Abstract
Heparan sulfate (HS) is an essential component of cell surface and matrix proteoglycans (HS-PGs) that include syndecans and perlecan. Because of their unique structural features, the HS chains are able to specifically interact with signaling proteins–including bone morphogenetic proteins (BMPs)-via their HS-binding domain, regulating protein availability, distribution and action on target cells. Hereditary Multiple Exostoses (HME) is a rare pediatric disorder linked to germline heterozygous loss-of-function mutations in EXT1 or EXT2 that encode Golgi-resident glycosyltransferases responsible for HS synthesis, resulting in a systemic HS deficiency. HME is characterized by cartilaginous/bony tumors-called osteochondromas or exostoses- that form within perichondrium in long bones, ribs and other elements. This review examines most recent studies in HME, framing them in the context of classic studies. New findings show that the spectrum of EXT mutations is larger than previously realized and the clinical complications of HME extend beyond the skeleton. Osteochondroma development requires a somatic “second hit” that would complement the germline EXT mutation to further decrease HS production and/levels at perichondrial sites of osteochondroma induction. Cellular studies have shown that the steep decreases in local HS levels: derange the normal homeostatic signaling pathways keeping perichondrium mesenchymal; cause excessive BMP signaling; and provoke ectopic chondrogenesis and osteochondroma formation. Data from HME mouse models have revealed that systemic treatment with a BMP signaling antagonist markedly reduces osteochondroma formation. In sum, recent studies have provided major new insights into the molecular and cellular pathogenesis of HME and the roles played by HS deficiency. These new insights have led to the first ever proof-of-principle demonstration that osteochondroma formation is a druggable process, paving the way toward the creation of a clinically-relevant treatment.
Keywords: Heparan Sulfate, Heparan sulfate proteoglycans, Hereditary Multiple Exostoses, EXT1, EXT2, Signaling proteins and pathways, Drug treatment
Introduction
Heparan sulfate (HS) is a versatile and variable glycosaminoglycan and is a critical component of cell surface and matrix proteoglycans (HSPGs) that include syndecans, glypicans and perlecan [1–4]. HS owes its complexity and character to a biosynthetic pathway that is carried out in the endoplasmic reticulum and Golgi and involves a series of multiple and sequential steps coordinated with synthesis and routing of different proteoglycan core proteins. HS synthesis initiates with the attachment of xylose to prescribed serine residues specific for each proteoglycan core protein and is followed by attachment of two galactose residues and glucuronic acid to form the tetrasaccharide linkage region [1–3]. Chain polymerization starts with the addition of a N-acetyl-D-glucosamine (GlcNAc) residue to the linkage region by the enzyme EXTL2 or EXTL3 ( ) and proceeds with the sequential and systematic addition of glucuronic acid (GlcA) and GlcNAc residues by a Golgi-resident enzyme complex composed of EXT1 and EXT2, eliciting repetitive disaccharide units and eventually producing chains with an overall size of 20 to 25 kDa. While these steps are ongoing, the growing chains are subjected to a series of concurrent structural modifications that start with elimination of certain acetyl groups from GlcNAc clusters and their replacement with sulfate by the action of members of the N-deacetylase-N-sulfotransferase family. The modifications continue with epimerization of some D-glucuronic acid residues to L-iduronic acid and sulfation at positions C2, C6 or C3 around glucosamine and glucuronic/iduronic rings by O-sulfotransferase family members. Such multiple serial biosynthetic and modification steps result in chains with diverse and unique sulfation and sugar modification patterns within 6–12 sugar residue-long segments flanked by segments of largely unmodified and un-sulfated sugars [3]. Additional processing of HS chains can occur at the cell surface by the action of the endosulfatases SULF1 and SULF2 that remove 6-O sulfate groups [5, 6]. The HS chains can also be trimmed and digested to various extents by the activity of extracellular heparanase [7, 8]. Because the intracellular and cell surface biosynthetic and modifying enzymes are expressed in tissue- and developmental-specific manners as are the proteoglycan core proteins, the overall output and structural characteristics of HS and HSPGs significantly differ in different tissues and cell types and are modulated at distinct prenatal and postnatal stages and processes [3, 4, 9, 10].
Members of the bone morphogenetic protein (BMP), hedgehog, fibroblast growth factor (FGF) and Wnt signaling protein families are essential regulators of multiple and critical developmental and postnatal processes that include progenitor cell recruitment, cell differentiation, tissue morphogenesis and homeostasis, and organogenesis [11–13]. A common and telling feature of most of these proteins is that they possess a specific domain recognizing and interacting with HS [14, 15]. The HS-binding domains contain several basic residues (Arg and/or Lys) spaced by hydropathic amino acids, have an average pI > 11, and vary greatly in length, organization and structure in different proteins [16, 17], though these features appear to be strictly conserved and may thus be important for specificity of HS-protein interactions [14]. Reciprocally, the 6–12 sugar residue-long sulfated and modified segments along the HS chains may have preferential protein-selective binding characteristics [3]. The resulting protein-HS interactions have many functions, but a key one is to regulate, limit and delimit protein distribution, diffusion, gradient formation, turn-over, bioavailability and action on target cells and tissues [9, 10]. This was originally and prominently demonstrated by the analysis of Drosophila embryos bearing loss-of-function mutations in tout-velu, the homologue of vertebrate Ext1 [18, 19]. The mutant embryos exhibited an abnormal and broad distribution of hedgehog protein and ectopic expression of its receptor Patched and were affected by developmental abnormalities, including defective segmentation polarity. Chondrocytes in zebrafish mutants lacking dackel (the homologue of vertebrate Ext2) exhibited abnormal growth, lost polarity and invaded surrounding tissues, thus behaving as cells in osteochondromas [20]. A subsequent study in mice bearing a hypomorphic Ext1 mutation showed that tissues including the growth plates in prenatal developing skeletal elements contained about 10% of normal HS levels and displayed a broader and unruly distribution of Indian hedgehog (Ihh), associated with abnormalities in growth plate function and skeletal growth and elongation [21]. Loss-of-function mutations in EXT1 and EXT2 have also been detected in various forms of human cancers, likely causing abnormal cell behavior and proliferation via greater availability of growth factors and cytokines [22, 23]. Mutations in Glypican-6 core protein were found to cause recessive Omodysplasia and impairment of endochondral ossification [24]. Recently, homozygous missense mutations in EXTL3 were identified in patients within unrelated families with a neuro-immuno-skeletal dysplasia syndrome characterized by multiple defects, including lack of T cells, kyphosis, neural deficiencies, brachydactyly and/or craniofacial malformations [25–27]. In sum, a normal complement and physiologic expression and function of HS and HSPGs are clearly required for embryonic development, postnatal growth, tissue homeostasis and organ function, and abnormalities in those components, expression patterns and mechanisms can lead to disease.
Hereditary Multiple Exostoses and its diverse clinical complications
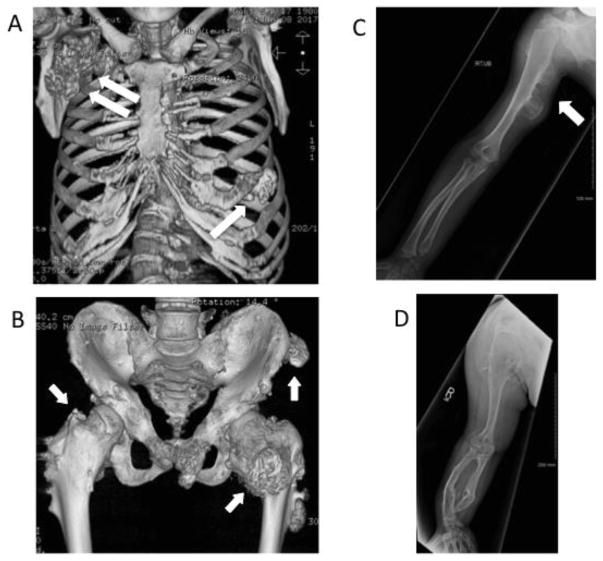
A disease most relevant to the present review article is Hereditary Multiple Exostoses (HME) that is associated with heterozygous loss-of-function mutations in EXT1 or EXT2 [28–31]. HME, also known as Multiple Osteochondromas (MO), is a rare orphan pediatric disorder with an incidence of about 1:50,000 individuals worldwide, without apparent geographic preference [32, 33]. The most obvious trait of the disease (from which its name) is the presence of benign cartilage-capped bony tumors called exostoses or osteochondromas that emerge within the perichondrium flanking the growth plates of long bones, vertebrae, ribs and cranial base in young and adolescent patients [34–37]. Many of the patients bear a large number of osteochondromas throughout their skeleton by the time their growth plates close at the end of puberty. Because of their large number, multiple anatomical locations and considerable size, the osteochondromas can, and do, cause a number of health problems, including skeletal deformities and bowing, joint ankylosis, skeletal growth disparities, and impingement of surrounding muscles, nerves and blood vessels. Fig. 1 provides vivid examples of osteochondroma burden and skeletal defects in HME patients, as revealed by computed tomography (CT) and X-ray imaging. Fig. 1A depicts the rib cage of a 37 yr-old HME patient exhibiting numerous osteochondromas along multiple ribs (Fig. 1A, arrow), infringing on mechanical function and neighboring tissues. A large mass made of soft tissues and calcified tissue was present on the upper right area near the clavicle (Fig. 1A, double arrow) that turned out to be a Grade 1 chondrosarcoma at pathology and was eventually resected by surgery. The pelvis of a 23 yr-old patient exhibited multiple osteochondromas located along the upper and lower iliac crest, pubic ramus and femurs (Fig. 1B). The pubic symphysis was likely fused by the osteochondromas, and the functioning of the left hip joint was compromised. Fig. 1C is from an 8 yr-old patient and reveals the presence of two large osteochondromas in posterior medial portion of the humerus (Fig. 1C, arrow), a small osteochondroma in distal radius, and bending of radius toward ulna. Fig. 1D illustrates the right arm from a 20 yr-old patient exhibiting significant bending of radius and ulna. The severe shortening of distal ulna created an ulna-minus wrist. The bulkiest osteochondromas were located in the distal shaft of radius and ulna and were present also in scapula and humerus.
Fig. 1.
Diagnostic CT and X-ray images obtained from HME patients. (A) This CT scan depicts the rib cage of a 37 yr-old HME patient exhibiting multiple osteochondromas along the ribs (arrow) and a large malignant chondrosarcoma Grade 1 on the upper right area near the clavicle (double arrow). The osteochondromas likely impinged on mechanical rib function, and the malignant tumor was eventually resected by surgery. (B) This CT scan shows the pelvis of a 23 yr-old patient exhibiting multiple osteochondromas along the upper and lower iliac crest (arrows), pubic ramus and femurs. The osteochondromas appear to fuse the pubic symphysis and their aggressive presence and size affected the left hip joint (arrow). (C) This X-ray image is from an 8 yr-old patient and reveals two large osteochondromas in posterior medial portion of humerus (arrow), a small osteochondroma in distal radius, and bending of radius toward ulna. (D) This X-ray image illustrates the right arm from a 20 yr-old patient displaying a marked degree of bending of radius and ulna. The severe shortening of distal ulna created an ulna-minus wrist. The bulkiest osteochondromas are located in the distal shaft of radius and ulna and are present also in scapula and humerus. All images were generously provided by the MHE Center at the Paley Orthopedic & Spine Institute, West Palm Beach, Florida.
Currently, surgery is the most common treatment for HME patients by which the most symptomatic, problematic and accessible osteochondromas are resected in order to ameliorate or correct major skeletal defects. However, osteochondromas may be difficult to reach or be located in potentially dangerous and delicate locations; as a consequence, many are often left in place, leading to long lasting problems and concerns [38]. in addition, osteochondromas undergo transformation in about 2–3% of patients and turn into chondrosarcomas (Fig. 1A) [39, 40]. Though rarely, these malignant cancers can become life threatening because of their typical resistance to chemo- or irradiation therapy, and their frequency appears to correlate with the number of benign osteochondromas left in place long term [39, 40].
Osetochondromas forming on the intracanal surface of the vertebrae are particularly dangerous and worrisome since they often impinge on, and affect the function of, the spinal cord and can cause nerve damage, physical weakness and progressive impediment of motion [41, 42]. Intracanal osetochondromas are seen in a significant number of HME patients and given their propensity to cause such problems, studies have recommended that all HME children be given routine MRI or CT screening and evaluation of their spine at presentation and consider surgical intervention [42]. A recent study focused on an additional possible complication of the spine in HME patients –scoliosis- that is not often included in assessment and natural history analyses of HME clinical symptoms [43]. The authors recruited 50 female and male HME patients with a mean age of 28.1 years (range 2–77 years) and clinically assessed them at presentation. Based on a disease severity classification that took into consideration deformities, functional limitation and number of osetochondroma-containing anatomical sites, the authors subdivided the patients into class I (least affected) to class III (most affected) groups. Clinically observable scoliosis was detected in about 70% of all patients exhibiting a mean primary curve of 15.3° and a mean minor curve of 10.6°, and spine severity encompassed King type I to type IV classification, but no type V [44]. Statistical calculations and analysis indicated that moderate scoliosis was significantly linked to disease severity and prevalent in class III patients, but was not related to overall number of osetochondromas, gender or limb deformities. Thus, mild to moderate scoliosis may be common among HME patients, but the data need to be verified in other and larger cohorts of patients before reaching conclusions about clinical significance and implications.
The multiple skeletal problems and physical difficulties associated with HME, including those described above, have long been known and appreciated, but more recent studies have focused attention to non-skeletal and often overlooked aspects of HME pathology. In a particularly attentive and revealing study [45], the authors contacted all the over 300 known patients with HME in the Netherlands and offered participation in an age-specific survey and questionnaire of: their daily function; school and professional activities; physical symptoms including pain; and overall well-being. Most adults and about 30% of the children completed the questionnaire, and data were assessed statistically by various methods. The majority of adult patients were employed, but a significant fraction of them reported to have changed jobs in connection to their own physical difficulties or a need to adapt to challenging work environments. A small number of patients were unable to hold jobs having been found to be medically unfit to work. Based on standard life quality assessment survey [46], the patients had low scores in several categories compared to control groups. About 50% of the children with HME described difficulties in school that affected and limited their participation in certain social activities and sports. Not surprisingly, both young and adult patients reported experiencing persistent pain. Data and insights in this study correlate well with those in a subsequent study of about 100 HME patients that included 57 adults and 42 children [47]. Together, these and other studies underline the pervasive and intrusive nature of HME with its negative effects on quality of life, social and personal well-being and activities, and even self-esteem. The studies call for closer and attentive clinical evaluation of HME patients, special attention to their physical and non-physical difficulties, health assessment over time, and specific remedies for diverse needs including early intervention.
EXT mutations and HME pathogenesis
As indicated above, the EXT1 and EXT2 proteins form Golgi-resident supermolecular complexes in which EXT1 exerts enzymatic function and EXT2 has a structural and scaffold role, with both proteins needed for efficient HS polymerization [48–50]. As a consequence, heterozygous mutations in either gene in HME patients lead to a systemic HS deficiency of about 50% [51]. This partial loss of HS has been found to be in itself detrimental to diverse physiologic mechanisms, including postprandial lipid clearance and metabolism and pancreatic beta-cell reserve functioning [52, 53]. However, a partial HS loss is not sufficient to cause multiple osteochondroma formation [54, 55]. As prescribed by Kundson Tumorigenesis Hypothesis [56], This process would require a somatic “second hit”, leading to further local decreases in HS synthesis and/or levels and causing resident cells to misbehave and turn tumorigenic [22]. Lines of evidence generated by several groups including ours have provided insights into these questions and have shed light on the spectrum of EXT mutations, nature of possible second hits, and consequences on osteochondroma formation and HME pathogenesis [57].
Genetic analyses conducted after those originally linking HME with EXT mutations [28, 30] have demonstrated that the spectrum of EXT mutations in this disease is quite broad and includes nonsense, missense, frame shift and splice-site mutations in EXT1 or EXT2 [31, 58, 59]. Mutations registered in the Multiple Osteochondroma Mutation Database (URL: http://medgen.uantwerpen.be/LOVDv.2.0/) exceeds 650 at present [58], but no definitive system is available to classify disease severity amongst HME patients and establish reliable genotype-phenotype correlations [60, 61]. This situation likely reflects the fact that clinically, HME is variable and can affect patients to divergent degrees, even within family members with the same EXT mutation and amongst unrelated patients with the same mutation [61]. In general however, patients with EXT1 mutations usually have a more severe clinical phenotype broadly and variously defined in terms of overall number of osteochondromas, skeletal deformations, anatomical sites affected, and physical difficulties and limitations [62]. The more severe phenotypes associated with EXT1 mutations possibly reflect its enzymatic function in HS synthesis compared to the structural roles played by EXT2.
HME does not have preferential geographical occurrence or different frequency around the world as pointed out above, and recent EXT mutation surveys in local populations in Japan, Poland, Italy and Spain have confirmed these basic important notions [63–66]. Not surprisingly, these studies have identified additional novel mutations in EXT1 or EXT2, broadening the mutation spectrum in HME to over 700. These studies raise anew long-standing questions regarding: how each mutation –and particularly the several distinct missense mutations- would interfere with EXT protein structure and function; whether and which of the mutations could lead to a dominant-negative effect on EXT complex function and HS synthesis; and which types of “second hits” could occur in HME patients and how they would relate to, and cooperate with, the germline heterozygous EXT mutations, eventually leading to steeper local HS loss and osteochondroma formation. To date, the best documented types of second hits observed in surgical retrieval human osteochondromas from HME patients are loss-of-heterozygosity (LOH), aneuploidy or other large genomic changes, all rendering local resident cells EXT1- or EXT2-null [54, 67–69]. To directly evaluate the significance of such genetic changes and their relevance to HME pathogenesis, several groups conducted mouse studies. Single heterozygous Ext1+/− or Ext2+/− mutant mice were found to be largely normal [50, 70], clearly showing that a partial decrease in Ext expression and HS synthesis is well tolerated and largely compatible with normal development and postnatal life. However, conditional loss of both Ext1 alleles in growth plate and/or perichondrium cells (accompanied by a steep decrease in HS levels) caused formation of multiple osteochondromas in long bones, ribs and vertebrae, thus mimicking the human HME phenotype and affirming the need for severe Ext/HS loss in disease initiation and progression [71–73]. In this regard, it is interesting to note that a few HME patients have been found to bear germline compound heterozygous mutations in both EXT1 and EXT2 [58, 59, 64]. It had remained unclear whether those patients would still require an additional second hit to develop HME or whether those inborn compound mutations would be sufficient. While this question is still pending, our own studies in mice have shown that compound heterozygous Ext1+/−;Ext2+/− mutant mice did develop multiple osteochondromas by 2 months of age [70], while companion single heterozygous mice did not as also shown previously [50]. Because both Ext1 and Ext2 are needed for HS synthesis, the compound heterozygous mice were expected to produce about 25% of HS compared to wild type mice, and our direct analyses of HS levels and distribution in mutant versus wild type growth plates confirmed that prediction. It is possible then that patients with germline compound heterozygous mutations in EXT1 and EXT2 may not require an additional second hit to develop a HME phenotype. Taken together, human and mouse studies carried out so far have established that single heterozygous mutations in EXT1 or EXT2 (and the resulting 50% reduction in systemic HS levels) are not sufficient to trigger a full-fledged HME phenotype. Rather, additional genetic changes are required that would cause a steep drop in HS levels, transform the phenotype of local resident cells, and provoke osteochondroma formation.
In this regard, it is of interest to further consider the distinct disease phenotype of patients with homozygous EXTL3 missense mutations identified recently [25–27]. As pointed out above, these patients suffer from various immune, neural and skeletal health problems and display a partial systemic HS deficiency (evaluated in serum and urine), but do not have typical symptoms of HME. Indeed, mutations in EXTL3 or EXTL2 have not been reported in HME patients [58]. It is likely then that the HS deficiency elicited by the homozygous missense EXTL3 mutations is moderate and insufficient to trigger HME and could also be compensated by expression of EXTL2 within growth plates and perichondrium where the development of osteochondromas occurs.
Mechanisms of osteochondroma development
The above studies have been quite important and illuminating regarding a need for additional somatic genetic changes, but did not experimentally test whether stochastic events such as LOH would indeed be sufficient to trigger HME. Also, the studies did not analyze how a severe drop in local HS levels would alter the phenotype of resident cells and trigger osteochondroma formation. The authors of a recent important study tackled the first question by creating transgenic mouse lines that contain head-to-head loxP sites flanking exon 2 in Ext1 (termed Ext1e2fl/e2fl) [71]. This genetic design was expected to yield a stochastic distribution of forward and inverted flanked segments after transient expression of Cre recombinase [74], leading to the generation of chimeric tissues that contained a low prevalence of mutant cells following inversion of both alleles. Cre expression was induced by including doxycycline in drinking water of compound Ext1e2fl/e2fl;Col2-rtTA-Cre mice from postnatal day 8 to day 15 (P8 to P15). Previous studies showed that Cre expression driven by Col2 regulatory sequences targets cells in both growth plate and neighboring perichondrium [75]. Numerous and large osteochondromas emerged near the knees and along the rib cage over the following 4 to 8 weeks in doxycycline-treated Ext1e2fl/e2fl;Col2-rtTA-Cre mice, but not in heterozygous Ext1e2fll+;Col2- rtTA-Cre mice or companions not given doxycycline. Loss of Ext1 enzymatic activity and HS synthesis in homozygous exon2-inverted tissues was confirmed by immunohistochemistry. Interestingly, Ext1e2fl/e2fl;Osterix-CreERT mice expressing CreERT in hypertrophic chondrocytes and bone cells [76] did not develop osteochondromas following tamoxifen treatment. This important study revealed that: stochastic loss of both Ext1 alleles is sufficient for, and does lead to, a HME-like phenotype; loss of one allele is not sufficient and is tolerated; and osteochondroma formation is ascribable to cells targeted by Col2- but not Osterix-driven Cre, pointing to specificity of cells involved in tumor formation.
These observations were extended in a concurrent and equally important study [72]. The authors relied on standard floxed Ext1 mice and mated them with Col2-CreERT mice [77], the latter previously assumed to express Cre recombinase only after tamoxifen treatment. However, the authors had observed a low level of leakiness by the Cre transgene without tamoxifen treatment, thus causing a random stochastic deletion of both Ext1 alleles in a small number of Col2-expressing cells. As pointed out above, the targeted cells included growth plate chondrocytes and adjacent perichondrial cells [75]. Numerous osteochondromas formed in long bone epiphyses and along the ribs in Ext1f/f;Col2-CreERT mice by one month of age, but not in heterozygous Ext1f/+;Col2-CreERT mice or those lacking the Cre transgene. Close histological inspection and analysis of each osteochondroma showed that the cells were a mixture of mutant and wild type cells, affirming the notion that mutant cells can recruit surrounding healthy cells into tumor formation and growth [67, 69]. Taken together, the above studies provided the most direct evidence to date that stochastic inactivation of both Ext alleles in a small number of cells is sufficient to elicit osteochondroma formation and leads to the development of a HME-like phenotype over time.
While providing important new information on HME molecular pathogenesis, the above studies did not examine aspects of HME cellular pathogenesis, and in particular the origin and nature of cells responsible for osteochondroma formation and what cellular mechanisms would be deranged as a consequence of local HS loss. At this regard, it is important to point out that osteochondromas were originally thought to derive from mutant growth plate chondrocytes (reviewed in [34]). However, we and other groups suggested that instead, osteochondromas originated from mesenchymal cells located in perichondrium [40, 55, 78] known to contain progenitors normally engaged in lateral appositional expansion of skeletal elements as well as tissue repair [79]. To directly test our hypothesis, we selectively targeted cells within perichondrium and asked whether conditional ablation of Ext1 alleles in those cells would be sufficient to provoke osteochondroma formation. We also asked what cellular mechanisms may be involved. We used Gdf5Cre mice that strongly express Cre recombinase in perichondrium flanking the epiphyseal portion of growth plates [78], being unique in that regard. Multiple osteochondroma-like masses did develop in Ext1f/f;Gdf5Cre mice over time that displayed a typical growth plate-like cartilaginous organization. A similar induction of osteochondroma formation occurred when long bone rudiments from E18.5 Ext1f/f mouse embryos reared in organ culture were treated with either adeno-Cre (to ablate Ext1) or Surfen, a HS antagonist drug [80]. These experiments revealed also that the onset of osteochondroma formation in vivo and in vitro was preceded by ectopic and excess BMP signaling (a major pro-chondrogenic pathway) within perichondrium as indicated by phosphorylated SMAD1/5/8 levels. In good correlation, we found that treatment of micromass cultures of Ext1f/f mouse embryo limb bud mesenchymal cells with adeno-Cre or Surfen caused: excessive chondrogenesis and cartilage nodule formation; overexpression of chondrogenic master and matrix genes; and cellular over-responsiveness to exogenous BMP proteins. Surface plasmon resonance assays indicated that interference with HS function by Surfen treatment reduced the physical association kinetics and interactions of recombinant BMP2 with immobilized HS chains. Taken together, our data strongly suggested that: osteochondromas originate from perichondrial progenitor cells; and loss of HS or interference with HS function provoke ectopic excessive BMP signaling in perichondrium, covert the phenotype of local progenitors from mesenchymal to chondrogenic, and induce osteochondroma formation. An important implication of these findings is that under physiologic circumstances, normal levels of HS and Ext expression would maintain the innate mesenchymal phenotype of perichondrial cells and prevent signaling by locally-expressed BMPs [81, 82]. It is important to note that while HS appears to normally restrain BMP signaling [83, 84], HS has the opposite effect on, and promotes, FGF signaling which is a major anti-chondrogenic pathway expressed in several non-cartilaginous tissues including perichondrium [85–87]. Thus, while boosting BMP signaling, the HS deficiency in HME could dampen FGF signaling, and this combination would alter the phenotype of perichondrial progenitors, induce chondrogenic differentiation and induce osteochondroma development (see model in Fig. 2).
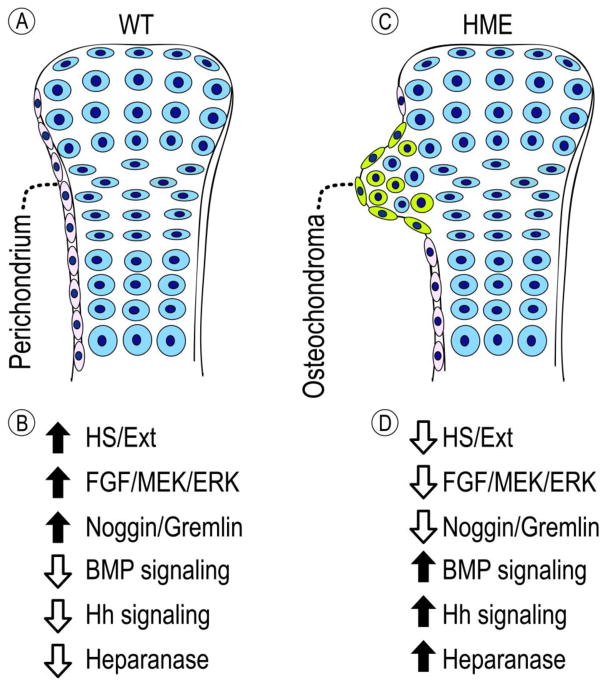
Fig. 2.
Schematic illustrating possible changes in regulatory steps that underlie osteochondroma formation. (A) In healthy wild-type (WT) circumstances, perichondrium (in off-white) would define the boundary of growth plate (in blue) in skeletal elements such as long bones. Resident perichondrial cells would be characterized by a normal mesenchymal phenotype and express traits including: a flat cell morphology; normal EXT expression and HS levels; strong anti-chondrogenic mechanisms, including FGF and ERK/MEK signaling and Noggin and Gremlin expression [98]; and low activity/expression of pro-chondrogenic mechanisms including BMP and hedgehog signaling and heparanase. (B) Up and down arrows depict the high and low levels of respective traits in normal perichondrium. (C) During the course of HME, LOH or other “second hits” would cause a steep and nearly complete loss of EXT expression and/or HS levels in local cells (in green) within perichondrium bordering the heterozygous EXT mutant growth plate. This would cause concerted decreases in anti-chondrogenic pathways and reciprocal increases in pro-chondrogenic pathways and heparanase expression in mutant cells (summarized in D), thus altering homeostatic signaling mechanisms and triggering differentiation of perichondria progenitors into round-shaped chondrocytes (in green). The growing osteochondromas would contain a mixture of mutant (green) and heterozygous (blue) cells, the latter being recruited into the osteochondroma forming process by mutant cells. The changes occurring in BMP, hedgehog and FGF signaling pathways and in expression of heparanase could each offer plausible therapeutic targets to inhibit osteochondroma inception and/or growth.
Therapeutic targets
In addition to their basic value and interest, the above data provided tantalizing clues and ideas about possible therapeutic targets. Given that ectopic BMP signaling in perichondrium precedes osteochondroma formation, we tested the possibility that systemic treatment with a BMP signaling antagonist could inhibit osteochondroma formation or growth [35]. To mimic HME pathogenic course more closely, we used an inducible mouse model of HME in which osteochondroma formation was first induced in juvenile mice during maximal skeletal growth. Thus, Ext1f/f;AggrecanCreER mice were injected with tamoxifen once at 5 weeks of age. On day 1 after tamoxifen injection, half of the mice were treated with the potent BMP signaling antagonist LDN-193189 (3 mg/kg) by daily IP injection [88], and the remaining half received daily injections of vehicle. After 6 weeks, the vehicle-treated mice exhibited multiple and large osteochondromas at every skeletal site examined, including long bones, ribs and cranial base. The tumors initially exhibited a typical cartilaginous growth plate-like organization and with time, acquired a bony stem in their proximal region linking them to the adjacent skeletal element. In mice treated with LDN-193189, however, osteochondroma formation was markedly reduced. Image analysis and quantification by micro-computed tomography and serial histological inspection showed that the decrease amounted to over 65% compared to vehicle-treated mice. No appreciable side effects of drug treatment were observed. Interestingly, when we examined early time points, we found that osteochondroma formation in vehicle-treated mice was preceded by ectopic excessive BMP signaling (as expected from our earlier studies) and a reciprocal decrease in phosphorylated ERK1/2 levels. The latter are major downstream effectors of FGF signaling [85, 89], providing evidence that osteochondroma formation is indeed preceded –and promoted-by opposite changes in BMP versus FGF signaling (see Fig. 2). Our data were the first to provide proof-of-principle evidence that osteochondorma formation is amenable to –and can be inhibited by- drug treatment [35]. Similar observations have just been reported by another group [90].
Studies have indicated that there is an inverse relationship between EXT and heparanase expression in several types of cancer cells [91–93]. On the surface, these observations appear paradoxical and counter-intuitive by suggesting that when HS production decreases as a result of decreased EXT expression or EXT mutations, the cells would up-regulate expression of heparanase to decrease the residual HS levels even further. In agreement with those cancer studies, osteochondromas resected from HME patients were originally shown to contain higher amounts of heparanase protein and RNA compared to normal cartilage from healthy donors [94]. We used immunohistochemistry on similar surgical retrieval specimens collected from HME patients at our Hospital and confirmed that heparanase was abundant and widespread in all osteochondroma chondrocytes, but was much lower in control growth plate chondrocytes (obtained from non-HME patients undergoing surgery for polydactyly). To test for mechanistic implications, we used recombinant human heparanase and found that it boosted chondrogenesis and BMP signaling in micromass cultures of mouse embryo limb bud mesenchymal cells. Notably, treatment of the cultures with bacterial heparitinase, Surfen or rhBMP2 up-regulated endogenous heparanase gene expression, pointing to positive regulatory loops between heparanase expression and response to decreasing HS levels or availability of HS-binding ligands. Notably also, we found that the potent heparanase inhibitor SST0001 [95] strongly inhibited chondrogenesis in the micromass cultures. Together, the data provide further evidence that heparanase is a potential important culprit in HME pathogenesis and thus, a potential therapeutic target. The data suggest also that an increase in heparanase expression could represent one of the “second hits” in HME pathogenesis.
Conclusions and perspectives
The above synopsis of current human and mouse HME literature demonstrates that there have been considerable advances in this biomedical research field over the last few years. Though the most obvious clinical trait of the disease are the osteochondromas and associated skeletal problems, HME can provoke additional difficulties and health problems for the patients that make the disease a syndrome. Regarding the molecular and cellular pathogenesis of osteochondroma formation, it is now well established that tumor formation requires steep decreases in EXT expression and/or HS levels. The former would result from LOH or other genetic changes, while the latter would be directly ascribable to: decreased EXT expression; compound heterozygous EXT1 and EXT2 mutations; or increases in heparanase expression. At the cellular levels, the steep local decrease in HS levels is likely to derange the fine balances amongst different resident signaling pathways within perichondrium; in particular, the HS loss would enhance BMP signaling and would decrease FGF signaling. As a result, the phenotype and lineage assignment of progenitors located in perichondrium would shift from mesenchymal to chondrogenic, promoting ectopic chondrogenesis and osteochondroma formation. As pointed out above, most members of key signaling protein families possess a specific HS-binding domain and under normal circumstances, the resulting interactions with HS and HSPGs regulate protein distribution and action. Hence, the steep local HS deficiency in HME could alter other signaling pathways in addition to BMP and FGF. Indeed, we showed a few years ago that mouse mutants expressing low HSPG family members in cartilage exhibited ectopic hedgehog signaling in perichondrium, followed by osteochondroma-like tissue formation [96]. Formation of these tumors would thus be the end result of a multiplicity of concurrent and opposing changes in signaling pathways (Fig. 2). These realizations and insights are not only important on their own, but have allowed the identification of plausible and possible therapeutic targets. Our direct demonstration that a drug-based inhibition with BMP signaling significantly decreases osteochondroma formation in HME model mice provides definitive proof-of-principle that HME is amenable to pharmacologic intervention [35]. Interference with BMP signaling by direct means –via drugs such as LDN-193189- or by indirect means -via drugs such as palovarotene [97]- should thus be considered as a possible treatment for HME deserving of clinical consideration and testing.
Highlights.
Heparan sulfate is a component of cell surface and matrix proteoglycans and regulates many developmental processes, including signaling protein distribution and action
Hereditary Multiple Exostoses is a rare disease caused by EXT mutations and heparan sulfate deficiency and is characterized by multiple growth plate-associated osteochondromas
This review assesses recent studies in the genetics of HME and the molecular and cellular pathogenesis of osteochondroma formation
Recent research advances have provided major new mechanistic insights and are paving the way toward a pharmacological treatment for osteochondroma prevention
Acknowledgments
The original studies in the author’s laboratory upon which this review is based were supported by the NIAMS grant R01AR061758. The content of this article is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health. I would like to express gratitude to the many colleagues participating in the studies, to collaborators providing reagents and mouse lines, and to Dr. E. Koyama in particular for contribution to Fig. 2 model. CT and X-ray images were generously provided by the MHE Center at the Paley Orthopedic & Spine Institute, West Palm Beach, Florida. Due to the concise nature of this review, not all relevant and deserving literature and authors could be cited. We would like to acknowledge the passionate efforts of the Multiple Hereditary Exostoses Research Foundation (http://www.mherf.org/), a private non-profit organization dedicated to the support of families and patients with HME and to advocating HME public awareness and biomedical research.
Abbreviations used
- HS
heparan sulfate
- HSPGs
heparan sulfate proteoglycans
- GlcNAc
N-acetyl-D-glucosamine
- GlcA
glucuronic acid
- HME
Hereditary Multiple Exostoses
- BMP
bone morphogenetic protein
- FGF
fibroblast growth factor
- LOH
loss of heterozygosity
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bishop JR, Schuksz M, Esko JD. Heparan sulphate proteoglycans fine-tune mammalian physiology. nature. 2007;446:1030–1037. doi: 10.1038/nature05817. [DOI] [PubMed] [Google Scholar]
- 2.Lamanna WC, Kalus I, Pavda M, Baldwin RJ, Merry CLR, Dierks T. The heparanome - The enigma of encoding and decoding heparan sulfate sulfation. J Biotechnology. 2007;129:290–307. doi: 10.1016/j.jbiotec.2007.01.022. [DOI] [PubMed] [Google Scholar]
- 3.Sarrazin S, Lamanna WC, Esko JD. Heparan sulfate proteoglycans. Cold Spring Harb Prospect Biol. 2011;3:a004952. doi: 10.1101/cshperspect.a004952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iozzo RV, Schaefer L. Proteoglycan form and function: a comprehensive nomenclature for proteoglycans. Matrix Biol. 2015;42:11–55. doi: 10.1016/j.matbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dhoot GK, Gustafsson MK, Ai X, Sun w, Standiford DM, Emerson CP. Regulation of Wnt signaling and embryo patterning by an extracellular sulfatase. Science. 2001;293:1663–1666. doi: 10.1126/science.293.5535.1663. [DOI] [PubMed] [Google Scholar]
- 6.Morimoto-Tomita M, Uchimura K, Werb Z, Hemmerich S, Rosen SD. Cloning and characterization of two extracellular heparin-degrading endosulfatases in mouse and human. J Biol Chem. 2002;277:49175–49185. doi: 10.1074/jbc.M205131200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fux L, Ilan N, Sanderson RD, Vlodavsky I. Heparanase: busy at the cell surface. Trends Biochem Sciences. 2009;34:511–519. doi: 10.1016/j.tibs.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vreys V, David G. Mammalian heparanase: what is the message? J Cell Mol Med. 2007;11:427–452. doi: 10.1111/j.1582-4934.2007.00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bulow HE, Hobert O. The molecular diversity of glycosaminoglycans shapes animal development. Annu Rev Cell Dev Biol. 2006;22:375–407. doi: 10.1146/annurev.cellbio.22.010605.093433. [DOI] [PubMed] [Google Scholar]
- 10.Lin X. Functions of heparan sulfate proteoglycans in cell signaling during development. Development. 2004;131:6009–6021. doi: 10.1242/dev.01522. [DOI] [PubMed] [Google Scholar]
- 11.Ornitz DM, Itoh N. The Fibroblast Growth Factor signaling pathway. WIREs Dev Biol. 2015;4:215–266. doi: 10.1002/wdev.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salazar VS, Gamer LW, Rosen V. BMP signaling in skeletal development, disease and repair. Nat Rev Endocrinology. 2016;12:203–221. doi: 10.1038/nrendo.2016.12. [DOI] [PubMed] [Google Scholar]
- 13.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 14.Billings PC, Pacifici M. Interactions of signaling proteins, growth factors and other proteins with heparan sulfate: mechanisms and mysteries. Connect Tissue Res. 2015;56:272–280. doi: 10.3109/03008207.2015.1045066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu D, Esko JD. Demystifying heparan sulfate-protein interactions. Annu Rev Biochem. 2014;83:129–157. doi: 10.1146/annurev-biochem-060713-035314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cardin AD, Weintraub HJ. Molecular modeling of protein-glycosaminoglycan interactions. Arterioschler Thromb Vasc Biol. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 17.Fromm JR, Hileman RE, Caldwell EE, Weiler JM, Linhardt RJ. Pattern and spacing of basic amino acids in heparin binding sites. Arch Biochem Biophys. 1997;343:92–100. doi: 10.1006/abbi.1997.0147. [DOI] [PubMed] [Google Scholar]
- 18.Bellaiche Y, The I, Perrimon N. Tout-velu is a Drosophila homologue of the putative tumor suppressor EXT-1 and is needed for Hh diffusion. Nature. 1998;394:85–88. doi: 10.1038/27932. [DOI] [PubMed] [Google Scholar]
- 19.The I, Bellaiche Y, Perrimon N. Hedgehog movement is regulated through tout-velu-dependent synthesis of a heparan sulfate proteoglycan. Mol Cell. 1999;4:633–639. doi: 10.1016/s1097-2765(00)80214-2. [DOI] [PubMed] [Google Scholar]
- 20.Clement A, Wiweger M, von der Hardt S, Rusch MA, Selleck S, Chien C-B, Roehl HH. Regulation of zebrafish skeletogenesis by ext2/dackel and papst1/pinscher. PLoS Genetics. 2008;4(7):E1000136. doi: 10.1371/journal.pgen.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koziel L, Kunath M, Kelly OG, Vortkamp A. Ext1-dependent heparan sulfate regulates the range of Ihh signaling during endochondral ossification. Dev Cell. 2004;6:801–813. doi: 10.1016/j.devcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Huegel J, Sgariglia F, Enomoto-Iwamoto M, Koyama E, Dormans JP, Pacifici M. Heparan sulfate in skeletal development, growth, and pathology: the case of Hereditary Multiple Exostoses. Dev Dyn. 2013;242:1021–1032. doi: 10.1002/dvdy.24010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Yang X, Yamagata S, Yamagata T, Sato T. Involvement of Ext and heparanase in migration of mouse FBJ osteosarcoma cells. Mol Cell Biochem. 2013;373:63–72. doi: 10.1007/s11010-012-1475-8. [DOI] [PubMed] [Google Scholar]
- 24.Campos-Xavier AB, Martinet D, Bateman J, Belluoccio D, Rowley L, Tan TY, Baxova A, Gutstavson K, Borochowitz ZU, Innes AM, Unger S, Backmann JS, Mittaz L, Ballhausen D, Superti-Furga A, Savarirayan R, Bonafe L. Mutations in the heparan-sulfate proteoglycan Glypican 6 (GPC6) impair endochondral ossification and cause recessive omodysplasia. A,m J Human Gen. 2009;84:760–770. doi: 10.1016/j.ajhg.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oud MM, Tujnenburg P, Hempel M, van Vlies N, Ren Z, Ferdinandusse S, Jansen MH, Santer R, Johannsen J, Bacchelli C, Alders M, Li R, Davis R, Dupuis L, Cale CM, Wanders RJ, Pals ST, Ocaka L, James C, Muller I, Lehmberg KST, Engels H, Williams HJ, Beales PL, Roepman R, Dias P, Brunner HG, Cobben JM, Hall C, Hartley T, Stabej P, Mendoza-Londono R, Davis EG, de Sousa SB, LD, Arts HH, Kuijpers TW. Mutations in EXTL3 cause neuro-immuno-skeletal dysplasia syndorme. Am J Hum Genet. 2017;100:281–296. doi: 10.1016/j.ajhg.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo L, Elcioglu NH, Mizumoto S, Wang Z, Noyan B, Albayrak HM, Yamada S, Matsumoto N, Miyake N, Nishimura G, Ikegawa S. Identification of biallelic EXTL3 mutations in a novel type of spondylo-epi-metaphyseal dysplasia. J Hum Genet. 2017;62:797–801. doi: 10.1038/jhg.2017.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Volpi S, Yamazaki Y, Brauer PM, van Rooijen E, Hayashida A, Slavotinek A, Kuehn HS, Di Rocco M, Rovolta C, Bortolomai I, Du L, Felgentreff K, de Bruin LO, Hayashida K, Freeman G, Marcovecchio GE, Caputer K, Rath P, Luche N, Hagedorn EJ, Buoncompagni A, Royer-Bertrand B, Giliani S, Poliani PL, Imberti L, Dobbs K, Poulain FE, Martini A, Manis J, Linhardt RJ, Bosticardo M, Rosenzweig SD, Lee HS, Puck JM, Zuniga-Pflucker jC, Zon L, Park PW, Superti-Furga A, Notarangelo LD. EXTL3 mutations cause skeletal dysplasia, immune deficiency, and developmental delay. J Exp Med. 2017;214:623–637. doi: 10.1084/jem.20161525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahn J, Ludecke HJ, Lindow S, Horton WA, Lee B, Wagner MJ, Horsthemke B, Wells DE. Cloning of the putative tumour suppressor gene for hereditary multiple exostoses (EXT1) Nat Genet. 1995;11:137–143. doi: 10.1038/ng1095-137. [DOI] [PubMed] [Google Scholar]
- 29.Cheung PK, McCormick C, Crawford BE, Esko JD, Tufaro F, Duncan G. Etiological point mutations in the hereditary multiple exostoses gene EXT1: a functional analysis of heparan sulfate polymerase activity. Am J Hum Genet. 2001;69:55–66. doi: 10.1086/321278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hecht JT, Hogue D, Strong LC, Hansen MF, Blanton SH, Wagner H. Hereditary multiple exostosis and chondrosarcoma: linkage to chromosome 11 and loss of heterozygosity for EXT-linked markers on chromosome 11 and 8. Am J Hum Genet. 1995;56:1125–1131. [PMC free article] [PubMed] [Google Scholar]
- 31.Wuyts W, Van Hul W. Molecular basis of multiple exostoses: mutations in the EXT1 and EXT2 genes. Hum Mutat. 2000;15:220–227. doi: 10.1002/(SICI)1098-1004(200003)15:3<220::AID-HUMU2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 32.Luckert Wicklund CL, Pauli RM, Johnson DR, Hecht JT. Natural history of Hereditary Multiple Exostoses. Am J Med Genet. 1995;55:43–46. doi: 10.1002/ajmg.1320550113. [DOI] [PubMed] [Google Scholar]
- 33.Schmale GA, Conrad EU, Raskind WH. The natural history of hereditary multiple exostoses. J Bone Joint Surg Am. 1994;76:986–992. doi: 10.2106/00004623-199407000-00005. [DOI] [PubMed] [Google Scholar]
- 34.Jones KB. Glycobiology and the Growth Plate: Current Concepts in Multiple Hereditary Exostoses. J Pediatr Orthop. 2011;31:577–586. doi: 10.1097/BPO.0b013e31821c7738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha R, Mundy C, Bechtold T, Sgariglia F, Ibrahim MM, Billings PC, Carroll K, Koyama E, Jones KB, Pacifici M. Unsuspected osteochondroma-like outgrowths in the cranial base of Hereditary Multiple Exostoses patients and modeling and treatment with a BMP antagonist in mice. PLoS Genetics. 2017;13:E1006742. doi: 10.1371/journal.pgen.1006742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon L. Hereditary multiple exostosis. J Bone Joint Surg. 1963;45B:292–304. [Google Scholar]
- 37.Stieber JR, Dormans JP. Manifestations of Hereditary Multiple Exostoses. J Am Acad Orthop Surg. 2005;13:110–120. doi: 10.5435/00124635-200503000-00004. [DOI] [PubMed] [Google Scholar]
- 38.Dormans JP. Pediatric Orthopaedics: Core Knowledge in Orthopaedics. Elsevier Mosby; Philadelphia: 2005. [Google Scholar]
- 39.Porter DE, Lonie L, Fraser M, Dobson-Stone C, Porter JR, Monaco AP, Simpson AH. Severity of disease and risk in malignant change in hereditary multiple exostoses. J Bone Joint Surg Br. 2004;86:1041–1046. doi: 10.1302/0301-620x.86b7.14815. [DOI] [PubMed] [Google Scholar]
- 40.Porter DE, Simpson AHRW. The neoplastic pathogenesis of solitary and multiple osteochondromas. J Pathol. 1999;188:119–125. doi: 10.1002/(SICI)1096-9896(199906)188:2<119::AID-PATH321>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 41.Ashraf A, Larson AN, Ferski G, Mielke CH, Wetjen NM, Guidera KJ. Spinal stenosis frequent in children with multiple hereditary exostoses. J Child Orthop. 2013;7:183–194. doi: 10.1007/s11832-013-0484-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roach JW, Klatt JWB, Faulkner ND. Involvement of the spine in patients with Multiple Hereditary Exostoses. J Bone Joint Surg. 2009;91:1942–1948. doi: 10.2106/JBJS.H.00762. [DOI] [PubMed] [Google Scholar]
- 43.Matsumoto Y, Matsumoto K, Harimaya K, Okada S, Doi T, Iwamoto Y. Scoliosis in patients with multiple hereditary exostoses. Eur Spine J. 2015;24:1568–1573. doi: 10.1007/s00586-015-3883-4. [DOI] [PubMed] [Google Scholar]
- 44.King HA, Moe JH, Bradford DS, Winter RB. The selection of fusion levels in thoracic idiopathic scoliosis. J Bone Joint Surg Am. 1983;65:1302–1312. [PubMed] [Google Scholar]
- 45.Goud AL, de Lange J, Scholtes VA, Bulstra SK, Ham SJ. Pain, physical and social functioning, and quality of life in individuals with multiple hereditary exostoses in The Netherlands: a national cohort study. J Bone Joint Surg Am. 2012;94:1013–1020. doi: 10.2106/JBJS.K.00406. [DOI] [PubMed] [Google Scholar]
- 46.Hays RD, Morales LS. The RAND-36 measure of health-related quality of life. Annu Med. 2001;33:350–357. doi: 10.3109/07853890109002089. [DOI] [PubMed] [Google Scholar]
- 47.Chhina H, Davis J, Alvarez CM. Health-related quality of life in people with hereditary Multiple Exostoses. J Pediatr Orthopaedics. 2012;32:210–214. doi: 10.1097/BPO.0b013e31823ee31c. [DOI] [PubMed] [Google Scholar]
- 48.Busse-Wicher M, Wicher KB, Kusche-Gullberg M. The exostosin family: proteins with many functions. Matrix Biol. 2014;35:25–33. doi: 10.1016/j.matbio.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Lin X, Wei G, Shi Z, Dryer L, Esko JD, Wells DE, Matzuk MM. Disruption of gastrulation and heparan sulfate biosynthesis in EXT1-deficient mice. Dev Biol. 2000;224:299–311. doi: 10.1006/dbio.2000.9798. [DOI] [PubMed] [Google Scholar]
- 50.Stickens D, Zak BM, Rougier N, Esko JD, Werb Z. Mice deficient in Ext2 lack heparan sulfate and develop exostoses. Development. 2005;132:5055–5068. doi: 10.1242/dev.02088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Anower-E-Khuda MF, Matsumoto K, Habuchi H, Morita H, Yokochi T, Shimizu K, Kimata K. Glycosaminoglycans in the blood of hereditary multiple exostoses patients: half reduction of heparan sulfate to chondroitin sulfate ratio and the possible diagnostic application. Glycobiology. 2013;23:865–876. doi: 10.1093/glycob/cwt024. [DOI] [PubMed] [Google Scholar]
- 52.Chen S, Wassenhove-McCarthy DJ, Yamaguchi Y, Holzman B, van Kuppevelt Th, Jenniskens G, Wijnhoven TJ, Woods AC, McCarthy KJ. Loss of heparan sulfate glycosaminoglycan assembly in podocytes does not lead to proteinuria. Kidney Int. 2008;74:289–299. doi: 10.1038/ki.2008.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mooij HL, BernelotMoens SJ, Gordts PL, Stanford KI, Foley EM, van den Boogert MA, Witjes JJ, Hassig HC, Tanck MW, van de Sande MA, Levels JH, Kstelein JJ, Stroes ES, Dallinga-Thie GM, Esko JD, Nieuwdorp M. Ext1 heterozygosity causes a modest effect on postprandial lipid clearance in humans. J Lipid Res. 2015;56:665–673. doi: 10.1194/jlr.M053504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bovee JV, Cleton-Jansen A-M, Wuyts W, Caethoven G, Taminiau AH, Bakker E, Hul WV, Cornelisse CJ, Hogendoorn PC. EXT-mutation analysis and loss of heterozygosity in sporadic and hereditary osteochondromas and secondary chondrosarcoma. Am J Hum Genet. 1999;65:689–698. doi: 10.1086/302532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hecht JT, Hayes E, Haynes R, Cole GC, Long RJ, Farach-Carson MC, Carson DD. Differentiation-induced loss of heparan sulfate in human exostosis derived chondrocytes. Differentiation. 2005;73:212–221. doi: 10.1111/j.1432-0436.2005.00025.x. [DOI] [PubMed] [Google Scholar]
- 56.Knudson AG. Hereditary cancer: two hits revisited. J Cancer Res Clin Oncol. 1996;122:135–140. doi: 10.1007/BF01366952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pacifici M. Hereditary Mutliple Exostoses: new insights into pathogenesis, clinical complications, and potential treatments. Curr Osteoporos Rep. 2017;15:142–152. doi: 10.1007/s11914-017-0355-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jennes I, Pedrini E, Zuntini M, Mordenti M, Balkassmi S, Asteggiano CG, Casey B, Bakker S, Sangiorgi L, Wuyts W. Multiple osteochondromas: mutation update and description of the multiple osteochondromas mutation database (MOdb) Hum Mutat. 2009;30:1620–1627. doi: 10.1002/humu.21123. [DOI] [PubMed] [Google Scholar]
- 59.Zuntini M, Pedrini E, Parra A, Sgariglia F, Gentile FV, Pandolfi M, Alberghini M, Sangiorgi L. Genetic models of osteochondroma onset and neoplastic progression: evidence for mechanisms alternative to EXT genes inactivation. Oncogene. 2010;29:3827–3834. doi: 10.1038/onc.2010.135. [DOI] [PubMed] [Google Scholar]
- 60.Alvarez C, Tredwell S, De Vera M, Hayden M. The genotype-phenotype correlation of hereditary multiple exostoses. Clin Genet. 2006;70:122–130. doi: 10.1111/j.1399-0004.2006.00653.x. [DOI] [PubMed] [Google Scholar]
- 61.Pedrini E, Jennes I, Tremosini M, Milanesi A, Mordenti M, Parra A, Sgariglia F, Zuntini M, Campanacci L, Fabbri N, Pignotti E, Wuyts W, Sangiorgi L. Genotype-phenotype correlation study in 529 patients with Hereditary Multiple Exostoses: identification of “protective” and “risk” factors. J Bone Joint Surg. 2011;93:2294–2302. doi: 10.2106/JBJS.J.00949. [DOI] [PubMed] [Google Scholar]
- 62.Clement ND, Porter DE. Hereditary multiple exostoses: anatomical distribution annd burdern of exostoses is dependent upon genotype and gender. Scottish Med J. 2014;59:34–44. doi: 10.1177/0036933013518150. [DOI] [PubMed] [Google Scholar]
- 63.Ciavarella M, Coco M, Baorda F, Stanziale P, Chetta M, Bisceglia L, Palumbo P, Bengala M, Raiteri P, Silengo M, Caldarini C, Facchini R, Lala R, Caveliere ML, De Brasi D, Pasini B, Zelante L, Guarnieri V, D’Agruma L. 20 novel point mutations and one large deletion in EXT1 and EXT2 genes: report of diagnostic screening in a large Italian cohort of patients affected by hereditary multiple exostosis. Gene. 2013;515:339–348. doi: 10.1016/j.gene.2012.11.055. [DOI] [PubMed] [Google Scholar]
- 64.Ishimaru D, Gotch M, Takayama S, Kosaki R, Matsumoto Y, Narimatsu H, Sato T, Kimata K, Akiyama H, Shimizu K, Matsumoto K. Large-scale mutational analysis in the EXT1 and EXT2 genes for Japanese patients with multiple osteochondromas. BMC Genetics. 2016;17:52. doi: 10.1186/s12863-016-0359-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jamsheer M, Socha M, Sowinska-Seidler A, Telega K, Trzeciak T, Latos-Bielenska A. Mutational screening of EXT1 and EXT2 genes in Polisj patients with hereditary multiple exostoses. J Applied Genet. 2014;55:183–188. doi: 10.1007/s13353-014-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarrion P, Sangorrin A, Urreizti R, Delgado A, Artuch R, Mantorell L, Armstrong J, Anton J, Torner F, Vilaseca MA, Nevado J, Lapunzina P, Asteggiano CG, Balcells S, Grinberg D. Mutations in the EXT1 and EXT2 genes in Spanish patients with multiple osteochondromas. Scientic Reports. 2013;3:1346. doi: 10.1038/srep01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bernard MA, Hall CE, Hogue DA, Cole WG, Scott A, Snuggs MB, Clines GA, Ludecke HJ, Lovett M, VWWB, Hecht JT. Diminshed levels of the putative tumor suppressor proteins EXT1 and EXT2 in exostosis chondrocytes. Cell Motil Cytoskeleton. 2001;48:149–162. doi: 10.1002/1097-0169(200102)48:2<149::AID-CM1005>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 68.Hameetman L, Szuhai K, Yavas A, Knijnenburg J, van Duin M, van Dekken H, Timiniau AH, Cleton-Jansen A-M, Bovee JV, Hogendoorn PC. The role of EXT1 in nonhereditary osteochondroma: identification of homozygous deletions. J Natl Cancer Inst. 2007;99:396–406. doi: 10.1093/jnci/djk067. [DOI] [PubMed] [Google Scholar]
- 69.Reijnders CM, Waaijer CJ, Hamilton A, Buddingh EP, Dijkstra SP, Ham J, Bakker E, Szuhai K, Karperien M, Hogendoorn PC, Stringer SE, Bovee JV. No haploinsufficiency but loss of heterozygosity for EXT in multiple osteochondromas. Am J Path. 2010;177:1946–1957. doi: 10.2353/ajpath.2010.100296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zak BM, Schuksz M, Koyama E, Mundy C, Wells DE, Yamaguchi Y, Pacifici M, Esko JD. Compound heterozygous loss of Ext1 and Ext2 is sufficient for formation of multiple exostoses in mouse ribs and long bones. Bone. 2011;48:979–987. doi: 10.1016/j.bone.2011.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jones KB, Piombo V, Searby C, Kurriger G, Yang B, Grabellus F, Roughley PJ, Morcuende JA, Buckwalter JA, Capechhi MR, VA, Sheffield VC. A mouse model of osteochondromagenesis from clonal inactivation of Ext1 in chondrocytes. Proc Natl Acad Sci USA. 2010;107:2054–2059. doi: 10.1073/pnas.0910875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Matsumoto K, Irie F, Mackem S, Yamaguchi Y. A mouse model of chondrocyte-specific somatic mutation reveals a role for Ext1 loss of heterozygosity in multiple hereditary exostoses. Proc Natl Acad Sci USA. 2010;107:10932–10937. doi: 10.1073/pnas.0914642107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sgariglia F, Candela ME, Huegel J, Jacenko O, Koyama E, Yamaguchi Y, Pacifici M, Enomoto-Iwamoto M. Epiphyseal abnormalities, trabecular bone loss and articular chondrocyte hypertrophy develop in the long bones of postnatal Ext1-deficient mice. Bone. 2013;57:220–231. doi: 10.1016/j.bone.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lam KP, Rajewsky K. Rapid elimination of mature autoreactive B cells demonstrated by Cre-induced change in B cell antigen receptor specificity in vivo. Proc Natl Acad Sci USA. 1998;95:13171–13175. doi: 10.1073/pnas.95.22.13171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ono N, Ono W, Nagasawa T, Kronenberg HM. A subset of chondrogenic cells provides early mesenchymal progenitors in growing bones. Nat Cell Biol. 2014;16:1157–1167. doi: 10.1038/ncb3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Maes C, Kobayashi A, Kronenberg HM. A novel transgenic mouse model to study the osteoblast lineage in vivo. Ann NY Acad Sci. 2007;1116:1490164. doi: 10.1196/annals.1402.060. [DOI] [PubMed] [Google Scholar]
- 77.Nakamura E, Nguyen M-T, Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreERT to assay temporal activity windows along the proximodistal limb skeleton. Dev Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- 78.Huegel J, Mundy C, Sgariglia F, Nygren P, Billings PC, Yamaguchi Y, Koyama E, Pacifici M. Perichondrium phenotype and border function are regulated by Ext1 and heparan sulfate in developing long bones: A mechanism likely deranged in Hereditary Multiple Exostoses. Dev Biol. 2013;377:100–112. doi: 10.1016/j.ydbio.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Colnot C, Lu C, Hu D, Helms JA. Distinguishing the contributions of the perichondrium, cartilage, and vascular endothelium to skeletal development. Dev Biol. 2004;269:55–69. doi: 10.1016/j.ydbio.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 80.Schuksz M, Fuster MM, Brown JR, Crawford BE, Ditto DP, Lawrence R, Glass CA, Wang LC, Tor Y, Esko JD. Surfen a small molecule antagonist of heparan sulfate. Proc Natl Acad Sci USA. 2008;105:13075–13080. doi: 10.1073/pnas.0805862105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bandyopadhyay A, Kubilus JK, Crochiere ML, Linsenmayer TF, Tabin CJ. Identification of unique molecular subdomains in the perichondrium and periosteum and their role in regulating gene expression in the underlying chondrocytes. Dev Biol. 2008;321:162–174. doi: 10.1016/j.ydbio.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Minina E, Schneider L, Rosowski M, Lauster R, Vortkamp A. Expression of Fgf and Tgfbeta signaling related genes during embryonic endochondral ossification. Gene Exp Patterns. 2005;6:102–109. doi: 10.1016/j.modgep.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 83.Ohkawara B, Iemura S, ten Dijke P, Ueno N. Action range of BMP is defined by its N-terminal basic amino acid core. Curr Biol. 2002;12:205–209. doi: 10.1016/s0960-9822(01)00684-4. [DOI] [PubMed] [Google Scholar]
- 84.Ruppert R, Hoffmann E, Sebald W. Human bone morphogenetic protein 2 contains a heparin-binding site which modifies its biological activity. Eur J Biochem. 1996;237:295–302. doi: 10.1111/j.1432-1033.1996.0295n.x. [DOI] [PubMed] [Google Scholar]
- 85.Buckland RA, Collinson JM, Graham E, Davidson DR, Hill RE. Antagonistic effects of FGF4 on BMP induction of apoptosis and chondrogenesis in the chick limb bud. Mech Dev. 1998;71:143–150. doi: 10.1016/s0925-4773(98)00008-2. [DOI] [PubMed] [Google Scholar]
- 86.Ornitz DM, Marie PJ. Fibroblast growth factor signaling in skeletal development and disease. Genes & Dev. 2015;29:1463–1486. doi: 10.1101/gad.266551.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yoon BS, Pogue R, Ovchinnikov DA, Yoshii I, Mishina Y, Behringer RR, Lyons KM. BMPs regulate multiple aspects of growth plate chondrogenesis through opposing actions of FGF pathways. Development. 2006;133:4667–4678. doi: 10.1242/dev.02680. [DOI] [PubMed] [Google Scholar]
- 88.Yu PB, Deng DY, et al. BMP type I receptor inhibition reduces heterotopic ossification. Nat Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Matsushita T, Chan YY, Kawanami A, Balmes G, Landreth GE, Murakami S. Extracellular signal-regulated kinase 1 (ERK1) and ERK2 play essential roles in osteoblast differentiation and in supporting osteoclastogenesis. Mol Cell Biol. 2009;29:5843–5857. doi: 10.1128/MCB.01549-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Inubushi T, Nazawa S, Matsumoto K, Irie F, Yamaguchi Y. Aberrant perichondrial BMP signaling mediates multiple osteochondromagenesis in mice. J Clin Inv Insight. 2017;2:E90049. doi: 10.1172/jci.insight.90049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hulett MD, Freeman C, Hamdorf BJ, Baker RT, Harris MJ, Parish CR. Cloning of mammalian heparanase: an important enzyme in tumor invasion and metastasis. Nat Med. 1999;5:183–187. doi: 10.1038/10525. [DOI] [PubMed] [Google Scholar]
- 92.Quiros RM, Rao G, Plate J, Harris JE, Brunn GJ, Platt JL, Gatuso P, Prinz RA, Xu X. Elevated serum heparanase-1 levels in patients with pancreatic carcinoma are associated with poor survival. Cancer. 2006;106:532–540. doi: 10.1002/cncr.21648. [DOI] [PubMed] [Google Scholar]
- 93.Zetser A, Bashenko Y, Miao HQ, Vlodavsky I, Ilan N. Heparanase affects adhesive and tumorigenic potential of human glioma cells. Cancer Res. 2003;63:7733–7741. [PubMed] [Google Scholar]
- 94.Trebicz-Geffen M, Robinson D, Evron Z, Glaser T, Fridkin M, Kollander Y, Vlodavsky I, Ilan N, Law KF, Cheah KSE, Chan D, Werner H, Nevo Z. The molecular and cellular basis of exostosis formation in hereditary multiple exostoses. Int J Exp Path. 2008;89:321–331. doi: 10.1111/j.1365-2613.2008.00589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ritchie JP, Ramani VC, Ren Y, Naggi A, Torri G, Casu B, Penco S, Pisano C, Camminati P, Tortoreto M, Zunino F, Vlodavsky I. SST0001, a chemically modified heparin, inhibits myeloma growth and angiogenesis via disruption of the heparanase/syndecan-1 shedding: a novel mechanism for stimulation of tumor growth and metastasis. Clin Cancer Res. 2011;17:1382–1393. doi: 10.1158/1078-0432.CCR-10-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koyama E, Young B, Nagayama M, Shibukawa Y, Enomoto-Iwamoto M, Iwamoto M, Maeda Y, Lanske B, Song B, Serra R, Pacifici M. Conditional Kif3a ablation causes abnormal hedgehog signaling topography, growth plate dysfunction, and excessive bone and cartilage formation during mouse skeletogenesis. Development. 2007;134:2159–2169. doi: 10.1242/dev.001586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shimono K, Tung W-E, Macolino C, Chi A, Didizian JH, Mundy C, Chandraratna RAS, Mishina Y, Enomoto-Iwamoto M, Pacifici M, Iwamoto M. Potent inhibition of heterotopic ossification by nuclear retinoic acid receptor-γ agonists. Nature Med. 2011;17:454–460. doi: 10.1038/nm.2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Nilsson O, Parker EA, Hedge A, Chau M, Barnes KM, Baron J. Gradients of bone morphogenetic protein-related gene expression across the growth plate. J Endocrinology. 2007;193:75–84. doi: 10.1677/joe.1.07099. [DOI] [PubMed] [Google Scholar]