ABSTRACT
The Epstein-Barr virus (EBV) oncoproteins latent membrane protein 1 (LMP1) and LMP2A constitutively activate multiple signaling pathways, and both have been shown to interact with cellular ubiquitin ligases and affect cellular ubiquitination. To detect the LMP1- and LMP2A-mediated effects on the global cellular proteome, epithelial cell lines expressing LMP1 or LMP2A were analyzed using label-free quantitative proteomics. To identify proteins whose ubiquitination is affected by the viral proteins, the cells were cultured in the presence and absence of deubiquitinase (DUB) and proteasome inhibitors. More than 7,700 proteins were identified with high confidence and considerably more proteins showed significant differences in expression in the presence of inhibitors. Few of the differentially expressed proteins with or without inhibitors were common between LMP1 and LMP2A, confirming that the viral proteins induce unique changes in cell expression and function. However, ingenuity pathway analysis (IPA) of the data indicated that LMP1 and LMP2A modulate many of the same cellular regulatory pathways, including cell death and survival, cell movement, and actin filament dynamics. In addition, various proteasome subunits, ubiquitin-specific peptidases and conjugating enzymes, vesicle trafficking proteins, and NF-κB and mitogen-activated protein kinase signaling proteins were affected by LMP1 or LMP2A. These findings suggest that LMP1 and LMP2A may commonly target critical cell pathways through effects on distinct genes, with many cellular proteins modified by ubiquitination and/or degradation.
IMPORTANCE
The Epstein-Barr virus proteins latent membrane protein 1 and 2 have potent effects on cell growth and signaling. Both proteins bind to specific ubiquitin ligases and likely modulate the cellular proteome through ubiquitin-mediated effects on stability and intracellular location. In this study, a comprehensive proteomic analysis of the effects of LMP1 and LMP2A revealed that both proteins affected proteasome subunits, ubiquitin-specific conjugases and peptidases, and vesical trafficking proteins. The data suggest that the effects of these proteins on the abundance and ubiquitination of cellular proteins are in part responsible for their effects on cell growth regulation.
INTRODUCTION
Epstein-Barr virus (EBV) is an important factor in the etiology of several major malignancies, including endemic Burkitt’s lymphoma (BL), nasopharyngeal carcinoma (NPC), Hodgkin disease (HD), and malignant lymphomas that arise in immunosuppressed or immunodeficient patients (1). The EBV proteins latent membrane proteins 1 and 2 (LMP1 and LMP2A) have both been shown to alter cell growth regulation and are considered the EBV oncogenes. LMP1 was initially identified as a viral oncogene, as it can transform rodent fibroblasts in vitro and is essential for B-cell transformation (2). LMP1 is expressed in multiple malignancies associated with EBV, including posttransplant lymphoma, Hodgkin disease, and nasopharyngeal carcinoma (NPC) (1). Many studies have shown that LMP1 has considerable effects on cellular biological properties and gene expression (2). Recent studies indicated that the carboxy-terminal activation domain 2 (CTAR2) is responsible for most of the LMP1-induced effects on cellular transcription through its activation of the canonical NF-κB pathway (3, 4). In contrast, CTAR1 activates the noncanonical NF-κB pathways but induces relatively few changes in transcription (4–6). However, it has profound effects on cellular biological properties and is sufficient for induction of epidermal growth factor receptor (EGFR), activation of phosphatidylinositol 3-kinase (PI3-kinase)/Akt, extracellular signal-regulated kinase (ERK), and rodent and epithelial cell transformation (6–8).
LMP2A is not essential for B-cell immortalization; however, it affects cell signaling and blocks signal transduction from the B-cell receptor (BCR) (9–12). The biological effects of LMP2A are subtle, but intriguing effects have been discerned in transgenic mice in which LMP2A expression enables B-cell survival in the absence of BCR signaling (12, 13). In transgenic mice that express both LMP1 and LMP2A in epithelial cells, LMP2A increased the development of squamous cell carcinoma (14). Additionally, LMP2A activates Akt in B lymphocytes and in normal, primary keratinocytes, resulting in the induction of the nuclear translocation of β-catenin and the activation of cellular promoters that are regulated by β-catenin (15–17). In organotypic raft cultures of the HaCat epithelial cell line LMP2A inhibited differentiation, and in the gastric carcinoma cell line HSC-39, LMP2A induced anchorage independence (18–20). Additionally, LMP2A inhibits the process of differentiation and lumen formation in the MCF10A cell line through inhibition of anoikis and induction of autophagy (21, 22).
Both LMP1 and LMP2A interact with cellular ubiquitin ligases that can affect both the levels and locations of cellular proteins. LMP1 is considered a constitutive member of the tumor necrosis factor receptor family and interacts with the same factors, TRAF1, -2, -3, -5, and -6, TRADD, and A20, most of which have ubiquitin ligase activity through a RING finger domain (23–26). LMP2A interacts with tyrosine kinases and with the NEDD 4 family of ubiquitin ligases (21, 27). Multiple studies have assessed the effects of LMP1 and LMP2A on cellular transcription; however, it is likely that both proteins have profound effects on the cellular proteome that contribute to their effects on cell growth regulation and transformation (28).
To identify proteins and pathways directly and indirectly affected by LMP1 and LMP2A through effects on protein levels, the individual cellular proteomes of cells expressing LMP1 or LMP2A were determined in the absence or presence of deubiquitinase and proteasome inhibitors N-ethylmaleimide and MG132, respectively. The use of deubiquitinase inhibitors would stabilize proteins whose ubiquitination is induced by the viral proteins, while the proteasome inhibitors would block their degradation. This approach would tentatively identify proteins that would otherwise be undetectable due to virus-mediated effects on ubiquitination and protein turnover. The ubiquitinated proteins include proteins whose levels are affected by the viral proteins through protein degradation but also would include proteins whose ubiquitination is induced by LMP1 or LMP2A to modulate cellular location or activity. The effects of these viral proteins on the proteome could have considerable functional outcomes in multiple aspects of cellular function, including signaling, endocytosis, and vesicle trafficking.
RESULTS
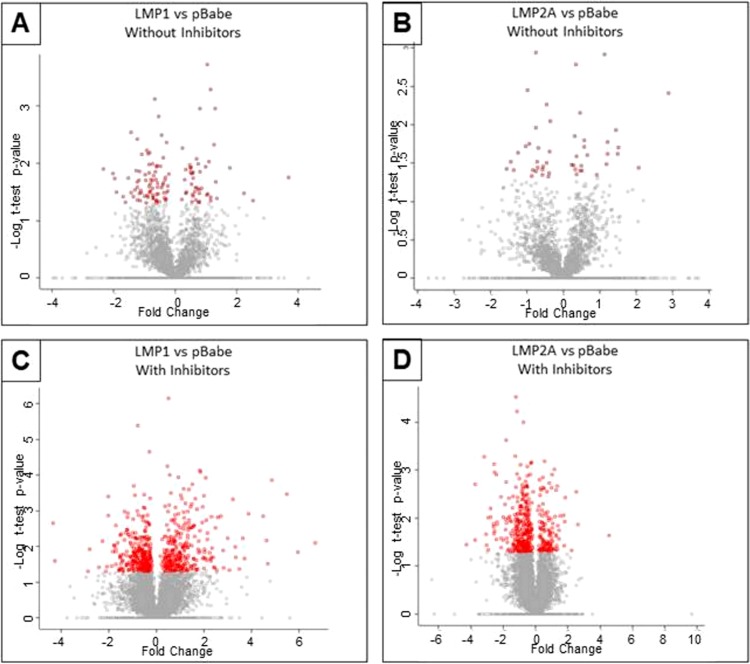
Equal amounts of cell lysates from MCF10 epithelial cells expressing LMP1, LMP2A, or pBabe empty vector control, incubated for 6 h with or without inhibitors, were fractionated by basic reverse-phase high-performance liquid chromatography (RP-HPLC) and were analyzed using LC-tandem mass spectrometry (LC-MS/MS). Two biological replicates with two technical replicates of each were analyzed using MaxQuant software (29). A total of 7,713 proteins were identified with 95% confidence limits with appropriate effects on abundance and distribution of increased and decreased expression (Fig. 1) in the presence of LMP1 or LMP2A. The addition of inhibitors consistently increased the number of proteins with significant differences from the pBabe control and between LMP1 and LMP2A (Table 1). In the absence of inhibitors, the LMP1-expressing cells showed increased expression of 49 proteins and decreased expression of 78 proteins, while LMP2A affected the abundance of 48 proteins, with 24 decreased and 24 increased. Expression of LMP1 with inhibitors affected 598 proteins (P < 0.05) in comparison to the pBabe-containing control cells, with an almost equal distribution of increased and decreased expression. In the LMP2A-expressing cells, 433 proteins (P < 0.05) were changed in comparison to the control cells; however, a considerably greater number of proteins were decreased relative to control cells, with 308 decreased and 125 increased. These data suggest that both LMP1 and LMP2A affect the cellular proteome through proteasome-mediated effects on levels of specific proteins. All P values and fold changes for each comparison are presented in Table S1 in the supplemental material).
FIG 1 .
Scatter plots (“volcano plots”) of t test P values against fold change between LMP1 and pBabe without inhibitors (A), LMP2A and pBabe without inhibitors (B), LMP1 and pBabe with inhibitors (C), and LMP2A and pBabe with inhibitors (D). Proteins with significant P values are indicated in red. The number of significantly changed proteins greatly increased with the addition of inhibitors.
TABLE 1 .
Abundance and distribution of significantly different proteins based on LC-MS/MS dataa
| Comparison | Inhibitors |
|||
|---|---|---|---|---|
| LMP1 |
LMP2 |
|||
| − | + | − | + | |
| No. of proteins with P < 0.05 | 127 | 598 | 48 | 433 |
| Increased vs pBabe | 49 | 286 | 24 | 125 |
| Decreased vs pBabe | 78 | 312 | 24 | 308 |
| Increased 2-fold or more | 15 | 136 | 9 | 29 |
| Decreased 2-fold or more | 23 | 70 | 5 | 107 |
Data reported are numbers of proteins for a given condition (in the presence [+] or absence [−] of inhibitors) that were found to have a significantly different abundance or distribution.
Proteomics data set. P values, fold changes, and gene names are provided for t test comparisons between LMP1 and pBabe without inhibitors, LMP2A and pBabe without inhibitors, LMP1 and pBabe with inhibitors, and LMP2A and pBabe with inhibitors. The results for each t test are presented on a separate tab. Download TABLE S1, PDF file, 1.9 MB (1.9MB, xls) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
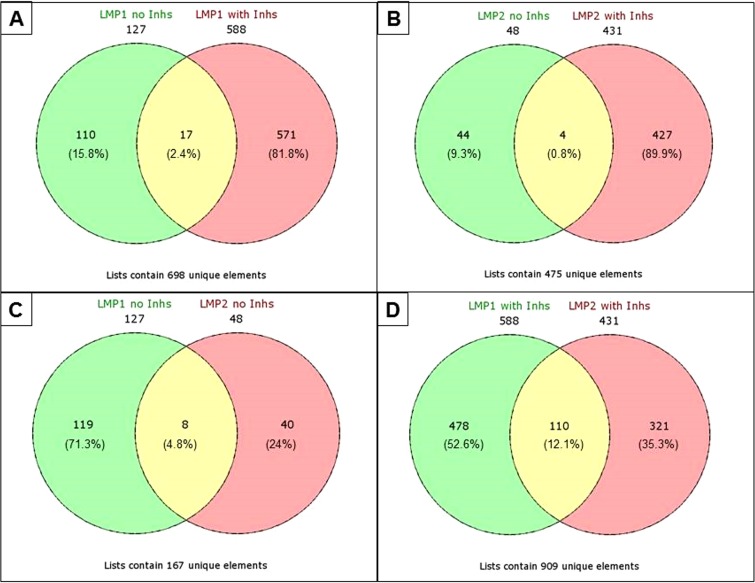
Comparison of the individual, significantly different proteins indicated little overlap between groups with and without inhibitors for either LMP1 (2.4%) or LMP2A (0.8%) (Fig. 2A and B; Table 2). The overlapping proteins were consistently affected in the same direction. There was also little overlap between genotypes (Fig. 2C and D), although with the addition of inhibitors the number of proteins in common between LMP1 and LMP2A increased to 110 (or 12%) from 8 (or 4.5%) proteins without inhibitors. This suggests that there is some overlap in their effects on the proteome through the ubiquitin pathway. Common proteins affected included Rab7A, TLR3, ATG2A, mitogen-activated protein kinase kinase 2 (MAP2K2), EIF3B and -3E, ribosomal protein L8 (RPL8) and RPL9, and integrin α5 (ITGA5) (Table 2). Additionally, despite the few overlapping individual significantly different proteins, further assessment of the data by use of the bioinformatics software Ingenuity Pathway Analysis (IPA) revealed substantial overlap in biological functions and signaling pathways between the study groups.
FIG 2 .
Venn diagrams displaying the overlap in the number of significantly different proteins between treatment groups. (A) LMP1-noInhs compared to LMP1+Inhs; (B) LMP2A-noInhs compared to LMP2A+Inhs; (C) LMP1-noInhs compared to LMP2A-noInhs; (D) LMP1+Inhs compared to LMP2A+Inhs. Proteins common to both groups are listed in Table 2.
TABLE 2 .
Significantly affected proteins altered in common between groups
| Comparison | Significantly different proteins in common between groups |
|---|---|
| LMP1-noInhs vs LMP1+Inhs | AARS, CA2, CALD1, CCDC47, CNN2, CSK, EIF4A1, ETHEd1, FARSB, HEBP2, LAMB1, MTHFD1, MVP, MYBBP1A, SARG, TBCE, TPD52L1 |
| LMP2-noInhs vs LMP2+Inhs | CHTOP, IGF2BP3, PML, RAB7A |
| LMP1-noInhs vs LMP2-noInhs | CS, EHD4, GARS, IAH1, NDRG1, SORD, SRSF5, ZC3H4 |
| LMP1+Inhs vs LMP2+Inhs | ABT1, ACLY, ACTR1A, ASMTL, ATG2A, ATP6V0D1, ATXN7L3, C9orf78, CAPZB, CCDC171, CCDC47, CHD1, CLIC2, CLIC3, CLPTM1L, COPS6, CSDA, CSNK1D, CTAGE5, DCAF4L2, DHX30, DOCK9, DUS3L, ECSIT, EFR3A, EFTUD2, EIF3B, EIF3E, EIF4A1, EIF4H, ENO1, EPPK1, EXOC3, FBXO7, FHL1, FHOD1, FSCN1, FUBP3, FXR1, G3BP1, G3BP2, GBAS, GBE1, GINS4, GRSF1, GTF2I, HDDC2, HDGFRP3, HEBP1, HELZ2, IL18, IPO9, ITGA5, KIF16B, LAMA5, LARP4, MAP2K2, MEAF6, MTERFD1, MYL6, NOLC1, NUDT19, P4HA2, PACSIN2, PGRMC1, PHGDH, PKD1, PPP1CC, PRMT7, PSMB1, PSMC2, PSMG1, RAB7A, RAP1GDS1, RBBP7, RMND1, RNF214, RPL8, RPL9, RPS6KC1, RRP7A, SCFD2, Selm, SERPINB11, SMARCAD1, SNRNP200, SNX24, SNX3, SPCS3, strap, TCEB2, THOC5, TLR3, TMEM63B, TMEM87A, TOMM5, TOMM7, TOR2A, UAP1, UBE3B, UBE4A, UFL1, USP38, UTP11L, VIM, WDR6, YIPF2, YIPF4, ZMYM2, ZNHIT2 |
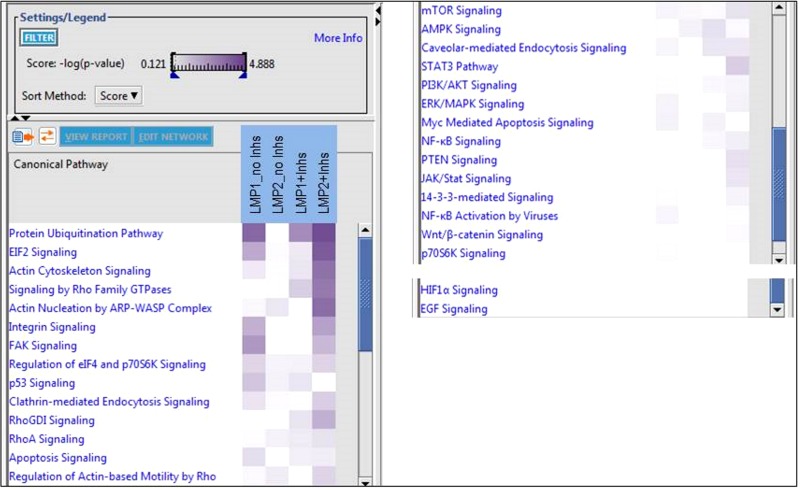
Identification of canonical pathways (Fig. 3) revealed major effects, with highly significant P values on the protein ubiquitination pathway in LMP1 with and without inhibitors and in LMP2A with inhibitors compared to the pBabe controls. Canonical pathways involved in EIF2 signaling, regulation of eIF4 and p70s6K signaling, actin cytoskeleton signaling, regulation of actin-based motility by Rho, integrin, FAK, PI3K/Akt, ERK/MAPK, and p53 signaling were all differentially regulated. Although not all of these pathways had a significant number of proteins associated with them to indicate activation or deactivation, the results did indicate that there were a number of significantly different proteins for these associated functions. Multiple pathways involved in vesicle formation and endocytosis, including Clathrin- and Caveolar-mediated endocytosis, were also differentially regulated (Fig. 3).
FIG 3 .
Analysis comparison of canonical pathways. A comparison of the canonical pathways associated with significantly different protein expression for each genotype, with or without inhibitors, was performed using IPA. The comparison was further sorted according to the P value of association (−log10) with a particular pathway and is represented by the intensity of each node in the heat map. A greater number of pathways were significantly associated with the addition of inhibitors, reflecting the increased number of significant changes in expression associated with these samples. In addition, the most significant pathway associated across almost all samples was the Protein ubiquitination pathway.
Analysis of diseases and biofunction identified many of the functions previously described for LMP1 based on significant Z scores (Fig. 4). A Z score greater than 2 strongly suggests activation or inhibition. Thus, cell death of tumor cells was considerably impaired in the LMP1 and LMP2A cells in the absence of inhibitors, while cell proliferation was activated in the LMP1 cells in the absence of inhibitors. Networks for cell death or proliferation and changes in migration of endothelial cells were significantly altered in LMP1 cells compared to the pBabe control. LMP1 without inhibitors also significantly decreased apoptosis but increased autophagy.
FIG 4 .
Analysis of the Diseases and bio-functions group. A comparison of the diseases and biological functions associated with significant expression changes for LMP1 and LMP2A, with or without inhibitors, versus the pBabe control, is shown. The scores are displayed on a scale from dark blue to orange, with dark blue representing the most negative scores or inactivation and orange representing the highest positive scores or activation. Only diseases and functions with a P value of <0.05 are displayed. Activation scores with an absolute value of 2 or more are considered significant. Again, an increase in the number of significant scores was found in samples where inhibitors were added.
The significant differences in diseases and biofunctions induced by the expression of LMP2A were also assessed using IPA. Many of the functions previously shown to be affected by LMP2A were identified in this analysis to be differentially regulated in the presence of inhibitors compared to the pBabe control. Binding of filaments was activated by LMP2A with inhibitors, while cell spreading was decreased (Fig. 4). In addition, potential new cellular functions predicted to be affected by LMP2A included RNA, DNA, and protein expression (expression of mRNA, translation of mRNA, expression of DNA, translation of mRNA, translation of RNA, and expression of RNA) (Fig. 4).
Overall, many of the pathways identified based on canonical pathways analysis have been previously described to be affected by LMP1 and LMP2A, including cell motility, cell junction signaling, actin cytoskeleton regulation, RhoA, PI3K/AKT, PTEN signaling, mTOR signaling, and p53 signaling, although not all of these had significant P values in each comparison. These data provided identification of novel protein targets that contribute to the effects mediated by LMP1 and LMP2A. Additionally, these data identified new effects on EIF2 signaling and regulation of eIF4 and p70s6K signaling. Analysis of canonical pathways identified new potential effects on mRNA processing, DNA methylation and transcriptional repression, cleavage and polyadenylation of pre-mRNA, and tRNA charging, which were all differentially regulated.
In order to simplify the description of results, the four comparison groups are referred to as LMP1-noInh (LMP1 versus pBabe without inhibitors), LMP2A-noInh (LMP2A versus pBabe without inhibitors), LMP1+Inh (LMP1 versus pBabe with inhibitors), and LMP2A+Inh (LMP2A versus pBabe with inhibitors), with specific attention on the protein ubiquitination pathway, vesicle formation and trafficking, transcription and translation, cytoskeleton cell movement and cell junctions, and pathways previously described for LMP1 and LMP2A (Tables 3 and 4; Tables S2, S3, S4, and S5).
TABLE 3 .
Proteins in the ubiquitination pathway identified via IPA
| Comparison and protein | Entrez gene name | P value | Fold change |
|---|---|---|---|
| LMP1 vs pBabe: no inhibitors (P = 1.92E−05) |
|||
| DNAJC7 | DnaJ (Hsp40) homologue, subfamily C, member 7 | 3.90E−02 | −1.852 |
| HSPA14 | Heat shock 70-kDa protein 14 | 1.98E−02 | −1.667 |
| HSPB1 | Heat shock 27-kDa protein 1 | 1.20E−02 | 3.427 |
| PSMD2 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 5.38E−03 | 1.171 |
| PSMD8 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 8 | 1.92E−02 | −1.728 |
| THOP1 | Thimet oligopeptidase 1 | 3.73E−02 | −1.982 |
| UBE2V1 | Ubiquitin-conjugating enzyme E2 variant 1 | 1.10E−03 | 1.739 |
| USP5 | Ubiquitin-specific peptidase 5 | 2.61E−02 | −1.490 |
| USP14 | Ubiquitin-specific peptidase 14 | 2.57E−02 | −2.353 |
| LMP2 vs pBabe: no inhibitors (P = 1) | |||
| None | |||
| LMP1 vs pBabe: with inhibitors (P = 2.35E−06) |
|||
| BAG1 | BCL2-associated athanogene | 2.96E−03 | 2.613 |
| BIRC3 | Baculoviral IAP repeat-containing 3 | 1.58E−02 | −1.760 |
| CUL1 | Cullin-1 | 3.57E−03 | −1.414 |
| DNAJC5 | DnaJ (Hsp40) homologue, subfamily C, member 5 | 2.41E−02 | −2.692 |
| HLA-B | Major histocompatibility complex, class I, B | 2.06E−02 | 4.993 |
| HSP90AA1 | Heat shock protein 90-kDa alpha (cytosolic), class A member 1 | 3.43E−02 | −1.335 |
| HSPA6 | Heat shock 70-kDa protein 6 (HSP70B′) | 5.54E−03 | −1.492 |
| HSPA8 | Heat shock 70-kDa protein 8 | 4.62E−02 | −2.934 |
| HSPA1A | Heat shock 70-kDa protein 1A | 2.34E−03 | −1.242 |
| PSMB1 | Proteasome (prosome, macropain) subunit, beta type, 1 | 1.08E−03 | 2.223 |
| PSMB8 | Proteasome (prosome, macropain) subunit, beta type, 8 | 9.89E−03 | 2.207 |
| PSMB10 | Proteasome (prosome, macropain) subunit, beta type, 10 | 1.03E−02 | 2.319 |
| PSMC2 | Proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 1.89E−02 | 1.127 |
| PSMD14 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 14 | 4.95E−02 | −1.390 |
| RBX1 | Ring box 1, E3 ubiquitin protein ligase | 2.30E−02 | −1.494 |
| TCEB1 | Transcription elongation factor B (SIII), polypeptide 1 | 4.49E−02 | −1.414 |
| TCEB2 | Transcription elongation factor B (SIII), polypeptide 2 | 1.33E−03 | −1.690 |
| UBE2I | Ubiquitin-conjugating enzyme E2I | 1.19E−02 | −1.485 |
| UBE2L3 | Ubiquitin-conjugating enzyme E2L 3 | 1.42E−02 | 4.031 |
| UBE3B | Ubiquitin protein ligase E3B | 6.61E−03 | 1.659 |
| UBE4A | Ubiquitination factor E4A | 1.94E−02 | −2.941 |
| USP38 | Ubiquitin-specific peptidase 38 | 3.60E−02 | −3.717 |
| LMP2 vs pBabe: with inhibitors (P = 4.26E−05) |
|||
| CDC34 | Cell division cycle 34 | 3.99E−02 | −2.274 |
| DNAJA1 | DnaJ (Hsp40) homologue, subfamily A, member 1 | 5.00E−02 | −1.763 |
| HSCB | HscB mitochondrial iron-sulfur cluster cochaperone | 4.67E−02 | −2.333 |
| PSMB1 | Proteasome (prosome, macropain) subunit, beta type, 1 | 1.73E−02 | 1.456 |
| PSMC2 | Proteasome (prosome, macropain) 26S subunit, ATPase, 2 | 2.62E−03 | 1.287 |
| PSMD2 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 2.70E−02 | −1.227 |
| TAP2 | Transporter 2, ATP-binding cassette, subfamily B | 3.16E−02 | −2.098 |
| TCEB2 | Transcription elongation factor B (SIII), polypeptide 2 | 1.27E−02 | −1.275 |
| TRAF6 | TNF receptor-associated factor 6, E3 ubiquitin protein ligase | 5.71E−03 | −1.943 |
| UBA1 | Ubiquitin-like modifier activating enzyme 1 | 2.85E−03 | −1.117 |
| UBE2C | Ubiquitin-conjugating enzyme E2C | 1.21E−02 | −5.242 |
| UBE2L6 | Ubiquitin-conjugating enzyme E2L 6 | 2.87E−02 | −2.430 |
| UBE3B | Ubiquitin protein ligase E3B | 4.58E−02 | 1.134 |
| UBE4A | Ubiquitination factor E4A | 9.86E−03 | −7.551 |
| USP38 | Ubiquitin-specific peptidase 38 | 2.81E−02 | −5.336 |
| USP48 | Ubiquitin-specific peptidase 48 | 8.21E−03 | −1.919 |
TABLE 4 .
Ubiquitination pathway proteins based on manual curation
| Comparison and protein | Entrez gene name | P value | Fold change |
|---|---|---|---|
| LMP1 vs pBabe: no inhibitors | |||
| VBP1 | Von Hippel-Lindau binding protein 1 | 1.50E−02 | −4.055781694 |
| CCDC50 (Ymer) | Coiled-coil domain containing 50 | 4.08E−02 | 2.574692252 |
| BAG2 | BCL2-associated athanogene 2 | 1.56E−02 | −1.975596065 |
| THOC1 (HPR1) | THO complex 1 | 7.01E−03 | −2.12573241 |
| LMP2 versus pBabe: no inhibitors | |||
| RPS25 | Ribosomal protein S25 | 3.99E−02 | 1.423354477 |
| SPG20 (Spartin) | Spastic paraplegia 20 (Troyer syndrome) | 1.20E−03 | 2.185767576 |
| LMP1 versus pBabe: with inhibitors | |||
| DCAF4L2 | DDB1- and CUL4-associated factor 4-like protein 2 | 3.00E−03 | 1.997755058 |
| FBXL18 | Isoform 4 of F-box/LRR-repeat protein 18 | 1.08E−02 | 2.490401919 |
| FBXO7 | F-box only protein 7 | 4.35E−02 | 1.868799777 |
| PSMG1 | Isoform 2 of proteasome assembly chaperone 1 | 2.51E−02 | −1.933470638 |
| SH3RF2 | Putative E3 ubiquitin-protein ligase SH3RF2 | 1.22E−02 | 1.739935853 |
| TRIM27 | Isoform beta of zinc finger protein RFP | 2.37E−02 | 1.677203226 |
| UBAP1 | Isoform 2 of ubiquitin-associated protein 1 | 5.99E−03 | 1.662662369 |
| UBE2K | Ubiquitin-conjugating enzyme E2 K | 1.86E−03 | −1.489748169 |
| UBR7 | Putative E3 ubiquitin-protein ligase UBR7 | 4.61E−02 | 0.364044384 |
| UFL1 | E3 UFM1-protein ligase 1 | 3.58E−02 | −1.27422905 |
| UFM1 | Ubiquitin-fold modifier 1 | 4.72E−02 | 1.622521302 |
| COPS6 | COP9 signalosome complex subunit 6 | 5.68E−03 | −1.935473811 |
| LMP2 vs pBabe: with inhibitors | |||
| ATXN3L | Putative ataxin-3-like protein | 2.32E−02 | 23.37327866 |
| ATXN7L3 | Ataxin-7-like protein 3 | 4.19E−02 | 1.367686338 |
| CBX4 | E3 SUMO-protein ligase CBX4 | 4.31E−02 | 1.424713914 |
| CCM2 | Isoform 2 of malcavernin | 1.69E−02 | −2.262188449 |
| COPS6 | COP9 signalosome complex subunit 6 | 3.01E−05 | −2.303932902 |
| FBXO7 | F-box only protein 7 | 1.37E−02 | 1.404454865 |
| PIAS3 | E3 SUMO-protein ligase PIAS3 | 1.35E−02 | 6.19446027 |
| POMP | Proteasome maturation protein | 4.47E−02 | −2.469462113 |
| PSMG1 | Isoform 2 of proteasome assembly chaperone 1 | 6.54E−03 | −1.961205514 |
| RNF114 | RING finger protein 114 | 4.65E−02 | −1.466037981 |
| RNF138 | E3 ubiquitin-protein ligase RNF138 | 2.37E−03 | 1.371846044 |
| SDCBP (MDA-9) | Isoform 3 of syntenin-1 | 2.26E−03 | −1.419647959 |
| UFL1 | E3 UFM1-protein ligase 1 | 2.88E−02 | −1.316652316 |
IPA data: vesicle formation and trafficking. The P value and fold change data for all proteins associated with vesicle formation and trafficking pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used for IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, data for other comparisons are also presented, unless they were from the manual curation. Download TABLE S2, PDF file, 0.11 MB (114.9KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: transcription and translation. The P value and fold change data for all proteins associated with transcription and translation pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, the data from other comparisons are also presented. Download TABLE S3, PDF file, 0.24 MB (239.6KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: cytoskeleton, cell movement, and cell junctions. The P value and fold change data for all proteins associated with cytoskeleton, cell movement, or cell junction pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, the data in other comparisons are also presented. Download TABLE S4, PDF file, 0.16 MB (161.3KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: other or known LMP1-LMP2A pathways. The P value and fold change data for all proteins associated with known LMP1- or LMP2A-associated pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) significant in one comparison, the data in other comparisons are also presented. Download TABLE S5, PDF file, 0.25 MB (252.2KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Protein ubiquitination pathway.
One of the top canonical pathways found by IPA was the protein ubiquitination pathway (Table 3). For LMP1-noInh, IPA found a significant number of proteins associated with the pathway (P = 1.92 × 10−5) (Table 3), some of which included DNAJC7 (downregulated 1.85-fold), the proteasome subunits PSMD2 (upregulated 1.17-fold) and PSMD8 (downregulated 1.73-fold), ubiquitin-specific peptidases USP5 and USP14 (downregulated 1.49- and 2.35-fold, respectively), and the E2 conjugase UBE2V1 (upregulated 1.74-fold). In contrast, for LMP2A-noInh, no proteins associated with the protein ubiquitination pathway were identified by IPA. However, further examination of the IPA data revealed a number of proteins with significantly different expression than their pBabe control that were not included by IPA but could be added to the protein ubiquitination pathway based on functions described in the literature. This manual curation (Table 4) identified RPS25 (upregulated 1.42-fold), which has been demonstrated to bind MDM2 and inhibit its E3 ubiquitin ligase activity, leading to a reduction of its p53 ubiquitylation and resulting in p53 stabilization (Table 4) (30). In addition, Spartin (SPG20) was significantly upregulated in LMP2A-expressing cells compared to the pBabe control (upregulated 2.19-fold). Spartin binds and activates ITCH, a major E3 ubiquitin ligase that binds to LMP2A and is responsible for LMP2A-induced turnover of the B-cell receptor (27, 31, 32).
IPA also found a significant number of proteins from the LMP1+Inh group that are associated with the protein ubiquitination pathway (P = 2.35 × 10−6) (Table 3). Some of these included the downregulation of a number of heat shock proteins, the E3 ubiquitin ligase component Cullin-1 (downregulated 1.414-fold), the proteasome subunits PSMB1 (upregulated 2.223-fold), PSMB8 (upregulated 2.207-fold), PSMB10 (upregulated 2.319-fold), PSMC2 (upregulated 1.127-fold), and PSMD14 (downregulated 1.390-fold), the E2 ubiquitin conjugases UBE2I (downregulated 1.485-fold) and UBE2L3 (upregulated 4.031-fold), the E3 ubiquitin ligase UBE3B (upregulated 1.659-fold), the ubiquitination factor UBE4A (downregulated 2.941-fold), the ubiquitin-specific peptidase USP38 (downregulated 3.717-fold), and the E3 ubiquitin ligase RBX1 (downregulated 1.494-fold). Manual curation (Table 4) also added two F-box proteins, FBXO7 (upregulated 1.869-fold) and FBXL18 (upregulated 2.490-fold), both substrate-recruiting subunits of Skp1-cullin1-FBP (SCF)-type E3 ubiquitin ligases (33), PSMG1 (downregulated 1.933-fold), E2 ubiquitin conjugase UBE2K (downregulated 1.490-fold), which is known to interact with RING finger proteins to modulate the ubiquitin linkage (34), and UFM1 (upregulated 1.623-fold), a ubiquitin-like protein.
For LMP2A+Inh, IPA revealed a significant number of proteins with functions associated with the protein ubiquitination pathway (P = 4.26 × 10−5) (Table 3). This group of proteins contained the E3 ubiquitin ligases TRAF6 (downregulated 1.943-fold) and UBE3B (upregulated 1.134-fold), the ubiquitin E2 conjugase enzymes UBE2C (downregulated 5.242-fold), UBE2L6 (downregulated 2.430-fold), and CDC34 (UBE2R1) (downregulated 2.274-fold), the proteasome subunits PSMB1 (upregulated 1.456-fold), PSMC2 (upregulated 1.287-fold), and PSMD2 (downregulated 1.227-fold), and the ubiquitin-specific peptidases USP38 (downregulated 5.336-fold) and USP48 (downregulated 1.919-fold). Manual curation (Table 4) added the E3-SUMO ligases CBX4 (upregulated 1.425-fold), which SUMOylates HNRNPK (35), and PIAS3 (upregulated 6.194-fold), which binds and stabilizes p53, inhibiting its ubiquitination by MDM2 and thereby regulating cell growth. In addition, manual curation also added FBOX7 (upregulated 1.404-fold), two proteasome component chaperones, POMP (downregulated 2.469-fold) and PSMG1 (downregulated 1.961-fold), and two RING finger proteins, RNF114 (downregulated 1.466-fold) and RNF138 (upregulated 1.372-fold), both of which act as E3 ubiquitin ligases. RNF114 interacts with A20 and modulates NF-κB activity (36). LMP2A has been shown to affect NF-κB activation by LMP1 through effects on TRAF2 (37). These proteomic study results suggest that effects on TRAF6 and RNF114 may be additional mechanisms through which LMP2A might affect the activity of LMP1.
Vesicle formation and trafficking.
IPA identified multiple proteins with functions in vesicle formation and trafficking pathways, including exocytosis, endocytosis, and autophagy. For LMP1-noInh, IPA identified a significant number of proteins in the “Autophagy of tumor cell lines” (P = 0.0126; Z score,1.987) and “Autophagy of cells” (P = 0.0279; z score, 1.334) categories in the Disease and Biofunctions group (Fig. 4; Table S2). Some of these proteins included Rab1A, calpain-1 (CAPN1), and EIF2AK2 (PKR). Manual curation added NDRG1 (N-myc downstream regulated 1 protein; upregulated 2.06-fold; P = 0.00019), which is a metastasis suppressor that inhibits stress-induced autophagy in cancer cells (38), CLIC4 (upregulated 1.73-fold; P = 0.048) which when downregulated enhances autophagy and endoplasmic reticulum (ER) stress-induced apoptosis (39), and VBP1 (von Hippel-Lindau-binding protein; downregulated 4.06-fold; P = 0.015), which mediates autophagy-dependent protein degradation of hMSH4, a MutS protein involved in regulating DNA double-strand break repair in homologous recombination (40) (Table S2).
Other proteins with known functions in vesical formation and trafficking, for example, Clathrin (CLTA), ARPC4, and Flottilin-2 (FLOT2), had significantly different expression in LMP1-expressing cells than in the pBabe control. However, the IPA pathways associated with these proteins did not contain a significant number of proteins (e.g., those from the groups for Viral entry via endocytic pathway, Clathrin-mediated endocytosis signaling, Caveolar-mediated endocytosis signaling, and Fc-gamma receptor phagocytosis). In addition, manual curation found a number of other proteins with functions in vesicle formation control (Table S2), including ATP6V1C1, a vacuolar ATPase subunit important for the acidification of intracellular components (41), SEC23B, a subunit of the COPII complex required for EGFR transport from the ER to the plasma membrane (42), EHD4, an Eps homology domain protein involved in the control of early endosome trafficking (43), and UBE2V1, an E2 Ub-conjugase that binds TRAF6 and ESCRT-0, a complex important for sorting of ubiquitinated cargo (44).
With LMP2A-noInh, IPA found a significant association with the Fusion of autophagosomes (P = 0.0154) and Fusion of late endosomes (P = 0.0176) categories, both of which only contained Rab7a from our data set, which was downregulated 1.678-fold (P = 0.04) (Table S2). Rab7A is a Rab family member of small RAS-related GTP-binding proteins that is important for regulating vesicle traffic from the late endosome to lysosomes and autolysosome formation (45). Rab7a was also associated with the Clathrin-mediated endocytosis signaling group (Table S2). Other differentially expressed proteins associated with IPA vesicle formation and trafficking pathways were VAMP3 (Fc-gamma receptor phagocytosis), SNX5 (macropinocytosis), and CHMP4B/VPS32 (Viral exit from cells) (Table S2). Manual curation also added a number of proteins to this functional category: MYOF and EHD4 (early endosome transport), which were downregulated, and RIN1 (ras effector protein capable of activating Rab5), NDRG1 (an inhibitor of stress induced autophagy), and SPG20 (Spartin), which were upregulated (Table S2) (46, 47). Spartin, which as described above activates Itch, also binds the ESCRT-III protein complex, which regulates vesicle trafficking and cargo sorting, primarily at the last stage of vesicle formation or budding. It has been shown that Spartin upregulation results in decreased EGFR degradation and decreased internalization, properties that would complement the EGFR induction by LMP1 (48).
IPA analysis of LMP1+Inh found 51 proteins with significant associations with 5 vesicle formation and trafficking pathways, including the groups Autophagy (P = 0.00219), Endocytosis (P = 2.29 × 10−4), Macropinocytosis (P = 0.01), Viral entry via endocytic pathway (P = 0.0437), and Caveolar-mediated endocytosis signaling (P = 0.0497) (Table S2). The Autophagy pathway contained 21 proteins in total, with 10 upregulated and 11 downregulated. Upregulated proteins included MAP1LC3B (LC3), RB1CC1, and ULK3, while downregulated proteins included Rab7a, eIF2S1, ATG2A, TBC1D4, and TBK1. ATG2A is an autophagy-related protein thought to regulate formation and dispersion from autophagosomes (49). Interestingly, TBK1 (TANK-binding kinase) is important in innate immunity and autophagy. Its activation is critically regulated by its cellular localization. The detection of this protein in the presence of inhibitors suggests that LMP1 may affect TBK localization and activation through effects on ubiquitination (50). Manual curation added ELMO2 (engulfment and cell motility protein 2) (upregulated 1.27-fold; P = 0.039), which is essential for phagocytosis and migration (47), TMEM59 (upregulated 1.53-fold; P = 0.037), which contains a novel ATG16L1-binding motif and promotes local activation of LC3 (also upregulated more than 4-fold). It has been demonstrated to bind ATG16L1 and mediate autophagy during Staphylococcus aureus infection (51). In addition, Rab8B, which is important for autophagosome maturation, was upregulated 1.9-fold (P = 0.034) (52) and WDR6 was downregulated 1.78-fold (P = 0.028). WDR6 is a negative regulator of amino acid starvation-induced autophagy (53).
Similarly, the Endocytosis pathway included 24 proteins in total, with 11 proteins upregulated and 13 downregulated. Upregulated proteins included ATP6V0D1, CLIC3, Ezrin (EZR), and RAC2, while downregulated proteins included Rab7a, SNX3, ATG2A, and AP1S1. Ezrin acts as a cross-linker between membranes and the cytoskeleton and is important for endocytosis, phagocytosis, and vesicle trafficking, enhancing cell motility and invasiveness. It is known that Ezrin phosphorylation is induced by LMP1-enhanced cell motility (54). Manual curation added another 18 proteins with Rab11FIP1, Rab GTPase-binding effector protein 2 (RabEP2), ATP6V1B1, TBC1D13, sorting nexin-24 (SNX24), and synaptogyrin-2 (SYNGR2) being upregulated. Rab11FIP1 is a Rab-interacting protein that is critical for trafficking into early endosomes (55). TBC1D13, a Rab35-specific GAP that contributes to GLUT4 trafficking, was also upregulated (56), while CCT8, NISCH, and VAMP5 were downregulated (Table S2).
For LMP2A+Inh, IPA revealed 32 proteins with significant associations with 3 vesicle formation and trafficking pathways (Table S2), Endocytosis (P = 2.63 × 10−4), Fusion of late endosomes (P = 0.0095), and Endocytosis by tumor cell lines (P = 0.013). The Endocytosis group contained Annexin-6 (ANXA6), which has been suggested to mediate the aggregation and fusion of endosomes during exocytosis in epithelial layers (57), as well as ATG2A, ATP6V1F, ATP6V1H, Rab7a, and SNX3, all of which were downregulated. Only ATP6V0D1 was upregulated. The Fusion of late endosomes group contained STX8, a syntaxin protein implicated in protein trafficking from early to late endosomes (58), Rab7a, and C20orf24 (RIP5), a Rab5 interaction protein (Table S2) (59). Other proteins associated via IPA with vesicle formation and trafficking pathways included MAP2K2, integrin α5, CHMP3 (VPS24), CD2AP, and VPS4A, which were significantly differentially expressed compared to the pBabe control (Table S2). CHMP3 is part of the ESCRT-III complex that recruits transmembrane proteins to lysosomes via the MVB, while VSP4 catalyzes ESCRT-III disassembly, reallocating the components to the cytoplasm for MVB sorting and contributing to herpesvirus envelopment (60). Manual curation added a number of significantly different proteins compared to the pBabe control with functions in vesicle formation and trafficking. Some of these included LAMTOR4, Rab1B, Rab38, EXOC3 (sec6), EXOC5, EXOC6B (sec15), and TMED4. LAMTOR4 recruits mTORC1 to lysosomes, leading to its activation, while Rab38 mediates protein trafficking to lysosomes (61, 62). EXOC3, EXOC5, and EXOC6B are components of the exocyst complex that targets exocytic vesicles to specific sites on the plasma membrane (63).
Transcription and translation.
IPA also revealed a number of overlapping pathways containing significant numbers of proteins with functions in transcription and translation (Table S3). For LMP1-noInh, IPA resulted in significant Fisher exact test scores in the pathways for EIF2 signaling (P = 4.65 × 10−3), tRNA charging (P = 1.18 × 10−2), translation of protein (P = 2.52 × 10−4), translation of mRNA (P = 7.45 × 10−5), expression of protein (P = 4.69 × 10−4), and processing of RNA (P = 9.51 × 10−3). In EIF2 signaling EIF2AK2, EIF3L, EIF4A1, and RPL12 were downregulated while EIF2B4 was upregulated. EIF2AK2, EIF2B4, and EIF3L were also included in the Translation of mRNA, Translation of protein, and Expression of protein pathways. All other proteins associated with these pathways are listed in Table S3. In contrast, for LMP2A-noInh, the only pathway IPA found with a significant number of proteins functioning in transcription and translation was the Cleavage and polyadenylation of pre-RNA pathway (P = 3 × 10−4). IPA found other pathways with LMP2A-noInh, but each of these had only one protein listed, ribosomal protein S25 (RPS25) in EIF2 signaling and eIF4-p70S6K signaling and glycyl-tRNA-synthetase (GARS) in tRNA charging.
With the addition of inhibitors, a greater number of transcription and translation pathways had significant scores with IPA. In addition, there were a greater number of differentially expressed proteins associated with these pathways, most of which were downregulated (Table S3). For instance, in LMP1+Inh, 13 out of 15 proteins associated with the EIF2 signaling pathway were downregulated. Among the proteins downregulated in LMP1+Inh were EIF2, -3, and -4 subunits and ribosomal proteins L8, L9, L31, and L32. In LMP2A+Inh, 10 out of 14 proteins were downregulated in the EIF2 signaling pathway, including EIF3, -4, and -5 subunits, as well as several ribosomal proteins. These proteins were also included in the eIF4 and p70s6K signaling, expression of mRNA, translation of mRNA, translation of RNA, expression of protein, and translation of protein pathways (Table S3).
Cytoskeleton, cell movement, and cell junctions.
With the addition of inhibitors, the number of cytoskeleton, cell movement, and cell junction pathways containing significant numbers of proteins identified via IPA increased dramatically in LMP1-expressing cells (Table S4). Without inhibitors, LMP1-expressing cells had only one significant pathway, Integrin signaling (P = 6.27 × 10−3), in which ARPC4, CAPN1, and RAP1B were downregulated while CAPNS1 and vinculin (VCL) were upregulated. Other pathways had proteins listed for LMP1-noInhs but their P values were not significant. In addition, for LMP2A-noInh there were no pathways with statistically significant numbers of proteins in these functional categories. With the addition of inhibitors to LMP2A-expressing cells, only the Rac signaling (P = 1.32 × 10−2) and Binding of filaments (P = 4.51 × 10−3) pathways contained significant numbers of proteins. In Rac signaling, six proteins were downregulated (ACTR2, BRK1, ITGA5, MAP2K2, NCKAP1, and RPS6KB1) and one was upregulated (NF-κB1), while in Binding of filaments all four proteins were downregulated (CSNK1D, FAS, FHL1, and VIM). It is intriguing that LMP2A would affect the expression of NF-κB1, a major component of the canonical NF-κB pathway that is activated by LMP1. This provides another mechanism for synergy between LMP1 and LMP2A.
In contrast, with LMP1+Inh cells the number of pathways IPA found with statistically significant numbers of proteins increased to 13 (Table S4). These pathways included Integrin signaling (P = 3.67 × 10−5), Actin cytoskeleton signaling (P = 9.58 × 10−6), Epithelial adherens junction signaling (P = 4.98 × 10−5), Sertoli cell-Sertoli cell junction signaling (P = 3.15 × 10−6), Tight junction signaling (P = 6.25 × 10−4), Rac signaling (P = 2.6 × 10−2), RhoA signaling (P = 6.52 × 10−3), Leucocyte extravasation signaling (P = 4.52 × 10−4), and ILK signaling (P = 5.18 × 10−3).
Pathways previously described for LMP1 and LMP2A (or other pathways).
A number of functional pathways previously described for LMP1 and LMP2A were also implicated by IPA (Table S5). In LMP1-noInh, the p53 signaling pathway contained a statistically significant number of proteins (P = 1.9 × 10−2), although only 3 proteins were listed. These were SERPINB5 and Stratifin (SFN), which were upregulated, and PCNA, which was downregulated. For LMP1+Inh, p53 signaling contained PERP (TP53 apoptosis effector) was downregulated 7.4-fold and TP53IP3 was upregulated 3.2-fold. LMP1+Inh also contained a significant number of proteins in the CDC42 signaling pathway (P = 1.67 × 10−2), with 8 of 10 proteins being upregulated. Some of these included IQGAP3, HLA-B, FDG1, and ARPC1A.
For LMP2A+Inh, the mTOR signaling (P = 1.33 × 10−3), PI3K/AKT signaling (P = 3.26 × 10−3), and PTEN signaling (P = 8.34 × 10−3) pathways contained significant numbers of proteins. In the mTOR signaling pathway, 9 of 11 proteins were downregulated and this included a number of EIF3 and EIF4 proteins. In the PI3K/AKT signaling pathway, 6 of 8 proteins were downregulated and also included INPP5K, MAP2K2, and PPP2R2A. For the PTEN signaling pathway, in which 5 of 7 proteins were downregulated, INPP5K, MAP2K2, and RPS6KB1 were also included (Table S5).
Several members of the NF-κB signaling pathway were identified, including NF-κB1 (P = 0.0162), and manual curation found NF-κB-activating protein (NKAP) (P = 0.0379) was significantly upregulated in LMP2A+Inh compared to its pBabe control. In addition, IKBKG was upregulated 1.742-fold in LMP1 with inhibitors (P = 5.03 × 10−3), while TANK-binding kinase (TBK1) was downregulated 1.305-fold (P = 0.0425) (Table S5). These data suggest that in the presence of LMP1, IKBKG had increased ubiquitination, which is in keeping with the activation of NF-κB by LMP1 mediated through effects on IKBKG.
Confirmation of protein expression changes associated with vesicle formation and trafficking.
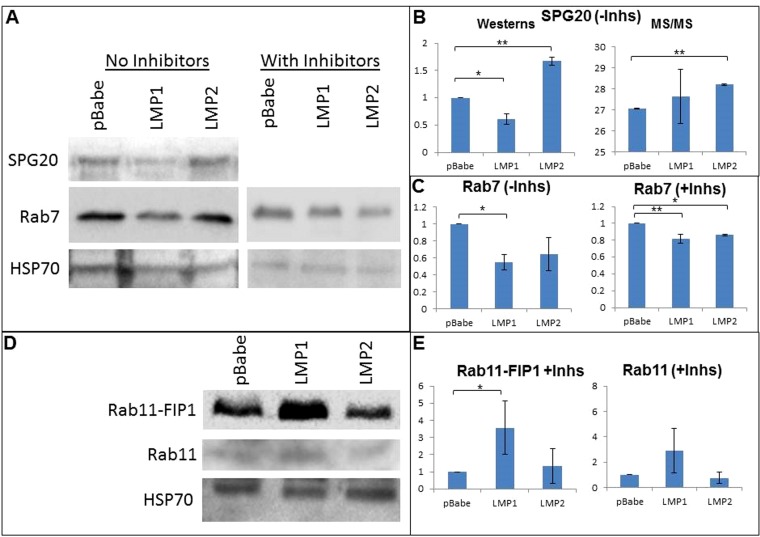
Several of the vesicle trafficking proteins altered with the expression of LMP1 or LMP2A were confirmed by Western transfer (Fig. 5). SPG20 (Spartin) expression was confirmed to be increased in LMP2A-noInh cells compared to the pBabe-noInh control, in agreement with the MS data (Fig. 5A and B). However, Spartin was significantly reduced in the LMP1-noInh cells compared to the pBabe-noInh control (P = 0.03), in contrast to the MS/MS data which indicated greater variance and were not significantly different (Fig. 5B). Western transfer for Rab7 expression without inhibitors was down in both LMP1 and LMP2A samples, although it was only significant with LMP1 expression (P < 0.05) (Fig. 5A and C). With the addition of inhibitors, Rab7 was significantly decreased by both LMP1 and LMP2A compared to their pBabe controls (P = 0.05 and 0.01, respectively). Western transfers also suggested that Rab11-FIP1 was significantly increased by LMP1 in the presence of inhibitors, in agreement with the MS/MS data. Interestingly, Rab11 was also shown to be increased based on immunoblotting, with significant variability (Fig. 5D and E).
FIG 5 .
Western transfers of SPG20, Rab7, Rab11FIP1, and Rab11. Protein expression changes were assessed for SPG20 and Rab7 in pBabe, LMP1 and LMP2A samples without inhibitors, and Rab7, Rab11FIP1, and Rab11 in pBabe, LMP1, and LMP2A samples with inhibitors. Western blots for SPG20 (A and B) confirmed its increased expression in LMP2A-noInh cells (P < 0.01). With LMP1-noInh cells, SPG20 was significantly reduced (P < 0.05), in contrast to the MS/MS data, which contained greater variance. Rab7 expression (A and C) without inhibitors was downregulated in both LMP1 and LMP2A samples, although this change was only significant with LMP1 expression (P < 0.05). With the addition of inhibitors, Rab7 levels in LMP1 and LMP2A cells were both significantly decreased compared to their pBabe controls (P = 0.05 and 0.01, respectively). Rab11FIP1 and Rab11 (D and E) were increased based on Western blot analysis in LMP1+Inh, but only Rab11-FIP1 in LMP1+Inh was significantly different (P < 0.05).
DISCUSSION
This is the first analysis of the effects of the EBV proteins LMP1 and LMP2A on the cellular proteome. As both proteins bind ubiquitin ligases, to identify proteins potentially regulated through this mechanism, the proteomes were also assessed in the presence of inhibitors of both ubiquitination and proteasome-mediated degradation. Not surprisingly, this analysis identified considerably more proteins in the presence of inhibitors that are apparently regulated by LMP1 or LMP2A through effects on ubiquitination or proteasomal processing. Inhibition did not necessarily increase the abundance of proteins, and considerable numbers of the affected proteins decreased in the presence of inhibitors. It is possible that these proteins are regulated at some level by LMP1 or LMP2A through effects on negative regulators of expression which are not degraded with proteasomal inhibition. Additionally, the inability to deubiquitinate proteins could also result in their degradation through standard lysosomal mechanisms.
However, the differences in the levels of expression suggest that LMP1 and LMP2A modulate the ubiquitination of these proteins. Both proteins also affected multiple ubiquitin-conjugating enzymes and peptidases, which could affect the specificity for ubiquitin targeting. It is known that LMP1 activates NF-κB through effects on the function of the canonical NF-κB inhibitor IκBα, and the data presented here indicate that it is highly ubiquitinated in the presence of LMP1 (64). Additionally, LMP1 also promotes the proteasome-mediated processing of NF-κB p100 into p52 in the noncanonical activation of NF-κB. Thus, it is not surprising that there would be little overlap between proteins regulated by NF-κB whose abundance is affected in the presence or absence of inhibitors. It is difficult to predict how the increase or decrease of specific proteins in the absence of deubiquitination and proteasomal degradation would modulate their activity in vivo. However, these data identified many proteins that are likely affected by LMP1 and LMP2A through ubiquitination and proteasomal turnover. Interestingly, multiple specific proteasome components were affected, and the effects of the viral proteins on these components may contribute to proteasome function and specificity.
The effects of LMP1 and LMP2A on cellular expression have previously largely been identified by changes in transcription. Although most of these data have been determined in EBV-infected lymphocytes, comparison of these data to existing transcriptional data identified several genes in common. Approximately 39% of genes modulated by LMP2A without inhibitors have been shown to be affected transcriptionally, while 29% with inhibitors were also identified transcriptionally (65, 66). Similarly, 28% of genes identified in transcriptional studies were identified in the proteome in the LMP1-expressing cells (3). For both LMP1 and LMP2A, the effects on protein abundance did not necessarily agree with effects identified on transcription, which is in keeping with some of the known mechanisms of transcriptional activation. However, comparison with the ENCODE data set indicated that for LMP1 and LMP2A with or without inhibitors, more than 80% of the proteins were genes previously identified in chromatin immunoprecipitation studies for relA (p65). Both LMP1 and LMP2A have known effects on NF-κB, and these findings indicate that both viral proteins affect this pathway directly as well as indirectly, with resulting effects on NF-κB binding.
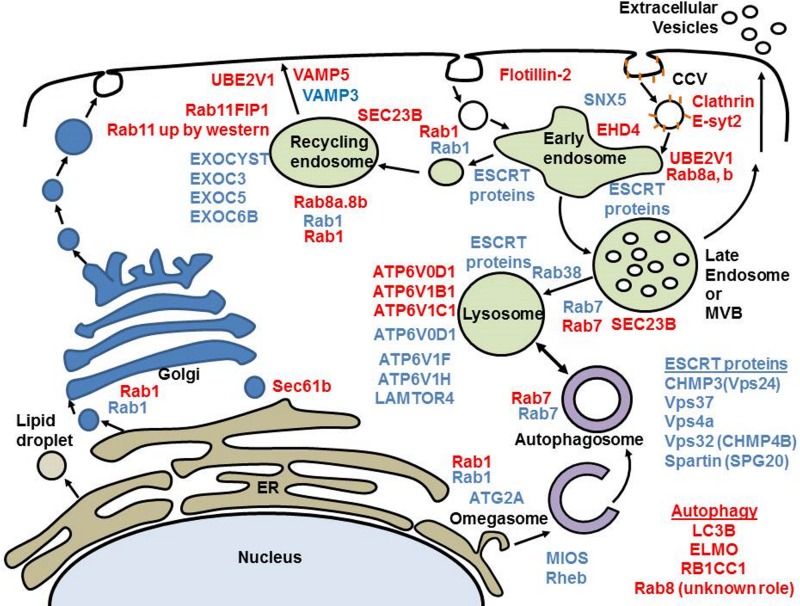
It is interesting to overlay the effects of both LMP1 and LMP2A on the pathways most clearly modulated. Evaluation of the effects on pathways involved in vesicle trafficking (Fig. 6) showed that LMP1 potentially affects the recycling endosome through activation of Rab11/Rab11FIP, while LMP2A potentially modulates activity of the exocyst complex. Additionally, both proteins potentially modulate lysosome activity through effects on ATP6V0D1 and other subunits of the proton pump V-ATPase. Interestingly, our analysis of pathways regulating autophagy predicts a greater effect of LMP1 on autophagosome-to-lysosome flux. It has previously been shown that autophagic flux is blocked in the final steps during the reactivation of EBV from latency. In this case, reduced Rab7 expression correlated with the lack of fusion or flux between lysosomes and autophagosomes (67). LMP1 has also been demonstrated to induce autophagy, increasing the levels of LC3II (MAP1LC3B) (68). In our experiments, LC3 was also significantly elevated and Rab7 significantly reduced in LMP1+Inh samples, with other proteins involved in autophagy, including RB1CC1, ELMO2, TMEM59, and Rab8B, also upregulated in LMP1+Inh samples. Thus, our results expand our knowledge of potential intermediaries regulating autophagosome-lysosome interactions influenced by LMP1.
FIG 6 .
Schematic of vesicle trafficking changes associated with LMP1 or LMP2A expression. Proteins significantly different with LMP1 expression are in red text, and proteins affected by LMP2A are in blue text. Only Rab1 and Rab7 were altered by both LMP1 and LMP2A expression. Although there are few proteins altered by both LMP1 and LMP2A, there is important overlap in the stages of vesicle trafficking predicted to be modulated by their individual expression. Both LMP1 and LMP2A affect the expression of different V-ATPase subunits potentially modulating lysosome activity, and both likely affect mTORC1 signaling via alternate intermediaries. In addition, LMP1 appears to affect the recycling endosome through Rab11/Rab11FIP1, while LMP2A affects the exocyst complex. CCV, Clathrin-coated vesicle; MVB, multivesicular body.
In these same MCF10A cells, we previously showed that LMP2A activates autophagy to prolong survival during the differentiation process of luminal formation (22). In contrast, the results presented here show that only Rab7 was downregulated in both LMP2A-Inh and LMP2A+Inh samples. This may reflect the considerable differences in growth conditions. The cells analyzed here were adherent cultures with the addition of serum, which likely inhibited the induction of autophagy and differentiation that occurs during luminal formation. However, significant changes in the LMP2A+Inh proteome were associated with vesicle trafficking and included a number of vacuolar ATPase subunits, exocyst subunits, Rab1, Rab38, VPS24 (CHMP3), VPS37B, and VPS4A. The majority of these proteins were downregulated, suggesting a dysregulation of vesicle trafficking not previously described with LMP2A.
The importance of virus-mediated effects on the ubiquitin system was clearly revealed in initial studies demonstrating that growth-transforming viral proteins such as E6 induced growth through specific effects and interactions with ubiquitination components, such as E6AP. The data presented here revealed that the EBV oncogenes also likely affect the abundance of many proteins through effects on ubiquitination. Additionally, these proteomics data suggest altered regulation of both endosomal vesicle trafficking and autophagosome-to-lysosome flux with the expression of LMP1 or LMP2A. The ability to affect vesicle formation and trafficking would also contribute to the regulation of many signaling complexes, such as those that are associated with endocytosed receptors or occur at the vesicle membrane. It will be important to further dissect how these virus-mediated effects on protein ubiquitination alter specific protein localization.
MATERIALS AND METHODS
Cell culture.
MCF10A breast epithelial cells expressing LMP1, LMP2A, or pBabe vector (21) were cultured in Dulbecco’s modified Eagle medium with F-12 medium (Gibco, Grand Island, NY) supplemented with 5% horse serum, 20 ng/ml recombinant human epidermal growth factor (rhEGF), 100 ng/ml cholera toxin, 500 µg/ml hydrocortisone, 10 µg/ml insulin, and 1% penicillin/streptomycin in a humidified growth chamber with 5% CO2. Each treatment/experimental group was plated in 6 replicates in 15 cm dishes. Treatment groups consisted of pBabe-, LMP1- or LMP2A-expressing cells untreated or incubated for 6 h with the DUB and proteasome inhibitors N-ethylmaleimide (2 µM final concentration) and MG-132 (10 µM final concentration), respectively.
Sample preparation.
Cells were lysed at 4°C in denaturing lysis buffer (8 M urea, 50 mM Tris [pH 7.5], 150 mM NaCl, 1 mM EDTA) with the addition of protease inhibitors (Complete EDTA-free; Roche) and DUB and proteasome inhibitors when applicable. The resulting lysates were centrifuged at 13,000 × g for 15 min at 4°C. Protein concentrations were estimated using a colorimetric protein assay (Pierce 660 nm; Thermo Scientific). For each treatment group, 5 mg of protein was reduced with 5 mM dithiothreitol (DTT) for 45 min at room temperature, followed by 10 mM N-ethylmaleimide for 30 min while protected from light. Samples were digested with 5 µg LysC (1:100 ratio; Promega) for 4 h at 37°C, then diluted to 2 M urea (final) by using 50 mM Tris (pH 7.5) and digested with 100 µg sequencing-grade trypsin (1:50 ratio; Promega) overnight at 37°C.
Peptide desalting and fractionation.
Following digestion, samples were acidified with formic acid (FA) to a final concentration of 0.5% and consequently desalted using a 500-mg C18 Sep-Pak Vac RC cartridge (Waters) according to the manufacturer’s instructions.
To increase the depth of coverage and quantitation of these lysates, the trypsin-digested desalted samples were fractionated using strong cation exchange (SCX) chromatography, collecting 32 fractions in each run. These fractions were then recombined noncontiguously into 8 final fractions per sample, such that each resulting fraction contained a less complex mixture of peptides with a maximum range of charges. Each fraction was subsequently analyzed by nano-liquid chromatography (nano-LC) run in-line with MS/MS identification of peptides. Samples were fractionated by basic RPLC on an XBridge 4.6- by 250-mm C18 column (Waters).
LC-MS/MS and label-free quantitative proteomics.
Samples were resuspended in 0.1% formic acid. Identification and quantitation of proteins were performed via RPLC-MS/MS on a two-dimensional nano-LC Ultra system coupled to an LTQ-Orbitrap Velos mass spectrometer (Thermo Scientific). Separation of the tryptic peptides was achieved with a 70-min linear gradient of 2 to 30% buffer B in buffer A at 200 nl/min, where buffer A was an aqueous solution of 0.1% formic acid and buffer B was a solution of 0.1% formic acid in acetonitrile (ACN).
Mass spectra were processed, and peptide identification was performed by using the Andromeda search engine found in MaxQuant software version 2.2.1. Peptides were identified using a peptide false-discovery rate (FDR) of 0.05 and a protein FDR of 0.01 with at least 2 unique peptides. Label-free quantitation (LFQ) was calculated by utilizing the measured area under the curve (AUC) of m/z and a retention time-aligned extracted ion chromatogram for each peptide. All 4 replicates (biological and technical) of each sample group were included in the LFQ experimental design, with protein-level quantitation and normalization performed using unique and razor peptide features. Statistical analysis was performed using Perseus (version 1.4.1.3), and the differences in expression profiles were further analyzed using Ingenuity Pathway Analysis software.
ACKNOWLEDGMENT
This work was supported by Public Health grants CA32979 and CA19014 to N.R.-T. from the National Cancer Institute.
Footnotes
Citation DeKroon RM, Gunawardena HP, Edwards R, Raab-Traub N. 2018. Global proteomic changes induced by the Epstein-Barr virus oncoproteins latent membrane protein 1 and 2A. mBio 9:e00959-18. https://doi.org/10.1128/mBio.00959-18.
REFERENCES
- 1.Raab-Traub N. 2007. EBV-induced oncogenesis, chapter 55. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom. [PubMed] [Google Scholar]
- 2.Kieff E, Rickinson AB. 2001. Epstein-Barr virus and its replication, p 2511–2573. In Knipe DM, Howley PM. (ed.), Fields virology, 4th ed., vol II Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 3.Gewurz BE, Mar JC, Padi M, Zhao B, Shinners NP, Takasaki K, Bedoya E, Zou JY, Cahir-McFarland E, Quackenbush J, Kieff E. 2011. Canonical NF-κB activation is essential for Epstein-Barr virus latent membrane protein 1 TES2/CTAR2 gene regulation. J Virol 85:6764–6773. doi: 10.1128/JVI.00422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards RH, Marquitz AR, Raab-Traub N. 2015. Changes in expression induced by Epstein-Barr virus LMP1-CTAR1: potential role of bcl3. mBio 6:e00441-15. doi: 10.1128/mBio.00441-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luftig M, Yasui T, Soni V, Kang MS, Jacobson N, Cahir-McFarland E, Seed B, Kieff E. 2004. Epstein-Barr virus latent infection membrane protein 1 TRAF-binding site induces NIK/IKK alpha-dependent noncanonical NF-κB activation. Proc Natl Acad Sci U S A 101:141–146. doi: 10.1073/pnas.2237183100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Everly DN Jr, Mainou BA, Raab-Traub N. 2004. Induction of Id1 and Id3 by latent membrane protein 1 of Epstein-Barr virus and regulation of p27/Kip and cyclin-dependent kinase 2 in rodent fibroblast transformation. J Virol 78:13470–13478. doi: 10.1128/JVI.78.24.13470-13478.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller WE, Mosialos G, Kieff E, Raab-Traub N. 1997. Epstein-Barr virus LMP1 induction of the epidermal growth factor receptor is mediated through a TRAF signaling pathway distinct from NF-κB activation. J Virol 71:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mainou BA, Everly DN Jr, Raab-Traub N. 2005. Epstein-Barr virus latent membrane protein 1 CTAR1 mediates rodent and human fibroblast transformation through activation of PI3K. Oncogene 24:6917–6924. doi: 10.1038/sj.onc.1208846. [DOI] [PubMed] [Google Scholar]
- 9.Miller CL, Burkhardt AL, Lee JH, Stealey B, Longnecker R, Bolen JB, Kieff E. 1995. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity 2:155–166. doi: 10.1016/S1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 10.Miller CL, Lee JH, Kieff E, Longnecker R. 1994. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci U S A 91:772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Speck P, Kline KA, Cheresh P, Longnecker R. 1999. Epstein-Barr virus lacking latent membrane protein 2 immortalizes B cells with efficiency indistinguishable from that of wild-type virus. J Gen Virol 80:2193–2203. doi: 10.1099/0022-1317-80-8-2193. [DOI] [PubMed] [Google Scholar]
- 12.Caldwell RG, Wilson JB, Anderson SJ, Longnecker R. 1998. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity 9:405–411. doi: 10.1016/S1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 13.Swanson-Mungerson M, Bultema R, Longnecker R. 2006. Epstein-Barr virus LMP2A enhances B-cell responses in vivo and in vitro. J Virol 80:6764–6770. doi: 10.1128/JVI.00433-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shair KH, Bendt KM, Edwards RH, Nielsen JN, Moore DT, Raab-Traub N. 2012. Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) and LMP2A function cooperatively to promote carcinoma development in a mouse carcinogenesis model. J Virol 86:5352–5365. doi: 10.1128/JVI.07035-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morrison JA, Klingelhutz AJ, Raab-Traub N. 2003. Epstein-Barr virus latent membrane protein 2A activates beta-catenin signaling in epithelial cells. J Virol 77:12276–12284. doi: 10.1128/JVI.77.22.12276-12284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scholle F, Bendt KM, Raab-Traub N. 2000. Epstein-Barr virus LMP2A transforms epithelial cells, inhibits cell differentiation, and activates Akt. J Virol 74:10681–10689. doi: 10.1128/JVI.74.22.10681-10689.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Swart R, Ruf IK, Sample J, Longnecker R. 2000. Latent membrane protein 2A-mediated effects on the phosphatidylinositol 3-kinase/Akt pathway. J Virol 74:10838–10845. doi: 10.1128/JVI.74.22.10838-10845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison JA, Raab-Traub N. 2005. Roles of the ITAM and PY motifs of Epstein-Barr virus latent membrane protein 2A in the inhibition of epithelial cell differentiation and activation of β-catenin signaling. J Virol 79:2375–2382. doi: 10.1128/JVI.79.4.2375-2382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukuda M, Longnecker R. 2007. Epstein-Barr virus latent membrane protein 2A mediates transformation through constitutive activation of the Ras/PI3-K/Akt pathway. J Virol 81:9299–9306. doi: 10.1128/JVI.00537-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fotheringham JA, Mazzucca S, Raab-Traub N. 2010. Epstein-Barr virus latent membrane protein-2A-induced ΔNp63α expression is associated with impaired epithelial-cell differentiation. Oncogene 29:4287–4296. doi: 10.1038/onc.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fotheringham JA, Raab-Traub N. 2013. Epstein-Barr virus latent membrane protein 2 effects on epithelial acinus development reveal distinct requirements for the PY and YEEA motifs. J Virol 87:13803–13815. doi: 10.1128/JVI.02203-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fotheringham JA, Raab-Traub N. 2015. Epstein Barr-virus latent membrane protein 2 induces autophagy to prevent cell death. J Virol 89:6940–6944. doi: 10.1128/JVI.03371-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. 1995. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell 80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 24.Izumi KM, Kieff ED. 1997. The Epstein-Barr virus oncogene product latent membrane protein 1 engages the tumor necrosis factor receptor-associated death domain protein to mediate B lymphocyte growth transformation and activate NF-κB. Proc Natl Acad Sci U S A 94:12592–12597. doi: 10.1073/pnas.94.23.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fries KL, Miller WE, Raab-Traub N. 1999. The A20 protein interacts with the Epstein-Barr virus latent membrane protein 1 (LMP1) and alters the LMP1/TRAF1/TRADD complex. Virology 264:159–166. doi: 10.1006/viro.1999.9980. [DOI] [PubMed] [Google Scholar]
- 26.Miller WE, Cheshire JL, Raab-Traub N. 1998. Interaction of tumor necrosis factor receptor-associated factor signaling proteins with the latent membrane protein 1 PXQXT motif is essential for induction of epidermal growth factor receptor expression. Mol Cell Biol 18:2835–2844. doi: 10.1128/MCB.18.5.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda A, Caldwell RG, Longnecker R, Ikeda M. 2003. Itchy, a Nedd4 ubiquitin ligase, downregulates latent membrane protein 2A activity in B-cell signaling. J Virol 77:5529–5534. doi: 10.1128/JVI.77.9.5529-5534.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longnecker R, Kieff ED, Cohen JI. 2012. Epstein-Barr virus, p 1898–1959. In Fields virology, 6th ed., vol VI Lippincott, Williams & Wilkins, Philadephia, PA. [Google Scholar]
- 29.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. [DOI] [PubMed] [Google Scholar]
- 30.Zhang X, Wang W, Wang H, Wang MH, Xu W, Zhang R. 2013. Identification of ribosomal protein S25 (RPS25)-MDM2-p53 regulatory feedback loop. Oncogene 32:2782–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards TL, Clowes VE, Tsang HT, Connell JW, Sanderson CM, Luzio JP, Reid E. 2009. Endogenous Spartin (SPG20) is recruited to endosomes and lipid droplets and interacts with the ubiquitin E3 ligases AIP4 and AIP5. Biochem J 423:31–39. doi: 10.1042/BJ20082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang M, Veselits M, O’Neill S, Hou P, Reddi AL, Berlin I, Ikeda M, Nash PD, Longnecker R, Band H, Clark MR. 2007. Ubiquitinylation of Ig beta dictates the endocytic fate of the B cell antigen receptor. J Immunol 179:4435–4443. doi: 10.4049/jimmunol.179.7.4435. [DOI] [PubMed] [Google Scholar]
- 33.Nelson DE, Randle SJ, Laman H. 2013. Beyond ubiquitination: the atypical functions of Fbxo7 and other F-box proteins. Open Biol 3:130131. doi: 10.1098/rsob.130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christensen DE, Brzovic PS, Klevit RE. 2007. E2-BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol 14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 35.Pelisch F, Pozzi B, Risso G, Muñoz MJ, Srebrow A. 2012. DNA damage-induced heterogeneous nuclear ribonucleoprotein K SUMOylation regulates p53 transcriptional activation. J Biol Chem 287:30789–30799. doi: 10.1074/jbc.M112.390120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez MS, Egaña I, Lopitz-Otsoa F, Aillet F, Lopez-Mato MP, Dorronsoro A, Lobato-Gil S, Sutherland JD, Barrio R, Trigueros C, Lang V. 2014. The RING ubiquitin E3 RNF114 interacts with A20 and modulates NF-κB activity and T-cell activation. Cell Death Dis 5:e1399. doi: 10.1038/cddis.2014.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guasparri I, Bubman D, Cesarman E. 2008. EBV LMP2A affects LMP1-mediated NF-κB signaling and survival of lymphoma cells by regulating TRAF2 expression. Blood 111:3813–3820. doi: 10.1182/blood-2007-03-080309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sahni S, Bae DH, Lane DJ, Kovacevic Z, Kalinowski DS, Jansson PJ, Richardson DR. 2014. The metastasis suppressor, N-myc downstream-regulated gene 1 (NDRG1), inhibits stress-induced autophagy in cancer cells. J Biol Chem 289:9692–9709. doi: 10.1074/jbc.M113.529511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhong J, Kong X, Zhang H, Yu C, Xu Y, Kang J, Yu H, Yi H, Yang X, Sun L. 2012. Inhibition of CLIC4 enhances autophagy and triggers mitochondrial and ER stress-induced apoptosis in human glioma U251 cells under starvation. PLoS One 7:e39378. doi: 10.1371/journal.pone.0039378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu Y, Her C. 2013. VBP1 facilitates proteasome and autophagy-mediated degradation of MutS homologue hMSH4. FASEB J 27:4799–4810. doi: 10.1096/fj.13-235127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng S, Deng L, Chen W, Shao J, Xu G, Li YP. 2009. Atp6v1c1 is an essential component of the osteoclast proton pump and in F-actin ring formation in osteoclasts. Biochem J 417:195–203. doi: 10.1042/BJ20081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naslavsky N, Caplan S. 2005. C-terminal EH-domain-containing proteins: consensus for a role in endocytic trafficking, EH? J Cell Sci 118:4093–4101. doi: 10.1242/jcs.02595. [DOI] [PubMed] [Google Scholar]
- 43.Scharaw S, Iskar M, Ori A, Boncompain G, Laketa V, Poser I, Lundberg E, Perez F, Beck M, Bork P, Pepperkok R. 2016. The endosomal transcriptional regulator RNF11 integrates degradation and transport of EGFR. J Cell Biol 215:543–558. doi: 10.1083/jcb.201601090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ. 2000. Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103:351–361. doi: 10.1016/S0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 45.Guerra F, Bucci C. 2016. Multiple roles of the small GTPase Rab7. Cells 5:34. doi: 10.3390/cells5030034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bernatchez PN, Acevedo L, Fernandez-Hernando C, Murata T, Chalouni C, Kim J, Erdjument-Bromage H, Shah V, Gratton JP, McNally EM, Tempst P, Sessa WC. 2007. Myoferlin regulates vascular endothelial growth factor receptor-2 stability and function. J Biol Chem 282:30745–30753. doi: 10.1074/jbc.M704798200. [DOI] [PubMed] [Google Scholar]
- 47.Blume JJ, Halbach A, Behrendt D, Paulsson M, Plomann M. 2007. EHD proteins are associated with tubular and vesicular compartments and interact with specific phospholipids. Exp Cell Res 313:219–231. doi: 10.1016/j.yexcr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 48.Miller WE, Earp HS, Raab-Traub N. 1995. The Epstein-Barr virus latent membrane protein 1 induces expression of the epidermal growth factor receptor. J Virol 69:4390–4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Velikkakath AK, Nishimura T, Oita E, Ishihara N, Mizushima N. 2012. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol Biol Cell 23:896–909. doi: 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Helgason E, Phung QT, Dueber EC. 2013. Recent insights into the complexity of TANK-binding kinase 1 signaling networks: the emerging role of cellular localization in the activation and substrate specificity of TBK1. FEBS Lett 587:1230–1237. doi: 10.1016/j.febslet.2013.01.059. [DOI] [PubMed] [Google Scholar]
- 51.Boada-Romero E, Letek M, Fleischer A, Pallauf K, Ramón-Barros C, Pimentel-Muiños FX. 2013. TMEM59 defines a novel ATG16L1-binding motif that promotes local activation of LC3. EMBO J 32:566–582. doi: 10.1038/emboj.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ao X, Zou L, Wu Y. 2014. Regulation of autophagy by the Rab GTPase network. Cell Death Differ 21:348–358. doi: 10.1038/cdd.2013.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T, Tooze SA. 2012. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J 31:1931–1946. doi: 10.1038/emboj.2012.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Endo K, Kondo S, Shackleford J, Horikawa T, Kitagawa N, Yoshizaki T, Furukawa M, Zen Y, Pagano JS. 2009. Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene 28:1725–1735. doi: 10.1038/onc.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schafer JC, McRae RE, Manning EH, Lapierre LA, Goldenring JR. 2016. Rab11-FIP1A regulates early trafficking into the recycling endosomes. Exp Cell Res 340:259–273. doi: 10.1016/j.yexcr.2016.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davey JR, Humphrey SJ, Junutula JR, Mishra AK, Lambright DG, James DE, Stöckli J. 2012. TBC1D13 is a RAB35 specific GAP that plays an important role in GLUT4 trafficking in adipocytes. Traffic 13:1429–1441. doi: 10.1111/j.1600-0854.2012.01397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Diego I, Schwartz F, Siegfried H, Dauterstedt P, Heeren J, Beisiegel U, Enrich C, Grewal T. 2002. Cholesterol modulates the membrane binding and intracellular distribution of annexin 6. J Biol Chem 277:32187–32194. doi: 10.1074/jbc.M205499200. [DOI] [PubMed] [Google Scholar]
- 58.Thoreau V, Bergès T, Callebaut I, Guillier-Gencik Z, Gressin L, Bernheim A, Karst F, Mornon JP, Kitzis A, Chomel JC. 1999. Molecular cloning, expression analysis, and chromosomal localization of human syntaxin 8 (STX8). Biochem Biophys Res Commun 257:577–583. doi: 10.1006/bbrc.1999.0503. [DOI] [PubMed] [Google Scholar]
- 59.Vitale G, Alexandrov K, Ullrich O, Horiuchi H, Giner A, Dobson C, Baykova O, Gournier H, Stenmark H, Zerial M. 1995. The GDP/GTP cycle of Rab5 in the regulation of endocytotic membrane traffic. Cold Spring Harb Symp Quant Biol 60:211–220. doi: 10.1101/SQB.1995.060.01.024. [DOI] [PubMed] [Google Scholar]
- 60.Kharkwal H, Smith CG, Wilson DW. 2016. Herpes simplex virus capsid localization to ESCRT-VPS4 Complexes in the presence and absence of the large tegument protein UL36p. J Virol 90:7257–7267. doi: 10.1128/JVI.00857-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Seto S, Tsujimura K, Koide Y. 2011. Rab GTPases regulating phagosome maturation are differentially recruited to mycobacterial phagosomes. Traffic 12:407–420. doi: 10.1111/j.1600-0854.2011.01165.x. [DOI] [PubMed] [Google Scholar]
- 63.Wu B, Guo W. 2015. The exocyst at a glance. J Cell Sci 128:2957–2964. doi: 10.1242/jcs.156398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cahir-McFarland ED, Carter K, Rosenwald A, Giltnane JM, Henrickson SE, Staudt LM, Kieff E. 2004. Role of NF-κB in cell survival and transcription of latent membrane protein 1-expressing or Epstein-Barr virus latency III-infected cells. J Virol 78:4108–4119. doi: 10.1128/JVI.78.8.4108-4119.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Portis T, Dyck P, Longnecker R. 2003. Epstein-Barr virus (EBV) LMP2A induces alterations in gene transcription similar to those observed in Reed-Sternberg cells of Hodgkin lymphoma. Blood 102:4166–4178. doi: 10.1182/blood-2003-04-1018. [DOI] [PubMed] [Google Scholar]
- 66.Shair KH, Raab-Traub N. 2012. Transcriptome changes induced by Epstein-Barr virus LMP1 and LMP2A in transgenic lymphocytes and lymphoma. mBio 3:e0288-12. doi: 10.1128/mBio.00288-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Granato M, Santarelli R, Farina A, Gonnella R, Lotti LV, Faggioni A, Cirone M. 2014. Epstein-Barr virus blocks the autophagic flux and appropriates the autophagic machinery to enhance viral replication. J Virol 88:12715–12726. doi: 10.1128/JVI.02199-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee DY, Lee J, Sugden B. 2009. The unfolded protein response and autophagy: herpesviruses rule! J Virol 83:1168–1172. doi: 10.1128/JVI.01358-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Proteomics data set. P values, fold changes, and gene names are provided for t test comparisons between LMP1 and pBabe without inhibitors, LMP2A and pBabe without inhibitors, LMP1 and pBabe with inhibitors, and LMP2A and pBabe with inhibitors. The results for each t test are presented on a separate tab. Download TABLE S1, PDF file, 1.9 MB (1.9MB, xls) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: vesicle formation and trafficking. The P value and fold change data for all proteins associated with vesicle formation and trafficking pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used for IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, data for other comparisons are also presented, unless they were from the manual curation. Download TABLE S2, PDF file, 0.11 MB (114.9KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: transcription and translation. The P value and fold change data for all proteins associated with transcription and translation pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, the data from other comparisons are also presented. Download TABLE S3, PDF file, 0.24 MB (239.6KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: cytoskeleton, cell movement, and cell junctions. The P value and fold change data for all proteins associated with cytoskeleton, cell movement, or cell junction pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) that was significant in one comparison, the data in other comparisons are also presented. Download TABLE S4, PDF file, 0.16 MB (161.3KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
IPA data: other or known LMP1-LMP2A pathways. The P value and fold change data for all proteins associated with known LMP1- or LMP2A-associated pathways in the IPA analysis are shown. In each pathway, the significant data for each comparison (LMP1-noInhs, LMP2A-noInhs, LMP1+Inhs, LMP2A+Inhs) are presented, with significant P values highlighted following the convention used by IPA. Green cells indicate significantly downregulated levels versus pBabe, and red cells indicate significantly upregulated levels versus pBabe. For each protein (or gene) significant in one comparison, the data in other comparisons are also presented. Download TABLE S5, PDF file, 0.25 MB (252.2KB, pdf) .
Copyright © 2018 DeKroon et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.