Abstract
Introduction
Melanoma is a deadly malignancy and the poor prognosis of patients with advanced disease is relatively poor. Recent studies indicate that long non-coding RNAs are involved in the pathogenesis of malignant melanoma. This study aims to investigate the role of the long non-coding RNA H19 in melanoma and to explore the underlying molecular mechanisms.
Materials and methods
The expression levels of H19 in clinical samples and melanoma cells were determined by quantitative real-time PCR. The cell growth and cell metastasis were assessed by Cell Counting Kit 8, cell invasion and wound healing assays. Cell apoptosis and cell cycle were determined by flow cytometry. Protein levels were determined by Western blotting assay.
Results
H19 was highly expressed in melanoma tissues compared to normal adjacent skin tissues, and the tissue expression level of H19 from melanoma patients with metastasis was significantly higher than that from patients without distant metastasis. In addition, the high expression of H19 in melanoma tissues was associated with advanced tumor invasion and TNM stage, distal metastasis, lymph node metastasis and shorter overall survival in patients with melanoma. The in vitro functional assays showed that knockdown of H19 inhibited cell growth, invasion and migration and also induced cell apoptosis as well as G0/G1 arrest in melanoma cells. Further quantitative real-time PCR and Western blot experiments showed that knockdown of H19 differentially regulated the epithelial–mesenchymal transition (EMT)-related gene expressions and reversed EMT in melanoma cell lines. Knockdown of H19 suppressed in vivo tumor growth and modulated the expressions of EMT-related genes in nude mice.
Conclusion
The results from this study suggest that upregulation of H19 contributes to melanoma development and progression.
Keywords: melanoma, lncRNAs, H19, apoptosis, invasion and migration, epithelial–mesenchymal transition
Introduction
Long non-coding RNAs (lncRNAs) are non-protein coding transcripts with more than 200 nucleotides in length.1 In the last decade, lncRNAs have been intensively studied for their diverse roles in cellular processes including chromatin remodeling, transcription, post-transcriptional processing and intracellular trafficking.2 Due to the diverse cellular functions of lncRNAs, studies have focused on the roles of lncRNAs in disease etiology, particularly in the pathogenesis of cancer.1 For example, aberrant lncRNA HOX transcript antisense intergenic RNA expression has been suggested as a positive biomarker for the prognosis of patients with carcinoma of digestive systems.3 Another line of evidence from the meta-analysis showed that the lncRNA HOXA transcript at the distal tip served as a novel predictor of lymph node metastasis and survival in human cancer.4 The lncRNA nuclear enriched abundant transcript 1 was found to promote pancreatic cancer progression via negatively modulating miR-506-3p expression.5 The lncRNA maternally expressed gene 3 contributes to the epigenetic regulation of epithelial–mesenchymal transition (EMT) in lung cancer cell lines.6 Therefore, these data suggest that lncRNAs play important roles in the pathogenesis of tumor malignancy.
Melanoma is the most aggressive type of skin cancers that originate from melanocytes, and melanoma accounts for approximately 70% death of skin cancer patients.7 In comparison to the stable or declining rates of most other types of cancers, new cases of and deaths due to melanoma are expected to rise.8 Though various treatments have been employed for treating melanoma, the most effective ways to treat melanoma were early diagnosis and surgery intervention.9 However, a substantial part of melanoma patients had distant metastasis, which results in poor prognosis with a 5-year survival rate of about 20%.9 Up to date, the exact molecular mechanisms underlying melanoma development, particularly metastasis, were largely unknown. Providing the important roles of lncRNA in the cancer pathogenesis, the functions of lncRNAs in melanoma development were explored in several studies. lncRNA antisense non-coding RNA in the INK4 locus (ANRIL) was found to trigger efficient therapeutic efficacy by reprogramming the aberrant INK4 hub in melanoma.10 Chen et al showed that the lncRNA growth arrest-specific 5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo.11 lncRNA urothelial cancer-associated 1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma.12 Recently, the lncRNA H19 has been extensively studied for its role in cancer development including gastric cancer, colorectal cancer, breast cancer, pancreatic cancer, cervical cancer and so on.13 However, the role of H19 in melanoma development has been not reported.
In the present study, the expression levels of H19 in clinical sample tissues were examined in patients with melanoma, and the potential prognostic role of H19 was also explored. Furthermore, in vitro functional roles of H19 in melanoma cell lines were examined by Cell Counting Kit 8 (CCK-8) assay, cell invasion assay, wound healing assay as well as flow cytometry. Whether H19 had an effect on EMT was also explored by qRT-PCR and Western blot assays. The in vivo knockdown effects of H19 on tumor growth and EMT were examined in a nude mice xenograft model.
Materials and methods
Clinical sample tissues
In the present study, 82 patients with melanoma were included in this study. All the patients were recruited from the Affiliated Wuxi No 4 People’s Hospital of Jiangnan University. Melanoma tissues and adjacent normal skin tissues were obtained from patients who underwent surgery at the Affiliated Wuxi No 4 People’s Hospital of Jiangnan University. No patient received chemotherapy or any other form of therapy before surgical resection. The clinicopathological parameters of all the patients are shown in Table 1. The study was performed in accordance with the institutional ethical guidelines, and the use of human skin tissues was approved by the Ethics Committee of the Affiliated Wuxi No 4 People’s Hospital of Jiangnan University. Written informed consent was obtained from all the patients, and the study was conducted according to the principles expressed in the Declaration of Helsinki. All collected samples were snap frozen in liquid nitrogen and then stored at −80°C for further experimentation.
Table 1.
The correlation between lncRNA H19 expression and clinicopathological parameters of patients with melanoma
| Parameters | Number of patients
|
P-value | |
|---|---|---|---|
| Low H19 expression (n) | High H19 expression (n) | ||
| Sex | 0.191 | ||
| Male | 21 | 28 | |
| Female | 19 | 14 | |
| Age (years) | 0.261 | ||
| <60 | 24 | 20 | |
| ≥60 | 16 | 22 | |
| Invasion | 0.0128 | ||
| T0–T2 | 22 | 11 | |
| T3–T4 | 18 | 31 | |
| TNM stage | 0.0076 | ||
| 0–I | 26 | 14 | |
| II–IV | 14 | 28 | |
| Distal metastasis | 0.0153 | ||
| No | 25 | 15 | |
| Yes | 15 | 27 | |
| Lymph node metastasis | 0.0481 | ||
| No | 22 | 14 | |
| Yes | 18 | 28 | |
Abbreviation: lncRNA, long non-coding RNA.
Cell culture
The melanoma cell lines including CHL-1, UACC904, A-375 and 1205Lu were obtained from American Type Culture collection (Manassas, VA, USA). Cells were maintained in DMEM medium supplemented with 10% fetal bovine serum (FBS; Gibco, Thermo Fisher Scientific, Carlsbad, CA, USA) in a humidified atmosphere at 37°C with 5% CO2. Human epidermal melanocyte (HEM) cells were obtained from Type Culture Collection of the Chinese Academy of Sciences (Shanghai, People’s Republic of China), and cells were maintained in the Melanocyte Growth Medium (Promo-Cell, Shanghai, People’s Republic of China) in a humidified atmosphere at 37°C with 5% CO2.
Oligonucleotides of siRNAs and cell transfection
The siRNAs (siRNA1 and siRNA2) for knockdown of H19 and the scramble siRNA for negative control were purchased from RiboBio (Guangzhou, People’s Republic of China). The constructed H19-overexpressing vectors (pcDNA3.1-H19) and pcDNA3.1 empty vectors were obtained from GenePharma (Shanghai, People’s Republic of China). For siRNA and vector transfection, cells were seeded into six-well plates at a density of 5 × 104 cells/well to reach about 50%–80% confluence for transfection. The transient transfection was performed by using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. At 24 h after transfection, cells were processed for further experimentation.
RNA extraction and quantitative real-time PCR (qRT-PCR)
The RNAs from tissue samples and cells were extracted by using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. Gene expression were determined by SYBR green qRT-PCR assay (Takara, Dalian, People’s Republic of China), and GAPDH was used as an internal control. The relevant gene expression levels were calculated by using 2−ΔΔCT method.
Cell growth assay
Cell growth was assessed by using CCK-8 assay (Dojindo Molecular Technologies, Rockville, MD, USA) according to the manufacturer’s protocol. Briefly, cells were seeded in a 96-well plate and transfected with corresponding siRNAs. At 0, 24, 48 and 72 h after transfection, 10 μL of CCK-8 solution was added to each well and incubated for 1 h. The OD values were detected at a wavelength of 450 nm using a microplate reader (Bio-Rad, Hercules, CA, USA).
Invasion assay
Cell invasion assay was carried out by using the Transwell insert chamber coated with Matrigel (BD Biosciences, San Jose, CA, USA). Briefly, the transfected cells (A-375 and 1205Lu) were re-suspended in a 100 μL serum-free medium at a density of 1 × 105 cells/mL and were added to the upper chamber. The lower chamber was supplied with 500 μL of DMEM medium supplemented with 10% FBS. After culturing for 24 h, the cells on the top surface of the insert were removed. Cells that invaded into the lower side of the membrane were fixed with methanol and stained with 20% Giemsa. The number of invaded cells was then counted under a light microscope.
Wound healing assay
After transfection, cells (A-375 and 1205Lu) were cultured in a six-well plate for 24 h until 90% of the base was filled. Wound line was created by scratching the plates with a 10 μL micropipette tip. Then, the cells were cultured for another 24 h. The migration distance was measured after images were taken under a light microscope. The migration distance of the control group was taken as 100% to assess cellular migration and compared with that of the experimental group.
Cell apoptosis and cell cycle analysis
Flow cytometry was performed to assess cell apoptosis and cell cycle distribution.
For cell apoptosis analysis, at 24 h after transfection, cells (A-375 and 1205Lu) were collected by trypsinization and washed with ice-cold PBS. Then, cells were stained with Annexin V-FITC and propidium iodide (PI) working solution by using the Vybrant Apoptosis Assay Kit (Invitrogen) at room temperature for 5 min in the dark. A FACScan flow cytometer (BD Biosciences) was used for the analysis of cell apoptosis.
For cell cycle analysis, at 24 h after transfection, cells (A-375 and 1205Lu) were collected by trypsinization and washed with ice-cold PBS and fixed in ice-cold 70% methanol overnight. Then, cells (A-375 and 1205Lu) were stained with 20 μg/mL PI and 100 μg/mL RNase A in PBS for 15 min at room temperature. A FACScan flow cytometer (BD Biosciences) and CellQuest software (BD Biosciences) were used for the analysis (488 nm emission and 570 nm excitation).
RT2 Profiler™ PCR array
The Human EMT RT2 Profiler™ PCR array was performed by KangChen Bio-tech (Shanghai, People’s Republic of China). Briefly, RNA from cells (A-375 and 1205Lu) was extracted by using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. qRT-PCR array was done using Super Array PCR master mix on an ABI PRISM7900 instrument (Applied Biosystems, Foster City, CA, USA), and the expression levels were calculated by using 2−ΔΔCT method.
Western blotting
Total protein from cells (A-375 and 1205Lu) was extracted using the RIPA buffer. Then, the extracted samples were boiled, and equal amounts of total lysates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. The transferred membranes were then blocked in 5% skimmed milk at room temperature for 1 h and then incubated with anti-E-cadherin, anti-vimentin and anti-N-cadherin antibodies (Abcam, Cambridge, MA, USA) at 4°C overnight; β-actin was used for loading control. After being washed with Tween 20-PBS, membranes were incubated with appropriate HRP-conjugated secondary antibodies for 1 h. Specific bands were then visualized by using an ECL kit (Amersham Biosciences, Little Chalfont, UK).
Lentivirus packaging and stable cell lines
Lentivirus carrying H19_shRNA2 or shRNA control was designed and packaged by GeneChem (Shanghai, People’s Republic of China). Lentivirus was packaged in HEK-293T cells and collected from the medium supernatant. Stable cells lines were established by infecting lentivirus into 1205Lu cells and selected by puromycin.
Xenograft mouse model
All the animal experiments were approved by the Animal Ethics Committee of the Affiliated Wuxi No 4 People’s Hospital of Jiangnan University and were in accordance with experimental protocol of the Affiliated Wuxi No 4 People’s Hospital of Jiangnan University. Briefly, the 6-week-old female nude mice (each group had 6 animals) (Cancer Institute of the Chinese Academy of Medical Science, Shanghai, People’s Republic of China) were subcutaneously injected with 1205Lu cells (1 × 106) stably expressing H19_shRNA2 or control shRNA. The tumor volume (length × width2 × 0.5) in the nude mice was measured every 4 days for up to 32 days after injection. At 32 days after injection, the mice were sacrificed and tumors were excised. After tumor weight was measured, the tumors were snap frozen in liquid nitrogen and then stored at −80°C for further experimentation.
Statistical analysis
All the data were analyzed by using the SPSS Version 13.0 software and GraphPad Prism Version 5.0 software. The experimental data were expressed as mean ± SEM. Significant differences between different groups were analyzed by one-way ANOVA or Student’s t-test, as appropriate. The survival curves were plotted according to the Kaplan–Meier method and were analyzed by using the log-rank test. All the in vitro experiments were performed in triplicates. Differences between groups were considered to be statistically significant when P<0.05.
Results
H19 was upregulated in melanoma tissues and correlated with poor clinical characteristics
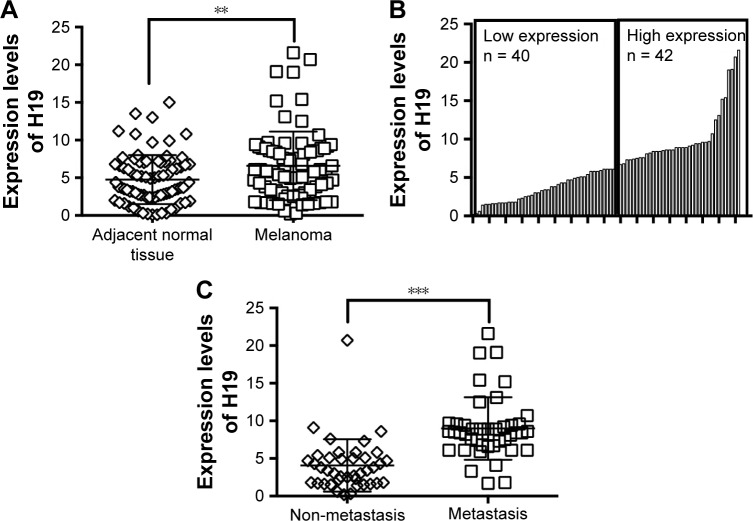
The expression levels of H19 in melanoma tissues and adjacent normal tissues from 82 patients were determined by qRT-PCR, and the results showed that the expression level of H19 melanoma tissues was significantly higher than that in adjacent normal tissues (Figure 1A). Based on H19 expression level in the melanoma tissues, the expression level of H19 was classified into low expression of H19 (n = 40) and high expression of H19 (n = 42) using median as the cutoff value (Figure 1B). Further analysis showed that the expression levels of H19 in melanoma tissues from patients with distal metastasis were significantly higher than those from patients without metastasis (Figure 1C). The correlation analysis between H19 expression and clinicopathological parameters is shown in Table 1, and high expression of H19 was correlated with advanced tumor invasion and TNM stage, distal metastasis and lymph node metastasis (Table 1). In addition, we also investigated if the expression level of H19 correlated with overall survival (OS) of patients with melanoma. Kaplan–Meier survival analysis showed that patients with high expression level of H19 in melanoma tissues had significantly shorter OS when compared to patients with low expression level of H19 in melanoma tissues (Figure 2).
Figure 1.
Differential expressions of H19 in clinical samples.
Notes: (A) The expression levels of H19 in adjacent normal tissues (n = 82) and melanoma tissues (n = 82) were determined by quantitative real-time PCR (qRT-PCR) assay. (B) The low expression of H19 (n = 40) and high expression of H19 (n = 42) in melanoma tissues were divided based on the median value of H19 expressions. (C) The expression levels of H19 in non-metastatic (n = 40) and distal metastatic (n = 42) melanoma tissues were compared. Significant differences between groups were shown as **P<0.01, ***P<0.001.
Figure 2.
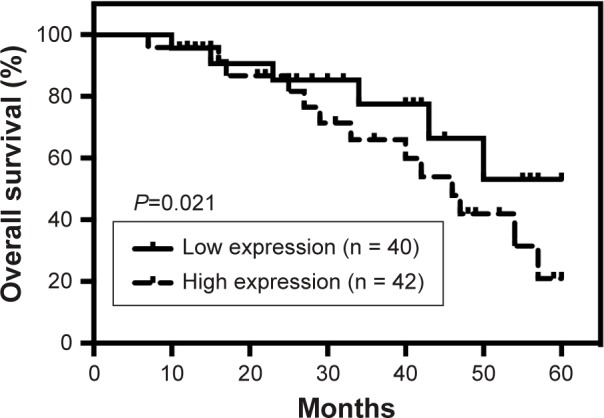
Kaplan–Meier survival analysis of high or low expression levels in melanoma tissues from cancer patients.
Note: The low expression (n = 40) and high expression (n = 42) of H19 were defined by the median of H19 expression.
The effect of H19 on cell growth, cell apoptosis and cell cycle in melanoma cell lines
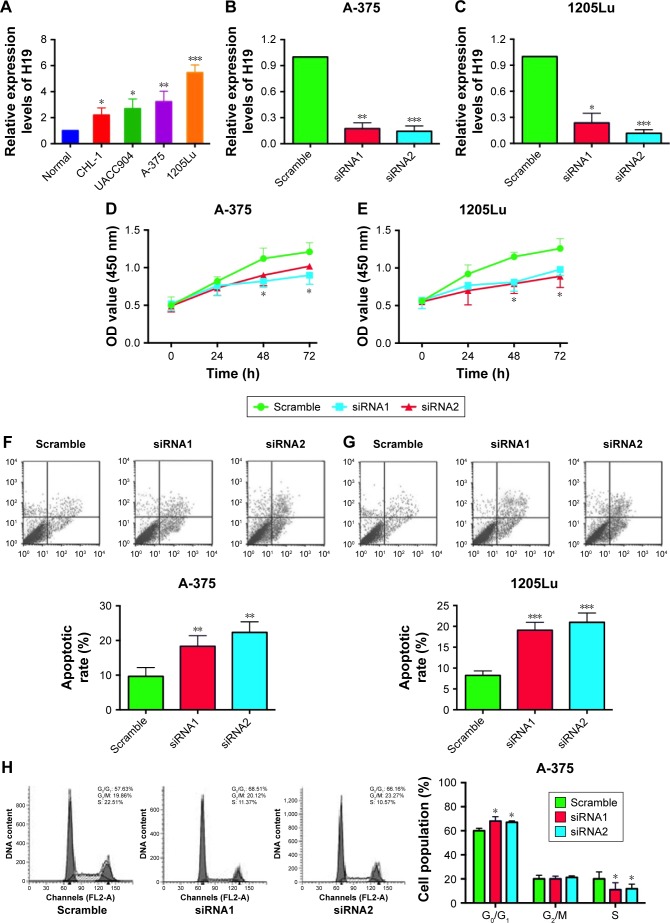
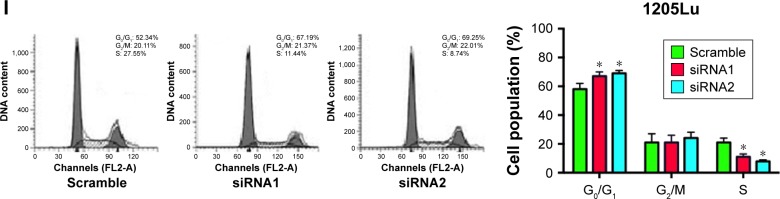
To further explore the underlying mechanisms of H19 in melanoma progression, a series of in vitro assays was performed in the melanoma cell lines. Firstly, we found that the expression level of H19 in melanoma cell lines including CHL-1, UACC904, A-375 and 1205Lu was significantly higher than that in HEM cells (Figure 3A) as determined by qRT-PCR. In order to perform the loss-of-function experiments, we transfected the A-375 cells and 1205Lu cells with H19 siRNAs, ie, siRNA1 and siRNA2, and H19 siRNA transfection significantly reduced the expression levels of H19 in A-375 cells and 1205Lu cells compared to cells transfected with scramble siRNA (Figure 3B and C). We then performed CCK-8 assay to examine the knockdown effect of H19 on cell growth in melanoma cell lines. As shown in Figure 3D and E, knockdown of H19 significantly reduced the cell growth in A-375 and 1205Lu cells as compared to cells transfected with scramble siRNA. The effects of H19 overexpression on cell proliferative ability was also examined in CHL-1 cells, and transfection with pcDNA3.1-H19 significantly increased H19 expression level and promoted cell growth of CHL-1 cells (Figure S1). Furthermore, flow cytometry experiments were performed to determine cell apoptotic rate and cell cycle. Knockdown of H19 significantly increased the cell apoptotic rate in A-375 and 1205Lu cells as compared to control group (Figure 3F and G). In the aspect of cell cycle, knockdown of H19 significantly increased the cell population at G0/G1 phase and correspondingly decreased the cell population at S phase; G2/M phase was not affected (Figure 3H and I).
Figure 3.
Knockdown of H19 inhibits cell growth, induces apoptosis and affects cell cycle in melanoma cell lines.
Notes: (A) The expression levels of H19 in the normal melanocytes, human epidermal melanocytes (HEM) and melanoma cells lines including CHL-1, UACC904, A-375 and 1205Lu were determined by quantitative real-time PCR (qRT-PCR). (B and C) Knockdown of H19 by siRNA1 and siRNA2 suppressed the expression levels of H19 in A-375 and 1205Lu cells. Cell Counting Kit 8 (CCK-8) assay was performed to determined cell growth, and H19 knockdown significantly inhibited cell growth in (D) A-375 cells and (E) 1205Lu cells. Flow cytometry analysis of cell apoptosis was performed in A-375 cell and 1205Lu cells, and knockdown of H19 induced cell apoptosis in (F) A-375 cells and (G) 1205Lu cells. For the cell cycle analysis, knockdown of H19 decreased cell population in G0/G1 phase and increased cell population in S phase as determined in (H) A-375 cells and (I) 1205Lu cells. All the experiments were performed in triplicates. Significant differences compared to Scramble control group were shown as *P<0.05, **P<0.01, ***P<0.001.
The effect of H19 on the metastatic ability of melanoma cell lines
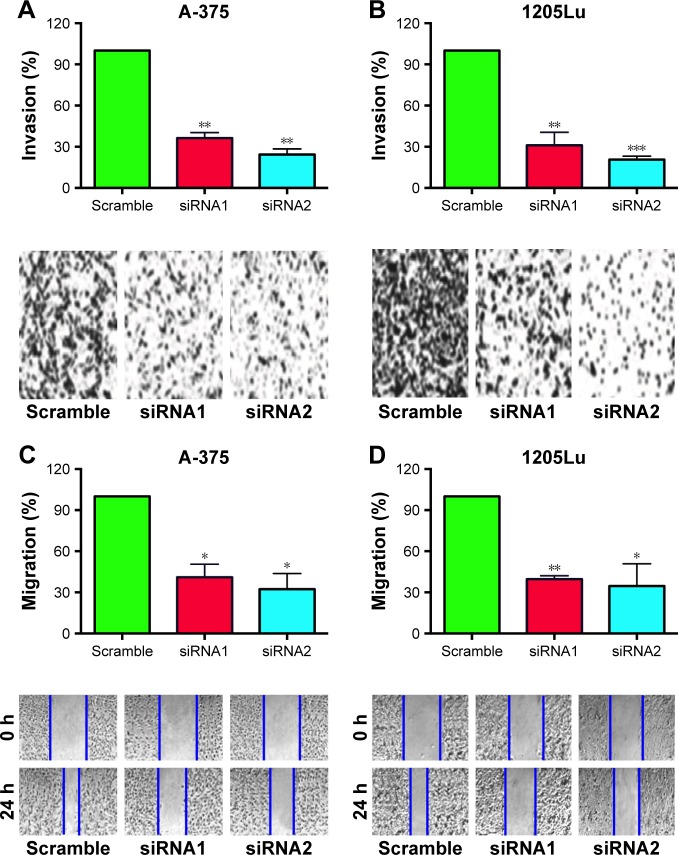
Transwell cell invasion and wound healing assays were performed to determine the effect of H19 on the metastatic ability in A-375 and 1205Lu cells. As shown in Figure 4A and B, the number of invaded cells in A-375 and 1205Lu cells transfected with H19 siRNAs was significantly reduced when compared to cells transfected with scramble siRNA (Figure 4A and B). For the wound healing assay, the results showed that cells transfected with H19 siRNAs had shorter migrating distance at 24 h as compared to cells transfected with scramble siRNA (Figure 4C and D).
Figure 4.
Knockdown of H19 inhibits cell invasion and migration in melanoma cell lines.
Notes: Knockdown of H19 reduced the number of invaded cells in (A) A-375 cells and (B) 1205Lu cells as determined by cell invasion assay. Knockdown of H19 reduced the migrating distance in (C) A-375 cells and (D) 1205Lu cells as determined by wound healing assay. All the experiments were performed in triplicates. Significant differences compared to Scramble control group were shown as *P<0.05, **P<0.01, ***P<0.001.
The effect of H19 on the EMT of melanoma cell lines
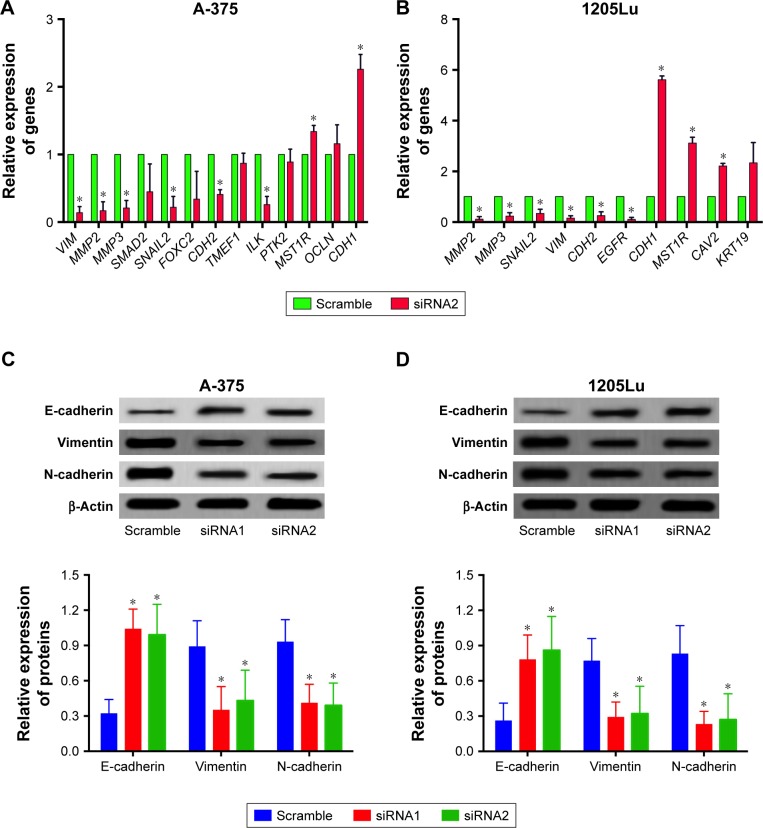
EMT is an important process for tumor invasion and metastasis. In this regard, we performed the RT2 Profiler™ PCR array to examine the effect of H19 on the differential expression of EMT-related genes. In this regard, we performed the RT2 ProfilerTM PCR array on the differential expression of EMT-related genes including cell surface receptors; cytoskeletal genes mediating cell adhesion, migration, motility and morphogenesis. The results showed that H19 knockdown significantly affected the expression levels of several key EMT-related markers, including E-cadherin, vimentin and N-cadherin. As shown in Table 2, knockdown of H19 decreased the expression levels of VIM, MMP2, MMP3, SMAD2, SNAIL2, FOXC2, CDH2, TMEF1, ILK and PTK2 and increased the expression levels of MST1R, OCLN and CDH1 in A-375 cells. The results from Table 3 showed that knockdown of H19 decreased the expression levels of MMP2, MMP3, SNAIL2, VIM, CDH2 and EGFR and increased the expression levels of CDH1, MST1R, CAV2 and KRT19 in 1205Lu cells. Further qRT-PCR experiments were performed to confirm the RT2 Profiler™ PCR results, and the qRT-PCR results showed that knockdown of H19 decreased the expression levels of VIM, MMP2, MMP3, SNAIL2, CDH2, TMEF1 and ILK and increased the expression levels of MST1R and CDH1 in A-375 cells (Figure 5A), and knockdown of H19 decreased the expression levels of MMP2, MMP3, SNAIL2, VIM, CDH2 and EGFR and increased the expression levels of CDH1, MST1R and CAV2 in 1205Lu cells (Figure 5B). Western blot results showed that knockdown of H19 increased the protein expression level of E-cadherin and decreased the protein expression levels of vimentin and N-cadherin in both A-375 cells and 1205Lu cells (Figure 5C and D).
Table 2.
Gene expressions of EMT-related genes (totally 84 genes were included for analysis) after H19 knockdown in A-375 cell lines
| Genes | Functional grouping | Fold change | P-value |
|---|---|---|---|
| VIM | Migration and motility | −7.83 | <0.001 |
| MMP2 | Extracellular matrix and cell adhesion | −7.12 | <0.001 |
| MMP3 | Extracellular matrix and cell adhesion | −6.54 | <0.001 |
| SMAD2 | Morphogenesis | −6.23 | <0.01 |
| SNAIL2 | Differentiation and development | −6.01 | <0.01 |
| FOXC2 | Differentiation and development | −5.67 | <0.01 |
| CDH2 | Extracellular matrix and cell adhesion | −5.34 | <0.01 |
| TMEF1 | Differentiation and development | −5.21 | <0.01 |
| ILK | Cell growth and proliferation | −5.11 | <0.01 |
| PTK2 | Extracellular matrix and cell adhesion | −4.02 | <0.01 |
| MST1R | Migration and motility | 6.12 | <0.001 |
| OCLN | Cell growth and proliferation | 5.13 | <0.01 |
| CDH1 | Extracellular matrix and cell adhesion | 4.17 | <0.01 |
Abbreviation: EMT, epithelial–mesenchymal transition.
Table 3.
Gene expressions of EMT-related genes (totally 84 genes were included for analysis) after H19 knockdown in 1205Lu cell lines
| Genes | Functional grouping | Fold change | P-value |
|---|---|---|---|
| MMP2 | Extracellular matrix and cell adhesion | −9.28 | <0.001 |
| MMP3 | Extracellular matrix and cell adhesion | −9.19 | <0.001 |
| SNAIL2 | Differentiation and development | −8.82 | <0.001 |
| VIM | Migration and motility | −7.43 | <0.01 |
| CDH2 | Extracellular matrix and cell adhesion | −6.12 | <0.01 |
| EGFR | Migration and motility | −4.1 | <0.01 |
| CDH1 | Extracellular matrix and cell adhesion | 7.88 | <0.001 |
| MST1R | Migration and motility | 5.14 | <0.01 |
| CAV2 | Cell growth and proliferation | 5.1 | <0.01 |
| KRT19 | Estrogen receptor signaling pathways | 4.27 | <0.01 |
Abbreviation: EMT, epithelial–mesenchymal transition.
Figure 5.
Differential expression of EMT-related markers and EMT-related genes in melanoma cell lines after H19 knockdown.
Notes: Knockdown of H19 reversed epithelial–mesenchymal transition (EMT) in (A) A-375 cells and (B) 1205Lu cells as determined by Western blot assay. Knockdown of H19 differentially affected the expression levels of EMT-related genes in (C) A-375 cells and (D) 1205Lu cells as determined by quantitative real-time PCR (qRT-PCR). All the experiments were performed in triplicates. Significant differences compared to Scramble control were shown as *P<0.05.
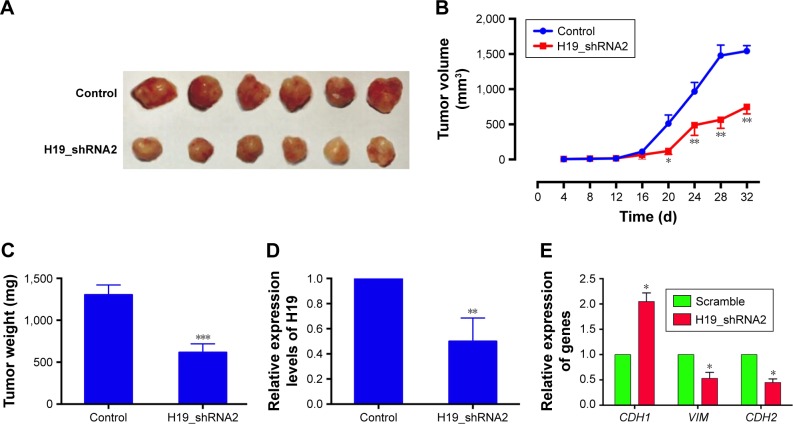
Knockdown of H19 suppressed the in vivo tumor growth in the nude mice
To further examine the effect of H19 on the in vivo tumor growth, we constructed the 1205Lu cells stably expressing H19 siRNA2 (H19_shRNA2) or the control shRNA (control). As shown in Figure 6A and B, the tumor volume was significantly decreased in the H19_shRNA2 group compared to the control group. The tumor weight in the H19_shRNA group was also lower than that in control group (Figure 6C). In addition, the expression level of H19 in the excised tumor from H19_shRNA2 group was significantly lower than that in control group (Figure 6D). The mRNA expression level of CDH1 was increased and the mRNA expression levels of VIM and CDH2 was decreased in the excised tumor form H19_ shRNA2 group as compared to control group (Figure 6E).
Figure 6.
Knockdown of H19 suppressed the in vivo tumor growth in the nude mice.
Notes: (A) Photographs of harvested tumors from the xenograft-transplanted nude mice were captured at 32 days after the injection of H19_shRNA2-overexpressing 1205Lu cells or control shRNA-expressing 1205Lu cells. (B) Tumor volume in the nude mice was measured every 4 days for a total of 32 days after injection. (C) Tumor weight was measured when the mice were sacrificed at 32 days after injection. (D) The expression levels of H19 and (E) the mRNA expression levels of CDH1, VIM and CDH2 from the excised tumors were determined by qRT-PCR. Each group had 6 animals. Significant differences compared to control group were shown as *P<0.05, **P<0.01, ***P<0.001.
Discussion
In the present study, we for the first time examined the role of H19 in melanoma development by studying clinical samples from melanoma patients as well as melanoma cell lines. The results showed that H19 was highly expressed in melanoma tissues compared to normal adjacent skin tissue. The tissue expression level of H19 from melanoma patients with metastasis was also significantly higher than that from patients without distal metastasis and correlated with advanced tumor invasion and TNM stage, distal metastasis and lymph node metastasis. In addition, the high expression of H19 in melanoma tissues was associated with shorter OS in patients with melanoma. The in vitro functional assays showed that knockdown of H19 inhibited cell proliferation, invasion and migration and also induced cell apoptosis as well as cell cycle arrest. Further qRT-PCR and Western blot experiments showed that knockdown of H19 differentially regulated the EMT-related gene expression and reversed EMT in melanoma cell lines. Knockdown of H19 suppressed the in vivo tumor growth in the nude mice. Collectively, the data suggest that upregulation of H19 contributes to melanoma development and progression.
The lncRNA H19 has been well studied in various types of cancer, and H19 is reported to act as an oncogene in prostate cancer, bladder cancer, gastric cancer and glioma.14–17 On the other hand, H19 was also found to be downregulated in hepatocellular cancer.18 The underlying mechanisms of H19 dysregulation are largely unknown. In the K562 leukemic cells, disruption of Bcr-Abl expression resulted in a decreased expression of c-Myc with simultaneously reduced levels of H19, and silencing c-Myc expression alone in K562 cells significantly decreased the level of H19, suggesting that the expression of H19 may be regulated by Bcr-Abl/c-Myc axis.19 The upregulation of H19 in melanoma may be due to the upregulation of the lncRNA HNF1A-AS1, as H19 was markedly inhibited by HNF1A-AS1 knockdown in oesophageal adenocarcinoma and human bladder cancer.20,21 The dysregulation of H19 may be also due to the alteration of gene methylation, as metformin induced H19 repression by altering DNA methylation in the endometrial cancer tissues.22 In our study, we found that H19 was upregulated in melanoma, which was consistent with most of the previous studies. The differential expressions of H19 in different cancer types may be the cancer-type specific. A meta-analysis from Liu et al showed that high expression of H19 predicted shorter OS in 9 types of cancers, and increased H19 was related to poor histological grades, positive lymph node metastasis and advanced clinical stage.13 In our study, we had similar findings that high expression of H19 predicted shorter OS in patients with melanoma and was associated with metastasis, which may suggest that H19 is a novel molecular marker for predicting melanoma.
For the in vitro studies, H19 was found to promote cell proliferation, cell invasion and migration in various types of cancer cell lines including esophageal squamous cell cancer cell lines, cervical cancer cell lines and colorectal cancer cell lines.23–25 Consistently, our results showed that knockdown of H19 significantly suppressed cell proliferation and cell invasion and migration, suggesting the oncogenic role of H19 in melanoma development. In addition, the flow cytometry experiments revealed that H19 knockdown induced cell apoptosis and cell cycle arrest, which is consistent with previous studies showing that H19 knockdown induced apoptosis and caused cell cycle arrest at G0/G1 phase in colorectal cancer cells, pancreatic cancer cells and gastric cancer cells.26–28
EMT is an important process correlated with the progression of many types of cancers and plays a key role in the dissemination of malignant cells from primary epithelial neoplasm and metastasis into the local tissue and blood vessels.29 Based on the clinical examinations and in vitro functional assays of this study, H19 was suggested to contribute to melanoma metastasis in cancer patients. In this regard, we considered to explore the potential correlation between H19 and EMT in melanoma. Our qRT-PCR array results revealed that knockdown of H19 differentially affected the expression profiles of EMT-related genes in melanoma cell lines, ie, A-375 and 1205Lu, and qRT-PCR examination for the most affected genes showed that several important EMT markers such as E-cadherin, N-cadherin and vimentin were significantly affected by H19 knockdown. Further Western blotting assay showed that knockdown of H19 increased the protein expression level of E-cadherin and decreased the protein expression level of N-cadherin and vimentin. In colon cancer, H19 was found to promote EMT by functioning as microRNA sponges.30 Ma et al also showed that H19 promotes pancreatic cancer metastasis by modulating HMGA2-mediated EMT.31 More importantly, downregulation of HNF1A-AS1 suppressed EMT in lung adenocarcinoma, suggesting that H19-mediated EMT process may be involved in HNF1A-AS1 in melanoma.32 Therefore, these findings may indicate that H19 promotes metastasis in melanoma via affecting the EMT processes. In addition, our in vivo data showed that knockdown of H19 suppressed tumor growth in the nude mice and also inhibited the EMT as indicated by changes in expression the EMT-related genes. However, the limitation of our study is the lack of in vivo tumor metastasis assay to evaluate knockdown effect of H19 on tumor metastasis in the nude mice.
Conclusion
Our results suggested that H19 may be an oncogenic lncRNA for tumor growth and metastasis in melanoma. The findings in the present study may highlight the important role of H19 in melanoma development, and H19 may represent a novel therapeutic target for melanoma in the future.
Supplementary material
Overexpression of H19 promoted CHL-1 cell proliferation.
Notes: (A) Transfection with H19-overexpressing vectors (pcDNA3.1-H19) significantly increased H19 expression in CHL-1 cells. (B) Cell Counting Kit 8 (CCK-8) assay was performed to determined cell growth, and H19 overexpression significantly promoted cell growth in CHL-1 cells. Significant differences compared to control group were shown as *P<0.05, **P<0.01.
Acknowledgments
The study was funded by the Basic Research Project of Nanjing Medical University (NMU201411C). The authors would like to thank Dr Zhang for the collecting human skin tissues.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Mol Cancer. 2016;15(1):62. doi: 10.1186/s12943-016-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao J. The functional role of long non-coding RNAs and epigenetics. Biol Proced Online. 2014;16:11. doi: 10.1186/1480-9222-16-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma G, Wang Q, Lv C, et al. The prognostic significance of HOTAIR for predicting clinical outcome in patients with digestive system tumors. J Cancer Res Clin Oncol. 2015;141(12):2139–2145. doi: 10.1007/s00432-015-1980-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, He A, Wang D, Liu Y, Huang W. Long noncoding RNA HOTTIP as a novel predictor of lymph node metastasis and survival in human cancer: a systematic review and meta-analysis. Oncotarget. 2017;8(8):14126–14132. doi: 10.18632/oncotarget.12981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang B, Liu C, Wu Q, et al. Long non-coding RNA NEAT1 facilitates pancreatic cancer progression through negative modulation of miR-506-3p. Biochem Biophys Res Commun. 2017;482(4):828–834. doi: 10.1016/j.bbrc.2016.11.120. [DOI] [PubMed] [Google Scholar]
- 6.Terashima M, Tange S, Ishimura A, Suzuki T. MEG3 long noncoding RNA contributes to the epigenetic regulation of epithelial-mesenchymal transition in lung cancer cell lines. J Biol Chem. 2017;292(1):82–99. doi: 10.1074/jbc.M116.750950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L, Yao L, Jiang Y. A novel integrative approach to identify lncRNAs associated with the survival of melanoma patients. Gene. 2016;585(2):216–220. doi: 10.1016/j.gene.2016.03.036. [DOI] [PubMed] [Google Scholar]
- 8.Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74(11):2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 9.Weinstein D, Leininger J, Hamby C, Safai B. Diagnostic and prognostic biomarkers in melanoma. J Clin Aesthet Dermatol. 2014;7(6):13–24. [PMC free article] [PubMed] [Google Scholar]
- 10.Xu S, Wang H, Pan H, et al. ANRIL lncRNA triggers efficient therapeutic efficacy by reprogramming the aberrant INK4-hub in melanoma. Cancer Lett. 2016;381(1):41–48. doi: 10.1016/j.canlet.2016.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Chen L, Yang H, Xiao Y, et al. LncRNA GAS5 is a critical regulator of metastasis phenotype of melanoma cells and inhibits tumor growth in vivo. Onco Targets Ther. 2016;9:4075–4087. doi: 10.2147/OTT.S98203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei Y, Sun Q, Zhao L, et al. LncRNA UCA1-miR-507-FOXM1 axis is involved in cell proliferation, invasion and G0/G1 cell cycle arrest in melanoma. Med Oncol. 2016;33(8):88. doi: 10.1007/s12032-016-0804-2. [DOI] [PubMed] [Google Scholar]
- 13.Peng J, Qi S, Wang P, et al. Meta-analysis of downregulated E-cadherin as a poor prognostic biomarker for cervical cancer. Future Oncol. 2016;12(5):715–726. doi: 10.2217/fon.15.332. [DOI] [PubMed] [Google Scholar]
- 14.Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281(16):3766–3775. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 15.Hua Q, Lv X, Gu X, et al. Genetic variants in lncRNA H19 are associated with the risk of bladder cancer in a Chinese population. Mutagenesis. 2016;31(5):531–538. doi: 10.1093/mutage/gew018. [DOI] [PubMed] [Google Scholar]
- 16.Hashad D, Elbanna A, Ibrahim A, Khedr G. Evaluation of the role of circulating long non-coding RNA H19 as a promising novel biomarker in plasma of patients with gastric cancer. J Clin Lab Anal. 2016;30(6):1100–1105. doi: 10.1002/jcla.21987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shi Y, Wang Y, Luan W, et al. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS One. 2014;9(1):e86295. doi: 10.1371/journal.pone.0086295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv J, Ma L, Chen XL, Huang XH, Wang Q. Downregulation of LncRNAH19 and MiR-675 promotes migration and invasion of human hepatocellular carcinoma cells through AKT/GSK-3β/Cdc25A signaling pathway. J Huazhong Univ Sci Technolog Med Sci. 2014;34(3):363–369. doi: 10.1007/s11596-014-1284-2. [DOI] [PubMed] [Google Scholar]
- 19.Guo G, Kang Q, Chen Q, et al. High expression of long non-coding RNA H19 is required for efficient tumorigenesis induced by Bcr-Abl oncogene. FEBS Lett. 2014;588(9):1780–1786. doi: 10.1016/j.febslet.2014.03.038. [DOI] [PubMed] [Google Scholar]
- 20.Feng Z, Wang B. Long non-coding RNA HNF1A-AS1 promotes cell viability and migration in human bladder cancer. Oncol Lett. 2018;15(4):4535–4540. doi: 10.3892/ol.2018.7878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang X, Song JH, Cheng Y, et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–890. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhong T, Men Y, Lu L, et al. Metformin alters DNA methylation genome-wide via the H19/SAHH axis. Oncogene. 2017;36(17):2345–2354. doi: 10.1038/onc.2016.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan D, Wu Y, Hu L, et al. Long noncoding RNA H19 is up-regulated in esophageal squamous cell carcinoma and promotes cell proliferation and metastasis. Dis Esophagus. 2017;30(1):1–9. doi: 10.1111/dote.12481. [DOI] [PubMed] [Google Scholar]
- 24.Iempridee T. Long non-coding RNA H19 enhances cell proliferation and anchorage-independent growth of cervical cancer cell lines. Exp Biol Med. 2017;242(2):184–193. doi: 10.1177/1535370216670542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohtsuka M, Ling H, Ivan C, et al. H19 noncoding RNA, an independent prognostic factor, regulates essential Rb-E2F and CDK8-β-catenin signaling in colorectal cancer. EBioMedicine. 2016;13:113–124. doi: 10.1016/j.ebiom.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han D, Gao X, Wang M, et al. Long noncoding RNA H19 indicates a poor prognosis of colorectal cancer and promotes tumor growth by recruiting and binding to eIF4A3. Oncotarget. 2016;7(16):22159–22173. doi: 10.18632/oncotarget.8063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ma L, Tian X, Wang F, et al. The long noncoding RNA H19 promotes cell proliferation via E2F-1 in pancreatic ductal adenocarcinoma. Cancer Biol Ther. 2016;17(10):1051–1061. doi: 10.1080/15384047.2016.1219814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu G, Xiang T, Wu QF, Wang WX. Long noncoding RNA H19-derived miR-675 enhances proliferation and invasion via RUNX1 in gastric cancer cells. Oncol Res. 2016;23(3):99–107. doi: 10.3727/096504015X14496932933575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh A, Settleman J. EMT, cancer stem cells and drug resistance: an emerging axis of evil in the war on cancer. Oncogene. 2010;29(34):4741–4751. doi: 10.1038/onc.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liang WC, Fu WM, Wong CW, et al. The lncRNA H19 promotes epithelial to mesenchymal transition by functioning as miRNA sponges in colorectal cancer. Oncotarget. 2015;6(26):22513–22525. doi: 10.18632/oncotarget.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ma C, Nong K, Zhu H, et al. H19 promotes pancreatic cancer metastasis by derepressing let-7’s suppression on its target HMGA2-mediated EMT. Tumour Biol. 2014;35(9):9163–9169. doi: 10.1007/s13277-014-2185-5. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overexpression of H19 promoted CHL-1 cell proliferation.
Notes: (A) Transfection with H19-overexpressing vectors (pcDNA3.1-H19) significantly increased H19 expression in CHL-1 cells. (B) Cell Counting Kit 8 (CCK-8) assay was performed to determined cell growth, and H19 overexpression significantly promoted cell growth in CHL-1 cells. Significant differences compared to control group were shown as *P<0.05, **P<0.01.