Abstract
Urbanization often substantially influences animal movement and gene flow. However, few studies to date have examined gene flow of the same species across multiple cities. In this study, we examine brown rats (Rattus norvegicus) to test hypotheses about the repeatability of neutral evolution across four cities: Salvador, Brazil; New Orleans, USA; Vancouver, Canada; and New York City, USA. At least 150 rats were sampled from each city and genotyped for a minimum of 15 000 genome-wide single nucleotide polymorphisms. Levels of genome-wide diversity were similar across cities, but varied across neighbourhoods within cities. All four populations exhibited high spatial autocorrelation at the shortest distance classes (less than 500 m) owing to limited dispersal. Coancestry and evolutionary clustering analyses identified genetic discontinuities within each city that coincided with a resource desert in New York City, major waterways in New Orleans, and roads in Salvador and Vancouver. Such replicated studies are crucial to assessing the generality of predictions from urban evolution, and have practical applications for pest management and public health. Future studies should include a range of global cities in different biomes, incorporate multiple species, and examine the impact of specific characteristics of the built environment and human socioeconomics on gene flow.
Keywords: urban evolution, population genomics, genetic diversity, brown rats, spatial autocorrelation, coancestry
1. Introduction
Urbanization is a rapidly accelerating human influence on the evolution of wildlife in and around cities. Genetic drift and gene flow have been the most commonly studied evolutionary processes in cities, probably because effective population sizes and dispersal are often reduced by urban development [1,2]. Urbanization thus often results in lower genome-wide diversity within populations [3], higher genetic differentiation between populations [4,5] and reduced connectivity across the landscape [6,7]. These impacts are most clearly seen in small-bodied organisms with limited mobility, whereas more mobile organisms may exhibit few negative effects from urbanization [8] or more subtle influences that produce heterogeneity in gene flow across urban environments [9]. It remains unclear, however, whether urbanization imposes a set of common pressures (e.g. arising from fragmentation) that give rise to parallel outcomes or whether local idiosyncrasies and structural differences between cities result in divergent outcomes. Further understanding of urban evolution requires that comparisons be drawn among cities using tools capable of resolving fine-scale variation in fundamental processes like genetic drift and gene flow [10,11].
The most useful model systems for examining the repeatability of evolutionary responses to urbanization occupy many cities around the world, are easy to sample in large numbers, have relatively short generation times, well-developed genomic resources, and strong ecological associations with urban habitat rather than just occasional presence in cities. The brown rat (Rattus norvegicus) meets all of these criteria, but its ecology and evolution in cities is poorly understood despite a long association with humans [12].
Recent global-scale genomic studies have uncovered some of the processes leading to establishment of brown rats in cities globally. Brown rats evolved in northern Asia, spread to Southeast Asia and slowly made their way through coastal routes to occupy all of Europe by the eighteenth century. Shortly after, ship activity associated with European imperialism spread the brown rat to nearly all coastal cities, followed by further spread to inland cities [13–15]. During their human-assisted migrations, rats evolved to exploit human resources and are now largely ‘anthrodependent’ [16]. Brown rats are one of the world's most destructive invasive vertebrates [17] and a chronic public health issue in many urban centres [18,19]. Rats carry many zoonotic pathogens of human concern (e.g. Bartonella, Leptospira, Seoul hantavirus) [20]. Pathogen distribution across urban rat populations is highly heterogeneous and probably influenced by patterns of rat dispersal on local and regional scales [21]. Eradication campaigns may even increase human pathogen risk by influencing rat movements or changing interactions among remaining rats [22]. Despite these concerns, our understanding of urban rat biology has lagged because pest control almost always takes precedence over research [23]. In this study, we use spatial population genomics to make insights about the operation of gene flow and drift in rat populations in four different cities.
Direct tracking or mark-recapture data are superior for characterizing rat movements, but nearly impossible to collect city-wide for wild rats owing to logistical challenges presented by urban environments [24]. Population genetic analyses provide a useful alternative, and have the added benefit of measuring gene flow. Previous behavioural ecology work indicated that rats can form relatively large social colonies composed of many relatives, and do not typically range farther than 150–200 m from their natal colony [25–27]. In dense populations, rats stay even closer to natal sites [28], although they are physically capable of long-distance movements [29–31]. Population genetic analyses using microsatellites confirmed short-distance movements (less than 150 m) of rats in Baltimore, USA, with occasional long distance dispersal [32]. Microsatellite-based analyses of rats in Salvador, Brazil reported genetic differentiation between human neighbourhoods separated by less than 100 m, largely because of major roadways and topographical slopes [33,34]. These results indicate that rat populations become genetically structured over fine spatial scales owing to landscape heterogeneity. However, the ability to detect fine-scale landscape effects may have been limited by lack of resolution in calculating genetic distance between individual rats [10].
To date, only a single study of urban rats has employed genome-wide single nucleotide polymorphisms (SNPs) to study population genomics of urban rats. This study of the brown rat population in Manhattan (New York City (NYC), USA) reported strong spatial autocorrelation in relatedness at distance classes less than 200 m, with detectable levels of autocorrelation up to 1400 m [9]. These results demonstrate that the closer rats are to each other, the more likely they are to be similar genetically because natal dispersal is limited, although occasional long distance dispersal was inferred. No evidence of sex-biased dispersal was detected among NYC rats, though several studies have inferred increased dispersal among male rats [33,35]. Despite continuous distribution and high connectivity among rats across Manhattan, there were also differences in genetic diversity and a genetic discontinuity that spanned a region characterized by commercial office buildings rather than residential areas (i.e. a resource desert). These results suggest that high relatedness over short distances is owing to limited dispersal, and fine-scale genetic structuring results from habitat heterogeneity in urban built environments. Determining whether this is a general phenomenon or idiosyncratic to NYC requires comparing rat populations across multiple cities. While examining the spatial distribution of genetic variation alone does not estimate the timing of divergence events or quantify the effect of landscape attributes on gene flow, these efforts establish hypotheses for landscape genetic or coalescent modelling to further understand migration-drift processes.
Here, we examine the repeatability of evolutionary outcomes in four cities: Salvador (SAL), Brazil; New Orleans (NOL), USA; Vancouver (VAN), Canada; and NYC, USA. These four cities are all ports in the Americas, but vary in climate from temperate (NYC, Vancouver) to subtropical (New Orleans) and tropical (Salvador). Processes of urbanization and rat invasion may be consistent worldwide, leading to similar population genetic outcomes despite climatic variability. Alternatively, differences in climate and city structure may influence drift and gene flow of invasive rat populations, particularly given that the brown rat is a cold-hardy species that originated in northern China. We employ much larger datasets and more powerful population genomic approaches than previous work on commensal rodents, constituting one of the first analyses of any vertebrate that compares evolutionary outcomes across multiple cities. We address the following specific questions:
(i) are spatial patterns and levels of genome-wide diversity similar across rat populations in different cities, or have idiosyncratic processes given rise to differences in diversity?
(ii) is spatial genetic structure from local isolation-by-distance consistently strong across geographically distinct urban landscapes? At what distance classes does drift outweigh the influence of local gene flow? and
(iii) do rat populations exhibit similar genetic structure and patterns of coancestry across cities? Do outcomes vary among cities in different climatic zones?
2. Material and methods
(a). Study sites and sampling
We studied patterns of neutral genetic variation within and between brown rat (Rattus norvegicus) populations in four coastal port cities: (i) SAL, Bahia, Brazil; (ii) NOL, Louisiana, USA; (iii) VAN, British Columbia, Canada; and (iv) NYC, New York, USA.
SAL is a tropical city with a large human population (2.9 mil total; 4187 km−2) that supports a patchwork of wealthier developed areas, dense lower income neighbourhoods (i.e. favelas), and natural areas. Rats (n = 150) were trapped and sampled between 2010 and 2017. Most samples were collected at the vertices of a grid array designed to maximize geographical coverage or through ongoing public health projects [36,37].
NOL is a subtropical city with a relatively low human population density (0.4 mil total; 858 km−2) that is subdivided by multiple waterways and contains large post-flood recovery areas with grassland habitat. Rats (n = 193) were sampled from 2014 to 2015 across 78 non-contiguous blocks spanning the city. Sites were chosen to represent different levels of income and flooding history for research on post-disaster re-assembly of coupled human-natural systems [38]. Full details are provided elsewhere [39].
VAN is a temperate city with a moderate population density (0.6 mil total; 5500 km−2). Rats (n = 615) were trapped from 2011 to 2012 from 32 contiguous city blocks within the Downtown-East neighbourhood, which has high rates of poverty and a significant homeless population. Tomahawk Rigid Traps (Tomahawk Live Trap, Hazelhurst, WI, USA, Model 102) were placed in alleyways bisecting each block and were pre-baited for one week, followed by two weeks of continuous trapping. Rats were trapped for a long-term study of urban rats with a focus on disease ecology, and detailed methods are reported elsewhere [40,41].
NYC experiences a temperate climate, and is dominated by the built environment and pervasive underground infrastructure (e.g. sewers, train tunnels) with an extremely high human population density (8.4 mil total; 10 800 km−2). Rats (n = 262) were sampled at the individual-level across the entire island of Manhattan from June 2014 to December 2015. Full details on this sampling and previous population genomic results are reported elsewhere [9].
(b). DNA sequencing, single nucleotide polymorphism genotyping and bioinformatics
We used an identical ddRADSeq approach to prepare libraries for genome-wide SNP genotyping, followed by bioinformatics processing, for all four cities. Briefly, we used Stacks v. 1.35 to demultiplex reads, then aligned reads to the Rnor v. 6.0 reference genome [42] and called SNPs using the pstacks, cstacks and sstacks scripts from Stacks v. 1.35. Details are in the electronic supplementary material, Methods.
(c). Population genomic analyses
We calculated four indices of genome-wide diversity for each dataset using the populations script in Stacks v. 1.35 for variant restriction site associated DNA (RAD) tags: expected heterozygosity (i.e. gene diversity), HE; observed heterozygosity, HO; nucleotide diversity, π; inbreeding coefficient, FIS. To understand the distribution of genetic diversity within each city, we used the sGD package [43] in R to calculate diversity (specifically HO, FIS, and allelic richness, Ar) for a genetic neighbourhood surrounding each individual based on a user-defined radius. For each city we used a radius reflecting the extent of local gene flow, specifically the distance at which our correlogram first indicated the lack of significant spatial autocorrelation (see below). We calculated the strength of isolation-by-distance within each city using a simple Mantel test between pairwise matrices of log-transformed Euclidean distances and pairwise genetic distances between all pairs of rats in the ecodist package within R. Each test was run with an alpha of 0.05 for 999 permutations.
To understand the extent of spatial genetic structure from local isolation-by-distance within each city's rat population, we created a Mantel correlogram using the ecodist R package. This analysis examines the extent of spatial autocorrelation between matrices of geographical and genetic distance across different distance classes. For NYC, SAL and NOL we used a 500 m lag size for ease of comparison. For VAN, we used a 100 m lag size because dense sampling allowed for interpretation at a smaller spatial scale. All correlograms were run for 999 permutations and generated 95% confidence intervals with 500 iterations of 90% bootstrapping.
To assess the population genetic structure of rats within each city we used four different analyses: (i) fineStructure; (ii) principal component analysis (PCA); (iii) discriminant analysis of principal components (DAPC); and (iv) Memgene. The fineStructure approach can use shared haplotypes along recombination blocks to estimate shared coancestry with dense genomic data, although with widely distributed individual loci, such as RADseq-derived SNPs used here, loci are essentially unlinked and the program captures allele frequency covariance [44]. While our data do not fully use fineStructure's ability to identify recent coancestry, it still provides a more complete visualization of population genetic structure compared to similar analyses. For each city we phased loci and imputed individual missing SNPs for loci on each chromosome using Fastphase [45]. Average missing data were low for all cities prior to imputation (SAL: 6.6%; NOL: 5.4%; VAN: 7.1%; NYC: 7.2%). Then we ran fineStructure with the unlinked model for 100 000 Markov chain Monte Carlo (MCMC) iterations, and 20 000 tree-building iterations, with a minimum of 500 SNPs and 10% of the genome used for expected maximization estimation. Finally, we inspected MCMC traces to confirm model convergence and created pairwise heatmap figures.
We created PCAs in R for each dataset using the PCs generated by fineStructure. Samples were mapped and labelled using a two-colour ramp corresponding to the scores of the first PC. We performed DAPC on each dataset using the adegenet R package [46]. We first ran find.clusters, retaining all PCs and choosing the K value corresponding to the lowest Bayesian information criterion. These group memberships were then used to assess spread of clusters in PC space using the dapc function. We chose to retain 40 PCs for NYC, SAL and NOL, and 50 PCs for VAN, based on the point of diminishing return in variance to avoid over-fitting [35].
Finally, we used the Memgene [47] package in R to describe processes that generate spatial neighbourhoods of genetically similar individuals. Memgene detects fine-scale patterns of differentiation and cryptic genetic structure [47]. This approach uses Moran's eigenvector maps (MEMs) to create orthogonal eigenvectors with sample coordinates, and then uses a regression framework to quantify the extent of genetic variation explained by each eigenvector. Samples are then mapped based on their eigenvector scores to describe levels of genetic differentiation across the landscape.
To confirm independence between the four sampled cities we used the ‘among-city’ SNP dataset (16 813 filtered SNPs; electronic supplementary material, Methods) and assessed the pattern of evolutionary clustering with DAPC, using an identical procedure as described above and retaining 40 PCs.
3. Results
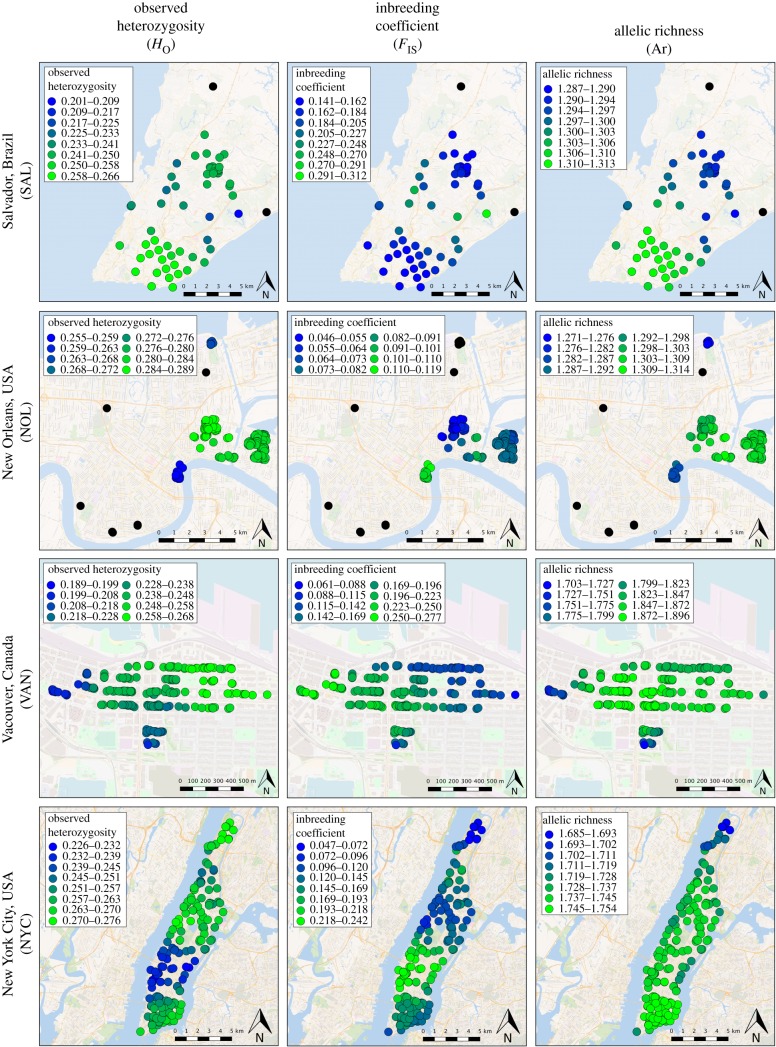
A minimum of 15 310 and maximum of 36 465 SNPs were called and included in downstream analyses for each city (electronic supplementary material, table S1). Despite variable sample sizes, heterozygosity and nucleotide diversity were similar across populations, but the inbreeding coefficient FIS was markedly higher among the VAN and SAL rats. Within each city, genetic diversity varied across different geographical regions as indicated by sGD (figure 1). In both NYC and SAL, heterozygosity was lower and inbreeding values were higher in the centre of the landscape, while in NOL the ‘French Quarter’ and ‘Gentilly’ neighbourhoods showed reduced diversity. Fine-scale sampling in VAN revealed gradual changes in diversity, with higher heterozygosity and lower inbreeding values in the North and East areas. Mantel tests for isolation-by-distance were significant in all cities (p = 0.001; electronic supplementary material, table S1) with the highest r for VAN (r = 0.71) and lowest for NYC (r = 0.30).
Figure 1.
Spatial patterns of genetic diversity within each city analysed using the software sGD. For each city, genetic diversity was calculated for the genetic neighbourhood surrounding each sample. The genetic neighbourhood size (i.e. radius) was determined by the distance at which spatial autocorrelation between genetic and geographical distance reached zero (figure 2). (Online version in colour.)
All four landscapes exhibited high autocorrelation of genetic distance at the shortest geographical distances in the Mantel correlogram (less than 500 m for SAL, NOL and NYC; less than 100 m for VAN; figure 2). Rats in NOL and NYC exhibited virtually no autocorrelation beyond 1500 m, but SAL rats were still positive up to 3000 m. Spatial autocorrelation was stronger among VAN rats at proximal distances (less than 100 m) and declined more rapidly compared to the other three cities, dropping below zero after 300 m.
Figure 2.
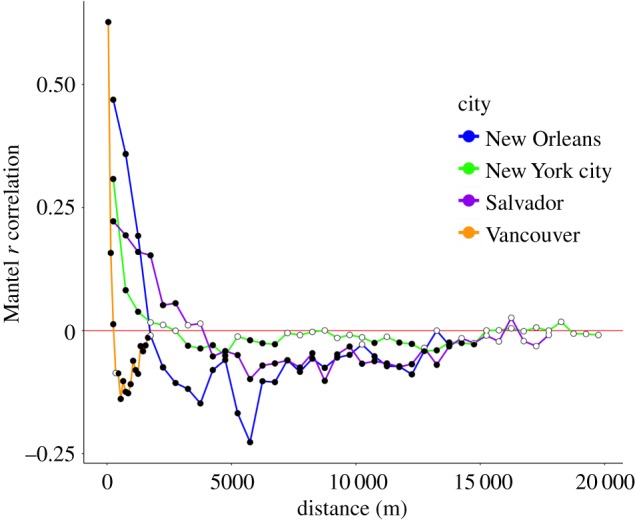
Mantel correlograms for brown rats in each of four studied cities. Filled points are statistically significant and hollow (i.e. white) points are not. For SAL, NYC and NOL each distance class is 500 m and for VAN each distance class is 100 m. (Online version in colour.)
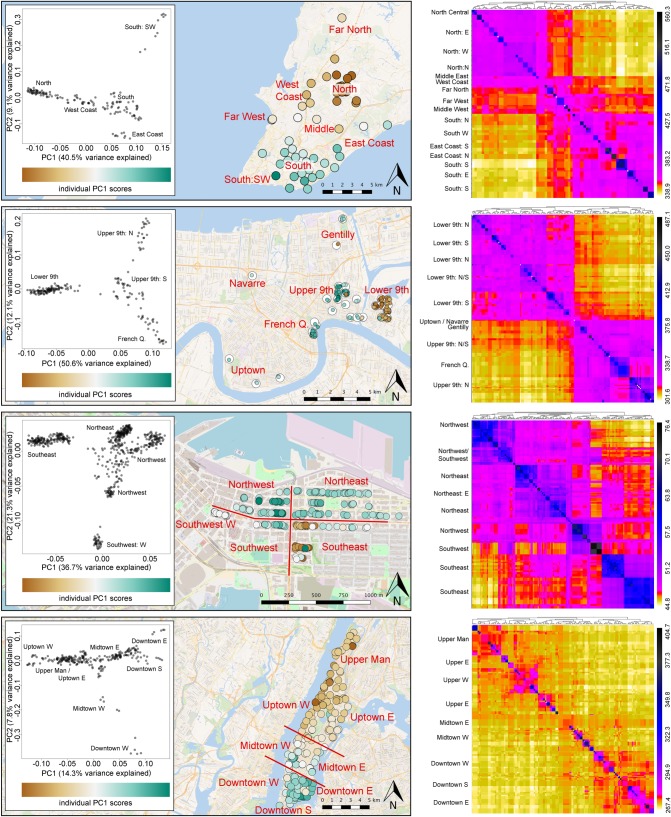
The fineStructure and PCA results indicated complex spatial patterns in all four cities, indicating connectivity at various spatial scales (figure 3). However, there are notable genetic discontinuities within each city that coincide with different urban features: a low density of human residential areas in NYC (i.e. resource desert), major waterways in NOL, and roadways in SAL and VAN (figure 3). The NYC fineStructure and PCA plots support the ‘Uptown’ and ‘Downtown’ clusters identified in previous analyses [9], but the variation is more clinal than in the other cities with lower coancestry across Manhattan.
Figure 3.
Population genetic structure for brown rats in four cities as described by a PCA and coancestry heatmap generated by fineStructure. For each city from left to right we show a PCA of within-city genetic variation, then map the scores of the first PC for each individual sample. On the right is a heatmap describing shared coancestry between all samples, shown with a high-contrast scale to maximize detail for less related individuals. Heatmap labels using suffixes N, S, W, E represent cardinal directions to indicate fine-scale differences within neighbourhoods. (Online version in colour.)
Evolutionary clustering described by DAPC differed among the urban landscapes in terms of the best supported K value, but all closely reflected the coancestry and PCA results (electronic supplementary material, figure S1). In NYC, K = 1 was best supported, but K = 2 cluster scenario also identified a North-South split that was previously identified by multiple analyses [9]. In SAL, K = 2 was best supported and indicated a split between rats in the Northern and Southern neighbourhoods of the city. In NOL, K = 2 was best supported, which suggested a division between rats east and west of a major canal. The K = 3 scenario in NOL identifies a third group occupying the ‘French Quarter’ neighbourhood (electronic supplementary material, figure S2). In VAN, the analyses identified a much higher number of clusters (K = 11) than estimated for the other three cities, probably owing to denser sampling of rats in a smaller area. We also identified two contrasting patterns in the spatial distribution of genetic variation. In the West and Central areas of the sampled neighbourhood, most city blocks contain a single evolutionary cluster that is unique to that location or shared only among a few nearby blocks, which probably represents colonial structure. In the North and East areas, two evolutionary clusters dominated over a dozen blocks. This area may lack stable, isolated colonies, probably experiencing higher gene flow or higher turnover among neighbouring rats than the West and Central areas.
Spatial patterns detected by Memgene (electronic supplementary material, figure S3) capture the same major genetic discontinuities identified in the PCA and fineStructure results, with some additional, more subtle patterns. The NYC analysis recovers the Uptown and Downtown split (8.3% variance explained). SAL rats exhibit a north-south split that coincides with a major roadway, with 20.2% of the variance explained by this first MEM axis. NOL rats also exhibit clear differentiation between rats east of the canal and those farther west (including the French Quarter) and north. Additionally, Memgene identifies subtle changes captured in the adjacent area west of the canal that correspond to changes in genetic diversity observed in sGD (47.9% variance explained). A major roadway in VAN also coincides with a genetic split between rats in the South-East area and most other rats (47.6% variance explained) from this single neighbourhood.
When assessing evolutionary differentiation among cities with DAPC, samples from all four cities clustered independently with no evidence of recent migration (electronic supplementary material, figure S4). SAL and VAN were clearly differentiated across the first discriminant function (DF), and both were separated from NOL and NYC by the second DF, which were most similar but separated by the third DF.
4. Discussion
(a). Are spatial patterns and levels of genome-wide diversity similar across rat populations in different cities?
Brown rat populations in four different coastal American cities exhibited similar patterns of genome-wide diversity despite differences in climate, human population density, land area and development patterns. In particular, HE and π were most similar among rat populations, while FIS values exhibited the largest differences. Cities with the highest human population density and levels of development, such as NYC, are expected to have higher rat density, but if there is a positive relationship between human and rat densities then it is not reflected in our results. Possible explanations are that all of these populations have achieved densities and connectivity that maintain high levels of genetic diversity. This outcome is predicted when local populations exhibit unique allele frequencies owing to genetic drift from isolation-by-distance processes [48]. These coastal rat populations were also derived from invasions from similar geographical sources, with the exception of VAN which probably had a more complex history of multiple invasions [13,15]. The VAN study site is also bordered by Canada's largest international shipping port, and thus may receive periodic migrants from diverse populations. Coalescent modelling of demographic histories of these populations may elucidate the source populations and whether similar numbers of rats (and genome-wide diversity) invaded each city.
All rat populations exhibited differences in genetic diversity within their respective urban landscapes (figure 1), suggesting that local conditions within cities alter the distribution of genetic variation. For example, within both NYC and SAL we observed reduced diversity across the centre of the landscape that may be owing to reduced effective population size and/or lower rates of gene flow. These differences in diversity, which can accrue over just a few city blocks (as seen in VAN), elucidate observed patterns of coancestry and clustering, but also indicate potential differences in adaptive potential across the urban landscape.
(b). Is isolation-by-distance consistently strong across geographically distinct urban landscapes? At what distance classes does drift outweigh the influence of local gene flow?
Landscape features such as rivers, buildings and other impervious surfaces generally restrict gene flow within and among urban populations of small vertebrates [6,7]. GPS tracking of many species also indicates that human-dominated environments reduce animal movements [2]. Thus, enhanced genetic drift and loss of genetic variation are among the most often reported results of urban evolution studies [1]. However, gene flow in some species is relatively unaffected [8]; many small mammals also sustain gene flow by using vegetated corridors within cities [49–51]. Invasive, anthrodependent species such as brown rats would be predicted to exhibit high dispersal rates in cities because they use urban infrastructure as habitat, but signatures of differential gene flow and drift between populations may still be detected if characteristics of different cities influence dispersal rates or distances.
Spatial autocorrelation of genetic and geographical distance among rats was similar across three cities, suggesting that isolation-by-distance and patterns of genetic distance within brown rat populations are driven by social factors and dispersal behaviour that do not vary widely. Our finding that rats are highly related within 500 m of one another used genome-wide SNPs for more accurate genetic distance estimation for this species than any previous study, but the overall conclusion is largely concordant with findings during the ‘golden age’ of rat field research in the mid-twentieth century [25,28]. There may be more variation among cities in the distance classes at which autocorrelation approaches zero owing to differential probabilities of long-distance dispersal [30], inbreeding, or the spatial scale at which rats were sampled. SAL rats exhibited greater autocorrelation at longer distance classes than the other cities, potentially owing to a more cohesive, panmictic population occupying a relatively smaller area than NOL or NYC, lack of seasonality and more consistent movements over time because of the tropical climate [52], or longer dispersal distances on average. The rapid dropoff in the VAN population is probably owing to very intense sampling of close relatives in a relatively small area, thus skewing genetic distances towards lower values and capturing sharp genetic breaks between nearby colonies.
(c). Do rat populations exhibit similar genetic structure and patterns of coancestry across cities?
Four conceptually different approaches, i.e. principal components, coancestry estimation in fineStructure, evolutionary clustering with DAPC, and Moran's eigenvector maps, identified the same spatial patterns of genetic variation across four rat populations. Our results indicate that genetic structure has emerged in all of these cities, though it remains unclear whether these patterns result from historical or recent events. Genetic breaks appear to coincide with canals, major roadways and resource deserts (i.e. neighbourhoods lacking in rubbish and harbourage), providing hypotheses for future landscape genetic modelling to quantify associations between these landscape features and genetic divergence. Roads, waterways and other urban barriers to dispersal are known to create pervasive genetic structure in native small rodents [3,4,53]. However, an invasive pest that occupies a broad range of urban conditions (limited primarily by access to water, rubbish and nesting/burrowing sites) might be expected to exhibit spatial genetic patterns driven only by isolation-by-distance. The fine-scale structuring of rats is more similar to native species that use certain types of urban infrastructure or substrates to maintain connectivity throughout cities, such as some lizards [7].
(d). Implications for pest management and public health
Our results on spatial autocorrelation and fine-scale genetic structure have the potential to improve management strategies for urban rats. The detection of genetic clusters on either side of rivers (NOL), roadways (SAL and VAN), or resource deserts (NYC) provide spatial targets for pest management that are less likely to be rapidly recolonized after control efforts. Roadways coincided with genetic breaks in the SAL and VAN populations, calling to question whether gene flow among rats may be limited over a scale of a few city blocks. Though further work is required to infer causality for such claims (e.g. landscape genetic modelling), identifying specific migration barriers caused by the built environment might allow for even finer-scale targeting of problematic infestations in single neighbourhoods. Estimates of spatial autocorrelation, particularly the geographical scale over which relatedness approaches zero, can also be used to identify the geographical scale of areas that need to be addressed to limit reinfestation from neighbouring rat colonies. Identification of genetic clusters, and knowledge of appropriately-sized buffers between them, could be powerful tools to guide city-wide rat mitigation.
Public health studies have identified many physical and social features that are associated with rat complaints by human residents, and it is likely that these same features positively influence rat movements through urban landscapes. For example, proximity to parkland (e.g. burrowing sites) and subway lines, density of restaurants and residential units, older or vacant housing, and high poverty neighbourhoods are all associated with active rat presence in NYC [54,55], and with proximity to waterways in Amsterdam, the Netherlands [56]. The midtown neighbourhood of NYC is associated with lower density of residential units and reduced rat activity and coincides with the soft boundary between two genetic clusters, suggesting the possibility that reduced gene flow and resulting genetic structure may be influenced by anthropogenic landscape factors [9]. However, rat densities may fluctuate seasonally and from block to block, thus complicating predictions about where and how quickly genetic structure will develop [40]. Similarly, the prevalence of rat-borne pathogens within and across cities is highly heterogeneous and generally not well understood [20,39], though genetic structure may help explain urban pathogen distributions. While geographical and genetic distance between colonies were not associated with pathogen communities among rat colonies in NYC [57], our results on the genetic structure of VAN rats are concordant with previously unexplained distributions of leptospirosis [39]. This finding suggests that gene flow among urban rats influences the prevalence of rat-borne pathogens.
(e). Limitations of this study
While successful at assessing genome-wide diversity, spatial autocorrelation and genetic structure of multiple rat populations, the analyses presented here have some limitations. Although identification of genetic breaks suggests ways landscape features may impact gene flow, we are limited in the conclusions we can draw regarding the timing of events or the mechanisms of gene flow without additional analyses. Sample collection in these four cities was not coordinated from the beginning; each project was originally initiated to address questions specific to each city, primarily regarding public health concerns (with the exception of the NYC study which has an explicit evolutionary focus [9]). Thus, some patterns differed between VAN and the other cities because rats were sampled more intensely over a much smaller scale. This sampling resulted in very different spatial autocorrelation and much greater overall coancestry between rats in VAN than the other three cities. Rats in SAL, NYC and NOL were sampled at roughly similar scales, but the trapping strategies deviated slightly, particularly in NOL where rats were sampled at clustered trapping stations. The spatial information for NOL rats was thus not as fine-grained as in the other cities, but given the similarity across cities this difference probably had little impact on the results.
(f). Conclusions and future directions
Overall, this study was successful at answering several questions about the generality of drift and gene flow across urban rat populations. Urban rats exhibit moderate to high genome-wide diversity. Local genetic drift as measured by spatial autocorrelation analyses typically does not operate much beyond 500 m owing to short dispersal distances in urban rats. However, long-distance dispersal is possible and occurs at low frequency. Major landscape features such as rivers and roadways may restrict gene flow, resulting in spatial genomic structure in urban rat populations, as might neighbourhoods that are resource deserts for rats. This study is the first, to our knowledge, to examine population genomic responses of a vertebrate to urbanization across multiple cities. Such replicated studies are crucial to assessing the generality of predictions from urban evolution [1] and landscape genetics [11]. Very few studies have examined urban evolution outside of temperate North American or European cities, but here our inclusion of tropical (SAL) and subtropical (NOL) cities allowed us to understand the generality of urban evolutionary responses in an invasive species to a degree otherwise unachievable. Future studies should include a range of global cities in different biomes, and incorporate multiple species with carefully chosen characteristics to test hypotheses about urban drift and gene flow. Although we were able to make qualitative statements about the influence of different types of neighbourhoods on rat gene flow, future efforts implementing coalescent approaches or landscape genetic modelling have the potential to significantly improve our understanding of how human socioeconomics and other attributes of the social landscape influence spatial population genomics of rats and other species. Overall, we highlight the importance of comparative approaches for understanding trends of differentiation and diversity in urban wildlife populations and examining repeatability of evolutionary outcomes in environments dominated by anthropogenic impact.
Supplementary Material
Acknowledgements
We thank two reviewers and the editors for their thoughtful comments leading to an improved manuscript. We thank T. Barr'e, A. Gulachenski, T. Madera, J. Park, A. Peterson, S. Piper, H. Rahn, G. Silveira, and the Vancouver Area Network of Drug Users for assistance in the field or laboratory.
Ethics
Collection of rats for this study adhered to all local guidelines. Approval in New York City was granted by the Fordham University Institutional Animal Care and Use Committee (IACUC; Protocol JMS-16-04); by the University of British Columbia's Animal Care Committee (A14-0265) in Vancouver; by the Tulane University IACUC (Protocol no.0451) in New Orleans; and by the IACUC committees at Yale University (protocol no. 2012-11498) and the Oswaldo Cruz Foundation (protocol no. 003/2012) for work in Salvador, Brazil.
Data accessibility
Illumina reads are available on the NCBI SRA, accession PRJNA414893. SNP genotypes are available on the Dryad digital repository (http://dx.doi.org/10.5061/dryad.kg1kd5s) [58].
Authors' contributions
M.C., J.M.-S. and J.L.R. designed the study, analysed data and wrote the manuscript; M.C., K.A.B., B.M.G. and J.L.R. conducted the laboratory work; J.M.-S., M.J.B., K.A.B., A.C., F.C., C.G.H. and J.L.R. obtained funding; all authors edited the manuscript and contributed samples.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by National Science Foundation grant nos. DEB-1457523 and DBI-1531639 to J.M.-S., BCS-1313703 to M.J.B., and ERI 1738789 to J.L.R. Vancouver sampling was supported by NSERC grant no. 2015-05058 to D. Patrick, and CIHR grant no. MOP-119530 and CGV-104833 to C.G.H. Salvador sampling was supported by National Institutes of Health R01-TW009504 and the Wellcome Trust 102330/Z/13/Z.
References
- 1.Johnson MT, Munshi-South J. 2017. Evolution of life in urban environments. Science 358, eaam8327 ( 10.1126/science.aam8327) [DOI] [PubMed] [Google Scholar]
- 2.Tucker MA, et al. 2018. Moving in the Anthropocene: global reductions in terrestrial mammalian movements. Science 359, 466–469. ( 10.1126/science.aam9712) [DOI] [PubMed] [Google Scholar]
- 3.Munshi-South J, Zolnik CP, Harris SE. 2016. Population genomics of the Anthropocene: urbanization is negatively associated with genome-wide variation in white-footed mouse populations. Evol. Appl. 9, 546–564. ( 10.1111/eva.12357) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gortat T, Rutkowski R, Gryczyńska A, Pieniążek A, Kozakiewicz A, Kozakiewicz M. 2015. Anthropopressure gradients and the population genetic structure of Apodemus agrarius. Conserv. Genet. 16, 649–659. ( 10.1007/s10592-014-0690-0) [DOI] [Google Scholar]
- 5.Lourenço A, Álvarez D, Wang IJ, Velo-Antón G. 2017. Trapped within the city: integrating demography, time since isolation and population-specific traits to assess the genetic effects of urbanization. Mol. Ecol. 26, 1498–1514. ( 10.1111/mec.14019) [DOI] [PubMed] [Google Scholar]
- 6.Unfried TM, Hauser L, Marzluff JM. 2013. Effects of urbanization on song sparrow (Melospiza melodia) population connectivity. Conserv. Genet. 14, 41–53. ( 10.1007/s10592-012-0422-2) [DOI] [Google Scholar]
- 7.Beninde J, Feldmeier S, Werner M, Peroverde D, Schulte U, Hochkirch A, Veith M. 2016. Cityscape genetics: structural vs. functional connectivity of an urban lizard population. Mol. Ecol. 25, 4984–5000. ( 10.1111/mec.13810) [DOI] [PubMed] [Google Scholar]
- 8.Noreen AME, Niissalo MA, Lum SKY, Webb EL. 2016. Persistence of long-distance, insect-mediated pollen movement for a tropical canopy tree species in remnant forest patches in an urban landscape. Heredity 117, 472–480. ( 10.1038/hdy.2016.64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Combs M, Puckett EE, Richardson J, Mims D, Munshi-South J. 2017. Spatial population genomics of the brown rat (Rattus norvegicus) in New York City. Mol. Ecol. 27, 83–98. ( 10.1111/mec.14437) [DOI] [PubMed] [Google Scholar]
- 10.Attard CRM, Beheregaray LB, Möller LM. 2017. Genotyping-by-sequencing for estimating relatedness in non-model organisms: avoiding the trap of precise bias. Mol. Ecol. Resour. 18, 381–390. ( 10.1111/1755-0998.12739) [DOI] [PubMed] [Google Scholar]
- 11.Richardson JL, Brady SP, Wang IJ, Spear SF. 2016. Navigating the pitfalls and promise of landscape genetics. Mol. Ecol. 25, 849–863. ( 10.1111/mec.13527) [DOI] [PubMed] [Google Scholar]
- 12.Feng AYT, Himsworth CG. 2014. The secret life of the city rat: a review of the ecology of urban Norway and black rats (Rattus norvegicus and Rattus rattus). Urban Ecosyst. 17, 149–162. ( 10.1007/s11252-013-0305-4) [DOI] [Google Scholar]
- 13.Puckett EE, et al. 2016. Global population divergence and admixture of the brown rat (Rattus norvegicus). Proc. R. Soc. B 283, 20161762 ( 10.1098/rspb.2016.1762) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Armitage PL. 1993. Commensal rats in the New World, 1492–1992. Biologist 40, 174–178. [Google Scholar]
- 15.Puckett EE, Munshi-South J. 2018. Brown rat demography reveals pre-commensal structure in eastern Asia prior to expansion into Southeast Asia during the Song dynasty. bioRxiv, 249862 ( 10.1101/249862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hulme-Beaman A, Dobney K, Cucchi T, Searle JB. 2016. An ecological and evolutionary framework for commensalism in anthropogenic environments. Trends Ecol. Evol. 31, 633–645. ( 10.1016/j.tree.2016.05.001) [DOI] [PubMed] [Google Scholar]
- 17.Jones HP, et al. 2016. Invasive mammal eradication on islands results in substantial conservation gains. Proc. Natl Acad. Sci. USA 113, 4033–4038. ( 10.1073/pnas.1521179113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Firth C, et al. 2014. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal rattus norvegicus in New York City. mBio 5, e01933-14 ( 10.1128/mBio.01933-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pepin M, Dupinay T, Zilber A-L, Mcelhinney L.. 2016. Of rats and pathogens : pathogens transmitted by urban rats with an emphasis on hantaviruses. CAB Rev. 11, 1–3. ( 10.1079/PAVSNNR201611019) [DOI] [Google Scholar]
- 20.Himsworth CG, Parsons KL, Jardine C, Patrick DM. 2013. Rats, cities, people, and pathogens: a systematic review and narrative synthesis of literature regarding the ecology of rat-associated zoonoses in urban centers. Vector-Borne Zoonotic Dis. 13, 349–359. ( 10.1089/vbz.2012.1195) [DOI] [PubMed] [Google Scholar]
- 21.Himsworth CG, et al. 2015. An investigation of Bartonella spp., Rickettsia typhi, and Seoul hantavirus in rats (Rattus spp.) from an inner-city neighborhood of Vancouver, Canada: is pathogen presence a reflection of global and local rat population structure? Vector Borne Zoonotic Dis. Larchmt. N 15, 21–26. ( 10.1089/vbz.2014.1657) [DOI] [PubMed] [Google Scholar]
- 22.Lee M, Byers KA, Donovan C, Bidulka J, Stephen C, Patrick DM, Himsworth CG. 2018. Effects of culling on Leptospira interrogans carriage by rats. Emerg. Infect. Dis. 24, 356–360. ( 10.3201/eid2402.171371) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsons MH, Banks PB, Deutsch MA, Corrigan RF, Munshi-South J. 2017. Trends in urban rat ecology: a framework to define the prevailing knowledge gaps and incentives for academia, pest management professionals (PMPs) and public health agencies to participate. J. Urban Ecol. 3, 376 ( 10.1093/jue/jux005) [DOI] [Google Scholar]
- 24.Byers KA, Lee MJ, Donovan CM, Patrick DM, Himsworth CG. 2017. A novel method for affixing global positioning system (GPS) tags to urban Norway rats (Rattus norvegicus): feasibility, health impacts and potential for tracking movement. J. Urban Ecol. 3, e68496 ( 10.1093/jue/jux010) [DOI] [Google Scholar]
- 25.Davis DE. 1953. The characteristics of rat populations. Q. Rev. Biol. 28, 373–401. ( 10.1086/399860) [DOI] [PubMed] [Google Scholar]
- 26.Heiberg A-C, Sluydts V, Leirs H. 2012. Uncovering the secret lives of sewer rats (Rattus norvegicus): movements, distribution and population dynamics revealed by a capture–mark–recapture study. Wildl. Res. 39, 202–219. ( 10.1071/WR11149) [DOI] [Google Scholar]
- 27.Costa F, Richardson JL, Dion K, Mariani C, Pertile AC, Burak MK, Childs JE, Ko AI, Caccone A. 2016. Multiple paternity in the Norway rat, Rattus norvegicus, from urban slums in Salvador, Brazil. J. Hered. 107, 181–186. ( 10.1093/jhered/esv098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis DE, Emlen JT, Stokes AW. 1948. Studies on home range in the brown rat. J. Mammal. 29, 207–225. ( 10.2307/1375387) [DOI] [Google Scholar]
- 29.Glass GE. 1989. Comparative ecology and social interactions of Norway rat (Rattus norvegicus) populations in Baltimore, Maryland. In Occasional Papers of the Museum of Natural History, University of Kansas, vol. 130, 33 p. Lawrence, KS: University of Kansas Publications. [Google Scholar]
- 30.Taylor KD, Quy RJ. 1978. Long distance movements of a common rat (Rattus norvegicus) revealed by radio-tracking. Mammalia 42, 63–72. ( 10.1515/mamm.1978.42.1.63) [DOI] [Google Scholar]
- 31.Glass GE, Klein SL, Norris DE, Gardner LC. 2016. Multiple paternity in urban Norway rats: extended ranging for mates. Vector-Borne Zoonotic Dis. 16, 342–348. ( 10.1089/vbz.2015.1816) [DOI] [PubMed] [Google Scholar]
- 32.Gardner-Santana L, Norris D, Fornadel C, Hinson E, Klein S, Glass G. 2009. Commensal ecology, urban landscapes, and their influence on the genetic characteristics of city-dwelling Norway rats (Rattus norvegicus). Mol. Ecol. 18, 2766–2778. ( 10.1111/j.1365-294X.2009.04232.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kajdacsi B, et al. 2013. Urban population genetics of slum-dwelling rats (Rattus norvegicus) in Salvador, Brazil. Mol. Ecol. 22, 5056–5070. ( 10.1111/mec.12455) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Richardson JL, et al. 2017. Using fine-scale spatial genetics of Norway rats to improve control efforts and reduce leptospirosis risk in urban slum environments. Evol. Appl. 10, 323–337. ( 10.1111/eva.12449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Desvars-Larrive A, et al. 2017. Population genetics, community of parasites, and resistance to rodenticides in an urban brown rat (Rattus norvegicus) population. PLoS ONE 12, e0184015 ( 10.1371/journal.pone.0184015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hagan JE, et al. 2016. Spatiotemporal determinants of urban leptospirosis transmission: four-year prospective cohort study of slum residents in Brazil. PLoS Negl. Trop. Dis. 10, e0004275 ( 10.1371/journal.pntd.0004275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Costa F, et al. 2014. Influence of household rat infestation on Leptospira transmission in the urban slum environment. PLoS Negl. Trop. Dis. 8, e3338 ( 10.1371/journal.pntd.0003338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rael RC, Peterson AC, Ghersi BM, Childs J, Blum MJ. 2016. Disturbance, reassembly, and disease risk in socioecological systems. EcoHealth 13, 450–455. ( 10.1007/s10393-016-1157-1) [DOI] [PubMed] [Google Scholar]
- 39.Peterson AC, et al. 2017. Rodent-borne Bartonella infection varies according to host species within and among cities. EcoHealth 14, 771–782. ( 10.1007/s10393-017-1291-4) [DOI] [PubMed] [Google Scholar]
- 40.Himsworth CG, Jardine CM, Parsons KL, Feng AYT, Patrick DM. 2014. The characteristics of wild rat (Rattus spp.) populations from an inner-city neighborhood with a focus on factors critical to the understanding of rat-associated zoonoses. PLoS ONE 9, e91654 ( 10.1371/journal.pone.0091654) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Himsworth CG, et al. 2013. Ecology of Leptospira interrogans in Norway rats (Rattus norvegicus) in an inner-city neighborhood of Vancouver, Canada. PLoS Negl. Trop. Dis. 7, e2270 ( 10.1371/journal.pntd.0002270) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbs RA, et al. 2004. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature 428, 493–521. ( 10.1038/nature02426) [DOI] [PubMed] [Google Scholar]
- 43.Shirk AJ., Cushman SA. 2011. sGD: software for estimating spatially explicit indices of genetic diversity. Mol. Ecol. Resour. 11, 922–934. ( 10.1111/j.1755-0998.2011.03035.x) [DOI] [PubMed] [Google Scholar]
- 44.Lawson DJ, Hellenthal G, Myers S, Falush D. 2012. Inference of population structure using dense haplotype data. PLoS Genet. 8, e1002453 ( 10.1371/journal.pgen.1002453) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Scheet P, Stephens M. 2006. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644. ( 10.1086/502802) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jombart T, Ahmed I. 2011. Adegenet 1.3-1: new tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. ( 10.1093/bioinformatics/btr521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galpern P, Peres-Neto PR, Polfus J, Manseau M. 2014. MEMGENE: spatial pattern detection in genetic distance data. Methods Ecol. Evol. 5, 1116–1120. ( 10.1111/2041-210X.12240) [DOI] [Google Scholar]
- 48.Nunney L. 2016. The effect of neighborhood size on effective population size in theory and in practice. Heredity 117, 224 ( 10.1038/hdy.2016.76) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munshi-South J. 2012. Urban landscape genetics: canopy cover predicts gene flow between white-footed mouse (Peromyscus leucopus) populations in New York City. Mol. Ecol. 21, 1360–1378. ( 10.1111/j.1365-294X.2012.05476.x) [DOI] [PubMed] [Google Scholar]
- 50.Goldingay RL, Harrisson KA, Taylor AC, Ball TM, Sharpe DJ, Taylor BD. 2013. Fine-scale genetic response to landscape change in a gliding mammal. PLoS ONE 8, e80383 ( 10.1371/journal.pone.0080383) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson A, Fenton B, Malloch G, Boag B, Hubbard S, Begg G. 2016. Urbanisation versus agriculture: a comparison of local genetic diversity and gene flow between wood mouse Apodemus sylvaticus populations in human-modified landscapes. Ecography 39, 87–97. ( 10.1111/ecog.01297) [DOI] [Google Scholar]
- 52.Panti-May JA, et al. 2016. A two-year ecological study of Norway rats (Rattus norvegicus) in a Brazilian urban slum. PLoS ONE 11, e0152511 ( 10.1371/journal.pone.0152511) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Noel S, Ouellet M, Galois P, Lapointe F-J. 2007. Impact of urban fragmentation on the genetic structure of the eastern red-backed salamander. Conserv. Genet. 8, 599–606. ( 10.1007/s10592-006-9202-1) [DOI] [Google Scholar]
- 54.Walsh MG. 2014. Rat sightings in New York City are associated with neighborhood sociodemographics, housing characteristics, and proximity to open public space. PeerJ 2, e533 ( 10.7717/peerj.533) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson S, Bragdon C, Olson C, Merlino M, Bonaparte S. 2016. Characteristics of the built environment and the presence of the Norway rat in New York City: results from a neighborhood rat surveillance program, 2008–2010. J. Environ. Health 78, 22–29. [PubMed] [Google Scholar]
- 56.van Adrichem MH, Buijs JA, Goedhart PW, Verboom J. 2013. Factors influencing the density of the brown rat (Rattus norvegicus) in and around houses in Amsterdam. Vis. Nat. 56, 77. [Google Scholar]
- 57.Angley LP, Combs M, Firth C, Frye MJ, Lipkin I, Richardson JL, Munshi-South J. 2017. Spatial variation in the parasite communities and genomic structure of urban rats in New York City. Zoonoses Public Health 65, e113–e123. ( 10.1111/zph.12418) [DOI] [PubMed] [Google Scholar]
- 58.Combs M, Byers KA, Ghersi BM, Blum MJ, Caccone A, Costa F, Himsworth CG, Richardson JL, Munshi-South J. 2018. Data from: Urban rat races: spatial population genomics of brown rats (Rattus norvegicus) compared across multiple cities Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.kg1kd5s) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Combs M, Byers KA, Ghersi BM, Blum MJ, Caccone A, Costa F, Himsworth CG, Richardson JL, Munshi-South J. 2018. Data from: Urban rat races: spatial population genomics of brown rats (Rattus norvegicus) compared across multiple cities Dryad Digital Repository. (http://dx.doi.org/10.5061/dryad.kg1kd5s) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
Illumina reads are available on the NCBI SRA, accession PRJNA414893. SNP genotypes are available on the Dryad digital repository (http://dx.doi.org/10.5061/dryad.kg1kd5s) [58].