Abstract
Bacterial resistance to antibiotics has made it necessary to resort to antibiotics that have considerable toxicities. Here, we show that the cyclic 9-amino acid peptide CARGGLKSC (CARG), identified via phage display on Staphylococcus aureus (S. aureus) bacteria and through in vivo screening in mice with S. aureus-induced lung infections, increases the antibacterial activity of CARG-conjugated vancomycin-loaded nanoparticles in S. aureus-infected tissues and reduces the needed overall systemic dose, minimizing side effects. CARG binds specifically to S. aureus bacteria but not Pseudomonas bacteria in vitro, selectively accumulates in S. aureus-infected lungs and skin of mice but not in non-infected tissue and Pseudomonas-infected tissue, and significantly enhances the accumulation of intravenously injected vancomycin-loaded porous silicon nanoparticles bearing the peptide in S. aureus-infected mouse lung tissue. The targeted nanoparticles more effectively suppress staphylococcal infections in vivo relative to equivalent doses of untargeted vancomycin nanoparticles or of free vancomycin. The therapeutic delivery of antibiotic-carrying nanoparticles bearing peptides targeting infected tissue may help combat difficult-to-treat infections.
Introduction
Staphylococcus aureus is one of the most common pathogens causing invasive infections, both in health care settings and the community.1–3 S. aureus infections can be severe and life threatening, and include abscesses, endocarditis, pneumonia, toxic shock syndrome, and sepsis. Methicillin-resistant S. aureus (MRSA), in particular, is of great current concern and represents a significant public health problem.4 The resistance of MRSA to the commonly used and relatively non-toxic first-line antibiotics makes it necessary to treat serious infections with antibiotics such as vancomycin that are kept in reserve or avoided because of their toxicity. These antibiotics can have serious adverse effects including thrombophlebitis, fever, kidney damage, epidermal necrolysis, and a hypersensitivity reaction known as red man syndrome, potentially resulting in toxic symptoms worse than the infection.5–7 In the United States, about half a million people acquire a staphylococcal infection annually and the healthcare costs related to these infections exceed $14 billion per year.8, 9 Therefore, there is an urgent need to develop better therapies. One solution is to find new and safer anti-bacterial drugs. Alternatively, existing last-resort antibiotics could be selectively delivered to the infected site, in order to increase drug potency locally and reduce side effects elsewhere.
One approach to reduce the toxicity of antibiotics is to improve their pharmacokinetics, particularly by packaging the antibiotics into nanoparticles.10–14 Nanoparticle formulations can prolong the half-life of the antibiotic payload and serve as a sustained release system, reducing the frequency of administration and improving therapeutic index. Active targeting of an antibiotic to bacteria in an infected tissue is another strategy that can be used to increase the therapeutic index of antibiotics. In recent years, rapid advances have been made in targeted delivery systems, but most of the advances in this area have been in cancer treatment.15–19 In the context of infectious disease, a rifampicin derivative has been coupled to an anti-MRSA antibody to treat systemic MRSA infections in a mouse model.20 The conjugate was more effective than the free antibiotic. More importantly, the targeted therapeutic appeared to reach intracellular bacteria, which otherwise could have been sheltered from the antibiotic to become a source for rekindling of the infection.
The combination of nanoparticle delivery with specific tissue targeting provides a particularly powerful means of improving drug delivery. Here we develop a targeted nanoparticle system to improve the delivery of antibiotics. The nanoparticles in this system have a biocompatible porous silicon core that provides a high loading capacity for drugs and a readily modified surface to accommodate targeting groups.21–26 To develop targeting devices for the nanoparticles we employed in vivo screening of phage-displayed peptide libraries to identify peptides that recognize sites of bacterial infection. This method has been found to be a powerful and unbiased method to discover peptides that specifically home to other diseased tissues, such as tumors, atherosclerotic plaques, and tissue injuries.27–30 Here we show that it lends itself to infection targeting as well.
Results
Identification of S. aureus targeting peptides by phage display
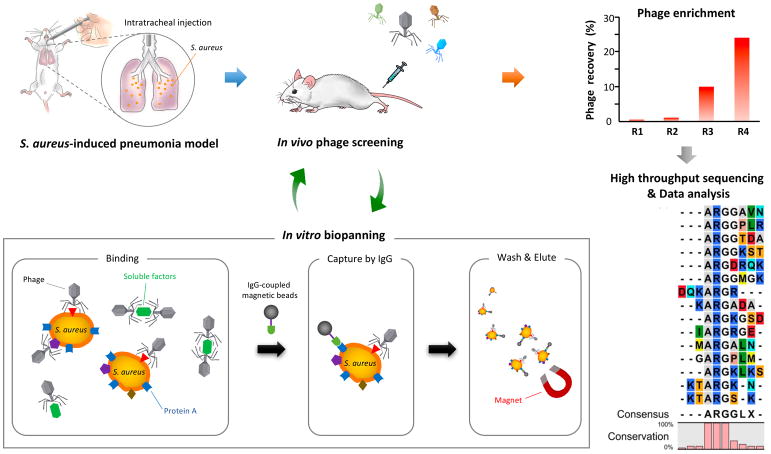
To isolate peptides that specifically bind to S. aureus-infected tissues, we screened a phage-displayed peptide library in a S. aureus pulmonary infection model (Fig. 1).31 The screen consisted of an in vivo round on infected lungs, a round of in vitro screening on cultured S. aureus bacteria to eliminate healthy tissue binders, and another in vivo round on infected lungs. High throughput sequencing and consensus motif analysis of the recovered phage pool revealed a dominant shared sequence motif, ARG or ARGG among the peptides encoded by the phages (Fig. 1). A consensus sequence was used to generate a synthetic 9-amino acid cyclic peptide, named CARG (sequence: CARGGLKSC).
Figure 1. Schematic representation of the peptide library screening by in vivo phage display in S. aureus-induced pneumonia model.
Lung infection was generated by direct inoculation of S. aureus into the lungs by intratracheal surgery. Forty-eight −72 h after the bacterial inoculation, a CX7C-peptide library was screened by in vivo phage display. After 3 in vivo rounds, the enriched phage pool was subjected to in vitro biopanning on cultured S. aureus. The in vitro phage pool was further screened in vivo in infected animals. The graph shows the enrichment obtained in 4 rounds of in vivo biopanning. The recovered phage pools were subjected to high throughput sequencing and the data were analyzed for dominant binding motifs.
The CARG peptide, synthesized with a 5(6)-carboxyfluorescein label (FAM-CARG), bound S. aureus bacteria in vitro, whereas a control peptide, with the same overall structure and charge (~ +2) (sequence: CNREARGRC) showed no binding (Fig. 2a). Further analysis of the binding by super-resolution microscopy showed that the peptide binds to the surface of bacteria (Fig. 2b). We also tested the ability of the CARG peptide to recognize a clinical isolate of methicillin-resistant S. aureus (MRSA). The FAM-CARG strongly bound to the MRSA strain USA300-0114 (Supplementary Fig. S1), a community-associated strain responsible for outbreaks of staphylococcal skin and soft tissue infections in the USA.32 Moreover, the FAM-CARG peptide also bound to Streptococcus pyogenes in culture (Supplementary Fig. S2), suggesting binding to other gram-positive strains.
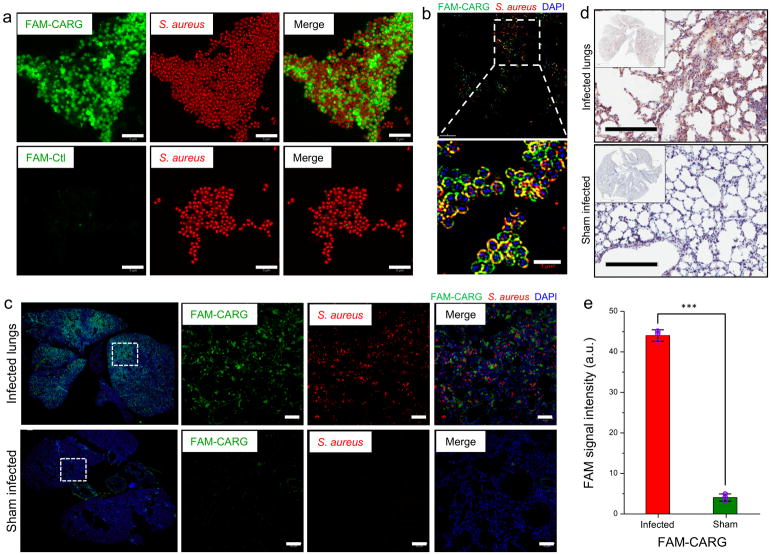
Figure 2. CARG peptide shows selective binding to cultured S. aureus in vitro and homes to infected lungs in vivo.
(a) Binding of FAM-labeled CARG peptide to cultured S. aureus in vitro (top row). The control peptide (FAM-Ctl) shows no bacterial binding (bottom row). Scale bar: 5 μm. (b) Super-resolution microscopy images of S. aureus after incubation with FAM-CARG peptide (green). Staphylococci visualized with an antibody are red, and bacterial nucleoid is stained with DAPI (blue). Scale bar: 3 μm. (c) S. aureus-induced lung infection was generated by intratracheal inoculation of S. aureus into the lungs in surgery. At 48–72 h post-infection, FAM-CARG was intravenously injected via the tail vein and allowed to circulate for 30 min. Lungs and other organs were harvested after perfusion and fixed for histological analysis. Sham-operated control mice underwent the same surgical procedure without S. aureus inoculation. Immunofluorescence microscopy of lung sections from mice with (top) or without (bottom) S. aureus infection. FAM-CARG (green), S. aureus (red) and cell nuclei stained with DAPI (blue). Images representative of at least five sections from each lung (n = 3–5 mice per group) are shown. Scale bar: 50 μm. (d) Immunohistochemical staining for the FAM label reveals intense signal (brown) from FAM-CARG in S. aureus-infected lungs as compared to healthy lungs. Sections were counterstained with DAPI (blue). Scale bar: 200 μm. (e) FAM integrated intensity was quantified using ImageJ (mean ± S.D., P < 0.001, two-tailed Student’s t test, n = 3; A. U. is arbitrary units).
In agreement with the in vitro binding results, intravenously injected CARG peptide homed to S. aureus-infected lungs, accumulating at the site of infection (Fig. 2c and d). There was only negligible homing to the healthy lungs of mice with a sham infection. Quantification of the FAM fluorescence signal showed that CARG accumulated in the infected regions of the lungs at 10-fold higher levels than in healthy lungs (Fig. 2e). Intravenously injected FAM-CARG peptide strongly homed to and accumulated at the infected tissue in a S. aureus skin infection model, and there was no detectable accumulation in uninfected skin (Fig. 3a and b). The control peptide for CARG showed essentially no homing to the S. aureus-infected lungs (Supplementary Fig. S3a).
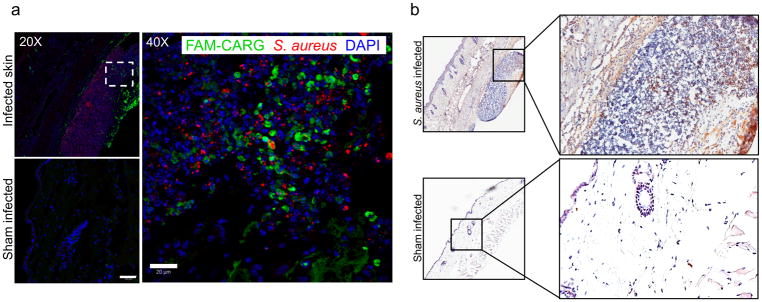
Figure 3. CARG is highly specific in targeting S. aureus infections.
(a–b) Skin infection in mice was generated by subcutaneous inoculation of S. aureus bacterial suspension. The sham-infected control mice received subcutaneous injection of PBS. At 72 h post-infection, FAM-CARG was intravenously injected via the tail vein and allowed to circulate for 30 min to home to the skin abscesses. (a) Immunofluorescence microscopy of skin sections from mice with (top) or without (bottom) S. aureus infection at low (20X) magnification. High (40X) magnification of CARG homing to infected skin section (Dashed square site). FAM-CARG (green), S. aureus (red) and cell nuclei stained with DAPI (blue). Representative images of at least five skin sections from each group (n = 3 mice per group) are shown. Scale bar: 50 μm (low magnification); 20 μm (high magnification). (b) Imunohistochemical staining for the FAM label reveals intense signal (brown) from FAM-CARG in S. aureus-infected skin as compared to healthy skin. Sections were counterstained with DAPI (blue). Scale bar: 200 μm.
We also tested a linearized version of the CARG peptide, where the terminal cysteines are replaced with alanines (sequence: AARGGLKSA). This linear ARG peptide showed negligible homing to the infected lungs (Supplementary Fig. S3b). Finally, we showed that CARG does not recognize cultured Pseudomonas aeruginosa bacteria or lung tissue infected with these bacteria (Supplementary Fig. S3c). We also tested a Pseudomonas-binding peptide (sequence: EKRTKSRLM; ref 45) on S. aureus-infected lungs. This peptide, which shows strong binding to P. aeruginosa cultures in vitro and homes to Pseudomonas lung infection in vivo (Supplementary Fig. S4a), did not show any homing to S. aureus-infected lungs (Supplementary Fig. S4b). These results show that CARG recognizes S. aureus infections in different tissues, and suggest that it is highly specific for S. aureus with some level of specificity to at least streptococci among the category of gram-positive bacteria.
CARG partially co-localizes with S. aureus and inflammatory leukocytes at sites of infection
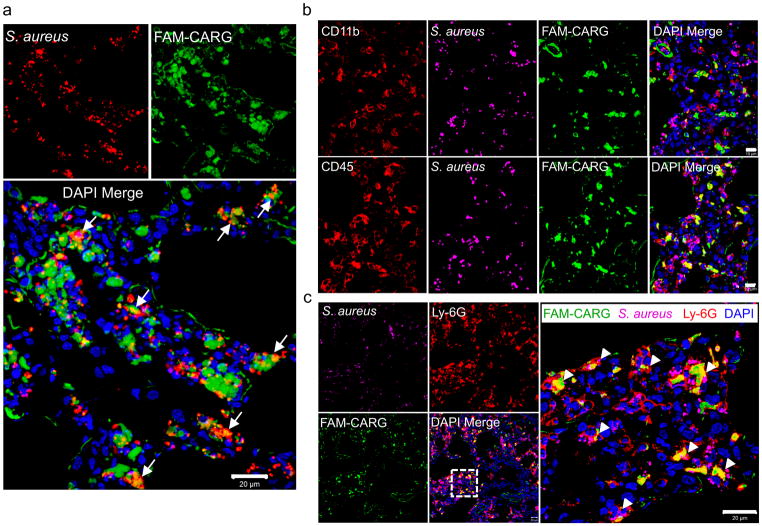
S. aureus is generally an extracellular pathogen, but it is also capable of invading and surviving inside mammalian phagocytic cells responsible for bacterial clearance.33–36 The intracellular localization provides the bacteria protection against the host immune system and antibiotics, leading to recurrent infections.37, 38 To determine whether the CARG peptide targets the bacteria and enters into host cells harboring intracellular bacteria, we intravenously injected S. aureus-infected mice with FAM-CARG and found that the injected peptide partially colocalized with the bacteria (Fig. 4a) and cells harboring intracellular bacteria (Fig. S5) in infected lung tissue. Quantitative colocalization analysis of the confocal immunofluorescence images showed 21.5 ± 2.2% colocalization of the FAM-CARG signal with the bacterial immune staining in infected lungs. Polymorphonuclear leukocytes (neutrophils) are a primary cellular host defense mechanism against S. aureus infections, and S. aureus abscesses are mostly composed of these cells.39 S. aureus-infected lungs were rich in inflammatory leukocytes (Fig. 4b), and the cells with engulfed bacteria were primarily neutrophils (Fig. 4c), as they expressed the neutrophil-specific marker Ly-6G.40 Analysis of CARG homing to S. aureus-infected skin sections also showed a co-localization of CARG with leukocytes at sites of infection (Supplementary Fig. S6). Thus, CARG-mediated targeting of antibiotics to neutrophils that contain intracellular S. aureus could potentially be harnessed to eradicate persistent infections.40
Figure 4. CARG peptide zeroes in on S. aureus bacteria at sites of infection and co-localizes with host phagocytic cells containing intracellular bacteria.
(a) The homing of FAM-labeled CARG to S. aureus infection was studied as described in Fig. 2. The FAM signal (green) shows strong homing and uptake in regions of Staphylococcus-infection (red). Cell nuclei are stained with DAPI (blue). The merged confocal image shows co-localization of the CARG peptide with bacteria at the infection site (white arrows). Scale bar: 20 μm. (b) Infected lung sections stained for CD11b (red, top row), CD45 (red, bottom row), S. aureus (magenta, pseudocolor), FAM (green) and nuclei (blue). The CARG peptide co-localizes with CD45-expressing leukocytes. Scale bar: 10 μm. (c) Infected lung sections stained for the neutrophil marker Ly-6G (red), S. aureus (magenta; pseudocolor), FAM (green) and nuclei (blue). The high magnification confocal image shows uptake and co-localization of the CARG peptide in neutrophils with engulfed bacteria (white arrowheads). Scale bar: 20 μm Representative images of at least five sections from infected lungs (n = 3–5 mice) are shown.
CARG-conjugated nanoparticles as a carrier of therapeutics to sites of S. aureus infection
The accumulation of FAM-CARG at sites of infection showed that CARG is capable of delivering a low molecular weight compound to infected tissue. This conclusion is based on the fact that FAM is in the same size range as a typical low molecular weight drug. We next examined the ability of CARG to deliver a nanoparticle (NP) payload by testing 20-nm CARG-coated silver NPs (CARG-AgNPs) on RAW 264.7 mouse macrophages. The RAW 264.7 cells are representative of activated macrophages, and this cell line has been used in earlier S. aureus infection studies.41 When incubated in media containing CARG-AgNPs and S. aureus, both the bacteria and the CARG-NPs, presumably as a complex, are taken up more effectively than control AgNPs, at least in the time frame we used. This result indicates that CARG-AgNPs are taken up by macrophages along with bacteria (Supplementary Fig. S7a). In vivo, intravenously administered CARG-AgNPs showed 8-fold greater accumulation relative to non-targeted FAM-AgNPs in S. aureus-infected lung tissue, and there was little accumulation in sham infected healthy lungs of either NPs (Supplementary Fig. S7b and 7c). The pattern of homing and localization of CARG-AgNPs was in good agreement with the FAM-CARG results. These findings demonstrate that the CARG peptide can target a nanoparticle to S. aureus-induced infections.
To investigate the translational potential of CARG for targeted drug delivery, we examined CARG-mediated delivery of the antibiotic vancomycin in the S. aureus lung infection model. The antibiotic was incorporated into porous silicon nanoparticles (pSiNPs) (mean diameter: ~ 180 nm) using a self-sealing chemical process involving calcium silicate formation that physically traps the drug payload without chemically altering its structure (Supplementary Fig. S8).42 In the present case the process loaded 12% by mass of vancomycin. The pSiNPs dissolved and the drug payload was released gradually over a 12 h period under simulated in vivo conditions, and the antibiotic retained 100% of its activity upon release (Supplementary Fig. S9). Although the drug release kinetics may be different in vivo, the results suggest delayed release of active antibiotic from the pSiNPs. Next, we modified the vancomycin-loaded pSiNPs with the CARG peptide for active targeting into infected tissue (Fig. 5a–d).
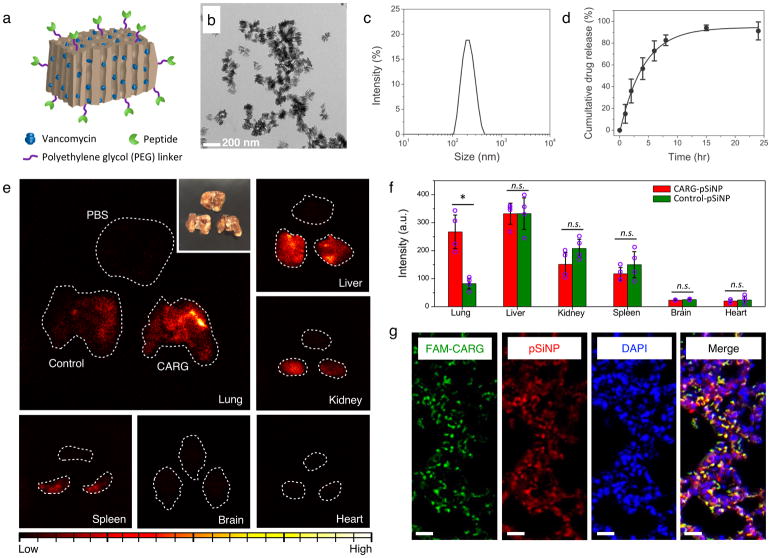
Figure 5. Targeted in vivo delivery of nanoparticles to S. aureus-infected lungs.
(a) A schematic illustration of therapeutic nanoparticle system consisting of pSiNP, chemotherapeutic agent, and homing peptide. (b) Transmission electron microscope (TEM) image of vancomycin-loaded pSiNP prepared by self-sealing chemistry. (c) Hydrodynamic size distribution of the nanoparticles measured by dynamic light scattering. Data are presented as mean ± standard deviation (n=3). (d) Release profile of vancomycin payload from the nanoparticles in phosphate-buffered saline at 37°C. (e) Time-gated luminescence images of pSiNPs in major organs harvested from mice after 1 hr of circulation (λex: 500 nm). Nanoparticles were intravenously injected to infected mice 24 h after intratracheally introduced S. aureus infection. White dashed line designates the outer boundary of each organ. Control nanoparticles were pSiNPs grafted with polyethylene glycol only (no targeting peptide). Corresponding volume (50 μL) of phosphate-buffered saline (PBS) was injected to mice intravenously as another negative control. Inset: White light photograph of lung tissues corresponding to the time-gated luminescence image. (f) Biodistribution of nanoparticles obtained from the luminescence intensity in each organ (n = 4 mice per group; * p < 0.01; n.s. means not significant, from two-tailed Student’s t test; Data are presented as mean ± standard deviation; a.u. is arbitrary units). Note that there is significantly greater accumulation of CARG-pSiNPs compared with control pSiNPs in infected lung tissue, but not in other organs. (g) Confocal fluorescence microscope images of infected lung tissue of mice injected with CARG-pSiNP. Green channel, CARG peptide labeled with FAM; red, intrinsic photoluminescence of pSiNPs; and blue, DAPI nuclear stain. Scale bar: 20 μm.
Time-gated luminescence imaging of the long-lived photoluminescence signal from the pSiNPs23 (Fig. 5e and f) showed that intravenously injected CARG-conjugated pSiNPs (CARG-pSiNPs) accumulated in infected lungs at markedly higher levels than control-pSiNPs (Fig. 5f), and the CARG-pSiNPs spread into the extravascular sites in infected, but not normal, lungs (Fig. 5g and Supplementary Fig. S10). Low, but detectable signal intensities from control-pSiNPs were observed in infected lungs and some CARG-pSiNPs were found in normal lungs (Supplementary Fig. S10). This background may be caused by NP uptake into the mononuclear phagocyte system (MPS) in the lungs.43, 44 We further characterized the percent injected dose per gram (%ID/g) of elemental silicon in different organs 6 hours after pSiNP administration in S. aureus-infected mice by inductively coupled plasma-optical emission spectroscopy (ICP-OES). Biodistribution in healthy mice was also analyzed for comparison. As shown in supplementary Fig. S11a, there was significantly higher accumulation of CARG-pSiNPs in the infected lungs than of control pSiNPs (> 4-fold). Accumulation of CARG-pSiNPs in the lungs of healthy animals did not differ from control pSiNPs. Thus, the substantial amount of the CARG-pSiNPs in the infected lungs (7% of the %ID/g) is attributable to CARG targeting. Overall, these results establish the specific targeting of staphylococcal infection by CARG-pSiNPs.
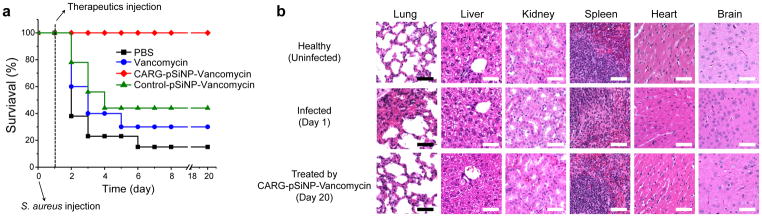
In our infection model, intratracheally introduced S. aureus causes serious pneumonia, resulting in 67% mortality (n=9 mice) between 24–48 h post-infection, and only 11 % survival by day 6. Treatment of infected mice with intravenous injections of the CARG-targeted vancomycin-pSiNPs one-day post-infection resulted in 100% recovery and long-term survival, whereas only 33% survival was seen when an equivalent dose (3 mg/kg) of free vancomycin was administered (Fig. 6a; p < 0.001 and S12a). At higher doses of free vancomycin (9 mg/kg and 15 mg/kg), the survival rates were 70–80% (n=5–7, Supplementary Fig. S12a and 12b) and 100% (n=3–5, Supplementary Fig. S12a and S13), respectively. Therefore, higher doses of vancomycin were progressively more effective. In a separate study, shown in Fig. S12b, CARG-pSiNP-vancomycin (1 mg/kg; n=5) was 100% effective, whereas 9 mg/kg of free vancomycin was again only partially effective (Figs. S12a and 12b). Thus, vancomycin delivered in CARG-pSiNPs is about 10-fold more effective than free vancomycin, and complete survival in our S. aureus infection model required at least a ~ 10-fold higher dose of free vancomycin than was delivered in the CARG-pSiNPs. We also evaluated the in vivo toxicity of the pSiNPs. Healthy mice injected with vancomycin-loaded pSiNPs or the free antibiotic at a therapeutically effective dose (3 mg/kg) showed normal aspartate aminotransferase and alanine transaminase levels (Table S1), indicating lack of toxicity to the liver, the organ that non-specifically takes up nanoparticles. Tissue histology supported the conclusion that the infection had completely resolved in the lungs of surviving treated mice (Fig. 6b), with no infectious foci or toxic damage observed in any of the major organs.
Figure 6. Targeted, infection-homing nanoparticles enhance antibiotic therapy.
(a) Survival rate (n = 9) of mice after intratracheal inoculation with 5 × 107 colony forming units (CFU) of S. aureus. Groups of mice received the following therapeutics intravenously 24 hours after the bacterial inoculation: free vancomycin, CARG-pSiNP-vancomycin, and control-pSiNP-vancomycin. The therapeutic compounds were pre-suspended in PBS prior to injection into the tail vein. The amount of vancomycin administered was adjusted to be the same in each therapeutic (3 ± 0.2 mg/kg). (b) Hematoxylin and eosin (H&E)-stained tissue sections from infected mice, showing that intratracheal injection of S. aureus only causes lung infection, whereas other organs show no detectable changes. Note that the infected lung lesions are resolved by treatment with vancomycin-loaded CARG-pSiNP (n=3; representative images are shown). Scale bar: 50 μm.
Discussion
We report here on a peptide that recognizes S. aureus bacteria and selectively accumulates at the sites of staphylococcal infection in mouse models. We show that this peptide, CARG, enhances the accumulation at sites of infection of systemically administered payloads with chemistries ranging from a drug-sized molecule to nanoparticles. We also show that porous silicon nanoparticles loaded with vancomycin and targeted with the CARG peptide are several times more effective than free vancomycin in eradicating a staphylococcal lung infection. This approach may have therapeutic advantages in situations that require the use of last-resort antibiotics to deal with difficult-to-treat infections, such as ones caused by MRSA.
We used in vivo phage display in our screens for peptides that recognize staphylococcal infections to increase the likelihood that the resulting peptides are relevant to real-life infections. Screening phage libraries on cultured bacterial cells readily yields peptides that bind to bacteria.45 However, the environment strongly influences bacterial gene expression, and it is entirely possible that a prominent surface component of cultured bacteria is not expressed, or is only a minor component in vivo. By combining screening for peptide homing to infected tissue in vivo with selection on cultured bacteria, we sought to ascertain that our peptides would be specific for the bacteria as they exist in the infected tissue, and that the peptide could reach the bacteria in the tissue. The CARG peptide we obtained fulfills these criteria.
The screening results and the CARG variations tested so far allow some conclusions regarding the structure function relationships. As shown in Figure 1, there is some conservation of the residues close to the ARG motif, and the ARG motif is positioned close to the center or toward the N-terminus of the peptide. The lack of bacterial binding by the control peptide suggests that the ARG motif cannot be adjacent to the C-terminal cysteine. The loss of activity upon linearization of the peptide shows that the cyclic structure is important for the activity, likely because the ARG motif is presented in an active conformation.
Several experiments demonstrated that the CARG peptide is highly specific for staphylococcal infections. First, the peptide bound to cultured S. aureus (two different strains, one a MRSA strain) and not to P. aeruginosa bacteria. Second, intravenously injected CARG accumulated in staphylococcal lung and skin infections, and not in normal tissues. Third, the peptide largely co-localized with the bacteria in the infected tissue. Fourth, CARG did not recognize lung tissue infected with P. aeruginosa. Thus, CARG appears suitable for specific targeting of S. aureus in infected tissues. Of note, CARG peptide accumulates at sites in infected tissue even where there are no bacteria detectable by antibody stain. A likely explanation for this distribution pattern is that CARG is targeting a bacterial component which is either derived from disintegrated bacteria, or which is shed from the surface of intact bacteria. This bacterial component then binds to host tissue and is targeted by CARG. The target is clearly a bacterial surface component because CARG strongly labels the bacterial surface in vitro. That the peptide targeting is not limited to intact bacteria is an advantage because it allows more peptide and payload to be targeted into the infected tissue than would be the case if the peptide only recognized intact bacteria.
The CARG peptide accumulated in host cells containing intracellular bacteria, colocalizing with the bacteria. This is a potentially important property of CARG, because the intracellular location protects bacteria against the host immune system and against antibiotics, which enables long-term colonization of the host, causing persistent and recurrent infections.37, 38 S. aureus is generally considered an extracellular pathogen. However, there is substantial evidence to the effect that these bacteria can survive intracellularly.33, 46, 47 The fact that CARG can reach intracellular bacteria suggests that CARG-mediated delivery of antibiotics has the potential of facilitating the eradication of bacteria from intracellular locations.
Antibiotics loaded into nanoparticles have found clinical use.48 The nanoparticle vehicle in these products is typically a liposome. The inorganic, non-liposomal pSiNPs used for antibiotic delivery in the present work have several attractive features: they are biocompatible and degrade in the body into silicic acid, which is naturally found in human tissues and efficiently excreted from the body through the urine.22 They have a high loading capacity for therapeutic payloads, which are released upon dissolution of the nanostructure, and this release can be controlled.22, 49–51 Finally, pSiNPs, and nanoparticles in general, are well suited for targeting with peptides because the avidity effect provided by the multivalent presentation on the NP surface compensates for the relatively low affinity of peptides for their target.27
Our results illustrate the advantages of targeted pSiNPs as a delivery vehicle for antibiotics. Promising gains in efficacy have been obtained by targeting antibiotics to bacteria.10, 11 Our delivery system combines the pSiNPs and targeting approaches. The CARG-coated pSiNPs gave an increase in vancomycin efficacy that was at least ~ 10-fold. The “last resort” antibiotics have typically been assigned to that status because, while effective, they are also toxic. In addition, some antibiotics are often poorly soluble and therefore difficult to administer.52 Delivery in pSiNPs has mitigated both of these problems,17, 18, 21, 24, 42 and targeted pSiNPs add further gains by lowering the total dose needed--thus reducing toxic side effects while increasing efficacy of the treatment.
We analyzed the biodistribution of pSiNPs by time-gated luminescence imaging and ICP-OES. The time-gated method images the intrinsic, long-lived photoluminescence from the pSiNP carrier and eliminates tissue autofluorescence,23 and allows the payload to be reliably quantified in intact organs. The quantitation of %ID/g of elemental silicon in different organs measured by ICP-OES showed a significantly higher accumulation of CARG-pSiNPs in the infected lungs than of control pSiNPs (> 4-fold). Accumulation of CARG-pSiNPs in the lungs of healthy animals did not differ from control pSiNPs. Thus, the substantial amount of the CARG-pSiNPs in the infected lungs is attributable to CARG targeting. Our biodistribution data are in good agreement with a previous analysis of silicon nanoparticle biodistribution.22 The elemental analysis and photoluminescence (PL) data are also in good agreement, although elemental analysis showed somewhat higher silicon content in the spleen. This is possibly due to rapid biodegradation of the pSiNPs in the spleen, or PL quenching by the spleen microenvironment. The data indicate that targeted delivery increases the amount of silicon delivered to infected lungs, translating to more efficacious delivery of nanoparticle-loaded vancomycin, in agreement with the enhanced antibacterial activity we observed. In contrast, targeted and control pSiNPs gave similar silicon levels in normal organs (Fig. S11a). Thus, the silicon biodistribution data indicate that for a given vancomycin dose in pSiNPs, the targeted pSiNPs provided improved efficacy with a comparable systemic drug burden.
The tendency of nanoparticles to clear through the liver raises the possibility of liver toxicity, which would not necessarily be seen with the free drug. Vancomycin is generally not toxic to the liver,53–55 and we saw no evidence of acute liver toxicity in this study. Even if the antibiotic payload is toxic to the liver, the reduced dose made possible by the addition of the targeting peptide component may compensate for the selective liver accumulation of the carrier NPs. It should be noted that clearance through the liver and spleen results in substantial NP accumulation in these organs. While no acute toxicity was noted in this study, extended administration of antibiotic-loaded NPs to treat chronic infections will require long-term safety studies.
In summary, we have developed a targeted drug delivery system that is based on selection of a peptide capable of recognizing and reaching an infectious agent in infected tissues, a biocompatible nanoparticle vehicle with a high loading capacity for an antibiotic, and the use of the peptide to concentrate the nanoparticles at the site of infection. Substantial efficacy gains in antibiotic treatment of a staphylococcal pneumonia mouse model validate the technology, which may be broadly applicable to other infectious diseases.
Materials and Methods
Bacterial strains
Staphylococcus aureus subsp. aureus (ATCC 25923), Methicillin-Resistant Staphylococcus aureus (MRSA USA300-0114; ATCC BAA-1756), and Pseudomonas aeruginosa (ATCC 10145) strains procured from the American Type Culture Collection (ATCC) were used. For inoculation experiments, subcultures were grown from freezer stocks in Brain Heart Infusion (BHI) agar plates and incubated at 37°C overnight. All subsequent liquid subcultures were derived from colonies isolated from these plates.
Animal models of infection
All animal experiments were conducted under an approved protocol of the Institutional Animal Care and Use Committee of Sanford Burnham Prebys Medical Discovery Institute. To create a lung infection model, we inoculated S. aureus bacteria intratracheally.31 The bacteria were grown overnight in BHI media with constant aeration. The cultures were diluted in BHI media by a factor of 100, and grown at 37°C until the mid-log phase. The Staphylococci were centrifuged and the pelleted bacteria were washed three times with cold, sterile phosphate-buffered saline (PBS) and finally suspended in 0.5 ml PBS. An aliquot of the culture was plated in BHI agar plates, and resulting colonies were counted (colony-forming units; CFU).
Male BALB/c mice (8–10 weeks old; Envigo) were anesthetized with 4% isoflurane (Aerrane; Baxter, UK) and secured on a surgery board angled at 60°. The fur on the neck was shaved, and the skin was disinfected with betadine and alcohol wipes. A 1 cm longitudinal incision was made over the trachea, the overlying muscle was retracted, and the trachea exposed. A tuberculin syringe fitted with a short-bevel 26-gauge needle was used to introduce 30 μL of bacterial suspension (107 CFU) into the trachea. Application of pressure to the abdomen of the mouse to deflate the lungs and its removal concurrent with the introduction of inoculum into the trachea assured inspiration of the inoculum into the lungs. The incision was closed with sutures and the mice were placed on a warming pad to recover from anesthesia before being placed back into their cage. A similar method was used in the P. aeruginosa pneumonia model.
A skin infection model was obtained by inoculating 8–10 week-old male BALB/c mice under isoflurane anesthesia with a subcutaneous injection in shaved right flank of 107 CFU S. aureus in 50 μL of PBS. This method produces consistent dermonecrotic abscesses within 2 days.56 The weights of all infected mice were recorded prior to inoculation and at 24-hour intervals post-infection during the course of disease.
In vivo phage display
In vivo phage screening of peptide libraries was carried out as described using mice with S. aureus-induced lung infection.57 The T7 library with the general structure of CX7C (C= cysteine, X= any amino acid) was injected 48 h post-infection and allowed to circulate for 30 min, after which the mice were perfused with 15–20 ml of warm PBS through the heart and tissues were collected for phage recovery. After three in vivo rounds of screening, the phage pool recovered was subjected to one round of in vitro biopanning on cultured S. aureus bacteria culture in vitro, followed by another round of in vivo screening. The phage pools obtained after the in vitro and the fourth in vivo rounds were subjected to high throughput sequencing using Ion Torrent (Thermo Fisher Scientific).
Peptide synthesis
Peptides were amidated on the C-terminus and synthesized on a microwave-assisted automated peptide synthesizer (Liberty; CEM, Matthews, NC) following Fmoc/tertiary butyl strategy on rink amide MBHA resin with HBTU (N,N,N′,N′-Tetramethyl-O-(1H-benzotriazol-1-yl) uronium hexafluorophosphate) activator, (or alternatively, O-(Benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate), collidine activator base and 5% piperazine for deprotection. When included, fluorescein was incorporated during the synthesis at the N-terminus of the sequence as 5(6)-carboxyfluorescein with a spacer, 6-aminohexanoic acid, separating the fluorophore and the sequence. Cleavage using a 95% trifluoroacetic acid (TFA) followed by purification on C18 reverse phase column using Acetonitrile water mixtures containing 0.1% TFA gave peptides with 90–95% purity. The peptides were analyzed by matrix assisted laser desorption mass spectrometry (MALDI).
Peptide targeting to infection sites and immunofluorescence
Animals with pneumonia were intravenously injected with 100 nmol of FAM-labeled synthetic peptide, at 24–48 h post-infection, and the peptide was allowed to circulate for 30–60 minutes. Following terminal cardiac perfusion with PBS, lungs and other organs were isolated, fixed in 4% (w/v) paraformaldehyde (PFA) in PBS overnight, washed with PBS, cryo-protected in sucrose solution [30% (w/v) in PBS] overnight, and processed through OCT embedding. Ten-μm sections were cut and analyzed by fluorescence microscopy. The primary antibodies for immunofluorescence were rabbit anti-Staphylococcus aureus, (ab20920, Abcam); rabbit anti-Fluorescein (A889, Invitrogen); rabbit anti-Pseudomonas (ab68538, Abcam); rat anti-mouse CD11b (550282, BD Pharmingen); rat anti-mouse CD45 (550539, BD Pharmingen); rat anti-mouse Ly-6G (551459, BD Pharmingen). These antibodies were incubated in diluted (1%) blocking buffer overnight at dilutions 1:100 or 1:200 at 4°C, the sections were washed with PBS-T and incubated with secondary antibodies diluted 1:500 or 1:1000, in 1% blocking buffer for one hour at room temperature. Subsequently sections were washed with PBS-T, counterstained with DAPI (1ug/mL) in PBS for 10 minutes, washed with PBS, mounted using mounting media (Molecular Probes, Life Technologies), and examined using a confocal microscope (Zeiss LSM-710).
Preparation of porous silicon nanoparticles
Porous silicon nanoparticles (pSiNPs) were prepared by electrochemical etch of crystalline, (100)-oriented silicon wafers (p-type, boron-doped, ~ 1 mΩ cm resistivity) as described previously.58, 59 In brief, the silicon wafer was anodized in an electrolyte composed of 3:1 (v:v) of 48 % aqueous HF:ethanol. The etching waveform consisted of a square wave in which a lower current density of 50 mA/cm2 was applied for 1.8 sec, followed by a higher current density pulse of 400 mA/cm2 for 0.36 sec. The pulsed waveform was repeated for 140 cycles, and the resulting porous silicon nanostructure was lifted-off from the silicon substrate by application of a current density of 3.7 mA/cm2 for 250 sec in an electrolyte consisting of 1:30 (v:v) of 48 % aqueous HF:ethanol. The freestanding porous silicon films were then fractured into nanoparticles of mean diameter 170 nm by ultrasonication in deionized water overnight.
Preparation of vancomycin-loaded porous silicon nanoparticles
Vancomycin was loaded and trapped in the pSiNPs following a published procedure previously used to load siRNA.42 An aqueous dispersion of pSiNPs (1 mg/mL in deionized water) was added to an aqueous solution of vancomycin (0.5 mg/mL) in a 1:1 (v:v) ratio, and ultrasonicated for 10 min. An equal volume of aqueous calcium chloride solution (2 M) was then added to the mixture, which was agitated (< 5 min) and ultrasonicated in an ice bath for 2 hr. Drug-loaded nanoparticles were purified by centrifugation and washing steps with deionized water, 70% ethanol, and 100 % ethanol, sequentially. To analyze the vancomycin loading capacity, the supernatants from centrifugation were collected and assayed for free vancomycin by measuring absorbance at 280 nm. The mass loading was verified by spectrophotometric quantification (λmax = 280 nm) of vancomycin released from the nanoparticles.
Coupling of peptide to the vancomycin-loaded porous silicon nanoparticles
One mL aliquot of pSiNPs (1 mg/mL in ethanol) was treated with 20 μL of 3-(ethoxydimethyl)-propylamine silane by vortexing overnight at room temperature. The amine-terminated nanoparticles were rinsed with ethanol and water, and then further functionalized by either MAL-PEG-SCM (maleimide-polyethylene glycol-succinimidyl carboxy methyl ester, MW: 5000) or mPEG-SCM (methoxy PEG-succinimidyl carboxy methyl ester, MW: 5000).23 The maleimide-activated nanoparticles were then mixed with the CARG peptide solution (0.5 mg/ml in water, 1 mL) and vortexed for 2 hr to conjugate the peptide via the free cysteine residue at the terminal group of the peptide (Supplementary Fig. S14).60 Peptide coupling was quantified and confirmed by measuring the fluorescence of the FAM label of the peptide.
Characterization of the porous silicon nanoparticles
Hydrodynamic size of the nanoparticles was measured by dynamic light scattering (DLS, Zetasizer ZS90, Malvern Instruments). Transmission electron microscopy (TEM) images were obtained with a JEOL-1200 EX II. Photoluminescence spectra (λex: 365 nm) were monitored using a spectrometer (QE Pro, Ocean Optics) with a 460 nm long-pass filter. Quantification of vancomycin was conducted by measuring absorbance at 280 nm with a spectrophotometer (NanoDrop 2000, Thermo Scientific) based on the standard curve of vancomycin.
In vitro antibacterial activity test
S. aureus bacteria were cultured overnight to achieve stable growth kinetics, and diluted bacteria were then separated to individual tubes. Bacterial growth and minimum inhibitory concentration of vancomycin were determined using optical density measurements.
In vivo antibiotic activity test
Six-to eight-week-old male BALB/c mice were used to evaluate in vivo anti-infective activity and to assess possible toxicity of the targeted nanoparticles. The S. aureus pneumonia model was generated by intratracheal inoculation as described above. S. aureus bacteria were introduced at a lethal dose of 5 × 107 CFU, which had been determined from a prior dose escalation study assessing the survival rate of mice 48 h after challenge. At this lethal dose, 67 % (n = 9) of mice died between 24 – 48 h post-infection, and 89 % of infected animals died within 6 days. After 24 h of infection, the mice received an intravenous injection of either PBS, free vancomycin, CARG-pSiNP-vancomycin and PEG-pSiNP-vancomycin (Control-pSiNP-vancomycin). The mice were observed over 20 days to evaluate survival. In order to compare antibacterial efficacy of each system, a separate dosing study involving free vancomycin (in the range 0.1–15 mg/kg) was performed to determine the survival rate for the infection protocol.
Quantitative in vivo biodistribution analysis
CARG-targeted or control pSiNPs (dose of pSiNPs: 20 mg/kg) and vehicle control (PBS) were injected intravenously into S. aureus-infected mice (24 h post-infection) and healthy mice. Nanoparticles were allowed to circulate for 6 h, and then the mice were sacrificed by cardiac perfusion with PBS under anesthesia. Major organs such as brain, heart, lung, liver, kidney, and spleen were collected, weighed, homogenized in a solution of PBS (0.3 mL), aqueous HNO3 (0.25 mL, ~15.7 M) and H2O2 (0.05 mL, 30 %), and digested for 2 days. The digested slurry was centrifuged (14,000 rpm, 10 min) and the supernatant was filtered using a syringe filter (Millex-HV Syringe Filter Unit, 0.45 μm), and then diluted with 2 % (v/v) HNO3 solution in water to prepare a final volume of 2 mL. Each solution was analyzed by inductively coupled plasma-optical emission spectroscopy (ICP-OES, Perkin Elmer Optima 3000DV). Silicon (Si) standard solutions (1, 0.5, 0.1, and 0.01 μg/mL) were used to calibrate Si content by dilution of a stock Si solution (1000 μg/mL) with 2 % aqueous HNO3.
Statistical analysis
All data represent mean value ± standard deviation. The significance analyses were performed using Statistica 8.0 or GraphPad Prism software, employing a one-way ANOVA or two-tailed heteroscedastic Student’s t test as detailed in the figure legends.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.
Supplementary Material
Acknowledgments
We thank Dr. Venkata Ramana Kotamraju for peptide synthesis and Joolz Ward for technical assistance. This work was supported by the Defense Advanced Research Projects Agency (DARPA) under Cooperative Agreement HR0011-13-2-0017, the National Science Foundation grant No. CBET-1603177 and in part by the National Institutes of Health, through contract No. R01 AI132413-01. The content of the information within this document does not necessarily reflect the position or the policy of the Government. This research was also supported in part by a grant from the Basic Science Research Program through the Korea NRF funded by the Ministry of Education (Grant No. 2017R1D1A1B03035525), a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Korea (Grant No. HI14C1090), and by the European Union through the European Regional Development Fund (Project No. 2014-2020.4.01.15-0012).
Footnotes
Author contributions
S.H. performed phage display screening and validated the Staphylococcus-binding peptide. J.J., G.B.B. and D.K. designed, synthesized, and characterized nanoparticles, performed in vitro experiments. S.H., J.J., G.B.B., Z.G.S., J.K., and B.K. performed and analyzed in vivo experiments. S.H., A.P.M., T.M and T.T performed Ion Torrent data analysis. S.C. and S.G. identified the Pseudomonas-binding peptide. S.H., J.J., G.B.B., D.K., B.K., J.K., M.J.S., and E.R. discussed and analyzed data. S.H., J.J., M.J.S., and E.R. conceived the project, and wrote the manuscript. All authors read and approved the manuscript.
Supplementary Information including associated results is available online.
Competing financial interests
M.J.S is a scientific founder of Spinnaker Biosciences, Inc., and has an equity interest in the company. Although the R01 AI132413-01 grant has been identified for conflict of interest management based on the overall scope of the project and its potential benefit to Spinnaker Biosciences, Inc., the research findings included in this particular publication may not necessarily relate to the interests of Spinnaker Biosciences, Inc. The terms of this arrangement have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
All the other authors declare no competing financial interests.
References
- 1.Moran GJ, et al. Methicillin-Resistant S. aureus Infections among Patients in the Emergency Department. New England Journal of Medicine. 2006;355:666–674. doi: 10.1056/NEJMoa055356. [DOI] [PubMed] [Google Scholar]
- 2.Klevens RM, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 3.Dantes R, et al. National burden of invasive methicillin-resistant Staphylococcus aureus infections, United States, 2011. JAMA Intern Med. 2013;173:1970–1978. doi: 10.1001/jamainternmed.2013.10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moellering RC. MRSA: the first half century. Journal of Antimicrobial Chemotherapy. 2012;67:4–11. doi: 10.1093/jac/dkr437. [DOI] [PubMed] [Google Scholar]
- 5.de Hoog M, Mouton JW, van den Anker JN. Vancomycin: pharmacokinetics and administration regimens in neonates. Clin Pharmacokinet. 2004;43:417–440. doi: 10.2165/00003088-200443070-00001. [DOI] [PubMed] [Google Scholar]
- 6.Pai MP, Mercier RC, Koster SA. Epidemiology of vancomycin-induced neutropenia in patients receiving home intravenous infusion therapy. Ann Pharmacother. 2006;40:224–228. doi: 10.1345/aph.1G436. [DOI] [PubMed] [Google Scholar]
- 7.Sorrell TC, Collignon PJ. A prospective study of adverse reactions associated with vancomycin therapy. J Antimicrob Chemother. 1985;16:235–241. doi: 10.1093/jac/16.2.235. [DOI] [PubMed] [Google Scholar]
- 8.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu Rev Microbiol. 2010;64:143–162. doi: 10.1146/annurev.micro.112408.134309. [DOI] [PubMed] [Google Scholar]
- 9.Inoshima I, et al. A Staphylococcus aureus pore-forming toxin subverts the activity of ADAM10 to cause lethal infection in mice. Nat Med. 2011;17:1310–1314. doi: 10.1038/nm.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Radovic-Moreno AF, et al. Surface Charge-Switching Polymeric Nanoparticles for Bacterial Cell Wall-Targeted Delivery of Antibiotics. ACS Nano. 2012;6:4279–4287. doi: 10.1021/nn3008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto-Alphandary H, Andremont A, Couvreur P. Targeted delivery of antibiotics using liposomes and nanoparticles: research and applications. International Journal of Antimicrobial Agents. 2000;13:155–168. doi: 10.1016/s0924-8579(99)00121-1. [DOI] [PubMed] [Google Scholar]
- 12.Courtney CM, et al. Photoexcited quantum dots for killing multidrug-resistant bacteria. Nat Mater. 2016;15:529–534. doi: 10.1038/nmat4542. [DOI] [PubMed] [Google Scholar]
- 13.Gao W, Thamphiwatana S, Angsantikul P, Zhang L. Nanoparticle approaches against bacterial infections. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology. 2014;6:532–547. doi: 10.1002/wnan.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morones-Ramirez JR, Winkler JA, Spina CS, Collins JJ. Silver Enhances Antibiotic Activity Against Gram-Negative Bacteria. Science Translational Medicine. 2013;5:190ra181. doi: 10.1126/scitranslmed.3006276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Low PS, Henne WA, Doorneweerd DD. Discovery and development of folic-acid-based receptor targeting for imaging and therapy of cancer and inflammatory diseases. Acc Chem Res. 2008;41:120–129. doi: 10.1021/ar7000815. [DOI] [PubMed] [Google Scholar]
- 16.Kukowska-Latallo JF, et al. Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]
- 17.Kell AJ, et al. Vancomycin-modified nanoparticles for efficient targeting and preconcentration of Gram-positive and Gram-negative bacteria. ACS Nano. 2008;2:1777–1788. doi: 10.1021/nn700183g. [DOI] [PubMed] [Google Scholar]
- 18.Simón-Gracia L, et al. iRGD peptide conjugation potentiates intraperitoneal tumor delivery of paclitaxel with polymersomes. Biomaterials. 2016;104:247–257. doi: 10.1016/j.biomaterials.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugahara KN, et al. Coadministration of a Tumor-Penetrating Peptide Enhances the Efficacy of Cancer Drugs. Science. 2010;328:1031–1035. doi: 10.1126/science.1183057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehar SM, et al. Novel antibody–antibiotic conjugate eliminates intracellular S. aureus. Nature. 2015;527:323–328. doi: 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- 21.Sailor MJ, Park JH. Hybrid Nanoparticles for Detection and Treatment of Cancer. Advanced Materials. 2012;24:3779–3802. doi: 10.1002/adma.201200653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park JH, et al. Biodegradable luminescent porous silicon nanoparticles for in vivo applications. Nat Mater. 2009;8:331–336. doi: 10.1038/nmat2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joo J, et al. Gated Luminescence Imaging of Silicon Nanoparticles. ACS Nano. 2015;9:6233–6241. doi: 10.1021/acsnano.5b01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Secret E, et al. Antibody-Functionalized Porous Silicon Nanoparticles for Vectorization of Hydrophobic Drugs. Advanced Healthcare Materials. 2013;2:718–727. doi: 10.1002/adhm.201200335. [DOI] [PubMed] [Google Scholar]
- 25.Serda RE, et al. Cellular Association and Assembly of a Multistage Delivery System. Small. 2010;6:1329–1340. doi: 10.1002/smll.201000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tasciotti E, et al. Mesoporous silicon particles as a multistage delivery system for imaging and therapeutic applications. Nat Nano. 2008;3:151–157. doi: 10.1038/nnano.2008.34. [DOI] [PubMed] [Google Scholar]
- 27.Ruoslahti E. Peptides as Targeting Elements and Tissue Penetration Devices for Nanoparticles. Advanced Materials. 2012;24:3747–3756. doi: 10.1002/adma.201200454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laakkonen P, Porkka K, Hoffman JA, Ruoslahti E. A tumor-homing peptide with a targeting specificity related to lymphatic vessels. Nat Med. 2002;8:751–755. doi: 10.1038/nm720. [DOI] [PubMed] [Google Scholar]
- 29.Sugahara KN, et al. Tissue-Penetrating Delivery of Compounds and Nanoparticles into Tumors. Cancer Cell. 2009;16:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mann AP, et al. A peptide for targeted, systemic delivery of imaging and therapeutic compounds into acute brain injuries. Nat Commun. 2016;7 doi: 10.1038/ncomms11980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeMaria TF, Kapral FA. Pulmonary infection of mice with Staphylococcus aureus. Infect Immun. 1978;21:114–123. doi: 10.1128/iai.21.1.114-123.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.King MD, et al. EMergence of community-acquired methicillin-resistant staphylococcus aureus usa 300 clone as the predominant cause of skin and soft-tissue infections. Annals of Internal Medicine. 2006;144:309–317. doi: 10.7326/0003-4819-144-5-200603070-00005. [DOI] [PubMed] [Google Scholar]
- 33.Thwaites GE, Gant V. Are bloodstream leukocytes Trojan Horses for the metastasis of Staphylococcus aureus? Nature reviews. Microbiology. 2011;9:215–222. doi: 10.1038/nrmicro2508. [DOI] [PubMed] [Google Scholar]
- 34.Gresham HD, et al. Survival of Staphylococcus aureus inside neutrophils contributes to infection. J Immunol. 2000;164:3713–3722. doi: 10.4049/jimmunol.164.7.3713. [DOI] [PubMed] [Google Scholar]
- 35.Anwar S, Prince LR, Foster SJ, Whyte MKB, Sabroe I. The rise and rise of Staphylococcus aureus: laughing in the face of granulocytes. Clinical & Experimental Immunology. 2009;157:216–224. doi: 10.1111/j.1365-2249.2009.03950.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garzoni C, Kelley WL. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol. 2009;17:59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Proctor RA, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nature reviews. Microbiology. 2006;4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 38.Foster TJ. Immune evasion by staphylococci. Nature reviews. Microbiology. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi SD, Malachowa N, DeLeo FR. Pathogenesis of Staphylococcus aureus Abscesses. The American Journal of Pathology. 2015;185:1518–1527. doi: 10.1016/j.ajpath.2014.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daley JM, Thomay AA, Connolly MD, Reichner JS, Albina JE. Use of Ly6G-specific monoclonal antibody to deplete neutrophils in mice. J Leukoc Biol. 2008;83:64–70. doi: 10.1189/jlb.0407247. [DOI] [PubMed] [Google Scholar]
- 41.Abed N, et al. An efficient system for intracellular delivery of beta-lactam antibiotics to overcome bacterial resistance. Sci Rep. 2015;5:13500. doi: 10.1038/srep13500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kang J, et al. Self-Sealing Porous Silicon-Calcium Silicate Core–Shell Nanoparticles for Targeted siRNA Delivery to the Injured Brain. Advanced Materials. 2016;28:7962–7969. doi: 10.1002/adma.201600634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anselmo AC, et al. Delivering Nanoparticles to Lungs while Avoiding Liver and Spleen through Adsorption on Red Blood Cells. ACS Nano. 2013;7:11129–11137. doi: 10.1021/nn404853z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choi HS, et al. Rapid translocation of nanoparticles from the lung airspaces to the body. Nat Biotech. 2010;28:1300–1303. doi: 10.1038/nbt.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Carnazza S, Foti C, Gioffrè G, Felici F, Guglielmino S. Specific and selective probes for Pseudomonas aeruginosa from phage-displayed random peptide libraries. Biosensors and Bioelectronics. 2008;23:1137–1144. doi: 10.1016/j.bios.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 46.Rogers DE, Tompsett R. The survival of staphylococci within human leukocytes. The Journal of experimental medicine. 1952;95:209–230. doi: 10.1084/jem.95.2.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garzoni C, Kelley WL. Return of the Trojan horse: intracellular phenotype switching and immune evasion by Staphylococcus aureus. EMBO molecular medicine. 2011;3:115–117. doi: 10.1002/emmm.201100123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allen TM, Cullis PR. Liposomal drug delivery systems: From concept to clinical applications. Advanced Drug Delivery Reviews. 2013;65:36–48. doi: 10.1016/j.addr.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 49.Joo J, et al. Porous silicon-graphene oxide core-shell nanoparticles for targeted delivery of siRNA to the injured brain. Nanoscale Horizons. 2016;1:407–414. doi: 10.1039/C6NH00082G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, et al. Fabrication of a Multifunctional Nano-in-micro Drug Delivery Platform by Microfluidic Templated Encapsulation of Porous Silicon in Polymer Matrix. Advanced Materials. 2014;26:4497–4503. doi: 10.1002/adma.201400953. [DOI] [PubMed] [Google Scholar]
- 51.Shen J, et al. High Capacity Nanoporous Silicon Carrier for Systemic Delivery of Gene Silencing Therapeutics. ACS Nano. 2013;7:9867–9880. doi: 10.1021/nn4035316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattacharjee M. Chemistry of antibiotics and related drugs. Springer Science+Business Media; New York, NY: 2016. [Google Scholar]
- 53.Farber BF, Moellering RC. Retrospective study of the toxicity of preparations of vancomycin from 1974 to 1981. Antimicrobial Agents and Chemotherapy. 1983;23:138–141. doi: 10.1128/aac.23.1.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elting LS, et al. Mississippi mud in the 1990s. Cancer. 1998;83:2597–2607. doi: 10.1002/(sici)1097-0142(19981215)83:12<2597::aid-cncr27>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 55.Wilhelm MP. Vancomycin. Mayo Clinic Proceedings. 1991;66:1165–1170. doi: 10.1016/s0025-6196(12)65799-1. [DOI] [PubMed] [Google Scholar]
- 56.Wardenburg JB, Schneewind O. Vaccine protection against Staphylococcus aureus pneumonia. The Journal of experimental medicine. 2008;205:287–294. doi: 10.1084/jem.20072208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Teesalu T, Sugahara KN, Ruoslahti E. In: Methods in Enzymology. Wittrup KD, Gregory LV, editors. Vol. 503. Academic Press; 2012. pp. 35–56. [DOI] [PubMed] [Google Scholar]
- 58.Qin Z, Joo J, Gu L, Sailor MJ. Size Control of Porous Silicon Nanoparticles by Electrochemical Perforation Etching. Particle & Particle Systems Characterization. 2014;31:252–256. [Google Scholar]
- 59.Joo J, Cruz JF, Vijayakumar S, Grondek J, Sailor MJ. Photoluminescent Porous Si/SiO2 Core/Shell Nanoparticles Prepared by Borate Oxidation. Advanced Functional Materials. 2014;24:5688–5694. [Google Scholar]
- 60.Cai W, Chen X. Preparation of peptide-conjugated quantum dots for tumor vasculature-targeted imaging. Nature protocols. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information.