Abstract
Advances in gene discovery for neurodevelopmental disorders have identified SCN2A dysfunction as a leading cause of infantile seizures, autism spectrum disorder, and intellectual disability. SCN2A encodes the neuronal sodium channel NaV1.2. Functional assays demonstrate strong correlation between genotype and phenotype. This insight can help guide therapeutic decisions and raises the possibility that ligands that selectively enhance or diminish channel function may improve symptoms. The well-defined function of sodium channels makes SCN2A an important test-case for investigating the neurobiology of neurodevelopmental disorders more generally. Here, we discuss the progress made, through the concerted efforts of a diverse group of academic and industry scientists as well as policy advocates in understanding and treating SCN2A-related disorders.
Keywords: autism spectrum disorder, epilepsy, sodium channel, neurodevelopmental disorder, developmental delay, neurodevelopment, intellectual disability, NaV1.2
The role of SCN2A in neurodevelopmental disorders
Our understanding of the genetic causes of neurodevelopmental disorders has progressed dramatically in the past decade, raising hope for better treatments by illuminating the biological mechanisms of disease. In that time, disruption of the gene SCN2A has been identified as a prominent cause of a wide range of neurodevelopmental disorders, including autism spectrum disorder (ASD), intellectual disability (ID), and infantile-onset seizures (before the first year of life) of varying severity. Moreover, and notably, the consequences of disease-associated variants on protein function can predict the nature and severity of the resulting phenotype.
SCN2A encodes the neuronal voltage-gated sodium channel NaV1.2, which is involved in the initiation and propagation of action potentials in a range of neuron classes. Critically, from a therapeutic perspective, ion channels such as NaV1.2 are “druggable” targets, and small molecule compounds can be developed to directly and selectively increase or decrease channel function as the basis for potential treatments. Thus, the well-defined biology describing NaV1.2 function, the strong genotype-phenotype correlations found within a genetically well-defined patient population, and the channel’s potential as a target for therapeutics all make SCN2A a leading candidate for focused efforts to understanding and treating neurodevelopmental disorders. Following an SCN2A-dedicated workshop organized by the Simons Foundation Autism Research Initiative (SFARI) that convened academic scientists, representatives from biotechnology and pharmaceutical companies, as well as funders and family advocacy organizations, we summarize the current state of research, future directions, and consider potential opportunities and challenges in the NaV1.2 field.
The SCN2A gene
The Sodium Channel, Voltage-Gated, Type II, Alpha (SCN2A) gene is located on the positive strand of chromosome 2 (2q24.3) in humans, between the sodium channel gene SCN3A and the nuclear protein gene CSRNP3. The sodium channel genes SCN1A and SCN9A are within 1,000 kbp downstream. The SCN2A mRNA transcript contains 27 exons that encode a 2,005 amino acid protein called NaV1.2. Due to the large last coding exon, only stop codons in the first 1,591 amino acids would be expected to induce nonsense-mediated decay (Figure 1A) [1]. There are two major developmentally regulated splice isoforms of NaV1.2 that use mutually exclusive copies of the fifth coding exon and differ by one amino acid at position 209: Asparagine (Asn/N; termed 5N) vs. Aspartic acid (Asp/D; 5A) [2,3] (Figure 1A).
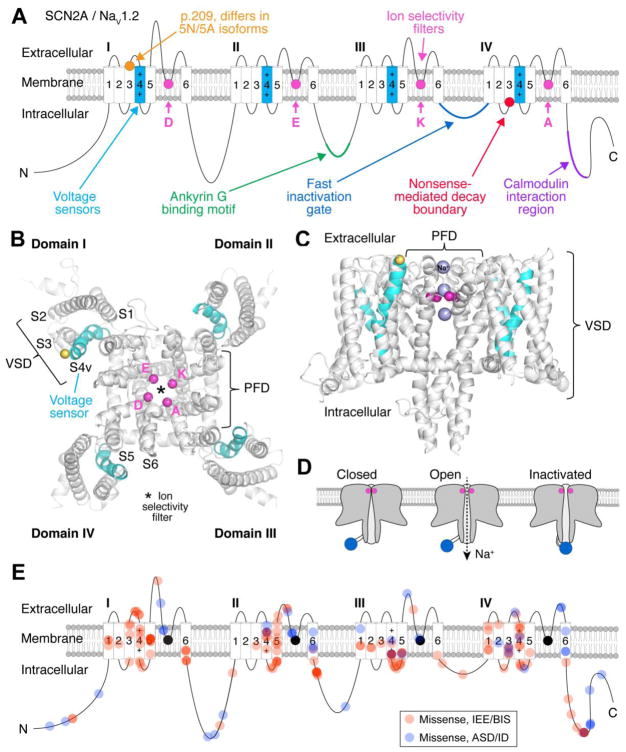
Figure 1. Overview of SCN2A/NaV1.2.
A) NaV1.2 resides in the cell membrane and is a pseudo-heterotetramer composed of four domains, called I-IV. Each domain contains a voltage sensor (cyan) and a pore loop with a ring of amino acids (DEKA) making up the ion selectivity filter (pink). The cytoplasmic loop between domains III and IV is the fast inactivation gate (dark blue, see D). Two isoforms are known, differing by one amino acid at p.209 (orange) [2]. The channel binds to Ankyrin G (ANK3), which anchors it to the membrane (green) [79], and interacts with calmodulin, which likely has a regulatory role (purple) [80]. Stop codons before the nonsense mediated decay boundary (red) are predicted to prevent protein translation, while those after this boundary may not.
B) Nav1.2 homology model of the transmembrane region, top view. Each domain is labeled. For domain I, transmembrane segments 1–6 are labeled S1–S6. S1–S4v represents the voltage sensing domain (VSD). The voltage sensor is labeled S4v and is colored cyan. S5–S6 represents the pore forming domain (PFD). The Cα carbons from each residue in the DEKA motif are represented as pink spheres. The Cα carbon of the residue that differs between the neonatal and adult isoforms is represented as a yellow sphere. The selectivity filter is represented by an asterisk. Model [81] is based on the NaV1.7 chimeric crystallographic structure [61].
C) Nav1.2 homology model of the transmembrane region, side view. Purple spheres represent sodium passing through the selectivity filter of NaV1.2.
D) NaV1.2 is closed at resting potential. Depolarization opens the channel, allowing sodium flux, after which the channel is blocked by the fast inactivation gate (blue). As the membrane voltage returns to rest the channel resets through hyperpolarization.
E) Location of missense variants observed in individuals with infantile epileptic encephalopathy and benign (familial) infantile seizures (red) and autism spectrum disorder/intellectual disability (blue). Increased color intensity represents multiple variants in proximity.
Panels A and E are based on previously published illustrations [26].
The structure and function of SCN2A / NaV1.2
SCN2A encodes the neuronal sodium channel NaV1.2, which is one of several sodium channels involved in initiation and propagation of action potentials in neurons. NaV1.2 is one of four sodium channel paralogs expressed throughout the central nervous system, along with NaV1.1 (SCN1A), NaV1.3 (SCN3A), and NaV1.6 (SCN8A). Sodium channels are pseudo-heterotetrameric protein structures consisting of four highly similar domains, termed I, II, III, and IV (Figure 1). Each domain contains 6 membrane-spanning segments S1-S6 (Figure 1) [4] with S1-S4v forming the voltage-sensing domains and S5-S6 forming the pore loops and DEKA selectivity filter. The positively charged 4th segment, called the voltage sensor or S4v, is sensitive to charge differences between the intracellular and extracellular sides of the membrane. When the intracellular electrical potential becomes less negative, the four S4v segments initiate conformational changes in the channel that lead to channel activation. Activation opens the central ion selectivity pore, permitting electrical conduction by way of sodium flux into the cell (Figure 1D). The inactivation gate then physically occludes the pore on the intracellular side, a shift in conformation termed “fast inactivation”. From here, the membrane must hyperpolarize to release the inactivation gate and recover from inactivation. While exceptions have been noted, sodium channels typically must go through all three steps to be available for subsequent activation [5].
These steps of activation, conductance, inactivation, and recovery from inactivation vary across sodium channel isoforms, both in kinetics and membrane voltage sensitivity. This is due to small differences in the amino acid sequence of the channels. Similarly, missense variants can conceivably alter the biophysical properties of the channel, especially if such variants are in parts of the protein that are important for sensing voltage (e.g., within the voltage sensing domain) or permitting ion flux (e.g., within the pore domain) (Figure 1). Furthermore, variants at essential sites for interactions with the beta subunit (e.g. SCN1B), other auxiliary proteins (e.g. calmodulin, fibroblast growth factor homologous factors), modulatory systems, or scaffolding proteins can all impact NaV1.2 activity [6]. How these variants can affect different aspects of channel function is described in detail below.
Developmental and cell-type specific patterns of NaV1.2 expression across brain regions
NaV1.2 is widely expressed throughout the human central nervous system, but not in peripheral tissues [7,8]. In cortical structures, NaV1.2 is co-expressed with NaV1.6 predominately in excitatory, glutamatergic neurons. Inhibitory interneurons largely express NaV1.1 instead of NaV1.2 [9,10], though NaV1.2 channels have been identified in some multipolar and bouquet-class interneurons in mouse [11]. NaV1.2 may co-express with NaV1.1 in the somatostatin-expressing interneuron subclass, though this observation is not consistent between human and mouse tissue [11,12]. Similar distributions across excitatory and inhibitory cells are found in the hippocampus [10,11,13].
The 5N and 5A splice isoforms of SCN2A are developmentally regulated, with 5N co-expressed with 5A in the fetal brain, eventually transitioning to neurons predominantly expressing 5A after the second postnatal week in mouse [2,14]. These isoforms influence channel voltage dependence; results indicate that the adult 5A encodes a channel that activates at lower depolarization voltages, resulting in increased neuronal activity [3,15].
In addition to alternative splicing, the subcellular distribution of NaV1.2 changes markedly over development in excitatory pyramidal cells. Early on, NaV1.2 is the only sodium channel isoform expressed in the axon initial segment (AIS) [3,16,17], an axonal subcompartment located proximal to the soma that is the site of action potential initiation [18]. In humans, this early developmental period spans from the late second trimester, when the cortex is beginning to form, to 1-2 years of age. At this time, NaV1.2 in the distal AIS, as well as the rest of the axon, is largely replaced by NaV1.6 (SCN8A), which has a lower voltage threshold for activation. Consequently, the distal AIS becomes the site of action potential initiation in mature neurons [19,20]. NaV1.2 is at this point restricted to the region of AIS most proximal to the soma, where it functions as a reserve pool of channels, right next to the cell body. Once action potentials are initiated in the AIS by NaV1.6, NaV1.2 is thought to help action potentials backpropagate into the somatodendritic compartment [21]. These backpropagating action potentials could then influence many functions, including activity dependent gene transcription, synaptic integration, and synaptic plasticity.
Another prominent site of NaV1.2 expression is the cerebellar cortex [22]. Here, NaV1.2 channels are co-expressed with NaV1.6 in unmyelinated axons of excitatory granule cells that form parallel fiber synapses on Purkinje neurons. In contrast to the developmental loss of NaV1.2 in cortical myelinated axons, NaV1.2 expression persists throughout development in the cerebellum, with an apparent peak in density within the second and third postnatal weeks in mice [22,23]. This suggests that NaV1.2 serves different roles in mature neurons in the neocortex and cerebellum.
SCN2A-mediated disorders
Variants in SCN2A are primarily associated with three distinct disorders [24,25]: 1) infantile epileptic encephalopathy (IEE), characterized by infantile-onset seizures, before 12 months of age, followed by neurodevelopmental delay; 2) benign (familial) infantile seizures (BIS), characterized by infantile-onset seizures, before 12 months of age, that resolve by 2 years of age without overt long-term neuropsychiatric sequelae; and 3) autism spectrum disorder/intellectual disability (ASD/ID), characterized by global developmental delay, particularly of social and language milestones. Up to a third of children in the ASD/ID category identified to date also develop childhood-onset seizures (≥12mths).
While most children with SCN2A variants fit into one of these three categories, there are some exceptions, including epileptic encephalopathy with choreoathetoid movements [26,27], benign infantile seizures with late-onset episodic ataxia [28–31], childhood-onset epileptic encephalopathy [32], and schizophrenia [33,34]. Further work is required to understand how these disorders relate to the other three distinct categories and the role of cerebellar NaV1.2 in ataxia, hypotonia, and other symptoms.
The 276 known SCN2A cases identified to date (Table S1) probably represent only a fraction of SCN2A cases, as more than 400 are predicted to be born each year in the USA alone (Table 1). Widespread adoption of genetic testing in IEE, ASD, and ID is expected to increase the frequency of known SCN2A cases to hundreds per year, similar to the incidence of SCN1A-mediated Dravet Syndrome at 6.4 per 100,000 births [35]. We would also expect the incidence of loss of function cases to be ~5-fold higher than gain of function cases (Table 1). At present the number of known SCN2A cases is roughly equal between IEE and ASD/ID, likely reflecting differences in the adoption of genetic testing.
Table 1.
Frequency of SCN2A-related disorders in the general population and literature
| IEE | BIS | ASD/ID | Total | |
|---|---|---|---|---|
| Cases per100,000 births | 120 [71,72] | 20 [71,72] | 2,500 [73] | NA |
| Population frequency | 0.12% | 0.02% | 2.5% | NA |
| Percent of cases with SCN2A variant | 1.2% [74] | 11% [75] | 0.3% [76] | NA |
| Estimate of SCN2A-related cases per 100,000 births | 1.2% of 120 = 1.4 | 11% of 20 = 2.2 | 0.3% of 2,500 = 7.5 | 11.1 |
| Expected SCN2A-mediated disorders born each year in the USA | 56 cases | 88 cases | 298 cases | 442 cases from IEE, BIS, and ASD/ID |
| Year SCN2A association documented | 2009 [56] | 2002 [77] | 2012 [78] | NA |
| Published variants | 107 cases | 35 families | 89 cases | 276 cases from all phenotypes |
Children with SCN2A-associated infantile epileptic encephalopathy
While epileptic encephalopathy frequently presents with seizures in the neonatal period, with some families retrospectively reporting rhythmic movements in utero, it is likely that seizure onset after three months occurs in 20%-40% of cases [24,25]. Of note, age of onset is often similar in individuals with the same genetic variant. Seizure type varies widely between individuals, with most experiencing more than one type of seizure. Some of the children with the earliest age of seizure onset are classified as having Ohtahara syndrome, based on tonic seizures and electroencephalography (EEG) burst suppression, or epilepsy of infancy with migrating focal seizures (EIMFS). In contrast, onset after three months often meets criteria for West Syndrome, with infantile spasms and a hypsarrhythmia pattern on EEG, evolving to Lennox-Gastaut syndrome with multiple seizure types as the child gets older [25]. Development varies from moderate to profound global developmental delay, often with comorbid hypotonia, microcephaly, cerebral and/or cerebellar atrophy, and cortical visual impairment. Some children also have movement disorders, including dystonia, chorea, or episodic ataxia.
Children with SCN2A-associated autism spectrum disorder/intellectual disability
In children with ASD/ID, early development is generally unremarkable up to about 6 months of age, followed by moderate motor delay and pronounced verbal delay. About a third of documented cases develop seizures, often between the age of 18 months and 4 years. These seizures are associated with more severe developmental delay and sometimes regression, particularly if the seizures are severe. Importantly, seizure prevalence is likely overestimated in the currently identified population since genetic screening is more common in the presence of syndromic features, such as seizures.
While children with SCN2A-associated ASD/ID do not exhibit a distinctive syndrome, several behavioral characteristics seem to be shared (unpublished observations, scn2a.org family group). Children are often described as placid and contented and enjoy physical contact with caregivers. They tend to engage in repetitive actions, including chewing objects, hand gestures, and roaming. Children rarely initiate social interactions and the response to social cues from others is slow. Eye contact is rarely made, and hand contact with people or novel objects is avoided. Motor skills are usually delayed to a lesser degree than social skills. Some children have signs of cerebellar involvement, including an unsteady or ataxic gait, clumsiness, and hypotonia. A case report described a muted response to pain and other aversive stimuli [36], which fits with parents’ experiences, though some report increased sensitivity to heat. Co-morbidities include cortical visual impairment, disrupted sleep, gastrointestinal disturbances, and uncoordinated oral movements.
Towards a model of SCN2A pathophysiology
An integrated analysis of genetic and electrophysiological data led to a model to account for the three disorders associated with SCN2A (Figure 2) [24]. Variants associated with greater NaV1.2 channel activity (i.e., gain of function) lead to IEE and BIS, while those associated with diminished channel activity (i.e., loss of function) lead to ASD/ID (Figure 2). Further, the degree to which the gain of function variants potentiate NaV1.2 activity distinguishes IEE from BIS, with severe variants leading to IEE and milder variants leading to BIS. This model is supported by several strands of evidence, including: 1) in vitro electrophysiology; 2) the restriction of protein truncating variants, resulting in loss of function, to ASD/ID cases; 3) shared symptoms between individuals with recurrent missense variants (Table S1); and 4) the clustering of IEE/BIS variants around the voltage sensor domain of the channel while the ASD/ID missense variants cluster near the pore loop regions (Figure 1E). The subsequent publication of 71 novel SCN2A patients from Europe [25] further support the model proposed in Ben-Shalom et al.. [26].
Figure 2. Current model of SCN2A pathophysiology.
Gain of function (GoF) variants (left) potentiate glutamatergic neuronal excitability, leading to infantile-onset seizure phenotypes, whereas loss of function (LoF) variants (right) impair excitability, leading to ASD and/or intellectual disability [24]. Seizure severity is correlated with the degree of GoF, with variants that cause the most excitability leading to infantile epileptic encephalopathy (IEE), while milder variants lead to benign (familial) infantile seizures (BIS) that resolve around 1–2 years of age without apparent neurological sequelae. The threshold that distinguishes IEE and BIS variants may be related to the degree of compensation of neuronal excitability by NaV1.6, which replaces NaV1.2 in generating action potentials and has a lower threshold for activation than wild type NaV1.2.
As with any model, there are likely to be exceptions. At least one has come to light: the variant K1422E alters the third residue in the DEKA ion selectivity filter (Figure 1) and is predicted to produce both loss of function and gain of function effects [37,38]. A child with this variant developed seizures at 13 months with developmental regression and features of ASD [39].
In summary, genotype and phenotype appear to be strongly correlated in disorders associated with variations of SCN2A. Identification of additional variants, their functional characterization and consistent phenotyping, should refine this model further, provide additional insights into SCN2A pathology and NaV1.2 function, and point to new potential therapeuticinterventions [40].
Linking disorders to changes in channel function
Several hotspots within the NaV1.2 protein are enriched with disorder-associated variants, including S4v-S5 in the voltage sensing domain, the intracellular N- and C-terminal domains, and the pore loops around the ion selectivity filter (Figure 1E). To date, only about 20 of the hundreds of disorder-associated variants have been electrophysiologically assessed (summarized in [24,25]). Nevertheless, some patterns are beginning to emerge, with variants within particular subdomains often evoking similar changes to channel biophysical properties (Table 2).
Table 2.
Electrophysiology and biophysics of SCN2A variants
| Effect on NaV1.2 channel |
Mutation type |
Common location in channel (if mutliple cases observed) |
GoF | Example | PMID | LoF | Exam ple |
PMID |
|---|---|---|---|---|---|---|---|---|
| Protein Truncatio n | Nonsense, canonical splice sites, frameshift insertion/deletions and large deletions | Anywhere in first 1,591 amino acids or 4,772 nucleotides | N/A | N/A | N/A | Mutations that prevent translation of one copy of the channel | C959X | 22495306; 28256214 |
| Gene duplication | Large duplication | Entire channel | Additional copies of channel translated | chr2:165798 270-166304847 duplication |
2.7E+ 07 | N/A | N/A | N/A |
| Non-conducting | Missense | Often clustered in the pore loop | N/A | N/A | N/A | Prevents sodium flux through channel | R379H | 25363760; 28256214 |
| Activation (voltage dependence) | Missense | Often clustered near the voltage sensor | Hyperpolarizing shifts (i.e., channel opens more readily at voltages near Vrest) | I1473M | 2E+07 | Depolarizin g shifts (more difficult to open channels for any given voltage) | D82G | 28256214 |
| Inactivation (voltage dependence) | Missense | Often clustered near base of channel, especially in intracellular region that links transmembrane domain 4 and 5. | Depolarizing shifts (i.e., more channels available for activation at resting membrane potential) | L1330F | 1.7E+07 | Hyperpolarizing shifts (fewer channels available for activation) | P1622S | 28379373 |
| Activation (kinetics) | Missense | Similar to voltage dependence | Faster channel activation | L1563V | 1.7E+07 | Slower channel activation | R1319Q | 17021166 |
| Fast Inactivation (kinetics) | Missense | Typically interacting with the “Ball and chain” inactivation gate between subdomain s III and IV, but can also be associated with voltage sensing components | Slower channel inactivation | F1597L | 2.8E+07 | Faster channel inactivation | D12N | 28256214 |
| Slow inactivation (kinetics) | Missense | Similar to voltage dependence | Faster recovery from inactivation | V261M | 2E+07 | Slower recovery from inactivation | Unknown | |
| Persistent current | Missense | Usually related to shifts in activation and inactivation that increase the size of the window current | Excess sodium flux at rest | M252V | 2.1E+07 | Unknown | N/A | N/A |
| Omega current | Missense | Arginine residues lining S4 transmembrane domain, may result in a mix of GoF and LoF aspects | Unknown | N/A | N/A | Unknown | N/A | N/A |
| Ion selectivity | Missense | Ion selectivity filter - formed by the apposition of D, E, K, and A residues, one from each repeat | Channel seems to have gained a persistent calcium current when at rest. | K1422E | 1313551; 9035373 | Overall conductance is reduced when active and non-selective for cations | K1422E | 1313551; 9035373 |
| Trafficking | Missense | Unknown, but Ankyrin G binding domain is likely | Unknown | N/A | N/A | Unknown | N/A | N/A |
| Channel regulation | Missense | C-terminus calmodulin regulatory domain | Unknown | N/A | N/A | Disrupted Ca-dependent regulation of voltage dependence | R1902C | 15316014 |
While each aspect of NaV1.2 function is listed separately in Table 2, it is quite common for one point variant to alter multiple channel properties. In some cases, the concerted changes to voltage dependence and kinetics are consistent, permitting easy interpretation of whether the variant enhances or decreases channel activity (i.e., gain- and loss-of-function, respectively). In other cases, a mix of what appear to be gain- and loss-of-function effects are observed, and it is more difficult to understand how neuronal excitability is affected. Compartmental modeling can provide insight into the net effect on neuronal excitability in such conditions [24]. Even with these efforts, it can be difficult to place certain variants into “gain” or “loss” categories. For example, some IEE-associated variants hyperpolarize the voltage dependence of activation to such a degree that neurons may end up being less excitable over time, as sodium channels may be activated at rest, but the neuron never hyperpolarizes to allow the same channels to recover from inactivation. This variant is characterized as a “gain-of-function” variant when describing channel biophysics, but this description may not hold at the level of overall neuronal excitability. Similarly, a variant at the ion selectivity filter (e.g., p.K1422E) can have mixed effects on excitability. This variant converts the channel into a non-selective cation channel with increased persistent, Ca2+-dominated current and reduced, non-selective conductance when activated [37,38]. Moreover, some variants can lead to an “omega current”, which stems from ion permeation through the S4v region when the central pore the channel is closed [41]. Developing models that capture net neuronal excitability effects in such cases is quite difficult, especially considering potential downstream effects on Ca2+-mediated cell signaling.
SCN2A homology in animal models and other channels
Voltage-gated sodium channels (NaV) are derived from a calcium channel (CaV3) early in the evolution of animals [42–44] with the DEKA ion selectivity filter in NaV1 being derived from the ancestral DEEA filter (Figure 3, Table 3). Of note, the variant p.K1422E, described earlier (Table 2), reverts the NaV1.2 filter to the ancestral state. The ten voltage-gated sodium channels in humans are derived from this NaV1 channel (Figure 3) [45–47] with different model organisms reflecting stages in this differentiation (Table 3). NaV1.2 retains a high degree of homology to both NaV1.1 and NaV1.3 and moderate homolog to NaV1.4, NaV1.5, NaV1.6, and NaV1.7 (Figure 3B). Loss of function variants of NaV1.1 cause Dravet syndrome [48], generalized epilepsy with febrile seizures plus [49], and have also been associated with ASD [50,51]. NaV1.5 is expressed in the heart, and its disruption can lead to cardiac arrhythmias, including heart block, long QT, Brugada syndrome, and cardiomyopathy [52]. Therapies aimed at modifying SCN2A/NaV1.2 function will need to be selective for NaV1.2 to avoid off target activity with these other sodium channels within both the brain and the periphery to prevent potential side effects.
Figure 3. Homology of SCN2A across model organisms and voltage-gated sodium channels.
A) Voltage-gated sodium channels (NaV) are derived from T-type voltage-gated calcium channels (CaV3) [42,43]. In fruit flies the ancestral NaV2 channel, with the ion selectivity filter composed of the four amino acids DEEA, co-exists with the NaV1 channel with the DEKA filter. All NaV channels in humans are derived from this NaV1 DEKA filter, while the NaV2 filter has been lost [45,46]. A series of gene duplication and differentiation events give rise to the ten channels seen in humans and reflected in the proximity of similar channels in the genome [45,46].
B) Homology between the 10 NaV channels in humans. A more intense shade of blue indicates higher homology. *Zebrafish have two copies of each of these four NaV channels due to relatively recent genome duplication. CNS: Central nervous system; PNS: Peripheral nervous system; GTEx: Genotype-Tissue Expression Project (www.gtexportal.org).
Table 3.
SCN2A homologues in model organisms
| Species | Number of NaV channel isoforms | Closest human NaV1.2homologue | Percent homology to human NaV1.2 |
|---|---|---|---|
| Yeast (Saccharomyces cerevisiae) | 0 | N/A | N/A |
| Worm (Caenorhabditis elegans) | 0 | N/A | N/A |
| Fruit Fly (Drosophila melanogaster) | 2 | Para / Para | 70% |
| Zebrafish (Danio rerio) | 4x2* | Scn1lab / Nav1.1Lb | 86% |
| Frog (Xenopus Tropicalis) | 6 | Scn2a / Nav1.2 | 85% |
| Chicken (Gallus gallus) | 9 | SCN2A / NaV1.2 | 93% |
| Mouse (Mus musculus) | 10 | Scn2a / NaV1.2 | 98% |
| Rat (Rattus norvegicus) | 10 | Scn2a / NaV1.2 | 98% |
| Rhesus (Macaca mulatta) | 10 | Scn2a / NaV1.2 | 99.9% |
| Human (Homo sapiens) | 10 | SCN2A / NaV1.2 | 100% |
Zebrafish have two copies of each of these four NaV channels due to relatively recent genome duplication.
Existing treatments
Seizures associated with SCN2A variants often cannot be controlled even with the use of multiple anti-epileptic drugs (AEDs). However, for infants with postnatal-onset seizures (≤3 months), non-selective sodium channel blockers, such as phenytoin and carbamazepine, are more effective [25], as would be expected based on a gain of function variant [24]. Of note, this is the opposite of current guidelines for empiric neonatal seizure treatment [53] and is a compelling reason for rapid genetic testing for this condition [54,55]. For children with ASD/ID and childhood-onset seizures (≥12 months), the opposite is true, with non-sodium channel inhibiting AEDs offering the best options, including levetiracetam, benzodiazepines, and valproate. For seizures starting between 3 months and 12 months there are limited SCN2A-specific data, so standard AED guidelines are recommended [53]. However, the gain of function nature of these variants suggest sodium channel inhibitors would be more efficacious. Some reports have described clinical improvement with less mainstream medications, including lidocaine in children with infantile epileptic encephalopathy [56,57], ethosuximide for late-onset seizures [25], and acetazolamide for episodic ataxia [28,29], though data on the generalizability of these effects are limited. This effort to think broadly about medications parallels new and emerging therapies that provide benefit in Dravet syndrome, such as bromides, fenfluramine, and cannabidiol [58]. Whether such medications would provide benefit with SCN2A-associated late onset seizure remains unknown. When considering such treatments, it is critical to remember that, at least in cortical regions, NaV1.1 channels affected in Dravet are more commonly expressed in inhibitory neurons [59], and that loss of function with NaV1.1 may have opposing effects on brain networks compared to NaV1.2 loss of function in excitatory neurons.
At present, no SCN2A-specific recommendations can be made for the treatment of other symptoms. Standard management for children with global developmental delay, ASD, and co-morbid disorders should be followed.
Future treatments
There is a clear unmet medical need for improved treatment options for SCN2A-associated disorders. Based on the current model of SCN2A pathology (see Figure 2), restoring NaV1.2 channel function to the “normal” range may provide therapeutic benefit for individuals with both classes of disorders (gain and loss of function). Since there is precedent for small molecule compounds that either enhance or diminish ion channel activity [60–62], it is conceivable to develop drugs for modulating NaV1.2 activity to treat IEE and ASD/ID. However, this approach has potential associated challenges. First, the high degree of conservation (Figure 3B) between SCN2A/NaV1.2 and other voltage-gated sodium channels, including SCN5A/NaV1.5 that can induce cardiac arrhythmias if disrupted, demonstrates that high selectivity for NaV1.2 will be essential to reduce side effects. Second, the SCN2A pathology will likely require normalizing the NaV1.2 channel current while avoiding overly enhancing or decreasing NaV1.2 channel activity that could result in IEE or ASD/ID symptoms, respectively. In addition, biophysical characterization of unknown patient-specific variants may be necessary to confirm the appropriate therapeutic intervention, especially in cases where mixes of GoF and LoF characteristics are present (Table 2).
An additional consideration is the reversibility of the symptoms. SCN2A expression begins early in gestation [8], and channel dysfunction may alter brain development. In keeping with this concern, computational modeling of loss of function variants suggests that neuronal excitability is restored in the mature brain as NaV1.6 replaces NaV1.2 as the critical channel underlying action potential generation at two years of age [24]. Despite this proposed functional compensation in action potential initiation, symptoms of ASD/ID continue beyond this age. It will therefore be critical to understand the physiological changes that persist within the brain in individuals with loss of function variants and whether these changes can be rescued by restoring NaV1.2 function. Similarly, the encephalopathy in gain of function variants may be secondary to seizures and tractable to treatment, alternatively seizures may simply be one feature of a more complex neurodevelopmental disorder that begins in utero.
To address these questions, and to understand where and when therapeutic interventions can occur, more advanced experimental tools must be applied. Transgenic rodent models, such as mice, rats, and voles, provide the opportunity to assay neurons in the context of the brain to capture non-autonomous, circuit-level, and behavioral effects. Understanding how alterations in NaV1.2 function affect developing and mature neuronal integration, and then understanding how this affects network function, is key to enable effective interventions. Further, experiments equivalent to the studies of symptom reversibility in MECP2 knockout mice [63] are critical for both loss and gain of function phenotypes.
Beyond rodent models, additional models and approaches can be brought to bear. Patient-derived induced pluripotent stem cell (iPSC)-based approaches including neuronal cultures [64,65] and organoids [66,67] hold great promise in this regard, as they would allow researchers to probe for the functional effects of SCN2A genetic variants on many levels including neuronal excitability, developmental progression and network behavior. Importantly, this can be done in human neurons, under physiological expression levels of the channel and under each patient’s unique genetic background. Further, studies in non-human primates may provide a better understanding of how therapeutics that prove beneficial in rodent models translate to more complex systems [68].
Towards personalized therapeutics
To develop treatments for SCN2A-mediated symptoms beyond epilepsy it is critical to advance the understanding of both gain and loss of function phenotypes. More specifically, observational studies are needed to characterize the natural course of the diseases and to identify endpoints for future interventional trials. Analysis of data from publications and clinical cohorts has already provided insights into genotype-phenotype relationships and appropriate antiepileptic drugs [24,25]. Further progress will require collaboration between scientists and families, including the participation of families in the FamilieSCN2A Foundation (www.scn2a.org) and the associated SCN2A patient registry (https://simonsvipconnect.org). There is room for further development of the tools for collecting and analyzing SCN2A patient data including greater data consistency, longitudinal, rather than cross sectional, phenotypes [69], and detailed records of the medications used and their efficacy and side effects. Furthermore, biomarkers that directly reflect the disease pathophysiology and could serve as surrogate marker for efficacious treatments – optimally translating to pre-clinical animal models – would be of great utility. To this end, electroencephalography, which directly measures electrophysiological activity, may well reflect NaV1.2 dysfunction. Alongside these phenotype data, it will be necessary to perform functional analyses and develop a database to record these data and outcomes (Table 2), possibly in concert with homologous sodium channels (Figure 3). The implementation of methods for performing high-throughput analysis of the impact of variants on channel function would accelerate this process considerably.
Concluding remarks
SCN2A variants can lead to at least two severe disorders, with gain of function leading to infantile-onset seizures and encephalopathy, and loss of function leading to autism spectrum disorder and/or intellectual disability. Normalization of NaV1.2 function has great potential to yield therapeutic benefit in both conditions, though the issues of therapeutic window and selectivity to other sodium channels must be addressed. Concurrent development of such therapies, alongside basic science to understand the neurological impact and reversibility of these variants, is essential (see Outstanding Questions). The results of these endeavors will have direct implications for families affected by SCN2A variants. In line with the principle that rare genetic disorders may inform therapeutic strategies for more common disorders [70], we hope that insights gleaned from SCN2A may also have wider implications for epileptic encephalopathy, autism spectrum disorder, and intellectual disability.
Outstanding Questions Box.
What is the contribution of NaV1.2 to neuronal processing across brain regions?
In which cell classes, and in which cellular compartments, are NaV1.2 channels expressed? How is their expression developmentally regulated across cell classes and within compartments?
How do alterations in NaV1.2 function produce particular phenotypes within these cells and brain regions?
Do changes in NaV1.2 exert cell-autonomous effects, or is the resulting dysfunction more of a circuit-level phenomenon stemming from interactions between cells?
To what extent does the etiology of SCN2A-associated disorders overlap with other neurodevelopmental disorders?
What is the full range of symptomatology in SCN2A-associated disorders? And do patients with loss of function mutations truly have an increased rate of seizures, or is this an ascertainment bias in the current data?
The current SCN2A genotype-phenotype model does not explain choreoathetoid movements, episodic ataxia, or late-onset epileptic encephalopathy, and provides only limited insight into the variation in symptom severity or treatment response. Consistent genetic, functional, and phenotype data for individuals with SCN2A-associated disorders would help refine this model.
Why do sodium channel blockers improve seizure control only in some of the individuals with SCN2A-associated early onset seizures? Why don’t sodium channel blockers improve seizure control in SCN2A-associated late onset seizures?
What are the therapeutic windows for treating SCN2A-associated disorders? Are there critical periods of neuronal development in which intervention must occur?
Supplementary Material
Highlights.
Dysfunction in SCN2A has been recently recognized as a major cause of neurodevelopmental disorders (NDD), including epilepsy, intellectual disability (ID), and autism spectrum disorder (ASD).
SCN2A encodes a neuronal sodium channel, NaV1.2. In contrast to many other NDD-linked genes, there is a well-established body of literature on sodium channel function and dysfunction.
Loss of NaV1.2 function contributes to ASD and ID, whereas gain of function contributes to early onset epilepsy. Such strong and bidirectional genotype-phenotype correlation is rare in brain disorders.
Sodium channel function can be enhanced or suppressed using pharmacology. This may allow for future, targeted treatment of SCN2A-associated disorders.
The neuropathophysiology revealed by investigating SCN2A may provide insight into the underlying biology of NDDs more generally.
Acknowledgments
This work was supported by grants from the Simons Foundation (SFARI #402281, SJS; #513133, KJB; #491201, ALG; # 349984, GP), the NIH (NIMH#100047, RAB), and the Stanley Center for Psychiatric Research at the Broad Institute (AJC, JRC, DL and FFW). We thank Markus Wolff and Katrine Johannesen for their help in combining the lists of published SCN2A variants (Table S1).
Glossary
- Action potential
an electrical signal used by neurons to transmit information from one neuron to the next. Action potentials are mediated largely by the opening of channels that allow positive sodium ions into the cell, “depolarizing” the “membrane potential,” then the opening of potassium channels that allow positive ions to leave the cell, “hyperpolarizing” the membrane potential. Variants in sodium channels that mediate action potentials can affect their initiation or propagation.
- Action potential threshold
the voltage at which a neuron will generate an action potential. This value is dynamic and depends on the sodium channel isoforms expressed within the neuron and the recent electrical activity of the neuron. Threshold can also be affected by the number of channels expressed on the neuron membrane.
- Brugada syndrome
A heart rhythm disorder that can lead to ventricular arrhythmias and sudden death. It is frequently caused by SCN5A mutations.
- Depolarize/Hyperpolarize/Repolarize
The opening of channels that allow positive ions into the cell, such as sodium channels, are said to “depolarize” the membrane potential, as they bring the membrane potential closer to 0 mV, or past 0 mV into the positive range. Conversely, ion channels that allow positive ions to leave the cell (e.g. potassium channels) “hyperpolarize” the membrane potential away from 0 mV and back towards resting membrane potential. This is often called “repolarization,” especially in the context of action potential generation.
- Dravet syndrome
An epileptic encephalopathy that presents with prolonged seizures, often alternating hemiconvulsions provoked by fever, in the first year of life. The interictal EEG and development are normal at onset, but signs of regression appear in the second year of life and are often accompanied by convulsive status epilepticus, myoclonic seizures, and EEG abnormalities. The majority of cases are caused by loss of function SCN1A mutations, which encodes NaV1.1, a sodium channel expressed in inhibitory interneurons.
- EEG
Electroencephalography. Recording of cortical electrical activity by a net of electrodes placed on the scalp.
- Exon
A segment of a gene that is not removed during splicing; these often contain the information to determine the order of amino acids in a protein.
- Expression
The amount of RNA produced from a gene in a given tissue or cell.
- Isoform
Versions of the RNA from a gene, often due to the inclusion of different exons; also called alternative splicing.
- Gain of function
When a genetic variant in an ion channel makes the channel more excitable. One example is when a variant hyperpolarizes the voltage required for channel opening.
- Gene
A region of DNA that is transcribed into RNA, e.g. SCN2A. For protein coding genes, this RNA is spliced, keeping exons and removing introns, then translated to determine the order of amino acids in a protein (e.g. NaV1.2) by a ribosome.
- Generalized epilepsy with febrile seizures plus (GEFS+)
The definition of this syndrome, previously known as “Generalized” Epilepsy with Febrile Seizures Plus, has changed over the past 10 years since its initial description. Currently, this is considered a childhood onset familial syndrome that encompasses several phenotypes, characterized by a spectrum of severity, that include combinations of febrile seizures, focal seizures, and generalized seizures.
- Lennox-Gastaut syndrome
This severe epileptic encephalopathy has typical onset in early childhood (3–5 years old) and is characterized by multiple seizure types (nocturnal tonic seizures and atonic seizures are typical) refractory to treatments, diffuse “slow” spike-and-wave and paroxysmal fast activity (EEG pattern), and intellectual disability.
- Loss of function
When a genetic variant in an ion channel lowers ion channel excitability, or destroys channel function altogether. Examples of this include “nonsense mediated decay,” a block of the pore, or changes in voltage dependence that make it harder for a channel to open.
- Membrane potential
The voltage differential between the inside and outside of a neuron’s membrane. Neurons that are not actively generating action potentials typically have a “resting membrane potential” between −60 to −90 mV (millivolts).
- Missense
A DNA variant that alters a single amino acid in a protein. This can have no impact on function, impair function, enhance function, or create new effects.
- Nonsense-mediated decay
A process that destroys RNA if there is a premature stop codon, preventing a protein being made.
- Ohtahara Syndrome
Also known as Early Infantile Epileptic Encephalopathy (EIEE), this severe epileptic encephalopathy has typical onset ≤3 months old and is characterized by intractable tonic seizures and burst suppression on EEG. Patients with EIEE may evolve into either West of Lennox-Gastaut syndromes with age.
- Protein truncating variant
A DNA variant that results in a premature stop codon, disrupting that copy of the gene. This includes nonsense, canonical splice site, and frameshift variants.
- West Syndrome
Also known as Infantile Spasms (IS), this severe epileptic encephalopathy has typical onset between 3–12 months old and is characterized by the classic triad of infantile spasms (seizure type), hypsarhythmia (EEG pattern), and developmental regression. Patients with IS may evolve into Lennox-Gastaut syndrome with age.
Footnotes
Disclosures: AJC has consulted for Praxis Precision Medicines. ALG is a member of a CNS scientific advisory board for Otsuka Pharmaceuticals. At the time of the workshop, TSO was an employee of F. Hoffmann-La Roche, Ltd. DH, JFH, and OK are full-time employees of F. Hoffmann-La Roche, Ltd. JRE is an employee and shareholder Xenon Pharmaceuticals. SP is a shareholder of RogCon Biotechnology and Praxis Precision Medicines.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 2.Kasai N, et al. Genomic structures of SCN2A and SCN3A - Candidate genes for deafness at the DFNA16 locus. Gene. 2001;264:113–122. doi: 10.1016/s0378-1119(00)00594-1. [DOI] [PubMed] [Google Scholar]
- 3.Gazina EV, et al. “Neonatal” Nav1.2 reduces neuronal excitability and affects seizure susceptibility and behaviour. Hum Mol Genet. 2015;24:1457–1468. doi: 10.1093/hmg/ddu562. [DOI] [PubMed] [Google Scholar]
- 4.De Lera Ruiz M, Kraus RL. Voltage-Gated Sodium Channels: Structure, Function, Pharmacology, and Clinical Indications. J Med Chem. 2015;58:7093–7118. doi: 10.1021/jm501981g. [DOI] [PubMed] [Google Scholar]
- 5.Armstrong CM. Na channel inactivation from open and closed states. Proc Natl Acad Sci U S A. 2006;103:17991–17996. doi: 10.1073/pnas.0607603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang TN, et al. Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci. 2014;17:240–7. doi: 10.1038/nn.3626. [DOI] [PubMed] [Google Scholar]
- 7.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catterall WA, et al. NaV1.1 channels and epilepsy. J Physiol. 2010;588:1849–59. doi: 10.1113/jphysiol.2010.187484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogiwara I, et al. Nav1.1 localizes to axons of parvalbumin-positive inhibitory interneurons: a circuit basis for epileptic seizures in mice carrying an Scn1a gene mutation. J Neurosci. 2007;27:5903–5914. doi: 10.1523/JNEUROSCI.5270-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamagata T, et al. Nav1.2 is expressed in caudal ganglionic eminence-derived disinhibitory interneurons: Mutually exclusive distributions of Nav1.1 and Nav1.2. Biochem Biophys Res Commun. 2017;491:1070–1076. doi: 10.1016/j.bbrc.2017.08.013. [DOI] [PubMed] [Google Scholar]
- 12.Tian C, et al. Molecular identity of axonal sodium channels in human cortical pyramidal cells. Front Cell Neurosci. 2014;8:297. doi: 10.3389/fncel.2014.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorincz A, Nusser Z. Molecular Identity of Dendritic Voltage-Gated Sodium Channels. Science. 2010;328:906–909. doi: 10.1126/science.1187958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gazina EV, et al. Differential expression of exon 5 splice variants of sodium channel alpha subunit mRNAs in the developing mouse brain. Neuroscience. 2010;166:195–200. doi: 10.1016/j.neuroscience.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Xu R, et al. A childhood epilepsy mutation reveals a role for developmentally regulated splicing of a sodium channel. Mol Cell Neurosci. 2007;35:292–301. doi: 10.1016/j.mcn.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Boiko T, et al. Functional specialization of the axon initial segment by isoform-specific sodium channel targeting. TL - 23. J Neurosci. 2003;23:2306–2313. doi: 10.1523/JNEUROSCI.23-06-02306.2003. VN-r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Osorio N, et al. Differential targeting and functional specialization of sodium channels in cultured cerebellar granule cells. J Physiol. 2005;569:801–816. doi: 10.1113/jphysiol.2005.097022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bender KJ, Trussell LO. The physiology of the axon initial segment. Annu Rev Neurosci. 2012;35:249–265. doi: 10.1146/annurev-neuro-062111-150339. [DOI] [PubMed] [Google Scholar]
- 19.Kole MHP, et al. Action potential generation requires a high sodium channel density in the axon initial segment. Nat Neurosci. 2008;11:178–186. doi: 10.1038/nn2040. [DOI] [PubMed] [Google Scholar]
- 20.Kole MHP, Stuart GJ. Signal Processing in the Axon Initial Segment. Neuron. 2012;73:235–247. doi: 10.1016/j.neuron.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Hu W, et al. Distinct contributions of Na(v)1.6 and Na(v)1.2 in action potential initiation and backpropagation. Nat Neurosci. 2009;12:996–1002. doi: 10.1038/nn.2359. [DOI] [PubMed] [Google Scholar]
- 22.Martínez-Hernández J, et al. Polarised Localisation of the Voltage-Gated Sodium Channel Nav1.2 in Cerebellar Granule Cells. The Cerebellum. 2013;12:16–26. doi: 10.1007/s12311-012-0387-1. [DOI] [PubMed] [Google Scholar]
- 23.Liao Y, et al. Molecular correlates of age-dependent seizures in an inherited neonatal-infantile epilepsy. Brain. 2010;133:1403–1414. doi: 10.1093/brain/awq057. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Shalom R, et al. Opposing effects on NaV1.2 function underlie differences between SCN2A variants observed in individuals with autism spectrum disorder or infantile seizures. Biol Psychiatry. 2017;82:1–9. doi: 10.1016/j.biopsych.2017.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wolff M, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. 2017 doi: 10.1093/brain/awx054. [DOI] [PubMed] [Google Scholar]
- 26.Samanta D, Ramakrishnaiah R. De novo R853Q mutation of SCN2A gene and West syndrome. Acta Neurol Belg. 2015 doi: 10.1007/s13760-015-0454-8. [DOI] [PubMed] [Google Scholar]
- 27.Kobayashi Y, et al. High prevalence of genetic alterations in early-onset epileptic encephalopathies associated with infantile movement disorders. Brain Dev. 2016;38:285–292. doi: 10.1016/j.braindev.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 28.Liao Y, et al. SCN2A mutation associated with neonatal epilepsy, late-onset episodic ataxia, myoclonus, and pain. Neurology. 2010;75:1454–1458. doi: 10.1212/WNL.0b013e3181f8812e. [DOI] [PubMed] [Google Scholar]
- 29.Leach EL, et al. Episodic ataxia associated with a de novo SCN2A mutation. Eur J Paediatr Neurol. 2016;20:772–776. doi: 10.1016/j.ejpn.2016.05.020. [DOI] [PubMed] [Google Scholar]
- 30.Schwarz N, et al. Mutations in the sodium channel gene SCN2A cause neonatal epilepsy with late-onset episodic ataxia. J Neurol. 2016;263:334–343. doi: 10.1007/s00415-015-7984-0. [DOI] [PubMed] [Google Scholar]
- 31.Johannesen KM, et al. Letter to the editor: confirming neonatal seizure and late onset ataxia in SCN2A Ala263Val. J Neurol. 2016;263:1459–1460. doi: 10.1007/s00415-016-8149-5. [DOI] [PubMed] [Google Scholar]
- 32.Howell KB, et al. SCN2A encephalopathy: A major cause of epilepsy of infancy with migrating focal seizures. Neurology. 2015;85:958–66. doi: 10.1212/WNL.0000000000001926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carroll LS, et al. Mutation screening of SCN2A in schizophrenia and identification of a novel loss-of-function mutation. Psychiatr Genet. 2016;26:60–5. doi: 10.1097/YPG.0000000000000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu YW, et al. Incidence of Dravet Syndrome in a US Population. Pediatrics. 2015;136:e1310–e1315. doi: 10.1542/peds.2015-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tavassoli T, et al. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med Genet. 2014;15:35. doi: 10.1186/1471-2350-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heinemann SH, et al. Calcium channel characteristics conferred on the sodium channel by single mutations. Nature. 1992;358:417–421. doi: 10.1038/356441a0. [DOI] [PubMed] [Google Scholar]
- 38.Schlief T, et al. Pore properties of rat brain II sodium channels mutated in the selectivity filter domain. Eur Biophys J. 1996;25:75–91. doi: 10.1007/s002490050020. [DOI] [PubMed] [Google Scholar]
- 39.Sundaram SK, et al. SCN2A mutation is associated with infantile spasms and bitemporal glucose hypometabolism. Pediatr Neurol. 2013;49:46–49. doi: 10.1016/j.pediatrneurol.2013.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.George AL. Lessons learned from genetic testing for channelopathies. Lancet Neurol. 2014;13:1068–1070. doi: 10.1016/S1474-4422(14)70123-1. [DOI] [PubMed] [Google Scholar]
- 41.Sokolov S, et al. Ion permeation through a voltage- sensitive gating pore in brain sodium channels having voltage sensor mutations. Neuron. 2005;47:183–189. doi: 10.1016/j.neuron.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 42.Anderson PAV, Greenberg RM. Phylogeny of ion channels: Clues to structure and function. Comp Biochem Physiol - B Biochem Mol Biol. 2001;129:17–28. doi: 10.1016/s1096-4959(01)00376-1. [DOI] [PubMed] [Google Scholar]
- 43.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol. 2001;63:871–94. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- 45.Widmark J, et al. Differential evolution of voltage-gated sodium channels in tetrapods and teleost fishes. Mol Biol Evol. 2011;28:859–871. doi: 10.1093/molbev/msq257. [DOI] [PubMed] [Google Scholar]
- 46.Liebeskind BJ, et al. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci U S A. 2011;108:9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moran Y, et al. Evolution of voltage-gated ion channels at the emergence of Metazoa. J Exp Biol. 2015;218:515–525. doi: 10.1242/jeb.110270. [DOI] [PubMed] [Google Scholar]
- 48.Claes L, et al. De Novo Mutations in the Sodium-Channel Gene SCN1A Cause Severe Myoclonic Epilepsy of Infancy. Am J Hum Genet. 2001;68:1327–1332. doi: 10.1086/320609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Escayg A, et al. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343–345. doi: 10.1038/74159. [DOI] [PubMed] [Google Scholar]
- 50.Berkvens JJL, et al. Autism and behavior in adult patients with Dravet syndrome (DS) Epilepsy Behav. 2015;47:11–16. doi: 10.1016/j.yebeh.2015.04.057. [DOI] [PubMed] [Google Scholar]
- 51.Han S, et al. Autistic-like behaviour in Scn1a+/ mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–390. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George ALJ. Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilmshurst JM, et al. Summary of recommendations for the management of infantile seizures: Task Force Report for the ILAE Commission of Pediatrics. Epilepsia. 2015;56:1185–1197. doi: 10.1111/epi.13057. [DOI] [PubMed] [Google Scholar]
- 54.Berg AT, et al. Early-Life Epilepsies and the Emerging Role of Genetic Testing. JAMA Pediatr. 2017;171:863–871. doi: 10.1001/jamapediatrics.2017.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shellhaas RA, et al. Profile of neonatal epilepsies: Characteristics of a prospective US cohort. Neurology. 2017;89:893–899. doi: 10.1212/WNL.0000000000004284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ogiwara I, et al. De novo mutations of voltage-gated sodium channel alphaII gene SCN2A in intractable epilepsies. Neurology. 2009;73:1046–1053. doi: 10.1212/WNL.0b013e3181b9cebc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Foster LA, et al. Pediatric Neurology Infantile Epileptic Encephalopathy Associated With SCN2A Mutation Responsive to Oral Mexiletine. Pediatr Neurol. 2017;66:108–111. doi: 10.1016/j.pediatrneurol.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 58.Wirrell EC. Treatment of Dravet Syndrome. Can J Neurol Sci. 2016;43:S13–S18. doi: 10.1017/cjn.2016.249. [DOI] [PubMed] [Google Scholar]
- 59.Lorincz A, Nusser Z. Cell-type-dependent molecular composition of the axon initial segment. J Neurosci. 2008;28:14329–14340. doi: 10.1523/JNEUROSCI.4833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McCormack K, et al. Voltage sensor interaction site for selective small molecule inhibitors of voltage-gated sodium channels. PNAS. 2013;110:E2724–32. doi: 10.1073/pnas.1220844110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahuja S, et al. Structural basis of Nav1.7 inhibition by an isoform-selective small-molecule antagonist. Science (80- ) 2015;350:aac5464. doi: 10.1126/science.aac5464. [DOI] [PubMed] [Google Scholar]
- 62.Frederiksen K, et al. A small molecule activator of Nav1.1 channels increases fast-spiking interneuron excitability and GABAergic transmission in vitro and has anti-convulsive effects in vivo. Eur J Neurosci. 2017;46:1887–1896. doi: 10.1111/ejn.13626. [DOI] [PubMed] [Google Scholar]
- 63.Guy J, et al. Reversal of Neurological Defects in a Mouse Model of Rett Syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ichida JK, Kiskinis E. Probing disorders of the nervous system using reprogramming approaches. EMBO J. 2015;34:1456–1477. doi: 10.15252/embj.201591267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muffat J, et al. CNS disease models with human pluripotent stem cells in the CRISPR age. Curr Opin Cell Biol. 2016;43:96–103. doi: 10.1016/j.ceb.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 66.Quadrato G, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Birey F, et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Izpisua Belmonte JC, et al. Brains, Genes, and Primates. Neuron. 2015;86:617–631. doi: 10.1016/j.neuron.2015.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bernier R, et al. Developmental trajectories for young children with 16p11.2 copy number variation. Am J Med Genet Part B Neuropsychiatr Genet. 2017;174:367–380. doi: 10.1002/ajmg.b.32525. [DOI] [PubMed] [Google Scholar]
- 70.Zhao Z, et al. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. Am J Hum Genet. 2006;79:514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ronen GM, et al. The epidemiology of clinical neonatal seizures in Newfoundland: A population-based study. J Pediatr. 1999;134:71–75. doi: 10.1016/s0022-3476(99)70374-4. [DOI] [PubMed] [Google Scholar]
- 72.Gaily E, et al. Incidence and outcome of epilepsy syndromes with onset in the first year of life: A retrospective population-based study. Epilepsia. 2016;57:1594–1601. doi: 10.1111/epi.13514. [DOI] [PubMed] [Google Scholar]
- 73.Christensen DL, et al. Prevalence and Characteristics of Autism Spectrum Disorder Among Children Aged 8 Years - Autism and Developmental Disabilities Monitoring Network, 11 Sites, United States, 2012. Morb Mortal Wkly report Surveill Summ. 2016;65:1–23. doi: 10.15585/mmwr.ss6503a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.EuroEPINOMICS-RES Consortium et al. De Novo Mutations in Synaptic Transmission Genes Including DNM1 Cause Epileptic Encephalopathies. Am J Hum Genet. 2014;95:360–370. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zara F, et al. Genetic testing in benign familial epilepsies of the first year of life: Clinical and diagnostic significance. Epilepsia. 2013;54:425–436. doi: 10.1111/epi.12089. [DOI] [PubMed] [Google Scholar]
- 76.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–1233. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Heron SE, et al. Sodium-channel defects in benign familial neonatal-infantile seizures. Lancet. 2002;360:851–852. doi: 10.1016/S0140-6736(02)09968-3. [DOI] [PubMed] [Google Scholar]
- 78.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bouzidi M, et al. Interaction of the Nav1.2a subunit of the voltage-dependent sodium channel with nodal ankyrinG. In vitro mapping of the interacting domains and association in synaptosomes. J Biol Chem. 2002;277:28996–29004. doi: 10.1074/jbc.M201760200. [DOI] [PubMed] [Google Scholar]
- 80.Mori M, et al. Novel interaction of the voltage-dependent sodium channel (VDSC) with calmodulin: Does VDSC acquire calmodulin-mediated Ca2+-sensitivity? Biochemistry. 2000;39:1316–1323. doi: 10.1021/bi9912600. [DOI] [PubMed] [Google Scholar]
- 81.Campbell A, et al. Improving our structural understanding of Nav1.2 missense mutations. [Accessed: 12.-Mar-2018];FamilieSCN2A Foundation Conference. 2016 [Online]. Available: www.scn2a.org/2016presentations.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.