Abstract
DEAD box proteins are putative RNA helicases that function in all aspects of RNA metabolism, including translation, ribosome biogenesis, and pre-mRNA splicing. Because many processes involving RNA metabolism are spatially organized within the cell, we examined the subcellular distribution of a human DEAD box protein, DDX1, to identify possible biological functions. Immunofluorescence labeling of DDX1 demonstrated that in addition to widespread punctate nucleoplasmic labeling, DDX1 is found in discrete nuclear foci ∼0.5 μm in diameter. Costaining with anti-Sm and anti-promyelocytic leukemia (PML) antibodies indicates that DDX1 foci are frequently located next to Cajal (coiled) bodies and less frequently, to PML bodies. Most importantly, costaining with anti-CstF-64 antibody indicates that DDX1 foci colocalize with cleavage bodies. By microscopic fluorescence resonance energy transfer, we show that labeled DDX1 resides within a Förster distance of 10 nm of labeled CstF-64 protein in both the nucleoplasm and within cleavage bodies. Coimmunoprecipitation analysis indicates that a proportion of CstF-64 protein resides in the same complex as DDX1. These studies are the first to identify a DEAD box protein associating with factors involved in 3′-end cleavage and polyadenylation of pre-mRNAs.
INTRODUCTION
DEAD box proteins are a family of putative RNA helicases found in all cellular organisms and in some viruses. They are characterized by eight conserved amino acid motifs, including the core DEAD (Asp-Glu-Ala-Asp) motif involved in ATP hydrolysis and coupling of ATPase and RNA helicase activity (Pause and Sonenberg, 1992). At least 14 human DEAD box proteins have been identified to date, summarized in the DExH/D protein family database (Jankowsky and Jankowsky, 2000). DEAD box proteins are thought to modulate RNA secondary structure in all cellular processes involving RNA, including transcription, pre-mRNA processing, ribosome biogenesis, RNA export, translation initiation, and RNA degradation (Schmid and Linder, 1992; de la Cruz et al., 1999). Although many of the biological functions of the prokaryotic and lower eukaryotic DEAD box proteins have been identified, DEAD box proteins in higher eukaryotes remain largely uncharacterized.
DDX1 is a human DEAD box protein that was identified by differential screening of a cDNA library enriched in transcripts present in two retinoblastoma (RB) cell lines: Y79 and RB522A (Godbout and Squire, 1993). The 2.7-kb DDX1 transcript encodes a protein with a predicted molecular mass of 82.4 kDa (Godbout et al., 1998). In addition to the eight conserved DEAD box family motifs, DDX1 also contains a region with homology to heterogeneous nuclear ribonucleoprotein U (hnRNP U) (Godbout et al., 1994). HnRNP U or scaffold attachment factor A, a protein located in the nuclear matrix, has recently been shown to function as a repressor of RNA polymerase II elongation by inhibiting transcription factor TFIIH-mediated carboxyl-terminal domain phosphorylation (Kim and Nikodem, 1999). Interestingly, the domain common to both hnRNP U and DDX1 was found to mediate binding of TFIIH to the RNA polymerase II holoenzyme.
The DDX1 gene has been mapped to chromosome 2p24, ∼400 kb telomeric to the proto-oncogene MYCN (Amler et al., 1996; Kuroda et al., 1996; Noguchi et al., 1996; Pandita et al., 1997). MYCN, a member of the myc family of transcription factors, is amplified and overexpressed in approximately one-third of all neuroblastoma (NB) tumors as well as in <10% of RB tumors (Brodeur et al., 1984; Seeger et al., 1985; Sakai et al., 1988; Doz et al., 1996). DDX1 is coamplified with MYCN and overexpressed in a subset of NB and RB cell lines and tumors (Godbout and Squire, 1993; Squire et al., 1995). Preliminary studies have shown that NB patients with amplification of both DDX1 and MYCN have a worse prognosis than do patients with only the MYCN gene amplified (Squire et al., 1995; George et al., 1996). The role of DDX1 in the tumorigenic process is not known, although it is predicted to involve RNA binding and the modulation of RNA secondary structure.
The eukaryotic cell nucleus is spatially organized into many subnuclear domains, including the nucleolus, Cajal (coiled) bodies, splicing factor compartments (speckles), promyelocytic leukemia (PML) nuclear bodies, Sam68 nuclear bodies (SNBs), and cleavage bodies (Schul et al., 1998; Misteli, 2000). A number of these structures have been implicated in RNA metabolism. For example, splicing factor compartments are thought to supply pre-mRNA splicing factors (Fu and Maniatis, 1990) as well as poly(A) binding protein II (Krause et al., 1994) and hyperphosphorylated RNA polymerase II (Bregman et al., 1995) to neighboring RNA transcription/processing sites. Cajal bodies, first discovered in 1903 by light microscopy, contain components involved in pre-mRNA splicing, rRNA processing, and histone mRNA 3′ end maturation (Matera, 1999; Gall, 2000). PML bodies are frequently found adjacent to Cajal bodies and contain proteins involved in RNA processing as well as apoptosis (Seeler and Dejean, 1999; Ruggero et al., 2000). Also located next to Cajal bodies and sometimes PML bodies, cleavage bodies contain proteins involved in pre-mRNA 3′-end cleavage and polyadenylation as well as the transcription factors TFIIE and TFIIF (Schul et al., 1996; Gall, 2000). Recently, the DEAD box protein Gemin3 has been shown to be associated with the spinal muscular atrophy gene product SMN in nuclear bodies called gems (Charroux et al., 1999). Gems are similar in size and commonly associate with Cajal bodies (Matera and Frey, 1998; Carvalho et al., 1999).
Previous cellular fractionation experiments with anti-DDX1 polyclonal antibodies revealed that DDX1 is primarily a nuclear protein, although it is also abundant in the cytoplasm of DDX1-amplified cells (Godbout et al., 1998). To determine the subnuclear localization of DDX1, we examined DDX1 distribution by confocal and digital imaging microscopy. DDX1 was found to be widely distributed throughout the nucleus; however, it was also located within nuclear foci of 0.5 μm diameter. These foci frequently costained with anti-CstF-64, previously reported to recognize cleavage bodies. Both coimmunoprecipitation and fluorescence resonance energy transfer (FRET) analyses suggest that CstF-64 and DDX1 are in proximity to each other. We propose that, like CstF-64, DDX1 may play a role in the processing of pre-mRNAs.
MATERIALS AND METHODS
Cell Culture
HeLa (human cervical carcinoma cells), GM38 (normal human lung fibroblasts), SKN (human neuroblastoma cells), COS-7 (transformed African green monkey kidney fibroblasts), HISM human intestinal smooth muscle cells, T24 (human bladder carcinoma cells), MRC-5 (normal human embryo lung fibroblasts), 293 (transformed human kidney cells), C3H 10T1/2 (mouse embryo fibroblasts), NIH 3T3 (mouse embryo fibroblasts), and Indian muntjac fibroblasts were grown on glass coverslips at 37°C in a 6% CO2 atmosphere in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal calf serum and antibiotics (100 U/ml penicillin and 100 μg/ml streptomycin [Invitrogen, Carlsbad, CA]). Human retinoblastoma cells RB778, RB893, RB522A, and RB(E)-2 were grown in suspension with the same medium described above. The cells were allowed to adhere to poly-l-lysine-coated coverslips for 30 min. All cells were used at 40–70% confluence. RNA was labeled by adding 2 mM 5′-fluorouridine (FU) (Sigma, St. Louis, MO) to cells at 37°C for 5, 10, or 15 min as previously described (Boisvert et al., 2000).
Immunofluorescence Labeling
Cells adhering to coverslips were fixed in 1% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min and permeabilized for 5 min in 0.5% Triton X-100/PBS. Alternatively, cells were fixed and permeabilized in −20°C 1:1 methanol/acetone for 10 min. Cells were incubated for 1 h in blocking buffer (3% bovine serum albumin in PBS) containing the primary antibody, rinsed in PBS, and incubated in blocking buffer containing the secondary antibody. Coverslips were mounted onto slides with glycerol containing 1 mg/ml p-phenylenediamine + 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI).
Antibodies and Immunoprecipitation
Rabbit polyclonal DDX1 antiserum (batch 2923) was produced using the use of nondenatured recombinant DDX1 (amino acids 1–186). To verify antibody specificity, GST-DDX1 was bacterially expressed and purified with the use of glutathione-Sepharose 4B (Godbout et al., 1998). Glutathione S-transferase (GST) was cleaved with thrombin and DDX1 (0 or 5 μg) was spotted onto nitrocellulose. The filters were incubated in blocking buffer and subsequently in primary antibody (anti-DDX1 or unrelated rabbit polyclonal anti-aldehyde dehydrogenase) at the dilution used for immunofluorescence labeling. After an overnight incubation at 4°C, the unbound antibody was used to stain fixed and permeabilized cells for immunofluorescence detection.
The following primary antibodies were used in this study: rabbit polyclonal p80 coilin (Andrade et al., 1993), mouse monoclonal Y12 against the Sm antigens (Pettersson et al., 1984), mouse monoclonal CstF-64 (Takagaki et al., 1990), mouse monoclonal Sam68 (Santa Cruz Biotechnology, Santa Cruz, CA), mouse monoclonal SC-35 (ATCC hybridoma 2031), mouse monoclonal 5E10 against PML bodies (Stuurman et al., 1992), and mouse monoclonal bromodeoxyuridine, which recognizes halogenated UTP (Sigma). Secondary antibodies included Alexa 488 goat anti-mouse, Alexa 594 goat anti-rabbit (Cedarlane Laboratories, Oakville, ON, Canada), and Cy3 donkey anti-rabbit (Jackson Immunoresearch, West Grove, PA).
For the coimmunoprecipitation experiments, we used rabbit polyclonal antiserum (batch 2910) generated with the use of denatured recombinant DDX1 (amino acids 1–186) (Godbout et al., 1998). The antibody was precleared with protein A-agarose beads and incubated for 3 h with nuclear extracts prepared from HeLa cells (Dignam et al., 1983). Immune complexes were precipitated with protein A-agarose beads and run on an SDS polyacrylamide gel. Proteins were transferred to nitrocellulose membrane by electroblotting, and Western blot analysis was carried out with anti-DDX1 antibody, anti-CstF-64 antibody, and anti-Sam68 antibody.
Affinity Purification of Anti-DDX1 Antibody
One milligram of glutathione-Sepharose 4B-purified GST-DDX1 was buffer exchanged into 1 ml of coupling buffer (0.2 M NaHCO3, 0.5 M NaCl; pH 8.3) with the use of P30 centricons (Millipore, Bedford, MA). The recombinant DDX1 was then applied to a 1-ml HiTrap N-hydroxysuccinimide activated Sepharose column (Amersham Pharmacia Biotech, Piscataway, NJ) and incubated for 30 min at room temperature. Deactivation and washing of the column was performed with alternate washes of 0.5 M ethanolamine, 0.5 M NaCl (pH 8.3) and 0.1 M acetate, 0.5 M NaCl (pH 4.0). The column was equilibrated with Tris-buffered saline and 1 ml of anti-DDX1 antiserum from batch 2923 was diluted 10-fold with Tris-buffered saline and applied to the GST-DDX1 affinity column at a flow rate of 1 ml/min and allowed to recirculate for 18 h at 4°C. The column was washed with 85% buffer A (10 mM Tris-HCl, 20% glycerol; pH 8.0) and 15% buffer B (buffer A + 1 M NaCl). Nonspecifically bound proteins were eluted with 50% buffer A and 50% buffer B. Antibodies were eluted with 100 mM glycine pH 3.0 and fractions neutralized to pH 8.0 with 1 M Tris-HCl, pH 11.0. The anti-DDX1 antibodies were concentrated with the use of P30 centricons and buffer-exchanged with buffer A in a final volume of 1 ml.
Light Microscopy
Images of single-labeled cells were collected on a Zeiss-Axioplan II microscope (Carl Zeiss, Thornwood, NY) with a cooled charge-coupled device camera (Cooke Corporation, Auburn Hills, MI). All double-labeled cells were viewed on a Zeiss LSM 510 confocal microscope with a plan apochromat 63×/1.4 oil immersion lens. Argon and helium-neon (HeNe) lasers were sequentially used to scan at wavelengths 488 and 543 nm, respectively. A UV laser was used to excite DAPI-stained cells. The image stacks were three-dimensionally reconstructed by maximum likelihood projections using the Zeiss LSM 510 image analysis software. To measure the three-dimensional distances between structures, the centers of each foci were marked and the Zeiss LSM 510 software calculated the distance based on pixel dimensions. For colocalization experiments, a minimum of 20 cells was three-dimensionally reconstructed and examined.
Where indicated, images were deconvolved with the use of Softworks 2.5 software (Applied Precision Inc., Issaquah, WA) to remove out-of-focus information. The required point spread functions were generated as recommended by the manufacturer. The deconvolution program is based on a constrained iterative algorithm developed in the laboratory of John Sedat (University of California, San Francisco, San Francisco, CA).
To determine the likelihood of nuclear bodies randomly colocalizing with DDX1 foci, we used a method published by Grande et al. (1996) that takes into account total cell volume and the size of the foci in question as well as their abundance. This calculation is based on the following equation: p = (4/3)π(d)3·n·m/v where p is the probability, d is the distance between the centers of adjacent structures, n and m are the average number of each structure per nucleus, and v is the volume of the nucleus in cubic micrometers.
Microscopic Fluorescence Resonance Energy Transfer
HeLa cells were fixed, permeabilized, and labeled for DDX1, CstF-64, PML, Sm, and p80 coilin as described above, with the exception that the coverslips were mounted in glycerol without p-phenylenediamine. The donors, anti-CstF-64, anti-PML, and anti-Sm antibodies, were labeled with Alexa 488 goat anti-mouse secondary antibody. The acceptors, anti-DDX1 and anti-p80 coilin antibodies, were labeled with Cy3 donkey anti-rabbit secondary antibody. Cells were observed with a Zeiss LSM 510 microscope, as described above, except that all images were collected with the pinhole apertures wide open. Alexa 488 was excited with a 488-nm Argon laser and detected with a 505–550-nm band-pass filter. Cy3 was excited with a 543-nm HeNe laser and detected with a 560-nm long-pass filter. The FRET image was collected with the 488-nm Argon laser for excitation and the 560-nm long-pass filter for detection. Photodestruction of the acceptor was done with the 543-nm HeNe laser.
FRET was determined on a pixel-by-pixel basis with the three-filter set method as described by Youvan et al. (1997). The background noise of every channel in all images was calculated by averaging four regions of 20 pixels outside of the cell. Subtracting the background value plus twice the SD produced corrected images. The final FRET image was calculated according to the following equation: final FRET = FRET − (FRET·donor ratio + FRET·acceptor ratio), where the donor and acceptor ratios are the amount of respective fluorophores contributing to the FRET intensity (calculated with the use of single labeled donor and acceptor slides). The Cy3 did not exhibit any excitation by the Argon laser at 488 at the current filter and laser settings. The final FRET images were corrected for background as described above with the use of Metamorph 4.5 software (Universal Imaging, Downingtown, PA). All images were rescaled identically in Adobe Photoshop 6.0 (Adobe Systems, Mountain View, CA) using the CstF-64 image to define the maximum value.
RESULTS
Subcellular Localization of DDX1
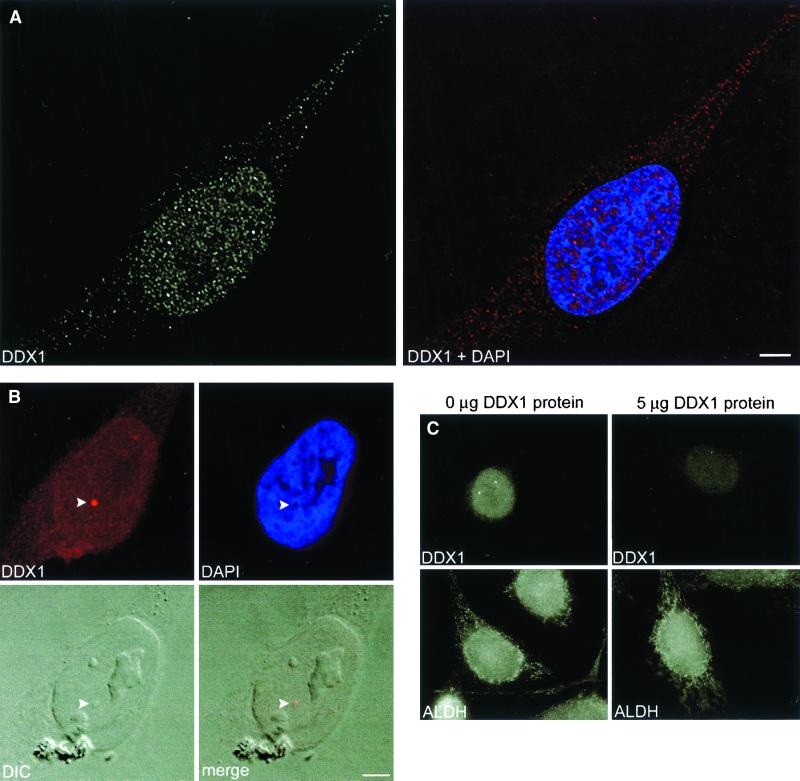
The subcellular distribution of DDX1 in HeLa cells was examined by indirect immunofluorescence using anti-DDX1 antiserum (2923). We observed predominant staining in the nucleus, although there was some signal in the cytoplasm as well (Figure 1A). These results are in agreement with previous cellular fractionation experiments (Godbout et al., 1998). In the nucleus, DDX1 was present in a granular/punctate pattern throughout most of the nucleoplasm, as well as in a few brightly labeled discrete foci. DDX1 was generally depleted in the nucleolus. Simultaneous staining with DAPI precluded a general association with chromatin. Approximately 90% of HeLa cells contained at least 1 DDX1 focus and 70% of these foci were visible by digital interference contrast (Figure 1B). The number of foci in each cell varied from 0 to 15, with an average of 4.7, each ∼0.5 μm in diameter. To ensure that DDX1 foci were not due to an artifact caused by cell fixation in paraformaldehyde, we also fixed cells with methanol/acetone. DDX1 foci were observed regardless of the fixative used.
Figure 1.
Immunofluorescence labeling of DDX1. HeLa cells were immunolabeled with anti-DDX1 polyclonal antibody (2923). (A) General staining pattern of DDX1 and its distribution relative to chromatin (DAPI). DDX1 is widely expressed in the nucleus, where it is found in both foci and nucleoplasm. The DDX1 signal has a punctate appearance in both the nucleoplasm and cytoplasm. Nuclear foci are shown in red in the merged (DDX1 + DAPI) image. The cells were observed with a Zeiss Axioplan II microscope equipped with a cooled, charge-coupled device camera; the image collected was deconvolved with the use of Softworks 2.5 software. (B) DDX1 foci are often visible by digital interference contrast (DIC) (indicated by the arrowhead). The image was collected with a Zeiss LSM 510 confocal laser scanning microscope. (C) Anti-DDX1 antibody is specifically competed by purified DDX1. Either anti-DDX1 polyclonal antibody (2923) or anti-ALDH polyclonal antibody was incubated overnight with either 0 or 5 μg of nitrocellulose-bound purifed DDX1 protein. The unbound fraction was then used to indirectly label HeLa cells. The image was collected with a Zeiss Axioplan II microscope. The DDX1 signal is greatly reduced after antibody adsorption to 5 μg of DDX1 protein, whereas the ALDH signal is unaffected. Bar, 5 μm.
To verify that anti-DDX1 antiserum specifically recognized DDX1, we carried out competition experiments with bacterially expressed DDX1 protein as described in MATERIALS AND METHODS. As shown in Figure 1C, the DDX1 signal was specifically reduced with 5 μg of DDX1 protein. No alteration in signal intensity was obtained with anti-aldehyde dehydrogenase (ALDH) antibody in the presence of DDX1 protein. Antibody specificity was further documented with anti-DDX1 antibody that had been affinity-purified with a DDX1-Sepharose column. Western blot analysis demonstrated that the affinity-purified antibody recognized a predominant band at 90 kDa, representing the main form of DDX1 (our unpublished data). Immunofluorescence staining with purified anti-DDX1 antibody gave a pattern indistinguishable from that of the nonpurified antiserum.
We examined 15 cell lines or cultures, including normal and transformed cells from different species, to determine the prevalence of nuclear DDX1 foci. Ten cell lines contained discrete nuclear DDX1 foci, including HeLa, GM38, 293, T24, NIH 3T3, RB778, MRC5, SKN, COS-7, and RB522A (which overexpresses DDX1 as the result of having amplified copies of the gene). Five cell lines contained DDX1 protein but no discrete foci, including Indian muntjac fibroblasts, C3H 10T1/2, HISM, RB(E)-2, and RB893. The presence of DDX1 foci in the normal lung fibroblast culture GM38 indicates that these foci are not restricted to transformed cells. These results suggest that DDX1 foci are nuclear bodies commonly found in a wide variety of cells.
DDX1 Foci Are Adjacent to PML and Cajal Bodies
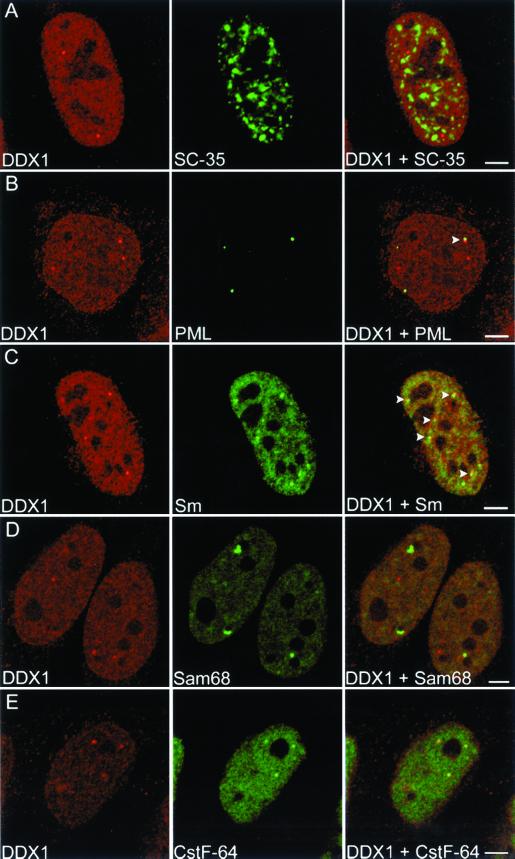
A number of nuclear bodies have been identified to date. Among the best defined are the splicing factor compartment (speckle), the PML oncogenic domain, and the Cajal body. Within the eukaryotic nucleus, the SC-35 antibody recognizes ∼20–50 speckles that contain splicing factors, hyperphosphorylated RNA polymerase II, and several polyadenylation factors (Fu and Maniatis, 1990; Schul et al., 1998). Double labeling with SC-35 and DDX1 antibodies demonstrated that DDX1 foci are not components of splicing factor compartments (Figure 2A).
Figure 2.
Subnuclear localization of DDX1 with respect to other nuclear bodies. HeLa cells were immunolabeled with anti-DDX1 antibody (2923) and either anti-SC-35 (A), anti-PML (B), anti-Sm (C), anti-Sam68 (D), or anti-CstF-64 (E) antibodies. Adjacent localization of a DDX1 focus and PML body is indicated by the arrowhead in B, merged image. Adjacent localization of DDX1 foci and Cajal bodies are indicated by the arrowheads in C, merged image. All 3 DDX1 foci colocalize with cleavage bodies in E, merged image. The images were collected with the use of a Zeiss LSM 510 confocal laser scanning microscope. Bar, 5 μm.
PML bodies, also known as PML oncogenic domains, are spherical structures 0.2–1.0 μm in diameter that are present in ∼10–30 copies per cell (Ruggero et al., 2000). Based on the growing list of proteins that localize to PML bodies, a variety of roles have been proposed for these structures, including gene regulation, control of cell growth, cellular differentiation, and apoptosis (Seeler and Dejean, 1999). Cajal bodies are spherical structures of ∼0.5 μm in diameter, which are often located next to PML bodies (Lamond and Carmo-Fonseca, 1993). First observed in the light microscope by Ramon y Cajal (1903), Cajal bodies appear as a tangle of electron-dense threads 0.1–1.0 μm in diameter (Monneron and Bernhard, 1969). The number of Cajal bodies, although variable, is usually one to five per cell (Lamond and Carmo-Fonseca, 1993). Cajal bodies contain many components required for the transcription and processing of pol I, pol II, and pol III RNAs and have been hypothesized to function as transcriptosome assembly sites (Gall, 2000).
We investigated whether there was a relationship between DDX1 foci and PML bodies. HeLa cells were costained with a mouse monoclonal antibody to the PML protein and anti-DDX1 antibody. Nuclear foci were observed with both these antibodies with an average of 12.8 PML bodies and 4.8 DDX1 foci per cell (Figure 2B and Table 1). The image stacks were three-dimensionally reconstructed to reveal the subnuclear distribution of the two foci. As shown in Figure 2B, DDX1 foci do not colocalize with PML bodies; however, 18% of foci were found next to PML bodies (Table 1). The three-dimensional distance between the DDX1 foci and PML bodies located adjacent to each other ranged from 0.35 to 0.6 μm.
Table 1.
Nuclear localization of PML, Cajal, and cleavage bodies with respect to DDX1 foci
| NB | Avg. no. of NBs per cella | Ave. no. of DDX1 foci per cell | % of DDX1 foci colocalized with NBsb | % of DDX1 foci partially colocalized with NBsc | % of DDX1 foci adjacent to NBsd |
|---|---|---|---|---|---|
| PML | 12.8 (30) | 4.8 | 0.0 (0.0) | 0.0 (0.0) | 18.0 (6.7) |
| Cajal bodies | 3.0 (65) | 4.4 | 1.1 (1.6) | 4.6 (6.8) | 27.9 (41.0) |
| Cleavage bodies | 4.6 (50) | 4.9 | 65.0 (70.0) | 0.0 (0.0) | 1.2 (1.3) |
Parentheses indicate number of cells analyzed.
Parentheses indicate percentage of NBs colocalizing with DDX1 foci.
Parentheses indicate percentage of NBs partially colocalizing with DDX1 foci.
Parentheses indicate percentage of NBs adjacent to DDX1 foci.
With the use of a method published by Grande et al. (1996) (described in MATERIALS AND METHODS), we determined the likelihood that nuclear bodies would randomly colocalize with DDX1 foci. Based on our calculations, the probability of a randomly positioned PML body pairing with a randomly positioned DDX1 focus is 1 event in 40 nuclei. We observed pairing in 15 out of 30 nuclei, indicating that there is a relationship between a fraction of DDX1 foci and PML bodies.
Next, we studied the subnuclear distribution of DDX1 foci in relation to Cajal bodies. HeLa cells were double-labeled with anti-DDX1 antibody and mouse monoclonal anti-Sm, an antibody that recognizes mature snRNPs in the nucleoplasm and in Cajal bodies (Lerner et al., 1981). In these experiments, there was an average of 3.0 Cajal bodies and 4.4 DDX1 foci per cell. Although only 1% of DDX1 foci colocalized with Cajal bodies, 4.6% of DDX1 foci partially overlapped with Cajal bodies and 28% of DDX1 foci were adjacent to them (Figure 2C, Video 1, and Table 1). Three-dimensional distances ranged from 0.2 μm for partially overlapping structures to 0.4 μm for adjacent structures. If the distribution of these two nuclear bodies is random, we would expect adjacent localization in 1 in 500 nuclei. The close association observed in 40 of 64 nuclei indicates a nonrandom relationship between DDX1 foci and Cajal bodies.
There was significant variation in the number of nuclear bodies observed from cell to cell. We therefore tested whether there was an overall correlation between the number of DDX1 foci and the number of either PML or Cajal bodies in any one cell. Linear regression analysis indicated positive correlation between the number of DDX1 foci and the number of PML bodies (p = 0.012) and Cajal bodies (p = 0.027). Previous studies have shown that fluctuations in PML body number occur during the cell cycle, with the highest number of PML bodies present during the G1-S phase transition (Koken et al., 1995; Terris et al., 1995). The general correlation between the number of DDX1 foci, PML bodies, and Cajal bodies may therefore reflect similar cell cycle variations.
DDX1 Foci Associate with Cleavage Bodies
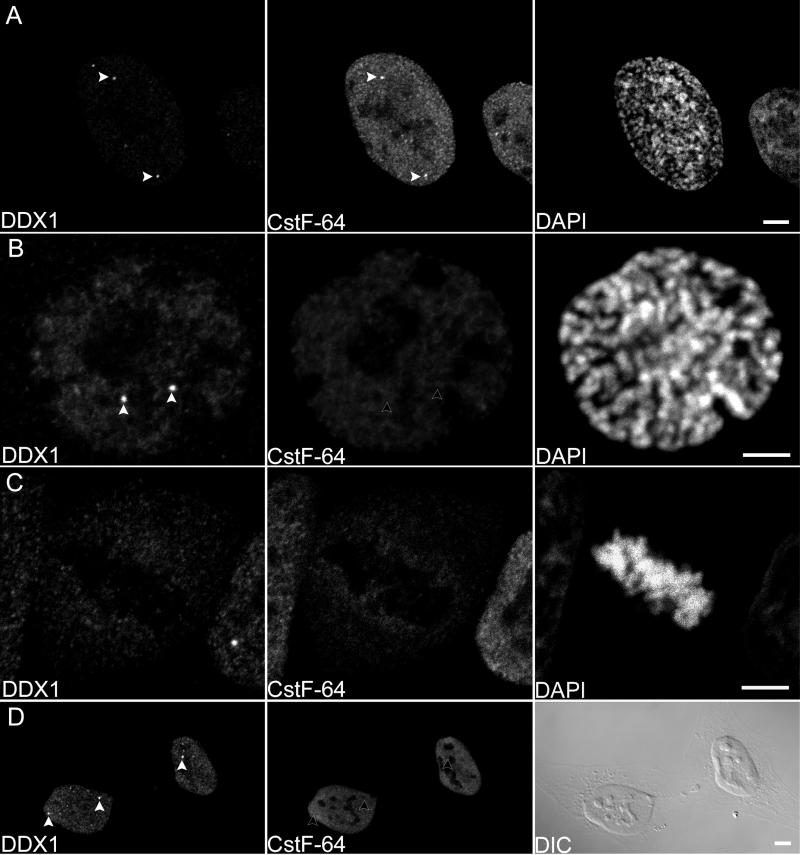
Other structures that have been found to localize adjacent to Cajal bodies include SNBs and cleavage bodies. SNBs contain Sam68, an RNA-binding protein (Wong et al., 1992) that serves as a possible adapter protein for Src kinases (Richard et al., 1995; Taylor et al., 1995), as well as SLM-1 and SLM-2, both of which are nuclear proteins that heterodimerize with Sam68 (Chen et al., 1999; Di Fruscio et al., 1999). Cleavage bodies contain the proteins CstF-64 and CPSF 100 kDa, both of which are involved in mRNA 3′ cleavage and polyadenylation (Schul et al., 1996) as well as the transcription factors TFIIE and TFIIF (Gall, 2000).
We costained HeLa cells with purified anti-DDX1 antibody and mouse monoclonal antibody to either Sam68 or CstF-64. DDX1 foci were never found to colocalize with SNBs (Figure 2D). Although HeLa cells were previously reported to contain elevated areas of CstF-64 staining rather than distinct cleavage bodies (Schul et al., 1996), we found that 90% of our HeLa cells had distinct cleavage bodies. When cells were double-labeled with anti-CstF-64 and anti-DDX1 antibodies, 65% of DDX1 foci colocalized with cleavage bodies and 1.2% showed adjacent (0.4 μm) localization (Figure 2E and Video 2). There was an average of 4.6 cleavage bodies and 4.9 DDX1 foci per cell (Table 1). Overall, 89% of cells that contained distinct DDX1 and CstF-64 foci appeared to have some foci that colocalized. These measurements only take into account distinct CstF-64 foci that colocalize with DDX1 foci. In cells with no distinct cleavage bodies, most DDX1 foci appeared to contain some CstF-64 protein. The distance between the three-dimensional CstF-64 and DDX1 foci was calculated to be 0.0 μm, strongly suggesting that DDX1 is found in the same structure as CstF-64.
Cleavage bodies have been reported to associate with Cajal bodies in a cell cycle-dependent manner in the T24 bladder carcinoma cell line; colocalization with Cajal bodies was predominantly observed in G1 phase cells, whereas cells in S phase displayed mostly adjacent localization (Schul et al., 1999). In the colon carcinoma line CaCo, cleavage bodies were found to colocalize with a Cajal body in only 1–5% of nuclei (Schul et al., 1996). In agreement with the latter, we observed 1 HeLa cell of 20 that showed colocalization of CstF-64 with p80 coilin, a protein found in Cajal bodies (our unpublished data). However, these Cajal bodies were weakly labeled compared with other Cajal bodies in the same cell. Overall, 45% of CstF-64 cleavage bodies were found adjacent (0.4 μm) to Cajal bodies, implying some spatial and/or functional relationship.
We analyzed additional cell lines with DDX1 foci to determine whether DDX1 was commonly enriched within cleavage bodies. COS-7, SKN, and GM38 cells were double-labeled with anti-DDX1 and anti-CstF-64 antibodies and the subnuclear distribution of the two proteins was analyzed. Although cleavage bodies were generally less distinct in these three cell lines compared with HeLa cells, we frequently observed colocalization of DDX1 foci and CstF-64 (our unpublished data). We therefore conclude that the DEAD box protein DDX1 is a common component of subnuclear domains previously identified as cleavage bodies.
Immunoprecipitation of DDX1 and CstF-64
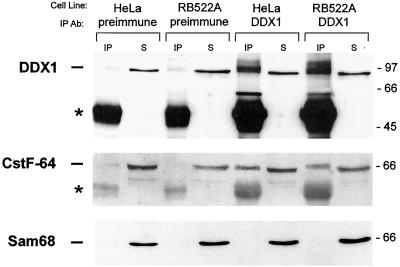
Coimmunoprecipitations were carried out to determine whether DDX1 and CstF-64 are located within the same complex. With anti-DDX1 antibody (2910), we were able to immunoprecipitate ∼5–8% of input DDX1 from both HeLa cells and RB522A cells (Figure 3, top, compare IP and S in the last four lanes). Considerably lower levels of DDX1 were immunoprecipitated with preimmune serum (lanes 1–4). A similar pattern was observed using anti-CstF-64 antibody, with >10-fold more CstF-64 coimmunoprecipitated with anti-DDX1 antibody than with preimmune serum (Figure 3, middle, compare lanes 1 and 3 with 5 and 7). The portion of CstF-64 that coimmunoprecipitated with DDX1 was relatively small, suggesting either a weak interaction between the two proteins or an association limited to a subset of DDX1 and CstF-64 proteins. To ensure that coimmunoprecipitation of CstF-64 with DDX1 was the result of specific interaction, the filter was immunostained with an antibody to the RNA binding protein, Sam68. As previously shown (Figure 2D), Sam68-containing nuclear bodies were never found to colocalize with DDX1 foci. Although Sam68 was abundant in both HeLa and RB522A nuclear extracts, there was no detectable Sam68 in any of the immunoprecipitation lanes (Figure 3, bottom).
Figure 3.
Coimmunoprecipitation of DDX1 and CstF-64. Nuclear extracts from HeLa cells and RB522A cells (1 mg) were precleared and incubated with 3 μl of either preimmune serum or anti-DDX1 antibody (2910). Complexes were precipitated with protein A-agarose beads and run on an 8% SDS-polyacrylamide gel. Proteins were transferred to nitrocellulose and detected with anti-DDX1 antibody (top), anti-CstF-64 antibody (middle), and anti-Sam68 antibody (bottom). For comparison, we have loaded 2.5% of the supernatant (S) from each immunoprecipitation reaction next to the immunoprecipitation (IP) lanes. IgG molecules are indicated by the asterisks. Size markers (in kDa) are indicated on the right.
Close Proximity of DDX1 and CstF-64 as Determined by Fluorescence Resonance Energy Transfer
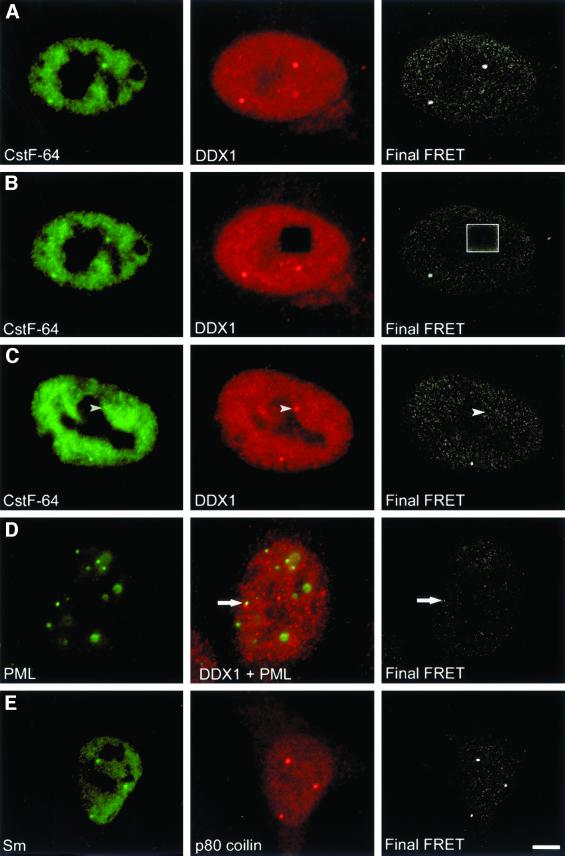
Fluorescence resonance energy transfer (FRET) is a technique that detects the radiationless transfer of energy from a fluorescent donor to an acceptor fluorophore. For energy transfer to occur, the two fluorophores must be in proximity (<10 nm) and their dipoles aligned in a specific orientation. FRET therefore achieves a resolution that is ∼25 times greater than that obtained by confocal imaging alone. To determine whether the CstF-64 and DDX1 proteins reside in close proximity within cleavage bodies, we analyzed the transfer of energy from CstF-64 labeled with Alexa 488 to DDX1 labeled with Cy3 using the three-filter set technique (Youvan et al., 1997; Schmid et al., 2001). In Figure 4, column 1 depicts donor fluorescence collected with a 505–550-nm filter. Column 2 represents acceptor fluorophore emission at 560 nm (except for Figure 4D, which contains both the acceptor and donor images). The final FRET image, displayed in column 3, is calculated from the FRET emission at 560 nm minus the contributions from both the donor and acceptor fluorophores (see MATERIALS AND METHODS). In Figure 4A, the FRET signal is detected in cleavage bodies and to some extent in the nucleoplasm. The photodestruction of Cy3 acceptor molecules efficiently quenches the FRET signal (Figure 4B). To ensure that FRET was indeed occurring within cleavage bodies, a cell was found which contained a DDX1 focus that was not paired with a cleavage body. In this instance, we observed no FRET at the focus site (Figure 4C, arrowhead). Therefore, CstF-64 may be entirely excluded from some DDX1 foci.
Figure 4.
DDX1 and CstF-64 undergo FRET in HeLa cells. HeLa cells were indirectly immunolabeled with anti-DDX1 antibody (2923) and either anti-CstF-64 (A–C) or anti-PML (D) antibody. The images were collected using the three-filter set method as described in MATERIALS AND METHODS. The first column represents the image obtained with the donor filters. The image in the second column was collected with the acceptor filters (except D, which represents the combined acceptor and donor signals). The third column represents the final FRET image processed to remove non-FRET components (as described in MATERIALS AND METHODS). (A) FRET was observed between CstF-64 and DDX1 in both the nucleoplasm and cleavage bodies. (B) After photodestruction of the acceptor molecule, FRET no longer occurred (boxed region). (C) Areas lacking CstF-64 foci (arrowhead) often did not demonstrate FRET. (D) Another subnuclear body, PML, demonstrated little FRET with DDX1. The arrow indicates the location of adjacent PML and DDX1 foci. (E) FRET was observed between two components of Cajal bodies, p80 coilin and Sm proteins. Bar, 5 μm.
As a negative control for FRET, we replaced the donor anti-CstF-64 antibody with anti-PML antibody. As indicated previously, 18% of DDX1 foci are found adjacent to PML bodies. There was little FRET between DDX1 and PML, implying that most of the labeled PML protein is situated at a distance >10 nm from labeled DDX1 (Figure 4D). To further validate the FRET technique, we labeled cells with anti-p80 coilin and anti-Sm antibodies. These antibodies recognize proteins that both localize within Cajal bodies. As shown in Figure 4E, FRET was observed between labeled p80-coilin and Sm proteins.
It is important to note that because we did our analysis with cells labeled with both primary and secondary antibodies, the distance between the structures may be greater than the (Förster) 10-nm distance. Nevertheless, these experiments represent a considerable improvement over traditional confocal microscopy resolution and suggest that DDX1 resides in proximity to CstF-64 within cleavage bodies and within the nucleoplasm.
DDX1 Foci Do not Accumulate Nascent RNA
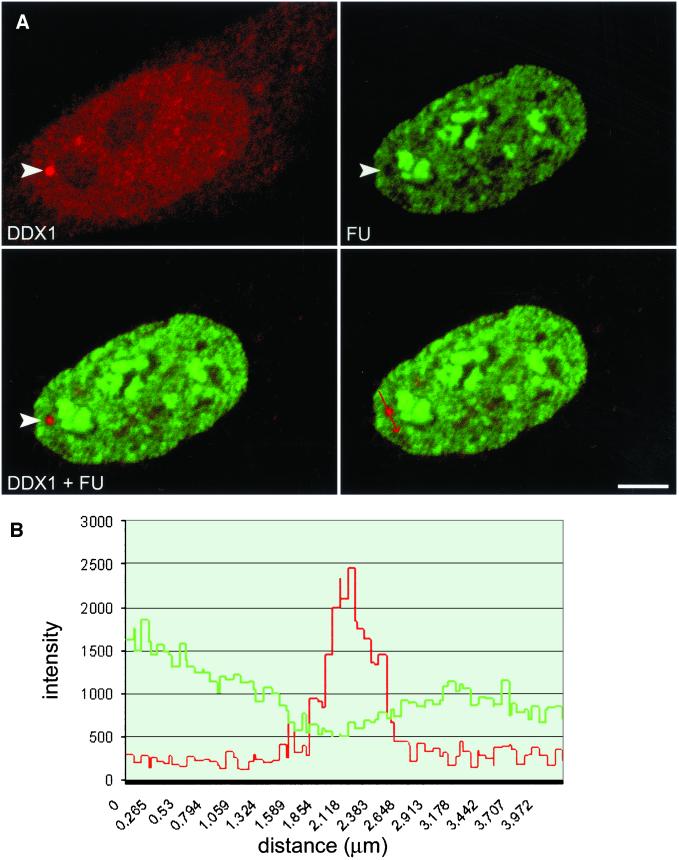
Previous studies in T24 bladder carcinoma cells have shown that ∼20% of cleavage bodies contain newly synthesized RNA. In addition, it is known that 18% of cleavage bodies do not colocalize with Cajal bodies in these cells (Schul et al., 1996). Because Cajal bodies do not contain nascent RNA (Moreno Diaz de la Espina et al., 1982; Raska, 1995; Schul et al., 1996), Schul et al. (1996) have postulated that all cleavage bodies not associated with Cajal bodies contain newly synthesized RNA. To determine whether cleavage bodies containing DDX1 have newly synthesized RNA, we examined DDX1 foci for the presence of nascent RNA by 5′-FU incorporation into HeLa cells. Cells were exposed to FU for 5, 10, or 15 min prior to fixation and analyzed by double-labeling with anti-DDX1 antibody and anti-bromodeoxyuridine antibody, which recognizes FU (Boisvert et al., 2000). The nuclear distribution of DDX1 and FU in cells labeled for 15 min is shown in Figure 5A. Although both FU and DDX1 are abundant throughout the nucleus, there does not appear to be any FU within DDX1 foci (Figure 5A, arrowhead). Similar results were observed at the earlier time points. To quantitate the intensity of the FU within the focus, an axis was drawn through a DDX1 focus (Figure 5A, arrow) and the signal intensities were plotted (Figure 5B and Video 3). There is an inverse relationship between intensity of the DDX1 signal and intensity of the FU signal, indicating that DDX1 foci do not accumulate nascent RNA. Three-dimensional analyses of 25 HeLa nuclei clearly demonstrate that DDX1 foci are not sites of active RNA transcription.
Figure 5.
DDX1 foci do not accumulate nascent RNA. (A) HeLa cells were incubated with FU for 15 min and stained with anti-DDX1 antibody (2923) and anti-bromodeoxyuridine antibody (FU). The arrowhead indicates a DDX1 focus. (B) The staining intensities of the region through a DDX1 foci (highlighted by the arrow in the bottom left panel) were profiled with the use of the Zeiss LSM 510 image software. The green line represents FU intensity, whereas the red line represents DDX1 intensity. Bar, 5 μm.
DDX1 during Cell Cycle
Previous publications indicate that cleavage bodies do not persist during mitosis in the T24 bladder carcinoma cell line (Schul et al., 1996, 1999). To investigate whether DDX1 foci follow the same pattern as cleavage bodies, HeLa cells were immunostained with anti-DDX1 antibody and the chromatin stained with DAPI. Stage of the cell cycle was determined by chromatin and cellular morphology. The majority of cells in prophase contained distinct DDX1 foci; however, the foci disappeared at metaphase (Figure 6, A–C). Cleavage bodies, as stained by anti-CstF-64, disappeared in late prophase, slightly earlier than DDX1 foci (Figure 6, A and B). DDX1 foci reappeared during telophase/early G1, concomitant with cytokinesis (our unpublished data). Cells in the G1 phase of the cell cycle (as detected by the presence of a midbody) usually contained at least one DDX1 focus, whereas cleavage bodies were not observed (Figure 6D). In contrast to DDX1 levels, which remained constant throughout mitosis, the overall CstF-64 signal was reduced during metaphase and anaphase (Figure 6C, compare the fluorescent intensities of the mitotic cell with the intensities of the surrounding cells).
Figure 6.
Disappearance of DDX1 and CstF-64 foci during mitosis. HeLa cells were immunolabeled with anti-DDX1 antibody (2923) and anti-CstF-64 antibody. The cells were mounted with DAPI to visualize the chromatin morphology. (A) Both DDX1 and CstF-64 foci are present during early prophase (indicated by the arrowheads). (B) Only DDX1 foci are present by late prophase (arrowheads). (C) Neither DDX1 nor CstF-64 foci can be detected during metaphase. (D) DDX1 foci reappear during G1 (arrowheads); however, CstF-64 foci are not observed. Although the overall DDX1 signal remains constant throughout mitosis, there is a decrease in CstF-64 signal. For example, compare the metaphase cell with adjacent cells in C. Bar, 5 μm.
DISCUSSION
Lower eukaryotic DEAD box proteins function in many aspects of RNA metabolism, including translation initiation, ribosome biogenesis, and pre-mRNA splicing. Because these functions occur in discrete areas of the cell, subcellular localization of higher eukaryote DEAD box proteins could lead to the identification of putative roles for these proteins. Based on this premise, we used confocal microscopy to examine the subcellular distribution of the DEAD box protein, DDX1, in HeLa cells. We established that the majority of DDX1 is distributed in a widespread punctate nucleoplasmic pattern, with lower levels in the cytoplasm. Interestingly, DDX1 was also found in discrete foci within the nucleus. There were an average of five DDX1 foci in HeLa cells with diameters of ∼0.5 μm. DDX1 protein was found in every cell line examined, with discrete DDX1 foci observed in 10 of 15 cell lines, including a variety of transformed cells as well as normal human fibroblasts. Taken together, these results support a primary role for DDX1 in nuclear, rather than cytoplasmic, RNA metabolism and suggest that many cell types have a nuclear domain that contains elevated concentrations of DDX1.
The eukaryotic nucleus is comprised of many distinct subdomains, each of which contains specific proteins believed to contribute to a functionally organized nucleus. Using a panel of antibodies previously shown to recognize specific nuclear subdomains, we found that DDX1 foci do not associate with SC-35 speckles or SNBs, but frequently reside adjacent to PML and Cajal bodies. Most importantly, we discovered that in HeLa, 65% of DDX1 foci colocalize with CstF-64-labeled cleavage bodies. Although there were no distinct cleavage bodies associated with the remaining DDX1 foci, the majority of these appeared to contain elevated levels of CstF-64. Colocalization of DDX1 foci and cleavage bodies was also observed in three other cell lines tested: COS-7, SKN, and GM38 cells. Analysis of the distribution pattern of DDX1 foci and cleavage bodies during the cell cycle indicates distinct patterns of disappearance and reappearance. DDX1 foci were visible until metaphase, whereas cleavage bodies disappeared during late prophase. Furthermore, only DDX1 foci were detected in the early G1 phase of the cell cycle. Unlike DDX1 protein levels, which remained constant throughout mitosis, CstF-64 levels decreased during prophase through to telophase, as previously reported (Schul et al., 1996). Based on these observations, we propose that DDX1 foci are identical to cleavage bodies and that the lack of colocalization observed in some cases may reflect cell cycle fluctuations in CstF-64. However, we cannot refute the possibility that DDX1 foci are entirely distinct nuclear subdomains, which can merge with cleavage bodies.
Because of the limits of resolution of the confocal microscope, proteins that appear to colocalize could be as far apart as 0.25 μm. We therefore used FRET, a technique based on the transfer of energy between molecules located within 10 nm of each other, to further study the proximity of DDX1 and CstF-64. This technique indicated that labeled DDX1 is within 10 nm of labeled CstF-64 in both cleavage bodies and within the nucleoplasm. Furthermore, coimmunoprecipitation experiments demonstrate that a fraction of CstF-64 is found within the same complex as DDX1. These results suggest a functional as well as a spatial relationship between these two proteins.
Within the nucleoplasm, CstF-64 is known to function as part of a heterotrimeric complex (CstF) in 3′-end pre-mRNA cleavage by binding a GU-rich element downstream of the AAUAAA consensus sequence (Wahle and Ruegsegger, 1999). The role of CstF-64 within cleavage bodies remains to be elucidated. Schul et al. (1999) have observed that in T24 bladder carcinoma cells undergoing DNA synthesis, cleavage bodies mainly exist independently of Cajal bodies and colocalize with replication-dependent histone gene clusters. These cleavage bodies were previously reported to contain newly transcribed RNA as measured by 5-bromo-uridine-triphosphate microinjection (Schul et al., 1996). Therefore, Schul et al. (1999) have hypothesized that cleavage bodies may play a role in histone mRNA synthesis. Because mammalian histone transcripts do not undergo typical 3′-end cleavage and polyadenylation (Dominski and Marzluff, 1999), the role of cleavage bodies in histone mRNA synthesis is not immediately obvious. Although our data do not refute the possibility that cleavage bodies may play a role in histone mRNA synthesis, three-dimensional analysis of 25 FU-labeled HeLa cells indicates that DDX1-containing cleavage bodies lack nascent RNA and, consequently, do not represent sites of active transcription. We therefore propose that cleavage bodies may function in facilitating the formation or regeneration of transcriptional complexes, as previously suggested for Cajal bodies (Gall, 2000). In support of this, we and others have observed that cleavage bodies frequently localize adjacent to Cajal bodies, which are known to contain a number of transcriptional and splicing/polyadenylation proteins (Gall et al., 1999). This juxtaposition suggests coordinate roles for these structures in RNA metabolism. Taking into consideration all observations with cleavage bodies to date, we believe that these structures play a general role in the transcription and processing of pre-mRNAs. Because histone gene transcription is particularly active during S phase, the observation of Schul et al. (1999) that histone gene clusters are associated with cleavage bodies would be consistent with a local requirement for elevated levels of factors involved in transcription and processing of histone genes.
The close proximity and possible interaction of DDX1 with CstF-64 suggests a role for DDX1 in 3′-end pre-mRNA cleavage and/or polyadenylation. Although a DEAD box protein has not yet been reported in the cleavage and polyadenylation pathway, there are numerous places where a protein involved in the modulation of RNA structure could play a role. For example, the CstF complex preferentially binds short RNA molecules, suggesting that RNA structure is important for binding (Takagaki and Manley, 1997). DDX1 could alter the secondary structure of pre-mRNAs by unwinding, or destabilizing double-stranded areas. DDX1 could also promote efficient binding of CstF to RNA substrates or could be involved in the recruitment of RNA. Because DDX1 is also found within the cytoplasm, it is possible that DDX1 plays a role in the export of mRNA from the nucleus. Although we have shown that DDX1 can bind to an in vitro transcribed RNA (our unpublished data), the substrate specificity or preference of this DEAD box protein remains to be determined.
In conclusion, we have shown that DDX1 is found in the nucleoplasm of all cell lines tested as well as in nuclear foci in 10 of 15 lines tested. These foci colocalize with cleavage bodies in all four cell lines examined. FRET analysis and coimmunoprecipitations suggest that DDX1 is in a complex with the pre-mRNA 3′-cleavage factor CstF-64. DDX1 therefore represents the first DEAD box protein associated with a cleavage and polyadenylation factor. Although it is possible that DDX1-containing foci represent distinct structures that frequently merge with cleavage bodies, our results are consistent with DDX1 foci and cleavage bodies representing the same dynamic structure with variable protein composition. In agreement with previous results in CaCo and T24, we found that cleavage bodies are often located adjacent to Cajal bodies in HeLa cells. Although the basis for this spatial association is not known, our findings reinforce the emerging view of an organized eukaryotic cell nucleus. The discovery of a DEAD box protein within cleavage bodies further documents the importance of this family of proteins in all processes involving RNA metabolism.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Joan Steitz for the anti-Sm Y12 antibody, Dr. Edward Chan for the anti-p80 coilin antibody, Dr. James Manley for the anti-CstF-64 antibody, and Dr. Roel van Driel for the anti-PML antibody (through Dr. David Bazett-Jones). We are grateful to Dr. John Hanson for help with the statistical analyses. This work was supported by the National Cancer Institute of Canada with funds from the Canadian Cancer Society.
Footnotes
REFERENCES
- Amler LC, Schürmann J, Schwab M. The DDX1 gene maps within 400 kbp 5′ to MYCN and is frequently coamplified in human neuroblastoma. Genes Chromosomes Cancer. 1996;15:134–137. doi: 10.1002/(SICI)1098-2264(199602)15:2<134::AID-GCC9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert F-M, Hendzel MJ, Bazett-Jones DP. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM, Seeger RC, Schwab M, Varmus HE, Bishop JM. Amplification of N-myc in untreated human neuroblastomas correlates with advanced disease stage. Science. 1984;224:1121–1124. doi: 10.1126/science.6719137. [DOI] [PubMed] [Google Scholar]
- Carvalho T, Almeida F, Calapez A, Lafarga M, Berciano MT, Carmo-Fonseca M. The spinal muscular atrophy disease gene product, SMN: a link between snRNP biogenesis and the Cajal (coiled) body. J Cell Biol. 1999;147:715–727. doi: 10.1083/jcb.147.4.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charroux B, Pellizzoni L, Perkinson RA, Shevchenko A, Mann M, Dreyfuss G. Gemin3: a novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J Cell Biol. 1999;147:1181–1193. doi: 10.1083/jcb.147.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Boisvert FM, Bazett-Jones DP, Richard S. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Di Fruscio M, Chen T, Richard S. Two novel Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc Natl Acad Sci USA. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Doz F, et al. N-MYC amplification, loss of heterozygosity on the short arm of chromosome 1 and DNA ploidy in retinoblastoma. Eur J Cancer. 1996;32A:645–649. doi: 10.1016/0959-8049(95)00626-5. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C. Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell. 1999;10:4385–4402. doi: 10.1091/mbc.10.12.4385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George RE, Kenyon RM, McGuckin AG, Malcolm AJ, Pearson AD, Lunec J. Investigation of co-amplification of the candidate genes ornithine decarboxylase, ribonucleotide reductase, syndecan-1 and a DEAD box gene, DDX1, with N-myc in neuroblastoma. Oncogene. 1996;12:1583–1587. [PubMed] [Google Scholar]
- Godbout R, Hale M, Bisgrove D. A human DEAD box protein with partial homology to heterogeneous nuclear ribonucleoprotein U. Gene. 1994;138:243–245. doi: 10.1016/0378-1119(94)90816-8. [DOI] [PubMed] [Google Scholar]
- Godbout R, Packer M, Bie W. Overexpression of a DEAD box protein (DDX1) in neuroblastoma and retinoblastoma cell lines. J Biol Chem. 1998;273:21161–21168. doi: 10.1074/jbc.273.33.21161. [DOI] [PubMed] [Google Scholar]
- Godbout R, Squire J. Amplification of a DEAD box protein gene in retinoblastoma cell lines. Proc Natl Acad Sci USA. 1993;90:7578–7582. doi: 10.1073/pnas.90.16.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grande MA, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort HT, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- Jankowsky E, Jankowsky A. The DEXH/D protein database. Nucleic Acids Res. 2000;28:333–334. doi: 10.1093/nar/28.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MK, Nikodem VM. HnRNP U inhibits carboxy-terminal domain phosphorylation by TFIIH and represses RNA polymerase II elongation. Mol Cell Biol. 1999;10:6833–6844. doi: 10.1128/mcb.19.10.6833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koken MH, Linares-Cruz G, Quignon F, Viron A, Chelbi-Alix MK, Sobczak-Thepot J, Juhlin L, Degos L, Calvo F, de The H. The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene. 1995;10:1315–1324. [PubMed] [Google Scholar]
- Krause S, Fakan S, Weis K, Wahle E. Immunodetection of poly(A) binding protein II in the cell nucleus. Exp Cell Res. 1994;214:75–82. doi: 10.1006/excr.1994.1235. [DOI] [PubMed] [Google Scholar]
- Kuroda H, White PS, Sulman EP, Manohar CF, Reiter JL, Cohn SL, Brodeur GM. Physical mapping of the DDX1 gene to 340 kb 5′ of MYCN. Oncogene. 1996;7:1561–1565. [PubMed] [Google Scholar]
- Lamond AI, Carmo-Fonseca M. The coiled body. Trends Cell Biol. 1993;3:198–204. doi: 10.1016/0962-8924(93)90214-l. [DOI] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway CA, Jr, Steitz JA. Monoclonal antibodies to nucleic acid-containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matera AG, Frey MR. Coiled bodies and gems: Janus or Gemini? Am J Hum Genet. 1998;63:317–321. doi: 10.1086/301992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Cell biology of transcription and pre-mRNA splicing: nuclear architecture meets nuclear function. J Cell Sci. 2000;113:1841–1849. doi: 10.1242/jcs.113.11.1841. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Moreno Diaz de la Espina S, Risueno MC, Medina FJ. Ultrastructural, cytochemical and autoradiographic characterization of coiled bodies in the plant cell nucleus. Biol Cell. 1982;44:229–238. [Google Scholar]
- Noguchi T, Akiyama K, Yokoyama M, Kanda N, Matsunaga T, Nishi Y. Amplification of a DEAD box gene (DDX1) with the MYCN gene in neuroblastomas as a result of cosegregation of sequences flanking the MYCN locus. Genes Chromosomes Cancer. 1996;15:129–133. doi: 10.1002/(SICI)1098-2264(199602)15:2<129::AID-GCC8>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- Pandita A, Godbout R, Zielenska M, Thorner P, Bayani J, Squire JA. Relational mapping of MYCN and DDXI in band 2p24 and analysis of amplicon arrays in double minute chromosomes and homogeneously staining regions by use of free chromatin FISH. Genes Chromosomes Cancer. 1997;20:243–252. doi: 10.1002/(sici)1098-2264(199711)20:3<243::aid-gcc4>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: the mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersson I, Hinterberger M, Mimori T, Gottlieb E, Steitz JA. The structure of mammalian small nuclear ribonucleoproteins. Identification of multiple protein components reactive with anti-(U1) ribonucleoprotein and anti-Sm autoantibodies. J Biol Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- Ramon y Cajal S. Un sencillo metodo de coloracion selectiva del reticulo protoplasmico y sus efectos en los diversos organos nerviosos de vertebrados y invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Raska I. Nuclear ultrastructures associated with the RNA synthesis and processing. J Cell Biochem. 1995;59:11–26. doi: 10.1002/jcb.240590103. [DOI] [PubMed] [Google Scholar]
- Richard S, Yu D, Blumer KJ, Hausladen D, Olszowy MW, Connelly PA, Shaw AS. Association of p62, a multifunctional SH2- and SH3-binding protein, with src-family tyrosine kinases, Grb2, and phospholiapse Cγ-1. Mol Cell Biol. 1995;15:186–197. doi: 10.1128/mcb.15.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Wang Z-G, Pandolfi PP. The puzzling multiple lives of PML and its role in the genesis of cancer. Bioessays. 2000;22:827–835. doi: 10.1002/1521-1878(200009)22:9<827::AID-BIES8>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- Sakai K, Tanooka H, Sasaki MS, Ejima Y, Kaneko A. Increase in copy number of N-myc in retinoblastomas in comparison with chromosome abnormality. Cancer Genet Cytogenet. 1988;30:119–126. doi: 10.1016/0165-4608(88)90099-4. [DOI] [PubMed] [Google Scholar]
- Schmid SR, Linder P. D-E-A-D protein family of putative RNA helicases. Mol Microbiol. 1992;6:283–291. doi: 10.1111/j.1365-2958.1992.tb01470.x. [DOI] [PubMed] [Google Scholar]
- Schmid JA, Scholze P, Kudlacek O, Freissmuth M, Singer EA, Sitte HH. Oligomerization of the human serotonin transporter and of the rat GABA transporter 1 visualized by fluorescence resonance energy transfer microscopy in living cells. J Biol Chem. 2001;276:3805–3810. doi: 10.1074/jbc.M007357200. [DOI] [PubMed] [Google Scholar]
- Schul W, de Jong L, van Driel R. Nuclear neighbors: the spatial and functional organization of genes and nuclear domains. J Cell Biochem. 1998;70:159–171. doi: 10.1002/(sici)1097-4644(19980801)70:2<159::aid-jcb2>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Schul W, Groenhout B, Koberna K, Takagaki Y, Jenny A, Manders EM, Raska I, van Driel R, de Jong L. The RNA 3′ cleavage factors CstF 64 kDa and CPSF 100 kDa are concentrated in nuclear domains closely associated with coiled bodies and newly synthesized RNA. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]
- Schul W, van der Kraan I, Matera AG, van Driel R, de Jong L. Nuclear domains enriched in RNA 3′-processing factors associate with coiled bodies and histone genes in a cell cycle-dependent manner. Mol Biol Cell. 1999;10:3815–3824. doi: 10.1091/mbc.10.11.3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeger RC, Brodeur GM, Sather H, Dalton A, Siegel SE, Wong KY, Hammond D. Association of multiple copies of the N-myc oncogene with rapid progression of neuroblastomas. N Engl J Med. 1985;313:1111–1116. doi: 10.1056/NEJM198510313131802. [DOI] [PubMed] [Google Scholar]
- Seeler JS, Dejean A. The PML nuclear bodies: actors or extras? Curr Opin Genet Dev. 1999;3:362–367. doi: 10.1016/s0959-437x(99)80054-9. [DOI] [PubMed] [Google Scholar]
- Squire JA, Thorner PS, Weitzman S, Maggi JD, Dirks P, Doyle J, Hale M, Godbout R. Co-amplification of MYCN and a DEAD box gene (DDX1) in primary neuroblastoma. Oncogene. 1995;10:1417–1422. [PubMed] [Google Scholar]
- Stuurman N, de Graaf A, Floore A, Josso A, Humbel B, de Jong L, van Driel R. A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. JCell Sci. 1992;101:773–784. doi: 10.1242/jcs.101.4.773. [DOI] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL. RNA recognition by the human polyadenylation factor CstF. Mol Cell Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Anafi M, Pawson T, Slalloway D. Functional interaction between c-src and its mitotic target, Sam68. J Biol Chem. 1995;270:10120–10124. doi: 10.1074/jbc.270.17.10120. [DOI] [PubMed] [Google Scholar]
- Terris B, Baldin V, Dubois S, Degott C, Flejou JF, Henin D, Dejean A. PML nuclear bodies are general targets for inflammation and cell proliferation. Cancer Res. 1995;55:1590–1597. [PubMed] [Google Scholar]
- Wahle E, Ruegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- Wong G, Muller O, Clark R, Conroy L, Moran MF, Polakis P, McCormick F. Molecular cloning and nucleic acid binding properties of the GAP-associated tyrosine phosphoprotein p62. Cell. 1992;69:551–558. doi: 10.1016/0092-8674(92)90455-l. [DOI] [PubMed] [Google Scholar]
- Youvan DC, Silva CM, Petersen J, Bylina EJ, Coleman WJ, Dilworth MR, Yang MM. Calibration of fluorescence resonance energy transfer in microscopy using genetically engineered GFP derivatives on nickel chelating beads. Biotechnology. 1997;3:1–18. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.