Abstract
Background
Despite advancements in the treatment of primary and metastatic breast cancer, many patients lack a durable response to these treatments. Patients with triple-negative breast cancer (TNBC) and human epidermal growth factor receptor 2(HER2)-positive breast cancer who do not have a pathological complete response (pCR) after neoadjuvant chemotherapy (NACT) have a very poor prognosis. Tumor-infiltrating lymphocytes (TILs) have been identified as a predictive marker for pCR after NACT in TNBC and HER2-positive breast cancer. These patient populations could also be suitable for novel treatment strategies including neoepitope-based therapies. This work analyses the effect of TILs on the pCR in neoadjuvantly treated patients in the TILGen study and presents the procedures aimed at establishing neoepitope-based therapies in this study.
Methods
Neoadjuvantly treated HER2-positive and TNBC patients were eligible for the presented analysis concerning the association between TILs and pCR. A total of 146 patients could be identified within the TILGen study. TILs were evaluated as percentage of stromal tumor tissue in core biopsies at primary diagnosis. The phenotype ‘lymphocyte-predominant breast cancer’ (LPBC) was associated with pCR by logistic regression adjusted for estrogen receptor status, progesterone receptor status, HER2 status, age at diagnosis, and grading.
Results
LPBC was seen in 24 (16.4%) patients. In this patient group, 66.7% achieved a pCR, while the pCR rate was 32.8% in patients with a low TIL count. The adjusted odds ratio was 6.60 (95% confidence interval 2.02–21.56; p < 0.01).
Conclusion
TILs are a strong predictor of pCR in TNBC and HER2-positive breast cancer patients. Implications for the use of this information including the effect on prognosis might help to identify patients most likely to benefit from a neoepitope-based therapy approach.
Keywords: Tumor-infiltrating lymphocytes, Response, Neoadjuvant chemotherapy, TILGen study
Introduction
Tumor microenvironment and immune cell interactions play a major role in breast cancer progression and therapy response [1]. Several studies have demonstrated an effector cell activation towards inflammation which is mediated by an antitumor response of the host [2].
However, the majority of breast cancer cells evade immune cell recognition by several immune escape mechanisms such as increased expression of immune checkpoint inhibitors, decreased major histocompatibility complex (MHC) class I expression or recruitment of tumor-associated regulatory cells, e.g., regulatory T cells (Tregs) [3, 4]. The tumor immune microenvironment is therefore crucial for both tumor progression and regression [5]. Immunogenicity varies between the 4 intrinsic molecular subtypes of breast cancer [6], which are represented in clinical routine by the hormone receptor(HR)-positive luminal A- and B-like subtypes (estrogen receptor(ER)- or progesterone receptor(PR)-positive immunohistochemistry (IHC)), human epidermal growth factor receptor 2(HER2)-positive breast cancers, and triple-negative breast cancers (TNBC). TNBC and HER2-positive subtypes show an elevated immunogenicity, indicated by a higher density of tumor-infiltrating lymphocytes (TILs) in the tumor environment [7]. Furthermore, the presence of TILs in pretreated TNBC and HER2-positive breast cancers is associated with improved overall and disease-free survival [8]. Additionally, high amounts of TILs are associated with pathological complete response (pCR) after neoadjuvant chemotherapy (NACT), regardless of the intrinsic breast cancer subtype [9, 10, 11]. In order to further facilitate this immunogenic potential and to minimize the systemic burdens of chemotherapies, targeted immunotherapies have been the focus of current research programs [12]. Immune checkpoint inhibitors targeting the programmed cell death 1 receptor (PD-1), its ligands (PD-L1 and PD-L2), the cytotoxic T-lymphocyte antigen-4 (CTLA-4), or indoleamine-pyrrole-2,3-dioxygenase (IDO) are Food and Drug Administration(FDA)-approved drugs for various cancer types (e.g., breast and lung cancer or melanoma) [12, 13, 14, 15, 16]. PD-1 and PD-L1 are physiologically expressed on immunocompetent cells including T cells and are overexpressed on TILs and breast cancer cells [17]. Breast cancer patients with PD-1-positive TILs or PD-L1-positive tumor cells have a worse clinical outcome [18, 19, 20]. PD-1 inhibitors like pembrolizumab or nivolumab block the PD-1 pathway and thus are able to reactivate the antitumor response within the breast cancer microenvironment [21, 22, 23]. Immune-targeted treatment using such immune checkpoint inhibitors relies on the overexpression of the target genes in malignant cells or immune cells, but can still affect healthy cells [24]. In order to reduce these side effects on healthy cells, a specific cancer cell-based immunotherapeutic approach would be necessary. Therefore, the identification of suitable molecular targets expressed solely by the tumor cells has to be of high priority [25]. Tumor-specific antigens (TSAs) or so-called neoepitopes seem to be an attractive target appropriate for cancer-specific therapy [26]. TSAs/neoepitopes mainly result from nonsynonymous somatic mutations. Furthermore, immunocompetent cells are able to recognize TSAs/neoepitopes as being foreign [27]. It was shown that TILs, especially cytotoxic T cells, recognize TSAs/neoepitopes in breast cancer and are able to induce antitumor responses in mice [28]. However, the knowledge about somatic mutations causing tumor regression is limited [29] and might be unique for each patient.
The aim of the presented work was to obtain first data from the iMODE-B/TILGen study by exploring the association between TILs from breast cancer core biopsies and pCR in order to confirm TILs as a pCR marker, and to describe the further study aims of the TILGen study.
Patients and Methods
Patient Selection
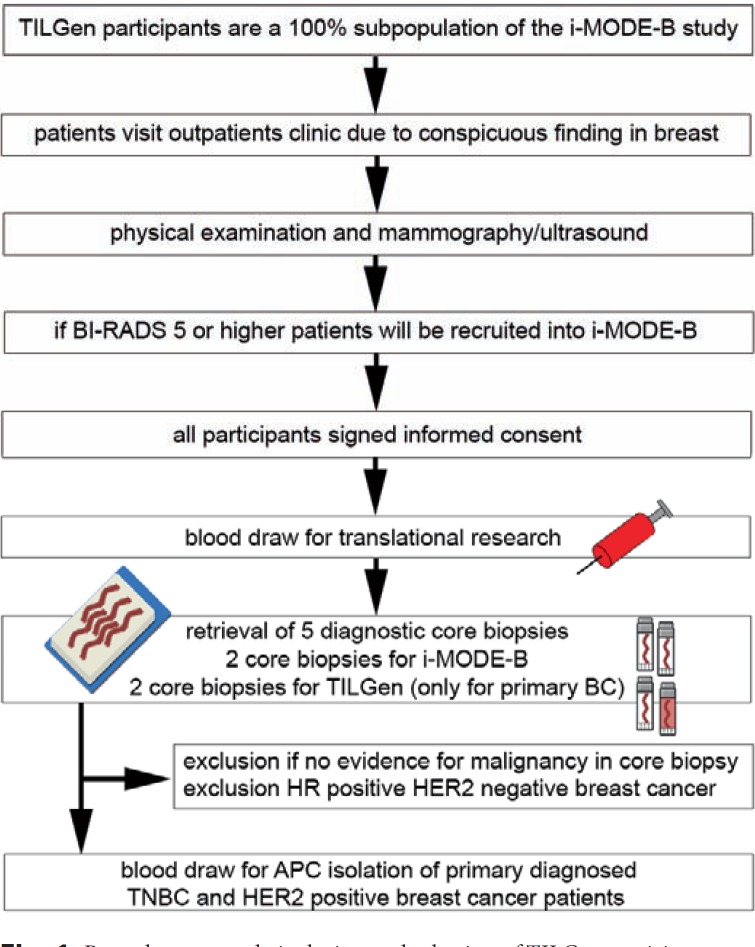
TILGen (TILs and genomics) is a predefined substudy of the iMODE-B (imaging and molecular detection of breast cancer) study. iMODE-B is concerned with molecular markers at the time of breast cancer diagnosis or progression, molecular detection, and imaging detection of breast cancer. The TILGen study focuses on the identification of antigen-specific TILs in TNBC and HER2-positive breast cancer patients in order to identify immunogenic targets that could help to improve cancer immunotherapy. A description of the full procedures of TILGen study inclusion is given in figure 1.
Fig. 1.
Procedures at study inclusion and selection of TILGen participants.
This analysis focuses on the association between TILs at primary diagnosis of breast cancer and pCR after NACT. Patients were eligible for inclusion in the iMODE-B study if an indication existed for a diagnostic biopsy because of a suspicious breast lesion. From May 2015 to April 2017, a total of 587 participants were recruited into the iMODE-B study in an outpatient clinic setting. Patients were excluded in the following hierarchical order: 49 patients were never biopsied at our study site, 169 were not diagnosed with breast cancer (controls as per iMODE-B protocol), and 31 patients were biopsied not with primary breast cancer. Therefore, the iMODE-B study population comprised 338 patients diagnosed with breast cancer at primary diagnosis. Of those, 192 patients were excluded because they were not treated with NACT. Thus, the final study population consisted of 146 patients treated with NACT within the TILGen study (fig. 2).
Fig. 2.

Patient selection table of the iMODE-B study for tumor-infiltrating lymphocyte (TIL) analysis.
All patients gave written informed consent, and the study was approved by the ethics committee of the University of Erlangen-Nuremberg (no. 4514).
Clinical Data
Patient data was collected prospectively, as required by the certification process of the German Cancer Society and by the German Society for the Study of Breast Diseases (Deutsche Gesellschaft für Senologie) [30]. A web-based database was used for documentation as previously described [31, 32]. Accordingly, each breast cancer case is prospectively documented, including patient and tumor characteristics, detailed treatment data, and epidemiological data. Follow-up treatments and disease characteristics were collected for up to 10 years after the primary diagnosis. All histological tumor data, such as tumor size, axillary lymph node status, grading, ER status, PR status, and HER2 status were documented. As part of the continuous certification process, the quality of the data was audited annually. Data obtained from the above-described collection and audit processes were used in the analyses presented here.
Histopathological Assessment
All of the histopathological parameters used in this study were directly documented from the original pathology reports after being reviewed by 2 investigators. IHC staining was performed with an automated staining module with supplied antibodies against ER, PR, HER2, and Ki67 according to manufacturer's instruction manual (BenchMark ULTRA IHC/ISH Staining Module, Ventana Medical Systems, Tucson, AZ, USA). ER, PR, and HER2 status as well as grading were determined on pre-therapeutic formalin-fixed paraffin-embedded (FFPE) breast cancer core biopsies. ER and PR protein expression was determined via IHC. Positive staining for ER and PR was defined as ≥1% [33]. HER2 was assessed with IHC for protein expression, and, if necessary, reflex testing with chromogenic in-situ hybridization (CISH) was conducted to proof HER2 positivity. HER2 gene amplification was determined by staining of the centromeres of the corresponding chromosome 17 with a staining kit for HER2 (ZytoDot, 2C SPEC HER2/CEN17, Zyto Vision Ltd., Bremerhaven, Germany). HER2 positivity was defined as IHC score 3+ alone or 2+ with positive HER2 CISH [34]. Proliferation was assessed with the proliferation marker Ki67. The cut-off for Ki67 was defined as >20% [35]. Grading before NACT was determined according to Elston and Ellis [36]. Patients with a ypT0 ypN0 assessment after NACT were considered to have pCR according to Sinn et al. [37].
Evaluation of TILs in Core Biopsies
Assessment of TILs followed the recommendations of the International TILs Working Group 2014, which are elaborately described by Salgado et al. [38]. For each case, we used 1 hematoxylin/eosin slide of FFPE preoperative breast core biopsy material. After reviewing the tumor cells, the tumor area and the stromal compartment of the tumor were defined. All mononuclear cells, but no polynuclear leukocytes, were evaluated as percentage per stromal tumor tissue area and only within tumor borders of the invasive carcinoma. Necrosis, regressive fibrosis/hyalinization, and inflammation around intraductal tumor components and normal lobules were excluded from the scoring area. The percentage of stromal TILs was averaged over the complete stromal tumor area and given as continuous variable but without any decimal place [38]. Lymphocyte-predominant breast cancer (LPBC) was defined as >50% stromal TILs versus non-LPBC (≤50% stromal TILs) according to Hendry et al. [39] and Swisher et al. [40].
Statistical Analysis
The influence of the amount of TILs (LPBC vs. non-LPBC) on pCR was investigated using logistic regression analyses. A simple logistic regression analysis with pCR as outcome and the amount of TILs as predictor was performed in order to obtain an unadjusted odds ratio (OR) with 95% confidence interval (CI) for LPBC versus non-LPBC. An adjusted OR for the amount of TILs was calculated using a multiple logistic regression analysis with the amount of TILs as predictor and additionally well-known predictors of pCR: age at diagnosis (continuous), clinical staging (cT, cT1 vs. cT2-cT4), grading (G1 and G2 vs. G3), ER (positive vs. negative), PR (positive vs. negative), and HER2 (positive vs. negative).
All tests were 2-sided, and a p value of <0.05 was regarded as statistically significant. Calculations were carried out using the R system for statistical computing (version 3.0.1).
Results
Patient Characteristics
The patient population for this analysis comprised 146 neoadjuvantly treated breast cancer patients, of which 56 (38.4%) achieved pCR. Patient characteristics according to pCR status are shown in table 1. The average age was 54.9 ± 13.1 years in the non-pCR and 52.9 ± 12.6 years in the pCR group. Known predictors of pCR were associated with it in the expected way. In ER-positive patients, 25.3% achieved a pCR, whereas 60% of patients with ER-negative breast cancer had pCR. PR-negative breast cancer patients responded to NACT with pCR in 54.1% of cases; for PR positivity, only 16.4% were responders. In patients positive for HER2, 58% achieved pCR. In patients with HER2-negative breast cancer, the pCR rate was 28.1%. All other distributions are shown in table 1.
Table 1.
Patient and tumor characteristics by pathological complete response (pCR) status
| Characteristic | No pCR | pCR |
|---|---|---|
| Patients (n = 146), n (%) | 90 (61.6) | 56 (38.4) |
| Age, mean ± SD, years | 54.9 ± 13.1 | 52.9 ± 12.6 |
| ER, n (%) | ||
| Negative (n = 55) | 22 (40) | 33 (60) |
| Positive (n = 91) | 68 (74.7) | 23 (25.3) |
| PR, n (%) | ||
| Negative (n = 85) | 39 (45.9) | 46 (54.1) |
| Positive (n = 61) | 51 (83.6) | 10 (16.4) |
| HER2, n (%) | ||
| Negative (n = 96) | 69 (71.9) | 27 (28.1) |
| Positive (n = 50) | 21 (42) | 29 (58) |
| cT, n (%) | ||
| cT1 (n = 56) | 34 (60.7) | 22 (39.3) |
| cT2–cT4 (n = 90) | 56 (62.2) | 34 (37.8) |
| Grading, n (%) | ||
| G1–G2 (n = 41) | 32 (78.1) | 9 (21.9) |
| G3 (n = 105) | 58 (55.3) | 47 (44.7) |
| LPBC, n (%) | ||
| 0–50% (n = 122) | 82 (67.2) | 40 (32.8) |
| 51–100% (n = 24) | 8 (33.3) | 16 (66.7) |
SD = Standard deviation; ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2; cT = clinical staging; LPBC = lymphocyte-predominant breast cancer.
TILs in Core Biopsies
A total of 24 (16.4%) samples had an LPBC phenotype. Patient characteristics according to LPBC status are shown in table 2. Higher LPBC rates were found in HR-negative samples (ER 27.3%, PR 22.3%) compared to HR-positive tumors (ER 9.9%, PR 8.2%). 14% of HER2-positive and 17.7% of HER2-negative tumors could be grouped into LPBC. Interestingly, no cT1 tumor showed an LPBC phenotype, while 36.7% of all tumors staged cT2 or higher did. Finally 20% of tumors with grading 3 compared to 7.3% with grading 1 and 2 were in the LPBC group.
Table 2.
Correlation of lymphocyte-predominant breast cancer (LPBC) with patient and tumor characteristics
| Characteristic | Non-LPBC | LPBC |
|---|---|---|
| Patients (n = 146), n (%) | 122 (83.6) | 24 (16.4) |
| Age, mean ± SD, years | 54.1 ± 12.8 | 54.3 ± 13.9 |
| ER, n (%) | ||
| Negative (n = 55) | 40 (72.2) | 15 (27.3) |
| Positive (n = 91) | 82 (90.1) | 9 (9.9) |
| PR, n (%) | ||
| Negative (n = 85) | 66 (77.6) | 19 (22.3) |
| Positive (n = 61) | 56 (91.8) | 5 (8.2) |
| HER2, n (%) | ||
| Negative (n = 96) | 79 (82.3) | 17 (17.7) |
| Positive (n = 50) | 43 (86) | 7 (14) |
| cT, n (%) | ||
| cT1 (n = 56) | 56 (100) | 0 (0) |
| cT2–cT4 (n = 90) | 66 (73.3) | 24 (36.7) |
| Grading, n (%) | ||
| G1–G2 (n = 41) | 38 (92.7) | 3 (7.3) |
| G3 (n = 105) | 84 (80) | 21 (20) |
SD = Standard deviation; ER = estrogen receptor; PR = progesterone receptor; HER2 = human epidermal growth factor receptor 2; cT = clinical staging.
pCR Rates with Regard to LPBC
A total of 66.7% of the patients with LPBC showed a pCR, while pCR in non-LPBC patients was 32.8% (fig. 3). Logistic regression analysis indicated that the amount of TILs was independently associated with pCR. The likelihood of a pCR was higher in LPBC patients than in non-LPBC patients. The unadjusted OR was 4.1 (95% CI 1.6–10.4; p < 0.01) and the adjusted OR was 6.6 (95% CI 2.0–21.6; p < 0.01).
Fig. 3.
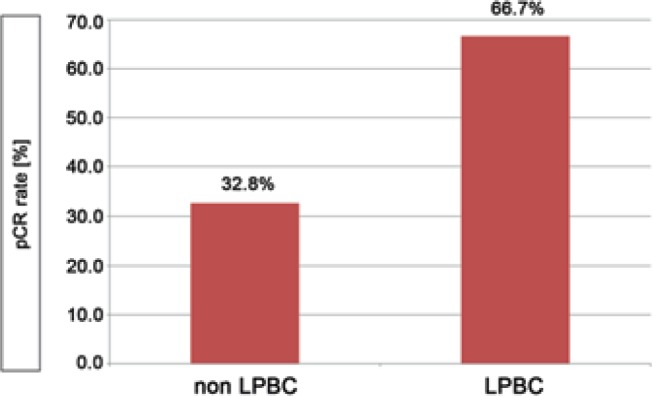
Pathological complete response (pCR) rates in non-lymphocyte-predominant breast cancer (non-LPBC) versus LPBC. pCR after neoadjuvant chemotherapy was defined according to Sinn et al. [37].
Discussion
In this investigation, we found a higher pCR rate in patients with tumors that had an LPBC phenotype (pCR rate 66.7%). In patients without an LPBC phenotype, the pCR rate was 32.8%. This effect was independent of ER, PR, HER2, grading, age at diagnosis, and clinical tumor staging with an OR of 6.6 (95% CI 2.0–21.6).
Our findings are in concordance with the data from the GeparSixto trial in which a high amount of TILs (LPBC: stromal TILs ≥60%) was associated with pCR independent of the tumor subtype (pCR rate: LPBC 59.9% vs. non-LPBC 33.8%). In addition, Dieci et al. [41] also reported pCR rates of 64.7% in LPBC (defined as ≥60% stromal TILs) compared to non-LPBCs with pCR rates of 25%. Our results further underline the use of stromal TILs as a predictive marker for NACT of primary breast cancer. Other studies confirmed that TILs were a strong predictor of pCR in HER2-positive breast cancer and TNBC and to a lesser degree in HR-positive breast cancers [42]. For example, in a meta-analysis, TILs in neoadjuvantly treated HER2-positive breast cancer were analyzed [43]. The OR for pCR was 2.46–2.88 depending on the treatment given [43]. In 2 German trials, LPBC phenotype (≥60% TILs) was predictive for pCR with an adjusted OR of 1.87 [44]. These ORs were lower than the OR in our total cohort (OR 6.6). However, in our cohort, only about one third of the cases were HER2-positive. We did not perform an analysis according to molecular subgroup because the sample size would have been too low to make robust statements. However, in concordance with the above-mentioned studies, we were able to observe that the majority of analyzed tumor samples were HR-negative or HER2-positive with regard to pCR and LPBC phenotype (table 2).
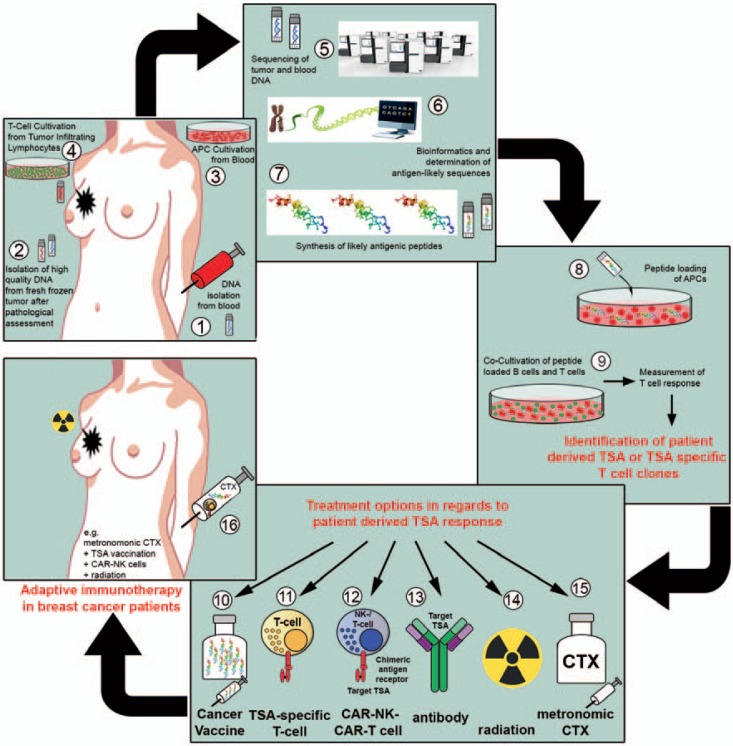
The biomaterials and data concerning therapy response and prognosis of the TILGen study will be used as a resource in the future to identify immunotherapy-related targets or pathways that might help with cancer therapies for individual patients, such as TSAs/neoepitopes [45]. The vision of the TILGen study is therefore to identify TSAs/neoepitopes which could be drivers of immunotherapies for these patients. An outline of how this could be achieved is shown in fig. 4.
Fig. 4.
Objective and strategy of the TILGen study and therapy concept. The overall objective of the TILGen study is the identification of tumor-specific antigens (TSAs)/neoepitopes as well as TSA-specific T cells, and the implementation of TSA/neoepitope-specific targeted therapy. The workflow with each working step is illustrated in numbers (1–16): 1) blood sample collection for isolation of germline DNA; 2) collection of fresh frozen breast cancer core biopsies for isolation of tumor DNA and RNA; 3) isolation and cultivation of antigen-presenting cells (APCs); 4) T-cell expansion out of tumor-infiltrating lymphocytes (TILs) from core biopsy; 5) whole genome sequencing; 6) determination of likely antigenic sequences by comparing germline and tumor DNA; 7) synthesis of likely antigenic peptides; 8) loading of antigenic peptides on APCs; 9) co-cultivation of peptide-loaded APCs and isolated tumor T cells to measure T cell response and identify TSAs and TSA-specific T cell clones; 10) production of cancer vaccine with regard to TSA; 11) expansion of TSA-specific T cells; 12) production of chimeric antigen receptor (CAR) natural killer cells (NK) or CAR T cells; 13) production of TSA-specific humanized antibodies; 14) low-dose radiation therapy; 15) metronomic chemotherapy; 16) treatment option with e.g. metronomic chemotherapy in combination with TSA vaccination, TSA-specific T cells, and radiation.
Dramatic improvements in the quality and speed of genomic sequencing and analysis as a clinical diagnostic tool for individual patients combined with the innovations propelling immuno-oncology are paving the way for a new era of truly personalized cancer treatment. At the heart of this new hope is the newfound ability to rapidly identify and target tumor cells with specific DNA mutations unique to each cancer patient. The products of mutated genes encoding altered proteins are so-called ‘neoepitopes’ and serve as a molecular address to direct and redirect immune cells in the killing of cancer cells and to procure long-term immunity. Neoepitopes are defined as unique mutations found specifically in a patient's tumor (not normal tissue) that can be targeted by the immune system to attack the tumor with minimal off-target toxicity.
Identifying neoepitopes for each patient is made possible by high-throughput whole genome or exome sequencing and by the direct comparison of abnormal tumor DNA with the patient's own normal DNA. The former widens the search for drugable targets (neoepitopes) among the >99% of genome deemed untargetable or unimportant by panel sequencing. The latter reduces the significantly high false-positive error rates associated with tumor-only sequencing techniques [46]. Precision in individualizing treatments targeting neoepitopes further requires confirmation of the expression of mutated genes, thus avoiding another potential pitfall of false-positive errors and the potential for the altered protein to induce immunogenicity. As an example, an expressed neoepitope, once confirmed to be presented to immune effector cells such as cytotoxic T cells by antigen-presenting cells, is a molecular address that may be delivered by an immunogenic vehicle, much like vaccination against a pathogen. One such vehicle is the adenovirus which may be engineered to express within its DNA many neoepitopes, and upon injection can locally infect dendritic cells which then present an identified neoepitope to the immune effector cells. Despite great promise, the use of adenovirus or other foreign delivery vehicles remains hindered due to the pre-existence or the induction of neutralizing antibodies against them in the patient's immune system.
This limitation has been overcome e.g. by engineered adenoviruses, which are capable of safely vaccinating and re-vaccinating against hundreds of neoepitopes and tumor-associated antigens despite pre-existing immunity against adenovirus [47]. Remarkable results have thus far been published demonstrating the delivery of tumor-associated antigens by this engineered adenovirus in a cohort of late-stage colorectal cancer patients [48].
The use of neoepitopes to guide treatment personalization is a mere extension of the in-practice use of advanced genomics and quantitative proteomics to identify and calculate the expression levels of TSAs. For example, sequencing genomic amplification in and quantifying proteomic alteration of the tumor-associated antigen HER2 has added significant precision to the use of HER2-targeting therapies such as trastuzumab [49]. Conversely, a TSA may initially be discovered as a novel expression antigen but reclassified due to its existence in large subsets of cancer patients. One such TSA is brachyury, a developmental protein found to be abnormally expressed during the epithelial-mesenchymal transition in chordoma and other cancers [50]. Using a heat-killed yeast platform designed to evade immune neutralization while inducing an immunogenic response against its payload, a phase I trial of yeast-brachyury resulted in specific T-cell immune responses in the majority of the trial's 34 patients despite heavy pretreatment and metastatic disease, and stable disease in 2 chordoma patients and 1 colorectal cancer patient [51]. A randomized phase II study using this platform is ongoing for chordoma patients. Similarly, the yeast platform has been used to engender T-cell responses against mutant KRAS in cancer patients, which despite their common appearance in multiple cancers can be thought of as neoepitopes given that a KRAS mutation is the result of a single nucleotide change in the KRAS gene which results in the coding of a novel (non-self) protein [52].
One of the challenges with neoepitope discovery and targeting will be the management of Big Data: teraflops of compute resources in a cloud environment to generate terabytes of sequencing data, including whole genome and/or whole exome sequencing, RNA sequencing, and molecular modeling of immune presentation of neoepitopes [53]. To meet the demands of heterogeneity, analysis and long-term storage of data from multiple biopsies for each patient is a further challenge. These activities require compute and storage under HIPAA (Health Insurance Portability and Accountability Act), and high-speed and large-bandwidth connectivity for rapidly transiting sequence data from sequencing labs to supercompute/cloud environments such that derivation and delivery of neoepitope targeting platforms is enabled in actionable time for each patient. These challenges require significant infrastructure and resources, and are overcome in private, dedicated Big Data supercompute clouds interconnected by dedicated fiber infrastructure capable of transporting terabytes of data at terabits per second. The latter are implemented for the benefit of financial trading markets, and now have been retrofitted to meet the needs of sequencing analysis and neoepitope discovery.
These perspectives are the background to further developing the TILGen study into a feeder mechanism for immunotherapeutic trials that explore the usage of vaccines, chimeric antigen receptor (CAR) T cells, CAR natural killer cells, and other targeted therapeutics as part of personalized medicine, which might mean the development of therapeutics on a n = 1 basis.
In summary, the confirmation of TILs as an independent marker for pCR is a first promising result from the iMODE-B/TILGen study. As patients without a pCR have a very unfavorable prognosis, this population may benefit from an improved immune response to the tumor and from a neoepitope-based therapy approach. To what extent information about TILs and chemotherapy response can be used to predict response to neoepitope-based therapies remains to be investigated. Also patients in the advanced therapeutic setting who have failed to respond to primary breast cancer treatment could be candidates for such a therapy. Which combination therapies (low-dose chemotherapy and radiation, checkpoint inhibition, other immune-supportive therapies) are best to combine with such a treatment will also be the aim of future research.
Disclosure Statement
PAF has received honoraria from Amgen, Celgene, Roche, Novartis, Pfizer, and Novartis. MPL received honoraria from Pfizer, Roche, MSD, Hexal, Novartis, Lilly, AstraZeneca, TEVA, Celgene, Eisai, and medac. SR is an employee of NantOmics LLC. NN received consultancy honoraria from Janssen-Cilag and travel support from Novartis. All other authors declare that they have no conflict of interest.
Acknowledgement
The authors want to thank all patients who participated in this study as well as coworkers Judith Bausenwein, Edith van der Meijden, Sonja Oeser, and Silke Landrith. This study was funded in part (AM, AK, and PAF) by the IZKF program of the University Hospital Erlangen as well as the Emerging Fields Initiative BIG-THERA (DD, MWB, PAF).
References
- 1.Jiang X, Shapiro DJ. The immune system and inflammation in breast cancer. Mol Cell Endocrinol. 2014;382:673–682. doi: 10.1016/j.mce.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Finn OJ. Immuno-oncology: understanding the function and dysfunction of the immune system in cancer. Ann Oncol. 2012;23((suppl 8)):viii6–9. doi: 10.1093/annonc/mds256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domschke C, Schneeweiss A, Stefanovic S, et al. Cellular immune responses and immune escape mechanisms in breast cancer: determinants of immunotherapy. Breast Care (Basel) 2016;11:102–107. doi: 10.1159/000446061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang M, Zhang C, Song Y, et al. Mechanism of immune evasion in breast cancer. Onco Targets Ther. 2017;10:1561–1573. doi: 10.2147/OTT.S126424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeNardo DG, Coussens LM. Inflammation and breast cancer. Balancing immune response: crosstalk between adaptive and innate immune cells during breast cancer progression. Breast Cancer Res. 2007;9:212. doi: 10.1186/bcr1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denkert C. The immunogenicity of breast cancer - molecular subtypes matter. Ann Oncol. 2014;25:1453–1455. doi: 10.1093/annonc/mdu235. [DOI] [PubMed] [Google Scholar]
- 7.Adams S, Gray RJ, Demaria S, et al. Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol. 2014;32:2959–2966. doi: 10.1200/JCO.2013.55.0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams S, Goldstein LJ, Sparano JA, et al. Tumor infiltrating lymphocytes (TILs) improve prognosis in patients with triple negative breast cancer (TNBC) Oncoimmunology. 2015;4:e985930. doi: 10.4161/2162402X.2014.985930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denkert C, Loibl S, Noske A, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi R, Tanaka M, Yano A, et al. Tumor-infiltrating lymphocytes are important pathologic predictors for neoadjuvant chemotherapy in patients with breast cancer. Hum Pathol. 2012;43:1688–1694. doi: 10.1016/j.humpath.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 11.Denkert C, von Minckwitz G, Darb-Esfahani S, et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: a pooled analysis of 3,771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018;19:40–50. doi: 10.1016/S1470-2045(17)30904-X. [DOI] [PubMed] [Google Scholar]
- 12.Migali C, Milano M, Trapani D, et al. Strategies to modulate the immune system in breast cancer: checkpoint inhibitors and beyond. Ther Adv Med Oncol. 2016;8:360–374. doi: 10.1177/1758834016658423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dempke WCM, Fenchel K, Uciechowski P, Dale SP. Second- and third-generation drugs for immuno-oncology treatment - the more the better? Eur J Cancer. 2017;74((suppl C)):55–72. doi: 10.1016/j.ejca.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Tomioka N, Azuma M, Ikarashi M, et al. The therapeutic candidate for immune checkpoint inhibitors elucidated by the status of tumor-infiltrating lymphocytes (TILs) and programmed death ligand 1 (PD-L1) expression in triple negative breast cancer (TNBC) Breast Cancer. 2018;25:34–42. doi: 10.1007/s12282-017-0781-0. [DOI] [PubMed] [Google Scholar]
- 15.Bi WW, Zhang WH, Yin GH, et al. Analysis of indoleamine 2–3 dioxygenase (IDO) and EGFR co-expression in breast cancer tissue by immunohistochemistry. Asian Pac J Cancer Prev. 2014;15:5535–5538. doi: 10.7314/apjcp.2014.15.14.5535. [DOI] [PubMed] [Google Scholar]
- 16.Lux MP, Janni W, Hartkopf AD, et al. Update breast cancer 2017 - implementation of novel therapies. Geburtshilfe Frauenheilkd. 2017;77:1281–1290. doi: 10.1055/s-0043-122885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dong H, Zhu G, Tamada K, Chen L. B7-H1, a third member of the B7 family, co-stimulates T-cell proliferation and interleukin-10 secretion. Nat Med. 1999;5:1365. doi: 10.1038/70932. [DOI] [PubMed] [Google Scholar]
- 18.Muenst S, Schaerli AR, Gao F, et al. Expression of programmed death ligand 1 (PD-L1) is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2014;146:15–24. doi: 10.1007/s10549-014-2988-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghebeh H, Mohammed S, Al-Omair A, et al. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: correlation with important high-risk prognostic factors. Neoplasia. 2006;8:190–198. doi: 10.1593/neo.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muenst S, Soysal SD, Gao F, et al. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res Treat. 2013;139:667–676. doi: 10.1007/s10549-013-2581-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimino-Mathews A, Foote JB, Emens LA. Immune targeting in breast cancer. Oncology (Williston Park) 2015;29:375–385. [PubMed] [Google Scholar]
- 22.Emens LA. Breast cancer immunotherapy: facts and hopes. Clin Cancer Res. 2017 doi: 10.1158/1078-0432.CCR-16-3001. DOI: 10.1158/1078-0432.CCR-16-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu B, Song Y, Liu D. Recent development in clinical applications of PD-1 and PD-L1 antibodies for cancer immunotherapy. J Hematol Oncol. 2017;10:174. doi: 10.1186/s13045-017-0541-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Postow MA. Managing immune checkpoint-blocking antibody side effects. Am Soc Clin Oncol Educ Book. 2015:76–83. doi: 10.14694/EdBook_AM.2015.35.76. [DOI] [PubMed] [Google Scholar]
- 25.Kitazawa Y, Fujino M, Wang Q, et al. Involvement of the programmed death-1/programmed death-1 ligand pathway in CD4+CD25+ regulatory T-cell activity to suppress alloimmune responses. Transplantation. 2007;83:774–782. doi: 10.1097/01.tp.0000256293.90270.e8. [DOI] [PubMed] [Google Scholar]
- 26.Cheever MA, Allison JP, Ferris AS, et al. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu Y-C, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol. 2016;28:22–27. doi: 10.1016/j.smim.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang X, Kim S, Hundal J, et al. Breast cancer neoantigens can induce CD8(+) T-cell responses and antitumor immunity. Cancer Immunol Res. 2017;5:516–523. doi: 10.1158/2326-6066.CIR-16-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Capietto AH, Jhunjhunwala S, Delamarre L. Characterizing neoantigens for personalized cancer immunotherapy. Curr Opin Immunol. 2017;46:58–65. doi: 10.1016/j.coi.2017.04.007. [DOI] [PubMed] [Google Scholar]
- 30.Beckmann MW, Brucker C, Hanf V, et al. Quality assured health care in certified breast centers and improvement of the prognosis of breast cancer patients. Onkologie. 2011;34:362–367. doi: 10.1159/000329601. [DOI] [PubMed] [Google Scholar]
- 31.Hein A, Gass P, Walter CB, et al. Computerized patient identification for the EMBRACA clinical trial using real-time data from the PRAEGNANT network for metastatic breast cancer patients. Breast Cancer Res Treat. 2016;158:59–65. doi: 10.1007/s10549-016-3850-8. [DOI] [PubMed] [Google Scholar]
- 32.Fasching PA, Brucker SY, Fehm TN, et al. Biomarkers in patients with metastatic breast cancer and the PRAEGNANT Study Network. Geburtshilfe Frauenheilkd. 2015;75:41–50. doi: 10.1055/s-0034-1396215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hammond ME, Hayes DF, Dowsett M, et al. Arch Pathol Lab Med. Vol. 134. American Society of Clinical Oncology: College of American Pathologists; 2010. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version) pp. e48–e72. [DOI] [PubMed] [Google Scholar]
- 34.Bethune GC, Veldhuijzen van Zanten D, MacIntosh RF, et al. Impact of the 2013 American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 (HER2) testing of invasive breast carcinoma: a focus on tumours assessed as ‘equivocal’ for HER2 gene amplification by fluorescence in-situ hybridization. Histopathology. 2015;67:880–887. doi: 10.1111/his.12723. [DOI] [PubMed] [Google Scholar]
- 35.Coates AS, Winer EP, Goldhirsch A, Panel Members Tailoring therapies - improving the management of early breast cancer: St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2015. Ann Oncol. 2015;26:1533–1546. doi: 10.1093/annonc/mdv221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 37.Sinn HP, Schmid H, Junkermann H, et al. Histologic regression of breast cancer after primary (neoadjuvant) chemotherapy (Article in German) Geburtshilfe Frauenheilkd. 1994;54:552–558. doi: 10.1055/s-2007-1022338. [DOI] [PubMed] [Google Scholar]
- 38.Salgado R, Denkert C, Demaria S, International TILs Working Group 2014 the evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015;26:259–271. doi: 10.1093/annonc/mdu450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hendry S, Salgado R, Gevaert T, et al. Assessing tumor-infiltrating lymphocytes in solid tumors: a practical review for pathologists and proposal for a standardized method from the International Immunooncology Biomarkers Working Group: Part 1: Assessing the host immune response, TILs in invasive breast carcinoma and ductal carcinoma in situ, metastatic tumor deposits and areas for further research. Adv Anat Pathol. 2017;24:235–251. doi: 10.1097/PAP.0000000000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swisher SK, Wu Y, Castaneda CA, et al. Interobserver agreement between pathologists assessing tumor-infiltrating lymphocytes (TILs) in breast cancer using methodology proposed by the International TILs Working Group. Ann Surg Oncol. 2016;23:2242–2248. doi: 10.1245/s10434-016-5173-8. [DOI] [PubMed] [Google Scholar]
- 41.Dieci MV, Prat A, Tagliafico E, et al. Integrated evaluation of PAM50 subtypes and immune modulation of pCR in HER2-positive breast cancer patients treated with chemotherapy and HER2-targeted agents in the CherLOB trial. Ann Oncol. 2016;27:1867–1873. doi: 10.1093/annonc/mdw262. [DOI] [PubMed] [Google Scholar]
- 42.Mao Y, Qu Q, Zhang Y, et al. The value of tumor infiltrating lymphocytes (TILs) for predicting response to neoadjuvant chemotherapy in breast cancer: a systematic review and meta-analysis. PLoS One. 2014;9:e115103. doi: 10.1371/journal.pone.0115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Solinas C, Ceppi M, Lambertini M, et al. Tumor-infiltrating lymphocytes in patients with HER2-positive breast cancer treated with neoadjuvant chemotherapy plus trastuzumab, lapatinib or their combination: a meta-analysis of randomized controlled trials. Cancer Treat Rev. 2017;57:8–15. doi: 10.1016/j.ctrv.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 44.Ingold Heppner B, Untch M, Denkert C, et al. Tumor-infiltrating lymphocytes: a predictive and prognostic biomarker in neoadjuvant-treated HER2-positive breast cancer. Clin Cancer Res. 2016;22:5747–5754. doi: 10.1158/1078-0432.CCR-15-2338. [DOI] [PubMed] [Google Scholar]
- 45.McGranahan N, Furness AJ, Rosenthal R, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones S, Anagnostou V, Lytle K, et al. Personalized genomic analyses for cancer mutation discovery and interpretation. Sci Transl Med. 2015;7:283ra53. doi: 10.1126/scitranslmed.aaa7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morse MA, Chaudhry A, Gabitzsch ES, et al. Novel adenoviral vector induces T-cell responses despite anti-adenoviral neutralizing antibodies in colorectal cancer patients. Cancer Immunol Immunother. 2013;62:1293–1301. doi: 10.1007/s00262-013-1400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Balint JP, Gabitzsch ES, Rice A, et al. Extended evaluation of a phase 1/2 trial on dosing, safety, immunogenicity, and overall survival after immunizations with an advanced-generation Ad5 (E1-, E2b-)-CEA(6D) vaccine in late-stage colorectal cancer. Cancer Immunol Immunother. 2015;64:977–987. doi: 10.1007/s00262-015-1706-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nuciforo P, Thyparambil S, Aura C, et al. High HER2 protein levels correlate with increased survival in breast cancer patients treated with anti-HER2 therapy. Mol Oncol. 2016;10:138–147. doi: 10.1016/j.molonc.2015.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vujovic S, Henderson S, Presneau N, et al. Brachyury, a crucial regulator of notochordal development, is a novel biomarker for chordomas. J Pathol. 2006;209:157–165. doi: 10.1002/path.1969. [DOI] [PubMed] [Google Scholar]
- 51.Heery CR, Singh BH, Rauckhorst M, et al. Phase I trial of a yeast-based therapeutic cancer vaccine (GI-6301) targeting the transcription factor brachyury. Cancer Immunol Res. 2015;3:1248–1256. doi: 10.1158/2326-6066.CIR-15-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chaft JE, Litvak A, Arcila ME, et al. Phase II study of the GI-4000 KRAS vaccine after curative therapy in patients with stage I-III lung adenocarcinoma harboring a KRAS G12C, G12D, or G12V mutation. Clin Lung Cancer. 2014;15:405–410. doi: 10.1016/j.cllc.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 53.Tresp V, Overhage JM, Bundschus M, et al. Going digital: a survey on digitalization and large-scale data analytics in healthcare. Proc IEEE. 2016;104:2180–2206. [Google Scholar]