Abstract
The digestive methods employed by amphioxus (Branchiostoma)—both intracellular phagocytic digestion and extracellular digestion—have been discussed since 1937. Recent studies also show that epithelial cells lining the Branchiostoma digestive tract can express many immune genes. Here, in Branchiostoma belcheri, using a special tissue fixation method, we show that some epithelial cells, especially those lining the large diverticulum protruding from the gut tube, phagocytize food particles directly, and Branchiostoma can rely on this kind of phagocytic intracellular digestion to obtain energy throughout all stages of its life. Gene expression profiles suggest that diverticulum epithelial cells have functional features of both digestive cells and phagocytes. In starved Branchiostoma, these cells accumulate endogenous digestive and hydrolytic enzymes, whereas, when sated, they express many kinds of immune genes in response to stimulation by phagocytized food particles. We also found that the distal hindgut epithelium can phagocytize food particles, but not as many. These results illustrate phagocytic intercellular digestion in Branchiostoma, explain why Branchiostoma digestive tract epithelial cells express typical immune genes and suggest that the main physiological function of the Branchiostoma diverticulum is different from that of the vertebrate liver.
Keywords: phagocytic intracellular digestion, Branchiostoma, digestive tract, diverticulum, gut, gene expression
1. Introduction
Every organism requires energy to survive, and most heterotrophic unicellular organisms phagocytize food particles directly and break them down via intracellular digestion [1]. Phagocytic intracellular digestion is the evolutionary cornerstone of both the digestive and immune mechanisms of multicellular animals [2–6]. Currently, however, phagocytosis in deuterostomes is seen primarily as an immune process, and research into it focuses on the biological mechanisms used to eliminate pathogens, clear up cell debris and present antigens [7]. It is commonly accepted that animals, especially deuterostomes, digest food using only extracellular digestion, which is the process of breaking down large food particles into small, water-soluble absorbable molecules by digestive enzymes in the lumen of a digestive organ [8]. In 1937, however, van Weel observed that, in addition to general extracellular digestion, the digestive tract of Branchiostoma, especially its protruding part, the diverticulum, can phagocytize food particles directly. He, therefore, suggested that both extracellular and intracellular digestion can occur in Branchiostoma [9]. Nevertheless, because autolysis by intracellular digestive enzymes tends to occur during sample fixation, phagocytic intracellular digestion in the Branchiostoma diverticulum is difficult to observe when using the typical fixatives used for light and election microscopy [10–14]. In addition, because the Branchiostoma diverticulum has been regarded as the homologue of the vertebrate liver for more than 150 years—since it was first hypothesized by Müller in 1844 [15]—it is difficult to accept that the living animal, like the chordate ancestor, mostly could use its ‘liver’ to ‘eat’ food directly [10,12].
Recent studies of the Branchiostoma digestive tract focus on tracing the origin of the vertebrate immune mechanism [16–27], but neglect its original digestive function. For instance, though variable region-containing chitin-binding proteins (VCBPs) have been shown to play important roles in the adaptive-like immune response [16–19] in Branchiostoma, there has been little investigation into why the VCBP genes are expressed in endoderm-derived epithelial cells of the digestive tract but not in mesoderm-derived professional immune cells. Consequently, determining the biological characteristics of the epithelial cells in the digestive tract, especially those in the diverticulum, is key to understanding the role of phagocytic intracellular digestion in Branchiostoma, and could shed light on the origins of the vertebrate immune mechanism and liver.
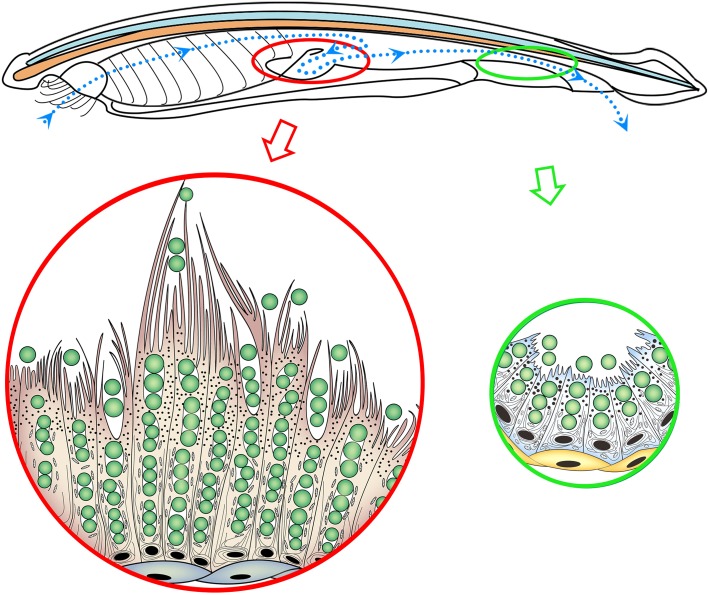
Here, by using an improved method of tissue fixation for transmission electron microscopy (TEM), we show that B. belcheri epithelial cells, especially those in the diverticulum, can phagocytize food particles directly (figure 1). The gene expression profile of these cells reveals that they can support both digestive and immune-like functions by expressing many kinds of immune genes and varying types and amounts of digestive enzymes. Interestingly, qRT-PCR results demonstrate that the gene expression profile of the cells can present two different states: after 3 days of starvation, they accumulate many kinds of digestive enzymes, as in vertebrate digestive cells, whereas, when the animal is sated, they also express immune genes in response to stimulation by exogenous food particles. Our results shed light on the context in which phagocytic intracellular digestion occurs in Branchiostoma, and suggest why the epithelial cells of the Branchiostoma digestive tract can express immune genes.
Figure 1.
Phagocytic epithelia of the Branchiostoma digestive tract. The epithelial cells of the diverticulum and hindgut can phagocytize food particles directly, and the phagocytic capability of diverticulum cells is greater than that of the hindgut epithelial cells.
2. Material and methods
(a) Animal and embryo procurement
Adult B. belcheri were collected from Zhanjiang Bay, Guangdong, China, during the summer breeding season (20 June–10 July, 2008–2015), and fed with cultured mixed algal powder including Spirulina platensis, Isochrysis galbana, Chlorella vulgaris and diatoms, etc., while being kept at 28–32°C at the Beihai Marine Station of Nanjing University, Guangxi, China. Wild samples are assumed to be sated because of the plentiful algal resources in Zhanjiang Bay [28]. Gametes were acquired by heat stimulation at 35–37°C, and then fertilized in filtered seawater. The embryos and larvae were cultured at 30 ± 2°C.
(b) Transmission electron microscopy and scanning electron microscopy
We sought a way to improve sample fixation so as to be able to visualize phagocytic intracellular digestion in the Brachiostoma digestive system. We found that initial fixation including glutaraldehyde is insufficient to fix the phagocytic epithelial cells of the B. belcheri digestive tract. Here, we used MEMPLA (0.1 mol l−1 morpholinopropanesulfonic acid, 2 mmol l−1 EGTA, 1 mmol l−1 MgSO4, 4% paraformaldehyde, 0.5 mol l−1 NaCl, pH 7.5, freshly mixed) to fix the B. belcheri digestive tract at 4°C for 12 h, prior to transfer to 2.5% glutaraldehyde in 0.02 mol l−1 PBS at 4°C. Other steps for TEM viewing and scanning electron microscopy (SEM) experiment were as previously described [29–31].
(c) The construction, sequencing and analysis of the full-length cDNA transcriptome library
Diverticula were dissected from 1-year-old adults and pooled from 100 individuals to ensure that enough RNA could be obtained (greater than 1 mg) for a full-length cDNA transcriptome library (hereafter ‘Full’). The total RNA was isolated with TRIzol LS Reagent (Thermo Fisher Scientific, 10296028) and treated with DNase I (Thermo Fisher Scientific, 18068015). The high-quality mRNA was isolated with a FastTrack MAG Maxi mRNA Isolation Kit (Thermo Fisher Scientific, K1580-02). To ensure that the results of Full reflect the natural state of diverticulum epithelial cells, we used the ‘technical route without normalization’ when constructing the library. The construction of Full was performed with a SuperScript Full-Length Library Construction Kit II (Thermo Fisher Scientific, A13268) with pDONR222 Vector according to the manufacturer's instructions (electronic supplementary material, figure S6).
The recombination pDONR222 vectors were sequenced with the Sanger method in a Thermo Fisher Scientific 3730xl DNA Analyzer using M13 forward primer; 15 000 vectors were sequenced, and 14 976 efficient expressed sequence tags (ESTs) were obtained. The redundancy statistic of the sequencing results is approximately 70% and reached the plateau phase. All the genes with Clustered-EST counts ≥10% were sequenced throughout from 5′ end to poly(A)+.
The unigenes were analysed using ORF finding, expression profile analysis, NCBI, SWISSPROT, KEGG, COG, Interpro and GO (electronic supplementary material, figure S7 and tables S3–S6). The annotated tissue-specific genes were verified by conserved homologous sequence using ClustalW and neighbour-joining trees using Mega6 with 1000 bootstraps.
(d) qRT-PCR assays
The qRT-PCR assays for determining the gene expression profile of the diverticulum epithelial cells include all the tissue-specific unigenes in Full (electronic supplementary material, table S2). From 1-year-old adults, we dissected the empty epithelial cells of the diverticulum (ED), natural sated epithelial cells of the diverticulum (SD), natural sated gut (SG), empty gut (EG), as well as pharyngeal bar (PB) and notochord (NT) cells as contrast samples. The empty samples were from animals that were starved for 3 days to ensure there were no residual algal cells in the epithelial cells of the digestive tract and that the gut had time to excrete all faeces completely.
The total RNA isolation was processed in the same way as the construction of Full. The two-step reverse transcript reaction Mixes I and II were prepared for the qRT-PCR reaction as follows. Mix I is a 12 µl system for 2 µg total RNA (adjusted to 5 µl), 0.5 µl of 50 µmol l−1 Oligo (dT) Primer (Thermo Fisher Scientific, AM5730G); 0.5 µl of Random Primer (Thermo Fisher Scientific, 48190011); 1 µl of 10 mmol l−1 dNTP Mix (Thermo Fisher Scientific, 18427013) and 5 µl of Nuclease-Free Water (Thermo Fisher Scientific, AM9938). Mix II is a 20 µl system consisting of 1 µl of SuperScript III RT at 200 U µl−1 (Thermo Fisher Scientific, 18080093), 4 µl of 5X First-Strand Buffer (Thermo Fisher Scientific, 18080093), 2 µl of 0.1 mol l−1 DTT (Thermo Fisher Scientific, 18080093); 1 µl of ribonuclease inhibitor at 40 U µl−1 (Thermo Fisher Scientific, 10777019) and 12 µl of Mix I. We kept Mix I at 65°C for 5 min, then put it on ice for 1 min and kept Mix II at 25°C for 5 min, 50°C for 1 h, 70°C for 15 min and on ice for 1 min. This cDNA can be stored at −20°C for six months. The PCR reaction is a 20 µl system performed according to the experimental standard of the manufacturer using the SYBR Green (Thermo Fisher Scientific, S7567) method.
The primers were designed online using Primer3 and NCBI Primer-BLAST, and the melt temperature (Tm) was set at 60 ± 0.5°C. All the primers were screened by electrophoresis and PCR product sequencing. The PCR products were purified with an E.Z.N.A. Ultra-Sep Gel Extraction Kit (Omega Bio-tek, D2510-01) and inserted into pMD18-T Vector (TaKaRa, 6011), then sequenced with the Sanger method using M13 forward primer. Matlab (MathWorks, USA) was used to process the raw qRT-PCR data by 2−ΔΔCt to output histograms (figures 4 and 5; electronic supplementary material, figures S9–S11, S12c), and the two-tailed Student's t-tests were used to assess statistical significance (data represent mean ± s.d.; *p ≤ 0.05, **p < 0.01, ***p < 0.001).
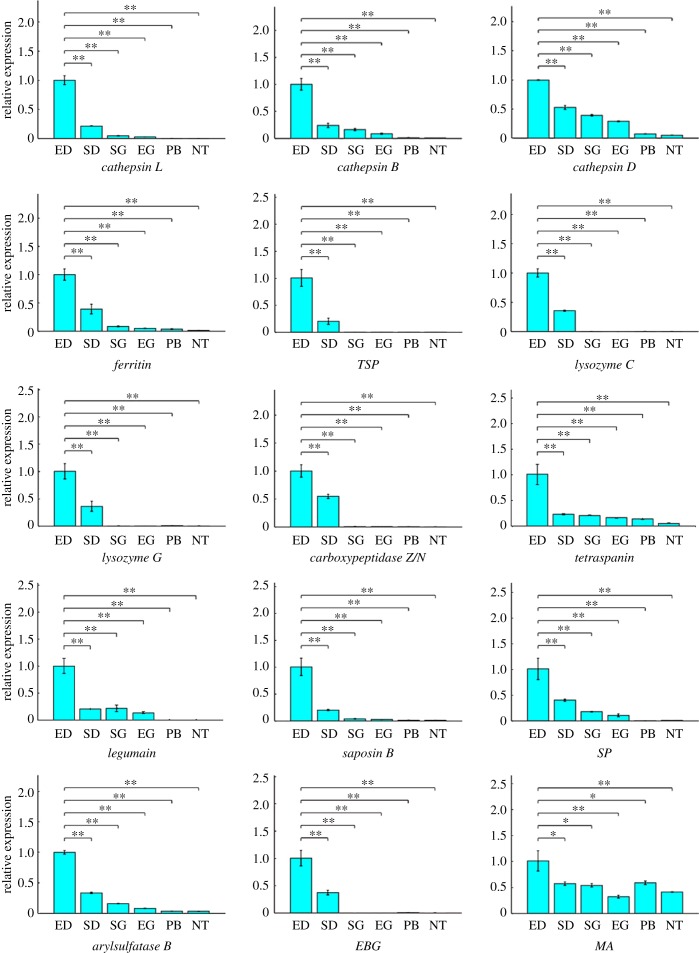
Figure 4.
The most highly expressed genes of diverticulum phagocytic epithelial cells after 3 days of starvation. qRT-PCR results reveal that these cells accumulate endogenous digestive enzymes when starved, ensuring that they are prepared to degrade phagocytized food particles. 18S was used as the internal reference gene. Data represent mean ± s.d. Two-tailed Student's t-tests were used to assess statistical significance. *p ≤ 0.05, **p < 0.01, ***p < 0.001. ED, empty diverticulum; SD, sated diverticulum; SG, sated gut; EG, empty gut; PB, pharyngeal bars; NT, notochord; TSP, trypsin-like serine protease; SP, subtilisin-like protease; EBG, endo-beta-1,4-glucanase; MA, methionine adenosyltransferase.
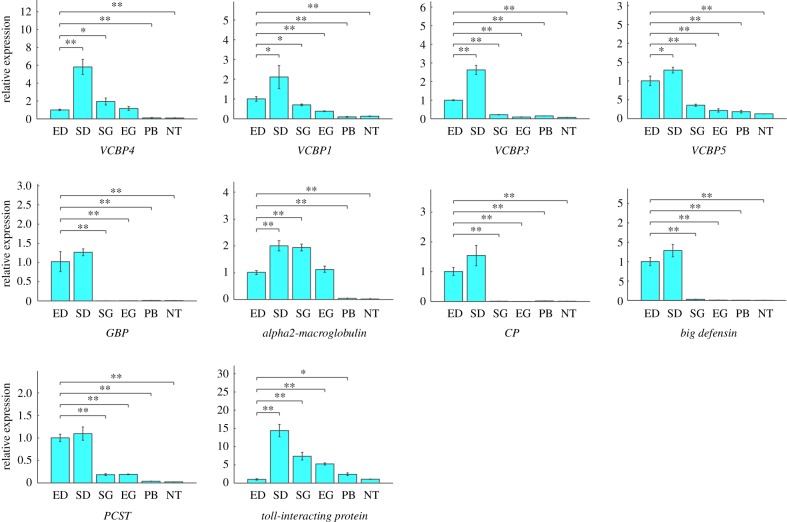
Figure 5.
The most highly expressed genes of the diverticulum phagocytic epithelial cells in the natural sated state. The Gram-negative bacteria-binding protein, alpha2-macroglobulin, chitotriosidase 1-like protein, big defensin and proprotein convertase subtilisin/kexin type 1 genes are highly expressed in the sated state relative to reference gene (18S). Data represent mean ± s.d. Two-tailed Student's t-tests were used to assess statistical significance. *p ≤ 0.05, **p < 0.01, ***p < 0.001. ED, SD, SG, EG, PB and NT: see figure 4. GBP, Gram-negative bacteria-binding protein; CP, chitotriosidase 1-like protein; PCSP, proprotein convertase subtilisin/kexin type 1.
(e) Suppression subtractive hybridization construction, sequencing and analysis
Suppression subtractive hybridization (SSH)-D and SSH-G were constructed as previously described [32] using a CloneMiner II cDNA Library Construction Kit (Thermo Fisher Scientific, A11180) and pMD18-T Vector (TaKaRa, 6011). SSH-D has 26 100 CFUs and a recombination rate of over 96%. SSH-G has 9360 CFUs and a recombination rate of over 96%. The methods of sample dissection, RNA isolation, sequencing and bioinformatics analysis (electronic supplementary material, tables S7–S10) are same as those used for Full.
(f) In situ hybridization
The samples were fixed in MEMPLA at room temperature for 1 h or at 4°C for 12 h, prior to storage in 75% ethanol at −20°C. The in situ hybridizations were performed as described in [33] and in [34] with some modifications. The templates were amplified from Full by 5′ or 3′ UTR and were inserted into pGEM-T Easy Vector Systems II (Promega, A1380), and then were amplified with M13 primers (forward: CGCCAGGGTTTTCCCAGTCACGAC; reverse: AGCGGATAACAATTTCACACAGGA). The digoxigenin (DIG)-labelled sense and antisense riboprobes were synthesized with T7 or Sp6 RNA polymerase (Roche, 11175025910) according to the manufacturer's instructions. The labelled riboprobes were precipitated with 4M LiCl/100% ethanol (40 µl/1000 µl) for 12 h. The hybridization experiment was performed in Netwell inserts (Corning, 3479). To reduce the background noise, the probe concentration was decreased to 0.1–0.2 ng µl−1 and the hybridization was performed at 60–65°C for 12–16 h. The post-hybridization washes were solution A (50% deionized formamide, 5× SSC and 0.1% Tween20) for 30 min × 3 at hybridization temperature, solution B (50% deionized formamide, 2× SSC and 0.1% Tween20) for 30 min × 2 at hybridization temperature, solution C (50% deionized formamide, 1× SSC and 0.1% Tween20) for 30 min × 2 at hybridization temperature, solution D (0.2× SSC and 0.1% Tween 20) for 30 min at 37°C and NaPBSTw (0.02 mol l−1 PBS and 0.1% Tween 20) for 10 min × 3 at room temperature on a rocker gently, with the omission of RNase treatments. The anti-DIG-AP (Roche, 11093274910) was visualized with 0.5× NBT/BCIP working solution (Roche, 11681451001) at 2–4°C for 30 min to 14 days. In order to inhibit endogenous alkaline phosphatase in the digestive tract, levamisole (Sigma, T1512) was added to NBT/BCIP working solution at a concentration of 0.48 mg ml−1 [35].
The samples were photographed with a stereoscopic microscope (ZEISS SteREO Discovery v. 20, Berlin, Germany). Images were combined using Adobe Photoshop CS6.
(g) Histology
Haematoxylin–eosin stain was used as previously described [16]. The samples were photographed with a DP72 CCD on IX81 microscope (Olympus, Tokyo, Japan). Images were acquired and processed using cellSens software (Olympus, Tokyo, Japan). Images were combined using Adobe Photoshop CS6.
3. Results
(a) Branchiostoma diverticulum epithelial cells can phagocytize food particles directly
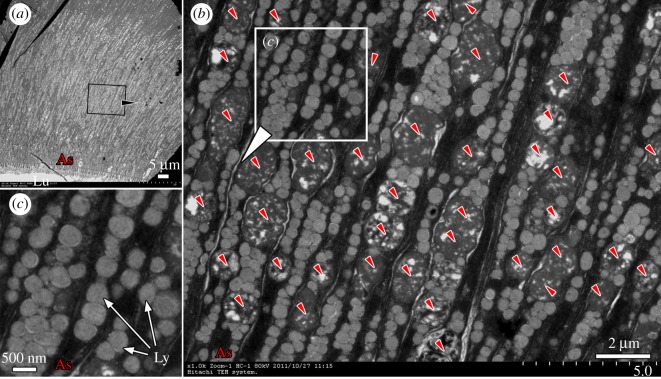
Branchiostoma filters food particles from the water with its oral cirri and pharyngeal bars. These selected food particles then flow through the oesophagus before being sorted again by the ileo-colon ring, which directs digestible food particles, such as small algal cells, to the lumen of diverticulum [36] (figure 2a). Interestingly, aside from being subject to normal extracellular digestion [8,10], the algal cells in the diverticulum lumen can also be phagocytized directly by diverticulum epithelial cells (figure 2b,c; electronic supplementary material, figure S1). These cells can phagocytize algal cells into their phagosomes (figure 2c,e; electronic supplementary material, figure S1c). They can even phagocytize more than one algal cell at once (figure 2c). The phagocytized algal cells are transported to the inner cytoplasm away from the apical side by phagosomes, waiting for further degradation (figure 2d,e; electronic supplementary material, figure S2). Importantly, the phagocytic function occurs in almost all of these cells (figure 2d; electronic supplementary material, figures S1c and S2). These cells produce large numbers of lysosomes to support their intracellular digestive function, especially after 24 h of starvation (figure 3).
Figure 2.
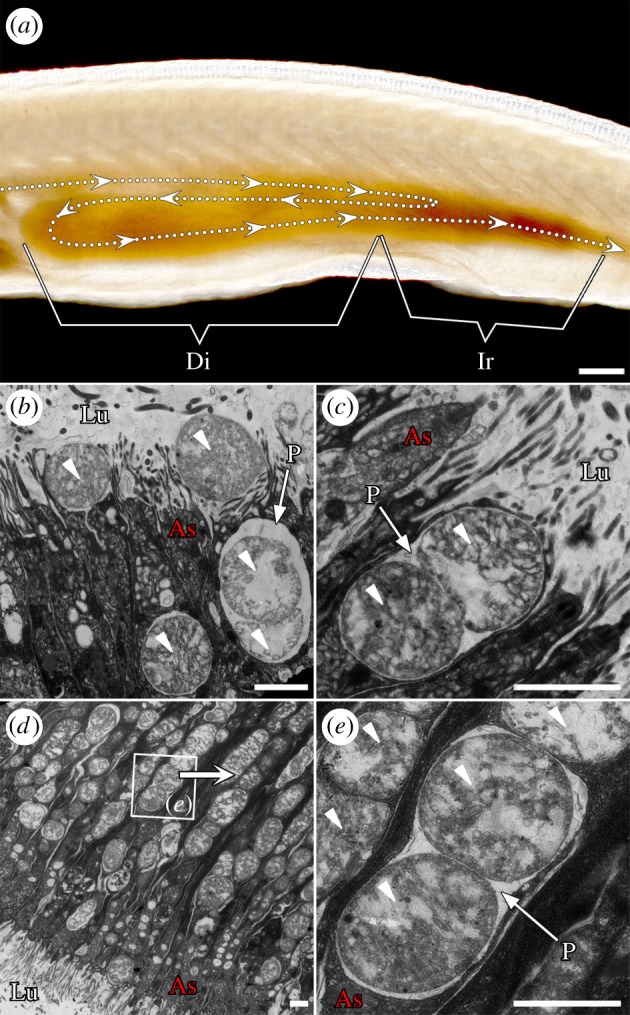
Branchiostoma diverticulum epithelial cells can phagocytize algal cells directly. (a) Food particles are transported into the diverticulum, as the arrows indicate. The ileo-colon ring sorts small and digestible cells into the diverticulum lumen. (b,c) Diverticulum epithelial cells phagocytize food cells into phagosomes directly. (d,e) Phagocytized algal cells are transported to the upper cytoplasm by phagosomes. Almost all diverticulum epithelial cells exhibit a phagocytic function (d). The small arrowheads mark the algal cells. Di, diverticulum; Ir, ileo-colon ring; Lu, lumen; As, apical side; P, phagosome. Scale bars: 100 µm (a); 2 µm (b–e).
Figure 3.
The phagocytic epithelial cells of the diverticulum produce large numbers of lysosomes to support their intracellular digestive function. (a) Cross section of diverticulum. (b, c) After 24 h of starvation, large numbers of lysosomes from 200 to 500 nm in diameter appear in the cytoplasm. The small arrowheads in (b) mark the degrading algal cells. Ly, lysosomes; Lu, lumen; As, apical side.
We also discovered that, just after the larval mouth opens at about 36 h, the epithelial cells of the diverticulum primordium can immediately begin to phagocytize algal cells (electronic supplementary material, figure S3), suggesting that phagocytic intracellular digestion occurs throughout all Branchiostoma life stages. Additionally, the ileo-colon ring is crucial because it can control the size of the food particles that enter the diverticulum: various sizes of algal cells can be found in the region of the ileo-colon ring (electronic supplementary material, figure S4), whereas the algal cells in the diverticulum lumen are mainly around 2 µm in diameter or less (figure 2b-e; electronic supplementary material, figures S1 and S2).
(b) The gene expression profile of diverticulum epithelial cells
In order to explore how the diverticulum epithelial cells deal with the phagocytized food particles, we constructed a full-length cDNA transcriptome library (abbreviation: Full; GenBank Accession Number: LIBEST_028542) to investigate the gene expression profile of these cells in a natural sated state (see Material and methods; electronic supplementary material, figures S5–S7). The genome of B. belcheri (Zhanjiang) has not been completely sequenced, and the reported genomic data from B. belcheri from Zhanjiang (studied in this paper), B. belcheri from Xiamen and B. floridae have obvious polymorphisms [37–40], so it is difficult to get precise transcriptome results from single NGS (next generation sequencing) techniques using non-mapping bioinformatic analysis based on the resulting shorter shotgun sequencing reads [39]. Thus, we discuss the analysis of individual intracellular digestive genes determined using the traditional Sanger method on a plasmid-based full-length cDNA library below.
The gene expression profile of the diverticulum epithelial cells overlaps with the patterns seen in both the digestive cells and phagocytes of vertebrates. The overwhelming majority of expressed gene markers belong to three families: cathepsin, ferritin and trypsin (table 1), and these three gene families occupy 10.57% of total ESTs and 67.79% of total annotated tissue-specific genes. The cathepsins are typical lysosomal proteases and, except for type K, most of them play major roles in intracellular protein hydrolysis [20]. In diverticulum epithelial cells, the major expressed cathepsins are cathepsin L and B (table 1), which are cysteine proteases, and cathepsin D, which is expressed at low levels, is aspartyl protease. Ferritin is an indispensable intracellular protein that acts as a buffer to maintain the balance of iron [41]. The intracellular iron store can prevent the spread of infective agents by impeding their proliferation [42]. The genes expressed at a moderate level are lysozyme, lipase, VCBPs and carboxypeptidase Z/N (table 1). Other genes expressed at low levels are tetraspanin, legumain, saposin B, subtilisin-like protease, arylsulfatase B, Gram-negative bacteria-binding protein, endo-beta-1,4-glucanase, alpha2-macroglobulin, methionine adenosyltransferase, plasminogen, chitotriosidase 1-like protein, big defensin, peroxiredoxin V, proprotein convertase subtilisin/kexin type 1 and Toll-interacting protein (table 1). Among these genes, lysozyme, VCBPs [16–19], tetraspanin, Gram-negative bacteria-binding protein, alpha2-macroglobulin [24], chitotriosidase 1-like protein [43], big defensin [16] and Toll-interacting protein are typical immune genes, and the other genes, except for methionine adenosyltransferase, are the main digestive or hydrolytic genes of both digestive cells and phagocytes. The in situ hybridization experiments show that all the genes mentioned are expressed in diverticulum epithelial cells (electronic supplementary material, figure S8).
Table 1.
Gene expression profile of diverticulum phagocytic epithelial cells.
| gene | EST counts (per cent of total ESTs) |
|---|---|
| cathepsinab | 582 (3.89%) |
| cathepsin L | 440 (2.94%) |
| cathepsin B | 132 (0.88%) |
| cathepsin D | 10 (0.07%) |
| ferritinb | 504 (3.37%) |
| trypsin-like serine proteasea | 497 (3.32%) |
| lysozymeac | 112 (0.75%) |
| lysozyme C | 92 (0.61%) |
| lysozyme G | 20 (0.13%) |
| pancreatic lipase-like proteina | 80 (0.53%) |
| VCBPsc | 79 (0.53%) |
| VCBP4 | 58 (0.40%) |
| VCBP1 | 11 (0.07%) |
| VCBP3 | 6 (0.04%) |
| VCBP5 | 4 (0.03%) |
| carboxypeptidase Z/Na | 74 (0.49%) |
| tetraspaninc | 52 (0.35%) |
| legumainab | 47 (0.31%) |
| saposin Ba | 43 (0.29%) |
| subtilisin-like proteasea | 42 (0.28%) |
| arylsulfatase Ba | 36 (0.24%) |
| gram-negative bacteria-binding proteinc | 28 (0.19%) |
| endo-beta-1,4-glucanasea | 25 (0.17%) |
| alpha2-macroglobulinc | 18 (0.12%) |
| methionine adenosyltransferase | 18 (0.12%) |
| plasminogena | 16 (0.11%) |
| chitotriosidase 1-like proteinac | 11 (0.07%) |
| big defensinc | 7 (0.05%) |
| peroxiredoxin Vb | 5 (0.03%) |
| proprotein convertase subtilisin/kexin type 1a | 5 (0.03%) |
| Toll-interacting proteinc | 1 (0.01%) |
aDigestive or hydrolytic genes.
bGenes concerned with immune reactions.
cTypical immune genes.
(c) Two different gene expression states of diverticulum epithelial cells
Next, to reveal the spatiotemporal pattern of gene expression of diverticulum epithelial cells, we compare the differences in gene expression between cells in empty (after 3 days of starvation) and sated states using qRT-PCR assays. In order to determine the results of the qRT-PCR assay more precisely, we selected both 18S and cytoplasmic actin as the reference genes [44].
The qRT-PCR assays show that cathepsin L, B and D, ferritin, trypsin-like serine protease, lysozymes C and G, carboxypeptidase Z/N, tetraspanin, legumain, saposin B, subtilisin-like protease, arylsulfatase B, endo-beta-1,4-glucanase and methionine adenosyltransferase are highly expressed in the empty state relative to both reference genes (figure 4; electronic supplementary material, figure S9). This gene expression profile indicates that the cells are prepared to degrade phagocytized food particles. It also reveals that VCBP4, 1, 3, 5, Gram-negative bacteria-binding protein, alpha2-macroglobulin, chitotriosidase 1-like protein, big defensin, proprotein convertase subtilisin/kexin type 1 and Toll-interacting protein are highly expressed in the natural sated state, relative to both reference genes (figure 5; electronic supplementary material, figure S10), indicating that phagocytized food particles can stimulate the innate immune-like response [16–19]. Moreover, our findings show that, though pancreatic lipase-like protein, plasminogen and peroxiredoxin V are highly expressed in diverticulum phagocytic epithelial cells, it is difficult to estimate whether they are highly expressed in either the empty or sated states because of the inconsistency caused by the two different reference genes, as one gives a high expression ratio and the other gives a low one (electronic supplementary material, figure S11). Plasminogen is the precursor of plasmin, which is a serine protease and can dissolve fibrin [45], whereas peroxiredoxin V has antioxidative and cytoprotective functions during oxidative stress triggered by immunogens [46]. The results of the qRT-PCR assays reveal that Branchiostoma phagocytic epithelial cells present different gene expression states in unfed versus sated states.
(d) Gut epithelial cells can also phagocytize food particles
In addition to the epithelial cells in the diverticulum, we also found that the epithelial cells lining the hindgut can phagocytize small algal cells (figure 6). The phagocytic capability of these cells is apparently more limited than that of diverticulum epithelial cells (figure 6b,c) because of their shorter length; however, this limited intracellular digestion still improves the utilization rate of food particles. Two lines of evidence show that the diverticulum is a more specialized digestive organ than the hindgut (figure 4; electronic supplementary material, table S1). First, the qRT-PCR results in figure 4 show higher expression for many digestive and hydrolytic enzymes. In fact, among all the annotated functional genes, only elastase I is obviously expressed more in the gut than in the diverticulum [47] (electronic supplementary material, figure S12). Second, the suppression subtractive hybridization (SSH) results reveal that the gene expression profiles of the diverticulum and gut epithelial cells are similar, but that the diverticulum can express greater numbers of functional genes than the gut. SSH technology allows for the PCR-based amplification of the cDNA fragments that differ between control (driver) and experimental (tester) transcriptomes [32]. That is, the diverticulum-to-gut SSH (SSH-D; the tester is the diverticulum transcriptome and the driver is the gut transcriptome) contains 5182 efficient ESTs (GenBank accession number: LIBEST_028557), and the gut-to-diverticulum SSH (SSH-G; the tester is the gut transcriptome and the driver is the diverticulum transcriptome) contains 3367 efficient ESTs (GenBank accession number: LIBEST_028556).
Figure 6.
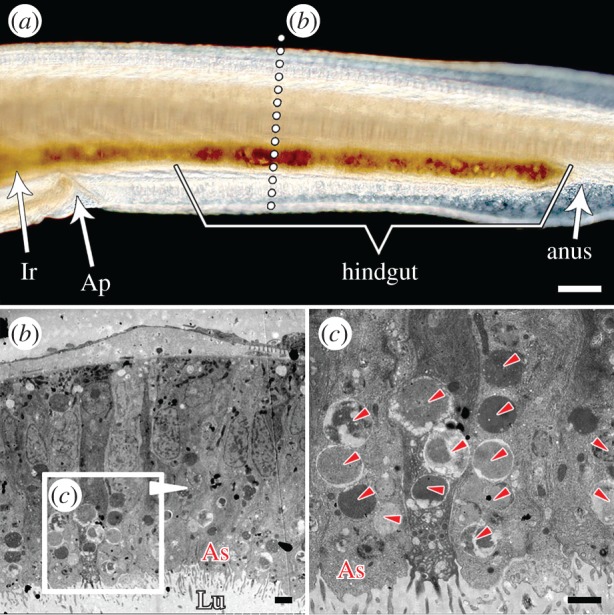
Gut epithelial cells outside the diverticulum can phagocytize. The epithelial cells in the hindgut can also phagocytize algal cells (c), however, the phagocytic capability of these cells seems more limited than that of diverticulum epithelial cells, which are longer and more numerous (figure 2). The small arrowheads in (c) mark the algal cells. Ap, atriopore; Ir, ileo-colon ring; Lu, lumen; As, apical side. Scale bars: 100 µm (a); 2 µm (b,c).
4. Discussion
(a) Food particles are exogenous immunogens to Branchiostoma phagocytic epithelial cells
The phagocytic epithelial cells of the Branchiostoma digestive tract are exposed not only to food resources, such as algal cells, but also to large quantities of exogenous, even pathogenic, single-celled organisms, such that ‘eating’ food can provoke a long-term immune-like response to protect intracellular and whole-body homeostasis. The phagocytic epithelial cells deal with this immune-like stimulation by expressing immune genes. Among these immune genes, lysozyme can present in the cytoplasmic granules to destroy Gram-positive bacteria [41], VCBPs can present an adaptive-like immune character by exhibiting high levels of polymorphism [16–19], Gram-negative bacteria-binding protein can bind to lipopolysaccharides to elicit immune responses and alpha2-macroglobulin can prevent cell lysis by inactivating various proteinases [24]. In addition, big defensin is a small cysteine-rich peptide that can bind to the cell membranes of bacteria and form pore-like defects that can lead to the lethal efflux of cytoplasmic ions and nutrients [16]. The functions of these typical immune genes and large quantities of other endogenous degrading enzymes ensure that the phagocytic epithelial cells can not only degrade phagocytized food particles sufficiently, but also maintain their cellular integrity.
(b) The physiological function of the Branchiostoma diverticulum differs from that of the vertebrate liver
Ever since Müller [15], the Branchiostoma diverticulum has been regarded as the homologue of the vertebrate liver. However, our findings demonstrate that the cell type, tissue structure and gene expression profile of the Branchiostoma diverticulum are quite different from those of the vertebrate liver. Furthermore, hepatic products homologous to those in vertebrates, such as albumin, alpha-fetoprotein, aspartate aminotransferase, alanine aminotransferase and miR-122, were not found in the Branchiostoma diverticulum through the tissue-specific expressions of hallmark genes—in fact, albumin, alpha-fetoprotein and miR-122, are absent from the Branchiostoma genome [22,37–40,48]. In summary, all of the aforementioned results suggest that the main physiological functions of the diverticulum are different from those of the liver.
(c) Phagocytic intracellular digestion in the Branchiostoma diverticulum cannot directly confirm the absence of liver in last common chordate
It would be reckless to infer directly that the invertebrate chordate lacked a liver just because Branchiostoma performs phagocytic intracellular digestion in its diverticulum. On the one hand, the determination of the homology between the diverticulum and liver needs to be supported by more evidence from evolutionary developmental biology investigations into the developmental gene regulatory networks of at least the liver, and preferably the whole endoderm [49–57]. On the other hand, because Branchiostoma cannot embody every feature of the ancestral chordate, and its diverticulum is possibly a specialized structure reflecting only Brachiostoma's own evolutionary history or lifestyle, studies of echinoderms, hemichordates, urochordates and ammocoetes (larval lampreys) are indispensable to investigating this question [58]. Furthermore, determining the evolutionary histories of animal characteristics cannot be done well without direct palaeontological evidence [59–61], so the question of whether the last common chordate had a liver or not will remain open until new fossil evidence that provides a clear answer can be found [62].
5. Conclusion
The results of this study confirm the existence of phagocytic intracellular digestion in Branchiostoma and explain why epithelial cells in the Branchiostoma digestive tract express a number of typical immune genes. The epithelial cells of the Branchiostoma digestive tract can phagocytize food particles, such as algal cells, directly. The phagocytic capability of diverticulum epithelial cells is greater than that of hind gut epithelial cells. The major physiological function of the Branchiostoma diverticulum is also different from that of the vertebrate liver.
Supplementary Material
Acknowledgements
We thank L. Holland, N. Holland and P. Holland for technical and scientific guidance, E. Davidson, J.-B. Caron and M. Cui for scientific guidance, M. Zhu, J. Lu and Z. Gai for scientific guidance and drafting the manuscript, and K. Zaret and D. Melton for scientific guidance on the embryonic patterning, organogenesis and origin of the vertebrate liver and pancreas.
Ethics
All sample collecting and processing was performed according to the local laws governing the welfare of invertebrate animals and was approved by the Southeast University (SEU) ethical committee.
Data accessibility
The full-length cDNA transcriptome library of Branchiostoma diverticulum: GenBank accession number, LIBEST_028542. The diverticulum-to-gut SSH: GenBank accession number, LIBEST_028557. The gut-to-diverticulum SSH: GenBank accession number, LIBEST_028556. Data are available at: https://datadryad.org/handle/10255/3/workflow?workflowID=128155&stepID=reviewStep&actionID=reviewAction.
Authors' contributions
C.H., J.-Y.C. and Z.L. conceived the project and wrote the paper. C.H., Z.H. and B.H. performed the electron microscopy experiments. C.H. and X.L. performed the WMISH experiments. R.G., T.T. and N.L. performed the bioinformatics analyses. C.H. and Q.Z. performed the histology experiments. C.H., T.H., Y.Z., Y.L. and Y.H. edited the paper. All authors discussed and commented on the data.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by the National Natural Science Foundation of China (6507030081) and the National Science and Technology Major Project of China (6307030004).
References
- 1.Kodama Y, Fujishima M. 2014. Symbiotic Chlorella variabilis incubated under constant dark conditions for 24 hours loses the ability to avoid digestion by host lysosomal enzymes in digestive vacuoles of host ciliate Paramecium bursaria. FEMS Microbial. Ecol. 90, 946–955. ( 10.1111/1574-6941.12448) [DOI] [PubMed] [Google Scholar]
- 2.Tauber AI. 2003. Metchnikoff and the phagocytosis theory. Nat. Rev. Mol. Cell Biol. 4, 897–901. ( 10.1038/nrm1244) [DOI] [PubMed] [Google Scholar]
- 3.Kaufmann SH. 2008. Immunology's foundation: the 100-year anniversary of the Nobel Prize to Paul Ehrlich and Elie Metchnikoff. Nat. Immunol. 9, 705–712. ( 10.1038/ni0708-705) [DOI] [PubMed] [Google Scholar]
- 4.Underhill DM, Goodridge HS. 2012. Information processing during phagocytosis. Nat. Rev. Immunol. 12, 492–502. ( 10.1038/nri3244) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Underhill DM, Gordon S, Imhof BA, Núñez G, Bousso P. 2016. Élie Metchnikoff (1845–1916): celebrating 100 years of cellular immunology and beyond. Nat. Rev. Immunol. 16, 651–656. ( 10.1038/nri.2016.89) [DOI] [PubMed] [Google Scholar]
- 6.Gordon S. 2016. Phagocytosis: the legacy of Metchnikoff. Cell 166, 1065–1068. ( 10.1016/j.cell.2016.08.017) [DOI] [PubMed] [Google Scholar]
- 7.Wynn TA, Chawla A, Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496, 445–455. ( 10.1038/nature12034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stumpp M, et al. 2015. Evolution of extreme stomach pH in bilateria inferred from gastric alkalization mechanisms in basal deuterostomes. Sci. Rep. 5, 10421 ( 10.1038/srep10421) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Weel PB. 1937. Die Ernahrungsbiologie von Amphioxus lanceolatus. Pubbl staz zool Napoli. 6, 221–272. [Google Scholar]
- 10.Barrington EJW. 1937. The digestive system of Amphioxus (Branchiostoma) lanceolatus. Phil. Trans. R. Soc. Lond. B 228, 269–311. ( 10.1098/rstb.1937.0013) [DOI] [Google Scholar]
- 11.van Weel PB. 1974. Hepatopancreas? Com. Biochem. Physiol. A Comp. Physiol. 47, 1–9. ( 10.1016/0300-9629(74)90044-9) [DOI] [PubMed] [Google Scholar]
- 12.Ruppert EE. 1997. Hemichordata, chaetognatha, and the invertebrate chordates. In Microscopic anatomy of invertebrates (eds Harrison FW, Ruppert EE), pp. 349–504. New York, NY: Wiley. [Google Scholar]
- 13.Han Y, et al. 2010. The immune system of amphioxus provides insights into the ancestral structure of the vertebrate immune system. Dev. Comp. Immunol. 34, 791–796. ( 10.1016/j.dci.2010.03.009) [DOI] [PubMed] [Google Scholar]
- 14.Yang P, et al. 2014. Origin of the phagocytic respiratory burst and its role in gut epithelial phagocytosis in a basal chordate. Free Radic. Biol. Med. 70, 54–67. ( 10.1016/j.freeradbiomed.2014.02.007) [DOI] [PubMed] [Google Scholar]
- 15.Müller J. 1844. Über den Bau und die Lebenserscheinungen des Branchiostoma lubricum Costa, Amphioxus lanceolatus Yarrell Abh K Preuss Akad. Wiss. Berl.1844, 79–116.
- 16.Cannon JP, Haire RN, Litman GW. 2002. (Identification of diversified genes that contain immunoglobulin-like variable regions in a protochordate. Nat. Immunol. 3, 1200–1207. ( 10.1038/ni849) [DOI] [PubMed] [Google Scholar]
- 17.Dishaw LJ, et al. 2008. Genomic complexity of the variable region-containing chitin-binding proteins in amphioxus. BMC Genet. 9, 78 ( 10.1186/1471-2156-9-78) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dishaw LJ, et al. 2010. The basis for haplotype complexity in VCBPs, an immune-type receptor in amphioxus. Immunogenetics 62, 623–631. ( 10.1007/s00251-010-0464-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dishaw LJ, et al. 2011. A role for variable region-containing chitin-binding proteins (VCBPs) in host gut–bacteria interactions. Proc. Natl Acad. Sci. USA 108, 16 747–16 752. ( 10.1073/pnas.1109687108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uinuk-Ool TS, et al. 2003. Phylogeny of antigen-processing enzymes: cathepsins of a cephalochordate, an agnathan and a bony fish. Scand. J. Immunol. 58, 436–448. ( 10.1046/j.1365-3083.2003.01322.x) [DOI] [PubMed] [Google Scholar]
- 21.Liang Y, Zhang S, Lun L, Han L. 2006. Presence and localization of antithrombin and its regulation after acute lipopolysaccharide exposure in amphioxus, with implications for the origin of vertebrate liver. Cell Tissue Res. 323, 537–541. ( 10.1007/s00441-005-0088-x) [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Zhang S, Li L. 2009. A transferrin-like homolog in amphioxus Branchiostoma belcheri: identification, expression and functional characterization. Mol. Immunol. 46, 3117–3124. ( 10.1016/j.molimm.2009.06.001) [DOI] [PubMed] [Google Scholar]
- 23.Yuan S, et al. 2009. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-kappaB pathway via MyD88. Mol. Immunol. 46, 2348–2356. ( 10.1016/j.molimm.2009.03.022) [DOI] [PubMed] [Google Scholar]
- 24.Liang Y, Pan A, Zhang S, Zhang Y, Liu M. 2011. Cloning, distribution and primary immune characteristics of amphioxus alpha-2 macroglobulin. Fish Shellfish Immunol. 31, 963–969. ( 10.1016/j.fsi.2011.08.014) [DOI] [PubMed] [Google Scholar]
- 25.Chao Y, Fan C, Liang Y, Gao B, Zhang S. 2012. A novel serpin with antithrombin-like activity in Branchiostoma japonicum: implications for the presence of a primitive coagulation system. PLoS ONE 7, e32392 ( 10.1371/journal.pone.0032392) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dishaw LJ, Haire RN, Litman GW. 2012. The amphioxus genome provides unique insight into the evolution of immunity. Brief Funct. Genomics 11, 167–176. ( 10.1093/bfgp/els007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang S, et al. 2016. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell 166, 102–114. ( 10.1016/j.cell.2016.05.032) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li X, Li B, Sun X. 2014. Effects of a coastal power plant thermal discharge on phytoplankton community structure in Zhanjiang Bay, China. Mar. Pollut. Bull. 81, 210–217. ( 10.1016/j.marpolbul.2013.08.006) [DOI] [PubMed] [Google Scholar]
- 29.Lacalli TC, West JE. 1986. Ciliary band formation in the doliolaria larva of Florometra. I. The development of normal epithelial pattern. J. Embryol. Exp. Morphol. 96, 303–323. [PubMed] [Google Scholar]
- 30.Hirakow R, Kajita N. 1990. An electron microscopic study of the development of amphioxus Branchiostoma belcheri tsingtaoensis cleavage. J. Morphol. 203, 331–344. ( 10.1002/jmor.1052030308) [DOI] [PubMed] [Google Scholar]
- 31.Koop D, Holland LZ, Setiamarga D, Schubert M, Holland ND. 2011. Tail regression induced by elevated retinoic acid signaling in amphioxus larvae occurs by tissue remodeling, not cell death. Evol. Dev. 13, 427–435. ( 10.1111/j.1525-142X.2011.00501.x) [DOI] [PubMed] [Google Scholar]
- 32.Diatchenko L, et al. 1996. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc. Natl Acad. Sci. USA 93, 6025–6030. ( 10.1073/pnas.93.12.6025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holland LZ, Holland PWH, Holland ND. 1996. Revealing homologies between body parts of distantly related animals by in situ hybridization to developmental genes: amphioxus versus vertebrates. In Molecular zoology: advances, strategies and protocols (eds Ferraris JD, Palumbi SR), pp. 267–282. New York, NY: Wiley-Liss. [Google Scholar]
- 34.Holland PWH. 1999. Whole-mount in situ hybridisation to amphioxus embryos. In Molecular embryology: methods and protocols (eds Sharpe PT, Mason IJ), pp. 703–706. Totowa, NJ: Humana. [Google Scholar]
- 35.Naser IB, et al. 2009. Analysis of a repulsive axon guidance molecule, draxin, on ventrally directed axon projection in chick early embryonic midbrain. Dev. Biol. 332, 351–359. ( 10.1016/j.ydbio.2009.06.004) [DOI] [PubMed] [Google Scholar]
- 36.Young JZ. 1981. The life of vertebrates, 3rd edn Oxford, UK: Clarendon Press. [Google Scholar]
- 37.Putnam NH, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064–1071. ( 10.1038/nature06967) [DOI] [PubMed] [Google Scholar]
- 38.Holland LZ, et al. 2008. The amphioxus genome illuminates vertebrate origins and cephalochordate biology. Genome Res. 18, 1100–1111. ( 10.1101/gr.073676.107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang S, et al. 2014. Decelerated genome evolution in modern vertebrates revealed by analysis of multiple lancelet genomes. Nat. Commun. 5, 5896 ( 10.1038/ncomms6896) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yue JX, et al. 2016. Conserved noncoding elements in the most distant genera of cephalochordates: the Goldilocks principle. Genome Biol. Evol. 8, 2387–2405. ( 10.1093/gbe/evw158) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, et al. 2010. Lysosomal proteolysis is the primary degradation pathway for cytosolic ferritin and cytosolic ferritin degradation is necessary for iron exit. Antioxid. Redox Signal. 13, 999–1009. ( 10.1089/ars.2010.3129) [DOI] [PubMed] [Google Scholar]
- 42.Honarmand Ebrahimi K, Bill E, Hagedoorn PL, Hagen WR. 2012. The catalytic center of ferritin regulates iron storage via Fe(II)-Fe(III) displacement. Nat. Chem. Biol. 8, 941–948. ( 10.1038/nchembio.1071) [DOI] [PubMed] [Google Scholar]
- 43.Kurt YG, et al. 2009. Serum chitotriosidase enzyme activity in patients with Crimean-Congo hemorrhagic fever. Clin. Chem. Lab. Med. 47, 1543–1547. ( 10.1515/CCLM.2009.347) [DOI] [PubMed] [Google Scholar]
- 44.Kusakabe R, Satoh N, Holland LZ, Kusakabe T. 1999. Genomic organization and evolution of actin genes in the amphioxus Branchiostoma belcheri and Branchiostoma floridae. Gene 227, 1–10. ( 10.1016/S0378-1119(98)00608-8) [DOI] [PubMed] [Google Scholar]
- 45.Miyata T, Iwanaga S, Sakata Y, Aoki N. 1982. Plasminogen Tochigi: inactive plasmin resulting from replacement of alanine-600 by threonine in the active site. Proc. Natl Acad. Sci. USA 79, 6132–6136. ( 10.1073/pnas.79.20.6132) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu L, et al. 2017. Molecular structure and functional characterization of the peroxiredoxin 5 in Procambarus clarkii following LPS and Poly I:C challenge. Fish Shellfish Immunol. 71, 28–34. ( 10.1016/j.fsi.2017.09.072) [DOI] [PubMed] [Google Scholar]
- 47.Sun T, Zhang S, Ji G. 2010. Identification and expression of an elastase homologue in Branchiostoma belcheri with implications to the origin of vertebrate pancreas. Mol. Biol. Rep. 37, 3303–3309. ( 10.1007/s11033-009-9916-5) [DOI] [PubMed] [Google Scholar]
- 48.Wang Y, Wang H, Li M, Gao Z, Zhang S. 2015. Identification, expression and regulation of amphioxus G6Pase gene with an emphasis on origin of liver. Gen. Comp. Endocrinol. 214, 9–16. ( 10.1016/j.ygcen.2014.12.021) [DOI] [PubMed] [Google Scholar]
- 49.Zaret KS. 2008. Genetic programming of liver and pancreas progenitors: lessons for stem-cell differentiation. Nat. Rev. Genet. 9, 329–340. ( 10.1038/nrg2318) [DOI] [PubMed] [Google Scholar]
- 50.Grapin-Botton A, Melton DA. 2000. Endoderm development: from patterning to organogenesis. Trends Genet. 16, 124–130. ( 10.1016/S0168-9525(99)01957-5) [DOI] [PubMed] [Google Scholar]
- 51.Holland LZ. 2015. Genomics, evolution and development of amphioxus and tunicates: the Goldilocks principle. J. Exp. Zool. B Mol. Dev. Evol. 324, 342–352. ( 10.1002/jez.b.22569) [DOI] [PubMed] [Google Scholar]
- 52.Brooke NM, Garcia-Fernàndez J, Holland PW. 1998. The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature 392, 920–922. ( 10.1038/31933) [DOI] [PubMed] [Google Scholar]
- 53.Ferrier DE, Holland PW. 2002. Ciona intestinalis ParaHox genes: evolution of Hox/ParaHox cluster integrity, developmental mode, and temporal colinearity. Mol. Phylogenet. Evol. 24, 412–417. ( 10.1016/S1055-7903(02)00204-X) [DOI] [PubMed] [Google Scholar]
- 54.Fröbius AC, Seaver EC. 2006. ParaHox gene expression in the polychaete annelid Capitella sp. I. Dev. Genes Evol. 216, 81–88. ( 10.1007/s00427-005-0049-0) [DOI] [PubMed] [Google Scholar]
- 55.Arnone MI, et al. 2006. Genetic organization and embryonic expression of the ParaHox genes in the sea urchin S. purpuratus: insights into the relationship between clustering and colinearity. Dev. Biol. 300, 63–73. ( 10.1016/j.ydbio.2006.07.037) [DOI] [PubMed] [Google Scholar]
- 56.Ikuta T, et al. 2013. Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava. BMC Evol. Biol. 13, 129 ( 10.1186/1471-2148-13-129) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fritsch M, Wollesen T, Wanninger A. 2016. Hox and ParaHox gene expression in early body plan patterning of polyplacophoran mollusks. J. Exp. Zoo. B Mol. Dev. Evol. 326, 89–104. ( 10.1002/jez.b.22671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swalla BJ, Smith AB. 2008. Deciphering deuterostome phylogeny: molecular, morphological and palaeontological perspectives. Phil. Trans. R. Soc. B. 363, 1557–1568. ( 10.1098/rstb.2007.2246) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Holland ND, Chen J. 2001. Origin and early evolution of the vertebrates: new insights from advances in molecular biology, anatomy, and palaeontology. BioEssays 23, 142–151. ( 10.1002/1521-1878(200102)23:2%3C142::AID-BIES1021%3E3.0.CO;2-5) [DOI] [PubMed] [Google Scholar]
- 60.Mallatt J, Chen J. 2003. Fossil sister group of craniates: predicted and found. J. Morphol. 258, 1–31. ( 10.1002/jmor.10081) [DOI] [PubMed] [Google Scholar]
- 61.Feinberg TE, Mallatt JM. 2016. The ancient origins of consciousness: how the brain created experience. Cambridge, MA: MIT Press. [Google Scholar]
- 62.Conway Morris S, Caron J-B. 2014. A primitive fish from the Cambrian of North America. Nature 512, 419–422. ( 10.1038/nature13414) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The full-length cDNA transcriptome library of Branchiostoma diverticulum: GenBank accession number, LIBEST_028542. The diverticulum-to-gut SSH: GenBank accession number, LIBEST_028557. The gut-to-diverticulum SSH: GenBank accession number, LIBEST_028556. Data are available at: https://datadryad.org/handle/10255/3/workflow?workflowID=128155&stepID=reviewStep&actionID=reviewAction.