Abstract

The compartmentalization of cell-free gene expression systems in liposomes provides an attractive route to the formation of protocells, but these models do not capture the physical (crowded) environment found in living systems. Here, we present a microfluidics-based route to produce monodisperse liposomes that can shrink almost 3 orders of magnitude without compromising their stability. We demonstrate that our strategy is compatible with cell-free gene expression and show increased protein production rates in crowded liposome protocells.
In recent years, there has been a significant research effort to produce synthetic cell-like compartments including liposomes,1 fatty acid vesicles,2 polymersomes,3 proteinosomes,4 water-in-oil droplets5 and coacervate droplets6 as protocells to study key aspects of living systems such as compartmentalization,5,7 replication,8 or metabolism.6b,9 Liposomes have been used to produce protocells for studying in vitro transcription and translation (IVTT),1a,1b cell division8b,10 and spatially confined catalysis.11 However, little progress has been made in generating protocells with adaptive performances: tunable volumes and surface areas in response to environmental changes.
Cells regulate their surface area and volume through membrane folding, invagination, or vesicle fusion and fission.12 Recent studies have demonstrated that fusion of micelles into protocell membranes,13 or in situ generation of lipids inside membranes,14 can lead to slow membrane growth, and lipid bilayers can generate lipid tubes or vesicles to balance hypertonic osmotic pressure.12b,15 However, there is no method to robustly swell and shrink liposome protocells without compromising their stability. Here, we propose a new route to regulate protocell volumes via incorporation of artificial oil-based organelles–lipid droplets (LDs). LDs are present in some cells as oil droplets with a lipid core surrounded by a phospholipid monolayer, and store lipids for energy metabolism and membrane synthesis.16 Here. we designed an oil-based lipid reservoir attached to liposomes to regulate the volume of protocells. The artificial LD allows collection or supply of lipids from/to the bilayer membrane as osmotic pressure fluctuates, resulting in shrinking or swelling of the protocells, opening up opportunities to study dynamic cellular features in vitro.
We used microfluidic emulsification to produce monodisperse water-in-oil-in-water (W/O/W) double emulsion droplets as templates (Figures 1a, S1 in Supporting Information (SI)), which subsequently undergo partial dewetting (Figure 1a). The dewetting process is determined by the spreading coefficient defined as Si = γjk – (γij + γik), where γij is the interfacial tension between fluids i and j.17 Only if Sw1 < 0, So < 0 and Sw2 < 0, will the emulsion templates form a stable partial dewetting configuration (see SI Experimental Section for details of the typically used inner, middle and outer fluids).17a The success of partial dewetting relies on careful control of the concentration of surfactant F-68 which tunes the interfacial energies. Without addition of F-68, the emulsion templates will keep the core–shell structure due to a positive So (Figures 1a,b, first panel).1b When F-68 is added into W2, γw1w2 and γow2 will decrease and So becomes negative, triggering the dewetting process. A lipid bilayer is formed via combining two lipid monolayers at the two water–oil interfaces (Figures 1a and S1). Higher concentrations of F-68 promote the formation of more bilayer until an intact liposome is generated (C(F-68) > 0.2 wt %),1b as F-68 in W2 reduces γw1w2 and γow2, which make the adhesion energy ΔF (ΔF = γow2 + γw1w2 – γow1)18 smaller, resulting in less contact area between oil droplet and liposome. Other combinations of liquids and surfactants reported in the literature enable tuning of the structures of double emulsion droplets17,20 and these also appear promising for making vesicles by the partial dewetting method.
Figure 1.
(a) Schematics and (b) confocal images of diverse configurations of protocells prepared from partial dewetting of W/O/W emulsion templates with 0.00–0.20 wt % F-68 in the W2 phase. (c) Reconstructed confocal image showing the 3D structure of protocells. (d) As-formed model protocells with different-sized oil organelles (in green). Scale bars, 100 μm.
Our approach yields excellent control over the liposomal structures by adjusting concentrations of F-68 or the flow rates (Figure 1b,c). As the concentrations of F-68 increase from 0%, 0.01%, 0.02%, 0.075% to 0.20%, the bilayer area gradually increases from 0%, 41%, 49%, 72% to 84% of the surface area of inner droplets (D = 69 μm), respectively. Moreover, the sizes of attached oil droplets can be easily tuned (Figure 1d), and complex structures with multiple compartments (so-called multisomes19) were also easily prepared in our method (Figure S2). Importantly, the as-formed liposomes are stable (Figures S3b,c), with no obvious loss in numbers after storage for 4 days (Figure S4).
Next, we studied the swelling and shrinking of the liposomes (Figure 2). As Figure 2a–c shows, when the protocells consisting of 0.05 wt % PEG were dispersed in a hypertonic solution of 2 M sucrose, they rapidly lost water and shrunk to balance the osmotic difference (Movies S1, S2). We did not observe any lipid spots, vesicles or tubule formation in the bilayers during the shrinking process, indicating the extra lipids from the bilayers were collected into the attached oil droplets. Remarkably, the membrane surface and volume are only 1/77 and 1/670, respectively, of their initial states (Figure S5, Movie S2). In contrast, liposomes without LDs collapsed and burst immediately under the same conditions (Figure S6). Inversely, LDs also allow the supply of lipids to the bilayers to induce membrane growth (Figure 2d,e), because a negative pressure outside the liposomes reverses the water flux, leading to the growth of surface area and volume. The shrinking process is reversible, as demonstrated in Figure S7. Liposomes with an inner phase of 1.7 wt % PEG and 170 mM sucrose were shrunk from 72 to 47 μm in 750 mM sucrose solution with 0.05 wt % F-68 added. Subsequently, an aqueous solution of 0.05 wt % F-68 was carefully added and the liposomes swelled to 60 μm.
Figure 2.
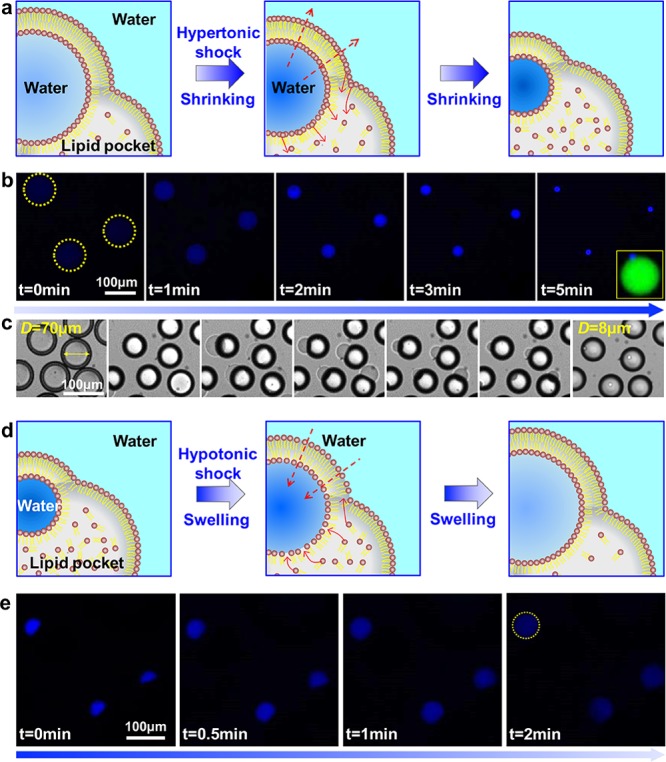
(a–c) Schematics, confocal and optical images of the shrinking process of protocells in response to hypertonic shock. (d,e) Schematics and confocal images of the swelling process of protocells when in hypotonic solution. Scale bars, 100 μm.
To verify the lipid exchange between the bilayers and the LDs, we performed fluorescence recovery after photobleaching (FRAP) experiments via labeling the bilayers and the inner water droplets with two different fluorophores (Figure 3a, see SI for details). As Figure 3b,c shows, the fluorescence of the whole bilayers recovered gradually in about 2 min after photobleaching (Movie S3), but the fluorescence in the interior did not recover due to shortage of dye supply (Figure S9). This experiment directly demonstrates the successful lipid exchange between the bilayers and the attached LDs.
Figure 3.

(a) Schematics of the FRAP experiments. (b) Confocal images of the recovery of fluorescence of bilayers after photobleaching. Insets in panel b showing the fluorescence recovery in the inner water droplet. (c) Fluorescence intensity–distance profiles along the line in panel b. Scale bars, 50 μm.
Cells are densely packed with macromolecules (total macromolecule concentrations in excess of 300 g·L–1 in E. coli),21 which influences biochemical kinetics.22 However, no method enables the production of liposomes with levels of crowding found in cells, because high concentrations of macromolecules are too viscous to encapsulate. To solve this issue and to reconstitute a realistic cell-like internal environment, we encapsulated cell lysate (60 g·L–1), nucleoids into E. coli lipid liposomes of 88 μm in diameter (see SI for details). We then shrunk them to 54 μm in diameter to form protocells with a concentration of macromolecules at about 260 g·L–1 (Figure 4a,b). To illustrate the dense interior, FRAP experiments were performed to probe the diffusion of enhanced green fluorescent protein (eGFP) encapsulated. As Figure 4c shows, the fluorescence recovers within 1 s in liposomes before shrinking, while it takes more than 40 s to recover after shrinking, which demonstrates the crowded interior of the protocells.
Figure 4.

(a,b) Illustration and images of macromolecularly crowded protocells. Inset in panel b1 is the sample before shrinking. (c) Fluorescence recovery of eGFP in liposomes before (upper sequence) and after (lower sequence) shrinking after photobleaching. (d) Expression profiles of mRFP in normal and shrunk liposomes and confocal images of liposomes after expression for 6 h. Scale bars: 50 μm in panels b, c; 100 μm in panel d.
We then performed IVTT in both crowded and noncrowded protocells to investigate the influence of crowded interiors on gene expression. We encapsulated a mix of cell lysate, feeding buffers and plasmids coding for monomeric red fluorescent protein (mRFP) (total concentration is about 40 g L–1) into l-α-phosphatidylcholine (eggPC) liposomes (see SI Experimental Section for details), then collected them into two containers (one with hypertonic solution, the other without) to form crowded protocells (diameter: 48 μm) and noncrowded protocells (diameter: 91 μm) (Figure 4d). In crowded protocells, the concentration of the interior solution increases approximately 6.8 times, yielding a concentration of IVTT mix of about 272 g L–1. The expression of mRFP in shrunk liposomes is notably enhanced compared to expression in normal liposomes which is very slow and barely detectable after 6 h (Movie S4). We postulate that the rate enhancement in gene expression is not only due to increased concentration of key components such as DNA or ribosomes in the IVTT mixture but also because of the molecularly crowded interior.5,6c The slightly increased ratio of the surface to volume of the bilayer membrane is not expected to alter gene expression significantly.23
To extend the technological scope for constructing protocells with more synthetic complexity, we induced complex coacervation of the cell lysates to create subcompartments in protocells. Complex coacervation is a form of liquid–liquid phase separation of oppositely charged polyelectrolytes, and provides powerful means of membrane-free compartmentalization. Coacervation has been explored extensively in protocell models for the construction of artificial cells or organelles.6,7 The phase separation of cell lysates to form crowded coacervates has been accomplished in water-in-oil droplets recently,6c but it has not been demonstrated in biological vesicles because of the use of concentrated salt solution (as high as 6 M) and fragile nature of vesicles. To address this problem, we shrunk the liposomes containing cell lysate, feeding buffers and 8 g·L–1 PEG via multistep osmotic shocks (Figure 5a, see SI Experimental Section for details). Meanwhile, plasmids coding for eGFP were also encapsulated into the protocells to perform IVTT. As Figures 5b,c and S10 show, shrinking the volume of liposomes induced coacervate droplet formation with cell lysate and PEG in the liposomes (Movie S5), due to the phase transition of salt and PEG as well as partitioning of cell lysate into the PEG phase. Notably, the expressed eGFP also prefers to partition into the innermost coacervate droplet (bright core in Figure 5b).6c
Figure 5.

(a,b) Coacervate formation in liposomes induced by decrease of volume. (c) Optical images of liquid–liquid phase separation process in protocells. Scale bars, 50 μm.
In summary, we have presented a novel design to regulate membrane area and volume of microfluidically prepared liposomes by exploiting the oil droplet as a reservoir which collects or supplies lipids from/to the bilayer membrane during shrinking or swelling. We demonstrated cell-free gene expression in cell-like conditions inside shrunk liposomes. Control over liposome volume may find use in research as diverse as maintaining artificial intracellular conditions for homeostasis,24 tuning biochemical reaction rates on the basis of membrane curvature,25 preparing monodisperse sub-micrometer-sized vesicles (Figure S11), and performing protein crystallization and growth.26
Acknowledgments
This work was supported by The Netherlands Organization for Scientific Research (NWO, TOPPUNT grant 718.014.001, the Ministry of Education, Culture and Science (Gravity programme, 024.001.035). N.-N. Deng acknowledges funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 659907.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.8b03123.
Experimental section (PDF)
Shrinking process of the protocells via hypertonic shock (AVI)
Increase of the concentration of fluorescent molecules in protocells during shrinking (AVI)
Recovery of fluorescence of the bilayers after photobleaching (AVI)
In vitro transcription and translation in crowded and noncrowded protocells (AVI)
Complex coacervation of cell lysates inside protocell (AVI)
The authors declare no competing financial interest.
Supplementary Material
References
- a Noireaux V.; Libchaber A. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 17669–17674. 10.1073/pnas.0408236101. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Deng N.-N.; Yelleswarapu M.; Huck W. T. S. J. Am. Chem. Soc. 2016, 138, 7584–7591. 10.1021/jacs.6b02107. [DOI] [PubMed] [Google Scholar]; c Deshpande S.; Caspi Y.; Meijering A. E. C.; Dekker C. Nat. Commun. 2016, 7, 10447. 10.1038/ncomms10447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy S. S.; Schrum J. P.; Krishnamurthy M.; Tobe S.; Treco D. A.; Szostak J. W. Nature 2008, 454, 122–125. 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino C.; Kim S.-H.; Horsfall L.; Abbaspourrad A.; Rosser S. J.; Cooper J.; Weitz D. A. Angew. Chem., Int. Ed. 2012, 51, 6416–6420. 10.1002/anie.201201443. [DOI] [PubMed] [Google Scholar]
- Li M.; Harbron R. L.; Weaver J. V. M; Binks B. P.; Mann S. Nat. Chem. 2013, 5, 529–536. 10.1038/nchem.1644. [DOI] [PubMed] [Google Scholar]
- Hansen M. M. K.; Meijer L. H. H.; Spruijt E.; Maas R. J. M.; Rosquelles M. V.; Groen J.; Heus H. A.; Huck W. T. S. Nat. Nanotechnol. 2016, 11, 191–197. 10.1038/nnano.2015.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Koga S.; Williams D. S.; Perriman A. W.; Mann S. Nat. Chem. 2011, 3, 720–724. 10.1038/nchem.1110. [DOI] [PubMed] [Google Scholar]; b Strulson C. A.; Molden R. C.; Keating C. D.; Bevilacqua P. C. Nat. Chem. 2012, 4, 941–946. 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]; c Sokolova E.; Spruijt E.; Hansen M. M. K.; Dubuc E.; Groen J.; Chokkalingam V.; Piruska A.; Heus H. A.; Huck W. T. S. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 11692–11697. 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Deng N.-N.; Huck W. T. S. Angew. Chem., Int. Ed. 2017, 56, 9736–9740. 10.1002/anie.201703145. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Deng N.-N.; Yelleswarapu M.; Zheng L.; Huck W. T. S. J. Am. Chem. Soc. 2017, 139, 587–590. 10.1021/jacs.6b10977. [DOI] [PubMed] [Google Scholar]
- a Kurihara K.; Tamura M.; Shohda K.-i.; Toyota T.; Suzuki K.; Sugawara T. Nat. Chem. 2011, 3, 775–781. 10.1038/nchem.1127. [DOI] [PubMed] [Google Scholar]; b Terasawa H.; Nishimura K.; Suzuki H.; Matsuura T.; Yomo T. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 5942–5947. 10.1073/pnas.1120327109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Gardner P. M.; Winzer K.; Davis B. G. Nat. Chem. 2009, 1, 377–383. 10.1038/nchem.296. [DOI] [PubMed] [Google Scholar]; b Adamala K.; Szostak J. W. Science 2013, 342, 1098–1100. 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M.; Anderson D. E.; Erickson H. P. Science 2008, 320, 792–794. 10.1126/science.1154520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolinger P.-Y.; Stamou D.; Vogel H. J. Am. Chem. Soc. 2004, 126, 8594–8595. 10.1021/ja049023u. [DOI] [PubMed] [Google Scholar]
- a Morris C. E.; Homann U. J. Membr. Biol. 2001, 179, 79–102. 10.1007/s002320010040. [DOI] [PubMed] [Google Scholar]; b Staykova M.; Holmes D. P.; Read C.; Stone H. A. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 9084–9088. 10.1073/pnas.1102358108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Budin I.; Debnath A.; Szostak J. W. J. Am. Chem. Soc. 2012, 134, 20812–20819. 10.1021/ja310382d. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Zhu T. F.; Szostak J. W. J. Am. Chem. Soc. 2009, 131, 5705–5713. 10.1021/ja900919c. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Budin I.; Szostak J. W. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 5249–5254. 10.1073/pnas.1100498108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Hardy M. D.; Yang J.; Selimkhanov J.; Cole C. M.; Tsimring L. S.; Devaraj N. K. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 8187–8192. 10.1073/pnas.1506704112. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Exterkate M.; Caforio A.; Stuart M. C. A.; Driessen A. J. M. ACS Synth. Biol. 2018, 7, 153–165. 10.1021/acssynbio.7b00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.; Lipowsky R.; Dimova R. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 4731–4736. 10.1073/pnas.1015892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Fujimoto T.; Parton R. G. Cold Spring Harbor Perspect. Biol. 2011, 3, a004838. 10.1101/cshperspect.a004838. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Thiam A. R.; Farese R. V. Jr.; Walther T. C. Nat. Rev. Mol. Cell Biol. 2013, 14, 775–786. 10.1038/nrm3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Torza S.; Mason S. G. Science 1969, 163, 813–814. 10.1126/science.163.3869.813. [DOI] [PubMed] [Google Scholar]; b Shum H. C.; Santanach-Carreras E.; Kim J.-W.; Ehrlicher A.; Bibette J.; Weitz D. A. J. Am. Chem. Soc. 2011, 133, 4420–4426. 10.1021/ja108673h. [DOI] [PMC free article] [PubMed] [Google Scholar]; c Deng N.-N.; Wang W.; Ju X.-J.; Xie R.; Weitz D. A.; Chu L.-Y. Lab Chip 2013, 13, 4047–4052. 10.1039/C3LC50638J. [DOI] [PubMed] [Google Scholar]
- Thiam A. R.; Bremond N.; Bibette J. Phys. Rev. Lett. 2011, 107, 068301. 10.1103/PhysRevLett.107.068301. [DOI] [PubMed] [Google Scholar]
- Villar G.; Heron A. J.; Bayley H. Nat. Nanotechnol. 2011, 6, 803–808. 10.1038/nnano.2011.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarzar L. D.; Sresht V.; Sletten E. M.; Kalow J. A.; Blankschtein D.; Swager T. M. Nature 2015, 518, 520–524. 10.1038/nature14168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulton A. B. Cell 1982, 30, 345–347. 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- van den Berg J.; Boersma A. J.; Poolman B. Nat. Rev. Microbiol. 2017, 15, 309–318. 10.1038/nrmicro.2017.17. [DOI] [PubMed] [Google Scholar]
- Sakamoto R.; Noireaux V.; Maeda Y. T. Sci. Rep. 2018, 8, 7364. 10.1038/s41598-018-25532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhart A. E.; Adamala K. P.; Szostak J. W. Nat. Chem. 2016, 8, 448–453. 10.1038/nchem.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallah-Araghi A.; Meguellati K.; Baret J.-C.; Harrak A. E.; Mangeat T.; Karplus M.; Ladame S.; Marques C. M.; Griffiths A. D. Phys. Rev. Lett. 2014, 112, 028301. 10.1103/PhysRevLett.112.028301. [DOI] [PubMed] [Google Scholar]
- Shim J.-u.; Cristobal G.; Link D. R.; Thorsen T.; Fraden S. Cryst. Growth Des. 2007, 7, 2192–2194. 10.1021/cg700688f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



