Abstract
In recent decades there has been a significant shift in our understanding of Idiopathic Pulmonary fibrosis (IPF), a progressive and lethal disorder. While initially much of the mechanistic understanding was derived from hypotheses generated from animal models of disease, in recent decades new insights derived from humans with IPF have taken precedence. This is mainly because of the establishment of large collections of IPF lung tissues and patient cohorts, and the emergence of high throughput profiling technologies collectively termed ‘omics’ technologies based on their shared suffix. In this review we describe impacts of ‘omics’ analyses of human IPF samples on our understanding of the disease. In particular, we discuss the results of genomics and transcriptomics studies, as well as proteomics, epigenomics and metabolomics. We then describe how these findings can be integrated in a modified paradigm of human idiopathic pulmonary fibrosis, that introduces the ‘hallmarks of aging’ as a central theme in the IPF lung. This allows resolution of all the disparate cellular and molecular features in IPF, from the central role of epithelial cells, through the dramatic phenotypic alterations observed in fibroblasts and the numerous aberrations that inflammatory cells exhibit. We end with reiterating a call for renewed efforts to collect and analyze carefully characterized human tissues, in ways that would facilitate implementation of novel technologies for high resolution single cell omics profiling.
Keywords: Pulmonary Fibrosis, Mitochondria, Telomere, Senescence, Microbiome, Genomics, transcriptomics
INTRODUCTION
Idiopathic pulmonary fibrosis (IPF) is a chronic Interstitial Lung Disease (ILD) with an etiology that remains to be fully elucidated. The natural course of patients with IPF is variable, but survival is worse than many cancers and is usually 3–5 years from diagnosis[1]. The prevalence estimates in the USA vary between 1.25 and 63 cases per 100,000 population and its incidence is 0.22 −8.6 per 100.000 of population, making it the most common among idiopathic ILD[2]. Despite the approval of Pirfenidone and Nintedanib by the FDA, the only curative treatment is lung transplantation.
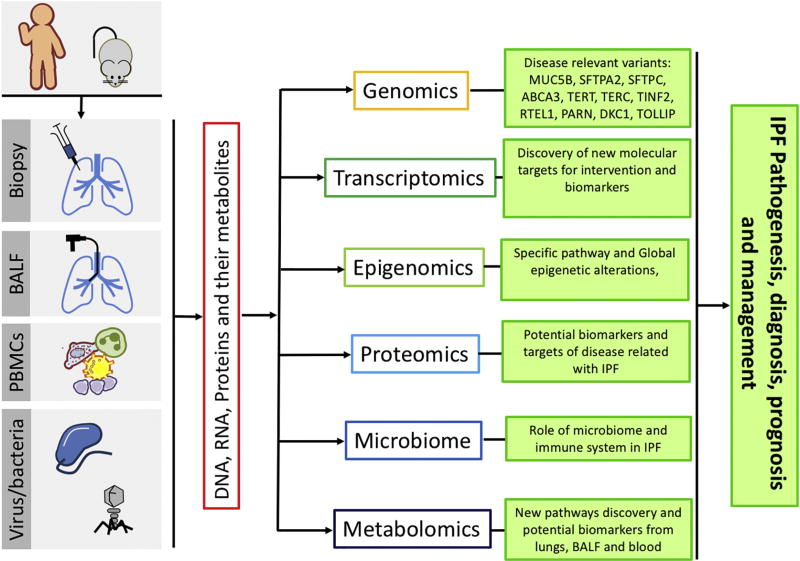
In the last decades, there has been significant progress understanding the molecular mechanisms of pulmonary fibrosis in animal models of disease, however understanding the exact molecular mechanism underlying the initiation and progression in humans are only now emerging. Traditionally, in the early decades of pulmonary fibrosis research, investigators formulated hypotheses based on experimental models, mostly the bleomycin model or on observations in other organ systems and then sought to validate them in humans, most frequently with low throughput protein profiling approaches in a limited number of samples. In the first decade of the 21st century, we observed a dramatic inversion of this process. The increased availability of well characterized human cells derived from tissues and the emergence of high throughput transcriptomic profiling technologies, led to novel hypotheses based on human observations that were followed by sophisticated molecular biology and mouse genetics techniques[3]. In the second decade of the 21st century, this trend expanded and matured, with a dramatic increase in the number of samples and application of high throughput molecular profiling technologies often referred to as “omics” technologies based on their common suffix. The lung genomics research consortium that provided parallel transcriptomic, epigenetic, genetic and microRNA profiles on hundreds of well characterized tissues[4–6], marked the start of this era. The discovery of IPF associated common genetic variants [7], the role of the microbiome [8], and the recent description of molecular and cellular phenotypes using single cell sequencing technologies [9] , are among the most recent and significant progress on the field (Figure 1). These studies provided a nearly chaotic molecular image of the lung with numerous molecular and cellular aberrations.
Figure 1.
Omics technologies and their effects in IPF research.
In this review, we describe what we have learned about pulmonary fibrosis from the application of omics technologies to human tissues, focusing mainly on the lung, but also mentioning other compartments reflective of the disease. We discuss how these findings fit and contribute to our perception of the central role of aging hallmarks in IPF, with a specific focus on telomere attrition and mitochondrial dysfunction.
Insights from OMICS analyses of human fibrotic lungs
Genomics
The tremendous progress in understanding the genetics of familial pulmonary fibrosis and sporadic idiopathic pulmonary fibrosis has been recently described [10]. Here we focus on the impact that those genetic findings have had on our understanding of the mechanisms of human pulmonary fibrosis.
Genetic studies of familial pulmonary fibrosis have been important in the identification of rare DNA sequence variants in coding and non-coding regions with significant mechanistic implications. They have identified variants in two broad categories: surfactant protein processing (SFTPA2, SFTPC, ABCA3 [11, 12]) and telomere maintenance and homeostasis machinery (TERT, TERC, TINF2, RTEL1, PARN, DKC1 [13–18]) gene mutations. These studies highlighted the central roles of endoplasmic reticulum (ER) stress and telomerase dysfunction in human pulmonary fibrosis. Elegant follow-up studies in genetically modified mice established the specific role of these mutations, as well as the general roles of ER stress [19, 20], and telomere attrition and telomerase dysfunction [21] as drivers of pulmonary fibrosis
Similarly, the application of genome wide association studies (GWAS), led to identification of common variants in both familiar pulmonary fibrosis and idiopathic pulmonary fibrosis with unprecedented reproducibility. The rs35705950 SNP in the promoter of MUC5B, a mucin expressed in airway epithelial cells, was associated with familial pulmonary fibrosis and IPF in multiple cohorts [22–29] and three large GWAS studies [7, 30, 31], accounting for ~30% of the risk for developing IPF by some estimates[10] . GWAS studies identified additional variants, including TOLLIP [31] and AKAP13 [30]. A retrospective analysis suggested that patients carrying the SNPs for TOLLIP, benefitted preferentially from treatment with N-Acetylcysteine in the PANTHER-IPF clinical trial [32] opening the door for future genetic variant driven clinical trials. Telomerase pathway variants were also associated with disease in these cohorts[7, 30, 31]. Thus, in pulmonary fibrosis, GWAS results led to identification of the role of bronchial epithelial cells, innate immunity and supported the role of the telomerase pathway.
Transcriptomics
The application of genome wide transcript profiling to human tissues in pulmonary fibrosis has had numerous impacts on IPF research. Conceptually, the unbiased transcriptomic analysis of human lungs shifted pulmonary fibrosis researchers from basing their hypotheses on model systems or biological plausibility-based hypotheses, to human lung based generated hypotheses, and led to identification of novel pathways and molecular targets. Traditionally, transcriptomic data can be classified as generating reductionist and mechanistic hypotheses, previously described as “cherry picking”, or generating global “systems level” conceptual observations[33].
Among the global discoveries that have been gleaned from transcriptomic data are: the discovery that matrix metalloproteases (MMPs) are active participants in pulmonary fibrosis [3, 34–36], the observation that developmental pathways are aberrantly activated in the IPF lungs [37], the observation that different classes of disease could be identified by gene expression patterns, allowing the recognition of distinct patient phenotypes [6, 38–40], the discovery of mitochondrial abnormalities in IPF and their implications [41, 42], and the extent of microRNA expression changes [43].
There have been numerous reductionist studies where investigators identified a gene differentially expressed in IPF lungs and utilized this information to develop a mechanistic insight. Sometimes investigators interested in a pathway, used publicly available datasets to support their hypotheses. Among the molecules identified as potential novel regulators of fibrosis are: MMP7 [3], OPN [44], IGBP3 and 5 [45, 46], WISP1 [47] , FKBP10 [48], PINK1 [41], RXFP1 [49], PTPN11 [50]. In most of these studies, combination of cell culture, genetically modified mice and other methods were included to make a plausible cause for their role in pulmonary fibrosis. The fact that these genes were indeed differentially expressed in human tissues, highlights the complexity of the regulatory networks in humans. Of particular interest in this context are microRNAs, small non-coding RNA molecules that regulate gene expression. It has been estimated that close to 10% of the microRNAs are changed in the IPF lung. Numerus microRNAs have been mechanistically related to pulmonary fibrosis in recent years. The most studied are: miR-21, let-7 and miR-29 family of microRNAs [43, 51–53].
Transcriptomics studies have also been utilized to prioritize biomarkers found in the blood stream, and to determine whether they were indicative of changes in the lung [54–56]. Direct analysis of peripheral blood mononuclear cells (PBMC), led to identification of a 52-gene expression signature that predicted more severe prognosis in patients with IPF [57], a finding later replicated in six independent cohorts [58].
The majority of these findings have been obtained from experiments utilizing microarrays and performed on bulk tissue. Studies using RNA-sequencing either in bulk [5], or at the single cell level [9] are emerging and will have a significant influence on the field.
Epigenomics
Changes in epigenetic regulation of specific genes have been reported in IPF [59–61, 62, 63], but several groups reported extensive methylation changes in IPF lungs [64, 65]. Comparison of IPF methylation patterns to lung cancer or control samples, revealed that IPF lungs displayed an intermediate methylation profile between lung cancer and control with 402 differentially methylated CpG islands overlapping between IPF and cancer. A later study [66] identified DNA methylation changes in IPF using comprehensive high-throughput arrays for relative methylation arrays (CHARM). This study focused on correlation of methylation and gene expression. Out of 2130 differentially methylated regions, 738 were associated with significant changes in gene expression and enriched for opposite changes between methylation and expression, suggesting a role for epigenetic regulation of gene expression in IPF.
To some extent, epigenomic studies did not lead to a wave of follow up studies. Critical questions regarding the cellular source of epigenetic changes, their relationship to patients’ fibrosis phenotypes and disease pathogenesis have not been answered. It is hoped that with the increased interest in aging related mechanisms in pulmonary fibrosis and the availability of advanced methodologies including single cell epigenomic analysis, a renewed interest will emerge [67, 68].
Microbiome
There has been a significant increase in information about the microbiome changes in IPF using state of the art next generation sequencing of bacterial 16r-rRNA Operational Taxonomic Units (OTU) clustering [69]. These new methods, more sensitive than the regular culture-dependent techniques, allow high-throughput analyses of numerous species. The majority of studies focused on the fibrotic lung microbiome in bronchoalveolar lavage (BAL) – a technique in which the content of the alveolar space is sampled by instilling fluid and suctioning it through a fiberoptic bronchoscope. Despite initial concerns about potential contaminations, the results have been reproducible and widely accepted[70, 71]. Two large studies identified correlations of changes in the microbiome with presence and progression of IPF [72, 73], as well connection with changes in peripheral blood gene expression[74, 75]. Changes in microbiome where observed in acute exacerbations [76] but not in patients treated with inhaled Interferon-γ [77]. While these associations do not necessarily imply causality, they do suggest a link between shifts in alveolar bacterial burden and IPF. Studies that directly assessed the microbiome of lung tissue, as opposed to BAL did not detect bacterial DNA in tissue samples obtained from patients with IPF competed to control lungs, questioning the true role of microbiome shifts in the lung[78].
At this stage, it is too early to determine whether the changes to the lung microbiome reported in BAL of IPF studies reflect basic pathogenetic mechanism, or are reflective of airway changes secondary to the remodeling. Experiments seeking to affect the lung microbiome in humans, and potentially studies assessing changes in fibrotic predisposition in gnotobiotic animals, will be required to resolve some of these questions.
Proteomics
Studies that applied targeted or global proteomic approaches to identify biomarkers or disease modifiers had a limited impact compared to transcriptomic, genomic or microbiome studies. Targeted proteomic approaches identified a peripheral blood protein signature that included MMP7 and MMP1 that distinguished patients with IPF from controls or other chronic lung diseases[54]. Later, applying a similar approach, the same group identified a peripheral blood protein signature that predicted outcome in IPF[79]. In both cases the investigators ended up with proteins that were increased in the human IPF lung, a comparison not performed in a later study that utilized a wider screening technique[80].
Comparison of IPF tissues with other interstitial lung disease revealed significant changes in proteins related to unfolded protein response, oxidative stress and DNA damage [81, 82] highlighting the extent of the aberrant expression of these proteins. While others revealed relatively predictable changes in proteins such as vimentin[83], a recent deep proteomic approach of fibrotic lung and skin led to the unexpected discovery of high levels of MZB1-positive plasma B cells in lung and skin fibrosis[84]. While the implications of this finding are not clear, they highlight the discovery potential of proteomic analysis of the lung, as well as the limitations of bulk analysis, as the major finding was a marker of infiltrating cells.
Metabolomics
Metabolomics analyses were, so far, rarely applied to the human fibrotic lung. A key study applied nuclear magnetic resonance spectroscopy to assess cellular metabolites in IPF and revealed elevations of Lactic acid and lactate dehydrogenase-5 (LDH5) in fibroblasts from IPF lungs[85]. Additional studies revealed alterations in glycolysis, glutathione biosynthesis, adenosine triphosphate degradation and ornithine aminotransferase pathways [86, 87]. Together with the evidence for metabolic reprogramming [88], mitochondrial dysfunction and extensive protein degradation in the IPF lung [35], these studies encourage a much wider, extensive and detailed studies of metabolomics in IPF.
Current insights into the pathogenesis of pulmonary fibrosis
The history of paradigm shifts in IPF can be described as a move from simple linear models of disease progression, highly influenced by tightly controlled animal models of disease, to more complex models based on information gleaned from the human lung. The initial inflammation driven fibrosis hypothesis that was mainly based on the bleomycin model of fibrosis, has been replaced by an epithelial primary injury paradigm. In this model, a primary chronic injury to epithelial cells, and a failure to repopulate them, leads to ongoing activation of fibroblasts, change in the phenotype of epithelial cells, and a self-perpetuating cycle of aberrant remodeling.
Two key human observations cemented this epithelial injury paradigm as probably the one that explains most features of IPF: First, the primary role of epithelial cells in IPF was solidified by the observation that the rs35705950 SNP, mostly associated with both familial pulmonary fibrosis and sporadic IPF in numerous cohorts [7, 22–31], is in the promoter of MUC5B, a gene encoding a mucin expressed in airway epithelial cells. Second, the complex and not necessarily all evil role of inflammation in IPF, was highlighted by the revelation that an immunosuppressive regimen recommended for patients with IPF was detrimental and associated with increased hospitalizations and mortality in a prospective randomized controlled study [89]. Previous smaller studies have repeatedly shown lack of an effect of use of steroids or other immunosuppressive medications in IPF [90, 91], and pulmonary fibrosis seems to develop independently of inflammation in animal models of fibrosis [92–95].
While these observations solidified the primary role of epithelial cells in IPF, they did not explain all of the molecular features of the human disease, this only happened when the hallmarks of aging were introduced to the IPF paradigm as described below.
Aging, the new wrinkle in IPF pathogenesis
To some extent, the connection of aging with IPF was in plain view of clinicians for at least three decades. Increased incidence and prevalence of IPF was consistently observed over the age of 65 and the disease is extremely rare below the age of 50[96–98]. However, only in the last decade this concept obtained molecular and genetic support. As mentioned, the IPF lung is dramatically altered at its cellular and molecular composition. Type I epithelial cells are lost and type II epithelial cells are stressed, exhibit senescence markers, shorter telomeres [99] and increased expression of senescence associated microRNAs such as mir-34 [100]. Airway basal cells infiltrate the lung. The extracellular matrix composition is changed and its stiffness increased. Fibroblasts exhibit myofibroblast characteristics, but also exhibit a senescent phenotype with metabolic reprogramming. Macrophages and other inflammatory cells are activated or changed. IPF lungs exhibit extensive molecular changes including local TGFB1 activation [101–103], WNT [104], SHH [105] pathways, the unfolded protein response [106], oxidative injury [107], epithelial injury and apoptosis[108], abnormal matrix deposition and stiffness [109, 110], fibroblast metabolic reprogramming [88, 111] and senescence [112], myofibroblast transdifferentiation [113, 114], metalloprotease activation and signaling [35, 115], changes in patterns of mRNA and microRNA expressions [6, 43] and epigenetic marks [116] . This nearly chaotic portrait of aberrant cell behavior in the IPF lungs became clearer when we consider that many of the hallmarks of aging [117] including telomere attrition, genomic instability, epigenetic alterations, mitochondrial dysfunction, cellular senescence, and altered intercellular communication are a present in the human IPF. Thus, aging, added to a genetic susceptibility and an environmental exposure, explains many of the molecular and cellular aberrations observed in the IPF lung. The majority of these attributes have been reviewed extensively recently [118], but to better explain the role of aging in IPF we will briefly describe the evidence for the role of telomeres and mitochondria in IPF.
Telomeres in IPF
Telomeres, the DNA repeats that protect the ends of linear chromosomes and, whose length is a measure of cellular age, emerged in IPF with the initial description of telomerase mutations in familial pulmonary fibrosis [14]. Since then, numerous mutations and variants in telomerase related proteins (TERT, TERC, TINF2, RTEL1, PARN, DKC1) have been identified in patients with familial and sporadic pulmonary fibrosis [13, 15–18, 119]. Shortened telomeres are found in the blood of patients with IPF as [13] and are predictive of worst outcome [120] before and after transplantation [121]. Short telomeres are also observed in the lungs of patients with IPF, in particular in type II alveolar epithelial cells [122]. Mice genetically engineered to have short telomeres or severe telomere dysfunction, develop pulmonary fibrosis [21, 99, 123]. This effect is evident only when telomerase dysfunction is induced in alveolar type II cells and not in collagen expressing cells [99], consistent with the centrality of epithelial cells in human IPF. In humans, telomere shortening is observed independently of the presence of telomerase mutations, in both sporadic IPF and familial pulmonary fibrosis [124], highlighting the potential universal importance of telomere shortening and dysfunction in fibrosis.
Mitochondrial Dysfunction
Multitude of circumstantial evidence point towards a potential role for mitochondrial dysfunction in IPF. Those include excessive epithelial cell apoptosis, fibroblast metabolic derangements and impaired autophagy and immune dysfunction (reviewed at [125]). However, direct evidence to mitochondrial dysfunction in human IPF lungs has been demonstrated only recently [41].
In IPF lungs, human alveolar type II epithelial cells exhibit accumulation of dysmorphic and swollen mitochondria. These findings are associated with decreased expression of PINK1, a regulator of mitochondrial homeostasis and mitophagy. PINK1-deficient mice exhibit predisposition to fibrosis and mimic the human mitochondrial phenotype [41]. We have recently demonstrated that delivery of thyroid hormone or sobetirome, small molecule thyroid hormone agonist devoid of cardiac or musculoskeletal side effects blunted fibrosis in mouse models of pulmonary fibrosis [126]. These effects were dependent on the presence of PARGC1A and PINK1, regulators of mitogenesis and mitophagy, suggesting that the antifibrotic effect of thyroid hormone was mediated through restoration of mitochondrial homeostasis. Thyroid hormone reversed mitochondrial injury induced by bleomycin both in-vivo in mice, and in-vitro in mouse and human primary cells [126].
Another intriguing piece of the puzzle emerges from analysis of free circuiting mitochondrial DNA (mtDNA) in patients with IPF [127]. Briefly, mtDNA concentrations are increased in BAL or plasma obtained from patients with IPF. Increased plasma mtDNA is associated with disease progression and decreased survival. In vitro, mtDNA release increased in response to conditions that mimic the lung environment in IPF (TGF-β1 or stiff matrix) and exposure of fibroblasts to mtDNA augmented their transdifferentiation to myofibroblasts. Taken together, these findings suggest a central role for mitochondrial dysfunction in the pathogenesis of pulmonary fibrosis.
Putting it all together: an integrated model of pulmonary fibrosis
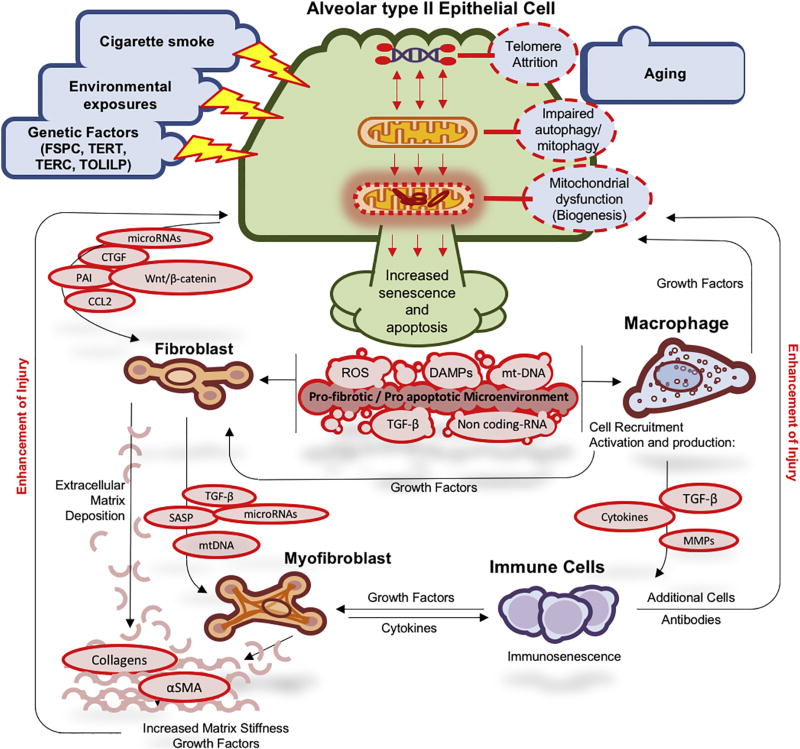
As previously mentioned, the introduction of the hallmarks of aging into the pulmonary fibrosis paradigm explains many of the features of the IPF lung. Telomere attrition and mitochondrial dysfunction become unifying themes that allow a cohesive model of the emergence, presence and progression of IPF (Figure 2). Basically, this model assumes a possible baseline genetic predisposition to injury, one that is not extremely detrimental to cells, otherwise it would not wait to be exhibited at the old age. Similarly, the model assumes a mild environmental injury, one that does not elicit an overwhelming inflammatory response, and thus, more often goes unnoticed clinically. The combination of the mild repeated injury, and the mild genetic predisposition lead to prolonged strain on the alveolar unit. In the alveolus, the cell that is more prone to this strain is the alveolar type II cell. This cell has been known to carry multiple functions critical to the integrity and function of the alveolar unit [128], including the metabolically intensive tasks of producing surfactant, serving as a progenitor for type I cells, and maintenance of fluid homeostasis. Indeed, type II cells are highly enriched with mitochondria [129, 130]. Thus, conditions that require enhanced replication because of loss of type I cells, or production of more surfactant (changes in microbiome, impaired mucus clearance), put a disproportionate metabolic and replicative stress on alveolar type II cells. The presence of relatively short telomeres or impaired telomerase function to begin with, could enhance this stress, because as cells proliferate, telomere attrition and uncapping leads to DNA damage response, downstream mitochondrial dysfunction, followed by release of ROS, mtDNA, DAMPs [131], and downstream activation of adjacent fibroblasts and macrophages creating a profibrotic microenvironment (Figure 2). If the fibroblasts also have relatively short telomeres, their response would potentially be abnormal and instead of proliferating normally and participating in a self-limited wound healing response, fibroblasts may develop a Senescence Associated Secretory Phenotype (SASP), characterized by growth arrest and secretion of extracellular matrix proteins such as fibronectin, proteases such as MMP-1, MMP-3 and MMP-10, PAI-1, IGFBPs [132], cytokines and chemokines [133], all known to be increased in IPF. SASP fibroblasts also exhibit metabolic derangements similar to what has been described in IPF and produce more ROS [134]. Macrophages will also be activated by the increase of DAMPs, cytokines and chemokines. It is also possible that in the presence of a systemic predisposition to aging, such as short telomeres or telomerase dysfunction, senescence will also emerge in macrophages and other lung inflammatory cells [135], a finding consistent with previous results in IPF patients [57, 136–138]. Therefore, instead of a self-resolving response to injury, the lung enters a vicious cycle, with increased matrix stiffness, enhanced local injury, accumulation of senescent fibroblasts and epithelial cells, a failure to resolve injury that leads to propagation of the disease to adjacent alveoli. While, many of the details of this model need to be worked out, the appeal of the model is that it allows integration of all the seemingly unrelated pieces of information gleaned from the application of OMICS technologies to the human lung.
Figure 2.
An updated paradigm of the Pulmonary Fibrosis Vicious Cycle – Prolonged injury from environmental exposure in the presence of genetic predisposition and abnormal aging lead to DNA damage response, downstream mitochondrial dysfunction, release of ROS, mtDNA, DAMPs, that activate both fibroblasts and macrophages, release of cytokines and growth factors, enhancement of injury which lead to sustained and self-propagating fibrosis.
Final remarks
In this review, we aimed to highlight the significant impacts that the application of omics technologies has had in our understanding of human pulmonary fibrosis. While never in isolation, the results led to novel hypotheses and sometimes provided the data to support and develop coherent conceptual models of disease. Publicly available datasets have allowed investigators to seek human correlates of their mechanistic findings. While we could not describe all of the findings, we chose to focus on the insights that supported a modified paradigm of pulmonary fibrosis: a disease driven by recurrent epithelial injury in the context of genetic predisposition and activated aging mechanisms. Understanding the central role of epithelial cells, telomere attrition and mitochondrial dysfunction, have been facilitated by collecting tissue and blood samples and carefully phenotype cohorts of patients with familial and sporadic IPF. In 2014, we published a call for an open access biorepository for IPF research [139]. While there has been a significant increase in the number of patients with IPF involved in research, with increased availability of DNA, RNA and peripheral blood samples, efforts to generate central repository of human lung tissues and cells from patients with IPF have not been renewed. We see this as a significant threat, because with the critical mass of genetic and genomic insights, the discovery, development and validation of novel therapies and biomarkers depends on us continuing to study human lungs and cells using novel approaches such as single cell profiling. Because if there is one lesson that we learned from our first two decades of applying Omics technologies to the human lung, is that we need to keep analyzing the human lung - this is where we will learn how to understand, diagnose and treat human pulmonary fibrosis.
Highlights.
Idiopathic Pulmonary Fibrosis (IPF) is a progressive, lethal and incurable disease
Genetic studies highlight ER stress, telomerase dysfunction, and epithelial injury.
Transcriptomics identified role of microRNAs and outcome predictive biomarkers in IPF
The lung microbiome, metabolome and proteome are substantially altered in IPF
The hallmarks of aging explain many of the omics findings in IPF lungs
Acknowledgments
The work was in part supported by US National Institute of Health (NIH) grants R01HL095397, R01HL127349 (N.K.), Pulmonary Fibrosis Foundation (PPF) Albert Rose Established Investigator Award 415245 (G.Y.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing financial interests
G.Y., and N.K., are inventors on a pending patent on use of thyroid hormone as an antifibrotic agent entitled: “Novel Methods of Treating or Preventing Fibrotic Lung Diseases” -OCR 6368 - 047162-7029P1 (00219). N.K. consulted Biogen Idec, Boehringer Ingelheim, Numedii, MMI, Pliant, Third Rock, Samumed and has an ongoing collaboration with MiRagen, all outside the submitted work.
References
- 1.Kim DS, Collard HR, King TE., Jr Classification and natural history of the idiopathic interstitial pneumonias. Proceedings of the American Thoracic Society. 2006;3:285–292. doi: 10.1513/pats.200601-005TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nalysnyk L, Cid-Ruzafa J, Rotella P, Esser D. Incidence and prevalence of idiopathic pulmonary fibrosis: review of the literature. European respiratory review : an official journal of the European Respiratory Society. 2012;21:355–361. doi: 10.1183/09059180.00002512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zuo F, Kaminski N, Eugui E, Allard J, Yakhini Z, Ben-Dor A, Lollini L, Morris D, Kim Y, DeLustro B, Sheppard D, Pardo A, Selman M, Heller RA. Gene expression analysis reveals matrilysin as a key regulator of pulmonary fibrosis in mice and humans. Proc Natl Acad Sci U S A. 2002;99:6292–6297. doi: 10.1073/pnas.092134099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Quackenbush J, Kaminski N, Schwartz DA, Spira A. Lung Genomics Research Consoritum Donwload Page. 2013 [Google Scholar]
- 5.Kusko RL, Brothers JF, 2nd, Tedrow J, Pandit K, Huleihel L, Perdomo C, Liu G, Juan-Guardela B, Kass D, Zhang S, Lenburg M, Martinez F, Quackenbush J, Sciurba F, Limper A, Geraci M, Yang I, Schwartz DA, Beane J, Spira A, Kaminski N. Integrated Genomics Reveals Convergent Transcriptomic Networks Underlying Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2016;194:948–960. doi: 10.1164/rccm.201510-2026OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Herazo-Maya JD, Kang DD, Juan-Guardela BM, Tedrow J, Martinez FJ, Sciurba FC, Tseng GC, Kaminski N. Integrative phenotyping framework (iPF): integrative clustering of multiple omics data identifies novel lung disease subphenotypes. BMC Genomics. 2015;16:924. doi: 10.1186/s12864-015-2170-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fingerlin TE, Murphy E, Zhang W, Peljto AL, Brown KK, Steele MP, Loyd JE, Cosgrove GP, Lynch D, Groshong S, Collard HR, Wolters PJ, Bradford WZ, Kossen K, Seiwert SD, du Bois RM, Garcia CK, Devine MS, Gudmundsson G, Isaksson HJ, Kaminski N, Zhang Y, Gibson KF, Lancaster LH, Cogan JD, Mason WR, Maher TM, Molyneaux PL, Wells AU, Moffatt MF, Selman M, Pardo A, Kim DS, Crapo JD, Make BJ, Regan EA, Walek DS, Daniel JJ, Kamatani Y, Zelenika D, Smith K, McKean D, Pedersen BS, Talbert J, Kidd RN, Markin CR, Beckman KB, Lathrop M, Schwarz MI, Schwartz DA. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat Genet. 2013;45:613–620. doi: 10.1038/ng.2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewitt RJ, Molyneaux PL. The respiratory microbiome in idiopathic pulmonary fibrosis. Annals of translational medicine. 2017;5:250. doi: 10.21037/atm.2017.01.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Mizuno T, Sridharan A, Du Y, Guo M, Tang J, Wikenheiser-Brokamp KA, Perl AT, Funari VA, Gokey JJ, Stripp BR, Whitsett JA. Single-cell RNA sequencing identifies diverse roles of epithelial cells in idiopathic pulmonary fibrosis. JCI insight. 2016;1:e90558. doi: 10.1172/jci.insight.90558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaur A, Mathai SK, Schwartz DA. Genetics in Idiopathic Pulmonary Fibrosis Pathogenesis, Prognosis, and Treatment. Front Med (Lausanne) 2017;4:154. doi: 10.3389/fmed.2017.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maitra M, Wang Y, Gerard RD, Mendelson CR, Garcia CK. Surfactant protein A2 mutations associated with pulmonary fibrosis lead to protein instability and endoplasmic reticulum stress. The Journal of biological chemistry. 2010;285:22103–22113. doi: 10.1074/jbc.M110.121467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou W, Wang Y. Candidate genes of idiopathic pulmonary fibrosis: current evidence and research. The application of clinical genetics. 2016;9:5–13. doi: 10.2147/TACG.S61999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz de Leon A, Cronkhite JT, Katzenstein AL, Godwin JD, Raghu G, Glazer CS, Rosenblatt RL, Girod CE, Garrity ER, Xing C, Garcia CK. Telomere lengths, pulmonary fibrosis and telomerase (TERT) mutations. PLoS One. 2010;5:e10680. doi: 10.1371/journal.pone.0010680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armanios MY, Chen JJ, Cogan JD, Alder JK, Ingersoll RG, Markin C, Lawson WE, Xie M, Vulto I, Phillips JA, 3rd, Lansdorp PM, Greider CW, Loyd JE. Telomerase mutations in families with idiopathic pulmonary fibrosis. N Engl J Med. 2007;356:1317–1326. doi: 10.1056/NEJMoa066157. [DOI] [PubMed] [Google Scholar]
- 15.Mushiroda T, Wattanapokayakit S, Takahashi A, Nukiwa T, Kudoh S, Ogura T, Taniguchi H, Kubo M, Kamatani N, Nakamura Y. A genome-wide association study identifies an association of a common variant in TERT with susceptibility to idiopathic pulmonary fibrosis. Journal of medical genetics. 2008;45:654–656. doi: 10.1136/jmg.2008.057356. [DOI] [PubMed] [Google Scholar]
- 16.Tsakiri KD, Cronkhite JT, Kuan PJ, Xing C, Raghu G, Weissler JC, Rosenblatt RL, Shay JW, Garcia CK. Adult-onset pulmonary fibrosis caused by mutations in telomerase. Proc Natl Acad Sci U S A. 2007;104:7552–7557. doi: 10.1073/pnas.0701009104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, Weissler J, Fitzgerald J, Kershaw C, Klesney-Tait J, Mageto Y, Shay JW, Ji W, Bilguvar K, Mane S, Lifton RP, Garcia CK. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nature genetics. 2015;47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathai SK, Pedersen BS, Smith K, Russell P, Schwarz MI, Brown KK, Steele MP, Loyd JE, Crapo JD, Silverman EK, Nickerson D, Fingerlin TE, Yang IV, Schwartz DA. Desmoplakin Variants Are Associated with Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2016;193:1151–1160. doi: 10.1164/rccm.201509-1863OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawson WE, Crossno PF, Polosukhin VV, Roldan J, Cheng DS, Lane KB, Blackwell TR, Xu C, Markin C, Ware LB, Miller GG, Loyd JE, Blackwell TS. Endoplasmic reticulum stress in alveolar epithelial cells is prominent in IPF: association with altered surfactant protein processing and herpesvirus infection. Am J Physiol Lung Cell Mol Physiol. 2008;294:L1119–1126. doi: 10.1152/ajplung.00382.2007. [DOI] [PubMed] [Google Scholar]
- 20.Lawson WE, Cheng DS, Degryse AL, Tanjore H, Polosukhin VV, Xu XC, Newcomb DC, Jones BR, Roldan J, Lane KB, Morrisey EE, Beers MF, Yull FE, Blackwell TS. Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc Natl Acad Sci U S A. 2011;108:10562–10567. doi: 10.1073/pnas.1107559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Povedano JM, Martinez P, Flores JM, Mulero F, Blasco MA. Mice with Pulmonary Fibrosis Driven by Telomere Dysfunction. Cell Rep. 2015;12:286–299. doi: 10.1016/j.celrep.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 22.Seibold MA, Wise AL, Speer MC, Steele MP, Brown KK, Loyd JE, Fingerlin TE, Zhang W, Gudmundsson G, Groshong SD, Evans CM, Garantziotis S, Adler KB, Dickey BF, du Bois RM, Yang IV, Herron A, Kervitsky D, Talbert JL, Markin C, Park J, Crews AL, Slifer SH, Auerbach S, Roy MG, Lin J, Hennessy CE, Schwarz MI, Schwartz DA. A common MUC5B promoter polymorphism and pulmonary fibrosis. N Engl J Med. 2011;364:1503–1512. doi: 10.1056/NEJMoa1013660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horimasu Y, Ohshimo S, Bonella F, Tanaka S, Ishikawa N, Hattori N, Kohno N, Guzman J, Costabel U. MUC5B promoter polymorphism in Japanese patients with idiopathic pulmonary fibrosis. Respirology (Carlton, Vic.) 2015;20:439–444. doi: 10.1111/resp.12466. [DOI] [PubMed] [Google Scholar]
- 24.Jiang H, Hu Y, Shang L, Li Y, Yang L, Chen Y. Association between MUC5B polymorphism and susceptibility and severity of idiopathic pulmonary fibrosis. Int J Clin Exp Pathol. 2015;8:14953–14958. [PMC free article] [PubMed] [Google Scholar]
- 25.Kishore A, Zizkova V, Kocourkova L, Petrkova J, Bouros E, Nunes H, Lostakova V, Muller-Quernheim J, Zissel G, Kolek V, Bouros D, Valeyre D, Petrek M. Association Study for 26 Candidate Loci in Idiopathic Pulmonary Fibrosis Patients from Four European Populations. Front Immunol. 2016;7:274. doi: 10.3389/fimmu.2016.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee MG, Lee YH. A meta-analysis examining the association between the MUC5B rs35705950 T/G polymorphism and susceptibility to idiopathic pulmonary fibrosis. Inflamm Res. 2015;64:463–470. doi: 10.1007/s00011-015-0829-6. [DOI] [PubMed] [Google Scholar]
- 27.Peljto AL, Selman M, Kim DS, Murphy E, Tucker L, Pardo A, Lee JS, Ji W, Schwarz MI, Yang IV, Schwartz DA, Fingerlin TE. The MUC5B promoter polymorphism is associated with idiopathic pulmonary fibrosis in a Mexican cohort but is rare among Asian ancestries. Chest. 2015;147:460–464. doi: 10.1378/chest.14-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vis JJ, Snetselaar R, Kazemier KM, ten Klooster L, Grutters JC, van Moorsel CH. Effect of Muc5b promoter polymorphism on disease predisposition and survival in idiopathic interstitial pneumonias. Respirology. 2016;21:712–717. doi: 10.1111/resp.12728. [DOI] [PubMed] [Google Scholar]
- 29.Zhu QQ, Zhang XL, Zhang SM, Tang SW, Min HY, Yi L, Xu B, Song Y. Association Between the MUC5B Promoter Polymorphism rs35705950 and Idiopathic Pulmonary Fibrosis: A Meta-analysis and Trial Sequential Analysis in Caucasian and Asian Populations. Medicine (Baltimore) 2015;94:e1901. doi: 10.1097/MD.0000000000001901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allen RJ, Porte J, Braybrooke R, Flores C, Fingerlin TE, Oldham JM, Guillen-Guio B, Ma SF, Okamoto T, John AE, Obeidat M, Yang IV, Henry A, Hubbard RB, Navaratnam V, Saini G, Thompson N, Booth HL, Hart SP, Hill MR, Hirani N, Maher TM, McAnulty RJ, Millar AB, Molyneaux PL, Parfrey H, Rassl DM, Whyte MKB, Fahy WA, Marshall RP, Oballa E, Bosse Y, Nickle DC, Sin DD, Timens W, Shrine N, Sayers I, Hall IP, Noth I, Schwartz DA, Tobin MD, Wain LV, Jenkins RG. Genetic variants associated with susceptibility to idiopathic pulmonary fibrosis in people of European ancestry: a genome-wide association study. Lancet Respir Med. 2017;5:869–880. doi: 10.1016/S2213-2600(17)30387-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Noth I, Zhang Y, Ma SF, Flores C, Barber M, Huang Y, Broderick SM, Wade MS, Hysi P, Scuirba J, Richards TJ, Juan-Guardela BM, Vij R, Han MK, Martinez FJ, Kossen K, Seiwert SD, Christie JD, Nicolae D, Kaminski N, Garcia JGN. Genetic variants associated with idiopathic pulmonary fibrosis susceptibility and mortality: a genome-wide association study. Lancet Respir Med. 2013;1:309–317. doi: 10.1016/S2213-2600(13)70045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldham JM, Ma SF, Martinez FJ, Anstrom KJ, Raghu G, Schwartz DA, Valenzi E, Witt L, Lee C, Vij R, Huang Y, Strek ME, Noth I, Investigators IP. TOLLIP, MUC5B, and the Response to N-Acetylcysteine among Individuals with Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2015;192:1475–1482. doi: 10.1164/rccm.201505-1010OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaminski N, Rosas IO. Gene expression profiling as a window into idiopathic pulmonary fibrosis pathogenesis: can we identify the right target genes? Proceedings of the American Thoracic Society. 2006;3:339–344. doi: 10.1513/pats.200601-011TK. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita CM, Dolgonos L, Zemans RL, Young SK, Robertson J, Briones N, Suzuki T, Campbell MN, Gauldie J, Radisky DC, Riches DW, Yu G, Kaminski N, McCulloch CA, Downey GP. Matrix metalloproteinase 3 is a mediator of pulmonary fibrosis. Am J Pathol. 2011;179:1733–1745. doi: 10.1016/j.ajpath.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pardo A, Selman M, Kaminski N. Approaching the degradome in idiopathic pulmonary fibrosis. Int J Biochem Cell Biol. 2008;40:1141–1155. doi: 10.1016/j.biocel.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 36.Yu G, Kovkarova-Naumovski E, Jara P, Parwani A, Kass D, Ruiz V, Lopez-Otin C, Rosas IO, Gibson KF, Cabrera S, Ramirez R, Yousem SA, Richards TJ, Chensny LJ, Selman M, Kaminski N, Pardo A. Matrix metalloproteinase-19 is a key regulator of lung fibrosis in mice and humans. Am J Respir Crit Care Med. 2012;186:752–762. doi: 10.1164/rccm.201202-0302OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Selman M, Pardo A, Kaminski N. Idiopathic pulmonary fibrosis: aberrant recapitulation of developmental programs? PLoS Med. 2008;5:e62. doi: 10.1371/journal.pmed.0050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang IV, Coldren CD, Leach SM, Seibold MA, Murphy E, Lin J, Rosen R, Neidermyer AJ, McKean DF, Groshong SD, Cool C, Cosgrove GP, Lynch DA, Brown KK, Schwarz MI, Fingerlin TE, Schwartz DA. Expression of cilium-associated genes defines novel molecular subtypes of idiopathic pulmonary fibrosis. Thorax. 2013;68:1114–1121. doi: 10.1136/thoraxjnl-2012-202943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selman M, Pardo A, Barrera L, Estrada A, Watson SR, Wilson K, Aziz N, Kaminski N, Zlotnik A. Gene expression profiles distinguish idiopathic pulmonary fibrosis from hypersensitivity pneumonitis. American journal of respiratory and critical care medicine. 2006;173:188–198. doi: 10.1164/rccm.200504-644OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Selman M, Carrillo G, Estrada A, Mejia M, Becerril C, Cisneros J, Gaxiola M, Perez-Padilla R, Navarro C, Richards T, Dauber J, King TE, Jr, Pardo A, Kaminski N. Accelerated variant of idiopathic pulmonary fibrosis: clinical behavior and gene expression pattern. PLoS One. 2007;2:e482. doi: 10.1371/journal.pone.0000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bueno M, Lai YC, Romero Y, Brands J, St Croix CM, Kamga C, Corey C, Herazo-Maya JD, Sembrat J, Lee JS, Duncan SR, Rojas M, Shiva S, Chu CT, Mora AL. PINK1 deficiency impairs mitochondrial homeostasis and promotes lung fibrosis. The Journal of clinical investigation. 2015;125:521–538. doi: 10.1172/JCI74942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu G, Tzouvelekis A, Wang R, Herazo-Maya JD, Ibarra GH, Srivastava A, de Castro JPW, DeIuliis G, Ahangari F, Woolard T, Aurelien N, Arrojo EDR, Gan Y, Graham M, Liu X, Homer RJ, Scanlan TS, Mannam P, Lee PJ, Herzog EL, Bianco AC, Kaminski N. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2017 doi: 10.1038/nm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pandit KV, Milosevic J. MicroRNA regulatory networks in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015;93:129–137. doi: 10.1139/bcb-2014-0101. [DOI] [PubMed] [Google Scholar]
- 44.Pardo A, Gibson K, Cisneros J, Richards TJ, Yang Y, Becerril C, Yousem S, Herrera I, Ruiz V, Selman M, Kaminski N. Up-regulation and profibrotic role of osteopontin in human idiopathic pulmonary fibrosis. PLoS Med. 2005;2:e251. doi: 10.1371/journal.pmed.0020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pilewski JM, Liu L, Henry AC, Knauer AV, Feghali-Bostwick CA. Insulin-like growth factor binding proteins 3 and 5 are overexpressed in idiopathic pulmonary fibrosis and contribute to extracellular matrix deposition. Am J Pathol. 2005;166:399–407. doi: 10.1016/S0002-9440(10)62263-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veraldi KL, Feghali-Bostwick CA. Insulin-like growth factor binding proteins-3 and −5: central mediators of fibrosis and promising new therapeutic targets. Open Rheumatol J. 2012;6:140–145. doi: 10.2174/1874312901206010140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Konigshoff M, Kramer M, Balsara N, Wilhelm J, Amarie OV, Jahn A, Rose F, Fink L, Seeger W, Schaefer L, Gunther A, Eickelberg O. WNT1-inducible signaling protein-1 mediates pulmonary fibrosis in mice and is upregulated in humans with idiopathic pulmonary fibrosis. The Journal of clinical investigation. 2009;119:772–787. doi: 10.1172/JCI33950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Staab-Weijnitz CA, Fernandez IE, Knuppel L, Maul J, Heinzelmann K, Juan-Guardela BM, Hennen E, Preissler G, Winter H, Neurohr C, Hatz R, Lindner M, Behr J, Kaminski N, Eickelberg O. FK506-Binding Protein 10, a Potential Novel Drug Target for Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2015;192:455–467. doi: 10.1164/rccm.201412-2233OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tan J, Tedrow JR, Dutta JA, Juan-Guardela B, Nouraie M, Chu Y, Trejo Bittar H, Ramani K, Biswas PS, Veraldi KL, Kaminski N, Zhang Y, Kass DJ. Expression of RXFP1 Is Decreased in Idiopathic Pulmonary Fibrosis. Implications for Relaxin-based Therapies. American journal of respiratory and critical care medicine. 2016;194:1392–1402. doi: 10.1164/rccm.201509-1865OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tzouvelekis A, Yu G, Lino Cardenas CL, Herazo-Maya JD, Wang R, Woolard T, Zhang Y, Sakamoto K, Lee H, Yi JS, DeIuliis G, Xylourgidis N, Ahangari F, Lee PJ, Aidinis V, Herzog EL, Homer R, Bennett AM, Kaminski N. SH2 Domain-Containing Phosphatase-2 Is a Novel Antifibrotic Regulator in Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017;195:500–514. doi: 10.1164/rccm.201602-0329OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montgomery RL, Yu G, Latimer PA, Stack C, Robinson K, Dalby CM, Kaminski N, van Rooij E. MicroRNA mimicry blocks pulmonary fibrosis. EMBO Mol Med. 2014;6:1347–1356. doi: 10.15252/emmm.201303604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pandit KV, Corcoran D, Yousef H, Yarlagadda M, Tzouvelekis A, Gibson KF, Konishi K, Yousem SA, Singh M, Handley D, Richards T, Selman M, Watkins SC, Pardo A, Ben-Yehudah A, Bouros D, Eickelberg O, Ray P, Benos PV, Kaminski N. Inhibition and role of let-7d in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2010;182:220–229. doi: 10.1164/rccm.200911-1698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu G, Friggeri A, Yang Y, Milosevic J, Ding Q, Thannickal VJ, Kaminski N, Abraham E. miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J Exp Med. 2010;207:1589–1597. doi: 10.1084/jem.20100035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rosas IO, Richards TJ, Konishi K, Zhang Y, Gibson K, Lokshin AE, Lindell KO, Cisneros J, Macdonald SD, Pardo A, Sciurba F, Dauber J, Selman M, Gochuico BR, Kaminski N. MMP1 and MMP7 as potential peripheral blood biomarkers in idiopathic pulmonary fibrosis. PLoS Med. 2008;5:e93. doi: 10.1371/journal.pmed.0050093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.DePianto DJ, Chandriani S, Abbas AR, Jia G, N’Diaye EN, Caplazi P, Kauder SE, Biswas S, Karnik SK, Ha C, Modrusan Z, Matthay MA, Kukreja J, Collard HR, Egen JG, Wolters PJ, Arron JR. Heterogeneous gene expression signatures correspond to distinct lung pathologies and biomarkers of disease severity in idiopathic pulmonary fibrosis. Thorax. 2015;70:48–56. doi: 10.1136/thoraxjnl-2013-204596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jia G, Chandriani S, Abbas AR, DePianto DJ, N’Diaye EN, Yaylaoglu MB, Moore HM, Peng I, DeVoss J, Collard HR, Wolters PJ, Egen JG, Arron JR. CXCL14 is a candidate biomarker for Hedgehog signalling in idiopathic pulmonary fibrosis. Thorax. 2017;72:780–787. doi: 10.1136/thoraxjnl-2015-207682. [DOI] [PubMed] [Google Scholar]
- 57.Herazo-Maya JD, Noth I, Duncan SR, Kim S, Ma SF, Tseng GC, Feingold E, Juan-Guardela BM, Richards TJ, Lussier Y, Huang Y, Vij R, Lindell KO, Xue J, Gibson KF, Shapiro SD, Garcia JG, Kaminski N. Peripheral blood mononuclear cell gene expression profiles predict poor outcome in idiopathic pulmonary fibrosis. Sci Transl Med. 2013;5:205ra136. doi: 10.1126/scitranslmed.3005964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herazo-Maya JD, Sun J, Molyneaux PL, Li Q, Villalba JA, Tzouvelekis A, Lynn H, Juan-Guardela BM, Risquez C, Osorio JC, Yan X, Michel G, Aurelien N, Lindell KO, Klesen MJ, Moffatt MF, Cookson WO, Zhang Y, Garcia JGN, Noth I, Prasse A, Bar-Joseph Z, Gibson KF, Zhao H, Herzog EL, Rosas IO, Maher TM, Kaminski N. Validation of a 52-gene risk profile for outcome prediction in patients with idiopathic pulmonary fibrosis: an international, multicentre, cohort study. Lancet Respir Med. 2017;5:857–868. doi: 10.1016/S2213-2600(17)30349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cisneros J, Hagood J, Checa M, Ortiz-Quintero B, Negreros M, Herrera I, Ramos C, Pardo A, Selman M. Hypermethylation-mediated silencing of p14(ARF) in fibroblasts from idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol. 2012;303:L295–303. doi: 10.1152/ajplung.00332.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanders YY, Tollefsbol TO, Varisco BM, Hagood JS. Epigenetic regulation of thy-1 by histone deacetylase inhibitor in rat lung fibroblasts. American journal of respiratory cell and molecular biology. 2011;45:16–23. doi: 10.1165/rcmb.2010-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, Peters-Golden M. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. The American journal of pathology. 2010;177:2245–2255. doi: 10.2353/ajpath.2010.100446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol. 2008;39:610–618. doi: 10.1165/rcmb.2007-0322OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coward WR, Feghali-Bostwick CA, Jenkins G, Knox AJ, Pang L. A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 2014;28:3183–3196. doi: 10.1096/fj.13-241760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rabinovich EI, Selman M, Kaminski N. Epigenomics of idiopathic pulmonary fibrosis: evaluating the first steps. American journal of respiratory and critical care medicine. 2012;186:473–475. doi: 10.1164/rccm.201208-1350ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanders YY, Ambalavanan N, Halloran B, Zhang X, Liu H, Crossman DK, Bray M, Zhang K, Thannickal VJ, Hagood JS. Altered DNA methylation profile in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2012;186:525–535. doi: 10.1164/rccm.201201-0077OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang IV, Pedersen BS, Rabinovich E, Hennessy CE, Davidson EJ, Murphy E, Guardela BJ, Tedrow JR, Zhang Y, Singh MK, Correll M, Schwarz MI, Geraci M, Sciurba FC, Quackenbush J, Spira A, Kaminski N, Schwartz DA. Relationship of DNA Methylation and Gene Expression in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2014;190:1263–1272. doi: 10.1164/rccm.201408-1452OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masser DR, Hadad N, Porter H, Stout MB, Unnikrishnan A, Stanford DR, Freeman WM. Analysis of DNA modifications in aging research. Geroscience. 2018 doi: 10.1007/s11357-018-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kelsey G, Stegle O, Reik W. Single-cell epigenomics: Recording the past and predicting the future. Science. 2017;358:69–75. doi: 10.1126/science.aan6826. [DOI] [PubMed] [Google Scholar]
- 69.Clarridge JE., 3rd Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clinical microbiology reviews. 2004;17:840–862. doi: 10.1128/CMR.17.4.840-862.2004. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, Bushman FD. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012;7:e42786. doi: 10.1371/journal.pone.0042786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim D, Hofstaedter CE, Zhao C, Mattei L, Tanes C, Clarke E, Lauder A, Sherrill-Mix S, Chehoud C, Kelsen J, Conrad M, Collman RG, Baldassano R, Bushman FD, Bittinger K. Optimizing methods and dodging pitfalls in microbiome research. Microbiome. 2017;5:52. doi: 10.1186/s40168-017-0267-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, Moore BB, White ES, Flaherty KR, Huffnagle GB, Martinez FJ. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: an analysis of the COMET study. The Lancet. Respiratory medicine. 2014;2:548–556. doi: 10.1016/S2213-2600(14)70069-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Molyneaux PL, Cox MJ, Willis-Owen SA, Mallia P, Russell KE, Russell AM, Murphy E, Johnston SL, Schwartz DA, Wells AU, Cookson WO, Maher TM, Moffatt MF. The role of bacteria in the pathogenesis and progression of idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2014;190:906–913. doi: 10.1164/rccm.201403-0541OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Molyneaux PL, Willis-Owen SAG, Cox MJ, James P, Cowman S, Loebinger M, Blanchard A, Edwards LM, Stock C, Daccord C, Renzoni EA, Wells AU, Moffatt MF, Cookson WOC, Maher TM. Host-Microbial Interactions in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017;195:1640–1650. doi: 10.1164/rccm.201607-1408OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang Y, Ma SF, Espindola MS, Vij R, Oldham JM, Huffnagle GB, Erb-Downward JR, Flaherty KR, Moore BB, White ES, Zhou T, Li J, Lussier YA, Han MK, Kaminski N, Garcia JGN, Hogaboam CM, Martinez FJ, Noth I, Investigators C-I. Microbes Are Associated with Host Innate Immune Response in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017;196:208–219. doi: 10.1164/rccm.201607-1525OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molyneaux PL, Cox MJ, Wells AU, Kim HC, Ji W, Cookson WO, Moffatt MF, Kim DS, Maher TM. Changes in the respiratory microbiome during acute exacerbations of idiopathic pulmonary fibrosis. Respir Res. 2017;18:29. doi: 10.1186/s12931-017-0511-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang J, Lesko M, Badri MH, Kapoor BC, Wu BG, Li Y, Smaldone GC, Bonneau R, Kurtz ZD, Condos R, Segal LN. Lung microbiome and host immune tone in subjects with idiopathic pulmonary fibrosis treated with inhaled interferon-gamma. ERJ Open Res. 2017;3 doi: 10.1183/23120541.00008-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kitsios GD, Rojas M, Kass DJ, Fitch A, Sembrat JC, Qin S, Veraldi KL, Gibson KF, Lindell K, Pilewski JM, Methe B, Li K, McDyer J, McVerry BJ, Morris A. Microbiome in lung explants of idiopathic pulmonary fibrosis: a case-control study in patients with end-stage fibrosis. Thorax. 2017 doi: 10.1136/thoraxjnl-2017-210537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Richards TJ, Kaminski N, Baribaud F, Flavin S, Brodmerkel C, Horowitz D, Li K, Choi J, Vuga LJ, Lindell KO, Klesen M, Zhang Y, Gibson KF. Peripheral blood proteins predict mortality in idiopathic pulmonary fibrosis. American journal of respiratory and critical care medicine. 2012;185:67–76. doi: 10.1164/rccm.201101-0058OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ashley SL, Xia M, Murray S, O’Dwyer DN, Grant E, White ES, Flaherty KR, Martinez FJ, Moore BB. Six-SOMAmer Index Relating to Immune, Protease and Angiogenic Functions Predicts Progression in IPF. PLoS One. 2016;11:e0159878. doi: 10.1371/journal.pone.0159878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Korfei M, Schmitt S, Ruppert C, Henneke I, Markart P, Loeh B, Mahavadi P, Wygrecka M, Klepetko W, Fink L, Bonniaud P, Preissner KT, Lochnit G, Schaefer L, Seeger W, Guenther A. Comparative proteomic analysis of lung tissue from patients with idiopathic pulmonary fibrosis (IPF) and lung transplant donor lungs. Journal of proteome research. 2011;10:2185–2205. doi: 10.1021/pr1009355. [DOI] [PubMed] [Google Scholar]
- 82.Korfei M, von der Beck D, Henneke I, Markart P, Ruppert C, Mahavadi P, Ghanim B, Klepetko W, Fink L, Meiners S, Kramer OH, Seeger W, Vancheri C, Guenther A. Comparative proteome analysis of lung tissue from patients with idiopathic pulmonary fibrosis (IPF), non-specific interstitial pneumonia (NSIP) and organ donors. Journal of proteomics. 2013;85:109–128. doi: 10.1016/j.jprot.2013.04.033. [DOI] [PubMed] [Google Scholar]
- 83.Ohara I, Aida S, Shimazaki H, Kobayashi H, Tsuda H, Toda T, Nakanishi K, Tamai S. Proteomic analysis in usual and nonspecific interstitial pneumonia. Histol Histopathol. 2014;29:377–386. doi: 10.14670/HH-29.377. [DOI] [PubMed] [Google Scholar]
- 84.Schiller HB, Mayr CH, Leuschner G, Strunz M, Staab-Weijnitz C, Preisendorfer S, Eckes B, Moinzadeh P, Krieg T, Schwartz DA, Hatz RA, Behr J, Mann M, Eickelberg O. Deep Proteome Profiling Reveals Common Prevalence of MZB1-Positive Plasma B Cells in Human Lung and Skin Fibrosis. American journal of respiratory and critical care medicine. 2017;196:1298–1310. doi: 10.1164/rccm.201611-2263OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kottmann RM, Kulkarni AA, Smolnycki KA, Lyda E, Dahanayake T, Salibi R, Honnons S, Jones C, Isern NG, Hu JZ, Nathan SD, Grant G, Phipps RP, Sime PJ. Lactic acid is elevated in idiopathic pulmonary fibrosis and induces myofibroblast differentiation via pH-dependent activation of transforming growth factor-beta. American journal of respiratory and critical care medicine. 2012;186:740–751. doi: 10.1164/rccm.201201-0084OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kang YP, Lee SB, Lee JM, Kim HM, Hong JY, Lee WJ, Choi CW, Shin HK, Kim DJ, Koh ES, Park CS, Kwon SW, Park SW. Metabolic Profiling Regarding Pathogenesis of Idiopathic Pulmonary Fibrosis. Journal of proteome research. 2016;15:1717–1724. doi: 10.1021/acs.jproteome.6b00156. [DOI] [PubMed] [Google Scholar]
- 87.Zhao YD, Yin L, Archer S, Lu C, Zhao G, Yao Y, Wu L, Hsin M, Waddell TK, Keshavjee S, Granton J, de Perrot M. Metabolic heterogeneity of idiopathic pulmonary fibrosis: a metabolomic study. BMJ open respiratory research. 2017;4:e000183. doi: 10.1136/bmjresp-2017-000183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xie N, Tan Z, Banerjee S, Cui H, Ge J, Liu RM, Bernard K, Thannickal VJ, Liu G. Glycolytic Reprogramming in Myofibroblast Differentiation and Lung Fibrosis. American journal of respiratory and critical care medicine. 2015;192:1462–1474. doi: 10.1164/rccm.201504-0780OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.N. Idiopathic Pulmonary Fibrosis Clinical Research. Raghu G, Anstrom KJ, King TE, Jr, Lasky JA, Martinez FJ. Prednisone, azathioprine, and N-acetylcysteine for pulmonary fibrosis. N Engl J Med. 2012;366:1968–1977. doi: 10.1056/NEJMoa1113354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Davies HR, Richeldi L, Walters EH. Immunomodulatory agents for idiopathic pulmonary fibrosis. Cochrane Database Syst Rev. 2003:CD003134. doi: 10.1002/14651858.CD003134. [DOI] [PubMed] [Google Scholar]
- 91.Wahidi MM, Ravenel J, Palmer SM, McAdams HP. Progression of idiopathic pulmonary fibrosis in native lungs after single lung transplantation. Chest. 2002;121:2072–2076. doi: 10.1378/chest.121.6.2072. [DOI] [PubMed] [Google Scholar]
- 92.Bonniaud P, Kolb M, Galt T, Robertson J, Robbins C, Stampfli M, Lavery C, Margetts PJ, Roberts AB, Gauldie J. Smad3 null mice develop airspace enlargement and are resistant to TGF-beta-mediated pulmonary fibrosis. J Immunol. 2004;173:2099–2108. doi: 10.4049/jimmunol.173.3.2099. [DOI] [PubMed] [Google Scholar]
- 93.Zhao J, Shi W, Wang YL, Chen H, Bringas P, Jr, Datto MB, Frederick JP, Wang XF, Warburton D. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. American journal of physiology. Lung cellular and molecular physiology. 2002;282:L585–593. doi: 10.1152/ajplung.00151.2001. [DOI] [PubMed] [Google Scholar]
- 94.Kaminski N, Allard JD, Pittet JF, Zuo F, Griffiths MJ, Morris D, Huang X, Sheppard D, Heller RA. Global analysis of gene expression in pulmonary fibrosis reveals distinct programs regulating lung inflammation and fibrosis. Proc Natl Acad Sci U S A. 2000;97:1778–1783. doi: 10.1073/pnas.97.4.1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, Rifkin DB, Sheppard D. The integrin alpha v beta 6 binds and activates latent TGF beta 1: a mechanism for regulating pulmonary inflammation and fibrosis. Cell. 1999;96:319–328. doi: 10.1016/s0092-8674(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 96.Raghu G, Chen SY, Hou Q, Yeh WS, Collard HR. Incidence and prevalence of idiopathic pulmonary fibrosis in US adults 18–64 years old. Eur Respir J. 2016;48:179–186. doi: 10.1183/13993003.01653-2015. [DOI] [PubMed] [Google Scholar]
- 97.Harari S, Madotto F, Caminati A, Conti S, Cesana G. Epidemiology of Idiopathic Pulmonary Fibrosis in Northern Italy. PLoS One. 2016;11:e0147072. doi: 10.1371/journal.pone.0147072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Raghu G, Chen SY, Yeh WS, Maroni B, Li Q, Lee YC, Collard HR. Idiopathic pulmonary fibrosis in US Medicare beneficiaries aged 65 years and older: incidence, prevalence, and survival, 2001–11. Lancet Respir Med. 2014;2:566–572. doi: 10.1016/S2213-2600(14)70101-8. [DOI] [PubMed] [Google Scholar]
- 99.Naikawadi RP, Disayabutr S, Mallavia B, Donne ML, Green G, La JL, Rock JR, Looney MR, Wolters PJ. Telomere dysfunction in alveolar epithelial cells causes lung remodeling and fibrosis. JCI insight. 2016;1:e86704. doi: 10.1172/jci.insight.86704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Disayabutr S, Kim EK, Cha SI, Green G, Naikawadi RP, Jones KD, Golden JA, Schroeder A, Matthay MA, Kukreja J, Erle DJ, Collard HR, Wolters PJ. miR-34 miRNAs Regulate Cellular Senescence in Type II Alveolar Epithelial Cells of Patients with Idiopathic Pulmonary Fibrosis. PloS one. 2016;11:e0158367. doi: 10.1371/journal.pone.0158367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tatler AL, Jenkins G. TGF-beta activation and lung fibrosis. Proceedings of the American Thoracic Society. 2012;9:130–136. doi: 10.1513/pats.201201-003AW. [DOI] [PubMed] [Google Scholar]
- 102.Goodwin A, Jenkins G. Role of integrin-mediated TGFbeta activation in the pathogenesis of pulmonary fibrosis. Biochemical Society transactions. 2009;37:849–854. doi: 10.1042/BST0370849. [DOI] [PubMed] [Google Scholar]
- 103.Henderson NC, Arnold TD, Katamura Y, Giacomini MM, Rodriguez JD, McCarty JH, Pellicoro A, Raschperger E, Betsholtz C, Ruminski PG, Griggs DW, Prinsen MJ, Maher JJ, Iredale JP, Lacy-Hulbert A, Adams RH, Sheppard D. Targeting of alphav integrin identifies a core molecular pathway that regulates fibrosis in several organs. Nature medicine. 2013;19:1617–1624. doi: 10.1038/nm.3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lehmann M, Baarsma HA, Konigshoff M. WNT Signaling in Lung Aging and Disease. Annals of the American Thoracic Society. 2016;13:S411–S416. doi: 10.1513/AnnalsATS.201608-586AW. [DOI] [PubMed] [Google Scholar]
- 105.Kugler MC, Joyner AL, Loomis CA, Munger JS. Sonic hedgehog signaling in the lung. From development to disease. American journal of respiratory cell and molecular biology. 2015;52:1–13. doi: 10.1165/rcmb.2014-0132TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kropski JA, Blackwell TS. Endoplasmic reticulum stress in the pathogenesis of fibrotic disease. The Journal of clinical investigation. 2018;128:64–73. doi: 10.1172/JCI93560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kurundkar A, Thannickal VJ. Redox mechanisms in age-related lung fibrosis. Redox Biol. 2016;9:67–76. doi: 10.1016/j.redox.2016.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Plataki M, Koutsopoulos AV, Darivianaki K, Delides G, Siafakas NM, Bouros D. Expression of apoptotic and antiapoptotic markers in epithelial cells in idiopathic pulmonary fibrosis. Chest. 2005;127:266–274. doi: 10.1378/chest.127.1.266. [DOI] [PubMed] [Google Scholar]
- 109.Herrera J, Henke CA, Bitterman PB. Extracellular matrix as a driver of progressive fibrosis. The Journal of clinical investigation. 2018;128:45–53. doi: 10.1172/JCI93557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tschumperlin DJ, Ligresti G, Hilscher MB, Shah VH. Mechanosensing and fibrosis. The Journal of clinical investigation. 2018;128:74–84. doi: 10.1172/JCI93561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bernard K, Logsdon NJ, Ravi S, Xie N, Persons BP, Rangarajan S, Zmijewski JW, Mitra K, Liu G, Darley-Usmar VM, Thannickal VJ. Metabolic Reprogramming Is Required for Myofibroblast Contractility and Differentiation. The Journal of biological chemistry. 2015;290:25427–25438. doi: 10.1074/jbc.M115.646984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Alvarez D, Cardenes N, Sellares J, Bueno M, Corey C, Hanumanthu VS, Peng Y, D’Cunha H, Sembrat J, Nouraie M, Shanker S, Caufield C, Shiva S, Armanios M, Mora AL, Rojas M. IPF lung fibroblasts have a senescent phenotype. American journal of physiology. Lung cellular and molecular physiology. 2017;313:L1164–L1173. doi: 10.1152/ajplung.00220.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Habiel DM, Hogaboam CM. Heterogeneity of Fibroblasts and Myofibroblasts in Pulmonary Fibrosis. Curr Pathobiol Rep. 2017;5:101–110. doi: 10.1007/s40139-017-0134-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bagnato G, Harari S. Cellular interactions in the pathogenesis of interstitial lung diseases. European respiratory review : an official journal of the European Respiratory Society. 2015;24:102–114. doi: 10.1183/09059180.00003214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pardo A, Cabrera S, Maldonado M, Selman M. Role of matrix metalloproteinases in the pathogenesis of idiopathic pulmonary fibrosis. Respiratory research. 2016;17:23. doi: 10.1186/s12931-016-0343-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Tzouvelekis A, Kaminski N. Epigenetics in idiopathic pulmonary fibrosis. Biochem Cell Biol. 2015;93:159–170. doi: 10.1139/bcb-2014-0126. [DOI] [PubMed] [Google Scholar]
- 117.Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mora AL, Rojas M, Pardo A, Selman M. Emerging therapies for idiopathic pulmonary fibrosis, a progressive age-related disease. Nat Rev Drug Discov. 2017;16:755–772. doi: 10.1038/nrd.2017.170. [DOI] [PubMed] [Google Scholar]
- 119.Dai J, Cai H, Zhuang Y, Wu Y, Min H, Li J, Shi Y, Gao Q, Yi L. Telomerase gene mutations and telomere length shortening in patients with idiopathic pulmonary fibrosis in a Chinese population. Respirology. 2015;20:122–128. doi: 10.1111/resp.12422. [DOI] [PubMed] [Google Scholar]
- 120.Dai J, Cai H, Li H, Zhuang Y, Min H, Wen Y, Yang J, Gao Q, Shi Y, Yi L. Association between telomere length and survival in patients with idiopathic pulmonary fibrosis. Respirology. 2015;20:947–952. doi: 10.1111/resp.12566. [DOI] [PubMed] [Google Scholar]
- 121.Newton CA, Kozlitina J, Lines JR, Kaza V, Torres F, Garcia CK. Telomere length in patients with pulmonary fibrosis associated with chronic lung allograft dysfunction and post-lung transplantation survival. J Heart Lung Transplant. 2017;36:845–853. doi: 10.1016/j.healun.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Snetselaar R, van Batenburg AA, van Oosterhout MFM, Kazemier KM, Roothaan SM, Peeters T, van der Vis JJ, Goldschmeding R, Grutters JC, van Moorsel CHM. Short telomere length in IPF lung associates with fibrotic lesions and predicts survival. PLoS One. 2017;12:e0189467. doi: 10.1371/journal.pone.0189467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Alder JK, Barkauskas CE, Limjunyawong N, Stanley SE, Kembou F, Tuder RM, Hogan BL, Mitzner W, Armanios M. Telomere dysfunction causes alveolar stem cell failure. Proc Natl Acad Sci U S A. 2015;112:5099–5104. doi: 10.1073/pnas.1504780112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Snetselaar R, van Moorsel CHM, Kazemier KM, van der Vis JJ, Zanen P, van Oosterhout MFM, Grutters JC. Telomere length in interstitial lung diseases. Chest. 2015;148:1011–1018. doi: 10.1378/chest.14-3078. [DOI] [PubMed] [Google Scholar]
- 125.Rangarajan S, Bernard K, Thannickal VJ. Mitochondrial Dysfunction in Pulmonary Fibrosis. Ann Am Thorac Soc. 2017;14:S383–S388. doi: 10.1513/AnnalsATS.201705-370AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yu G, Tzouvelekis A, Wang R, Herazo-Maya JD, Ibarra GH, Srivastava A, de Castro JPW, DeIuliis G, Ahangari F, Woolard T, Aurelien N, Arrojo EDR, Gan Y, Graham M, Liu X, Homer RJ, Scanlan TS, Mannam P, Lee PJ, Herzog EL, Bianco AC, Kaminski N. Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat Med. 2018;24:39–49. doi: 10.1038/nm.4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ryu C, Sun H, Gulati M, Herazo-Maya J, Chen Y, Osafo-Addo A, Brandsdorfer C, Winkler J, Blaul C, Faunce J, Pan H, Woolard T, Tzouvelekis A, Antin-Ozerkis DE, Puchalski JT, Slade M, Gonzalez AL, Bogenhagen DF, Kirillov V, Feghali-Bostwick C, Gibson K, Lindell K, Herzog RI, Dela Cruz CS, Mehal W, Kaminski N, Herzog EL, Trujillo G. Extracellular Mitochondrial DNA is Generated by Fibroblasts and Predicts Death in Idiopathic Pulmonary Fibrosis. American journal of respiratory and critical care medicine. 2017 doi: 10.1164/rccm.201612-2480OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Castranova V, Rabovsky J, Tucker JH, Miles PR. The alveolar type II epithelial cell: a multifunctional pneumocyte. Toxicol Appl Pharmacol. 1988;93:472–483. doi: 10.1016/0041-008x(88)90051-8. [DOI] [PubMed] [Google Scholar]
- 129.Cloonan SM, Choi AM. Mitochondria in lung disease. The Journal of clinical investigation. 2016;126:809–820. doi: 10.1172/JCI81113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Piantadosi CA, Suliman HB. Mitochondrial Dysfunction in Lung Pathogenesis. Annu Rev Physiol. 2017;79:495–515. doi: 10.1146/annurev-physiol-022516-034322. [DOI] [PubMed] [Google Scholar]
- 131.Birch J, Barnes PJ, Passos JF. Mitochondria, telomeres and cell senescence: Implications for lung ageing and disease. Pharmacol Ther. 2017 doi: 10.1016/j.pharmthera.2017.10.005. [DOI] [PubMed] [Google Scholar]
- 132.Mavrogonatou E, Pratsinis H, Papadopoulou A, Karamanos NK, Kletsas D. Extracellular matrix alterations in senescent cells and their significance in tissue homeostasis. Matrix Biol. 2017 doi: 10.1016/j.matbio.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 133.Ohtani N, Hara E. Roles and mechanisms of cellular senescence in regulation of tissue homeostasis. Cancer Sci. 2013;104:525–530. doi: 10.1111/cas.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chandrasekaran A, Idelchik M, Melendez JA. Redox control of senescence and age-related disease. Redox Biol. 2017;11:91–102. doi: 10.1016/j.redox.2016.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Frasca D. Senescent B cells in aging and age-related diseases: Their role in the regulation of antibody responses. Exp Gerontol. 2017 doi: 10.1016/j.exger.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huang Y, Ma SF, Vij R, Oldham JM, Herazo-Maya J, Broderick SM, Strek ME, White SR, Hogarth DK, Sandbo NK, Lussier YA, Gibson KF, Kaminski N, Garcia JG, Noth I. A functional genomic model for predicting prognosis in idiopathic pulmonary fibrosis. BMC Pulm Med. 2015;15:147. doi: 10.1186/s12890-015-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Gilani SR, Vuga LJ, Lindell KO, Gibson KF, Xue J, Kaminski N, Valentine VG, Lindsay EK, George MP, Steele C, Duncan SR. CD28 down-regulation on circulating CD4 T-cells is associated with poor prognoses of patients with idiopathic pulmonary fibrosis. PLoS One. 2010;5:e8959. doi: 10.1371/journal.pone.0008959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Feghali-Bostwick CA, Tsai CG, Valentine VG, Kantrow S, Stoner MW, Pilewski JM, Gadgil A, George MP, Gibson KF, Choi AM, Kaminski N, Zhang Y, Duncan SR. Cellular and humoral autoreactivity in idiopathic pulmonary fibrosis. J Immunol. 2007;179:2592–2599. doi: 10.4049/jimmunol.179.4.2592. [DOI] [PubMed] [Google Scholar]
- 139.White ES, Brown KK, Collard HR, Conoscenti CS, Cosgrove GP, Flaherty KR, Leff JA, Martinez FJ, Roman J, Rose D, Violette S, Kaminski N. Open-access biorepository for idiopathic pulmonary fibrosis. The way forward. Ann Am Thorac Soc. 2014;11:1171–1175. doi: 10.1513/AnnalsATS.201406-289OI. [DOI] [PMC free article] [PubMed] [Google Scholar]