Abstract
The pig is the large animal model of choice for study of nerve regeneration and wound repair. Availability of porcine sensory neural cells would conceptually allow for analogous cell-based peripheral nerve regeneration in porcine injuries of similar severity and size to those found in humans. After recently reporting that porcine (or pig) induced pluripotent stem cells (piPSCs) differentiate into neural rosette (NR) structures similar to human NRs, here we demonstrate that pig NR cells could differentiate into neural crest cells and other peripheral nervous system-relevant cell types. Treatment with either bone morphogenetic protein 4 or fetal bovine serum led to differentiation into BRN3A-positive sensory cells and increased expression of sensory neuron TRK receptor gene family: TRKA, TRKB, and TRKC. Porcine sensory neural cells would allow determination of parallels between human and porcine cells in response to noxious stimuli, analgesics, and reparative mechanisms. In vitro differentiation of pig sensory neurons provides a novel model system for neural cell subtype specification and would provide a novel platform for the study of regenerative therapeutics by elucidating the requirements for innervation following injury and axonal survival.
Keywords: : neural development, neural crest cells, sensory neural cells
Introduction
Mounting evidence indicates a disparity in the translation of neural injury and disease therapeutics developed in rodent models to clinical relevance in human patients (Mak et al., 2014). Due to these translational limitations, interest and use of porcine models of neural injury and disease continue to rise (Duberstein et al., 2014; Hughes et al., 2003; Platt et al., 2014; Suzuki et al., 2011; Swindle et al., 2012) as parallel modeling paradigms in porcine and human systems emerge. Anatomical parities with humans, including a gyrencephalic brain, similar white-to-gray matter ratios, and size, have made the pig a particularly useful model for both the central and peripheral nervous systems (PNSs) (Costine et al., 2015; Platt et al., 2014; Sullivan et al., 2015; Zurita et al., 2012).
Similarities with human injury, including spatial considerations, increased critical distance, and nerve diameter, make the pig a needed, more rigorous model organism for treatment assessment for nerve regeneration and wound repair (Middelkoop et al., 2004; Shen et al., 2015), especially after therapeutics touted in rodent models have failed to produce desirable human clinical outcomes (Moore et al., 2009).
Substantial improvements in treating peripheral nerve injuries have been made with nerve grafts and nerve transfers, but these therapies have considerable limitations, including availability of compatible donor material and insufficient functional and sensory recovery of denervated tissue (Korus et al., 2016; Liu et al., 2015). Novel stem cell-based therapeutic strategies have provided varying degrees of improvement in rodent models, but these studies have yet to lead to the development of viable therapeutic treatments for people [reviewed in (Korus et al., 2016)]. This has left nerve autograft as the current gold standard treatment in many instances despite inadequate recovery (Korus et al., 2016).
Human induced pluripotent stem cell-derived sensory neurons (iPSC-SNs) are a robust source of cells with the potential to overcome many of the limitations associated with cell therapies, as iPSC-SNs are highly scalable due to their rapid proliferation, and can undergo autologous derivation limiting immunogenic potential (Marmigere and Ernfors 2007; Nayagam and Minter 2012). However, these cells must be tested for efficacy and safety, which would be best done in a large animal model similar to humans such as the pig. Having analogous porcine sensory neural cells would serve as a means to determine how iPSC-SNs would respond in a cytotoxic neural injury environment and the ability of these cells to differentiate and integrate into diseased or damaged tissues in an allogeneic pig model.
We recently reported that both pig and human iPSCs go through similar processes as they differentiate into neural cells and respond to the same regionalization cues (Gallegos-Cardenas et al., 2015). The goal of the current study was to expand upon these parallels in early neural differentiation and investigate the prospect of differentiating porcine induced pluripotent stem cells (piPSCs) into PNS-relevant cell types. Pig neural rosettes (NRs) were positive for neural crest (NC) markers, therefore we hypothesized that this mixed neural population may contain a subpopulation capable of PNS differentiation after treatment with bone morphogenetic protein 4 (BMP4) or fetal bovine serum (FBS) (Hornbruch et al., 2005; Nayagam and Minter 2012).
In this study, we report the development of piPSC-derived TRKA-positive sensory neural-like cells (piPSC-SNs). Human iPSC-derived sensory neurons have been generated (Marmigere and Ernfors 2007; Nayagam and Minter 2012), but to test in an allogeneic model system, having analogous porcine sensory neural-like cells would serve as a means to determine how similar human and porcine cells are in response to sensory stimuli and reparative mechanisms.
Materials and Methods
Cell culture
Pig iPSC line POU5F1high/SSEA4low was generated as previously described (West et al., 2010; Yang et al., 2013). This line was less than 20 passages from derivation and continuously maintained on Matrigel (diluted 1:100 in Dulbecco's modified Eagle's medium—in DMEM/F12, BD Bioscience)-coated dishes in mTeSR1 (Stemcell Technologies) medium. Neural induction (NI) was performed by dissociating iPSCs in dispase and plated on Matrigel in mTESR1 at 12,500 cells/cm2 and cultured at 37°C under 5% CO2. After 24 hours, pig iPSCs were placed in chemically defined NI medium consisting of DMEM/F12, N2 (Gibco), 2 mM L-glutamine (Invitrogen), 50 U/mL penicillin, 50 μg/mL streptomycin (Invitrogen), and 10 ng/mL fibroblast growth factor-2 (bFGF, R&D System) replaced every day without passage for 15 days.
Following NR formation, cells were transferred to NIF medium (NI medium plus 10 ng/mL leukemia inhibitory factor (LIF, Millipore)). These cells reformed NRs after 7 days, which were disassociated manually and split at a ratio of 1:2. For immunostaining and flow cytometry assay, pig iPSC-derived NR passage 1, passage 3, and passage 7 were used. All experiments were replicated a minimum of three times.
Differentiation into peripheral sensory neural lineage
Pig iPSC-derived NR passage 7 cells were dissociated and seeded at 20,000 cells/cm2 on Matrigel-coated dishes and 4-well Permanox chamber slides (Lab-Tek). After 24 hours, cells were transferred into neural differentiation medium containing NI medium plus ANS (ArunA Biomedical) with/without BMP4 (10 ng/mL,) or FBS (5%, Hyclone). Cells in BMP4 treatment were transferred to FBS medium after 5 days. Medium was changed every other day and all cells were maintained under these conditions for 14 days.
Immunocytochemistry
Pig iPSCs and NRs were plated onto glass, four-well chamber slides and fixed with 4% paraformaldehyde (PFA, Electron Microscopy Sciences) for 15 min. Cells were permeabilized with 0.1% Triton X-100 and 1% polyvinylpyrrolidone (PVP, Sigma-Aldrich) in blocking solution (4% normal donkey serum, from Jackson Immuno Research). Primary antibodies were diluted in blocking solution and incubated at 4°C overnight. Primary antibodies used were p75 nerve grow factor receptor (p75, Abcam, 1:100), human natural killer 1 (HNK1, SIGMA, 1:100), PAX6 (Millipore, 1:1000), SOX1 (R&D System, 1:30), beta III-tubulin (βIII-TUB, Abcam, 1:200), brain-specific homeobox/POU domain protein 3A (BRN3A, Chemicon/Millipore, 1:100), glial fibrillary acidic protein (Dako, 1:400), and alpha smooth muscle actin (α-SMA, Millipore, 1:1000).
Primary antibodies were detected using a fluorescently conjugated secondary antibody, Alexa Fluor (Invitrogen, 1:1000), and incubated for 1 hour at room temperature. Fluorescence images were taken under a laser scanning confocal microscope (Zeiss LSM 710) and were recorded using LSM 710 Software.
Flow cytometry
Cells were fixed in 4% PFA for 10 min at room temperature. Following three washes in phosphate-buffered saline (PBS), cells were blocked in PBS containing 4% normal donkey serum for 45 min. Primary antibodies against p75 (1:50) and HNK1 (1:50) were incubated for 1 hour, followed by three washes in blocking solution. Primary antibodies were detected using fluorescently conjugated secondary antibody Alexa Flour 488 (1:500; Invitrogen) and Alexa Fluor 647 (1:500; Invitrogen). Cells were analyzed using Dakocytomation Cyan (DakoCytomation) and FlowJo cytometry analysis software (Tree Star).
RNA isolation, polymerase chain reaction, and quantitative real-time polymerase chain reaction
Total RNA was extracted from collected cells using the RNeasy mini kit (Qiagen; Germantown, MD) according to the manufacturer's protocol. To disrupt cellular membranes, cells were briefly sonicated immediately before RNA isolation. Isolated RNA was assessed using the NanoDrop 8000, (Thermo Scientific; Waltham, MA) and 500 ng of RNA was then reverse transcribed using the iScript cDNA (BioRad; Irvine, CA). For reverse transcriptase–polymerase chain reaction (RT-PCR) analysis, PCR amplification was performed using GoTaq Green master mix (Promega). Primers were designed using Primer-BLAST software or based on literature (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/cell).
PCR reactions were performed by initially denaturing cDNA at 95°C for 3 min, followed by 30–35 cycles (depending on the particular mRNA abundance) of denaturing at 95°C for 1 min, annealing temperatures at 53°C–60°C for 30 sec according to the primers, and polymerization at 72°C for 45 sec, followed by a 10-min extension at 72°C. Products were separated on 2% agarose gel and imaged using the Alpha Innotech HD2. Omission of transcriptase on cDNA sample during PCR served as negative control.
Using 25 ng of cDNA, quantitative RT-PCR (qRT-PCR) was performed for the following genes: RPL4, TUJ1, BRN3A, ISL1, PRPH, TRKA, TRKB, and TRKC. The primer sequences were developed using the PubMed database and purchased from Integrated DNA Technologies (San Diego, CA) (Supplementary Table S2). Sequencing was performed on the Prism 7900HT Sequence Detection System using SYBR Green master mix (Applied Biosystems; Carlsbad, CA). Biological and technical replicates were both run in triplicate, and the comparative Ct method was used for the purpose of data analysis (Livak and Schmittgen 2001). Data were normalized to the endogenous control and basal (day 0) gene expression to obtain ddCt values.
Statistics
Flow cytometry data were evaluated by one-way ANOVA (SAS® 9.3), followed by Tukey's LSD post hoc test to determine significant differences between groups. One-way ANOVA was used to determine effect sizes in qRT-PCR experiments. In cases of a significant treatment effect, between-group differences were measured using Fisher LSD (STATISTICA version 7.0; Tulsa, OK). Statistical significance was set at p < 0.05.
Results and Discussion
Originating cell lines do not express NC cell markers, but continual passage does increase NC marker p75, but not HNK1
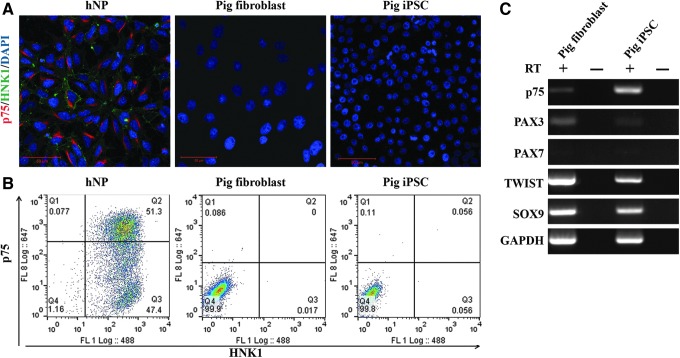
The original fibroblast population and piPSCs derived from them did not express either the low-affinity nerve growth factor receptor p75 or HNK1 proteins at a detectable level, (Fig. 1A, B), but did express p75 mRNA (Fig. 1C), indicating that translation of these NC cell-associated mRNAs is inhibited under these conditions. Both lines were positive for NC markers PAX3, TWIST, and SOX9, but negative for marker PAX7 (Fig. 1C).
FIG. 1.
Porcine fibroblasts nor derived piPSCs express neural crest cell markers. (A) Immunocytochemistry and flow cytometry (B) analysis reveal no protein expression of either p75 or HNK1, while RT-PCR (C) indicates the presence of p75 mRNA in piPSCs. Both fibroblasts and piPSCs were positive for TWIST and SOX9, both indicated in cell lineage determination and differentiation. HNK1, human natural killer 1; RT-PCR, reverse transcriptase–polymerase chain reaction.
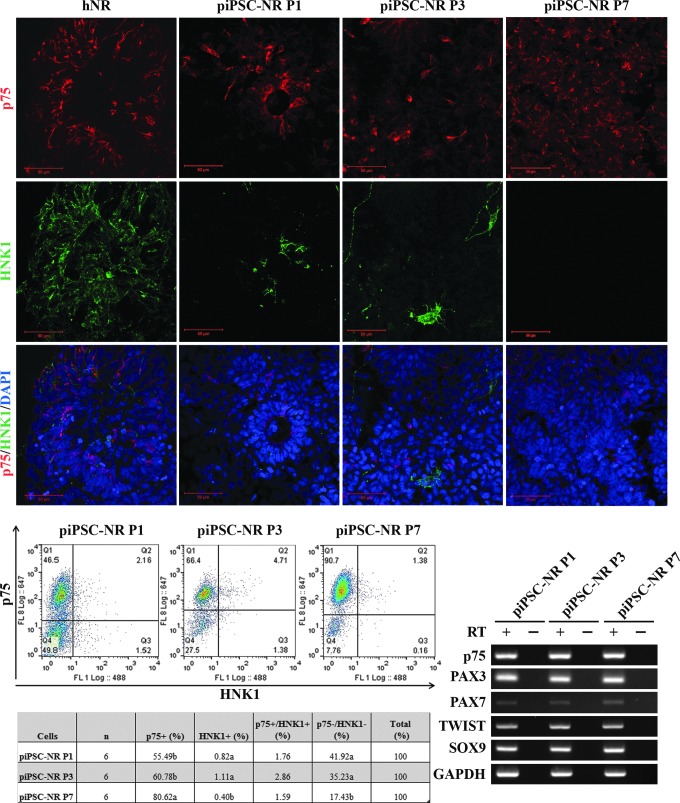
NR formation was induced as described previously (Gallegos-Cardenas et al., 2015) and cells were evaluated for the presence of NC marker-positive cells. The originating fibroblast and piPSC lines were negative for the NC markers upon initial evaluation and, after NR formation, were moved into differentiation medium containing either BMP4 (involved in NC induction) or FBS (Boisvert et al., 2015). At the NR stage, the nerve growth factor receptor p75 was detected at the protein level (Fig. 2A) in over 55% of cells and increased until present in over 80% of cells by P7 (Fig. 2 B, C). HNK1 expression, another marker for migrating NC cells (Giovannone et al., 2015), was also detectable (Fig. 2A–C), although at much lower levels (<3%), irrespective of passage (Fig. 2B).
FIG. 2.
Following NR formation, a subset of cells express neural crest markers. (A) While absent from the originating fibroblast and piPSC lines, expression of neural crest marker p75 is detectable after NR formation, and HNK1 is detectable, although in a smaller percentage of cells. Quantification by flow cytometry (B) and RT PCR (C) supports this analysis, with ∼55% of cells expressing p75 at P1 increasing to over 80% by P7; while HNK1 is detectable in the population, its expression remains under 3% with passage. (D) RT-PCR analysis indicates mRNA for PAX3 and PAX7 after NR formation and retained expression of TWIST and SOX9. NR, neural rosette.
Gene expression of TWIST, which regulates signaling downstream of BMP (Petryk et al., 2004), and SOX9, a transcription factor that regulates NC development (Cheung and Briscoe 2003), was retained through NR formation and passage (Fig. 2D). Taken together, these data indicate that this cell population is NC cell-like and potentially competent to differentiate into PNS-relevant cell types.
BMP4 or FBS treatment induces expression of peripheral neuron marker BRN3A
NC cells are a population of multipotent cells that contribute to a wide array of patterning processes in the developing embryo and may express marker proteins indicative of different lineages that include peripheral neurons. Due to the presence of a population of cells differentiating into p75-expressing cells, we hypothesized that this population was NC-like cells capable of differentiating into presumptive peripheral neurons. Cells were cultured under proneuronal conditions in the presence of BMP4, FBS, or control media without BMP4 or FBS, and cells were assessed for the presence of the neuronal protein TUJ1.
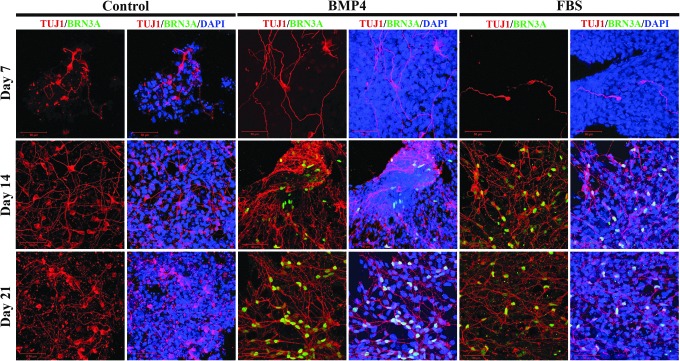
To evaluate the possibility of differentiating the resultant piPSCs into peripheral neuron-like cell types, we performed immunocytochemistry for BRN3A, (Fig. 3) a POU homeodomain transcription factor expressed in all sensory neurons. Cells treated with either BMP4 or FBS contained cell populations expressing BRN3A, while proneuronal conditions alone did not induce expression in piPSC NR cell cultures.
FIG. 3.
BMP4 or FBS treatment induces expression of peripheral neuron marker BRN3A. Expression of BRN3A is not detectable 7 days after transition into medium supplemented with either BMP4 or FBS, but is present after 14 days in differentiation medium, and further increased after 21 days. BMP4 appears to generate a more robust induction of BRN3A expression compared with FBS. Control piPSCs did not differentiate into cells expressing BRN3A in the absence of BMP4 or FBS. BMP4, bone morphogenetic protein 4; FBS, fetal bovine serum.
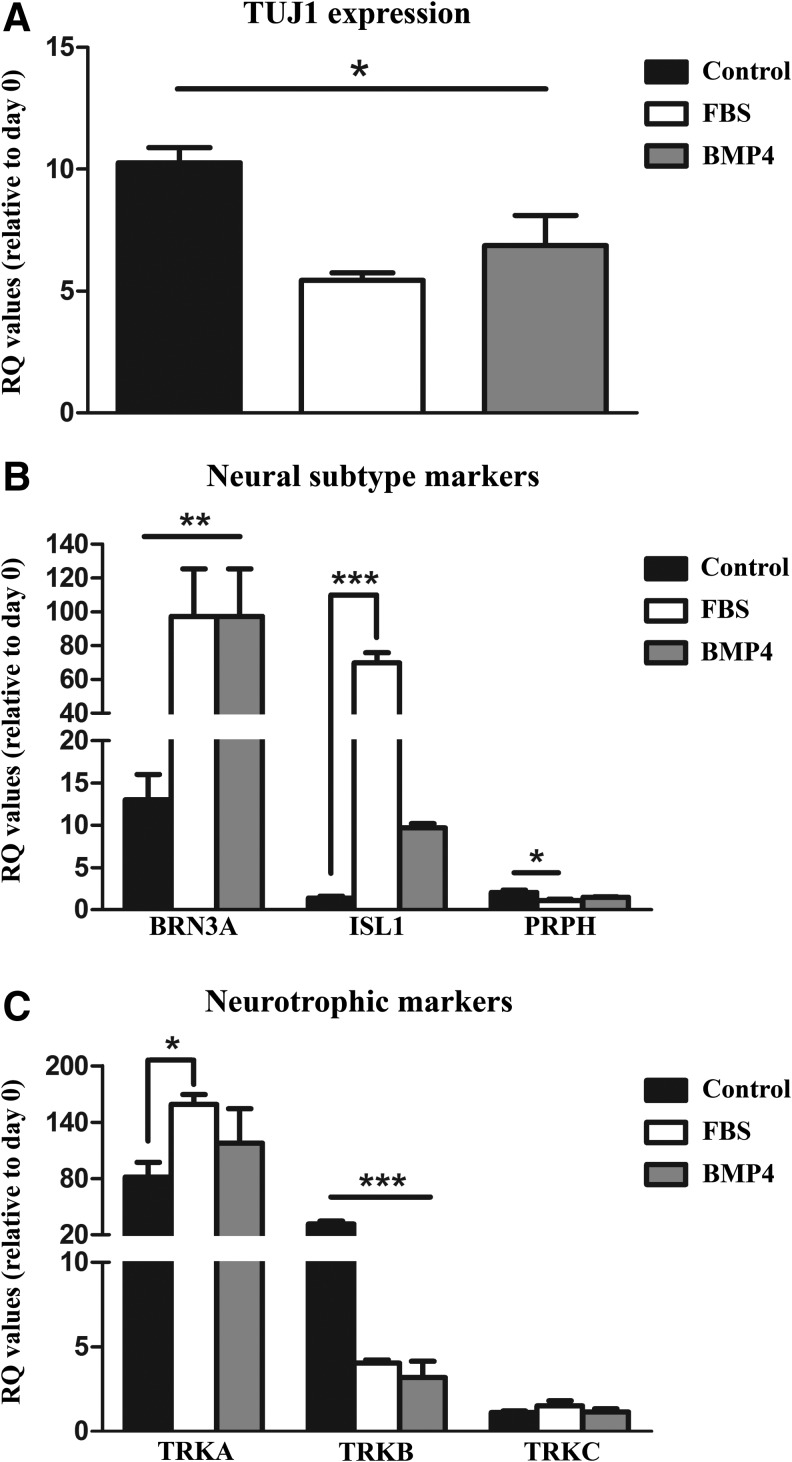
A significant effect of the differentiation medium was found (Fig. 4A–C) (p < 0.01). All differentiation media resulted in an increase in TUJ1 expression compared with immature day 0 cells (Fig. 4A) (p < 0.05). No difference was found between FBS and BMP4 treatment; however, the control differentiation protocol increased TUJ1 expression to a higher extent than the FBS and BMP4 treatments (Fig. 4A) (p < 0.05).
FIG. 4.
Gene expression for (A) TUJ1, (B) peripheral neural subtype markers (BRN3A, ISL1, and PRPH), and (C) neurotrophic markers (TRKA, TRKB, and TRKC). Cells were differentiated under the standard protocol (control), with FBS, or with BMP4 before assay. Relative quantification (RQ) values were determined from calculating dCT to RPL4 and ddCT from immature day 0 cells. Between-group differences are designated by *p < 0.05, **p < 0.01, and ***p < 0.001. Abbreviations: brain-specific homeobox/POU domain protein 3A (BRN3A), ISL LIM homeobox 1 (ISL1), peripherin 2 (PRPH), neuron-specific class III beta-tubulin (TUJ1), tropomyosin receptor kinase (TRKA, TRKB, TRKC).
Gene expression of peripheral neuron and subtype-specific markers was then assessed (BRN3A, ISL1, PRPH). BRN3A and ISL1 are homeodomain transcription factors found in most sensory neurons, and PRPH is an intermediate filament protein expressed largely by peripheral neurons (Marmigere and Ernfors 2007; Sun et al., 2008). The addition of BMP4 and FBS during differentiation robustly increased BRN3A expression compared with immature day 0 cells in addition to the control differentiation protocol (p < 0.01) (Fig. 4B). A similar significant treatment effect was observed with ISL1 expression (p < 0.001). FBS addition in particular greatly enhanced ISL1 transcript concentrations compared with both day 0 cells and control differentiation and BMP4-treated cells (p < 0.001).
While BMP4 treatments failed to reach significance in ISL1 transcript, a trend was observed between the treatment and day 0 and control cells (p = 0.08 and p = 0.09, respectively). An additional treatment effect was measured in the peripheral neural marker PRPH [p < 0.02; F (6.31, 3)]. The pan-neuronal control differentiation conditions increased the expression of PRPH compared with the immature day 0 cells and FBS-treated cells (p < 0.01). No other differences were found (Fig. 4B).
Differentiation of piPSCs increases gene expression of sensory neural markers
Peripheral sensory neurons comprise three distinct subtypes, which are distinguished by their functional subclass and characterized by their selective expression of one member of the TRK receptor gene family: TRKA, TRKB, and TRKC. Expression of these genes in sensory neuron subtypes dictates which sensory modalities sensory neurons are specialized to detect (Marmigere and Ernfors 2007). For instance, whether neurons detect pain sensation or detect limb movement is dictated by the presence of this peptide family. Differentiation resulted in higher expression (p < 0.05) of nociceptor and proprioceptor subclass of neurons as indicated by increased TRKA expression, regardless of the differentiation protocol (Fig. 4C).
Compared with pan-proneural differentiation, FBS treatment enhanced TRKA transcript levels (p < 0.05). A significant treatment effect was also measured in TRKB transcript (mechanoreceptor; p < 0.001). Differentiation under control conditions resulted in higher TRKB expression compared with day 0 undifferentiated NC cells, in addition to the FBS and BMP4 treatments (p < 0.001). No other between-group differences were found with TRKB. Furthermore, differentiation did not induce any changes in the expression of TRKC (Fig. 4C).
Taken together, these data indicate that like human iPSC populations, piPSCs can be differentiated into NRs, NC-like cells, and more specific lineages, including piPSC-SNs. Both BMP4 and serum are sufficient to induce BRN3A expression in both human and pig cells, although human cells can be differentiated more rapidly with small molecules (Chambers et al., 2012; Zhou et al., 2010). It is interesting that continued culture in the presence of BMP4 or FBS results in increased gene expression of TRKA, while decreasing expression of TRKB and TRKC expression did not change with treatment.
It is of note that we did not detect an SOX10-positive NC stage, which may indicate that these cells are more reminiscent of the progenitor population that gives rise to the trigeminal nociceptors that innervate the face (Ching and Kingham 2015). The findings do not support earlier findings that pig fibroblasts can directly differentiate into sensory neurons; we found that prosensory neural culture conditions induced BRN3A expression, suggesting that human and pig NC cells are responsive to similar sensory neuron in vitro differentiation cues, which others indicate as being a necessary step in the sensory neuron specification of NC cells (Blanchard et al., 2015). Taken together, these data indicate that piPSCs can generate sensory neuron-specific subtypes that recapitulate human iPSC in vitro differentiation into PNS sensory neurons.
Supplementary Material
Acknowledgment
This work was supported by the Georgia Research Alliance endowment.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Blanchsard J.W., Eade K.T., Szűcs A., Sardo V.L., Tsunemoto R.K., Williams D., Sanna P.P., and Baldwin K.K. (2015). Selective conversion of fibroblasts into peripheral sensory neurons. Nat Neurosci 18, 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisvert E.M., Engle S.J., Hallowell S.E., Liu P., Wang Z.-W., and Li X.-J. (2015). The specification and maturation of nociceptive neurons from human embryonic stem cells. Sci Rep 5, 16821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Qi Y., Mica Y., Lee G., Zhang X.J., Niu L., Bilsland J., Cao L., Stevens E., Whiting P., et al. (2012). Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat Biotechnol 30, 715–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung M., and Briscoe J. (2003). Neural crest development is regulated by the transcription factor Sox9. Development 130, 5681–5693 [DOI] [PubMed] [Google Scholar]
- Ching R.C., and Kingham P.J. (2015). The role of exosomes in peripheral nerve regeneration. Neural Regen Res 10, 743–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costine B.A., Missios S., Taylor S.R., McGuone D., Smith C.M., Dodge C.P., Harris B.T., and Duhaime A.C. (2015). The subventricular zone in the immature piglet brain: anatomy and exodus of neuroblasts into white matter after traumatic brain injury. Dev Neurosci 37, 115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duberstein K.J., Platt S.R., Holmes S.P., Dove C.R., Howerth E.W., Kent M., Stice S.L., Hill W.D., Hess D.C., and West F.D. (2014). Gait analysis in a pre- and post-ischemic stroke biomedical pig model. Physiol Behav 125, 8–16 [DOI] [PubMed] [Google Scholar]
- Gallegos-Cardenas A., Webb R., Jordan E., West R., West F.D., Yang J.Y., Wang K., and Stice S.L. (2015). Pig induced pluripotent stem cell-derived neural rosettes developmentally mimic human pluripotent stem cell neural differentiation. Stem Cells Dev 24, 1901–1911 [DOI] [PubMed] [Google Scholar]
- Giovannone D., Ortega B., Reyes M., El-Ghali N., Rabadi M., Sao S., and de Bellard M.E. (2015). Chicken trunk neural crest migration visualized with HNK1. Acta Histochem 117, 255–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbruch A., Ma G., Ballermann M.A., Tumova K., Liu D., and Cairine Logan C. (2005). A BMP-mediated transcriptional cascade involving Cash1 and Tlx-3 specifies first-order relay sensory neurons in the developing hindbrain. Mech Dev 122, 900–913 [DOI] [PubMed] [Google Scholar]
- Hughes G.C., Post M.J., Simons M., and Annex B.H. (2003). Translational physiology: porcine models of human coronary artery disease: implications for preclinical trials of therapeutic angiogenesis. J Appl Physiol 94, 1689–1701 [DOI] [PubMed] [Google Scholar]
- Korus L., Ross D.C., Doherty C.D., and Miller T.A. (2016). Nerve transfers and neurotization in peripheral nerve injury, from surgery to rehabilitation. J Neurol Neurosurg Psychiatry 87, 188–197 [DOI] [PubMed] [Google Scholar]
- Liu G.Y., Jin Y., Zhang Q., and Li R. (2015). Peripheral nerve repair: a hot spot analysis on treatment methods from 2010 to 2014. Neural Regen Res 10, 996–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., and Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- Mak I.W., Evaniew N., and Ghert M. (2014). Lost in translation: animal models and clinical trials in cancer treatment. Am J Transl Res 6, 114–118 [PMC free article] [PubMed] [Google Scholar]
- Marmigere F., and Ernfors P. (2007). Specification and connectivity of neuronal subtypes in the sensory lineage. Nat Rev Neurosci 8, 114–127 [DOI] [PubMed] [Google Scholar]
- Middelkoop E., van den Bogaerdt A.J., Lamme E.N., Hoekstra M.J., Brandsma K., and Ulrich M.M.W. (2004). Porcine wound models for skin substitution and burn treatment. Biomaterials 25, 1559–1567 [DOI] [PubMed] [Google Scholar]
- Moore A.M., Kasukurthi R., Magill C.K., Farhadi H.F., Borschel G.H., and Mackinnon S.E. (2009). Limitations of conduits in peripheral nerve repairs. Hand 4, 180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayagam B.A., and Minter R.L. (2012). A comparison of in vitro treatments for directing stem cells toward a sensory neural fate. Am J Otolaryngol 33, 37–46 [DOI] [PubMed] [Google Scholar]
- Petryk A., Anderson R.M., Jarcho M.P., Leaf I., Carlson C.S., Klingensmith J., Shawlot W., and O'Connor M.B. (2004). The mammalian twisted gastrulation gene functions in foregut and craniofacial development. Dev Biol 267, 374–386 [DOI] [PubMed] [Google Scholar]
- Platt S.R., Holmes S.P., Howerth E.W., Duberstein K.J.J., Dove C.R., Kinder H.A., Wyatt E.L., Linville A.V., Lau V.W., Stice S.L., et al. (2014). Development and characterization of a Yucatan miniature biomedical pig permanent middle cerebral artery occlusion stroke model. Exp Transl Stroke Med 6, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y.-I., Song H.-H.G., Papa A.E., Burke J.A., Volk S.W., and Gerecht S. (2015). Acellular hydrogels for regenerative burn wound healing: Translation from a Porcine Model. J Invest Dermatol 135, 2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan S., Eucker S., Gabrieli D., Bradfield C., Coats B., Maltese M., Lee J., Smith C., and Margulies S. (2015). White matter tract-oriented deformation predicts traumatic axonal brain injury and reveals rotational direction-specific vulnerabilities. Biomech Model Mechanobiol 14, 877–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Dykes I.M., Liang X., Eng S.R., Evans S.M., and Turner E.E. (2008). A central role for Islet1 in sensory neuron development linking sensory and spinal gene regulatory programs. Nat Neurosci 11, 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Yeung A.C., and Ikeno F. (2011). The representative porcine model for human cardiovascular disease. J Biomed Biotechnol 2011, 195483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindle M.M., Makin A., Herron A.J., Clubb F.J., and Frazier K.S. (2012). Swine as models in biomedical research and toxicology testing. Vet Pathol 49, 344–356 [DOI] [PubMed] [Google Scholar]
- West F.D., Terlouw S.L., Kwon D.J., Mumaw J.L., Dhara S.K., Hasneen K., Dobrinsky J.R., and Stice S.L. (2010). Porcine induced pluripotent stem cells produce chimeric offspring. Stem Cells Dev 19, 1211–1220 [DOI] [PubMed] [Google Scholar]
- Yang J.Y., Mumaw J.L., Liu Y., Stice S.L., and West F.D. (2013). SSEA4-positive pig induced pluripotent stem cells are primed for differentiation into neural cells. Cell Transplant 22, 945–959 [DOI] [PubMed] [Google Scholar]
- Zhou J., Su P., Li D., Tsang S., Duan E., and Wang F. (2010). High-efficiency induction of neural conversion in human ESCs and human induced pluripotent stem cells with a single chemical inhibitor of transforming growth factor beta superfamily receptors. Stem Cells 28, 1741–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurita M., Aguayo C., Bonilla C., Otero L., Rico M., Rodríguez A., and Vaquero J. (2012). The pig model of chronic paraplegia: a challenge for experimental studies in spinal cord injury. Prog Neurobiol 97, 288–303 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.