ABSTRACT
Mycobacterium tuberculosis releases membrane vesicles (MV) that modulate host immune responses and aid in iron acquisition, although they may have additional unappreciated functions. MV production appears to be a regulated process, but virR remains the only characterized genetic regulator of vesiculogenesis. Here, we present data supporting a role for the M. tuberculosis Pst/SenX3-RegX3 signal transduction system in regulating MV production. Deletion of pstA1, which encodes a transmembrane component of the phosphate-specific transport (Pst) system, causes constitutive activation of the SenX3-RegX3 two-component system, leading to increased protein secretion via the specialized ESX-5 type VII secretion system. Using proteomic mass spectrometry, we identified several additional proteins hyper-secreted by the ΔpstA1 mutant, including LpqH, an MV-associated lipoprotein. Nanoparticle tracking analysis revealed a 15-fold increase in MV production by the ΔpstA1 mutant. Both hyper-secretion of LpqH and increased MV release required RegX3 but were independent of VirR, suggesting that Pst/SenX3-RegX3 controls MV release by a novel mechanism. Prior proteomic analysis identified ESX-5 substrates associated with MV. We therefore hypothesized that MV release requires ESX-5 activity. We constructed strains that conditionally express eccD5, which encodes the predicted ESX-5 transmembrane channel. Upon EccD5 depletion, we observed reduced secretion of the ESX-5 substrates EsxN and PPE41, but MV release was unaffected. Our data suggest that ESX-5 does not affect vesicle production and imply that further characterization of the Pst/SenX3-RegX3 regulon might reveal novel mechanisms of M. tuberculosis vesicle biogenesis.
KEYWORDS: ESX secretion, type VII secretion, lipoproteins, tuberculosis, vesicle
IMPORTANCE
In Gram-negative bacteria, MV derived from the outer membrane have diverse functions in bacterial physiology and pathogenesis, and several factors regulating their production have been identified. Though Gram-positive bacteria and mycobacteria that lack an outer membrane also produce vesicles with described roles in pathogenesis, the mechanisms of MV biogenesis in these organisms remain poorly characterized. Defining mechanisms of MV biogenesis might yield significant insights into the importance of MV production during infection. In M. tuberculosis, only a single genetic element, virR, is known to regulate MV production. Our work reveals that the Pst/SenX3-RegX3 signal transduction system is a novel regulator of MV biogenesis that controls MV production by a mechanism that is independent of both VirR and activation of the specialized ESX-5 protein secretion system. Understanding which genes in the RegX3 regulon cause increased MV production might reveal novel molecular mechanisms of MV release.
INTRODUCTION
Virtually all Gram-negative bacteria produce membrane vesicles (MV) by passive budding from the outer membrane (1). For many years, it was assumed that Gram-positive bacteria and mycobacteria would not release MV due to the difficulty of transporting membranous material through their complex outer cell walls. In the last decade, however, MV production has been described for many of these organisms, including the mycobacteria (2), although the mechanisms of MV release are not well understood (3).
Mycobacterium tuberculosis, the etiologic agent of tuberculosis in humans, produces MV derived from the inner membrane that modulate the host immune response (2, 4). MV contain glycolipids, including lipoarabinomannan (LAM), that inhibit CD4+ T cell activation (4) and lipoproteins, such as LpqH, that act in a TLR2-dependent manner to elicit the production of inflammatory cytokines and chemokines, including interleukin 1β (IL-1β), IL-6, IL-12, tumor necrosis factor alpha (TNF-α), and CXCL1 (2). LpqH can also regulate CD4+ T cell activation and macrophage major histocompatibility complex (MHC) class II expression (5, 6). It is unclear whether these MV-induced responses are beneficial or detrimental to bacterial survival. In mice pretreated with MV via the intratracheal route, these inflammatory responses impaired control of subsequent M. tuberculosis infection (2). However, when MV were administered systemically via subcutaneous injection, they induced a level of protection similar to that produced by the Mycobacterium bovis BCG vaccine (7). Furthermore, an M. tuberculosis mutant that produces more MV was hyper-inflammatory, inducing greater production of TNF-α and IL-6 from infected primary human macrophages, and this mutant was attenuated compared to wild-type (WT) M. tuberculosis (8). M. tuberculosis MV-null mutants might be used to conclusively determine the role of either MV-induced inflammation or MV-mediated inhibition of T cell function in pathogenesis, but such mutants have yet to be described.
MV production is an active and regulated process, since MV release can be induced by iron limitation (9), but the mechanisms that control MV biogenesis remain largely uncharacterized. The only genetic element currently known to affect M. tuberculosis MV production is virR (locus tag rv0431); disruption of virR resulted in increased MV release (8). VirR is a cytosolic protein that associates with the inner membrane and was proposed to act as part of a higher-order complex to regulate MV production through an unknown mechanism. Genetic screens in the model Gram-negative organism Escherichia coli revealed that numerous unique mutations cause increased MV release (10, 11), suggesting that virR is unlikely to be the only regulator of MV production in M. tuberculosis.
In addition to producing MV, M. tuberculosis interacts with the host via several protein secretion systems. Among these are five specialized type VII secretion systems, collectively referred to as the ESX systems (12). ESX-1, -3, and -5 each play an important role in pathogenesis. ESX-1 mediates the escape of M. tuberculosis from the phagosome and helps activate the inflammasome (13, 14). Furthermore, ESX-1 is required to trigger type I interferon responses, which are dependent on the cytosolic DNA sensor cyclic GMP-AMP synthase (cGAS) (14, 15). ESX-3 is essential for bacterial growth; it plays a role in mycobactin-mediated iron acquisition (16) but also has an iron-independent role in virulence (17). ESX-5 is important for inducing the inflammatory response and IL-1β production, as well as causing caspase-dependent cell death (18). These responses likely aid bacterial survival within the host (18). EccD5, the protein predicted to form the ESX-5 transmembrane channel, is required for full pathogenicity in macrophages and severe combined immunodeficiency (SCID) mice (19–21). We recently reported that ESX-5 activity is regulated by the Pst/SenX3-RegX3 system in response to extracellular inorganic phosphate (Pi) availability (22).
The Pst (phosphate-specific transport) system is a high-affinity, low-velocity, ABC-type transport system. We have previously shown that the Pst system controls gene expression in response to extracellular Pi availability through an interaction with the SenX3-RegX3 two-component signal transduction system (23). This system, comprising a membrane-bound sensor histidine kinase (SenX3) and DNA binding response regulator (RegX3), is inhibited by the Pst system when Pi is abundant. When Pi is limiting, however, inhibition by the Pst system is relieved, leading to activation of SenX3-RegX3. Deletion of pstA1, which encodes a transmembrane component of the Pst system, causes constitutive activation of SenX3-RegX3 regardless of Pi availability (23). We demonstrated that in addition to controlling expression of genes involved in the Pi scavenging response, RegX3 directly binds and regulates transcription of esx-5 genes (22, 23). The ΔpstA1 mutant exhibits a RegX3-dependent increase in esx-5 transcription and hyper-secretion of the ESX-5 substrates PPE41 and EsxN (22).
Here, we describe that in addition to affecting protein secretion via ESX-5, the ΔpstA1 mutant hyper-secretes the lipoproteins LpqH and PstS1, which are released from M. tuberculosis in association with MV (2, 8, 24). The ESX-5 substrates EsxN and PPE41 have also previously been detected in association with MV by proteomic approaches (2, 24). We therefore hypothesized that the Pst/SenX3-RegX3 system regulates MV production and that MV release depends on the ESX-5 system. We show that while the ΔpstA1 mutant does hyper-secrete MV in a RegX3-dependent manner, MV release occurs independently of ESX-5 activity. Furthermore, our data indicate that MV hyper-secretion from the ΔpstA1 mutant is independent of VirR. Our data suggest that genes under the control of RegX3 impact MV release from ΔpstA1 bacteria by a novel mechanism.
RESULTS
The ΔpstA1 mutant hyper-secretes the lipoproteins LpqH and PstS1.
We previously demonstrated that ESX-5 secretion system activity is regulated in response to Pi availability by the Pst/SenX3-RegX3 signal transduction system (22). A ΔpstA1 mutant, in which the RegX3 response regulator is constitutively activated, hyper-secretes the ESX-5 substrates EsxN and PPE41 under standard Pi-replete culture conditions (22). We observed that the ΔpstA1 mutant has a secreted protein profile distinct from that of the wild-type (WT) control (Fig. 1A). To preliminarily characterize these differences in protein secretion, we selected four protein bands at molecular weights of approximately 75, 50, 37, and 25 kDa that appeared to be hyper-secreted by the ΔpstA1 mutant and subjected them to proteomic mass spectrometry (MS) analysis. Multiple proteins were identified in each band; the complete data are provided in Table S1 in the supplemental material.
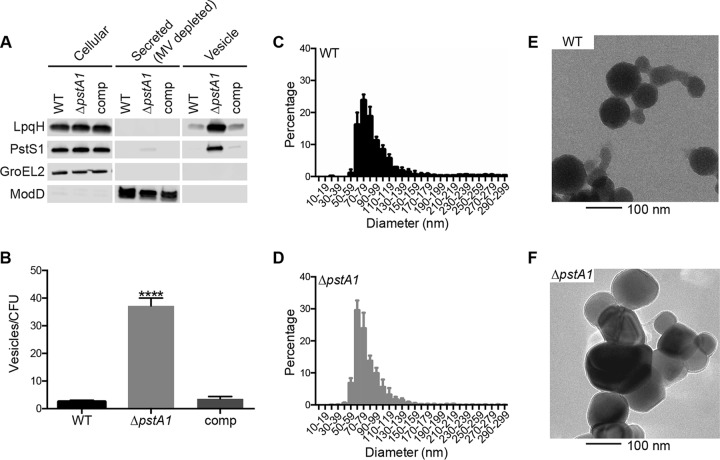
FIG 1 .
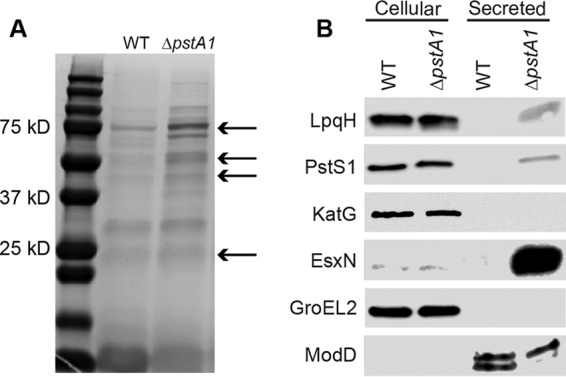
The ΔpstA1 mutant hyper-secretes the lipoproteins LpqH and PstS1. Wild-type M. tuberculosis Erdman (WT) and ΔpstA1 mutant cultures were grown for 5 days in Sauton’s complete medium without Tween 80. Cellular (10 µg) and secreted (5 µg) proteins were separated by SDS-PAGE. (A) Secreted proteins were stained with Imperial protein stain. Arrows indicate the bands selected for analysis by proteomic mass spectrometry. (B) Western blot analysis of proteins identified by mass spectrometry.
Proteomic mass spectrometry analysis of proteins hyper-secreted by the ΔpstA1 mutant. Proteins analyzed by Western blotting are highlighted in yellow. Download TABLE S1, XLSX file, 0.7 MB (685.3KB, xlsx) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We obtained antibodies against several proteins identified by mass spectrometry (LpqH, PstS1, and KatG) and tested secretion of these proteins by Western blotting. We detected increased secretion of the lipoproteins LpqH and PstS1 by the ΔpstA1 mutant compared to their levels in the WT control, though there was no change in the abundance of these proteins in the cellular fraction (Fig. 1B). As in our previous report, we observed increased secretion of the ESX-5 substrate EsxN from the ΔpstA1 mutant compared to its level in the WT control. EsxN was undetectable in the secreted WT fraction when 5 µg of protein was loaded, but we previously reported that EsxN secretion is induced at least 8-fold in the ΔpstA1 mutant (22). The catalase-peroxidase KatG was detected only in the cellular fraction of both strains, and as such, it was excluded from further analyses (Fig. 1B). GroEL2 served as a loading control for cell-associated protein and indicated that cellular lysis did not contaminate the secreted protein fraction. ModD, a protein secreted by the general Sec system, served as a control for equivalent loading of secreted proteins; we detected ModD as a doublet, consistent with its glycosylation (25). These data corroborated the mass spectrometry identification of LpqH and PstS1 as being hyper-secreted by the ΔpstA1 mutant.
The ΔpstA1 mutant overproduces membrane vesicles containing the lipoproteins LpqH and PstS1.
The LpqH and PstS1 lipoproteins were previously described to be associated with M. tuberculosis MV (2, 8, 24). Based on their lipid composition, mycobacterial MV are derived from the inner membrane and contain lipoproteins and other protein cargo (2, 24). Since both LpqH and PstS1 were hyper-secreted by the ΔpstA1 mutant, we sought to determine whether these proteins were MV associated. We isolated MV from culture filtrates by ultracentrifugation and performed Western blotting experiments to analyze the distribution of LpqH and PstS1. Extracellular LpqH and PstS1 were localized exclusively to the MV fraction, and both proteins were more abundant in MV released from the ΔpstA1 mutant than in the WT control (Fig. 2A). Complementation of the ΔpstA1 mutation restored the MV-associated release of LpqH and PstS1 to levels comparable to those in the WT control.
FIG 2 .
The ΔpstA1 mutant hyper-secretes membrane vesicles containing LpqH and PstS1. Cultures of wild-type M. tuberculosis Erdman (WT), the ΔpstA1 mutant, and the ΔpstA1 pMVpstA1 complemented strain (comp) were grown for 5 days in Sauton’s complete medium without Tween 80, and membrane vesicles were isolated. (A) Cellular proteins (5 µg), secreted proteins depleted of MV (5 µg), and MV suspension (20 µl) were analyzed by Western blotting to detect the indicated proteins. (B to D) Nanoparticle tracking analysis of culture supernatants. (B) Numbers of particles per milliliter were normalized to numbers of CFU per milliliter calculated from a control culture of each strain grown in Sauton’s complete medium with Tween 80. Data are means ± standard deviations for three independent cultures. ****, P < 0.0001. (C and D) Distribution of particle sizes from the WT and ΔpstA1 strains, binned in 10-nm increments. (E and F) Transmission electron micrographs of membrane vesicles purified and concentrated from culture supernatants of WT and ΔpstA1 cultures.
To quantify MV release, we conducted nanoparticle-tracking analysis (NTA) on culture supernatants collected from WT, ΔpstA1, and ΔpstA1 pMVpstA1 bacteria using a NanoSight instrument. We observed a statistically significant 15-fold increase in MV release from ΔpstA1 bacteria compared to that in WT bacteria but no difference in MV release between the WT and ΔpstA1 pMVpstA1 strains (Fig. 2B). The accompanying size analysis of MV isolated from WT and ΔpstA1 bacteria revealed that both strains produced particles that fell within the 40- to 250-nm-diameter range previously observed for M. tuberculosis MV (2) (Fig. 2C and D). To further confirm these results, purified MV from these strains were imaged via transmission electron microscopy (TEM). We observed vesicular structures with the predicted size of MV from both WT and ΔpstA1 mutant bacteria (Fig. 2E and F). Taken together, these data support the finding that the ΔpstA1 mutant releases an increased number of MV, and these vesicles are enriched for both the LpqH and PstS1 lipoproteins.
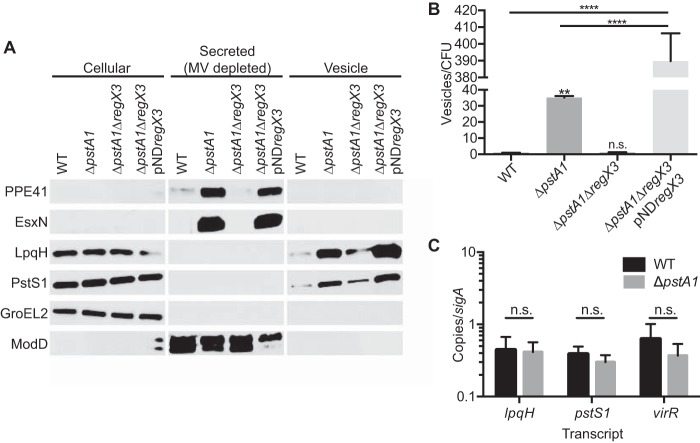
Overproduction of membrane vesicles by the ΔpstA1 mutant requires RegX3.
Many of the phenotypes that we have previously reported for the ΔpstA1 mutant are dependent on RegX3 (22, 23, 26, 27). To determine whether constitutive activation of RegX3 also contributes to overproduction of MV by the ΔpstA1 mutant, we performed additional Western blotting and NTA on the ΔpstA1 ΔregX3 and ΔpstA1 ΔregX3 pNDregX3 strains. Hyper-secretion of the ESX-5 substrates EsxN and PPE41 was abolished in the ΔpstA1 ΔregX3 mutant (Fig. 3A), as previously described (22). EsxN and PPE41 were detected only in the secreted protein fraction despite previous reports from proteomic analyses that these proteins are MV associated (2, 24). This is likely due to the lower sensitivity of Western blots than of proteomic mass spectrometry. Both LpqH and PstS1 were detected in the vesicle fraction from the ΔpstA1 ΔregX3 mutant but at reduced levels compared to those in the ΔpstA1 strain (Fig. 3A). In all cases, complementation of the ΔregX3 deletion with pNDregX3 restored hyper-secretion (Fig. 3A).
FIG 3 .
Increased release of membrane vesicles from the ΔpstA1 mutant requires RegX3. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, ΔpstA1 ΔregX3, and ΔpstA1 ΔregX3 pNDregX3 strains were grown for 5 days in Sauton’s complete medium without Tween 80. (A) Cellular proteins (5 µg), secreted proteins depleted of MV (5 µg), and MV suspension (20 µl) were separated and analyzed by Western blotting to detect the indicated proteins. (B) Nanoparticle tracking analyses of culture supernatants. The results were normalized to numbers of CFU per milliliter determined from a control culture grown in Sauton’s medium with Tween 80 and plated on 7H10 medium. Data are means ± standard deviations for three independent cultures. **, P < 0.01; ****, P < 0.0001. n.s., the difference was not significant. (C) The transcript abundance of lpqH, pstS1, and virR relative to that of sigA were determined by quantitative RT-PCR for the WT and ΔpstA1 strains grown to mid-logarithmic phase in 7H9 complete medium. Results are the means ± standard deviations for three independent experiments.
We again used NTA to quantify MV release from the regX3 deletion mutant and complemented strains. Size analysis revealed that all strains produced MV of the appropriate diameter, with the majority clustering between 50 and 100 nm (Fig. S1). As suggested by the reduced abundance of LpqH and PstS1 in the vesicle fraction from the ΔpstA1 ΔregX3 strain, NTA revealed that vesicle release from the ΔpstA1 ΔregX3 mutant returned to WT levels (Fig. 3B). When the regX3 deletion was complemented, we observed a significant increase in MV release of more than 10-fold compared to MV release in the ΔpstA1 strain. We hypothesize that this increase is due to overexpression of regX3 from the complementation vector, as previously reported (23). We observed no significant difference in abundance of the lpqH or pstS1 transcripts between the ΔpstA1 mutant and WT bacteria (Fig. 3C), consistent with no apparent change in LpqH or PstS1 production in the cellular protein fraction (Fig. 3A). These results demonstrate that increased release of LpqH and PstS1 associated with MV is not due to changes in their expression. Taken together, these results indicate that some other factor, or factors, regulated by RegX3 influence MV production.
The ΔpstA1 ΔregX3 and ΔpstA1 ΔregX3 pNDregX3 strains produce membrane vesicles. The ΔpstA1 ΔregX3 and ΔpstA1 ΔregX3 pNDregX3 strains were grown for 5 days in Sauton’s complete medium without Tween 80. Nanoparticle tracking analyses were performed on culture supernatants, and the resulting particle sizes were binned in 10-nm increments. Download FIG S1, PDF file, 0.5 MB (517.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
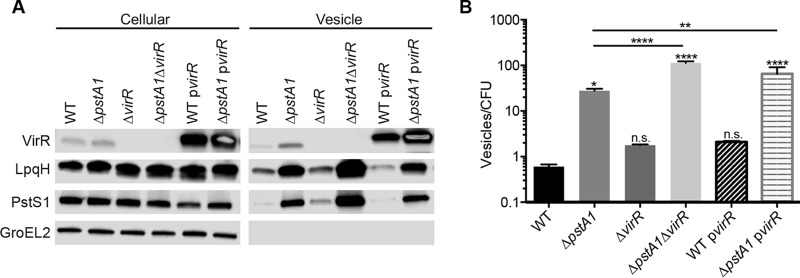
Increased vesicle production by ΔpstA1 bacteria is not dependent on VirR.
virR was previously implicated as a regulator of MV production; transposon disruption of virR led to increased MV biogenesis (8). We analyzed the transcription of virR to determine whether changes in virR expression contribute to increased MV release. While the difference was not statistically significant, we observed a 1.7-fold decrease in the virR transcript level in the ΔpstA1 mutant compared to that in the WT control (Fig. 3C). We investigated whether decreased virR transcript abundance could account for increased MV release from the ΔpstA1 strain. Western blotting revealed similar levels of VirR production in the cellular fractions of WT and ΔpstA1 bacteria, suggesting that the modest differences in virR transcript abundance do not affect VirR protein production (Fig. 4A). VirR associates with MV and is more abundant in MV from the ΔpstA1 strain, consistent with enhanced MV release (Fig. 4A).
FIG 4 .
Increased vesicle release from the ΔpstA1 mutant is independent of VirR. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, ΔvirR, ΔpstA1 ΔvirR, WT pvirR, and ΔpstA1 pvirR strains were grown for 5 days in Sauton’s complete medium without Tween 80. (A) Cellular proteins (5 µg) and MV suspension (20 µl) were analyzed by Western blotting to detect the indicated proteins. (B) Nanoparticle tracking analysis of culture supernatants. Numbers of particles per milliliter were normalized to numbers of CFU per milliliter determined from a control culture grown in Sauton’s medium with Tween 80 and plated on 7H10 medium. Data are means ± standard deviations for three independent cultures. The results of all statistical analyses are compared to WT levels unless otherwise indicated. *, P < 0.05; **, P < 0.01; ****, P < 0.0001.
We took two approaches to determine whether MV production by the ΔpstA1 mutant depends on virR. We overexpressed FLAG-tagged VirR (pvirR) (8) to restore virR expression. Western blots of the WT pvirR and ΔpstA1 pvirR strains revealed a dramatic overproduction of VirR (Fig. 4A). We also constructed a deletion in virR, which we predicted would not affect MV production by the ΔpstA1 mutant if both PstA1 and VirR act in the same MV biogenesis pathway. To avoid polarity on adjacent essential genes, we deleted amino acid residues 85 to 103 of VirR, which correspond to the core globular structure of the protein (8). Western blotting confirmed loss of VirR production in both the ΔvirR and the ΔpstA1 ΔvirR strains (Fig. 4A).
The overexpression of virR did not dramatically alter LpqH or PstS1 abundance in the MV fraction. However, VirR was much more abundant in MV from both the WT pvirR and the ΔpstA1 pvirR strains (Fig. 4A), indicating that VirR may become overrepresented in MV when it is overproduced. Both VirR overexpression strains had slightly increased MV production compared to that of the corresponding parental controls, but this was statistically significant only for the ΔpstA1 pvirR strain (Fig. 4B). Notably, ΔpstA1 pvirR bacteria still produced more MV than WT bacteria, demonstrating that virR overexpression does not rescue the MV hyper-secretion phenotype of the ΔpstA1 strain (Fig. 4B).
The ΔvirR strain had slightly more LpqH and PstS1 in the MV fraction than the WT control (Fig. 4A), suggesting increased MV production as previously reported (8). Both LpqH and PstS1 were also more abundant in the MV fraction from the ΔpstA1 ΔvirR mutant than in either single mutant or WT bacteria (Fig. 4A). NTA confirmed that the ΔvirR strain produced 3-fold-more MV than the WT strain, although this increase was not statistically significant (Fig. 4B). NTA also confirmed that the ΔpstA1 ΔvirR strain produced 4-fold-more MV than the ΔpstA1 mutant and 63-fold-more MV than the ΔvirR mutant, both significant increases (Fig. 4B). Taken together, these results indicate that overproduction of MV by the ΔpstA1 strain is independent of VirR. Furthermore, the mechanisms driving increased MV production in the ΔpstA1 and ΔvirR strains appear to synergize with each other, as a strain harboring deletions of both of these genes produced significantly more MV than either single mutant.
Construction of conditional eccD5 depletion mutants.
We previously demonstrated that the ΔpstA1 mutant exhibits increased ESX-5 activity that is dependent on the RegX3 response regulator (22). Furthermore, others have reported detection of ESX-5 substrates within MV (2, 24). We therefore sought to determine whether increased MV production by the ΔpstA1 mutant was due to increased ESX-5 activity. We attempted to delete eccD5, which encodes a core component of the ESX-5 secretion system that is predicted to form an inner membrane pore through which substrates are secreted (21, 28). We constructed a ΔeccD5 allelic-exchange vector and introduced it into both the WT Erdman and ΔpstA1 mutant strain backgrounds. After obtaining cointegrates with the plasmid integrated at the eccD5 locus, we screened sucrose-resistant colonies for the ΔeccD5 deletion by PCR. All the colonies screened (130 with the WT background, 169 with the ΔpstA1 background) retained the WT eccD5 allele, suggesting that eccD5 is essential for the replication of M. tuberculosis in the standard Middlebrook 7H9 and 7H10 media that we used.
To create strains for conditional expression of eccD5, we first introduced a plasmid (pTIC-eccD5) in which eccD5 transcription is under the control of a tetracycline-inducible (Tet-ON) promoter. We then attempted to delete the chromosomal copy of eccD5 in the WT pTIC-eccD5 and ΔpstA1 pTIC-eccD5 strains. All strain construction steps were conducted in the presence of anhydrotetracycline (ATc) to stimulate eccD5 transcription from pTIC-eccD5. We screened 8 and 16 sucrose-resistant, hygromycin (Hyg)-sensitive isolates in the WT and ΔpstA1 mutant backgrounds, respectively. Among these isolates, we identified 2 ΔeccD5 pTIC-eccD5 mutants and 2 ΔpstA1 ΔeccD5 pTIC-eccD5 mutants that we herein refer to as eccD5 Tet-ON and ΔpstA1 eccD5 Tet-ON, respectively. These data support the contention that eccD5 is essential for the replication of M. tuberculosis under standard in vitro culture conditions.
We analyzed the growth of the eccD5 Tet-ON and ΔpstA1 eccD5 Tet-ON strains compared to that of the parental controls in liquid medium and on agar plates. Both eccD5 Tet-ON strains had a growth defect in liquid medium compared to the growth of the respective parental control even in the presence of ATc to induce eccD5 transcription (Fig. S2A and B). The eccD5 Tet-ON strains also produced smaller colonies on agar plates regardless of ATc treatment (data not shown). Although eccD5 transcription was induced in the eccD5 Tet-ON strains in the presence of ATc, there was still detectable transcript in no-ATc controls (Fig. S2C), and we observed no difference in EccD5 protein abundance between treatment conditions (Fig. S2D). Additionally, PPE41 was secreted even in the absence of ATc (Fig. S2D). Overall, our data suggest that leaky expression from the Tet-inducible promoter in the absence of ATc is sufficient to support the normal activity of the ESX-5 secretion system in the eccD5 Tet-ON strains.
Residual eccD5 expression in uninduced eccD5 Tet-ON strains is sufficient for protein secretion through ESX-5. Kanamycin (15 µg/ml) was added to all eccD5 Tet-ON cultures to maintain plasmid integration. (A and B) Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains were inoculated in 7H9 complete medium at an OD600 of 0.05 and grown at 37°C with aeration. Anhydrotetracycline hydrochloride (ATc; 50 ng/ml) was added every 3 days to the indicated cultures. Growth was monitored by daily OD600 measurements. (C) The transcript abundance of eccD5 relative to that of sigA was determined by quantitative RT-PCR for the WT, ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains grown to mid-logarithmic phase in 7H9 complete medium with and without ATc. (D) The WT, ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains were grown for 5 days in Sauton’s complete medium without Tween 80. ATc (50 ng/ml) was added to the indicated cultures to induce eccD5. Equivalent amounts of cellular (5 µg) and secreted (5 µg) proteins were analyzed by Western blotting to detect the indicated proteins. Download FIG S2, PDF file, 0.8 MB (834.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We therefore turned to construction of strains harboring tetracycline-repressible eccD5 (eccD5 Tet-OFF). We cloned eccD5 under the control of the tetO-4C5G operator (29) on a vector that integrates at the same site as pTIC10a. This pGMCH eccD5 Tet-OFF vector was introduced into the eccD5 Tet-ON strains, and integrants in which pGMCH eccD5 Tet-OFF replaced pTIC-eccD5 by a plasmid swap were selected on 7H10 containing hygromycin. We recovered over 15-fold-more colonies from electroporation with pGMCH eccD5 Tet-OFF than with an empty hygromycin resistance (Hygr) plasmid control. The majority of pGMCH eccD5 Tet-OFF transformants were hygromycin resistant and kanamycin (Kan) sensitive, while isolates transformed with the empty Hygr plasmid were resistant to both hygromycin and kanamycin, indicating that they retained the pTIC-eccD5 plasmid. These data provide further evidence that eccD5 is essential for M. tuberculosis replication in vitro. Strains in which the pGMCH eccD5 Tet-OFF plasmid replaced pTIC-eccD5 were confirmed by PCR and designated eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF. We also attempted construction of dual-control strains in which both eccD5 transcriptional repression and EccD5 protein degradation would occur upon addition of ATc due to the addition of a DAS+4 degradation tag on EccD5 (29). However, attempts to introduce the plasmid encoding DAS-tagged EccD5 by plasmid swap produced <10 colonies, all of which were resistant to both hygromycin and kanamycin, suggesting that EccD5-DAS+4 is nonfunctional.
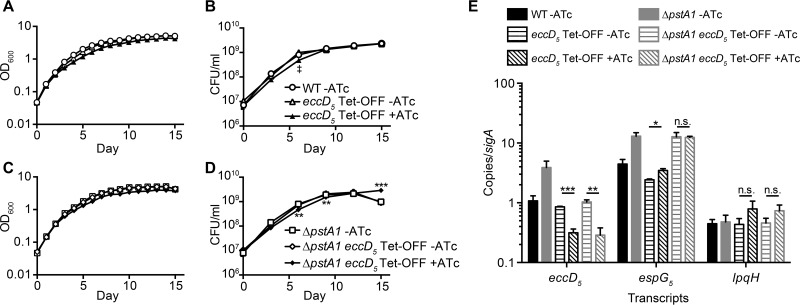
The eccD5 Tet-OFF strains in both the WT and ΔpstA1 backgrounds had statistically significant growth defects (P < 0.05) in liquid medium containing ATc beginning at day 3 compared to the growth of the no-drug controls (Fig. 5A and C). These defects were maintained throughout the incubation period until day 15, when they dropped just below the level of statistical significance. Similarly, we recovered fewer viable CFU from cultures of both eccD5 Tet-OFF strains at days 3 to 12 when ATc was added (Fig. 5B and D). These differences did not quite achieve statistical significance for the eccD5 Tet-OFF strain (Fig. 5B). Addition of ATc significantly reduced the growth of the ΔpstA1 eccD5 Tet-OFF strain at days 6 and 9, but by day 15, we observed a significant increase in the number of viable CFU recovered from these cultures (Fig. 5D). Both eccD5 Tet-OFF strains produced smaller colonies when grown on 7H10 medium with ATc (Fig. S3). Importantly, the addition of ATc did not affect the growth kinetics or colony morphology of the WT or ΔpstA1 parent strain (Fig. S3 and S4). Quantitative reverse transcription-PCR (qRT-PCR) confirmed that addition of ATc to the eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF strains resulted in significant 2.7-fold and 3.6-fold repression of eccD5, respectively (Fig. 5E). This repression did not dramatically alter the expression of other genes within the ESX-5 locus, such as espG5, or the expression of lpqH itself (Fig. 5E). To confirm that reduced eccD5 transcription altered EccD5 protein production and ESX-5 secretion system activity, we performed Western blotting. EccD5 production was reduced 2.4-fold and 3.5-fold in the eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF strains, respectively, in the presence of ATc (Fig. 6A). These results suggest that eccD5 expression is not fully repressed by ATc in the Tet-OFF strains and that sufficient EccD5 remains to support growth, even when the cultures contain ATc.
FIG 5 .
Incomplete repression of eccD5 transcription has moderate effects on growth. (A to D) Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, eccD5 Tet-OFF, and ΔpstA1 eccD5 Tet-OFF strains were inoculated in 7H9 complete medium at an OD600 of 0.05 and grown at 37°C with aeration. Anhydrotetracycline hydrochloride (ATc; 100 ng/ml) was added at day 0 and day 7 as indicated. Growth was monitored by daily OD600 measurements (A and C) and by plating serially diluted cultures on 7H10 medium to determine numbers of viable CFU per milliliter on days 0, 3, 6, 9, 12, and 15 (B and D). The key in panel B applies to panels A and B; the key in panel D applies to panels C and D. **, P < 0.01; ***, P < 0.001; ‡, P = 0.057. (E) The transcript abundances of eccD5, espG5, and lpqH relative to that of sigA were determined by quantitative RT-PCR for the WT, ΔpstA1, eccD5 Tet-OFF, and ΔpstA1 eccD5 Tet-OFF strains grown to mid-logarithmic phase in 7H9 complete medium with and without ATc. Results are the means ± standard deviations for three independent experiments. *, P = 0.0167, **; P = 0.0042; ***, P = 0.0005.
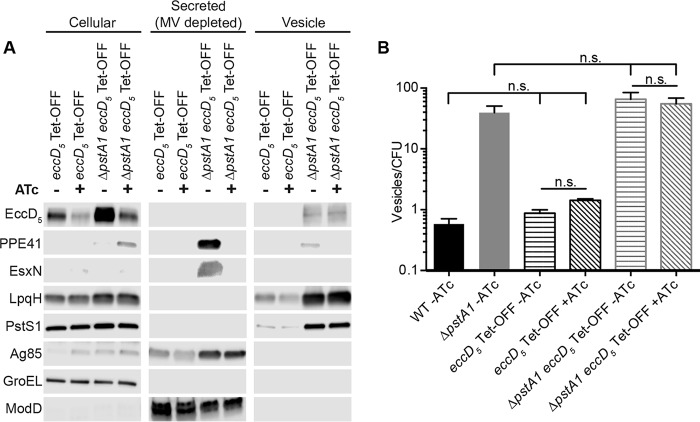
FIG 6 .
Depletion of EccD5 prevents ESX-5 secretion but does not affect membrane vesicle production. The eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF mutants were grown for 5 days in Sauton’s complete medium without Tween 80. Anhydrotetracycline hydrochloride (ATc; 100 ng/ml) was added as indicated to repress eccD5. (A) Cellular proteins (5 µg), secreted proteins depleted of MV (5 µg), and MV suspension (20 µl) were analyzed by Western blotting to detect the indicated proteins. (B) Nanoparticle tracking analysis of culture supernatants. Numbers of particles per milliliter were normalized to numbers of CFU per milliliter determined from a control culture of each strain grown with Tween 80 and plated on 7H10 complete medium. The data are means ± standard deviations for three independent cultures.
Repression of eccD5 results in small-colony morphology. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, eccD5 Tet-OFF, and ΔpstA1 eccD5 Tet-OFF strains were inoculated in 7H9 complete medium at an OD600 of 0.05. Cultures were serially diluted, and 10 µl of the indicated dilutions were spot plated on 7H10 complete medium with or without ATc (100 ng/ml). Download FIG S3, PDF file, 10.4 MB (10.6MB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ATc does not affect the growth of WT or ΔpstA1 bacteria. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1 strain were inoculated in 7H9 complete medium at an OD600 of 0.05 and grown at 37°C with aeration. Anhydrotetracycline hydrochloride (ATc; 100 ng/ml) was added at day 0 and day 7 as indicated. Growth was monitored by daily OD600 measurements (A and C) and by plating serially diluted cultures on 7H10 medium to determine numbers of viable CFU per milliliter on days 0, 3, 6, 9, 12, and 15 (B and D). Download FIG S4, PDF file, 0.5 MB (547.7KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We observed typical hyper-secretion of both EsxN and PPE41 from the ΔpstA1 eccD5 Tet-OFF strain in the absence of ATc, but secretion of these proteins was abrogated when ATc was added (Fig. 6A). Treatment of the ΔpstA1 eccD5 Tet-OFF strain with ATc also caused an increase in PPE41 associated with the cellular fraction (Fig. 6A), indicating that PPE41 becomes trapped within the cell when ESX-5 secretion is prevented. We were able to faintly detect PPE41 only in the MV fraction of the untreated ΔpstA1 eccD5 Tet-OFF strain, further supporting our finding that the majority of secreted EsxN and PPE41 is not MV associated. Other secreted proteins, such as the antigen 85 (Ag85) complex and ModD, were not affected by the repression of eccD5 (Fig. 6A). Taken together, these analyses confirm that the Tet-OFF strains function as expected and demonstrate that repression of eccD5 results in reduced secretion through ESX-5.
The ESX-5 secretion system is not required for membrane vesicle release.
We predicted that repression of eccD5 transcription in the ΔpstA1 eccD5 Tet-OFF strain would prevent MV hyper-secretion. MV were isolated from eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF strains as described above but with the addition of ATc to the indicated cultures during the final growth phase. Western blots for LpqH and PstS1 demonstrated that MV were hyper-secreted from the ΔpstA1 eccD5 Tet-OFF strain in similar amounts whether or not ATc was added to the cultures (Fig. 6A). NTA confirmed that there was no significant difference in the numbers of MV released between samples with and without ATc treatment or between the Tet-OFF strains and the corresponding parental control (Fig. 6B). These findings refute our initial hypothesis and instead indicate that ESX-5 secretion system activity is not required for MV release from M. tuberculosis.
DISCUSSION
M. tuberculosis produces MV, but the molecular mechanisms and regulatory processes controlling their release are poorly characterized. We demonstrate that the ΔpstA1 mutant hyper-secretes MV containing the lipoproteins LpqH and PstS1 in a RegX3-dependent manner. Because the ΔpstA1 mutant also exhibits a RegX3-dependent increase in the activity of the ESX-5 secretion system and ESX-5 substrates had previously been identified within MV, we initially hypothesized that MV release and ESX-5 activity were connected. However, the overproduction of MV from the ΔpstA1 eccD5 Tet-OFF strain was not affected by repressing ESX-5 secretion. Our data demonstrate that the Pst/SenX3-RegX3 system regulates the production of MV independently of ESX-5 activity.
Our data suggest that one or more factors regulated by RegX3 causes increased MV release from the ΔpstA1 mutant. Previous work implicated virR as a regulator of MV production (8), but we observed only modest, insignificant changes in virR expression in ΔpstA1 bacteria compared to its expression in the WT. Furthermore, deletion of virR in the ΔpstA1 background resulted in an even greater increase in MV production, suggesting that these genes promote MV production through separate mechanisms that are capable of synergizing with each other. Overexpression of virR in the ΔpstA1 background also resulted in a slight increase in MV production, providing further support for this hypothesis and suggesting that dramatic overproduction of VirR may also influence MV production. Together, these findings indicate that MV production is mediated by a novel mechanism in ΔpstA1 bacteria. Over 60 genes are dysregulated in the ΔpstA1 mutant in a RegX3-dependent manner (23), and any one or more of these genes may be involved in MV release. Our future studies will seek to identify the basis for increased MV release by the ΔpstA1 mutant to provide mechanistic insight into MV biogenesis.
By carefully examining the RegX3 regulon, we developed several hypotheses to explain increased MV production from the ΔpstA1 mutant. Several highly upregulated genes in the ΔpstA1 mutant are involved, or predicted to be involved, in the production of lipoproteins and glycolipids. One of the most upregulated genes, lppF, was overexpressed 20-fold in the ΔpstA1 mutant and is predicted to encode a lipoprotein (23). While LppF has not been detected in association with MV, several other lipoproteins, including LpqH, LppX, LprA, LprG, and LprF, are highly abundant (24). Additionally, rv0557 (mgtA) transcript levels are almost 14-fold higher in ΔpstA1 bacteria (23). The Corynebacterium glutamicum ortholog of this gene is required for production of a novel glycolipid, 1,2-di-O-C16/C18:1-(α-d-mannopyranosyl)-(1→4)-(α-d-glucopyranosyluronic acid)-(1→3)-glycerol (ManGlcAGroAc2), that contributes to the lipomannan (LM) pool (30). In M. tuberculosis, inactivation of rv0557 results in reduced cell wall LAM and LM (31). As both LAM and LM are incorporated in MV (2, 4), it is possible that changes in the production of these lipoglycans influence MV release. Two genes encoding putative acyltransferases (rv3027c and rv3026c) are also upregulated in the ΔpstA1 mutant 10- and 7-fold, respectively (23). These genes may contribute to increased lipid production, resulting in more MV biogenesis. Finally, rv1491c is also upregulated 3-fold in the ΔpstA1 mutant (23) and encodes a DedA family protein. In E. coli, deletion of genes encoding DedA family proteins causes cell division defects and altered membrane phospholipid composition (32). We hypothesize that Rv1491c has similar roles in M. tuberculosis membrane biogenesis and thus influences MV production. Additionally, Rv1491c is encoded near Rv1488, a putative membrane protein that interacts with VirR (8). It is possible that both Rv1491c and Rv1488 play roles in MV biogenesis. We intend to explore these hypotheses as part of our future studies.
We initially hypothesized that ESX-5 would be involved in MV release based in part on prior detection of the ESX-5 substrates PPE41 and EsxN in MV by mass spectrometry (2, 24), yet we were generally unable to detect these proteins in MV by Western blotting. We observed the majority of PPE41 and EsxN only in the secreted protein fraction and confirmed that their secretion requires EccD5. Our results indicate that PPE41 and EsxN are secreted primarily as soluble protein via ESX-5.
Our data also suggest that the ESX-5 core component EccD5 is essential for M. tuberculosis viability. We were unable to delete eccD5 unless a complementing copy of the gene was provided in trans, and we also could not efficiently replace the complementing copy of eccD5 with an empty vector. Similarly, both eccB5 and eccC5, which encode two additional core components of ESX-5, could not be deleted without a complementing copy of the appropriate gene provided in trans, suggesting that they are also essential for M. tuberculosis viability (33). Conserved ESX-5 components eccC5 and mycP5 also appear to be essential for the growth of the mycobacterial species M. marinum and M. bovis BCG (34). Others have reported deletion (20) or disruption (35) of eccD5, in contrast to our data. It is possible that these mutants were still viable due to compensatory mutations. Indeed, eccC5 could be deleted in M. marinum mutants deficient for production of the virulence lipid phthiocerol dimycocerosate (PDIM) (34), and spontaneous loss of PDIM production by M. tuberculosis has previously been reported (36). Overall, our data support a growing body of literature suggesting that ESX-5 secretion in general and the EccD5 core component in particular are essential for mycobacterial growth in vitro.
To circumvent the essentiality of EccD5, we generated conditional eccD5 Tet-OFF strains in both the WT and ΔpstA1 backgrounds. Addition of ATc to these strains repressed eccD5 transcription but caused only moderate growth defects either in liquid medium or on solid agar. EccD5 production was also decreased in both eccD5 Tet-OFF strains when ATc was added, although protein was still detectable by Western blotting. We hypothesize that the residual EccD5 produced by these strains was sufficient to support some growth. Nevertheless, depletion of EccD5 led to decreased ESX-5 activity, as indicated by reduced secretion of PPE41 and EsxN. Collectively, these data indicate that native EccD5 is necessary for M. tuberculosis viability and that lower eccD5 expression results in decreased growth and reduced protein secretion via ESX-5. Future work will use these eccD5 Tet-OFF strains to identify additional secreted substrates of the ESX-5 system and to investigate the role that ESX-5 secretion plays in pathogenesis.
MATERIALS AND METHODS
Strains and culture conditions.
M. tuberculosis Erdman and ΔpstA1, ΔpstA1 pMVpstA1, ΔpstA1 ΔregX3, and ΔpstA1 ΔregX3 pND-regX3 strains were previously described (23). Construction of the ΔvirR, ΔpstA1 ΔvirR, virR overexpression, eccD5 Tet-OFF, and ΔpstA1 eccD5 Tet-OFF strains are described below. Bacteria were routinely cultured in Middlebrook 7H9 liquid medium (Difco) supplemented with 10% albumin-dextrose-saline (ADS), 0.5% glycerol, and 0.1% Tween 80 or on Middlebrook 7H10 agar medium (Difco) supplemented with oleic acid-albumin-dextrose-catalase (OADC; BD Biosciences) and 0.5% glycerol. Strains containing the pvirR plasmid were grown in the presence of 50 µg/ml hygromycin B (Sigma). Frozen stocks were prepared by growing cultures to mid-logarithmic phase, adding glycerol to a 15% final concentration, and aliquoting for storage at −80°C.
Proteomic analysis.
M. tuberculosis WT and ΔpstA1 bacteria were grown in Sauton’s medium without Tween 80, and the secreted protein fractions were isolated as previously described (22). Total secreted proteins (10 µg) were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was fixed for 30 min in 40% ethanol-10% acetic acid and then stained with Imperial protein stain (Thermo Scientific) and destained in water. Bands of interest from three SDS-PAGE gel lanes of the ΔpstA1 mutant corresponding to 75, 50, 37, and 25 kDa were excised from the gel, pooled, cut into ~2- by 2-mm cubes, and processed by in-gel trypsin digestion. Gel slices were washed twice for 15 min at room temperature in wash buffer (1:1 acetonitrile-100 mM NH4HCO3) and then treated with 100% acetonitrile until the pieces shrank and turned white and semiopaque. Proteins were reduced with 10 mM dithiothreitol (DTT) in 10 mM NH4HCO3 for 1 h at 56°C and then alkylated by treatment with 55 mM iodoacetamide in 100 mM NH4HCO3 for 30 min at room temperature in the dark. Gel slices were washed twice with wash buffer and then washed briefly with 100% acetonitrile before rehydration in digestion buffer (50 mM NH4HCO3, 5 mM CaCl2, 12.5 ng/µl trypsin) at 4°C for 15 min, followed by overnight incubation in 50 mM NH4HCO3, 5 mM CaCl2 at 37°C. Peptides were extracted from the gel slices with 50% acetonitrile, 0.3% formic acid for 15 min and then with 80% acetonitrile, 0.3% formic acid for 15 min. Peptides were incubated at −80°C for 30 min, dried in a speed vac, and stored at −80°C prior to analysis by liquid chromatography-tandem mass spectrometry (LC/MS-MS) on an LTQ Orbitrap Velos mass spectrometer (Thermo, Fisher). The MS-MS data were compared with sequences in an M. tuberculosis proteomic database using Scaffold V4 software.
Purification of His6-tagged EccD51–131.
The portion of eccD5 encoding a predicted soluble cytoplasmic domain from residues 1 to 131 (EccD51–131) was amplified from M. tuberculosis Erdman genomic DNA by PCR with primers His-eccD5_F1 and His-eccD5_R1 and cloned into plasmid pET28b+ between the NdeI and HindIII restriction enzyme sites to generate pET28-His6EccD5, encoding EccD51–131 with an N-terminal His6 tag. The pET28-His6EccD5 plasmid was introduced into E. coli BL21(DE3), and the protein was purified by Ni2+-nitrilotriacetic acid (NTA) affinity chromatography (Qiagen) as previously described for PPE41-His6 (37, 38). Briefly, purified protein was bound to the column under native conditions in 20 mM HEPES buffer, 300 mM NaCl, pH 7.8, and eluted in 20 mM HEPES buffer, 500 mM NaCl, pH 7.8, containing 50 to 150 mM imidazole. Purified His6EccD5 was concentrated through a 5-kDa-cutoff Amicon Ultra centrifugal filtration unit (Millipore), and then contaminant proteins were removed by fast-protein liquid chromatography (FPLC) using a BioLogic DuoFlow apparatus (Bio-Rad). Protein was bound to an Enrich Sec 650 (Bio-Rad) column and eluted in phosphate-buffered saline (PBS) using an isocratic flow at a rate of 500 µl/min. Protein which eluted at 0.02 absorbance unit was collected and pooled.
Antiserum production.
Polyclonal antisera against purified His6-EccD51–131 were generated in rabbits by Pierce custom antibodies (Thermo Scientific) using TiterMax Gold adjuvant (Sigma). Polyclonal rabbit antisera against EsxN and PPE41 were previously described (38).
Preparation of membrane vesicles.
Thirty-milliliter M. tuberculosis cultures were grown in Sauton’s medium as previously described for analysis of secreted proteins (22). MV were isolated from culture supernatants as previously described, with minor modifications (2). Briefly, cells were pelleted by centrifugation (4,700 × g, 15 min, 4°C), and supernatants were sequentially filtered through 0.45-µm and 0.22-µm Millex syringe filters (Millipore). The supernatants were centrifuged (4,000 × g, 15 min, 4°C) to remove remaining cellular debris and then concentrated to approximately 1 ml using 100-kDa-cutoff Amicon Ultra centrifugal filtration devices (Millipore). The filtrates were further processed to concentrate secreted proteins using a 5-kDa-cutoff Amicon Ultra centrifugal filtration device, as previously described (22). The 100-kDa filter retentates were then centrifuged at 100,000 × g (75 min, 4°C) to pellet MV. The MV pellets were resuspended in 500 µl PBS containing Complete EDTA-free protease inhibitors (Roche).
Western blotting.
Cellular and secreted proteins were isolated from M. tuberculosis cultures grown in Sauton’s medium as previously described, with minor modifications (22). Briefly, bacteria were pelleted and culture supernatants were filter sterilized before MV were purified and secreted proteins were concentrated as described above. Pellets were resuspended in PBS containing Complete EDTA-free protease inhibitors (Roche) and subjected to bead beating to lyse the cells before they were filtered to remove intact cells. Cellular and secreted proteins were quantified via a bicinchoninic acid (BCA) assay (Pierce BCA protein assay kit; ThermoFisher). Five micrograms of cellular and secreted proteins was separated by SDS-PAGE and transferred to 0.2-µm nitrocellulose membranes (Bio-Rad) as previously described (22). The protein concentration in MV preparations was below the limit of detection of the BCA assay, so 20 µl of MV samples, corresponding to 1.2 ml of culture, was separated by SDS-PAGE. Membranes were blocked in PBS with 0.1% Tween 20 (PBST) containing 5% nonfat milk powder at room temperature for 1 to 2 h. Membranes were washed in PBST and then probed overnight at 4°C with primary antisera diluted in PBST containing 2.5% nonfat milk powder. Primary antisera were used at the following dilutions: mouse anti-KatG, 1:1,000; mouse anti-LpqH, 1:500; mouse anti-PstS1, 1:1,000; mouse anti-GroEL2, 1:1,000; rabbit anti-ModD, 1:25,000; rabbit anti-PPE41, 1:1,000; rabbit anti-EsxN 1:1,000; rabbit anti-EccD5, 1:1,000; and rabbit anti-VirR, 1:10,000. Membranes were again washed in PBST before incubation for 1 to 2 h at room temperature with the appropriate secondary antibody (either goat anti-rabbit or rabbit anti-mouse antibody conjugated to horseradish peroxidase; Sigma) diluted 1:30,000 (1:5,000 when used to detect VirR) in PBST containing 2.5% nonfat milk powder. Membranes were washed again in PBST, and bands were detected using SuperSignal West Pico chemiluminescent substrate (Thermo Scientific). Blots were imaged using an Odyssey Fc imaging system (LI-COR), and protein abundance was analyzed using Image Studio software (LI-COR).
Vesicle quantification.
Filter-sterilized culture supernatants from M. tuberculosis strains grown in Sauton’s medium without Tween 80 were analyzed with a NanoSight instrument to determine vesicle numbers and sizes. Culture filtrates were diluted in a 1-ml final volume of 1× PBS (Corning) to a concentration acceptable for analysis by the NanoSight NS300 (Malvern Instruments, United Kingdom). Triplicate videos of each sample were taken at 24.5°C in light scatter mode using the equipped 532-nm green laser and a syringe pump. Particle displacement was detected with a camera level of 14, and analyses were performed using NanoSight 3.0 or 3.1 software and a threshold of between 3 and 5. Triplicate video statistics were averaged for each sample. A control culture of each strain grown in Sauton’s medium with Tween 80 was serially diluted and plated on 7H10 medium in duplicate. Colonies were counted after 3 weeks of incubation at 37°C to determine numbers of CFU per milliliter, and the number of particles per milliliter was normalized to this value to determine the number of vesicles per CFU.
EM of purified vesicles.
All reagents were electron microscopy (EM) grade and were purchased from Electron Microscopy Supply Co. Transmission electron microscopy (TEM) was used to visualize MV purified from 600-ml cultures grown as described above. Formvar-coated copper grids were floated on 20-µl drops of purified MV solutions for 20 min. The grids were washed in water and then floated on 1% uranyl acetate for 30 s. The grids were washed again before being imaged on a FEI Tecnai Spirit Bio-Twin.
Construction of virR deletion and overexpression strains.
Sequences of all primers used for cloning and strain construction are provided in Table S2 in the supplemental material. A vector for the deletion of base pairs 252 to 306 of virR was constructed in the allelic-exchange plasmid pJG1100, which contains the aph (kanamycin [Kan] resistance), hyg (hygromycin [Hyg] resistance), and sacB (sucrose sensitivity) markers (36). Regions of the M. tuberculosis genome ~900 bp 5′ and 3′ of the deletion site were amplified by PCR with primers virR_F2/virR_R6 and virR_F6/virR_R3. The virR_R6 primer to amplify the 5′ region of virR was designed with an AvrII restriction site in frame with the virR start codon, and the virR_F6 primer used to amplify the 3′ region of virR was designed with an AvrII restriction site in frame with the stop codon. The resulting construct encodes a copy of virR lacking 54 bp. The PCR products were cloned into PCR2.1-TOPO (Invitrogen) and sequenced. The 5′ and 3′ homology regions were subsequently removed from the pCR2.1 vector by digestion with PacI/AvrII or AvrII/AscI, respectively, and cloned in pJG1100 digested with PacI/AscI. The resulting pJG-ΔvirR vector was confirmed by sequencing. Plasmids for the overexpression of FLAG-tagged virR (pvirR) were previously described (8) and generously provided by Carl Nathan.
Oligonucleotide primers used for cloning, strain construction, or qRT-PCR. Download TABLE S2, DOCX file, 0.1 MB (109.7KB, docx) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The pJG-ΔvirR and pvirR vectors were introduced into WT and ΔpstA1 bacteria by electroporation as previously described (23). Transformants containing pvirR were selected by plating them on complete 7H10 containing 50 µg/ml Hyg. The presence of pvirR was confirmed by PCR with the primer pair pDE43_F/pDE43_R. Transformants containing pJG-ΔvirR were selected by plating them on complete 7H10 containing 50 µg/ml Hyg and 15 µg/ml Kan. Integration of pJG-ΔvirR at the virR locus was confirmed using primers virR_F4/PJGR and PJGF/virR_R4. Transformants that had integrated the plasmid were grown in complete 7H9 medium before serial dilution and plating on complete 7H10 medium containing 2% sucrose to counterselect the pJG-ΔvirR plasmid. Sucrose-resistant isolates were screened by PCR with the primer pair virR_F4/virR_R4 to ensure loss of the pJG-ΔvirR plasmid, and the 54-bp deletion was confirmed by sequencing.
Cloning of eccD5 deletion and conditional expression vectors.
A vector for deletion of eccD5 was constructed in the allelic-exchange plasmid pJG1100. Regions of the M. tuberculosis genome ~800 bp 5′ and 3′ of eccD5 were amplified by PCR with primer pairs eccD5F1/eccD5R1 and eccD5F2/eccD5R2. The eccD5R1 primer to amplify the 5′ region was designed with an AvrII restriction site in frame with the eccD5 translation start codon. The eccD5F2 primer to amplify the 3′ region was designed with an AvrII restriction site in frame with the eccD5 stop codon. To avoid polarity of the final construct upon expression of mycP5, which overlaps the eccD5 gene at the 3′ end, the eccD5R2 primer was designed 5′ of the predicted mycP5 start codon. The resulting construct encodes the first 5 amino acids of EccD5 fused in frame to the last 16 amino acids of EccD5. The PCR products were cloned in pCR2.1-TOPO (Invitrogen) and sequenced. The 5′ and 3′ homology regions were subsequently removed from the pCR2.1 vector by digestion with PacI/AvrII or AvrII/AscI, respectively, and cloned in pJG1100 digested with PacI/AscI. The resulting pJG-ΔeccD5 vector was confirmed by sequencing.
For tetracycline-inducible expression, eccD5 was cloned in pTIC10a, a derivative of the integrative pTIC6 vector (39), which contains a codon-optimized Tet repressor expressed under the control of the constitutive mycobacterial groEL promoter, the aph Kan resistance marker, and the Psmyc-tetO mycobacterial promoter and Tet operator 5′ of the multicloning site. Full-length eccD5 was amplified from M. tuberculosis Erdman genomic DNA by PCR with primers Tet-eccD5F and Tet-eccD5R, cloned in pCR2.1-TOPO (Invitrogen), and sequenced. The eccD5 insert was removed by digestion with HindIII and EcoRI and cloned in similarly digested pTIC10a. The resulting pTIC-eccD5 vector was confirmed by sequencing.
A vector for tetracycline-repressible expression of eccD5 was constructed as previously described using Gateway vectors generously provided by Dirk Schnappinger (40). eccD5 was amplified using primers eccD5_P1, which adds an attB2 site upstream of the eccD5 promoter sequence, and eccD5_P4, which adds an attB3 site downstream of the eccD5 stop codon, before being cloned into the pDO23A entry vector via a BP Gateway reaction (Gateway BP clonase II enzyme mix; ThermoFisher) to create pEN23A-eccD5. The final pGMCH-T38S38-750-eccD5 vector was constructed via an LR Gateway reaction (Gateway LR clonase II enzyme mix; ThermoFisher) using plasmids pDE43-MCH, pEN41A-T38S38, pEN12A-P750, and pEN23A-eccD5 and confirmed by sequencing. A similar strategy was used to construct a vector for the expression of eccD5-DAS containing the DAS+4 degradation tag (29). eccD5-DAS was amplified using primers eccD5_P1, eccD5_P2, and eccD5_P3. The eccD5_P2 and eccD5_P3 primers were used in sequential PCRs to add the DAS+4 tag with a stop codon and introduce an attB3 site as described previously (40). Gateway reactions to clone eccD5-DAS into pDO23A and to construct the final pGMCH-T38S38-750-eccD5-DAS plasmid were performed as described above, and the final plasmid was confirmed by sequencing.
Construction of a conditional eccD5 depletion strain.
The pTIC-eccD5 vector was introduced into WT and ΔpstA1 mutant M. tuberculosis by electroporation as described previously (23), and transformants were selected by plating them on complete 7H10 agar containing 15 µg/ml Kan. The presence of the plasmid in transformants was verified by PCR with the primer pair pTseqF/Q95R1. The resulting WT pTIC-eccD5 and ΔpstA1 pTIC-eccD5 strains were subsequently electroporated with pJGΔeccD5, and transformants were selected on 7H10 agar containing 50 µg/ml Hyg and 15 µg/ml Kan. Integration of the pJG-ΔeccD5 vector at the eccD5 locus in transformants was confirmed by PCR using primers Q94F1/mycP5R1 and Rv1794_3′F/Q96R1 to detect integration via the 5′ and 3′ regions of homology, respectively. Isolates in which the plasmid integrated site specifically were then grown in 7H9 supplemented with 15 µg/ml Kan and 50 ng/ml anhydrotetracycline hydrochloride (ATc; Sigma) to induce eccD5 expression from pTIC-eccD5 and allow excision of pJGΔeccD5. The culture was serially diluted and plated on 7H10 agar containing 15 µg/ml Kan, 100 ng/ml ATc, and 2% sucrose to counterselect the pJGΔeccD5 plasmid. Sucrose-resistant isolates were patched to 7H10 medium containing 50 µg/ml Hyg to identify those that had excised pJGΔeccD5. Hyg-sensitive colonies were grown in 7H9 medium containing 15 µg/ml Kan and 50 ng/ml ATc and tested by PCR for the ΔeccD5 mutation using primers Q94F1 and Q96R1. These strains were designated eccD5 Tet-ON and ΔpstA1 eccD5 Tet-ON.
The pGMCH-T38S38-750-eccD5 vector was introduced into eccD5 Tet-ON and ΔpstA1 ΔeccD5 Tet-ON bacteria by electroporation, and transformants were selected on complete 7H10 medium containing 50 µg/ml Hyg. Transformants were patched onto 7H10 medium containing 50 µg/ml Hyg and 7H10 medium containing 15 µg/ml Kan. Colonies that grew in the presence of Hyg but not in the presence of Kan were screened by PCR to ensure that the strains harbored the hyg cassette (primers hygro_forward and hygro_reverse) and the pGMCH-eccD5 vector (primers pGMCH_D5_F and pGMCH_Rev) and had lost the pTIC10a-eccD5 vector via a plasmid swap (primers pTIC_KanR_For and pTIC_KanR_Rev). These strains were designated eccD5 Tet-OFF and ΔpstA1 eccD5 Tet-OFF.
Growth curves.
Frozen bacterial stocks were inoculated into complete 7H9 medium and grown to mid-logarithmic phase (optical density at 600 nm [OD600] = 0.6). Cultures were diluted to an OD600 of 0.05 in 7H9 medium with and without 100 ng/ml ATc, and OD600 measurements were taken daily for 15 days. Every 3 days, aliquots of each culture were serially diluted, plated on 7H10 agar, and incubated at 37°C for 3 to 4 weeks to determine numbers of viable CFU.
qRT-PCR.
Bacteria were inoculated into 7H9 medium at an OD600 of 0.1, and ATc (100 ng/ml) was added where indicated in the figures. Cultures were grown at 37°C with aeration to late-logarithmic phase (OD600s = 0.6 to 0.8) before extraction of RNA and reverse transcription to cDNA as previously described (22, 23). Quantitative PCR primers to amplify an internal region of genes of interest (eccD5, espG5, lpqH, virR, sigA) were designed with similar melting temperatures (58 to 60°C) using Primer Express software (Applied Biosystems) or ProbeFinder Assay Design software (Roche). Sequences of these primers can be found in Table S2. qRT-PCRs were carried out on a LightCycler 480 (Roche), and gene copy numbers were normalized to that of sigA as previously described (22).
Statistical analysis.
All statistical analyses were performed using GraphPad Prism6 software. P values of <0.05 were considered significant. Statistics for Fig. 2B, 3B, 4B, and 6B were calculated by ordinary one-way analysis of variance (ANOVA) with Holm-Sidak’s multiple-comparison test applied post hoc. Statistics for Fig. 3C, 5A, and 5B were calculated by unpaired t test between the indicated strains.
ACKNOWLEDGMENTS
We thank Dirk Schnappinger for providing the pDO23A, pDE43-MCH, pEN41A-T38S38, and pEN12A-P750 plasmids used to construct the eccD5 Tet-OFF plasmid, Carl Nathan for providing the pvirR overexpression plasmid and VirR primary antibody, Anthony Baughn for providing plasmids pTIC10a and pJT6a, Ryan Hunter for assistance with electron microscopy, Shannon Kordus for assistance with FPLC, Kelsey Binder for assistance with pTIC-eccD5 plasmid construction, and Alyssa Brokaw for constructing the pJGΔeccD5 plasmid. The following reagents were obtained through BEI Resources, NIAID, NIH: monoclonal anti-Mycobacterium tuberculosis GroEL2 (Rv0440), clone IT-70 (DCA4) (produced in vitro), catalog no. NR-13657; polyclonal anti-Mycobacterium tuberculosis Mpt32 (Rv1860) (antiserum, rabbit), catalog no. NR-13807; polyclonal anti-Mycobacterium tuberculosis antigen 85 complex (FbpA/Fbp/FbpC; Rv3804c, Rv1886c, and Rv0129c) (antiserum, rabbit), catalog no. NR-13800; monoclonal anti-Mycobacterium tuberculosis KatG (Rv1908c), clone IT-42 (HBT1) (produced in vitro), catalog no. NR-13791; monoclonal anti-Mycobacterium tuberculosis LpqH (Rv3763), clone IT-54 (produced in vitro), catalog no. NR-13792; and monoclonal anti-Mycobacterium tuberculosis PhoS1/PstS1 (Rv0934) clone IT-23 (TB71) (produced in vitro), catalog no. NR-13657.
This work was supported by an NIH Director’s New Innovator Award (1DP2AI112245 to A.D.T.) and institutional startup funds from the University of Minnesota (A.D.T. and L.T.B.). The purchase of the NanoSight NS300 was partially supported by an equipment grant from the University of Minnesota Foundation (L.T.B.). Parts of this work were carried out in the Characterization Facility, University of Minnesota, which receives partial support from the NSF through the MRSEC program.
Footnotes
Citation White DW, Elliott SR, Odean E, Bemis LT, Tischler AD. 2018. Mycobacterium tuberculosis Pst/SenX3-RegX3 regulates membrane vesicle production independently of ESX-5 activity. mBio 9:e00778-18. https://doi.org/10.1128/mBio.00778-18.
REFERENCES
- 1.Beveridge TJ. 1999. Structures of Gram-negative cell walls and their derived membrane vesicles. J Bacteriol 181:4725–4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, Camara C, Nosanchuk JD, Besra GS, Chen B, Jimenez J, Glatman-Freedman A, Jacobs WR Jr, Porcelli SA, Casadevall A. 2011. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest 121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown L, Wolf JM, Prados-Rosales R, Casadevall A. 2015. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 13:620–630. doi: 10.1038/nrmicro3480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Athman JJ, Sande OJ, Groft SG, Reba SM, Nagy N, Wearsch PA, Richardson ET, Rojas R, Boom WH, Shukla S, Harding CV. 2017. Mycobacterium tuberculosis membrane vesicles inhibit T cell activation. J Immunol 198:2028–2037. doi: 10.4049/jimmunol.1601199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lancioni CL, Li Q, Thomas JJ, Ding X, Thiel B, Drage MG, Pecora ND, Ziady AG, Shank S, Harding CV, Boom WH, Rojas RE. 2011. Mycobacterium tuberculosis lipoproteins directly regulate human memory CD4(+) T cell activation via Toll-like receptors 1 and 2. Infect Immun 79:663–673. doi: 10.1128/IAI.00806-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noss EH, Pai RK, Sellati TJ, Radolf JD, Belisle J, Golenbock DT, Boom WH, Harding CV. 2001. Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J Immunol 167:910–918. doi: 10.4049/jimmunol.167.2.910. [DOI] [PubMed] [Google Scholar]
- 7.Prados-Rosales R, Carreño LJ, Batista-Gonzalez A, Baena A, Venkataswamy MM, Xu J, Yu X, Wallstrom G, Magee DM, LaBaer J, Achkar JM, Jacobs WR Jr, Chan J, Porcelli SA, Casadevall A. 2014. Mycobacterial membrane vesicles administered systemically in mice induce a protective immune response to surface compartments of Mycobacterium tuberculosis. mBio 5:e01921-14. doi: 10.1128/mBio.01921-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rath P, Huang C, Wang T, Wang T, Li H, Prados-Rosales R, Elemento O, Casadevall A, Nathan CF. 2013. Genetic regulation of vesiculogenesis and immunomodulation in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 110:E4790–E4797. doi: 10.1073/pnas.1320118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prados-Rosales R, Weinrick BC, Piqué DG, Jacobs WR Jr, Casadevall A, Rodriguez GM. 2014. Role for Mycobacterium tuberculosis membrane vesicles in iron acquisition. J Bacteriol 196:1250–1256. doi: 10.1128/JB.01090-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McBroom AJ, Johnson AP, Vemulapalli S, Kuehn MJ. 2006. Outer membrane vesicle production by Escherichia coli is independent of membrane instability. J Bacteriol 188:5385–5392. doi: 10.1128/JB.00498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kulp AJ, Sun B, Ai T, Manning AJ, Orench-Rivera N, Schmid AK, Kuehn MJ. 2015. Genome-wide assessment of outer membrane vesicle production in Escherichia coli. PLoS One 10:e0139200. doi: 10.1371/journal.pone.0139200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majlessi L, Prados-Rosales R, Casadevall A, Brosch R. 2015. Release of mycobacterial antigens. Immunol Rev 264:25–45. doi: 10.1111/imr.12251. [DOI] [PubMed] [Google Scholar]
- 13.Houben D, Demangel C, van Ingen J, Perez J, Baldeón L, Abdallah AM, Caleechurn L, Bottai D, van Zon M, de Punder K, van der Laan T, Kant A, Bossers-de Vries R, Willemsen P, Bitter W, van Soolingen D, Brosch R, van der Wel N, Peters PJ. 2012. ESX-1-mediated translocation to the cytosol controls virulence of mycobacteria. Cell Microbiol 14:1287–1298. doi: 10.1111/j.1462-5822.2012.01799.x. [DOI] [PubMed] [Google Scholar]
- 14.Wassermann R, Gulen MF, Sala C, Perin SG, Lou Y, Rybniker J, Schmid-Burgk JL, Schmidt T, Hornung V, Cole ST, Ablasser A. 2015. Mycobacterium tuberculosis differentially activates cGAS- and inflammasome-dependent intracellular immune responses through ESX-1. Cell Host Microbe 17:799–810. doi: 10.1016/j.chom.2015.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Manzanillo PS, Shiloh MU, Portnoy DA, Cox JS. 2012. Mycobacterium tuberculosis activates the DNA-dependent cytosolic surveillance pathway within macrophages. Cell Host Microbe 11:469–480. doi: 10.1016/j.chom.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A 106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tufariello JM, Chapman JR, Kerantzas CA, Wong KW, Vilchèze C, Jones CM, Cole LE, Tinaztepe E, Thompson V, Fenyö D, Niederweis M, Ueberheide B, Philips JA, Jacobs WR Jr.. 2016. Separable roles for Mycobacterium tuberculosis ESX-3 effectors in iron acquisition and virulence. Proc Natl Acad Sci U S A 113:E348–E357. doi: 10.1073/pnas.1523321113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdallah AM, Bestebroer J, Savage ND, de Punder K, van Zon M, Wilson L, Korbee CJ, van der Sar AM, Ottenhoff TH, van der Wel NN, Bitter W, Peters PJ. 2011. Mycobacterial secretion systems ESX-1 and ESX-5 play distinct roles in host cell death and inflammasome activation. J Immunol 187:4744–4753. doi: 10.4049/jimmunol.1101457. [DOI] [PubMed] [Google Scholar]
- 19.Abdallah AM, Verboom T, Hannes F, Safi M, Strong M, Eisenberg D, Musters RJ, Vandenbroucke-Grauls CM, Appelmelk BJ, Luirink J, Bitter W. 2006. A specific secretion system mediates PPE41 transport in pathogenic mycobacteria. Mol Microbiol 62:667–679. doi: 10.1111/j.1365-2958.2006.05409.x. [DOI] [PubMed] [Google Scholar]
- 20.Bottai D, Di Luca M, Majlessi L, Frigui W, Simeone R, Sayes F, Bitter W, Brennan MJ, Leclerc C, Batoni G, Campa M, Brosch R, Esin S. 2012. Disruption of the ESX-5 system of Mycobacterium tuberculosis causes loss of PPE protein secretion, reduction of cell wall integrity and strong attenuation. Mol Microbiol 83:1195–1209. doi: 10.1111/j.1365-2958.2012.08001.x. [DOI] [PubMed] [Google Scholar]
- 21.Beckham KS, Ciccarelli L, Bunduc CM, Mertens HD, Ummels R, Lugmayr W, Mayr J, Rettel M, Savitski MM, Svergun DI, Bitter W, Wilmanns M, Marlovits TC, Parret AH, Houben EN. 2017. Structure of the mycobacterial ESX-5 type VII secretion system membrane complex by single-particle analysis. Nat Microbiol 2:17047. doi: 10.1038/nmicrobiol.2017.47. [DOI] [PubMed] [Google Scholar]
- 22.Elliott SR, Tischler AD. 2016. Phosphate starvation: a novel signal that triggers ESX-5 secretion in Mycobacterium tuberculosis. Mol Microbiol 100:510–526. doi: 10.1111/mmi.13332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tischler AD, Leistikow RL, Kirksey MA, Voskuil MI, McKinney JD. 2013. Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect Immun 81:317–328. doi: 10.1128/IAI.01136-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee J, Kim SH, Choi DS, Lee JS, Kim DK, Go G, Park SM, Kim SH, Shin JH, Chang CL, Gho YS. 2015. Proteomic analysis of extracellular vesicles derived from Mycobacterium tuberculosis. Proteomics 15:3331–3337. doi: 10.1002/pmic.201500037. [DOI] [PubMed] [Google Scholar]
- 25.Dobos KM, Swiderek K, Khoo KH, Brennan PJ, Belisle JT. 1995. Evidence for glycosylation sites on the 45-kilodalton glycoprotein of Mycobacterium tuberculosis. Infect Immun 63:2846–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramakrishnan P, Aagesen AM, McKinney JD, Tischler AD. 2015. Mycobacterium tuberculosis resists stress by regulating PE19 expression. Infect Immun 84:735–746. doi: 10.1128/IAI.00942-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Namugenyi SB, Aagesen AM, Elliott SR, Tischler AD. 2017. Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating Pst/SenX3-RegX3 phosphate sensing. mBio 8:e00494-17. doi: 10.1128/mBio.00494-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stoop EJ, Bitter W, van der Sar AM. 2012. Tubercle bacilli rely on a type VII army for pathogenicity. Trends Microbiol 20:477–484. doi: 10.1016/j.tim.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Kim JH, O’Brien KM, Sharma R, Boshoff HI, Rehren G, Chakraborty S, Wallach JB, Monteleone M, Wilson DJ, Aldrich CC, Barry CE III, Rhee KY, Ehrt S, Schnappinger D. 2013. A genetic strategy to identify targets for the development of drugs that prevent bacterial persistence. Proc Natl Acad Sci U S A 110:19095–19100. doi: 10.1073/pnas.1315860110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tatituri RV, Illarionov PA, Dover LG, Nigou J, Gilleron M, Hitchen P, Krumbach K, Morris HR, Spencer N, Dell A, Eggeling L, Besra GS. 2007. Inactivation of Corynebacterium glutamicum NCgl0452 and the role of MgtA in the biosynthesis of a novel mannosylated glycolipid involved in lipomannan biosynthesis. J Biol Chem 282:4561–4572. doi: 10.1074/jbc.M608695200. [DOI] [PubMed] [Google Scholar]
- 31.Torrelles JB, DesJardin LE, MacNeil J, Kaufman TM, Kutzbach B, Knaup R, McCarthy TR, Gurcha SS, Besra GS, Clegg S, Schlesinger LS. 2009. Inactivation of Mycobacterium tuberculosis mannosyltransferase pimB reduces the cell wall lipoarabinomannan and lipomannan content and increases the rate of bacterial-induced human macrophage cell death. Glycobiology 19:743–755. doi: 10.1093/glycob/cwp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doerrler WT, Sikdar R, Kumar S, Boughner LA. 2013. New functions for the ancient DedA membrane protein family. J Bacteriol 195:3–11. doi: 10.1128/JB.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Di Luca M, Bottai D, Batoni G, Orgeur M, Aulicino A, Counoupas C, Campa M, Brosch R, Esin S. 2012. The ESX-5 associated eccB5-eccC5 locus is essential for Mycobacterium tuberculosis viability. PLoS One 7:e52059. doi: 10.1371/journal.pone.0052059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ates LS, Ummels R, Commandeur S, van de Weerd R, Sparrius M, Weerdenburg E, Alber M, Kalscheuer R, Piersma SR, Abdallah AM, Abd El Ghany M, Abdel-Haleem AM, Pain A, Jiménez CR, Bitter W, Houben EN. 2015. Essential role of the ESX-5 secretion system in outer membrane permeability of pathogenic mycobacteria. PLoS Genet 11:e1005190. doi: 10.1371/journal.pgen.1005190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jiménez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol 86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 36.Kirksey MA, Tischler AD, Siméone R, Hisert KB, Uplekar S, Guilhot C, McKinney JD. 2011. Spontaneous phthiocerol dimycocerosate-deficient variants of Mycobacterium tuberculosis are susceptible to gamma interferon-mediated immunity. Infect Immun 79:2829–2838. doi: 10.1128/IAI.00097-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strong M, Sawaya MR, Wang S, Phillips M, Cascio D, Eisenberg D. 2006. Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barczak AK, Avraham R, Singh S, Luo SS, Zhang WR, Bray MA, Hinman AE, Thompson M, Nietupski RM, Golas A, Montgomery P, Fitzgerald M, Smith RS, White DW, Tischler AD, Carpenter AE, Hung DT. 2017. Systematic, multiparametric analysis of Mycobacterium tuberculosis intracellular infection offers insight into coordinated virulence. PLoS Pathog 13:e1006363. doi: 10.1371/journal.ppat.1006363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glover RT, Kriakov J, Garforth SJ, Baughn AD, Jacobs WR Jr.. 2007. The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J Bacteriol 189:5495–5503. doi: 10.1128/JB.00190-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schnappinger D, O’Brien KM, Ehrt S. 2015. Construction of conditional knockdown mutants in mycobacteria. Methods Mol Biol 1285:151–175. doi: 10.1007/978-1-4939-2450-9_9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomic mass spectrometry analysis of proteins hyper-secreted by the ΔpstA1 mutant. Proteins analyzed by Western blotting are highlighted in yellow. Download TABLE S1, XLSX file, 0.7 MB (685.3KB, xlsx) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The ΔpstA1 ΔregX3 and ΔpstA1 ΔregX3 pNDregX3 strains produce membrane vesicles. The ΔpstA1 ΔregX3 and ΔpstA1 ΔregX3 pNDregX3 strains were grown for 5 days in Sauton’s complete medium without Tween 80. Nanoparticle tracking analyses were performed on culture supernatants, and the resulting particle sizes were binned in 10-nm increments. Download FIG S1, PDF file, 0.5 MB (517.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Residual eccD5 expression in uninduced eccD5 Tet-ON strains is sufficient for protein secretion through ESX-5. Kanamycin (15 µg/ml) was added to all eccD5 Tet-ON cultures to maintain plasmid integration. (A and B) Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains were inoculated in 7H9 complete medium at an OD600 of 0.05 and grown at 37°C with aeration. Anhydrotetracycline hydrochloride (ATc; 50 ng/ml) was added every 3 days to the indicated cultures. Growth was monitored by daily OD600 measurements. (C) The transcript abundance of eccD5 relative to that of sigA was determined by quantitative RT-PCR for the WT, ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains grown to mid-logarithmic phase in 7H9 complete medium with and without ATc. (D) The WT, ΔpstA1, eccD5 Tet-ON, and ΔpstA1 eccD5 Tet-ON strains were grown for 5 days in Sauton’s complete medium without Tween 80. ATc (50 ng/ml) was added to the indicated cultures to induce eccD5. Equivalent amounts of cellular (5 µg) and secreted (5 µg) proteins were analyzed by Western blotting to detect the indicated proteins. Download FIG S2, PDF file, 0.8 MB (834.8KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Repression of eccD5 results in small-colony morphology. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1, eccD5 Tet-OFF, and ΔpstA1 eccD5 Tet-OFF strains were inoculated in 7H9 complete medium at an OD600 of 0.05. Cultures were serially diluted, and 10 µl of the indicated dilutions were spot plated on 7H10 complete medium with or without ATc (100 ng/ml). Download FIG S3, PDF file, 10.4 MB (10.6MB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ATc does not affect the growth of WT or ΔpstA1 bacteria. Wild-type M. tuberculosis Erdman (WT) and the ΔpstA1 strain were inoculated in 7H9 complete medium at an OD600 of 0.05 and grown at 37°C with aeration. Anhydrotetracycline hydrochloride (ATc; 100 ng/ml) was added at day 0 and day 7 as indicated. Growth was monitored by daily OD600 measurements (A and C) and by plating serially diluted cultures on 7H10 medium to determine numbers of viable CFU per milliliter on days 0, 3, 6, 9, 12, and 15 (B and D). Download FIG S4, PDF file, 0.5 MB (547.7KB, pdf) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotide primers used for cloning, strain construction, or qRT-PCR. Download TABLE S2, DOCX file, 0.1 MB (109.7KB, docx) .
Copyright © 2018 White et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.