of secreted and membrane‐bound molecules that play critical functions in diverse biological processes.1, 2, 3, 4, 5, 6, 7, 8, 9 SEMA1A/Fasciclin IV is the founding member of the semaphorin family and was first reported in 1992 as a regulator of axon branching in the growth cone of grasshopper embryos.10 To date, more than 20 semaphorin members have been discovered in viruses, invertebrates, and vertebrates.3, 5, 11 In addition to acting as guidance cues for axon growth in the nervous system, semaphorins have been implicated in many other systems, such as immune responses, tumor angiogenesis, bone development and homeostasis, and cardiovascular development.1, 2, 3, 4, 5, 6, 7, 8, 9
Semaphorins are categorized into 8 classes. Classes 1 and 2 are found in invertebrates, classes 3 to 7 are present in vertebrates, and class V semaphorins are found only in viruses.3, 12 All semaphorins contain a conserved domain of about 500 amino acids, termed the sema domain, which is located close to the N‐terminal end of the molecule13 (Figure 1). Crystal structures have revealed that the sema domain is composed of a 7‐bladed β‐propeller fold, arranged radially around a central axis. Each blade comprises a 4‐strand antiparallel β‐sheet.14, 15 With the exception of certain poxvirus semaphorins, other viral and all animal semaphorins contain a plexin‐semaphorin‐integrin domain in their extracellular regions, immediately to the C‐terminal of the sema domain.12, 16, 17 The plexin‐semaphorin‐integrin domain is a disulfide‐rich motif that is found in plexins, semaphorins, and integrins. In addition to the defined sema and plexin‐semaphorin‐integrin domains, semaphorins can be further distinguished by other specific additional motifs (Figure 1). For instance, classes 2, 3, 4, and 7 contain an immunoglobulin‐like domain, class 5 members contain 7 thrombospondin type 1 repeats, and class 3 members contain a basic domain. Some class V members contain an immunoglobulin‐like domain, but others do not.3, 12, 16 Semaphorins can be membrane anchored or secreted. Classes 1, 4, 5, and 6 are transmembrane proteins, whereas classes 2, 3, and V are secreted proteins. Class 7 proteins are glycosylphosphatidylinositol‐linked proteins.3, 11 Some transmembrane semaphorins, such as SEMA6D, also have secreted forms to act both locally and over a long distance.18, 19
Figure 1.
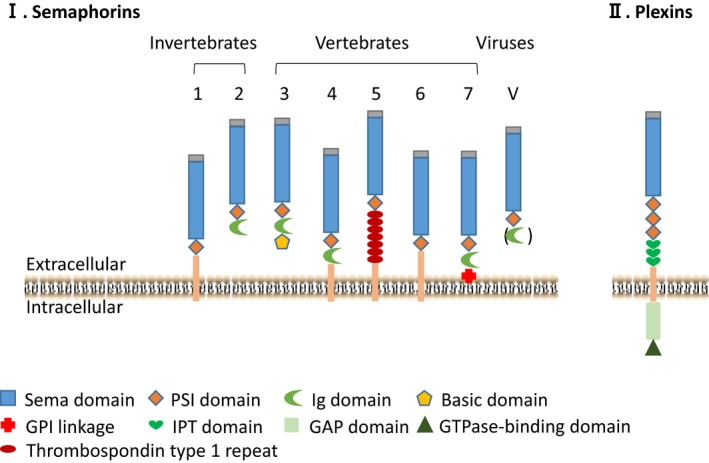
Semaphorins and plexins contain multiple functional domains.
Plexins are the primary receptors for semaphorins20, 21, 22 (Table 1). They are segregated into 4 subfamilies, including PLXNA1‐4, PLXNB1‐3, PLXNC, and PLXND.2, 6, 12, 23, 24 Similar to semaphorins, all plexins contain a sema domain at their N‐terminal ends (Figure 1). The extracellular domains of plexins also contain 3 plexin‐semaphorin‐integrin motifs and 3 to 6 immunoglobulin‐like plexin transcription factors domains. On the intracellular side, plexins are comprised of a rho GTPase‐binding domain and a segment GTPase‐activating protein domain.2, 6, 12, 23, 24 Recent structural studies have shown that semaphorins form a homodimer through intermolecular disulfide bridges to trigger plexin signaling. Semaphorins and plexins interact with each other through their respective sema domains in a “head‐to‐head” orientation. Two plexin molecules bind 1 semaphorin homodimer to form a bivalent 2:2 complex to mediate cell‐cell signaling. Proteolytic cleavage of the semaphorin dimer results in its dissociation to monomeric semaphorin, which can still bind plexin but fails to trigger signaling.11, 23, 25 The same semaphorin molecule can interact with different plexins, and different semaphorins can interact with the same plexins, adding to the complexity of semaphorin‐plexin signaling (Table 1). It is noteworthy that the biological activities of the semaphorin‐plexin interaction can be further modified by coreceptors. For example, class 3 semaphorins bind to neuropilin (NRP)/plexin complexes, which require NRP1 and/or NRP2 as coreceptors in the complexes.20, 21, 22 In another example, SEMA6D interacts with the PLXNA4/vascular endothelial growth factor (VEGFR2) receptor complex to stimulate endothelial cell migration in the outflow tract (OFT) region of chicken embryonic hearts, whereas interaction with the PLEXINA4/PTK7 complex inhibits endothelial cell migration in the ventricle region.18
Table 1.
| Semaphorins | Organisms | Plexins |
|---|---|---|
| SEMA1A | Invertebrate | PLXN A |
| SEMA1B | Invertebrate | PLXN A |
| SEMA2B | Invertebrate | PLXN B |
| SEMA3A | Vertebrate | PLXN A1, A2, A3, A4, D1 |
| SEMA3B | Vertebrate | PLXN A1, A2, A3, A4 |
| SEMA3C | Vertebrate | PLXN A1, A2, B1, D1 |
| SEMA3D | Vertebrate | PLXN D1 |
| SEMA3E | Vertebrate | PLXN B2, D1 |
| SEMA3F | Vertebrate | PLXN A1, A3, A4 |
| SEMA4A | Vertebrate | PLXN B1, B2, B3, D1 |
| SEMA4B | Vertebrate | PLXN B2 |
| SEMA4C | Vertebrate | PLXN B2 |
| SEMA4D | Vertebrate | PLXN B1, B2, C1 |
| SEMA4G | Vertebrate | PLXN B2 |
| SEMA5A | Vertebrate | PLXN A1, A3, B3 |
| SEMA5B | Vertebrate | PLXN A1, A3 |
| SEMA6A | Vertebrate | PLXN A2, A4 |
| SEMA6B | Vertebrate | PLXN A1, A2, A4 |
| SEMA6C | Vertebrate | PLXN A1 |
| SEMA6D | Vertebrate | PLXN A1, D1 |
| SEMA7A | Vertebrate | PLXN C1 |
| Poxvirus A39R | Virus | PLXN C1 |
Although plexins are the best‐studied semaphorin receptors, recent findings suggest that other proteins can also act as receptors by binding to the extracellular domain of semaphorins. For example, TIM2, which belongs to the Tim protein family and is characterized by expression on activated T cells and the presence of conserved immunoglobulin domain and mucin domains, is a receptor for SEMA4A.26 CD72 is a novel class of SEMA4D receptor on lymphocytes that belongs to the C‐type lectin family.27 Moreover, SEMA7A exerts an essential function by binding and stimulating monocytes through the α1β1 integrin receptor in both the nervous and immune systems.28
Brief Introduction of Cardiovascular Development
Cardiovascular development in embryos can be subdivided into heart development and vascular development. The heart is the first functional organ formed in mammals.9, 29, 30, 31, 32 Initially, the heart develops from clusters of progenitor cells, which coalesce to form the cardiac crescent and the heart tube.31, 32, 33 Subsequently, the heart tube undergoes elongation, looping, septation, remodeling, and maturation to form the final 4‐chambered organ.31, 32, 33 Multiple cell types, including myocardial, endocardial, epicardial, and neural crest cells (NCCs) act coordinately during complicated cardiogenesis in vertebrates. NCCs are a group of pluripotent cells that are generated at the edge of neural tubes. Cardiac NCCs migrate through pharyngeal arches 3, 4, and 6 to enter the distal region of the OFT, where they play a critical role in separation of the aorta root and pulmonary trunk.34, 35, 36 Some cardiac NCCs in the OFT region eventually become interstitial cells in semilunar valves. In addition, NCCs give rise to a group of smooth muscle cells in the pharyngeal arch arteries. In the proximal region of the OFT and atrioventricular (AV) canal region, a subset of endocardial cells respond to signals released from the myocardium and undergo epithelial‐mesenchymal transition (EMT) to become cushion mesenchymal cells during midgestation. The cellularized OFT and AV cushions facilitate unidirectional blood flow in embryos and are further remodeled into mature septa and valve structures.37, 38, 39, 40, 41 Most cushion mesenchymal cells become interstitial cells in valves. Vascular development is initiated with vasculogenesis. In this process, mesoderm‐derived angioblasts form a primitive vascular plexus,42 which is further developed into a mature blood vessel system to ensure proper blood supply for the whole body.43
Numerous signaling pathways and downstream transcription factors are required for normal cardiogenesis and blood vessel development.9, 31, 32, 33, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51 In the past 2 decades, accumulated studies have indicated that impaired semaphorin signaling results in various cardiovascular disorders during development and in multiple disease states. In this article we summarize recent discoveries regarding the function of semaphorins during mammalian cardiovascular development, with a primary focus on members of the SEMA3, SEMA5, and SEMA6 families, whose activities during cardiovascular development have been supported with gene inactivation studies in mice.
Versatile Activities of Semaphorins During Cardiovascular Development
Role of Class 3 Semaphorins During Cardiovascular Development
SEMA3A Role
SEMA3A regulates cardiac innervation patterning and is essential for heart rate control.52 Sema3A −/− mice lack a cardiac sympathetic innervation gradient, which leads to sinus bradycardia, abrupt sinus slowing, and stellate ganglia defects.52 In support of the direct role of SEMA3A in regulating innervation in cardiomyocytes, myocardial‐specific overexpression of Sema3A resulted in prolonged action potential duration, reduction of sympathetic innervation, and spontaneous ventricular arrhythmia.52 The role of SEMA3A in regulating sympathetic innervation is not limited to developing hearts. In a rat myocardial infarction model, overexpression of Sema3A in left stellate ganglion and myocardium reduces sympathetic reinnervation in the myocardial infarction border zone and the susceptibility to malignant arrhythmia.53, 54 In an independent gene inactivation study, the major cardiac phenotype of Sema3A homozygous mutant mice is postnatal right ventricle hypertrophy.55 The differential phenotypes reported in the literature are likely due to the specific knockout strategies and/or mouse strains used by different groups.
In support of the clinical relevance of SEMA3A function in rodent cardiomyocytes, addition of SEMA3A to cardiomyocytes derived from human‐induced pluripotent stem cells inhibited the Kv4.3 (Ito) channel, as observed in heterologous human embryonic kidney cells.56 Furthermore, several missense mutations in SEMA3A (R552C, R734W, and I334V) were shown to be associated with Brugada syndrome and unexplained cardiac arrest.56, 57 These mutations impaired the ability of SEMA3A to inhibit the Kv4.3 (Ito) channel.56
SEMA3A is also important for normal development of blood vessels. Knocking out Sema3A in mice led to abnormal patterning of anterior cardinal veins in the head and intersomitic vessels in the trunk region.58 The cranial blood vessels in mutants remain at the primitive capillary plexus stage and fail to remodel. The mouse vascular defects can be observed in the CD‐1 background but not in 129/Sv background, suggesting the involvement of other genetic factors in determining vascular phenotypes in Sema3A‐null mice. Severe defects in dorsal aorta development were also observed in zebrafish with sema3a either knocked down or overexpressed59; however, a similar phenotype was not reported in Sema3A mutant mice. A further mechanistic study showed that SEMA3A acts as a selective inhibitor of VEGF‐mediated angiogenesis via disruption of focal adhesion kinase/Src signaling and as a potent inducer of microvascular permeability via activation of NRP1.60
SEMA3C Roles
SEMA3C/PLXNA2 signaling and SEMA3C/NRP1 signaling are required for NCC development, which is essential for proper septation of the cardiac OFT.61, 62, 63 Using NCCs isolated from Hamburger Hamilton 10 chicken embryos, Toyofuku et al found that SEMA3C promoted NCC migration through PLXND1 and NRP1.64 Sema3C complete knockout mice are cyanotic and die shortly after birth from interruption of the aortic arch, persistent truncus arteriosus, and septation defects in the OFT.61 These morphological defects are likely caused by failure of NCCs to migrate into the proximal OFT.61 A recent study using a conditional gene inactivation approach indicated that SEMA3C expressed in NCCs activates NRP1 in endocardial cells of the OFT to promote EMT in OFT cushions,63 which are essential for proper OFT septation and semilunar valve formation. A recent study systematically examined the cis‐regulatory elements that control the proper expression of Sema3C in the OFT and pharyngeal arch regions.65 This group of researchers found that transcription factors FOXC1 and FOXC2 can directly bind the FOX binding sites in the enhancer region of Sema3C to promote its transcription in the OFT myocardium. In the pharyngeal arch region, expression of Sema3C is repressed by TBX1‐FGF8. This study strongly supports the idea that proper spatiotemporal expression of SEMA3C is essential for normal septation of the OFT.65
SEMA3C can also regulate blood vessel formation. It inhibits VEGF‐induced endothelial cell adhesion and migration through PLXND1 and NRP1 receptors in both in vitro and in vivo assays.66 Moreover, the local administration of SEMA3C into the vitreous body of a retinopathy of prematurity model prevents the formation of pathological retinal angiogenesis.66
SEMA3D Roles
Functions of SEMA3D during cardiovascular development have been found in multiple vertebrates. Knocking down expression of sema3D in zebrafish led to dysmorphic hearts with smaller ventricles, smaller atrium, and thickened myocardial wall.67 Endocardium was present in sema3D‐knockdown fish; however, AV valves and trabeculation were absent.
The function of SEMA3D in mammals appears to be different from that in zebrafish. Rather, mammalian SEMA3D provides repulsive cues to direct normal endothelial cell development. In vitro analyses using primary human umbilical vascular endothelial cells showed that exogenous SEMA3D inhibits endothelial cell migration and tube formation and that this activity requires PLXND1/NRP168, 69 and activation of the PI3K/AKT pathway.68 Inactivation of Sema3D in mice led to total anomalous pulmonary venous connection in which pulmonary veins abnormally enter the coronary sinus.70 These results suggest that signals provided by SEMA3D are particularly important for endothelial cells of pulmonary veins in vivo. Sema3D ‐/– mice can survive to adulthood but show severe cardiomegaly due to dilation of right atria and ventricles accompanied by left‐to‐right shunt, which is likely secondary to the total anomalous pulmonary venous connection defect.70 Furthermore, a point mutation (F602L) in SEMA3D was identified in a human patient with partial anomalous pulmonary venous connection.70
In addition to the loss‐of‐function allele, a gain‐of‐function allele was also identified in a human patient who carried a duplication of the 5′ half of SEMA3D.71 This patient displayed transposition of the great arteries, ventricular septal defect, and coarctation of the aorta. The authors speculated that migration of cardiac NCCs into the OFT is impaired in patients. However, a role of SEMA3D in NCCs has not been demonstrated using animal models.
SEMA3E Role
SEMA3E is a potent repulsive guidance cue for endothelial cells. Sema3E mRNA is robustly expressed in the caudal region of each somite in E11.5 mouse embryos from in situ hybridization analysis.72 Knocking out Sema3E led to disorganized intersomitic vessels, suggesting the essential role of SEMA3E in guiding intersomitic vessel formation and patterning. Further detailed examination of the Sema3E‐null mice revealed more vascular defects, including the paired dorsal aortas and fusion of a large plexus of blood vessels.73, 74 To further support SEMA3E as a repulsive cue for endothelial cells, electroporation of a SEMA3E expression plasmid into E3 chicken embryos reduced vessel formation in the area where SEMA3E was ectopically expressed.72
SEMA3E appears to act primarily through PLXND1 in blood vessels. Alkaline phosphatase‐tagged SEMA3E (AP‐SEMA3E) bound to COS cells expressing PLXND1, but not NRP1 or NRP2.72 AP‐SEMA3E bound to the blood vessels in sections from wild type but not PlnxD1 null embryos. Functional analysis showed that addition of SEMA3E caused collapse of PLXND1‐expressing COS cells. Unlike SEMA3D, SEMA3E‐mediated cytoskeletal reorganization does not require NRP1.68 Inactivation of PlxnD1 results in similar organizational defects in the somatic vasculature as observed in Sema3E‐null embryos.72
The Role of SEMA5A During Cardiovascular Development
SEMA5A is the only known member of the SEMA5 family for which there is clear genetic evidence to support its role during cardiovascular development. SEMA5A is a proangiogenic molecule that potently induces endothelial cell proliferation and migration and inhibits apoptosis.75, 76 Treatment of immortalized human dermal microvascular endothelial cells with SEMA5A significantly increased their migration. Furthermore, subcutaneous injection of SEMA5A‐containing matrigel enhanced blood vessel sprouting.75 Knocking out Sema5A in mice led to embryonic lethality between E11.5 and E12.5.77 A thorough examination of the cardiovascular system in mutants revealed that the number of secondary and tertiary branches of blood vessels in the cranial region was reduced, although the capillary network was not affected.77 No other cardiovascular defect was reported in mutant embryos. Therefore, the proangiogenic activity of SEMA5A is essential only in the cranial region in vivo. The major cause for the death of Sema5A‐null embryos remains unidentified, as the vascular defect in the cranial region is unlikely lethal to the embryo.
The Role of Class 6 Semaphorins During Cardiovascular Development
SEMA6A Role
In vitro analysis using human umbilical vascular endothelial cells showed that SEMA6A promotes endothelial cell survival and growth by modulating VEGFR2 signaling.78 The vascular defects in Sema6A‐null animals are limited to the eye. At the P4 stage, hyaloid vessels displayed a significantly reduced network complexity in mutant animals. Also at the same stage, the extension of the vascular network from the optic nerve to the periphery was also reduced in mutants. The vascular defects in eyes disappeared at the P8 stage.78 One likely reason is that other members of the semaphorin family are able to compensate for the loss of SEMA6A in mutant eyes at later stages. Sema3A is expressed in the vasculature of eyes during both embryonic and postnatal eye development79 and thus is a likely candidate.
SEMA6A inhibits migration of NCCs isolated from Hamburger Hamilton 10 chicken embryos, in contrast to SEMA3C, which stimulates NCC migration.64 However, no defect was observed in the OFT and pharyngeal arch arteries in Sema6A‐null mice. It is thus possible that SEMA6A exerts different activities in chicken and mammals.
SEMA6D Roles
The initial evidence indicating a role for SEMA6D during heart development came from chicken studies. It was found that SEMA6D could promote endocardial cell migration in the OFT region through the PLXNA1‐VEGFR2 receptor complex, whereas it inhibited endocardial cell migration in the ventricle region through the PLXNA1‐PTK7 receptor complex.18 In addition to acting on endocardial cells, SEMA6D can also regulate myocardial wall morphogenesis in chicken embryos through its cytoplasmic domain‐dependent reverse signaling.19 The chicken studies described above applied comprehensive loss‐of‐function and gain‐of‐function approaches; however, these activities of SEMA6D in chicken were not replicated in subsequent mouse studies.
We recently developed a novel conditional immortal AV cushion mesenchymal cell line, tsA58‐AVM, and used this line to identify Sema6D as the regulatory target of bone morphogenetic protein signaling in AV cushions.80 Conditional inactivation of Sema6D in endocardial cells of mouse embryos using the Nfatc1‐Cre driver led to hypocellular AV cushions at E9.25 and E9.5 due to reduced EMT in the AV canal region. Functional tests revealed that SEMA6D activates Rho through PLXNA1‐FARP1 to promote cushion mesenchymal cell formation in the AV canal80 (Figure 2). Thus, EMT by endocardial cells in the OFT and AV canal both rely on semaphorin signaling, with the OFT region needing SEMA3C and the AV canal region requiring SEMA6D. The AV cushion defect in Nfatc1‐Cre/Sema6D loxp/loxp embryos was resolved at a later stage (E10.5), likely due to the compensatory effect from increased expression of SEMA6C.80 Studies of double‐knockout mice of Sema6D and Sema6C are required to test this hypothesis. No defect in the OFT cushions or in the endocardial cells of ventricles was observed in Nfatc1‐Cre/Sema6D loxp/loxp embryos.80 No myocardial wall defect was reported in Sema6D complete knockout mice.81 The apparent difference in the functions of SEMA6D between chicken studies and mouse studies suggests that this cytokine has differential roles in the cardiovascular system in birds and mammals.
Figure 2.
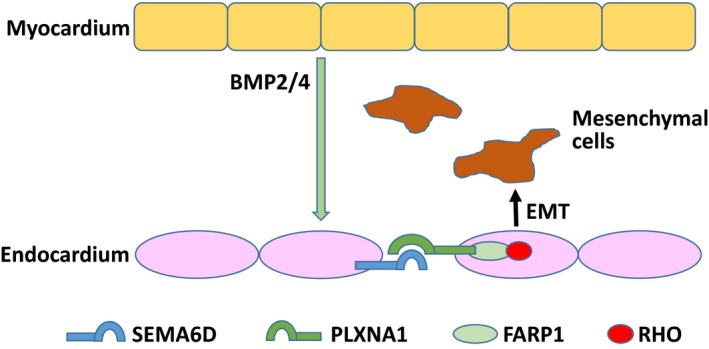
SEMA6D promotes EMT at AV cushions. In responding to BMP ligands released from the overlying myocardium (such as BMP2 and BMP4), expression of SEMA6D in endocardial cells is upregulated in the AV canal region of mouse embryos at E9.0–9.5. SEMA6D acts on adjacent endocardial cells through the PLXNA1/FARP1/RHO axis to promote cushion mesenchymal cell formation and migration in AV cushions. AV indicates atrioventricular; BMP, bone morphogenic protein; EMT, epithelial‐mesenchymal transition.
Summary
Multiple semaphorin molecules play an essential role in regulating cardiovascular development. This conclusion is supported by convincing mouse genetic evidence complemented with tests on other model systems including cell culture, zebrafish, and chicken studies (summarized in Table 2). The activities of semaphorin signaling are highly versatile and include regulation of NCC migration, endocardial cell EMT in the OFT and AV canal regions, cardiac innervation, myocardial wall morphogenesis, endothelial cell migration during blood vessel formation, and patterning of vessel networks.
Table 2.
Summary of Functions of Different Semaphorins During Cardiovascular Development
| Semaphorins | Cardiovascular Expression | Cardiovascular Defects in Mutants |
|---|---|---|
| SEMA3A | Trabecular zone of embryonic hearts, Purkinje fiber, vascular endothelia cells52, 58 |
Zebrafish: Abnormal dorsal aorta development59
Mouse: sinus bradycardia, abrupt sinus slowing, stellate ganglia defects, right ventricle hypertrophy, abnormal patterning of anterior cardinal veins and intersomitic vessels52, 55, 58 Human: Brugada syndrome56, 57 |
| SEMA3C | Neural crest cells, outflow tract myocardial cells61, 62, 63, 65 | Mouse: interruption of the aortic arch, persistent truncus arteriosus, septation defects in the outflow tract61, 62, 63 |
| SEMA3D | Mesocardial reflection and proepicardial organ in embryos, neural crest cells70 |
Zebrafish: dysmorphic hearts, absence of atrioventricular valves and trabeculation67
Mouse: total anomalous pulmonary venous connection, cardiomegaly70 Human: partial anomalous pulmonary venous connection, transposition of the great arteries, ventricular septal defect, coarctation of the aorta70, 71 |
| SEMA3E | Notochord, lateral plate mesoderm, caudal region of somites72, 73 | Mouse: disorganized intersomitic vessels, paired dorsal aortas, fusion of a large plexus of blood vessels72, 73, 74 |
| SEMA5A | Atrium septum, endocardial cells, cushion mesenchymal cells, mesoderm surrounding cranial vessels77 | Mouse: reduced number of secondary and tertiary branches of blood vessels in the cranial region77 |
| SEMA6A | Hyaloid vessels, retinal vessels78 | Mouse: reduced network complexity in hyaloid and retinal vessels at P4, defects resolved at P878 |
| SEMA6D | Myocardial, endocardial, cushion mesenchymal cells18, 19, 80 |
Chicken: altered endocardial cell migration, reduced myocardial wall trabeculation, small ventricle18, 19
Mouse: reduced cushion mesenchymal cell number at E9.5, defect resolved at later stages80 |
There remain some outstanding questions regarding the underlying mechanisms by which semaphorin signaling regulates cardiovascular development. We list 3 here. (1) The functions of semaphorins are highly cell‐type and/or tissue‐type dependent. In most cases it is unclear how such specificity is achieved. Detailed characterization of conditional gene‐inactivation mouse models, in which semaphorin/plexin genes are specifically inactivated in different cardiovascular cell types, would provide crucial clues to answer this question. (2) Semaphorins can activate many downstream effectors to accomplish their complex biological activities. Unlike many other signaling pathways, such as TGFβ/BMP signaling, there is no canonical pathway that is associated with semaphorin signaling. How the semaphorin/plexin complex on the cell surface selectively activates downstream cytoplasmic effectors in a context‐dependent fashion remains largely elusive. (3) Another critical question to be addressed is how semaphorin signaling interacts with other signaling pathways during cardiovascular development. Such interaction may occur at the cell surface through sharing (or competing for) the same coreceptors and/or in the cytoplasm through crosstalk between different cytoplasmic effectors.
Answering the above questions will help us design effective strategies to accurately and specifically modulate semaphorin signaling for therapeutic purposes. Recent studies have shown that tumor angiogenesis can be regulated by different semaphorins.4, 82 A better understanding of the molecular mechanism by which semaphorins regulate blood vessel formation may lead to identification of novel intervention targets for cancer treatment. Another potential area for translational research of semaphorin signaling is regenerative medicine. For example, semaphorin signaling is involved in generation of cushion mesenchymal cells in both the OFT and AV canal regions. These mesenchymal cells are precursors of septa and valves in mature hearts. Our knowledge of semaphorin signaling during OFT and AV cushion development may provide crucial guidance for us to differentiate pluripotent stem cells into valvular/septal cells for tissue repair. This area of research remains a blank in the literature.
In summary, semaphorins are versatile signaling molecules that regulate multiple aspects of cardiovascular development. Studies on semaphorin signaling are highly significant for both basic and translational research.
Sources of Funding
Research in the authors’ laboratory is supported by NIH R01 (R01HL095783), R03 (R03HD082634), and R21 (R21CA199586) grants awarded to Jiao.
Disclosures
None.
Acknowledgments
We regret that due to space limitations, the work of all of our colleagues could not be cited here. We thank the members of the Jiao laboratory for their comments and suggestions for the article.
J Am Heart Assoc. 2018;7:e008853 DOI: 10.1161/JAHA.118.008853.29776958
Contributor Information
Kexiang Liu, Email: kxliu64@hotmail.com.
Kai Jiao, Email: kjiao@uab.edu.
References
- 1. Tessier‐Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. 1996;274:1123–1133. [DOI] [PubMed] [Google Scholar]
- 2. Tamagnone L, Artigiani S, Chen H, He Z, Ming GI, Song H, Chedotal A, Winberg ML, Goodman CS, Poo M, Tessier‐Lavigne M, Comoglio PM. Plexins are a large family of receptors for transmembrane, secreted, and GPI‐anchored semaphorins in vertebrates. Cell. 1999;99:71–80. [DOI] [PubMed] [Google Scholar]
- 3. Goodman C, Kolodkin A, Luo Y, Püschel A, Raper J. Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97:551–552.10367884 [Google Scholar]
- 4. Neufeld G, Mumblat Y, Smolkin T, Toledano S, Nir‐Zvi I, Ziv K, Kessler O. The semaphorins and their receptors as modulators of tumor progression. Drug Resist Updat. 2016;29:1–12. [DOI] [PubMed] [Google Scholar]
- 5. Gurrapu S, Tamagnone L. Transmembrane semaphorins: multimodal signaling cues in development and cancer. Cell Adh Migr. 2016;10:675–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kruger RP, Aurandt J, Guan KL. Semaphorins command cells to move. Nat Rev Mol Cell Biol. 2005;6:789–800. [DOI] [PubMed] [Google Scholar]
- 7. Suzuki K, Kumanogoh A, Kikutani H. Semaphorins and their receptors in immune cell interactions. Nat Immunol. 2008;9:17–23. [DOI] [PubMed] [Google Scholar]
- 8. Kang S, Kumanogoh A. Semaphorins in bone development, homeostasis, and disease. Semin Cell Dev Biol. 2013;24:163–171. [DOI] [PubMed] [Google Scholar]
- 9. Epstein JA, Aghajanian H, Singh MK. Semaphorin signaling in cardiovascular development. Cell Metab. 2015;21:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kolodkin AL, Matthes DJ, O'Connor TP, Patel NH, Admon A, Bentley D, Goodman CS. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 1992;9:831–845. [DOI] [PubMed] [Google Scholar]
- 11. Siebold C, Jones EY. Structural insights into semaphorins and their receptors. Semin Cell Dev Biol. 2013;24:139–145. [DOI] [PubMed] [Google Scholar]
- 12. Yazdani U, Terman JR. The semaphorins. Genome Biol. 2006;7:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75:1389–1399. [DOI] [PubMed] [Google Scholar]
- 14. Love CA, Harlos K, Mavaddat N, Davis SJ, Stuart DI, Jones EY, Esnouf RM. The ligand‐binding face of the semaphorins revealed by the high‐resolution crystal structure of SEMA4D. Nat Struct Mol Biol. 2003;10:843–848. [DOI] [PubMed] [Google Scholar]
- 15. Antipenko A, Himanen J‐P, van Leyen K, Nardi‐Dei V, Lesniak J, Barton WA, Rajashankar KR, Lu M, Hoemme C, Püschel AW. Structure of the semaphorin‐3A receptor binding module. Neuron. 2003;39:589–598. [DOI] [PubMed] [Google Scholar]
- 16. Bork P, Doerks T, Springer TA, Snel B. Domains in plexins: links to integrins and transcription factors. Trends Biochem Sci. 1999;24:261–263. [DOI] [PubMed] [Google Scholar]
- 17. Gherardi E, Love CA, Esnouf RM, Jones EY. The sema domain. Curr Opin Struct Biol. 2004;14:669–678. [DOI] [PubMed] [Google Scholar]
- 18. Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Suto F, Kamei J, Aoki K, Yabuki M, Hori M, Fujisawa H, Kikutani H. Dual roles of Sema6D in cardiac morphogenesis through region‐specific association of its receptor, Plexin‐A1, with off‐track and vascular endothelial growth factor receptor type 2. Genes Dev. 2004;18:435–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Toyofuku T, Zhang H, Kumanogoh A, Takegahara N, Yabuki M, Harada K, Hori M, Kikutani H. Guidance of myocardial patterning in cardiac development by Sema6D reverse signalling. Nat Cell Biol. 2004;6:1204–1211. [DOI] [PubMed] [Google Scholar]
- 20. Negishi M, Oinuma I, Katoh H. Plexins: axon guidance and signal transduction. Cell Mol Life Sci. 2005;62:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fujisawa H. Discovery of semaphorin receptors, neuropilin and plexin, and their functions in neural development. J Neurobiol. 2004;59:24–33. [DOI] [PubMed] [Google Scholar]
- 22. He Z, Tessier‐Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90:739–751. [DOI] [PubMed] [Google Scholar]
- 23. Nogi T, Yasui N, Mihara E, Matsunaga Y, Noda M, Yamashita N, Toyofuku T, Uchiyama S, Goshima Y, Kumanogoh A. Structural basis for semaphorin signalling through the plexin receptor. Nature. 2010;467:1123–1127. [DOI] [PubMed] [Google Scholar]
- 24. Hota PK, Buck M. Plexin structures are coming: opportunities for multilevel investigations of semaphorin guidance receptors, their cell signaling mechanisms, and functions. Cell Mol Life Sci. 2012;69:3765–3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Klostermann A, Lohrum M, Adams RH, Puschel AW. The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. J Biol Chem. 1998;273:7326–7331. [DOI] [PubMed] [Google Scholar]
- 26. Kumanogoh A, Marukawa S, Suzuki K, Takegahara N, Watanabe C, Ch'ng E, Ishida I, Fujimura H, Sakoda S, Yoshida K, Kikutani H. Class IV semaphorin Sema4A enhances T‐cell activation and interacts with Tim‐2. Nature. 2002;419:629–633. [DOI] [PubMed] [Google Scholar]
- 27. Kumanogoh A, Watanabe C, Lee I, Wang X, Shi W, Araki H, Hirata H, Iwahori K, Uchida J, Yasui T, Matsumoto M, Yoshida K, Yakura H, Pan C, Parnes JR, Kikutani H. Identification of CD72 as a lymphocyte receptor for the class IV semaphorin CD100: a novel mechanism for regulating B cell signaling. Immunity. 2000;13:621–631. [DOI] [PubMed] [Google Scholar]
- 28. Suzuki K, Okuno T, Yamamoto M, Pasterkamp RJ, Takegahara N, Takamatsu H, Kitao T, Takagi J, Rennert PD, Kolodkin AL, Kumanogoh A, Kikutani H. Semaphorin 7A initiates T‐cell‐mediated inflammatory responses through α1β1 integrin. Nature. 2007;446:680–684. [DOI] [PubMed] [Google Scholar]
- 29. Schleich J‐M, Abdulla T, Summers R, Houyel L. An overview of cardiac morphogenesis. Arch Cardiovasc Dis. 2013;106:612–623. [DOI] [PubMed] [Google Scholar]
- 30. Sylva M, van den Hoff MJ, Moorman AF. Development of the human heart. Am J Med Genet A. 2014;164:1347–1371. [DOI] [PubMed] [Google Scholar]
- 31. Epstein JA. Cardiac development and implications for heart disease. N Engl J Med. 2010;363:1638–1647. [DOI] [PubMed] [Google Scholar]
- 32. Christoffels VM, Burch JB, Moorman AF. Architectural plan for the heart: early patterning and delineation of the chambers and the nodes. Trends Cardiovasc Med. 2004;14:301–307. [DOI] [PubMed] [Google Scholar]
- 33. Sizarov A, Ya J, de Boer BA, Lamers WH, Christoffels VM, Moorman AF. Formation of the building plan of the human heart. Circulation. 2011;123:1125–1135. [DOI] [PubMed] [Google Scholar]
- 34. Creazzo TL, Godt RE, Leatherbury L, Conway SJ, Kirby ML. Role of cardiac neural crest cells in cardiovascular development. Annu Rev Physiol. 1998;60:267–286. [DOI] [PubMed] [Google Scholar]
- 35. Maschhoff KL, Baldwin HS. Molecular determinants of neural crest migration. Am J Med Genet. 2000;97:280–288. [DOI] [PubMed] [Google Scholar]
- 36. Plein A, Fantin A, Ruhrberg C. Neural crest cells in cardiovascular development. Curr Top Dev Biol. 2015;111:183–200. [DOI] [PubMed] [Google Scholar]
- 37. Combs MD, Yutzey KE. Heart valve development: regulatory networks in development and disease. Circ Res. 2009;105:408–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lin CJ, Lin CY, Chen CH, Zhou B, Chang CP. Partitioning the heart: mechanisms of cardiac septation and valve development. Development. 2012;139:3277–3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Markwald RR, Norris RA, Moreno‐Rodriguez R, Levine RA. Developmental basis of adult cardiovascular diseases: valvular heart diseases. Ann N Y Acad Sci. 2010;1188:177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. [DOI] [PubMed] [Google Scholar]
- 41. Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Annu Rev Physiol. 2011;73:29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73–91. [DOI] [PubMed] [Google Scholar]
- 43. Risau W. Mechanisms of angiogenesis. Nature. 1997;386:671. [DOI] [PubMed] [Google Scholar]
- 44. Harvey RP. Patterning the vertebrate heart. Nat Rev Genet. 2002;3:544. [DOI] [PubMed] [Google Scholar]
- 45. Qian Y, Xiao D, Guo X, Chen H, Hao L, Ma X, Huang G, Ma D, Wang H. Multiple gene variations contributed to congenital heart disease via GATA family transcriptional regulation. J Transl Med. 2017;15:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. England J, Granados‐Riveron J, Polo‐Parada L, Kuriakose D, Moore C, Brook JD, Rutland CS, Setchfield K, Gell C, Ghosh TK. Tropomyosin 1: multiple roles in the developing heart and in the formation of congenital heart defects. J Mol Cell Cardiol. 2017;106:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wilting J, Christ B. Embryonic angiogenesis: a review. Naturwissenschaften. 1996;83:153–164. [DOI] [PubMed] [Google Scholar]
- 48. Drake CJ, Fleming PA. Vasculogenesis in the day 6.5 to 9.5 mouse embryo. Blood. 2000;95:1671–1679. [PubMed] [Google Scholar]
- 49. Tepper OM, Capla JM, Galiano RD, Ceradini DJ, Callaghan MJ, Kleinman ME, Gurtner GC. Adult vasculogenesis occurs through in situ recruitment, proliferation, and tubulization of circulating bone marrow‐derived cells. Blood. 2005;105:1068–1077. [DOI] [PubMed] [Google Scholar]
- 50. Chetty SC, Rost MS, Enriquez JR, Schumacher JA, Baltrunaite K, Rossi A, Stainier DY, Sumanas S. VEGF signaling promotes vascular endothelial differentiation by modulating ETV2 expression. Dev Biol. 2017;424:147–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Baltrunaite K, Craig MP, Desai SP, Chaturvedi P, Pandey RN, Hegde RS, Sumanas S. ETS transcription factors Etv2 and Fli1b are required for tumor angiogenesis. Angiogenesis. 2017;20:307–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ieda M, Kanazawa H, Kimura K, Hattori F, Ieda Y, Taniguchi M, Lee J‐K, Matsumura K, Tomita Y, Miyoshi S. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat Med. 2007;13:604–612. [DOI] [PubMed] [Google Scholar]
- 53. Chen RH, Li YG, Jiao KL, Zhang PP, Sun Y, Zhang LP, Fong XF, Li W, Yu Y. Overexpression of Sema3a in myocardial infarction border zone decreases vulnerability of ventricular tachycardia post‐myocardial infarction in rats. J Cell Mol Med. 2013;17:608–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yang L‐C, Zhang P‐P, Chen X‐M, Li C‐Y, Sun J, Hou J‐W, Chen R‐H, Wang Y‐P, Li Y‐G. Semaphorin 3a transfection into the left stellate ganglion reduces susceptibility to ventricular arrhythmias after myocardial infarction in rats. Europace. 2015;18:1886–1896. [DOI] [PubMed] [Google Scholar]
- 55. Behar O, Golden JA, Mashimo H, Schoen FJ, Fishman MC. Semaphorin III is needed for normal patterning and growth of nerves, bones and heart. Nature. 1996;383:525. [DOI] [PubMed] [Google Scholar]
- 56. Boczek NJ, Ye D, Johnson EK, Wang W, Crotti L, Tester DJ, Dagradi F, Mizusawa Y, Torchio M, Alders M. Characterization of SEMA3A‐encoded semaphorin as a naturally occurring Kv4.3 protein inhibitor and its contribution to Brugada syndrome. Circ Res. 2014;115:460–469. DOI: 10.1161/CIRCRESAHA.115.303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nakano Y, Chayama K, Ochi H, Toshishige M, Hayashida Y, Miki D, Hayes CN, Suzuki H, Tokuyama T, Oda N. A nonsynonymous polymorphism in semaphorin 3A as a risk factor for human unexplained cardiac arrest with documented ventricular fibrillation. PLoS Genet. 2013;9:e1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Serini G, Valdembri D, Zanivan S, Morterra G, Burkhardt C, Caccavari F, Zammataro L, Primo L, Tamagnone L, Logan M. Class 3 semaphorins control vascular morphogenesis by inhibiting integrin function. Nature. 2003;424:391–397. [DOI] [PubMed] [Google Scholar]
- 59. Shoji W, Isogai S, Sato‐Maeda M, Obinata M, Kuwada JY. Semaphorin3a1 regulates angioblast migration and vascular development in zebrafish embryos. Development. 2003;130:3227–3236. [DOI] [PubMed] [Google Scholar]
- 60. Acevedo LM, Barillas S, Weis SM, Göthert JR, Cheresh DA. Semaphorin 3A suppresses VEGF‐mediated angiogenesis yet acts as a vascular permeability factor. Blood. 2008;111:2674–2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Feiner L, Webber AL, Brown CB, Lu MM, Jia L, Feinstein P, Mombaerts P, Epstein JA, Raper JA. Targeted disruption of semaphorin 3C leads to persistent truncus arteriosus and aortic arch interruption. Development. 2001;128:3061–3070. [DOI] [PubMed] [Google Scholar]
- 62. Brown CB, Feiner L, Lu M‐M, Li J, Ma X, Webber AL, Jia L, Raper JA, Epstein JA. PlexinA2 and semaphorin signaling during cardiac neural crest development. Development. 2001;128:3071–3080. [DOI] [PubMed] [Google Scholar]
- 63. Plein A, Calmont A, Fantin A, Denti L, Anderson NA, Scambler PJ, Ruhrberg C. Neural crest–derived SEMA3C activates endothelial NRP1 for cardiac outflow tract septation. J Clin Invest. 2015;125:2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Toyofuku T, Yoshida J, Sugimoto T, Yamamoto M, Makino N, Takamatsu H, Takegahara N, Suto F, Hori M, Fujisawa H. Repulsive and attractive semaphorins cooperate to direct the navigation of cardiac neural crest cells. Dev Biol. 2008;321:251–262. [DOI] [PubMed] [Google Scholar]
- 65. Kodo K, Shibata S, Miyagawa‐Tomita S, Ong SG, Takahashi H, Kume T, Okano H, Matsuoka R, Yamagishi H. Regulation of Sema3c and the interaction between cardiac neural crest and second heart field during outflow tract development. Sci Rep. 2017;7:6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Yang WJ, Hu J, Uemura A, Tetzlaff F, Augustin HG, Fischer A. Semaphorin‐3C signals through Neuropilin‐1 and PlexinD1 receptors to inhibit pathological angiogenesis. EMBO Mol Med. 2015;7:1267–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sato M, Tsai H‐J, Yost HJ. Semaphorin3D regulates invasion of cardiac neural crest cells into the primary heart field. Dev Biol. 2006;298:12–21. [DOI] [PubMed] [Google Scholar]
- 68. Aghajanian H, Choi C, Ho VC, Gupta M, Singh MK, Epstein JA. Semaphorin 3d and semaphorin 3e direct endothelial motility through distinct molecular signaling pathways. J Biol Chem. 2014;289:17971–17979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hamm MJ, Kirchmaier BC, Herzog W. Sema3d controls collective endothelial cell migration by distinct mechanisms via Nrp1 and PlxnD1. J Cell Biol. 2016;215:415–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Degenhardt K, Singh MK, Aghajanian H, Massera D, Wang Q, Li J, Li L, Choi C, Yzaguirre AD, Francey LJ. Semaphorin 3d signaling defects are associated with anomalous pulmonary venous connections. Nat Med. 2013;19:760–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Sanchez‐Castro M, Pichon O, Briand A, Poulain D, Gournay V, David A, Caignec CL. Disruption of the SEMA3D gene in a patient with congenital heart defects. Hum Mutat. 2015;36:30–33. [DOI] [PubMed] [Google Scholar]
- 72. Gu C, Yoshida Y, Livet J, Reimert DV, Mann F, Merte J, Henderson CE, Jessell TM, Kolodkin AL, Ginty DD. Semaphorin 3E and plexin‐D1 control vascular pattern independently of neuropilins. Science. 2005;307:265–268. [DOI] [PubMed] [Google Scholar]
- 73. Meadows SM, Fletcher PJ, Moran C, Xu K, Neufeld G, Chauvet S, Mann F, Krieg PA, Cleaver O. Integration of repulsive guidance cues generates avascular zones that shape mammalian blood vessels. Circ Res. 2012;110:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Meadows SM, Ratliff LA, Singh MK, Epstein JA, Cleaver O. Resolution of defective dorsal aortae patterning in Sema3E‐deficient mice occurs via angiogenic remodeling. Dev Dyn. 2013;242:580–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sadanandam A, Rosenbaugh EG, Singh S, Varney M, Singh RK. Semaphorin 5A promotes angiogenesis by increasing endothelial cell proliferation, migration, and decreasing apoptosis. Microvasc Res. 2010;79:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Sadanandam A, Sidhu S, Wullschleger S, Singh S, Varney M, Yang C, Ashour A, Batra SK, Singh R. Secreted semaphorin 5A suppressed pancreatic tumour burden but increased metastasis and endothelial cell proliferation. Br J Cancer. 2012;107:501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fiore R, Rahim B, Christoffels VM, Moorman AF, Püschel AW. Inactivation of the Sema5a gene results in embryonic lethality and defective remodeling of the cranial vascular system. Mol Cell Biol. 2005;25:2310–2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Segarra M, Ohnuki H, Maric D, Salvucci O, Hou X, Kumar A, Li X, Tosato G. Semaphorin 6A regulates angiogenesis by modulating VEGF signaling. Blood. 2012;120:4104–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ko JA, Mizuno Y, Yanai R, Chikama T, Sonoda KH. Expression of semaphorin 3A and its receptors during mouse corneal development. Biochem Biophys Res Commun. 2010;403:305–309. [DOI] [PubMed] [Google Scholar]
- 80. Peng Y, Song L, Li D, Kesterson R, Wang J, Wang L, Rokosh G, Wu B, Wang Q, Jiao K. Sema6D acts downstream of bone morphogenetic protein signalling to promote atrioventricular cushion development in mice. Cardiovasc Res. 2016;112:532–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Takamatsu H, Takegahara N, Nakagawa Y, Tomura M, Taniguchi M, Friedel RH , Rayburn H, Tessier‐Lavigne M, Yoshida Y, Okuno T, Mizui M, Kang S, Nojima S, Tsujimura T, Nakatsuji Y, Katayama I, Toyofuku T, Kikutani H, Kumanogoh A. Semaphorins guide the entry of dendritic cells into the lymphatics by activating myosin II. Nat Immunol. 2010;11:594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Neufeld G, Mumblat Y, Smolkin T, Toledano S, Nir‐Zvi I, Ziv K, Kessler O. The role of the semaphorins in cancer. Cell Adh Migr. 2016;10:652–674. [DOI] [PMC free article] [PubMed] [Google Scholar]


