Abstract
Brain tumor initiating cells (BTICs), also known as cancer stem cells, hijack high-affinity glucose uptake active normally in neurons to maintain energy demands. Here we link metabolic dysregulation in human BTICs to a nexus between MYC and de novo purine synthesis, mediating glucose-sustained anabolic metabolism. Inhibiting purine synthesis abrogated BTIC growth, self-renewal and in vivo tumor formation by depleting intracellular pools of purine nucleotides, supporting purine synthesis as a potential therapeutic point of fragility. In contrast, differentiated glioma cells were unaffected by the targeting of purine biosynthetic enzymes, suggesting selective dependence of BTICs. MYC coordinated the control of purine synthetic enzymes, supporting its role in metabolic reprogramming. Elevated expression of purine synthetic enzymes correlated with poor prognosis in glioblastoma patients. Collectively, our results suggest that stem-like glioma cells reprogram their metabolism to self-renew and fuel the tumor hierarchy, revealing potential BTIC cancer dependencies amenable to targeted therapy.
Glioblastoma (World Health Organization grade IV glioma) is the most prevalent and malignant primary intrinsic brain tumor, with current therapies offering only palliation for patients1. Glioblastomas display intratumoral heterogeneity stemming from genetic and epigenetic aberrations. Functional diversity exists within this heterogeneity in the form of a cellular hierarchy maintained by self-renewing brain tumor initiating cells (BTICs)2. While BTICs recapitulate self-renewal and differentiation, partially mimicking neural precursor cells (NPCs)3, BTICs demonstrate distinguishing features, including increased frequency, proliferation, aberrant expression of stem and progenitor cell markers, and tumor formation. The importance of BTICs is underscored by their relative resistance to radiation and chemotherapy as compared to non-stem or differentiated glioma cells (DGCs)4,5. This resistance may contribute to tumor recurrence and relapse. BTICs also promote angiogenesis and tumor invasion, potentially limiting the efficacy of antiangiogenic agents6,7. Therefore, targeting BTICs may be important for improved outcomes for glioblastoma patients.
The normal brain potently extracts circulating glucose to provide energy required for organ function. Metabolic reprogramming, namely the Warburg effect, is recognized as a key hallmark of cancer, including brain tumors8. Tumor cells preferentially rely on glycolysis, rather than the mitochondrial oxidative phosphorylation used in nonmalignant cells, to generate ATP9,10. Although energetically inefficient, this phenomenon may provide cancer cells with the key ability to channel glycolytic intermediates into anabolic biosynthesis, enabling rapid tumor growth even under hypoxia. Metabolic reprograming in cancer is not simply a passenger in tumorigenesis, but may be an initiating event, as recurrent somatic mutations of metabolic enzymes have been reported10. Neomorphic mutations in isocitrate dehydrogenase 1 or 2 (IDH1 and IDH2, respectively) occur in most low-grade gliomas and recurrent glioblastomas11. The mutant IDH1 or IDH2 produces 2-hydroxyglutarate, an oncometabolite, generating global epigenetic alterations associated with tumor initiation and progression12.
Adaptation to the varied microenvironments in the brain ecosystem is essential to tumor maintenance and progression. Stressful conditions, such as hypoxia, low glucose or low pH, maintain BTICs13-15. We previously showed that BTICs upregulate GLUT3 (solute carrier family 2 (facilitated glucose transporter), member 3; SLC2A3), a neuronal survival mechanism, to compete for glucose in response to nutrient supply fluctuations13. The GLUT3 transporter has fivefold greater affinity for glucose than the ubiquitously expressed GLUT1 protein, creating an increased glucose influx and secured carbon flow in normal neurons and BTICs. Similar phenomena also exist in embryonic stem cells and induced pluripotent stem cells, suggesting similarity in signaling pathways between pluripotent stem cells and BTICs10,16. Glucose also induces resistance to targeted therapy through the promotion of protein acetylation17. Clinically, hyperglycemia and overexpression of GLUT3 predict shorter survival in glioblastoma patients13,18. The precise mechanism of how BTICs reprogram the metabolic machinery to sustain glucose influx, promote self-renewal and maintain tumorigenic capacity is unknown.
We traced the downstream carbon flow following glucose influx, using unbiased metabolomic profiling and in silico genomic discovery. Carbon influx in BTICs was used to sustain de novo purine synthesis, maintained by the core transcription factor MYC. As metabolic reprogramming represents a unique BTIC signature stemming from both intrinsic and extrinsic influences, this study reveals new metabolic nodes for improved glioblastoma therapeutics.
RESULTS
Combined metabolomic and genomic analyses reveal specific upregulation of de novo purine synthesis in BTICs
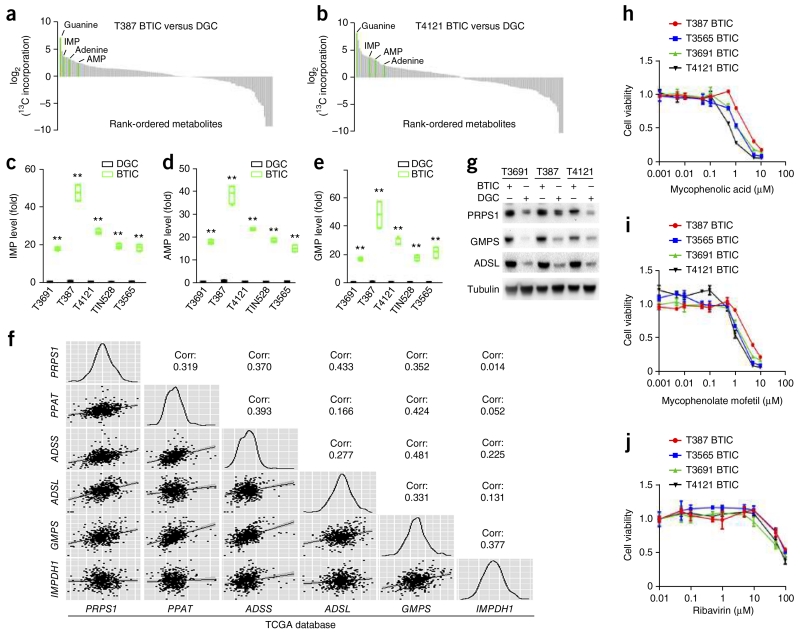
To determine the downstream metabolic reprogramming events associated with elevated glucose uptake in BTICs, we performed extensive molecular characterization of patient-derived BTIC xenograft models that we previously showed to contain functional cellular hierarchies13-15. Whole-exome sequencing, RNA sequencing (RNA-seq) and whole-genome DNA methylation analysis revealed that these models replicate key features of the genetics and transcriptional signatures of glioblastoma, when compared to data extracted from The Cancer Genome Atlas (TCGA) (Supplementary Fig. 1a–d). Using these models, we performed unbiased carbon tracing of two glioblastoma patient-derived BTICs and matched DGCs with [13C6]glucose, followed by nontargeted small molecule mass spectrometry. We scored each metabolite by calculating the relative 13C incorporation in BTICs over patient-matched DGCs. Among the top hits were metabolites in the de novo purine synthesis pathway, such as inosine 5′-monophosphate (IMP), adenosine 5′-monophosphate (AMP), adenine and guanine (Fig. 1a,b). However, the level of ribose 5-phosphate (R5P), the starting substrate of de novo purine synthesis, was not significantly different between BTICs and DGCs (data not shown). We thus hypothesized that the de novo purine pathway is preferentially used in BTICs. Using targeted small molecule mass spectrometry, we confirmed elevated levels of IMP, AMP and guanosine monophosphate (GMP) in BTICs in five patient-derived glioblastoma models (Fig. 1c–e). The level of hypoxanthine, a purine degradation product, was also elevated in BTICs (Supplementary Fig. 2a), diminishing the possibility that blockage in the purine degradation pathway causes the observed higher levels of IMP, AMP and GMP. Collectively, these results support the idea that one outcome of increased glucose intake of BTICs is shuttling of carbon atoms preferentially into de novo purine synthesis.
Figure 1.
BTICs upregulate de novo purine synthesis. (a,b) Waterfall plots of non-targeted small molecule mass spectrometry profiling differential levels of metabolites in BTICs and matched DGCs from two patient-derived glioblastoma specimens, T387 and T4121, treated with [13C6]glucose. Each identified small molecule was scored by its relative level of 13C incorporation in BTICs compared to that in DGCs. (c–e) Targeted mass spectrometry showed elevated levels of purine metabolites (IMP, AMP and GMP) using cell lysates of BTICs and matched DGCs from five patient-derived glioblastoma specimens (T3691, T387, T4121, TIN528 and T3565). The relative levels of these three metabolites normalized to those of DGCs are shown. Center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 4 independent experiments per group. (IMP: T3691, t(6) = 37.688; T387, t(6) = 20.709; T4121, t(6) = 21.137; TIN528, t(6) = 32.948; T3565, t(6) = 18.947. AMP: T3691, t(6) = 37.525; T387, t(6) = 18.385; T4121, t(6) = 24.088; TIN528, t(6) = 46.790; T3565, t(6) = 17.106. GMP: T3691, t(6) = 29.267; T387, t(6) = 9.347; T4121, t(6) = 16.995; TIN528, t(6) = 20.899; T3565, t(6) = 11.145.) (f) Pairwise correlation analysis of six representative genes in purine synthesis pathway. Plots indicate gene expression data from TCGA glioblastoma patients for PRPS1, PPAT, ADSS, ADSL, GMPS and IMPDH1. Correlation coefficient (R) values are shown. (g) BTICs upregulate protein levels of purine synthesis enzymes (PRPS1, GMPS and ADSL) relative to DGCs across human glioblastoma specimens T3691, T387 and 4121. Full-length western blots are presented in Supplementary Figure 15. (h) Sensitivity of four patient-derived BTIC models (T387, T3565, T3691 and T4121) to the purine synthesis inhibitor mycophenolic acid. (i) Sensitivity of four patient-derived BTIC models (T387, T3565, T3691 and T4121) to the purine synthesis inhibitor mycophenolate mofetil. (j) Sensitivity of four patient-derived BTIC models (T387, T3565, T3691 and T4121) to the purine synthesis inhibitor ribavirin. Data are presented as mean ± s.e.m.
To determine the molecular pathways involved in the increased BTIC purine synthesis, we assessed the gene expression pattern of core enzymes involved in purine synthesis in patient-derived samples from the TCGA glioblastoma data set19,20. PRPS1 (phosphoribosyl pyrophosphate synthetase 1) and PPAT (phosphoribosyl pyrophosphate amidotransferase) function in the main branch synthesizing IMP from R5P. Downstream of IMP, the pathway branches in two to synthesize AMP and GMP. Their syntheses are catalyzed, respectively, by ADSS (adenylosuccinate synthase) plus ADSL (adenylosuccinate lyase) and by IMPDH1 (IMP dehydrogenase 1) plus GMPS (guanine monophosphate synthase). The expression of genes encoding these six enzymes significantly and positively correlated with one another across 530 glioblastoma tumor samples analyzed (Fig. 1f). The correlation coefficient between ADSS and GMPS was as high as 0.481, supporting the coordinated regulation of genes involved in the purine synthesis pathway in glioblastoma specimens. Using immunoblot analysis, the expression levels of three purine synthesis enzymes, PRPS1, ADSL and GMPS—each representing an arm of the purine biosynthesis pathway—were significantly elevated in BTICs as compared to matched DGCs (Fig. 1g). As the most commonly used BTIC marker, CD133, is not universally informative for BTICs, we confirmed our findings using an independent surface marker for stemness in human glioblastoma, CD15 (SSEA-1)21. We derived matched populations of CD15+ and CD15− tumor cells from two fresh surgical glioblastoma specimens in the absence of any culture to avoid derivation bias. The expression levels of all six de novo purine biosynthetic pathway genes were upregulated in BTICs compared to those in DGCs (Supplementary Fig. 2b,c).
Elevated levels of purines and their biosynthetic enzymes in BTICs suggest that BTICs may be sensitive to inhibition of this pathway. Therefore, we treated a series of patient-derived BTIC models with three pharmacologic inhibitors of purine biosynthesis: mycophenolic acid, mycophenolate mofetil and ribavirin. BTIC proliferation was attenuated with mycophenolic acid and mycophenolate mofetil at submicromolar concentrations (Fig. 1h,i), whereas ribavirin inhibited BTIC proliferation at higher concentrations (Fig. 1j). Similar concentration ranges of these three inhibitors had no effect on DGC proliferation (Supplementary Fig. 3a–c), supporting a specific role of de novo purine synthesis in BTICs. Treatment of two BTIC models with 5 μM mycophenolic acid, 5 μM mycophenolate mofetil or 50 μM ribavirin all caused significant reduction in GMP, assessed with targeted mass spectrometry (Supplementary Fig. 3d–f). Further, treatment with an intermediate of inosine monophosphate production, AICAR (5-aminoimidazole-4-carboxamide ribonucleotide), an AMP analog, suppressed BTIC proliferation (Supplementary Fig. 3g–j). While AICAR is considered an activator of AMP kinase, recent reports support an AMP kinase–independent mechanism in glioblastoma22,23.
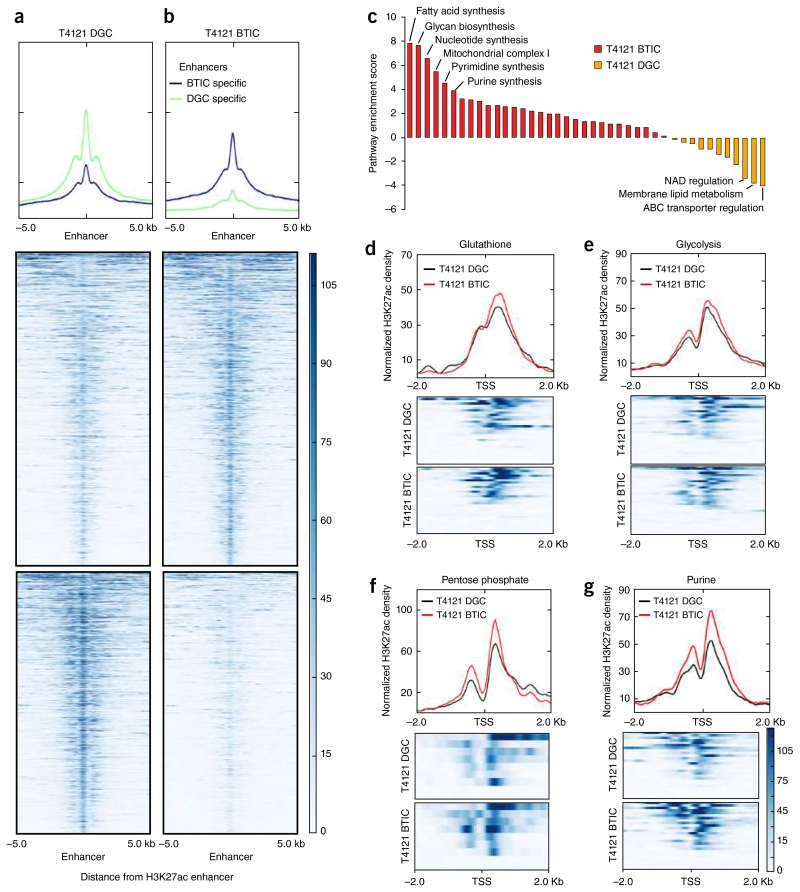
To evaluate the genomic upregulation of de novo purine synthesis pathway in BTICs, we performed genome-wide histone H3 Lys27 acetylation chromatin immunoprecipitation followed by deep sequencing (H3K27ac ChIP-seq) on a matched pair of BTICs and DGCs to map active enhancers and promoters. The differential chromatin regulation of H3K27ac levels at specific enhancers was compared between BTICs and DGCs (Fig. 2a,b). Unbiased enrichment analysis of all metabolic pathways identified the upregulation of H3K27ac at nucleotide synthesis pathway and particularly purine synthesis pathway genes in BTICs (Fig. 2c, Supplementary Fig. 4a and Supplementary Table 1). Focused analysis was performed at the promoter regions for genes involved in four key metabolic pathways: glutathione, glycolysis, pentose phosphate and purine pathways. Active chromatin (as demonstrated by H3K27 acetylation) for all four metabolic pathways was increased in BTICs compared to DGCs, with the strongest differential regulation in purine synthesis pathway genes (Fig. 2d–g). We then performed RNA-seq using the same matched pair of BTICs and DGCs to characterize the expression profiles. Analysis of the above four metabolic pathways revealed selective upregulation of purine synthesis pathway in BTICs (Supplementary Fig. 4b–e). In contrast, no consistent differential regulation was observed for the other three metabolic pathways. Our combined efforts of metabolomic profiling and genomic discovery demonstrate that BTICs specifically activate de novo purine synthesis, supporting purine biosynthesis as a potential point of sensitivity in resistant brain tumor populations.
Figure 2.
Genomic profiling reveals specific upregulation of de novo purine synthesis pathway in BTICs. (a,b) H3K27ac ChIP-seq coverage plots that illustrate (a) DGC-specific and (b) BTIC-specific enhancers in T4121 glioma cells. Heat maps are shown to depict H3K27ac signal, normalized to read depth, for ±5 kb surrounding enhancer peaks. Color scale indicates reads per kilobase per million mapped reads (RPKM). The y axis is also normalized H3K27ac read depth (RPKM). (c) Enrichment analysis of all metabolic pathways in BTICs (red) versus DGCs (orange) derived from differential H3K27 acetylation in the T4121 glioblastoma cell line. Pathway enrichment was assessed using single-sample gene set enrichment analysis comparing pathway enrichment scores between T4121 BTICs and DGCs. NAD, nicotinamide adenine dinucleotide; ABC, ATP-binding cassette. (d–g) H3K27ac ChIP-seq analysis of matched T4121 BTICs and DGCs for focused metabolic pathways. Transcriptional start sites (TSS) for selected metabolic genes were mapped for (d) glutathione, (e) glycolysis, (f) pentose phosphate and (g) purine metabolism.
Glycolytic activity modulates de novo purine synthesis in BTICs
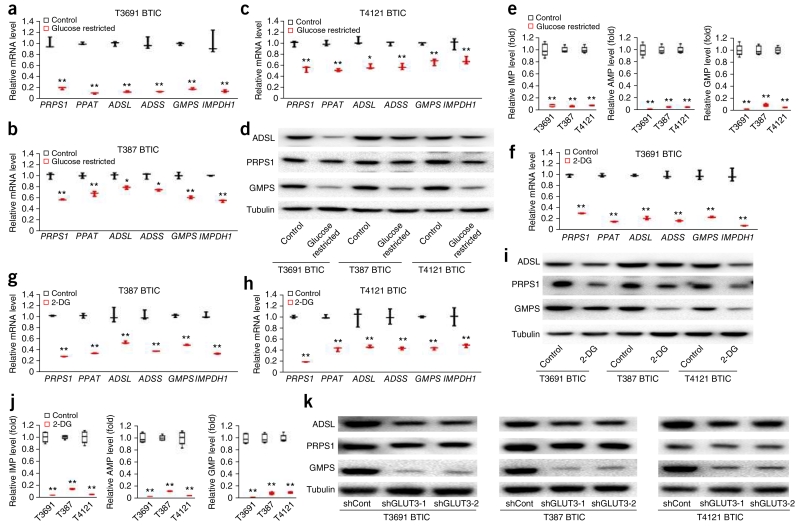
Preferential activation of purine synthesis in BTICs prompted us to investigate its metabolic significance in processing increased carbon flow through glycolysis. Standard cell culture conditions provide glucose at supraphysiologic levels to facilitate unrestricted proliferation, but these artificial conditions fail to replicate nutrient availability in tumors. To mimic tumor conditions, we grew BTICs with 0.45 g/L glucose, which is one-tenth of that in standard cell culture conditions, for 2 d. Short-term (48 h) glucose restriction of BTICs decreased mRNA levels of several molecular precursor markers, including SOX2, NES and OLIG2, and increased mRNA levels of differentiation markers, including GFAP and MBP (Supplementary Fig. 5a–c). In contrast, long-term glucose restriction for 7 d in BTICs induced a twofold increase in precursor markers of SOX2, NES and OLIG2, and significant decrease in differentiation markers of GFAP and MBPF, indicating metabolic reprogramming in BTIC in response to metabolic stress (Supplementary Fig. 5d–f), as is consistent with our previous report13. To evaluate the role of purine synthesis in maintaining BTIC stemness, we examined the effect of short-term glucose restriction of 48 h on mRNA levels of genes encoding all six purine synthetic enzymes. All six were decreased (Fig. 3a–c). This was also observed at the protein levels for PRPS1, ADSL and GMPS (Fig. 3d). Concordantly, levels of purine synthetic intermediates IMP, AMP and GMP were all decreased in BTICs under glucose-restricted conditions (Fig. 3e), establishing a tight connection between glucose uptake and downstream purine synthesis activity. We then used 2-deoxyglucose (2-DG), an inhibitor of glycolysis, to mimic glucose depletion under standard glucose conditions. Upon treatment with 10 mM 2-DG for 2 d, BTICs downregulated the transcription of genes involved in purine synthesis by 50–80% (Fig. 3f–h). Change of mRNA levels corresponded with decreased protein levels of PRPS1, ADSL and GMPS after 2-DG treatment (Fig. 3i) and the levels of purine intermediates, including IMP, AMP and GMP (Fig. 3j).
Figure 3.
Glycolytic activity modulates de novo purine synthesis in BTICs. (a–c) Effect of standard (4.5 g/L) and restricted (0.45 g/L) glucose conditions on mRNA levels of six purine synthesis pathway genes (PRPS1, PPAT, ADSL, ADSS, GMPS and IMPDH1) in T3691, T387 and T4121 BTICs. Two-tailed unpaired Student’s t-test; *P < 0.05; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T3691 (a): PRPS1, P = 0.0002, t(4) = 13.322; PPAT, P < 0.0001, t(4) = 53.492; ADSL, P < 0.0001, t(4) = 32.615; ADSS, P < 0.0001, t(4) = 42.225; GMPS, P < 0.0001, t(4) = 40.682; IMPDH1, P = 0.0021, t(4) = 7.025. T387 (b): PRPS1, P = 0.0066, t(4) = 9.452; PPAT, P = 0.0074, t(4) = 8.557; ADSL, P = 0.0320, t(4) = 4.487; ADSS, P = 0.0160, t(4) = 5.083; GMPS, P = 0.0038, t(4) = 8.158; IMPDH1, P = 0.0002, t(4) = 12.822. T4121 (c): PRPS1, P = 0.0015, t(4) = 8.907; PPAT, P = 0.0020, t(4) = 11.505; ADSL, P = 0.0100, t(4) = 7.569; ADSS, P = 0.0001, t(4) = 10.662; GMPS, P = 0.0056, t(4) = 6.886; IMPDH1, P = 0.0270, t(4) = 4.463.) (d) Protein levels of PRPS1, ADSL and GMPS under restricted glucose conditions (0.45 g/L) compared to standard conditions (4.5 g/L). (e) Levels of purine metabolites (IMP, AMP and GMP) under standard (4.5 g/L) and restricted (0.45 g/L) glucose conditions in T3691, T387 and T4121 BTICs. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 4 independent experiments per group. (IMP: T3691, P < 0.0001, t(6) = 19.866; T387, P < 0.0001, t(6) = 42.874; T4121, P < 0.0001, t(6) = 39.350. AMP: T3691, P < 0.0001, t(6) = 22.970; T387, P < 0.0001, t(6) = 34.374; T4121, P < 0.0001, t(6) = 34.989. GMP: T3691, P < 0.0001, t(6) = 24.155; T387, P < 0.0001, t(6) = 27.871; T4121, P < 0.0001, t(6) = 41.121.) (f–h) Treatment with 2-DG decreased the mRNA levels of six purine synthesis pathway genes (PRPS1, PPAT, ADSL, ADSS, GMPS and IMPDH1) in T3691, T387 and T4121 BTICs. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T3691: PRPS1, P < 0.0001, t(4) = 38.981; PPAT, P < 0.0001, t(4) = 45.247; ADSL, P < 0.0001, t(4) = 36.126; ADSS, P < 0.0001, t(4) = 20.815; GMPS, P = 0.0001, t(4) = 15.147; IMPDH1, P = 0.0002, t(4) = 13.439. T387: PRPS1, P < 0.0001, t(4) = 97.734; PPAT, P < 0.0001, t(4) = 26.615; ADSL, P = 0.0036, t(4) = 6.118; ADSS, P = 0.0002, t(4) = 12.486; GMPS, P < 0.0001, t(4) = 22.360; IMPDH1, P < 0.0001, t(4) = 20.446. T4121: PRPS1, P < 0.0001, t(4) = 50.270; PPAT, P < 0.0001, t(4) = 19.526; ADSL, P = 0.0051, t(4) = 5.572; ADSS, P = 0.0021, t(4) = 7.070; GMPS, P = 0.0002, t(4) = 13.103; IMPDH1, P = 0.0055, t(4) = 5.452. (i) Immunoblot analysis highlighting decreased protein levels of PRPS1, ADSL and GMPS under 2-DG treatment (0.45 g/L) compared to control. (j) Targeted mass spectrometry to examine the level of purine metabolites (IMP, AMP and GMP) under 2-DG or control treatment in T3691, T387 and T4121 BTICs. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 4 independent experiments per group. (IMP: T3691, t(6) = 26.947; T387, t(6) = 67.355; T4121, t(6) = 22.435. AMP: T3691, t(6) = 29.278; T387, t(6) = 47.672; T4121, t(6) = 20.124. GMP: T3691, t(6) = 28.602; T387, t(6) = 32.596; T4121, t(6) = 31.558.) (k) Immunoblot analysis highlighting decreased protein levels of PRPS1, ADSL and GMPS after shRNA-mediated GLUT3 knockdown. All full-length western blots are presented in Supplementary Figure 15. In e,j, center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum.
As the GLUT3 transporter mediates preferential glucose uptake in BTICs13, we examined the link between GLUT3 and purine synthesis activity. Using two independent short hairpin RNAs against the GLUT3 (SLC2A3) gene, we successfully knocked down GLUT3 expression by over 80% compared to control nontargeting shRNA (shCont) (Supplementary Fig. 5g). Upon GLUT3 depletion in BTICs, mRNA levels of purine synthesis pathway genes decreased (Supplementary Fig. 5h–j). Concordantly, GLUT3 knockdown reduced the protein levels of PRPS1, ADSL and GMPS (Fig. 3k).
As glucose is not the sole carbon source in brain and cancer metabolism, we further interrogated the role of additional metabolic sources such as glutamine and acetate in the context of glucose restriction. Induced glucose deprivation in two BTIC models caused loss of cell viability (Supplementary Fig. 5k,l). Glutamine partially rescued cell viability, while adding acetate showed no significant benefit (Supplementary Fig. 5k,l). Glutamine is a known nitrogen source for metabolic reprogramming in cancer cells24, so we analyzed the contribution of nitrogen from glutamine to synthesize purine in BTICs. Using a [15N2]glutamine tracer, we found that glutamine contributed nitrogen atoms to purine synthesis (Supplementary Fig. 6). These data confirm a link between GLUT3-mediated glucose influx and the functional activation of de novo purine synthesis in BTICs, creating critical metabolic support for BTIC maintenance.
The transcription factor MYC regulates purine synthesis pathway in BTICs
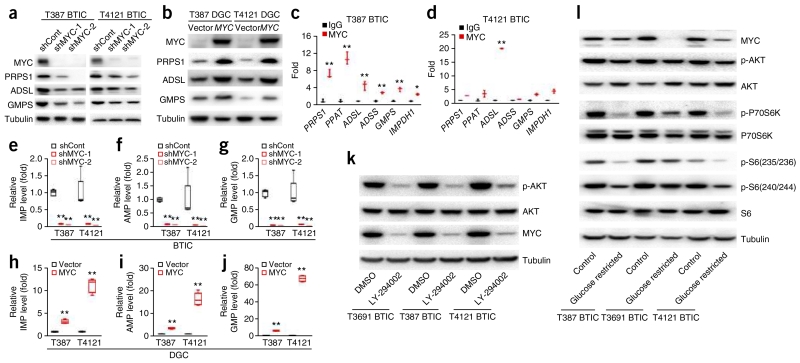
We and others previously showed that MYC is essential to the maintenance of BTIC self-renewal and tumorigenic capacity25,26. MYC regulates enzymes involved in basic cellular metabolism, supporting a central role of MYC in metabolic reprogramming in cancers27. Using two independent shRNAs targeting MYC, we found that silencing MYC in BTICs significantly reduced mRNA levels of all six purine biosynthetic enzymes and protein levels of PRPS1, ADSL and GMPS (Fig. 4a and Supplementary Fig. 7a–g). Reciprocally, MYC overexpression in DGCs increased mRNA levels of all six purine biosynthetic enzymes and protein levels of PRPS1, ADSL and GMPS (Fig. 4b and Supplementary Fig. 7h–n). Linking these findings to the stem-like status of BTICs, knocking down MYC decreased mRNA level of precursor markers, including SOX2, NES and OLIG2, and elevated levels of differentiation markers, such as GFAP and MBPF (Supplementary Fig. 7o,p). Concordantly, overexpression of MYC in DGCs caused a two- to sevenfold increase in precursor markers, including SOX2, NES and OLIG2, and a decrease in mRNA levels of differentiation markers GFAP and MBP (Supplementary Fig. 7q,r). To assess the direct transcriptional effect of MYC on purine synthesis gene targets, we performed chromatin immunoprecipitation of MYC protein in two BTIC models, followed by reverse transcription and quantitative PCR (qRT-PCR) with primer sets designed for the promoter regions of PRPS1, PPAT, ADSL, ADSS, GMPS and IMPDH1. Compared to control IgG, MYC was more likely to occupy the promoter regions of genes involved in purine synthesis in BTICs (Fig. 4c,d).
Figure 4.
Transcription factor MYC regulates de novo purine synthesis pathway enzymes in BTICs. (a) Knockdown of MYC in BTICs using two independent shRNAs decreased the protein levels of PRPS1 and GMPS in T387 and T4121 BTICs. Tubulin was used as a loading control. (b) Overexpression of exogenous MYC upregulated protein levels of purine synthesis enzymes PRPS1, ADSL and GMPS in T387 and T4121 DGCs. (c,d) Transcription factor MYC bound directly to promoter regions of enzymes in purine synthesis pathway in (c) T387 and (d) T4121 BTICs. Two-tailed unpaired Student’s t-test; *P < 0.05; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T387: PRPS1, P = 0.0075, t(4) = 5.379; PPAT, P = 0.0078, t(4) = 4.904; ADSL, P = 0.0072, t(4) = 6.054; ADSS, P = 0.0008, t(4) = 12.225; GMPS, P = 0.0009, t(4) = 8.949; IMPDH1, P = 0.0200, t(4) = 3.654. T4121: PRPS1, P = 0.0005, t(4) = 10.270; PPAT, P = 0.0026, t(4) = 7.175; ADSL, P < 0.0001, t(4) = 25.230; ADSS, P = 0.0260, t(4) = 3.427; GMPS, P = 0.0017, t(4) = 7.917; IMPDH1, P = 0.0086, t(4) = 6.138.) (e–g) Knockdown of MYC using two independent shRNAs decreased the level of purine metabolites in BTICs. Mass spectrometry was used to assess the levels of (e) IMP, (f) AMP and (g) GMP in BTICs. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 4 independent experiments per group. (IMP: T387, shRNA-1, P < 0.0001, t(6) = 24.084; shRNA-2, P < 0.0001, t(6) = 24.802; T4121, shRNA-1, P = 0.0008, t(6) = 4.653; shRNA-2, P = 0.0006, t(6) = 4.890. AMP: T387, shRNA-1, P < 0.0001, t(6) = 30.416; shRNA-2, P < 0.0001, t(6) = 31.323; T4121, shRNA-1, P = 0.0069, t(6) = 3.143; shRNA-2, P = 0.0057, t(6) = 3.272. GMP: T387, shRNA-1, P < 0.0001, t(6) = 20.868; shRNA-2, P < 0.0001, t(6) = 21.045; T4121, shRNA-1, P = 0.0003, t(6) = 5.393; shRNA-2, P = 0.0002, t(6) = 5.687.) (h–j) Overexpression of exogenous MYC upregulates the level of purine metabolites of (h) IMP, (i) AMP and (j) GMP in T387 and T4121 DGCs. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 4 independent experiments per group. (IMP: T387, t(6) = 11.949; T4121, t(6) = 15.436. AMP: T387, t(6) = 17.874; T4121, t(6) = 12.641. GMP: T387, t(6) = 23.280; T4121, t(6) = 45.919.) (k) Effect of PI3K inhibitor LY-294002 (5 μM for 48 h) on the levels of phospho-AKT (p-AKT), total AKT and MYC in three BTIC models (T3691, T387 and T4121). (l) Effect of restricted glucose conditions (0.45 g/L) and standard conditions (4.5 g/L) for 48 h on the levels of MYC, phospho-AKT, total AKT, phospho-p70 S6 kinase (p-P70S6K) (Thr389), total p70 S6 kinase (P70S6K), S6 phosphorylated on Ser235 or Ser236 (p-S6(235/236)), S6 phosphorylated on Ser240 or Ser244 (p-S6(240/244)) and total S6 in three BTIC models (T3691, T387 and T4121). All full-length western blots are presented in Supplementary Figure 15. In e–j, center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum.
We next examined the functional significance of MYC-regulated purine synthesis at the metabolic level. Knockdown of MYC in two BTIC models significantly decreased the absolute levels of IMP, AMP and GMP metabolites (Fig. 4e–g). Conversely, overexpressing MYC in two DGC models caused a dramatic increase in the absolute levels of IMP, AMP and GMP (Fig. 4h–j).
Metabolic crosstalk permits the integration of cell state and metabolic activity. Phosphatidylinositol-4,5-bisphosphate 3-kinase (PI3K) is a central signaling node that organizes the cellular response to the nutritional state, regulating survival and motility. To determine the function of PI3K in our findings, we treated three BTIC models with a PI3K inhibitor (LY294002), revealing attenuated activity of AKT, a downstream effector, and reduced MYC levels, suggesting that PI3K is an upstream regulator of MYC (Fig. 4k). Further, acute glucose deprivation of 48 h reduced the activation of metabolic stress indicators, including AKT, mammalian target of rapamycin (mTOR) and ribosomal protein S6 kinase (S6K), as well as MYC levels, confirming the signaling crosstalk between key metabolic pathways (Fig. 4l). Our findings indicate that in BTICs, MYC directs carbon flow into the synthesis of purine metabolites by directly activating the transcription of purine synthesis genes.
AMP and GMP synthesis is required for BTIC maintenance
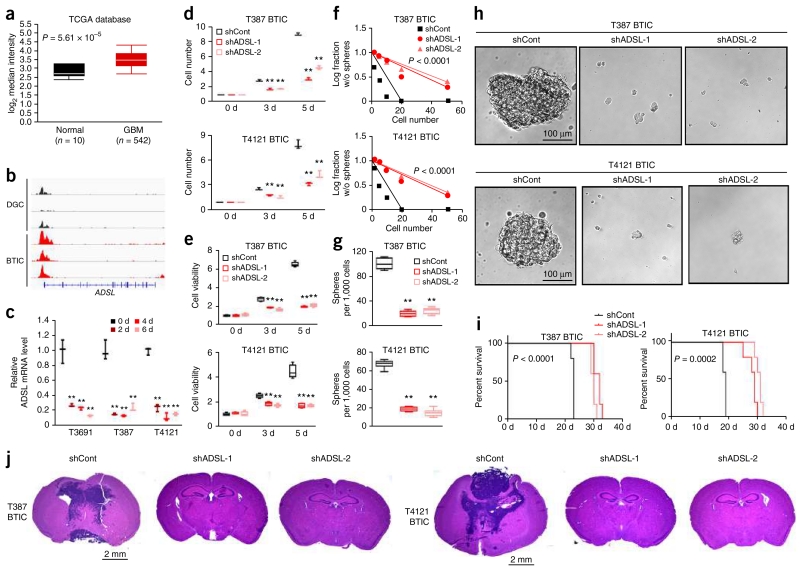
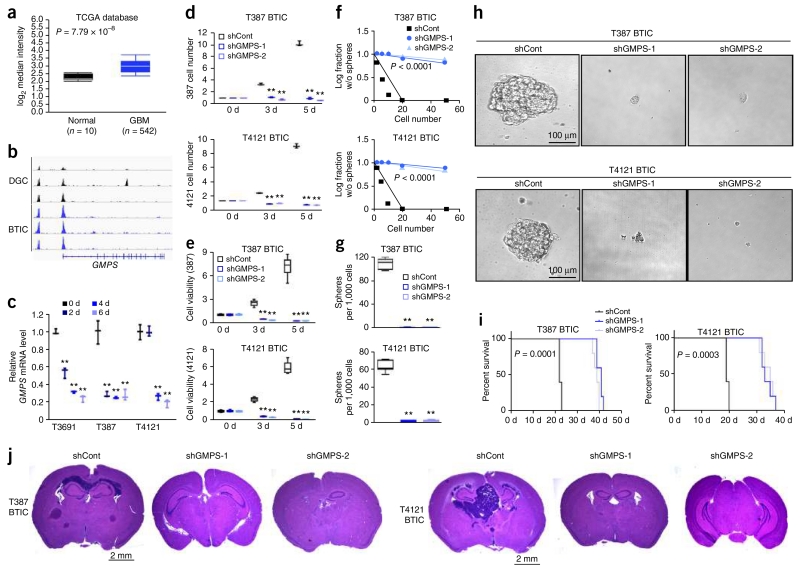
De novo purine synthesis pathway branches at the point of IMP: ADSS and ADSL catalyze the synthesis of AMP from IMP, while IMPDH and GMPS cooperate to synthesize GMP. To determine the functional significance of these two downstream synthetic pathways in BTIC maintenance, we assessed the roles of ADSL and GMPS expression and function in glioma (Figs. 5 and 6). Levels of ADSL and GMPS mRNA were significantly elevated in 542 samples from the TCGA glioblastoma data set as compared to normal brain tissues (Figs. 5a and 6a)19,20. Expression levels of ADSL, GMPS and other purine synthesis genes did not significantly differ across molecular subtypes of glioblastoma, nor did known precursor markers, including OLIG2, MYC and SOX2, demonstrate a consistent pattern among subtypes, suggesting that these mechanisms may be shared across glioblastoma (Supplementary Fig. 8a–c). We interrogated the promoter regulation of purine synthetic genes across three matched pairs of BTICs and DGCs derived from three independent glioblastoma patients28, revealing that increased activation, as measured by H3K27ac peak levels, was greater in BTICs (Figs. 5b and 6b and Supplementary Fig. 8d,e). We next investigated mRNA levels for purine synthetic genes during serum-induced differentiation in three different BTIC models, demonstrating that differentiation suppressed the expression of purine synthetic genes in BTICs by approximately 80% (Figs. 5c and 6c and Supplementary Fig. 8g,h).
Figure 5.
ADSL regulates BTIC growth and self-renewal. (a) Microarray-based gene expression levels of ADSL from 542 glioblastoma (GBM) specimens and 10 different specimens of normal brain tissues. Expression data were derived from the TCGA glioblastoma data set (P < 0.0001, t(550) = 6.095, two-tailed unpaired Student’s t-test). (b) H3K27ac ChIP-seq enrichment plot centered at the gene locus for ADSL. Enrichment is shown for three matched pairs of DGCs and BTICs from patient-derived glioblastoma specimens. H3K27ac ChIP-seq data were downloaded from NCBI Gene Expression Omnibus GSE54047. (c) qRT-PCR quantification of ADSL mRNA in BTICs and DGCs. BTICs derived from three primary human glioblastoma specimens (T3691, T387, T4121) were treated with serum to induce differentiation over a course of 2, 4 or 6 d. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T3691: 2 d, P = 0.0014, t(4) = 7.967; 4 d, P = 0.0011; t(4) = 8.381; 6 d, P = 0.0007, t(4) = 9.534. T387: 2 d, P = 0.0004, t(4) = 11.190; 4 d, P = 0.0003, t(4) = 11.454; 6 d, P = 0.0007, t(4) = 9.351. T4121: 2 d, P < 0.0001, t(4) = 19.147; 4 d, P < 0.0001, t(4) = 21.366; 6 d, P < 0.0001, t(4) = 27.708. (d) Two independent shRNAs targeting ADSL decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to a non-targeting control shRNA (shCont), as measured by direct cell number count. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T387: 3 d, shRNA-1, P = 0.0004, t(4) = 11.280; shRNA-2, P = 0.0001, t(4) = 14.945; 5 d, shRNA-1, P < 0.0001, t(4) = 42.426; shRNA-2, P < 0.0001, t(4) = 25.324. T4121: 3 d, shRNA-1, P = 0.0018, t(4) = 7.379; shRNA-2, P = 0.0018, t(4) = 7.391; 5 d, shRNA-1, P = 0.0001, t(4) = 14.215; shRNA-2, P = 0.0009, t(4) = 8.960.) (e) Two independent shRNAs targeting ADSL decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to shCont, as measured by cell-titer assay. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 6 independent experiments per group. (T387: 3 d, shRNA-1, P < 0.0001, t(10) = 9.842; shRNA-2, P < 0.0001, t(10) = 10.423; 5 d, shRNA-1, P < 0.0001, t(10) = 45.883; shRNA-2, P < 0.0001, t(10) = 35.452. T4121: 3 d, shRNA-1, P < 0.0001, t(10) = 7.161; shRNA-2, P < 0.0001, t(10) = 9.656; 5 d, shRNA-1, P < 0.0001, t(10) = 13.049; shRNA-2, P < 0.0001, t(10) = 13.473.) (f) In vitro extreme limiting dilution assays (ELDAs) demonstrate that knockdown of ADSL in T387 (top) and T4121 (bottom) BTICs decreases the frequency of neurosphere formation (T387, P < 0.0001; T4121, P < 0.0001 by ELDA analysis). (g) Knockdown of ADSL in T387 (top) and T4121 (bottom) BTICs decreases the number of spheres formed in ELDA per 1,000 cells seeded. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 6 independent experiments per group. (T387: shRNA-1, t(10) = 23.115; shRNA-2, t(10) = 21.493. T4121: shRNA-1, t(10) = 27.062; shRNA-2, t(10) = 23.890.) (h) Representative images of neurospheres derived from T387 (left) and T4121 (right) BTICs expressing shCont, shADSL-1 or shADSL-2. Scale bar, 100 μm. Each image is representative of at least 5 similar experiments. (i) Kaplan-Meier survival curves of immunocompromised mice bearing intracranial T387 (top) or T4121 (bottom) BTICs expressing shCont, shADSL-1 or shADSL-2. (T387, P < 0.0001; T4121, P = 0.0002 by log-rank test; n = 5 animals per group). (j) Representative images of hematoxylin and eosin stained sections of mouse brains collected on day 18 after transplantation of T387 (top) or T4121 (bottom) BTICs expressing shCont, shADSL-1 or shADSL-2. Scale bar, 2 mm. Each image is representative of at least 3 similar experiments. In a,e,g, center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum.
Figure 6.
GMPS regulates BTIC growth and self-renewal. (a) Microarray-based gene expression level of GMPS from 542 glioblastoma (GBM) specimens and 10 different specimens of normal brain tissues. Expression data were derived from the TCGA glioblastoma data set (P < 0.0001, t(550) = 10.818, two-tailed unpaired Student’s t-test). (b) H3K27ac ChIP-seq enrichment plot centered at the gene locus for GMPS. Enrichment is shown for three matched pairs of DGCs and BTICs from patient-derived glioblastoma specimens. H3K27ac ChIP-seq data were downloaded from NCBI Gene Expression Omnibus GSE54047. (c) qRT-PCR quantification of GMPS mRNA levels in BTICs and DGCs. BTICs derived from three primary human glioblastoma specimens (T3691, T387, T4121) were treated with serum to induce differentiation over a course of 2, 4 or 6 d. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T3691: 2 d, P = 0.0003, t(4) = 11.523; 4 d, P < 0.0001, t(4) = 35.939; 6 d, P < 0.0001, t(4) = 26.326. T387: 2 d, P = 0.0008, t(4) = 9.076; 4 d, P = 0.0006, t(4) = 9.748; 6 d, P = 0.0010, t(4) = 8.561. T4121: 2 d, P = 0.951, t(4) = 0.066; 4 d, P = 0.0018, t(4) = 10.669; 6 d, P = 0.0001, t(4) = 15.436.) (d) Two independent shRNAs targeting GMPS decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to a non-targeting control shRNA, as measured by direct cell number count. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. T387: 3 d, shRNA-1, P = 0.0051, t(4) = 13.914; shRNA-2, P = 0.0048, t(4) = 14.422; 5 d, shRNA-1, P = 0.0016, t(4) = 24.686; shRNA-2, P = 0.0012, t(4) = 27.858. T4121: 3 d, shRNA-1, P = 0.0003, t(4) = 56.569; shRNA-2, P = 0.0012, t(4) = 24.033; 5 d, shRNA-1, P = 0.0013, t(4) = 26.916; shRNA-2, P = 0.0014, t(4) = 26.845. (e) Two independent shRNAs targeting GMPS decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to a non-targeting control shRNA, as measured by cell-titer assay. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 6 independent experiments per group. (T387: 3 d, shRNA-1, P < 0.0001, t(10) = 13.910; shRNA-2, P < 0.0001, t(10) = 14.932; 5 d, shRNA-1, P < 0.0001, t(10) = 13.856; shRNA-2, P < 0.0001, t(10) = 13.841. T4121: 3 d, shRNA-1, P < 0.0001, t(10) = 20.981; shRNA-2, P < 0.0001, t(10) = 20.981; 5 d, shRNA-1, P < 0.0001, t(10) = 21.738; shRNA-2, P < 0.0001, t(10) = 21.924.) (f) In vitro extreme limiting dilution assays (ELDA) demonstrate that knockdown of GMPS in T387 (top) and T4121 (bottom) BTICs decreases the frequency of neurosphere formation (T387, P < 0.0001; T4121, P < 0.0001, by ELDA analysis). (g) Knockdown of GMPS in T387 (top) and T4121 (bottom) BTICs decreases the number of spheres formed in ELDA per 1,000 cells seeded. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 6 independent experiments per group. (T387: shRNA-1, t(10) = 32.575; shRNA-2, t(10) = 32.367. T4121: shRNA-1, t(10) = 26.678; shRNA-2, t(10) = 26.638.) (h) Representative images of neurospheres derived from T387 (left) and T4121 (right) BTICs expressing shCont, shGMPS-1 or shGMPS-2. Scale bar, 100 μm. Each image is representative of at least 5 similar experiments. (i) Kaplan-Meier survival curves of immunocompromised mice bearing intracranial T387 (top) or T4121 (bottom) BTICs expressing shCont, shGMPS-1 or shGMPS-2. (T387, P = 0.0001; T4121, P = 0.0003 by log-rank test; n = 5 animals per group). (j) Representative images of hematoxylin and eosin stained sections of mouse brains collected on day 18 after transplantation of T387 (top) or T4121 BTICs expressing shCont, shGMPS-1 or shGMPS-2. Scale bar, 2 mm. Each image is representative of at least 3 similar experiments. In a,e,g, center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum.
To determine the functional consequences of AMP and GMP synthesis in BTICs, we targeted the key enzymes in each synthetic pathway using a lentivirus-based vector to create independent, non-overlapping shRNAs targeting ADSL (shADSL-1 and shADSL-2) and GMPS (shGMPS-1 and shGMPS-2). The efficiency of ADSL and GMPS knockdown was confirmed at the protein level by immunoblotting (Supplementary Fig. 9a,b). Reducing the protein levels of either ADSL or GMPS significantly decreased BTIC growth and cell viability in two different models (Figs. 5d,e and 6d,e). The specificity of shRNA knockdown was confirmed by reconstituting ADSL and GMPS expression (Supplementary Fig. 9d,e), partially restoring the defect in cell viability of BTIC (Supplementary Fig. 9g,h,j,k). Knocking down ADSL or GMPS also reduced precursor markers SOX2, NES and OLIG2, and elevated differentiation markers GFAP and MBPF (Supplementary Fig. 9m,n,p,q). In contrast, purine synthesis is dispensable in DGCs, as targeting ADSL or GMPS expression had no effect on proliferation (Supplementary Fig. 10a,b).
Using in vitro limiting dilution assay, we demonstrated that knockdown of ADSL or GMPS significantly impaired self-renewal capacity, as measured by neurosphere formation (Figs. 5f and 6f). Sphere size, often considered a surrogate for proliferation, was significantly reduced in BTICs expressing shRNAs targeting ADSL or GMPS compared to shCont (Figs. 5g,h and 6g,h). We transplanted BTICs into the brains of immunocompromised mice to interrogate the effect of ADSL or GMPS in tumor initiation of BTICs in vivo. Mice bearing BTICs expressing shRNA targeting ADSL (shADSL-1 or shADSL-2) or GMPS (shGMPS-1 or shGMPS-2) demonstrated increased survival relative to shCont mice, establishing the role of ADSL and GMPS in maintaining tumorigenic capacity of BTICs in vivo (Fig. 5i,j and Fig. 6i,j). To determine the mechanism of these in vivo effects, we examined the protein levels of Ki-67, cleaved caspase-3 and SOX2, as markers for proliferation, apoptosis and stemness, respectively, using immunohistochemistry in the transduced xenograft tumor specimens. Upon knockdown of ADSL or GMPS, cleaved caspase-3 increased, while Ki-67 and SOX2 decreased (Supplementary Fig. 11a,b).
As cancer metabolism interacts with the immune system, we interrogated the roles of purine synthesis pathway in vivo using a murine glioma model in a syngeneic host. Specifically, we knocked down the expression of Adsl or Gmps in the mouse glioma 261 line (GL261) in an immunocompetent host. Knockdown efficiency was confirmed using qRT-PCR (Supplementary Fig. 12a,b). Intracranial implantation of GL261 cells expressing either control or gene-targeting shRNA was performed on MHC-matched C57BL/6J mice. Knocking down Adsl or Gmps increased the survival of immunocompetent mice bearing GL261 tumors (Supplementary Fig. 12d,e) and reduced tumor volume (Supplementary Fig. 12g,h). These results establish nonredundant dependency of BTICs on the downstream components of de novo purine synthesis pathway, specifically AMP and GMP synthesis, as essential to maintaining BTIC proliferation, self-renewal and tumorigenicity.
PRPS1 is required for BTIC maintenance
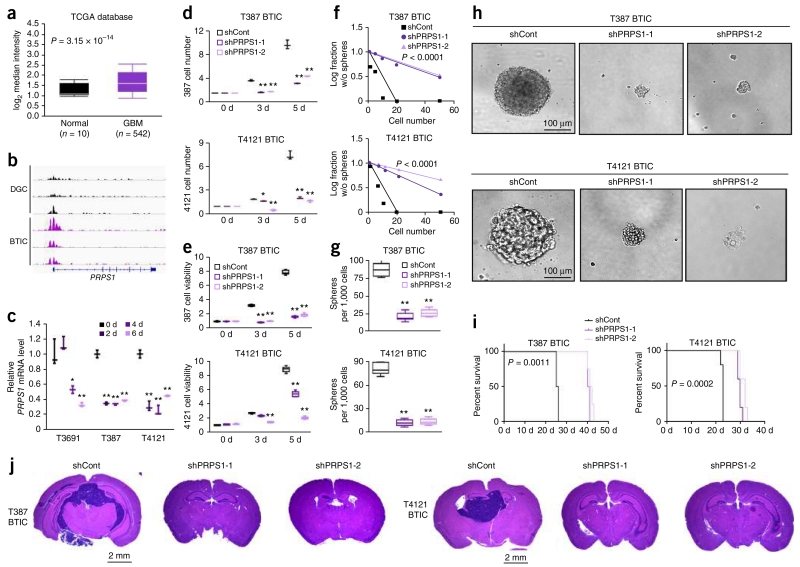
PRPS1 catalyzes the first step of de novo purine synthesis by adding pyrophosphate onto R5P. Using gene expression data of glioblastoma and normal brain tissue from TCGA19,20, we found that the PRPS1 gene was overexpressed in glioblastoma (Fig. 7a). Consistently, the promoter regions of PRPS1 and PPAT were active in BTICs, as shown by accumulation of H3K27ac at these locations in BTICs as compared to matched DGCs (Fig. 7b and Supplementary Fig. 8f)28. Induction of BTIC differentiation in three patient-derived glioblastomas significantly decreased PRPS1 and PPAT mRNA levels (Fig. 7c and Supplementary Fig. 8i). These data led us to hypothesize that the gene encoding PRPS1 is critical to BTIC maintenance.
Figure 7.
PRPS1 regulates BTIC growth and self-renewal. (a) Microarray-based gene expression level of PRPS1 from 542 glioblastoma (GBM) specimens and 10 different specimens of normal brain tissues. Expression data were derived from the TCGA glioblastoma data set (P = 0.0003, t(550) = 4.719, two-tailed unpaired Student’s t-test). (b) H3K27ac ChIP-seq enrichment plot centered at the gene locus for PRPS1. Enrichment is shown for three matched pairs of DGCs and BTICs from patient-derived glioblastoma specimens. H3K27ac ChIP-seq data were downloaded from NCBI Gene Expression Omnibus GSE54047. (c) qRT-PCR quantification of PRPS1 mRNA levels in BTICs and DGCs. BTICs derived from three primary human glioblastoma specimens (T3691, T387, T4121) were treated with serum to induce differentiation into DGCs, with time points taken at 2, 4 and 6 d. Two-tailed unpaired Student’s t-test; *P < 0.05; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. (T3691: 2 d, P = 0.3309, t(4) = 1.106; 4 d, P = 0.0109, t(4) = 4.487; 6 d, P = 0.0028, t(4) = 6.576. T387: 2 d, P = 0.0005, t(4) = 16.496; 4 d, P < 0.0001, t(4) = 22.363; 6 d, P < 0.0001, t(4) = 20.931. T4121: 2 d, P = 0.0022, t(4) = 6.946; 4 d, P = 0.0018, t(4) = 7.388; 6 d, P = 0.0092, t(4) = 4.671.) (d) Two independent shRNAs targeting PRPS1 decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to a non-targeting control shRNA, as measured by direct cell number count. Two-tailed unpaired Student’s t-test; **P < 0.01; n = 3 independent experiments per group. Data are presented as median ± s.e.m. T387: 3 d, shRNA-1, P = 0.0029, t(4) = 18.336; shRNA-2, P = 0.0030, t(4) = 17.441; 5 d, shRNA-1, P = 0.0002, t(4) = 63.302; shRNA-2, P = 0.0004, t(4) = 53.000. T4121: 3 d, shRNA-1, P = 0.0136, t(4) = 8.485; shRNA-2, P = 0.0062, t(4) = 12.611; 5 d, shRNA-1, P = 0.0088, t(4) = 10.590; shRNA-2, P = 0.0079, t(4) = 11.207. (e) Two independent shRNAs targeting PRPS1 decreased the growth of T387 (top) and T4121 (bottom) BTICs in comparison to a non-targeting control shRNA, as measured by cell-titer assay. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 6 independent experiments per group. (T387: 3 d, shRNA-1, t(10) = 40.496; shRNA-2, t(10) = 38.005; 5 d, shRNA-1, t(10) = 47.379; shRNA-2, t(10) = 42.150. T4121: 3 d, shRNA-1, t(10) = 7.316; shRNA-2, t(10) = 23.789; 5 d, shRNA-1, t(10) = 15.959; shRNA-2, t(10) = 39.988.) (f) In vitro extreme limiting dilution assays (ELDA) demonstrate that knockdown of PRPS1 in T387 (top) and T4121 (bottom) BTICs decreases the frequency of neurosphere formation. (T387, P < 0.0001; T4121, P < 0.0001 by ELDA analysis). (g) Knockdown of PRPS1 in T387 (top) and T4121 (bottom) BTICs decreases the number of neurospheres formed in ELDA per 1,000 cells seeded. Two-tailed unpaired Student’s t-test; **P < 0.0001; n = 6 independent experiments per group. (T387: shRNA-1, t(10) = 14.805; shRNA-2, t(10) = 13.952. T4121: shRNA-1, t(10) = 20.889; shRNA-2, t(10) = 20.584.) (h) Representative images of neurospheres derived from T387 (left) and T4121 (right) BTICs expressing shCont, shPRPS1-1 or shPRPS1-2. Scale bar, 100 μm. Each image is representative of at least 5 similar experiments. (i) Kaplan-Meier survival curves of immunocompromised mice bearing intracranial T387 (top) or T4121 (bottom) BTICs expressing shCont, shPRPS1-1 or shPRPS1-2. (T387, P = 0.0011; T4121, P = 0.0002 by log-rank test; n = 5 animals per group). (j) Representative images of hematoxylin and eosin stained sections of mouse brains collected on day 18 after transplantation of T387 (top) or T4121 BTICs expressing shCont, shPRPS1-1 or shPRPS1-2. Scale bar, 2 mm. Each image is representative of at least 3 similar experiments. In a,e,g, center line represents the median, lower and upper limits of the box represent the 25th and 75th percentiles, and whiskers show maximum and minimum.
We then developed two non-overlapping shRNAs to knock down PRPS1, designated shPRPS1-1 and shPRPS1-2. The efficiency of PRPS1 knockdown was confirmed at the protein level by immunoblotting (Supplementary Fig. 9c). Targeting PRPS1 significantly decreased the growth and cell viability of BTICs (Fig. 7d,e). The specificity of shRNA knockdown was confirmed by reconstituting the expression of PRPS1 (Supplementary Fig. 9f), which partially rescued the defect in cell viability of BTICs (Supplementary Fig. 9i,l). Knockdown of PRPS1 was also accompanied by a decrease in the precursor markers SOX2, NES and OLIG2 and elevation of the differentiation markers GFAP and MBP (Supplementary Fig. 9o,r). In contrast, targeting PRPS1 expression in matched DGCs did not alter cellular proliferation (Supplementary Fig. 10c), again supporting dispensability of purine biosynthesis in differentiated tumor cells.
Using in vitro limiting dilution assay, we found that PRPS1 knockdown resulted in a more than threefold reduction in the frequency of sphere formation (Fig. 7f). Sphere size was also significantly reduced in BTICs expressing shRNAs targeting PRPS1 compared to those expressing shCont (Fig. 7g,h). To examine the effect of PRPS1 in maintaining tumorigenic potential of BTICs in vivo, we transplanted BTICs expressing PRPS1-directed shRNAs or shCont into the brains of immunocompromised mice. Animals bearing BTICs expressing shRNA targeting PRPS1 (shPRPS1-1 or shPRPS1-2) demonstrated significantly delayed tumor formation and increased survival relative to controls (Fig. 7i,j). The in vivo impact of targeting PRPS1 was also assayed by measuring the protein levels of Ki-67, cleaved caspase-3 and SOX2 as markers of proliferation, apoptosis and stemness, respectively, using immunohistochemistry in the xenograft specimens. Upon PRPS1 knockdown, cleaved caspase-3 increased, while Ki-67 and SOX2 decreased (Supplementary Fig. 11c). To determine a potential interaction with immune function, we first validated in vivo tumorigenesis experiments using murine GL261 glioma model in C57BL/6J syngeneic immunocompetent mice. Efficiency of PRPS1 knockdown was confirmed using qRT-PCR (Supplementary Fig. 12c). Intracranial implantation of GL261 cells expressing either control or gene-targeting shRNA was performed on MHC-matched C57BL/6J mice. Knocking down Prps1 significantly increased the survival of immunocompetent mice bearing GL261 tumors (Supplementary Fig. 12f) and reduced tumor volume (Supplementary Fig. 12i). Taken together, our data support a role of the gene encoding purine synthesis enzyme PRPS1 in sustaining BTIC phenotype, as measured by proliferation, self-renewal and tumorigenicity.
Overexpression of de novo purine synthesis pathway genes is associated with poor prognosis in glioma patients
To generalize our findings of purine synthesis pathway dependence to glioma patients, we investigated glioma databases to examine gene expression of enzymes involved in purine synthesis19,20. Mapping purine synthetic genes across gliomas of all grades, purine synthetic genes were strongly enriched in non-CpG island methylator phenotype (non-CIMP) grade III and IV gliomas, without correlation to specific genetic lesions beyond IDH wild-type status (Supplementary Fig. 13a). These results were concordant with our functional data, as we selected BTIC models to represent each of the major TCGA transcriptional groups (data not shown). To interrogate the functional linkage between purine biology and tumor processes in silico, we performed a gene set enrichment analysis with a purine metabolic signature, finding cell cycle regulation as the dominant process, with linkage to mRNA metabolism, Wnt signaling and antigen processing (Supplementary Fig. 13b). Cell cycle progression would be expected to depend on purine availability, but we previously reported that BTICs and DGCs display similar cell cycle profiles29, suggesting that BTICs preferentially depend on de novo purine synthesis.
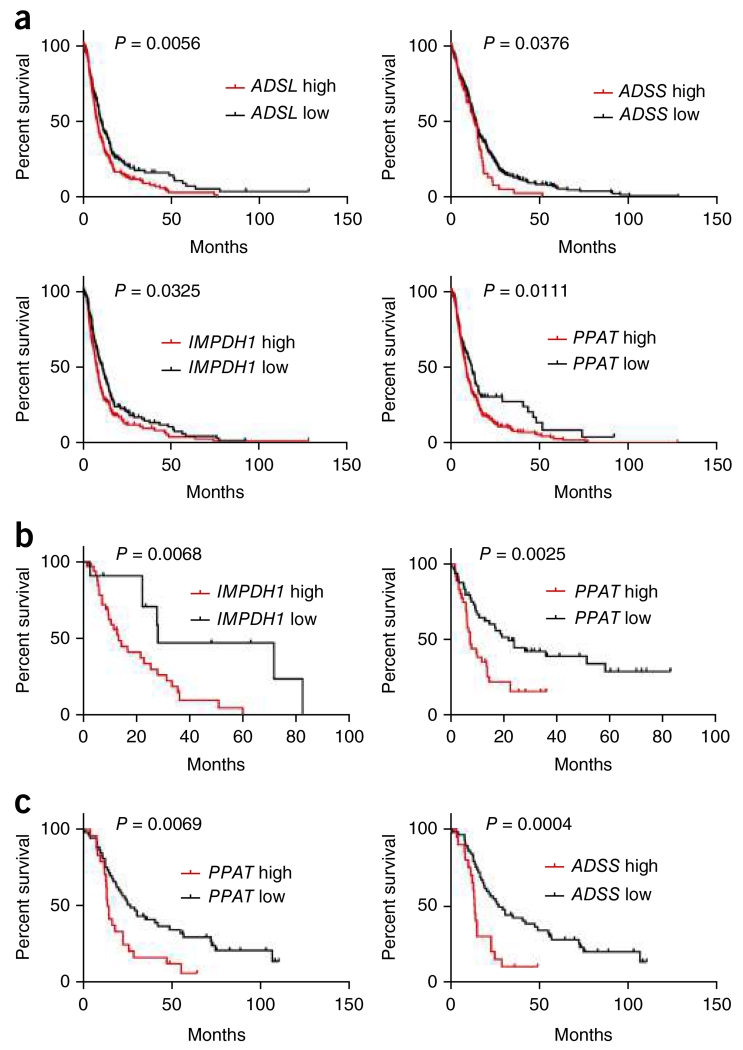
To determine the clinical relevance of purine biosynthesis, we interrogated the linkage between purine synthesis enzymes and patient outcome. Increased levels of four genes from de novo purine synthesis pathway, ADSL, ADSS, IMPDH1 and PPAT, were associated with poor prognosis in glioblastoma patients (Fig. 8a). PPAT is the second enzyme on the main branch that synthesizes IMP from R5P. Higher PPAT gene expression is associated with shorter survival in glioblastoma (P = 0.0111). Through retrospective analysis of the TCGA data set, increased expression of IMPDH1 (on the sub-branch synthesizing GMP from IMP) and both ADSL and ADSS (on the sub-branch synthesizing AMP from IMP) were correlated with worse prognosis in glioblastoma patients (IMPDH1: P = 0.0325; ADSL: P = 0.0056; ADSS: P = 0.0376).
Figure 8.
Overexpression of purine synthesis genes is associated with poor brain tumor patient survival. (a) Analysis of the TCGA glioblastoma data set indicates a significant correlation between reduced expressions of purine synthesis genes (PPAT, ADSL, ADSS and IMPDH1) and overall survival in all patients. Log-rank test. (ADSLlow, n = 256; ADSLhigh, n = 261; P = 0.0056. ADSSlow, n = 338; ADSShigh, n = 179; P = 0.0376. IMPDH1low, n = 279; IMPDH1high, n = 238; P = 0.0325. PPATlow, n = 382; PPAThigh, n = 135; P = 0.0111.) (b) Analysis of the Freije high-grade glioma data set indicates a significant inverse correlation between expression of purine synthesis genes (PPAT and IMPDH1) and overall survival in all patients. Log-rank test. (PPATlow, n = 49; PPAThigh, n = 37; P = 0.0068. IMPDH1low, n = 50; IMPDH1high, n = 36; P = 0.0025.) (c) Analysis of the Phillips high-grade glioma data set indicates a significant correlation between reduced expressions of purine synthesis enzymes (PPAT and ADSS) and overall survival in all patients. Log-rank test. (PPATlow, n = 53; PPAThigh, n = 24; P = 0.0069. ADSSlow, n = 57; ADSShigh, n = 20; P = 0.0004.)
To further assess the significance of purine synthesis pathway expression in brain tumor patients, we generated Kaplan-Meier survival curves in independent glioma data sets30-32, which include both low-grade glioma and glioblastoma specimens. Overexpression of PPAT was associated with significantly shorter survival among glioma patients in both the Freije (P = 0.0025) and Phillips (P = 0.0069) data sets (Fig. 8b,c). Higher expression of two other purine synthesis genes, IMPDH1 (P = 0.0066) and ADSS (P = 0.0004), was also correlated with worse survival in the Freije and Phillips data sets, respectively (Fig. 8b,c). Comprehensive analysis of each of the six de novo purine biosynthesis enzymes supports a general link between activation of purine metabolism and poor clinical outcome (Supplementary Fig. 13c–r).
DISCUSSION
Improved outcomes for brain cancer patients will require new therapeutic models. Using comprehensive metabolomic and genomic approaches in patient-derived glioma models, we found that BTICs activate de novo purine synthesis to maintain self-renewal, proliferation and tumor forming capacity (modeled in Supplementary Fig. 14). Purine synthesis starts with ribose 5-phosphate (R5P), the sugar backbone of the nucleotide, which is typically generated in normal cells by diverting glucose-6-phosphate into the oxidative pentose phosphate pathway (PPP)33. An important byproduct generated by the PPP is NADPH, which donates hydrogen in reductive anabolism, including lipid synthesis and cellular redox balance through the transformation of glutathione34. Cancer cells synthesize R5P through an alternative, nonoxidative branch of the PPP catalyzed by the concerted actions of transketolase and transaldolase35,36. This metabolic reprogramming facilitates production of purine nucleotides without creating excessive NADPH and perturbing redox balance in cancer cells.
Recent analysis of cell mass formation demonstrated that glucose, together with glutamine and serine, significantly contributed to the carbon sources of DNA and RNA, both of which are downstream from purine synthesis37. Carbon flow from glucose and serine in mouse embryonic fibroblasts into de novo purine synthesis through the mitochondrial tetrahydrofolate cycle is regulated by mTORC1 and ATF4 (ref. 38). In contrast, we did not observe a consistent activation of the PPP in BTICs (data not shown). Instead, BTICs upregulate the enzymes involved in purine synthesis (including PRPS1 and PPAT on the common branch to synthesize IMP, ADSL and ADSS on one side branch to synthesize AMP; and GMPS and IMPDH1 on the other side branch to synthesize GMP) to efficiently channel glucose-derived R5P into the production of basic purine nucleotides, including IMP, AMP and GMP. AMP and GMP serve as critical building blocks for DNA replication and for RNA production to support protein synthesis, collectively sustaining BTIC self-renewal and proliferative capacity. Further, cAMP and cGMP, synthesized from purine nucleotides through adenylate cyclase and guanylate cyclase, function as second messengers. Genetic perturbation of key enzymes, including ADSL, GMPS and PRPS1, significantly decreased the BTIC maintenance in both immunodeficient and immunocompetent mouse models. Our findings are consistent with a recent report of 13C-based glucose carbon tracing experiments in non-small-cell lung cancer patients during surgery, which showed heterogeneous glucose metabolism within these tumors39. Glucose was found to be the dominant fuel for energy and anabolism in the less perfused regions39, supporting the link between nutrient restriction and cellular reprogramming13.
Metabolic reprogramming in cancer cells is often executed through coordinated regulation of enzymes for a whole synthetic pathway, in contrast to the activation or inhibition of a single rate-limiting step enzyme found in untransformed cells33. By examining the TCGA data set and testing in patient-derived xenograft models, we found that the expression of purine synthesis enzymes in BTICs is regulated in a concerted manner in glioblastoma, indicating a potential upstream transcriptional regulator. In glioblastoma, we previously demonstrated that MYC is essential to the maintenance of the BTIC phenotype25. Here we interrogated the downstream targets of MYC in BTIC, showing that MYC directly activates the transcription of all six essential genes in the purine biosynthetic pathway. De novo purine synthesis is regulated by several mechanisms beyond MYC in cancer cells33,36. The PI3K–AKT signaling pathway promotes metabolic reprogramming in tumors and can modulate carbon flow into purine synthesis40. We previously showed that inhibition of this pathway is a specific therapeutic approach in targeting BTICs compared to differentiated progeny41, further highlighting a potential link between the PI3K–AKT pathway and purine synthesis in promoting tumor initiating phenotype. In the current study, we demonstrate that the PI3K–AKT pathway regulates MYC protein level, which in turn modulate the expression of purine synthesis enzymes in BTICs.
The preferential requirement of de novo purine synthesis in BTICs can be exploited therapeutically for glioblastoma treatment42. Inhibitors blocking purine synthesis could be used as an adjuvant therapy specifically targeting BTICs in combination with current regimens of radiation, temozolomide and bevacizumab. Two IMPDH inhibitors are already widely used in the clinic. Mycophenolate mofetil (CellCept) and mycophenolate sodium (Myfortic) are both potent immunosuppressants used in organ transplant and autoimmune diseases43,44. The active component, mycophenolic acid, inhibits IMPDH at nanomolar range by acting as the nicotinamide portion of the cofactor nicotinamide adenine dinucleotide45. Ribavirin, a broad-spectrum antiviral agent active against both DNA and RNA viruses, inhibits the activity of human IMPDH, which depletes cellular pool of GTP and dGTP to block viral replication46,47. Newer agents with greater potency inhibiting IMPDH, such as VX-497, are under clinical investigation as antivirals48. In addition to directly targeting the purine synthesis pathway, inhibitors to upstream regulatory mechanisms are also potential choices for anti-BTIC therapy. For example, BKM-120, a pan-class I PI3K inhibitor capable of penetrating the blood-brain barrier, is undergoing phase II clinical study for recurrent glioblastoma (NCT01339052)49,50. Taken together, the results of our study demonstrate the distinct requirement of de novo purine synthesis in BTIC maintenance and survival, an intratumoral population difficult to eradicate with current treatment strategies. Developing therapeutics against purine synthesis could therefore synergize with radiation and chemotherapy to achieve improved management of glioblastoma.
ONLINE METHODS
Isolation and culture of cells
Glioblastoma tissues were obtained from excess surgical materials from patients at the Cleveland Clinic after neuropathology review with appropriate consent, in accordance with an IRB-approved protocol. To prevent culture-induced drift, patient-derived xenografts were generated and maintained as a recurrent source of tumor cells for study. Immediately upon xenograft removal, a papain dissociation system (Worthington Biochemical) was used to dissociate tumors according to the manufacturer’s instructions. Cells were then cultured in Neurobasal medium (Life Technologies) supplemented with B27, l-glutamine, sodium pyruvate, 10 ng/ml basic fibroblast growth factor and 10 ng/ml epidermal growth factor (R&D Systems) for at least 6 h to recover expression of surface antigens. No marker is uniformly informative for BTICs, so we use a combination of functional criteria to validate BTICs. Both BTICs and DGCs were derived immediately after dissociation or after transient xenograft passage in immunocompromised mice using prospective sorting followed by assays to confirm stem cell marker expression, sphere formation and secondary tumor initiation. For experiments using matched BTIC and DGC cultures, we segregated AC133 marker-positive and marker-negative populations using CD133/1 antibody-conjugated magnetic beads (Miltenyi Biotech, Auburn, CA), as previously described4,13. The BTIC phenotype was validated by stem cell marker expression (OLIG2 and SOX2), functional assays of self-renewal (serial neurosphere passage), and tumor propagation using in vivo limiting dilution.
Plasmids and lentiviral transduction
Lentiviral clones expressing shRNAs directed against human ADSL (TRCN0000078271, TRCN0000078272), human GMPS (TRCN0000045939, TRCN0000045940), human PRPS1 (TRCN0000010123, TRCN0000010125), murine Adsl (TRCN0000120128, TRCN0000339620), murine Gmps (TRCN0000251162, TRCN0000251160), murine Prps1 (TRCN0000024886, TRCN0000024888), human MYC (TRCN0000039640, TRCN0000039642), human GLUT3 (TRCN0000043616, TRCN0000043615) or a control shRNA that has no targets in either human or mouse genome (shCont: SHC002) were obtained from Sigma-Aldrich (St. Louis, MO). shRNAs with non-overlapping sequences that had the best relative knockdown efficiency were used for all experiments. Lentiviral overexpression plasmid for human MYC was generated by cloning the ORF with the N-terminal Flag into the pCDH-MCS-T2A-Puro-MSCV vector (System Biosciences)51. Lentiviral particles were generated in 293FT cells in Neurobasal complete medium (Life Technologies) with cotransfection with the packaging vectors pCMV-dR8.2 dvpr and pCI-VSVG (Addgene) using standard calcium phosphate transfection.
Metabolic analysis of BTIC using mass spectrometry
For metabolomics screening experiments, pairs of BTIC and DGC cells were incubated with medium in which all glucose was replaced with [13C6]glucose (Cambridge Isotope Laboratories #CLM-1396-1) for 24 h before harvesting. Cell lysates were then subjected to untargeted mass spectrometry. For nitrogen tracing experiments, BTICs were incubated with medium in which all glutamine was replaced with 13N2-glutamine (Cambridge Isotope Laboratories #NLM-1328-PK) for 24 h before harvesting. For targeted mass spectrometry of purine synthesis intermediates, pure samples of AMP, GMP and IMP were purchased from Sigma-Aldrich, and the internal standard [13C5]AMP was purchased from Toronto Research Chemical, Inc. All solvents used were HPLC grade from Fisher Scientific. Cells were dried and then extracts of cell lysates were reconstituted with 180 μl water, 20 μl of internal standard (50 μM [13C5]AMP). After vortexing for 60 s, the reconstituted sample was centrifuged for 60 s using a Beckman Coulter centrifuge (Microfuge 22R, Brea, CA) at 12,000 r.p.m. at 4 °C for 10 min. After centrifugation, 140 μl of supernatant was transferred into a vial for analysis by LC/MS/MS. The Waters 2690 Module (Waters, Corp., Franklin, MA) HPLC system was directly interfaced with a triple-quadrupole mass spectrometer (Quattro Ultima, Waters Inc., Beverly, MA) using electrospray ionization in negative mode and multiple reaction monitoring. Peak areas in the LC/MS/MS chromatogram for each analyte and internal standard were generated using QuanLynx attached to Masslynx V4.1 software. Internal standard calibration curves were used to quantify the three monophosphate nucleotides in cell lysates.
Proliferation and neurosphere formation assay
Cell proliferation was measured using Cell-Titer Glow (Promega, Madison, WI). All data were normalized to day 0 and are presented as mean ± s.e.m. Neurosphere formation was measured by in vitro limiting dilution, as previously described13,52. Briefly, decreasing numbers of cells per well (50, 20, 10, 5 and 1) were plated into 96-well plates. Seven days after plating, the presence and number of neurospheres in each well were recorded. Extreme limiting dilution analysis was performed using software available at http://bioinf.wehi.edu.au/software/elda, as previously described13,52.
Quantitative RT-PCR
Total cellular RNA was isolated using Trizol reagent (Sigma-Aldrich), followed by reverse transcription into cDNA using the qScript cDNA Synthesis Kit (Quanta BioSciences). Real-time PCR was performed using an Applied Biosystems 7900HT cycler using SYBR-Green PCR Master Mix (Thermo Fisher Scientific). Sequences for gene-specific primer sets were as follows: human PRPS1 forward 5′-ATCTTCTCCGGTCCTGCTATT-3′ and reverse 5′-TGGTGACTACTACTGCCTCAAA-3′; human PPAT forward 5′-GATGGGAGTTCGGTGCCAA-3′ and reverse 5′-CAACGAAGGGC TGACAATTTTC-3′; human ADSL forward 5′-GCTGGAGGCGATCATGGTTC-3′ and reverse 5′-TGATAGGCAAACCCAATGTCTG-3′; human ADSS forward 5′-TGGGTATGCCACCTCAAAATG-3′ and reverse 5′-GCTCT GTAGGAAAGGCACCAATA-3′; human GMPS forward 5′-ATGGCTCTGT GCAACGGAG-3′ and reverse 5′-CCTCACTCTTCGGTCTATGACT-3′; human IMPDH1 forward 5′-CAGCAGGTGTGACGTTGAAAG-3′ and reverse 5′-AGCTCATCGCAATCATTGACG-3′; murine Prps1 forward 5′-ACTTATCCCAGAAAATCGCTGAC-3′ and reverse 5′-CCACACCCACTTTGAA CAATGTA-3′; murine Adsl forward 5′-AAAGCTGCGGGCATTATTCAT-3′ and reverse 5′-GCACGATCCTTAGCAAAGTCG-3′; murine Gmps forward 5′-GCCCTGTGCAACGGAGATT-3′ and reverse 5′-GCTCCTTCGTAATGGTGAGA AC-3′; human MYC forward 5′-GGCTCCTGGCAAAAGGTCA-3′ and reverse 5′-CTGCGTAGTTGTGCTGATGTH3′; and human 18S RNA forward 5′-AACCCGTTGAACCCCATT-3′ and reverse 5′-CCATCCAATCGGTAGTAGCG-3′.
Western blotting
Cells were collected and lysed in RIPA buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% NP-40; 50 mM NaF with protease inhibitors) and incubated on ice for 30 min. Lysates were centrifuged at 20,817g at 4 °C for 10 min and supernatant was collected. Protein concentration was determined using the Bradford assay (Bio-Rad Laboratories, Hercules, CA). Equal amounts of protein samples were mixed with SDS Laemmli loading buffer, boiled and electrophoresed using NuPAGE Bis-Tris Gels (Life Technologies), then transferred onto PVDF membranes (Millipore). Blocking was performed for 45 min using TBST supplemented with 5% non-fat dry milk and blotting performed with primary antibodies at 4 °C for 16 h. The following antibodies were used: ADSL (Abcam #ab154182), GMPS (Cell Signaling #14602), PRPS1 (Abcam #ab154721), MYC (N-262, Santa Cruz #sc-764), total AKT (Cell Signaling #4691), phospho-AKT (S473, Cell Signaling #9271), total S6 (Cell Signaling #2317), phospho-S6 (235/236, Cell Signaling #4858), phospho-S6 (240/242, Cell Signaling #5364), total p70 S6K (Cell Signaling #2708), phospho-p70 S6K (Cell Signaling #9234) and α-tubulin (Sigma #T6074). All antibody validation information is available in the product’s manual. For western blotting, the dilution was 1:500 for MYC antibody and 1:1,000 for all other antibodies.
Immunohistochemistry
Immunohistochemistry experiments were performed using specimens from xenografted tumors in mouse model. The following primary antibodies were used: cleaved caspase 3 (Cell Signaling #9664, 1:300), SOX2 (R&D #MAB2018, 1:200) and Ki-67 (DAKO #M7240, 1:300). Biotinylated secondary antibodies were used together with the Vectastain ABC kit and the DAB kit (Vector Laboratories) per manufacturer’s instructions. All antibody validation information is available in the product’s manual.
Chromatin immunoprecipitation (ChIP) assay
4 × 106 cells per condition were collected, and ChIP was performed using the EZ-ChIP chromatin immunoprecipitation kit (EMD Millipore #17-371) following the manufacturer’s protocol. Briefly, 5 μg MYC antibody (N-262, Santa Cruz #sc-764) or rabbit IgG was used for the immunoprecipitation of the DNA-protein immunocomplexes. Crosslinking was reversed by heating for 6 h at 65 °C, followed by digestion with proteinase K. The purified DNA was subjected to quantitative PCR using the following primer sets: human PRPS1 forward 5′-GAGGGTGCAGCTCTGTGACT-3′ and reverse 5′-CAGCCACTATCTATTGGGTC-3′; human PPAT forward 5′-TGCG GTACATCCAGCTGCGT-3′ and reverse 5′-TGGTTGGTGCTTACACCT TG-3′; human ADSL forward 5′-TAGCGACAGGTATAAATTCC-3′ and reverse 5′-TCTCCTGCCCTTGCTTTCCT-3′; human ADSS forward 5′-TCT CGCTTTCAAACTGGA-3′ and reverse 5′-TGTTTGTACTGCGAAATTCC-3′; human GMPS forward 5′-CTGCGGAGGGTATCTGAG-3′ and reverse 5′-CGCCGAATGGAGCAACAG-3′; human IMPDH1 forward 5′-GTCT GCATCCCCCAACCAAAG-3′ and reverse 5′-ACTGCTGCAGGCCGGCTAC-3′.
For ChIP-seq experiments, 5–10 mg of flash-frozen BTIC or DGC cells was subjected to ChIP using 5 mg of H3K27ac antibody per specimen (Abcam-AB4729). Enriched DNA was quantified using Picogreen (Invitrogen) and ChIP libraries were amplified and barcoded using the Thruplex DNA-seq library preparation kit (Rubicon Genomics) according to manufacturer recommendations. Following library amplification, DNA fragments of less than 1 kb were size selected using a 1.0% agarose gel, assessed using Bioanalyzer (Agilent Technologies) and sequenced using Illumina Hi-Seq 2000 100-bp single-end sequencing.
In vivo tumorigenesis
BTICs derived from human glioblastoma specimens were transduced with lentiviral vectors expressing shRNA targeting ADSL, GMPS or PRPS1, or expressing a non-targeting control shRNA (shCont) for the knockdown experiment. 36 h after infection, viable cells were counted and engrafted intracranially into NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, The Jackson Laboratory, Bar Harbor, ME) mice. For the immunocompetent model, murine glioma 261 (GL261) cells were transduced with lentiviral vectors expressing shRNA targeting murine Adsl, Gmps or Prps1, or expressing shCont. 36 h after infection, viable cells were counted and engrafted intracranially into syngeneic host stain C57BL/6J (The Jackson Laboratory). All mice experiments were performed under an animal protocol approved the Institutional Animal Care and Use Committee of the Cleveland Clinic. Healthy female mice of NSG or C57BL/6J background, 4–6 weeks old, were randomly selected and used in this study for intracranial injection. Animals were maintained until neurological signs were apparent, at which point they were sacrificed. Brains were collected and fixed in 4% formaldehyde, cryopreserved in 30% sucrose and then cryosectioned. Sections were stained with hematoxylin and eosin for histological analysis. In parallel survival experiments, animals were monitored until they developed neurological signs.
Statistical analysis
No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in previous publications4,13. A two-sided unpaired Student’s t-test was used to assess differences between groups. Data distribution was assumed to be normal, but this was not formally tested. Kaplan-Meier survival curves were generated using Prism software and a log-rank test was performed to assess statistical significance between groups. For all figures presented in box-and-whisker format, the center line represents the median and the lower and upper limits of the box represent the 25th and 75th percentiles. The maximum and minimum are connected to the center box through the vertical lines (whiskers). No animals or data points were excluded from the analyses for any reason. Blinding and randomization were performed in all experiments. Correlation between gene expression and patient survival was performed through analysis of TCGA, Freije, Phillips and REMBRANDT brain tumor data sets, downloaded from the TCGA data portal or NCBI GEO database. High and low expression groups were defined as above and below the mean expression level, respectively. A Supplementary Methods Checklist is available.
Data and code availability
The exome sequencing data are available from the NCBI SRA Database (accession code SRR5275896). The ChIP-seq data are available from Gene Expression Omnibus (accession code GSE95216). The data and analysis routines that support the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We appreciate mass spectrometry analysis by R. Zhang, flow cytometry assistance by C. Shemo and S. O’Bryant, the glioblastoma tissue provided by M. McGraw and the Cleveland Clinic Tissue Procurement Service, and the IN528 model from I. Nakano at Ohio State University. We thank T. Roberts, J. Suh, G. Narla and members of the Rich laboratory for discussions. This work was supported by funding from National Institutes of Health grants CA197718, CA154130, CA169117, CA171652, NS087913 and NS089272 (J.N.R.), CA184090, NS091080 (S.B.) and CA168997, CA193256, CA201963 (J.W.L.); the James S. McDonnell Foundation (J.N.R.); the Research Programs Committees of the Cleveland Clinic (J.N.R. and K.Y.); Clinical and Translational Science Collaborative of Cleveland grant UL1TR000439 from the National Center for Advancing Translational Sciences (J.N.R. and K.Y.); a P&F grant from NIH Resource Center for Stable Isotope Resolved Metabolomics (RC-SIRM) at University of Kentucky (J.N.R. and K.Y.); and an ENGAGE grant from the National Center for Regenerative Medicine (K.Y.).
Footnotes
Note: Any Supplementary Information and Source Data files are available in the online version of the paper.
AUTHOR CONTRIBUTIONS
X.W., K.Y. and J.N.R. designed the experiments, analyzed the data and wrote the manuscript with contributions from all authors. X.W., K.Y., Q.X., Q.W., S.C.M., C.G.H., T.E.M., W.Z., Z.H. and X.F. performed the experiments. K.Y., Y.S., L.J.Y.K., B.C.P., W.A.F., M.S., A.R., M.L.S. and T.H.H. performed database analyses. X.W., K.Y., Q.X., X.L. and J.W.L. performed metabolic experiments. S.B. provided scientific input and helped edit the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Stupp R. et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 2.Singh SK. et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 3.Gage FH, Temple S. Neural stem cells: generating and regenerating the brain. Neuron. 2013;80:588–601. doi: 10.1016/j.neuron.2013.10.037. [DOI] [PubMed] [Google Scholar]
- 4.Bao S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 5.Liu G. et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol. Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bao S. et al. Stem cell-like glioma cells promote tumor angiogenesis through vascular endothelial growth factor. Cancer Res. 2006;66:7843–7848. doi: 10.1158/0008-5472.CAN-06-1010. [DOI] [PubMed] [Google Scholar]
- 7.Cheng L. et al. Glioblastoma stem cells generate vascular pericytes to support vessel function and tumor growth. Cell. 2013;153:139–152. doi: 10.1016/j.cell.2013.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23:27–47. doi: 10.1016/j.cmet.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan H. et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Turcan S. et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flavahan WA. et al. Brain tumor initiating cells adapt to restricted nutrition through preferential glucose uptake. Nat. Neurosci. 2013;16:1373–1382. doi: 10.1038/nn.3510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z. et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hjelmeland AB. et al. Acidic stress promotes a glioma stem cell phenotype. Cell Death Differ. 2011;18:829–840. doi: 10.1038/cdd.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panopoulos AD. et al. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Res. 2012;22:168–177. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masui K. et al. Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc. Natl. Acad. Sci. USA. 2015;112:9406–9411. doi: 10.1073/pnas.1511759112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Derr RL. et al. Association between hyperglycemia and survival in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 2009;27:1082–1086. doi: 10.1200/JCO.2008.19.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brennan CW. et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu X. et al. Discrete mechanisms of mTOR and cell cycle regulation by AMPK agonists independent of AMPK. Proc. Natl. Acad. Sci. USA. 2014;111:E435–E444. doi: 10.1073/pnas.1311121111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo D. et al. The AMPK agonist AICAR inhibits the growth of EGFRvIII-expressing glioblastomas by inhibiting lipogenesis. Proc. Natl. Acad. Sci. USA. 2009;106:12932–12937. doi: 10.1073/pnas.0906606106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J. et al. c-Myc is required for maintenance of glioma cancer stem cells. PLoS One. 2008;3:e3769. doi: 10.1371/journal.pone.0003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng H. et al. p53 and Pten control neural and glioma stem/progenitor cell renewal and differentiation. Nature. 2008;455:1129–1133. doi: 10.1038/nature07443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suvà ML. et al. Reconstructing and reprogramming the tumor-propagating potential of glioblastoma stem-like cells. Cell. 2014;157:580–594. doi: 10.1016/j.cell.2014.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venere M. et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014;21:258–269. doi: 10.1038/cdd.2013.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freije WA. et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64:6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 31.Madhavan S. et al. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol. Cancer Res. 2009;7:157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phillips HS. et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Lane AN, Fan TW. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015;43:2466–2485. doi: 10.1093/nar/gkv047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wood T. Physiological functions of the pentose phosphate pathway. Cell Biochem. Funct. 1986;4:241–247. doi: 10.1002/cbf.290040403. [DOI] [PubMed] [Google Scholar]
- 35.Boros LG. et al. Transforming growth factor β2 promotes glucose carbon incorporation into nucleic acid ribose through the nonoxidative pentose cycle in lung epithelial carcinoma cells. Cancer Res. 2000;60:1183–1185. [PubMed] [Google Scholar]
- 36.Tong X, Zhao F, Thompson CB. The molecular determinants of de novo nucleotide biosynthesis in cancer cells. Curr. Opin. Genet. Dev. 2009;19:32–37. doi: 10.1016/j.gde.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hosios AM. et al. Amino acids rather than glucose account for the majority of cell mass in proliferating mammalian cells. Dev. Cell. 2016;36:540–549. doi: 10.1016/j.devcel.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ben-Sahra I, Hoxhaj G, Ricoult SJ, Asara JM, Manning BD. mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science. 2016;351:728–733. doi: 10.1126/science.aad0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hensley CT. et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang W. et al. The phosphatidylinositol 3-kinase/Akt cassette regulates purine nucleotide synthesis. J. Biol. Chem. 2009;284:3521–3528. doi: 10.1074/jbc.M806707200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eyler CE. et al. Brain cancer stem cells display preferential sensitivity to Akt inhibition. Stem Cells. 2008;26:3027–3036. doi: 10.1634/stemcells.2007-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat. Rev. Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- 43.Brazelton TR, Morris RE. Molecular mechanisms of action of new xenobiotic immunosuppressive drugs: tacrolimus (FK506), sirolimus (rapamycin), mycophenolate mofetil and leflunomide. Curr. Opin. Immunol. 1996;8:710–720. doi: 10.1016/s0952-7915(96)80090-2. [DOI] [PubMed] [Google Scholar]
- 44.D’Cruz DP, Khamashta MA, Hughes GR. Systemic lupus erythematosus. Lancet. 2007;369:587–596. doi: 10.1016/S0140-6736(07)60279-7. [DOI] [PubMed] [Google Scholar]
- 45.Sintchak MD. et al. Structure and mechanism of inosine monophosphate dehydrogenase in complex with the immunosuppressant mycophenolic acid. Cell. 1996;85:921–930. doi: 10.1016/s0092-8674(00)81275-1. [DOI] [PubMed] [Google Scholar]
- 46.Sidwell RW. et al. Broad-spectrum antiviral activity of virazole: 1-β-d-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science. 1972;177:705–706. doi: 10.1126/science.177.4050.705. [DOI] [PubMed] [Google Scholar]
- 47.Wray SK, Gilbert BE, Noall MW, Knight V. Mode of action of ribavirin: effect of nucleotide pool alterations on influenza virus ribonucleoprotein synthesis. Antiviral Res. 1985;5:29–37. doi: 10.1016/0166-3542(85)90012-9. [DOI] [PubMed] [Google Scholar]
- 48.Markland W, McQuaid TJ, Jain J, Kwong AD. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: a comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob. Agents Chemother. 2000;44:859–866. doi: 10.1128/aac.44.4.859-866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bendell JC. et al. Phase I, dose-escalation study of BKM120, an oral pan-class I PI3K inhibitor, in patients with advanced solid tumors. J. Clin. Oncol. 2012;30:282–290. doi: 10.1200/JCO.2011.36.1360. [DOI] [PubMed] [Google Scholar]
- 50.Wen PY, Lee EQ, Reardon DA, Ligon KL, Alfred Yung WK. Current clinical development of PI3K pathway inhibitors in glioblastoma. Neuro-oncol. 2012;14:819–829. doi: 10.1093/neuonc/nos117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fang X. et al. The zinc finger transcription factor ZFX is required for maintaining the tumorigenic potential of glioblastoma stem cells. Stem Cells. 2014;32:2033–2047. doi: 10.1002/stem.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie Q. et al. Mitochondrial control by DRP1 in brain tumor initiating cells. Nat. Neurosci. 2015;18:501–510. doi: 10.1038/nn.3960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The exome sequencing data are available from the NCBI SRA Database (accession code SRR5275896). The ChIP-seq data are available from Gene Expression Omnibus (accession code GSE95216). The data and analysis routines that support the findings of this study are available from the corresponding author upon reasonable request.