Abstract
Endophytic actinobacteria exist widely in plant tissues and are considered as a potential bioresource library of natural products. Tea plants play important roles in human health and in the lifestyles of Asians, especially the Chinese. However, little is known about the endophytic actinobacteria of tea plants. In this study, 16 actinobacteria of 7 different genera and 28 actinobacteria of 8 genera were isolated and analyzed by 16S rRNA gene sequencing from tea cultivars of Zijuan and Yunkang-10 (Camellia sinensis var. assamica), respectively. The diversity of actinobacteria species from Zijuan were higher in July than December (6 vs. 3 genera), but the diversity of species from Yunkang-10 were higher in December than July (7 vs. 3 genera). No actinobacteria isolates were obtained from any tea cultivar in September. Ten isolates from Yunkang-10 exhibited antimicrobial activity against at least one human pathogenic microorganism (Staphylococcus epidermidis, Shigella flexneri, and Escherichia coli), but none of the isolates from Zijuan exhibited antimicrobial activities. Fourteen strains were further exammined the genes of polyketide synthetase (PKS)-I and PKS-II and non-ribosomal peptide synthetase (NRPS). Brevibacterium sp. YXT131 from Yunkang-10 showed strong inhibitory activity against S. epidermidis, Sh. flexneri, and E. coli, and PKS-I and PKS-II and NRPS genes were obtained from the strain. In in vitro assays, extracts from 14 actinobacteria that were tested for antibiotic biosynthetic genes showed no inhibition of concanavalin A (ConA)-induced murine splenocyte proliferation. In in vivo assays, the crude extract of YXT131 modulated the immune response by decreasing the proinflammatory cytokines interleukin (IL)-12/IL-23 p40 and tumor necrosis factor (TNF)-α in the serum of mice. These results confirm that endophytic actinobacteria from tea plants might be an undeveloped bioresource library for active compounds.
Keywords: Camellia sinensis, endophytic actinobacteria, diversity, antimicrobial activity, immunomodulatory activities
Introduction
Actinobacteria are aerobic, gram-positive bacteria and are well-known producers of a vast array of secondary metabolites, including antibiotics, immunosuppressive agents, antitumor agents, and enzymes, many of which are of great importance to the pharmaceutical and agricultural industries (Saini et al., 2015; Landwehr et al., 2016; Salcedo et al., 2016; Yang et al., 2016). Endophytes are microorganisms that ubiquitously colonize the internal tissues of plants without causing any negative effects, and some endophytes are able to control plant pathogens and promote the growth of plants (Santoyo et al., 2016; Kandel et al., 2017). Although numerous species of actinobacteria occur in the soil, other microbial habitats, such as leaf litter and plants, are potential sources of actinobacteria for the isolation of biologically active compounds (Sardi et al., 1992; Takahashi and Omura, 2003; Gos et al., 2017). In recent years, endophytic actinobacteria have been isolated from many crop plants (such as wheat, rice, and potatoes) (Coombs and Franco, 2003; Sessitsch et al., 2004; Tian et al., 2007) and medicinal plants (Qin et al., 2009; Gos et al., 2017). Endophytic actinobacteria are a potential source for the production of secondary metabolites that are used in the direct antagonism of pests and diseases (Cao et al., 2005) as well as various natural products with antimicrobial, antitumor, and anti-infection activities (Qin et al., 2011; Gos et al., 2017). Endophytic actinobacteria can also confer salt tolerance to host plants and promote host-plant growth (Qin et al., 2017).
As a popular non-alcoholic beverage, tea and tea drinks play important roles in human health and lifestyle, such as by reducing cardiovascular mortality and treating digestive disorders (Yang C.S. et al., 2009; Persson et al., 2010; Begas et al., 2017). As the same plant species, the tea cultivars of Zijuan and Yunkang-10 belong to the taxonomic species of Camellia sinensis var. assamica, which originated from the Yunnan province of China (Wang et al., 2017; Zhu et al., 2017). The two closely related cultivars of Zijuan and Yunkang-10 are the major raw materials for Pu’er tea (a kind of dark tea) produced in Yunnan. Catechins are the major secondary metabolic products in tea leaves (especially in green tea), and polyphenols are known to confer health benefits (Khan and Mukhtar, 2007). Anthocyanin is another functional flavonoid with health benefits and is widely distributed in higher plants (e.g., vegetables, flowers, fruits, cereals, and tea plants) (Joshi et al., 2015; Fabroni et al., 2016; Chen et al., 2017). The total anthocyanin content in Zijuan has been reported to be almost three times that found in other Chinese tea cultivars (including Yunkang-10), and thus Zijuan is considered to be an anthocyanin-rich cultivar (Yang X.R. et al., 2009). However, only a few studies have observed on tea plant endophytes, and those studies have mainly been performed on endophytic fungi and for plant disease protection (Rabha et al., 2014). For example, the endophytic fungus Colletotrichum gloeosporioides from healthy tea plant leaves shows strong inhibitory activity against tea pathogens of Pestalotiopsis theae and Colletotrichum camelliae; the inhibitory factors may be the highly efficient fungal chitinase and protease (Rabha et al., 2014). Compared to endophytic fungi, endophytic actinobacteria have received almost no attention in tea plants, and the interactions between endophytic actinobacteria and tea plants have not been investigated. Based on recent research progress in other plants, the study of endophytic actinobacteria culture and bioactivity is of great significance for developing bioresource libraries. In this study, the endophytic actinobacteria from tea plants of Zijuan and Yunkang-10 were isolated at different seasons, and the bioactivities of antimicrobial and immunomodulatory were screened and further evaluated for the isolates.
Materials and Methods
Tea Sample Collection
Three-year-old tea plants of Zijuan and Yunkang-10 (Camellia sinensis var. assamica) were obtained from Dechang tea plantation (Shucheng China, 31°11’ N, 116°47’ E). The leaf samples of Zijuan and Yunkang-10 were collected on July 5, September 10, and December 5 of 2015. At the each time of sampling, 15 branches with the same growth and no pest damage were selected for each tea cultivar (considered as 1 sample). The healthy branches were placed in fully soaked floral foam and transported to the laboratory within 2 h for actinobacteria isolation; the isolation procedures were performed within 96 h.
Endophytic Actinobacteria Isolation
The leaf samples were pretreated following the method described by Qin et al. (2009) with minor modifications. The samples were air-dried for 48 h at room temperature and then washed with ultrasonic cleaning (160 W, 15 min). After drying, the samples were sterilized in the following order: a 6-min wash in 5% NaOCl, followed by a 10-min wash in 2.5% Na2S2O3, a 5-min wash in 70% ethanol, a 5-min wash in sterile water, and a final rinse in 10% NaHCO3 for 10 min. The sterilized tissues were imprinted on nutrient agar (NA, Difco) and tryptic soy agar (TSA, Difco), and incubated at 28°C for 2 weeks to ensure the sterilization effectiveness. After surface sterilization and thorough drying under aseptic conditions, the samples were cut up in a sterile mortar and ground to a homogenate, followed by dilution to 10-1 to 10-3 with sterile water. Aliquots of 200 μL of the dilutions were spread-plated onto a series of isolation media as indicated in Table 1 and incubated at 28°C for 2–3 weeks for actinobacteria cultivation. The pH of the selected media was adjusted to 7.2. Each isolation medium was amended with nalidixic acid (50 mg/L) and nystatin (100 mg/L) to prevent the growth of gram-negative bacteria and fungi. As colonies appeared on the plates, candidate colonies were observed and selected carefully according to phenotypic characteristics.
Table 1.
Culture medium composition for endophytic actinobacteria isolation.
| Medium | Composition (per 1000 mL) | Reference |
|---|---|---|
| GAUZE’s medium | 5 g K2HPO4, 20 g soluble starch, 0.5 g MgSO4⋅7H2O, 0.01 g FeSO4⋅7H2O, 1 g KNO3, 0.5 g NaCl, 18 g agar | Ma et al., 2017 |
| TWYE | 0.5 g K2HPO4, 0.25 g yeast extract, 18 g agar | Crawford et al., 1993 |
| YECD | 2 g K2HPO4, 0.3 g yeast extract, 0.3 g glucose, 18 g agar | Coombs and Franco, 2003 |
| Humic acid-vitamin agar (HV) | 0.02 g CaCO3, 0.5 g Na2HPO4, 0.5 g MgSO4⋅7H2O, 0.01 g FeSO4⋅7H2O, 1 g Humic acid, 1.7 g KCl, 0.5 mg VB6, 0.5 mg p-aminobenzoic acid, 0.5 mg riboflavin, 0.5 mg thiamine, 0.5 mg inositol, 0.5 mg pantothenic acid, 0.5 mg nicotinic acid, 0.25 mg biotin, 18 g agar | Hayakawa and Nonomura, 1989 |
| Glucose-Asparagine modified media (GA) | 1 g K2HPO4, 1 g asparagine, 0.01 g ZnSO4⋅7H2O, 0.01 g FeSO4⋅7H2O, 10 g glucose, 0.01 g MnCl2⋅4H2O, 18 g agar | Shirling and Gottlieb, 1966 |
DNA Extraction, 16S rRNA Gene Sequencing, and Phylogenetic Analysis
The obtained isolates were subjected to 16S rRNA gene sequence analysis for genus and species identification. The genomic DNA was extracted using the method of Li et al. (2007). The 16S rRNA gene of each isolate was amplified using primer pairs 27F and 1492R (Table 2), and polymerase chain reaction (PCR) amplification was carried out as described by Li et al. (2007). The reagents for PCR reaction were purchased from TaKaRa (Dalian, China). The PCR -products were separated by agarose gel electrophoresis, purified using QIA quick gel extraction kits (Qiagen, Hilden, Germany), then ligated into a pMD-19T vector (TaKaRa). Positive clones were screened further, and insert DNA sequencing was performed by Invitrogen (Shanghai, China) on an Applied Biosystems PRISM 3730 DNA sequencer.
Table 2.
Polymerase chain reaction (PCR) primers used in this study.
| Primer name | Sequence (5′—3′) | Target gene | Length (bp) | Reference |
|---|---|---|---|---|
| 27F | 5′-AGAGTTTGATCCTGGCTCAG-3′ | 16S rRNA | 1400–1500 | Li et al., 2007 |
| 1492R | 5′-ACGGTTACCTTGTTACGACTT-3′ | |||
| K1F | 5′-TSAAGTCSAACATCGGBCA-3′ | PKS-I | 1200–1400 | Ayuso-Sacido and Genilloud, 2005 |
| M6R | 5′-CGCAGGTTSCSGTACCAGTA-3′ | |||
| KSα | 5′-TSGCSTGCTTGGAYGCSATC-3′ | PKS-II | 600 | Metsa-Ketela et al., 1999 |
| KSβ | 5′-TGGAANCCGCCGAABCCTCT-3′ | |||
| A3F | 5′-GCSTACSYSATSTACACSTCSGG-3′ | NRPS | 700–800 | Ayuso-Sacido and Genilloud, 2005 |
| A7R | 5′-SASGTCVCCSGTSCGGTAS-3′ | |||
The 16S rRNA gene analysis was performed by BLAST searches in the National Center for Biotechnology Information database1 and EzBioCloud2. Multiple sequence alignment of selected 16S rDNA sequence was carried out using CLUSTAL_X (version 2.0) (Thompson et al., 1997), and a phylogenetic tree was constructed using MEGA v6.0 (Tamura et al., 2013). Distances (distance options according to the Kimura two-parameter model) (Kimura, 1980) and clustering were based on the neighbor-joining (Saitou and Nei, 1987) method. Bootstrap analysis based on 1000 resamplings was used to evaluate the topology of the neighbor-joining tree (Felsenstein, 1985). The 16S rRNA gene sequences of the 44 isolates have been deposited in GenBank under the accession numbers (MH298662–MH298705).
Detection of PKS-I, PKS-II, and NRPS Genes
Three sets of degenerate primers for amplification of the genes encoding polyketide synthases I and II (PKS-I and PKS-II) and non-ribosomal peptide synthetase (NRPS) were selected (Table 2), and amplification was carried out as recommended by Metsa-Ketela et al. (1999) and Ayuso-Sacido and Genilloud (2005). The reaction mixture contained 2.5 U of Taq DNA polymerase, 1 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, 2 μM each primer, and 5% dimethyl sulfoxide in a 50-μL reaction volume. A reaction mixture with no actinobacterial DNA template was used as a negative control. Thermocycling conditions consisted of one denaturation step of 94°C for 5 min, 30 amplification cycles of 94°C for 1 min, 57°C (for K1F–M6R and A3F–A7R) or 58°C (for KSα–KSβ) for 1 min, and 72°C for 2 min; and a final extension at 72°C for 5 min.
Active Compound Extraction and Bioactivity Evaluation
The endophytic isolates were cultured in GAUZE’s liquid medium at 28°C and 180 r/min (Rehacek, 1959). After 7–12 days of cultivation, the 100 mL culture broth was collected by centrifugation at 12,000 ×g for 10 min and extracted by 100 mL ethyl acetate for three times. The organic phase was evaporated under reduced pressure to yield a dry extract. The dry extract was resuspended by 5 mL sterile water and used for antimicrobial screening. The antimicrobial susceptibility was examined by placing antimicrobial testing disks (7 mm diameter) containing 25 μL test extract suspension onto LB plates (Mearns-Spragg et al., 1998). The tested plates were incubated at 37°C, and the diameters of the inhibition zones were measured after 24 h. A 25-μL volume of sterile water was used as a negative control. The pathogenic bacteria Staphylococcus epidermidis, Shigella flexneri, Escherichia coli, and Bacillus cereus were used as the indicator microorganisms for antimicrobial determination. The pathogenic microorganisms were obtained from the Institute of Quality and Standard for Agro-products, Zhejiang Academy of Agricultural Sciences.
Animal Experiments and Physiological Tests
Female BALB/c mice at 8–9 weeks of age were purchased from the Zhejiang Laboratory Animal Center (Hangzhou, China). The mice were maintained in pathogen-free conditions with standard laboratory chow and water ad libitum. Animal experiments were approved and performed in accordance with the guidelines of the Animal Care Committee of Zhejiang province, China. Dried extracts from the test endophytic actinobacteria were resuspended by saline with concentration of 2 mg/mL and injected i.p. 100 μL per mouse once daily for 2 weeks. The same volume (100 μL per mouse) of saline was given as a vehicle control. Three independent experiments were repeated, and in each experiment five mice were used. Clinical signs of poisoning were assessed and the weights of mice were recorded daily. Blood samples (∼500 μL per mouse) were taken from the retro-orbital venous plexus at the end of the experiment and incubated at 4°C for 30 min. Serum was collected by centrifugation at 4500 ×g for 10 min and stored at -20°C until analysis. Cytokines [interleukin-2 (IL-2), interleukin-6 (IL-6), the shared p40 subunit of IL-12 and IL-23 (IL-12/IL-23 p40), and tumor necrosis factor-alpha (TNF-α)] in the serum were determined with sandwich enzyme-linked immunosorbent assay (ELISA) kits according to the manufacturer’s instructions (Dakewe Biotech, Shenzhen, China).
Splenocyte Proliferation
Freshly isolated splenocytes were obtained from BALB/c mice and incubated in 200 μL RPMI 1640 with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 5 μg/mL of concanavalin A (ConA) in a humidified, 37°C, 5% CO2-containing incubator for 48 h in the presence or absence of extracts. Cyclosporin A (CsA) (500 ng/mL) was used as a positive control. Three independent experiments were repeated, and in each experiment five wells of splenocytes were used. RPMI 1640 medium was purchased from Gibco, Thermo Fisher Scientific (Waltham, MA, United States). ConA and CsA were purchased from Sigma (St. Louis, MO, United States). The cell number was determined by a Millipore Guava easyCyte 8HT Flow Cytometer (Millipore, Billerica, MA, United States).
Statistical Analysis
Data are presented as means ± standard deviation. Statistical analyzes were performed using Student’s t test. P < 0.05 was considered statistically significant.
Ethics Statement
This study was carried out in accordance with the guidelines of the Animal Care Committee of Zhejiang province, China (Government Decree No. 263). The protocol was approved by the Committee on the Ethics of Animal Experiments of Zhejiang Academy of Agricultural Science.
Results
Evaluation of Surface Sterilization
Surface sterilization is critical for the study of plant endophytic actinobacteria. In this study, the surface-sterilized leaves were examined by NA and TSA, and no microbial colony was observed after 2 weeks of incubation at 28°C. This indicated that the surface-sterilization protocol modified from Qin et al. (2009) was effective in removing phyllospheric microorganisms of tea plants.
Selective Isolation of Culturable Endophytic Actinobacteria From Zijuan and Yunkang-10
To obtain as many endophytic actinobacteria as possible, five selective isolation media were used simultaneously in this study (Table 1). Endophytic actinobacteria were isolated on all of five media. In total, 44 actinobacterial strains (28 from Yunkang-10 and 16 from Zijuan) were isolated from 3 samples of Zijuan and 3 samples of Yunkang-10 (Table 3).
Table 3.
The isolated endophytic actinobacteria of Zijuan and Yunkang-10.
| Tea cultivars | July |
September |
December |
|||
|---|---|---|---|---|---|---|
| Species | Number | Species | Number | Species | Number | |
| Zijuan | Brachybacterium sp. | 1 | Brachybacterium sp. | 2 | ||
| Brevibacterium sp. | 1 | Kocuria sp. | 3 | |||
| Kocuria sp. | 1 | Microbacterium sp. | 1 | |||
| Leucobacter sp. | 1 | |||||
| Micrococcus sp. | 4 | |||||
| Streptomyces sp. | 2 | |||||
| Total number | 10 | Total number | 0 | Total number | 6 | |
| Yunkang-10 | Brevibacterium sp. | 4 | Pseudarthrobacter sp. | 1 | ||
| Micrococcus sp. | 1 | Brachybacterium sp. | 1 | |||
| Mycobacterium sp. | 3 | Brevibacterium sp. | 6 | |||
| Kocuria sp. | 4 | |||||
| Microbacterium sp. | 5 | |||||
| Micrococcus sp. | 2 | |||||
| Saccharomonospora sp. | 1 | |||||
| Total number | 8 | Total number | 0 | Total number | 20 | |
Diversity of Endophytic Actinobacteria Analyzed by 16S rRNA Gene Sequencing
The endophytic actinobacteria obtained from Zijuan were distributed among 7 genera [i.e., Brachybacterium sp. (3 isolates), Brevibacterium sp. (1 isolate), Kocuria sp. (4 isolates), Leucobacter sp. (1 isolate), Micrococcus sp. (4 isolates), Microbacterium sp. (1 isolate), and Streptomyces sp. (2 isolates)] within the class actinobacteria (Figure 1). Among them, Brachybacterium sp. and Kocuria sp. were two mutual groups both isolated in July and December, while the others were only isolated in July or December, and no endophytic actinobacteria were obtained in September. The diversity of endophytic actinobacteria from the Zijuan cultivar was in the order of July (6 genera) > December (3 genera) > September (0 genera). The 28 endophytic actinobacteria isolated from Yunkan-10 were distributed among 8 genera [i.e., Brevibacterium sp. (10 isolates), Micrococcus sp. (3 isolates), Mycobacterium sp. (3 isolates), Pseudarthrobacter sp. (1 isolate), Brachybacterium sp. (1 isolate), Kocuria sp. (4 isolates), Microbacterium sp. (5 isolates), and Saccharomonospora sp. (1 isolate)] within the class actinobacteria (Figure 1). Isolates of Brevibacterium sp. and Micrococcus sp. were obtained from both July and December specimens, and isolates of Mycobacterium sp. were found only in July. The isolates from other genera were only obtained in December, while no endophytic actinobacteria were obtained in September. The diversity of endophytic actinobacteria from Yunkang-10 was in the order of December (7 genera) > July (3 genera) > September (0 genera). In comparing the two cultivars, isolates of Leucobacter sp. and Streptomyces sp. were endemic actinobacterial groups to the Zijuan cultivar, while Mycobacterium sp., Pseudarthrobacter sp., and Saccharomonospora sp. were endemic to Yunkang-10.
FIGURE 1.
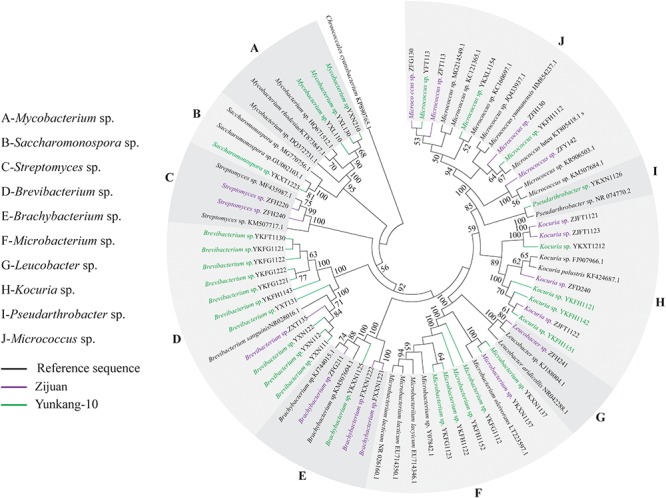
Phylogenetic analysis of endophytic actinobacteria in different seasons from Zijuan and Yunkang-10. The phylogenetic tree was constructed by using 73 16S rRNA gene sequences.
Endophyte Antimicrobial Activity and Sequencing of PKS and NRPS Genes
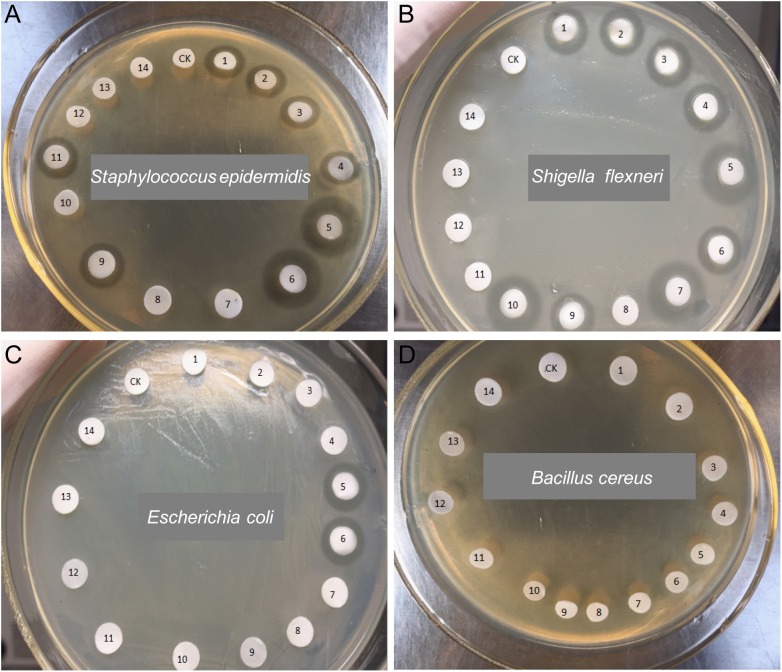
In antimicrobial screening test, several culture media indicated in Table 1 were preliminary evaluated, and GAUZE’s medium was the best one for the majority of actinobacteria isolates from Zijuan and Yunkang-10. The extracts from GAUZE’s medium showed no obvious antimicrobial difference from other media, but all the tested strains could be cultured by the GAUZE’s medium. The 44 isolates were screened for antimicrobial activities against the pathogenic bacteria S. epidermidis, Sh. flexneri, E. coli, and B. cereus. Ten of the 28 isolates (35.7%) from Yunkang-10 exhibited activity against at least one of the tested pathogenic microorganisms. Surprising, none of the 16 isolates from Zijuan showed obvious antimicrobial activity. A total of 14 isolates selected (10 antimicrobial positive strains and four negative strains) were selected for the determination of antibiotic biosynthetic gene sequences of PKS-I, PKS-II, and NRPS by PCR amplification using specific primer sets K1F–M6R, KSα–KSβ, and A3F–A7R, respectively (Table 4). As shown by primary screening, the inhibitory effect on Sh. flexneri was the most frequent detected antimicrobial activity in this study. Eight isolates were active against S. epidermidis, and 7 isolates were found to inhibit two or more pathogenic microorganisms. However, none of the isolates of this study exhibited activity against B. cereus. Two isolates, YXT131 and YKFG1221, which belong to the genus Brevibacterium, appeared to have a broad spectrum of antimicrobial activity (three pathogenic microorganisms). Brevibacterium sp. YXT131 exhibited high inhibitory effects against S. epidermidis, Sh. flexneri, and E. coli (Figure 2 and Table 5).
Table 4.
Endophytic actinobacteria isolated from Zijuan and Yunkang-10, and similarity values for 16S rRNA gene sequences.
| Isolate no. | Sequence length | Closest cultivated species | Similarity (%) | Query coverage (%) |
|---|---|---|---|---|
| YFT113 | 1345 | Micrococcus yunnanensis (FJ214355) | 99.33 | 93.0 |
| YXN120 | 1334 | Brevibacterium bullata (D12785) | 99.77 | 96.5 |
| YXN111 | 1441 | Brevibacterium celere (AY228463) | 99.44 | 97.8 |
| YXN112 | 1441 | Brevibacterium celere (AY228463) | 99.44 | 97.9 |
| YXT131 | 1395 | Brevibacterium celere (AY228463) | 99.78 | 96.1 |
| YKFG1221 | 1531 | Brevibacterium casei (X76564) | 99.72 | 100 |
| YKFG1121 | 1489 | Brevibacterium casei (X76564) | 99.24 | 100 |
| YKFG1122 | 1489 | Brevibacterium casei (X76564) | 99.31 | 100 |
| YKFT1130 | 1492 | Brevibacterium casei (X76564) | 99.72 | 100 |
| YKFH1122 | 1487 | Microbacterium lacticum (X77441) | 99.73 | 100 |
| YKFG1112 | 1487 | Microbacterium lacticum (X77441) | 99.59 | 100 |
| ZFY142 | 1428 | Micrococcus endophyticus (EU005372) | 97.88 | 99.0 |
| ZFG130 | 1366 | Micrococcus yunnanensis (FJ214355) | 99.19 | 94.5 |
| ZJFT1121 | 1459 | Kocuria marina (AY211385) | 99.86 | 100 |
FIGURE 2.
Antimicrobial activities of endophytic actinobacteria against pathogenic bacteria. Staphylococcus epidermidis (A), Shigella flexneri (B), Escherichia coli (C), and Bacillus cereus (D) were the tested strains. CK-Vehicle, 1-YFT113, 2-YXN120, 3-YXN111, 4-YXN112, 5-YXT131, 6-YKFG1221, 7-YKFG1121, 8-YKFG1122, 9-YKFT1130, 10-YKFH1122, 11-YKFG1112, 12-ZFY142, 13-ZFG130, 14-ZJFT1121.
Table 5.
Antimicrobial activities and PKS/NRPS genes of culturable actinobacteria from Zijuan and Yunkang-10.
| Isolate no. | Activitya against |
Presenceb of gene |
|||||
|---|---|---|---|---|---|---|---|
| S. epidermidis | Sh. flexneri | E. coli | B. cereus | PKS-I | PKS-II | NRPS | |
| YFT113 | ++ | ++ | - | - | + | - | - |
| YXN120 | ++ | ++ | - | - | + | - | - |
| YXN111 | ++ | ++ | - | - | + | - | - |
| YXN112 | + | ++ | - | - | + | - | - |
| YXT131 | +++ | +++ | ++ | - | + | + | + |
| YKFG1221 | +++ | + | ++ | - | + | + | + |
| YKFG1121 | - | +++ | - | - | + | + | - |
| YKFG1122 | - | - | - | - | - | - | - |
| YKFT1130 | + | + | - | - | + | + | - |
| YKFH1122 | - | ++ | - | - | - | - | + |
| YKFG1112 | + | - | - | - | - | - | + |
| ZFY142 | - | - | - | - | - | + | - |
| ZFG130 | - | - | - | - | + | - | - |
| ZJFT1121 | - | - | - | - | - | + | - |
aEstimated by measuring the diameter of the clear zone of growth inhibition. Symbols: -, no activity; +, ++, and +++, weak activity, moderate activity, and strong activity, respectively. b+, present; -, absent.
The PKS-I sequence was detected in 9 isolates (64.3%), while the PKS-II and NRPS sequences were detected in 6 and 4 of the 14 strains (Table 5 and Supplementary Figure S1), respectively. The isolates YXT131 and YKFG1221, which have broad spectrum antimicrobial activity, gave positive amplification products with PKS-I, PKS-II, and NRPS primers. The isolates from Yunkang-10 that exhibited antimicrobial activity against pathogenic microorganisms also gave positive amplification products for at least one of the PKS-I, PKS-II, and NRPS genes. The isolate YKFG1122 neither exhibited antimicrobial activity to the four tested pathogenic microorganisms nor provided any positive amplification products for the three biosynthetic genes. The three antimicrobial negative isolates from Zijuan still provided positive amplification products PKS-I or PKS-II.
Immunomodulatory Activity in Selected Actinobacteria
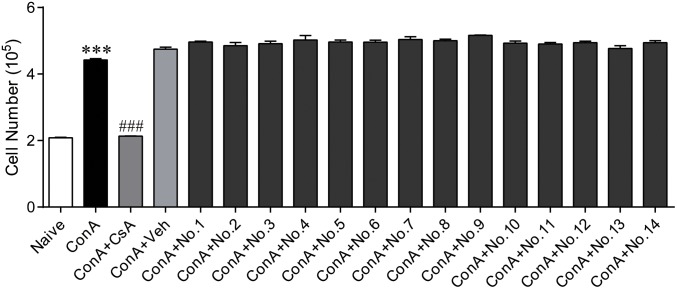
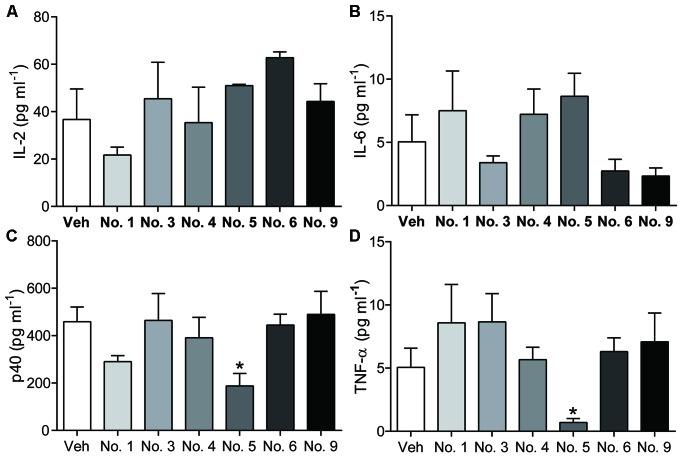
Chattopadhyay et al. (2012) showed that black tea has potential anti-inflammatory and immunomodulatory effects in animal models and in human peripheral mononuclear cells, and actinobacteria can produce secondary metabolites with immunosuppressive activity by suppressing cytokine expression and T cell proliferation (Yang et al., 2016). In in vitro splenocyte proliferation assays, extracts from 14 actinobacteria that were tested for antibiotic biosynthetic genes showed no inhibition of ConA-induced murine splenocyte proliferation (Figure 3), indicating that extracts from actinobacteria might not directly affect splenocyte proliferation. In the animal model, six isolates, YFT113, YXN111, YXN112, YXT131, YKFG1221, and YKFT1130, were examined for their potential immunomodulatory activity. Compared with the control group, BALB/c mice treated by fermentation extracts of the six isolates showed no obvious clinical signs of poisoning or other atypical signs throughout the trial. No significant differences in body weight were observed between control and fermentation extract-treated mice during the 2 weeks of testing (data not shown). CD4+ T cells are the key components of the adaptive immune system, and naïve CD4+ T cells can differentiate into effector T helper cell subsets (e.g., Th1, Th2, or Th17) by the coordinated functioning of distinct cytokines, including IL-6, IL-12, IL-23, and IL-2 (Murphy and Stockinger, 2010). TNF-α is a multifunctional cytokine that coordinates tissue homeostasis by regulating cytokine production, cell survival, and cell death (Annibaldi and Meier, 2018). In this study, no significant differences in IL-2 and IL-6 concentrations in serum were observed between control and fermentation extract-treated mice (Figures 4A,B). IL-12 and IL-23 are heterodimeric cytokines that share a common p40 subunit. To our surprise, the serum levels of IL-12/IL-23 p40 and TNF-α in Brevibacterium sp. YXT131 fermentation extract-treated mice were significantly lower than in the control group, but other fermentation extract-treated groups showed no significant differences (Figures 4C,D). These results indicated that the isolate YXT131 appeared to have immunosuppressive activity.
FIGURE 3.
Splenocyte proliferation in ConA-induced splenocytes treated with extracts of endophytic actinobacteria from Zijuan and Yunkang-10. Naive mouse splenocytes were stimulated with 5 μg/mL of ConA in the presence of different extracts, and 500 ng/mL CsA was set as a positive control. No.1-YFT113, No.2-YXN120, No.3-YXN111, No.4-YXN112, No.5-YXT131, No.6-YKFG1221, No.7-YKFG1121, No.8-YKFG1122, No.9-YKFT1130, No.10-YKFH1122, No.11-YKFG1112, No.12-ZFY142, No.13-ZFG130, 14-ZJFT1121. Data are means ± SD (n = 5), ∗∗∗p < 0.001 vs. naive, ###p < 0.001 vs. ConA.
FIGURE 4.
Cytokine levels in sera of mice treated by extracts of endophytic actinobacteria. IL-2 (A), IL-6 (B), IL-12/IL-23 p40 (C), and TNF-α (D) levels in serum were measured by ELISA. No.1-YFT113, No.3-YXN111, No.4-YXN112, No.5-YXT131, No.6-YKFG1221, No.9-YKFT1130. Data are means ± SD (n = 5), ∗p < 0.05 vs. vehicle.
Discussion
As described by previous studies, the natural characteristics and planting environment of Zijuan and Yunkang-10 are similar, and the two cultivars both originate from the Yunnan province of China (Wang et al., 2017; Zhu et al., 2017). The high anthocyanin content of Zijuan is one of the most important differences between the two cultivars (Yang X.R. et al., 2009). Previous studies have indicated that plants secondary metabolites such as alkaloids, phenolics, and terpenoids can interfere with cancer cells, bacteria, and fungi (Wink et al., 2012), and that anthocyanins act as antimicrobial agents of natural plant origin (Cisowska et al., 2011). The different anthocyanin content of Zijuan and Yunkang-10 may influence the microbial community, diversity and bioactivity of endophytic actinobacteria. In this study, the endophytic actinobacteria from Zijuan and Yunkang-10 were isolated and compared for their diversity and antimicrobial and immunomodulatory activities.
In our study, 44 isolates were isolated in July and December, but none of the endophytic actinobacteria was obtained in September. This result might be an incidental, alternatively, endophytic actinobacteria community in Zijuan and Yunkang-10 changed in September and is unculturable on these media. Many studies indicated that the endophytes of plant tissues [such as maple tree sap (Filteau et al., 2010), the buds of Scots pine trees (Pirttilä et al., 2005) and the grape endosphere (Baldan et al., 2014; Bulgari et al., 2014)] were shown to be sensitive to seasonal changes. Other than culture-dependent method, a variety culture independent methods including T-RFLP, PCR fingerprinting and 16S rRNA specific probes were used to investigate the seasonal community changes, and these results were consistent to the present culture-dependent investigation from tea plant (Shen and Fulthorpe, 2015).
In recent years, the antibiotics abuse has been resulted serious bacterial resistance, and become a heavy threat to public health. For instance, methicillin-resistant staphylococcal infections are an important cause of catheter-associated disease, and 75–90% among hospital isolates are S. epidermidis (Otto, 2009). Antibiotic resistance by Shigella species is also a global issue now (Lampl, 2015). E. coli and B. cereus are abundant in nature, and many factors make them a potential threat for the food industry (Gundogan and Avci, 2014). Endophytic actinobacteria are well-known producers of a vast array of secondary metabolites, including antibiotics. In our study, all of 44 isolates were screened for antimicrobial activities against S. epidermidis, Sh. flexneri, E. coli, and B. cereus. Ten isolates from Yunkang-10 exhibited antimicrobial activity against at least one of the tested pathogenic microorganisms, but none of the isolates from Zijuan showed obvious activity. The high inhibitory activity and broad antimicrobial spectrum of these tested strains suggested that the endophytic actinobacteria from tea cultivar Yunkang-10 are potential candidates for novel antimicrobial agents. For PKS-I, PKS-II, and NRPS screening, the antimicrobial activity results and the biosynthetic genes seemed to be positively correlated in isolates from Yunkang-10, but 3 isolates from Zijuan that showed no antimicrobial activity still provided positive amplification products for PKS-I or PKS-II. The 3 isolates from Zijuan that showed negative antimicrobial results in this study might produce the antimicrobial agents to other pathogenic microorganisms. The 6 isolates with a broad spectrum of antimicrobial activity and high inhibitory effects were then selected for potential immunomodulatory activities in in vivo test. The isolate Brevibacterium sp. YXT131, which have broad spectrum antimicrobial activity, gave positive amplification products with PKS-I, PKS-II, and NRPS primers, also exhibited high inhibitory effects of the serum levels of IL-12/IL-23 p40 and TNF-α. These results indicated that endophytic actinobacteria from Yunkang-10 might be an undeveloped bioresource library for active compounds.
Conclusion
In this study, we found that the endophytic actinobacterial communities in the tea cultivars Zijuan and Yunkang-10 were quite different. The isolates of Leucobacter sp. and Streptomyces sp. were endemic actinobacterial groups for the Zijuan cultivar, while Mycobacterium sp., Pseudarthrobacter sp., and Saccharomonospora sp. were endemic actinobacterial groups for Yunkang-10. Ten of the 28 isolates (35.7%) from Yunkang-10 exhibited activity against at least one of the tested pathogenic microorganisms, but none of the 16 isolates from Zijuan showed obvious antimicrobial activity. Brevibacterium sp. YXT131 and Brevibacterium sp. YKFG1221 from Yunkang-10 appeared to have broad-spectrum antimicrobial activity (against S. epidermidis, Sh. flexneri, and E. coli) and gave positive amplification products for the PKS-I, PKS-II, and NRPS genes. The crude extract from Brevibacterium sp. YXT131 showed no inhibition of ConA-induced splenocyte proliferation but decreased IL-12/IL-23 p40 and TNF-α levels in the serum of a mouse model, indicating that Brevibacterium sp. YXT131 had immunosuppressive activity. Endophytic actinobacteria from Yunkang-10 might be an undeveloped bioresource library for active compounds.
Author Contributions
JX, XW, and YZ conceived of and designed the experiments. WW, XW, FC, XY, and YL performed the experiments. WW drafted the manuscript. WW, YZ, JX, CW, and XC analyzed the data. JX and XW revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank LetPub for its linguistic assistance during the preparation of this manuscript.
Funding. This work was supported by grants from the Natural Science Foundation of Anhui Province (1608085QC57), the Natural Science Foundation of Zhejiang Province (LQ17C180001), and the Open Fund of State Key Laboratory of Tea Plant Biology and Utilization (SKLTOF20150102).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01304/full#supplementary-material
DNA gel electrophoresis of PKS-I, PKS-II, and NRPS. The PCR amplification products were resolved using electrophoresis in 1.5% agarose gels. M-Marker, 1-YFT113, 2-YXN120, 3-YXN111, 4-YXN112, 5-YXT131, 6-YKFG1221, 7-YKFG1121, 8-YKFG1122, 9-YKFT1130, 10-YKFH1122, 11-YKFG1112, 12-ZFY142, 13-ZFG130, 14-ZJFT1121.
References
- Annibaldi A., Meier P. (2018). Checkpoints in TNF-induced cell death: implications in inflammation and cancer. Trends Mol. Med. 24 49–65. 10.1016/j.molmed.2017.11.002 [DOI] [PubMed] [Google Scholar]
- Ayuso-Sacido A., Genilloud O. (2005). New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49 10–24. 10.1007/s00248-004-0249-6 [DOI] [PubMed] [Google Scholar]
- Baldan E., Nigris S., Populin F., Zottini M., Squartini A., Baldan B. (2014). Identification of culturable bacterial endophyte community isolated from tissues of Vitis vinifera “Glera”. Plant Biosyst. 148 508–516. 10.1080/11263504.2014.916364 [DOI] [Google Scholar]
- Begas E., Tsioutsiouliti A., Kouvaras E., Haroutounian S. A., Kasiotis K. M., Kouretas D., et al. (2017). Effects of peppermint tea consumption on the activities of CYP1A2, CYP2A6, Xanthine Oxidase, N-acetyltranferase-2 and UDP-glucuronosyltransferases-1A1/1A6 in healthy volunteers. Food Chem. Toxicol. 100 80–89. 10.1016/j.fct.2016.12.021 [DOI] [PubMed] [Google Scholar]
- Bulgari D., Casati P., Quaglino F., Bianco P. A. (2014). Endophytic bacterial community of grapevine leaves influenced by sampling date and phytoplasma infection process. BMC Microbiol. 14:198. 10.1186/1471-2180-14-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Qiu Z., You J., Tan H., Zhou S. (2005). Isolation and characterization of endophytic streptomycete antagonists of fusarium wilt pathogen from surface-sterilized banana roots. FEMS Microbiol. Lett. 247 147–152. 10.1016/j.femsle.2005.05.006 [DOI] [PubMed] [Google Scholar]
- Chattopadhyay C., Chakrabarti N., Chatterjee M., Mukherjee S., Sarkar K., Chaudhuri A. R. (2012). Black tea (Camellia sinensis) decoction shows immunomodulatory properties on an experimental animal model and in human peripheral mononuclear cells. Pharmacogn. Res. 4 15–21. 10.4103/0974-8490.91029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G., Liu H., Wei Q., Zhao H., Liu J., Yu Y. (2017). The acyl-activating enzyme PhAAE13 is an alternative enzymatic source of precursors for anthocyanin biosynthesis in petunia flowers. J. Exp. Bot. 68 457–467. 10.1093/jxb/erw426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cisowska A., Wojnicz D., Hendrich A. B. (2011). Anthocyanins as antimicrobial agents of natural plant origin. Nat. Prod. Commun. 6 149–156. [PubMed] [Google Scholar]
- Coombs J. T., Franco C. M. (2003). Isolation and identification of actinobacteria from surface-sterilized wheat roots. Appl. Environ. Microbiol. 69 5603–5608. 10.1128/AEM.69.9.5603-5608.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford D. L., Lynch J. M., Whipps J. M., Ousley M. A. (1993). Isolation and characterization of actinomycete antagonists of a fungal root pathogen. Appl. Environ. Microbiol. 59 3899–3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabroni S., Ballistreri G., Amenta M., Romeo F. V., Rapisarda P. (2016). Screening of the anthocyanin profile and in vitro pancreatic lipase inhibition by anthocyanin-containing extracts of fruits, vegetables, legumes and cereals. J. Sci. Food Agric. 96 4713–4723. 10.1002/jsfa.7708 [DOI] [PubMed] [Google Scholar]
- Felsenstein J. (1985). Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39 783–791. 10.1111/j.1558-5646.1985.tb00420.x [DOI] [PubMed] [Google Scholar]
- Filteau M., Lagacé L., LaPointe G., Roy D. (2010). Seasonal and regional diversity of maple sap microbiota revealed using community PCR fingerprinting and 16S rRNA gene clone libraries. Syst. Appl. Microbiol. 33 165–173. 10.1016/j.syapm.2010.02.003 [DOI] [PubMed] [Google Scholar]
- Gos F., Savi D. C., Shaaban K. A., Thorson J. S., Aluizio R., Possiede Y. M., et al. (2017). Antibacterial activity of endophytic actinomycetes isolated from the medicinal plant Vochysia divergens (Pantanal. Brazil). Front. Microbiol. 8:1642 10.3389/fmicb.2017.01642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundogan N., Avci E. (2014). Occurrence and antibiotic resistance of Escherichia coli, Staphylococcus aureus and Bacillus cereus in raw milk and dairy products in Turkey. Int. J. Dairy Technol. 67 562–569. 10.1111/1471-0307.12149 [DOI] [Google Scholar]
- Hayakawa M., Nonomura H. (1989). A new method for the intensive isolation of actinomycetes from soil. Actinomycetologica 3 95–104. 10.3209/saj.3_95 22901022 [DOI] [Google Scholar]
- Joshi R., Rana A., Gulati A. (2015). Studies on quality of orthodox teas made from anthocyanin-rich tea clones growing in Kangra valley, India. Food Chem. 176 357–366. 10.1016/j.foodchem.2014.12.067 [DOI] [PubMed] [Google Scholar]
- Kandel S. L., Firrincieli A., Joubert P. M., Okubara P. A., Leston N. D., McGeorge K. M., et al. (2017). An in vitro study of bio-control and plant growth promotion potential of Salicaceae endophytes. Front. Microbiol. 8:386. 10.3389/fmicb.2017.00386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan N., Mukhtar H. (2007). Tea polyphenols for health promotion. Life Sci. 81 519–533. 10.1016/j.lfs.2007.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980). A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16 111–120. 10.1007/BF01731581 [DOI] [PubMed] [Google Scholar]
- Lampl K. A. (2015). Antimicrobial resistance in Shigella species. Antimicrob. Food Saf. 7 119–135. 10.1016/B978-0-12-801214-7.00007-7 [DOI] [Google Scholar]
- Landwehr W., Wolf C., Wink J. (2016). Actinobacteria and myxobacteria-two of the most important bacterial resources for novel antibiotics. Curr. Top. Microbiol. Immunol. 398 273–302. 10.1007/82_2016_503 [DOI] [PubMed] [Google Scholar]
- Li W. J., Xu P., Schumann P., Zhang Y. Q., Pukall R., Xu L. H., et al. (2007). Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int. J. Syst. Evol. Microbiol. 57(Pt 7) 1424–1428. 10.1099/ijs.0.64749-0 [DOI] [PubMed] [Google Scholar]
- Ma G. Q., Xia Z. F., Wan C. X., Zhang Y., Luo X. X., Zhang L. L. (2017). Streptomyces kalpinensis sp. nov., an actinomycete isolated from a salt water beach. Int. J. Syst. Evol. Microbiol. 67 4892–4896. 10.1099/ijsem.0.002125 [DOI] [PubMed] [Google Scholar]
- Mearns-Spragg A., Bregu M., Boyd K. G., Burgess J. G. (1998). Cross-species induction and enhancement of antimicrobial activity produced by epibiotic bacteria from marine algae and invertebrates, after exposure to terrestrial bacteria. Lett. Appl. Microbiol. 27 142–146. 10.1046/j.1472-765X.1998.00416.x [DOI] [PubMed] [Google Scholar]
- Metsa-Ketela M., Salo V., Halo L., Hautala A., Hakala J., Mantsala P., et al. (1999). An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 180 1–6. 10.1016/S0378-1097(99)00453-X [DOI] [PubMed] [Google Scholar]
- Murphy K. M., Stockinger B. (2010). Effector T cell plasticity: flexibility in the face of changing circumstances. Nat. Immunol. 11 674–680. 10.1038/ni.1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto M. (2009). Staphylococcus epidermidis — the “accidental” pathogen. Nat. Rev. Microbiol. 7 555–567. 10.1038/nrmicro2182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson I. A., Persson K., Hagg S., Andersson R. G. (2010). Effects of green tea, black tea and Rooibos tea on angiotensin-converting enzyme and nitric oxide in healthy volunteers. Public Health Nutr. 13 730–737. 10.1017/S1368980010000170 [DOI] [PubMed] [Google Scholar]
- Pirttilä A. M., Pospiech H., Laukkanen H., Myllylä R., Hohtola A. (2005). Seasonal variations in location and population structure of endophytes in buds of Scots pine. Tree Physiol. 25 289–297. 10.1093/treephys/25.3.289 [DOI] [PubMed] [Google Scholar]
- Qin S., Feng W. W., Wang T. T., Ding P., Xing K., Jiang J. H. (2017). Plant growth-promoting effect and genomic analysis of the beneficial endophyte Streptomyces sp. KLBMP 5084 isolated from halophyte Limonium sinense. Plant Soil 416 117–132. 10.1007/s11104-017-3192-2 [DOI] [Google Scholar]
- Qin S., Li J., Chen H. H., Zhao G. Z., Zhu W. Y., Jiang C. L., et al. (2009). Isolation, diversity, and antimicrobial activity of rare actinobacteria from medicinal plants of tropical rain forests in Xishuangbanna, China. Appl. Environ. Microbiol. 75 6176–6186. 10.1128/AEM.01034-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S., Xing K., Jiang J. H., Xu L. H., Li W. J. (2011). Biodiversity, bioactive natural products and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89 457–473. 10.1007/s00253-010-2923-6 [DOI] [PubMed] [Google Scholar]
- Rabha A. J., Naglot A., Sharma G. D., Gogoi H. K., Veer V. (2014). In vitro evaluation of antagonism of endophytic colletotrichum gloeosporioides against potent fungal pathogens of Camellia sinensis. Indian J. Microbiol. 54 302–309. 10.1007/s12088-014-0458-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehacek Z. (1959). Isolation of actinomycetes and determination of the number of their spores in soil. Microbiol. USSR 28 220–225. [Google Scholar]
- Saini A., Aggarwal N. K., Sharma A., Yadav A. (2015). Actinomycetes: a source of lignocellulolytic enzymes. Enzyme Res. 2015:279381. 10.1155/2015/279381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4 406–425. 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Salcedo R. G., Olano C., Gomez C., Fernandez R., Brana A. F., Mendez C., et al. (2016). Characterization and engineering of the biosynthesis gene cluster for antitumor macrolides PM100117 and PM100118 from a marine actinobacteria: generation of a novel improved derivative. Microb. Cell Fact. 15:44. 10.1186/s12934-016-0443-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., Orozco-Mosqueda Mdel C., Glick B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183 92–99. 10.1016/j.micres.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Sardi P., Saracchi M., Quaroni S., Petrolini B., Borgonovi G. E., Merli S. (1992). Isolation of endophytic streptomyces strains from surface-sterilized roots. Appl. Environ. Microbiol. 58 2691–2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessitsch A., Reiter B., Berg G. (2004). Endophytic bacterial communities of field-grown potato plants and their plant-growth-promoting and antagonistic abilities. Can. J. Microbiol. 50 239–249. 10.1139/w03-118 [DOI] [PubMed] [Google Scholar]
- Shen S. Y., Fulthorpe R. (2015). Seasonal variation of bacterial endophytes in urban trees. Front. Microbiol. 6:427. 10.3389/fmicb.2015.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Takahashi Y., Omura S. (2003). Isolation of new actinomycete strains for the screening of new bioactive compounds. J. Gen. Appl. Microbiol 49 141–154. 10.2323/jgam.49.141 [DOI] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 302725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. D., Gibson T. J., Plewniak F., Jeanmougin F., Higgins D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25 4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., Cao L., Tan H., Han W., Chen M., Liu Y., et al. (2007). Diversity of cultivated and uncultivated actinobacterial endophytes in the stems and roots of rice. Microb. Ecol. 53 700–707. 10.1007/s00248-006-9163-4 [DOI] [PubMed] [Google Scholar]
- Wang W., Fu X. W., Dai X. L., Hua F., Chu G. X., Chu M. J., et al. (2017). Novel acetylcholinesterase inhibitors from Zijuan tea and biosynthetic pathway of caffeoylated catechin in tea plant. Food Chem. 237 1172–1178. 10.1016/j.foodchem.2017.06.011 [DOI] [PubMed] [Google Scholar]
- Wink M., Ashour M. L., El-Readi M. Z. (2012). Secondary metabolites from plants inhibiting ABC transporters and reversing resistance of cancer cells and microbes to cytotoxic and antimicrobial agents. Front. Microbiol. 3:130. 10.3389/fmicb.2012.00130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. S., Wang X., Lu G., Picinich S. C. (2009). Cancer prevention by tea: animal studies, molecular mechanisms and human relevance. Nat. Rev. Cancer 9 429–439. 10.1038/nrc2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X. R., Bao Y. X., Huang M. (2009). The botanical and quality characteristics of the tea cultivar “Zi-Juan” in Yunnan province. J. Tea 35 17–18. [Google Scholar]
- Yang Y., Yu L., Komaki H., Oku N., Igarashi Y. (2016). Absolute configuration of NFAT-133, an aromatic polyketide with immunosuppressive and antidiabetic activity from actinomycetes. J. Antibiot. 69 69–71. 10.1038/ja.2015.80 [DOI] [PubMed] [Google Scholar]
- Zhu M., Li N., Zhao M., Yu W., Wu J. L. (2017). Metabolomic profiling delineate taste qualities of tea leaf pubescence. Food Res. Int. 94 36–44. 10.1016/j.foodres.2017.01.026 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA gel electrophoresis of PKS-I, PKS-II, and NRPS. The PCR amplification products were resolved using electrophoresis in 1.5% agarose gels. M-Marker, 1-YFT113, 2-YXN120, 3-YXN111, 4-YXN112, 5-YXT131, 6-YKFG1221, 7-YKFG1121, 8-YKFG1122, 9-YKFT1130, 10-YKFH1122, 11-YKFG1112, 12-ZFY142, 13-ZFG130, 14-ZJFT1121.