Abstract
Background
Studies have shown that miR-17-5p plays an important role in the development of cancer. The aim of this meta-analysis was to quantitatively analyze the association of miR-17-5p with prognosis in various cancers.
Materials and methods
We searched the PubMed, EMBASE, Web of Science, and Cochrane library databases for relevant studies through August 2017. The prognostic data and clinico-pathological features of overall survival (OS) and disease-free survival (DFS) were extracted to investigate the association between miR-17-5p expression and tumor prognosis. In addition, odds ratios (ORs) were used to assess the correlations between miR-17-5p expression and clinicopathological characteristics.
Results
A total of ten studies were incorporated into this systematic review, and we found that high miR-17-5p expression can predict poor OS for malignancies (combined hazard ratio [HR]=1.87; 95% confidence interval [CI], 1.37–2.55; P=0.000) as well as poor DFS (combined HR=1.60; 95% CI, 1.05–2.44; P=0.027). Further subgroup analyses suggested that high miR-17-5p expression was related to poor OS in Asian patients (combined HR=1.92; 95% CI, 1.37–2.71; P=0.000) and the serum/plasma sample source subgroup (combined HR=2.13; 95% CI, 1.36–3.31; P=0.001). The combined OR indicated that the expression of miR-17-5p was associated with lymph node invasion (OR=1.28; 95% CI, 1.05–1.56; P=0.016) and venous invasion (OR=1.92; 95% CI, 1.40–2.63; P=0.000).
Conclusion
Elevated expression of miR-17-5p suggested a poor prognosis in cancer patients and may serve as a new tumor marker to monitor cancer development and progression.
Keywords: miR-17-5p, cancer, prognosis, meta-analysis
Introduction
Cancer has become a global public health problem, seriously endangering human life in recent decades.1 Early diagnosis of cancer greatly improves the survival of patients and their quality of life; however, there is no effective way to assess the prognosis of cancer patients based on clinical parameters. microRNAs (miRNAs), as a new type of biomarker, have gradually attracted the attention of many researchers.2 miRNAs are non-coding RNAs of approximately 18–25 nucleotides that facilitate messenger RNA (mRNA) degradation and inhibit translation by binding to the 3′-untranslated region of the mRNA.3
Recently, a growing amount of evidence has shown that miRNAs are engaged in diverse biological processes, including cellular growth, differentiation, proliferation, metastasis, invasion, migration, and apoptosis.4–8
The miR-17-92 cluster (which is situated at the human 13q31 and includes miR-17-3p, miR-17-5p, miR-19a, miR-18a, miR-19b, miR-20a, and miR-92-1) first attracted the attention of researchers after a series of reports linked these miRNAs to tumor formation and progression.9 Specifically, researchers have found that miR-17-5p is a crucial member of the miR-17 cluster family that is highly expressed in various cancers such as hepatocellular carcinoma (HCC),10,11 lung cancer (LC),12 colorectal cancer (CRC), 13 and other cancers.13–18
Furthermore, researchers have found that high expression of miR-17-5p is negatively correlated with overall survival (OS) and disease-free survival (DFS) in different kinds of cancers.10,11,19 However, some studies have reported contradictory results; for example, Liu et al20 observed that high expression of miR-17-5p could negatively regulate the invasion and migration of HCC cell lines. Until now, a systematic review of the prognostic value of miR-17-5p in human tumors has not been reported. Therefore, we performed this meta-analysis to explore the prognostic role of elevated miR-17-5p expression in various cancers.
Materials and methods
Literature search
To identify relevant studies, we searched the online PubMed, EMBASE, Web of Science, and the Cochrane library database through July 2017. The keywords for the search were as follows: “miR-17-5p”, “microRNA-17-5p”, “miRNA-17-5p”, “cancer”, “carcinoma”, “neoplasm”, and “tumor” in combination with “prognosis”. In addition, the references of identified articles were also explored for additional relevant articles. All of the articles were reviewed by three investigators independently, and any disagreements were settled by discussion.
Inclusion and exclusion criteria
Researchers first screened the title and abstract and then screened out articles by skimming the full text. An eligible study was selected if it was consistent with the following inclusion criteria: 1) the study was based on human research; 2) hazard ratios (HRs) and their 95% confidence intervals (CIs) could be extracted from the article; 3) all patients could be separated into high and low expression level groups based on miR-17-5p quantity; and 4) the study was written in English and its full text was available.
Exclusion criteria for the articles were as follows: 1) the study was conducted using animals and/or cell lines; 2) the study was a systematic review or case report; 3) the study was not written in English; 4) the study lacked significant data; and 5) the study was a repeat study.
Data extraction
Three investigators (Kong WH, Cheng YS, and Liang H) extracted the data and information from the selected studies. Any disagreements were resolved through a panel discussion. Extracted data from the eligible studies included cancer type, the first author’s surname, country, sample size, specimen source, test method, criteria for high miR-17-5p expression, rate of high miR-17-5p expression, follow-up period, age, gender, tumor depth, lymph node invasion, metastasis, vein invasion, histology differentiation, tumor size, and HRs with corresponding 95% CIs for OS, progress-free survival (PFS), and DFS.
Quality evaluation
We assessed the selected studies using the MOOSE method proposed by the Netherland Cochrane Center. The basic principles of this analysis included determining if 1) the number of patients was sufficient; 2) the design method was clear; 3) sufficient data were reported; 4) the results were clearly evaluated; 5) miR-17-5p measurement was described in detail; and 6) adequate follow-up time was available for each study.21 Studies that did not meet the above six requirements were excluded.
Statistical analysis
Statistical analyses of the HRs for OS, DFS, and odds ratios (ORs) for clinicopathological characteristics were computed using STATA and RevMan software. Heterogeneity was assessed using Q tests and I2 statistics (P<0.05 or I2>50% was deemed to be statistically heterogeneous). When the heterogeneity of the combined study was small (I2<50%, P<0.10), we used the fixed effect model for statistical analysis. Otherwise, a random effects model was used. Subgroup analysis was conducted to further explore the association of miR-17-5p with sample size, ethnicity, source, definition method, tumor type, and statistical mode for various carcinomas. We used Egger’s test, Begg’s test, and funnel plots to detect publication bias. Sensitive analysis was also implemented to evaluate the stabilization of the results. A P-value below 0.05 was deemed to be statistically significant.
Results
Literature search and study characteristics
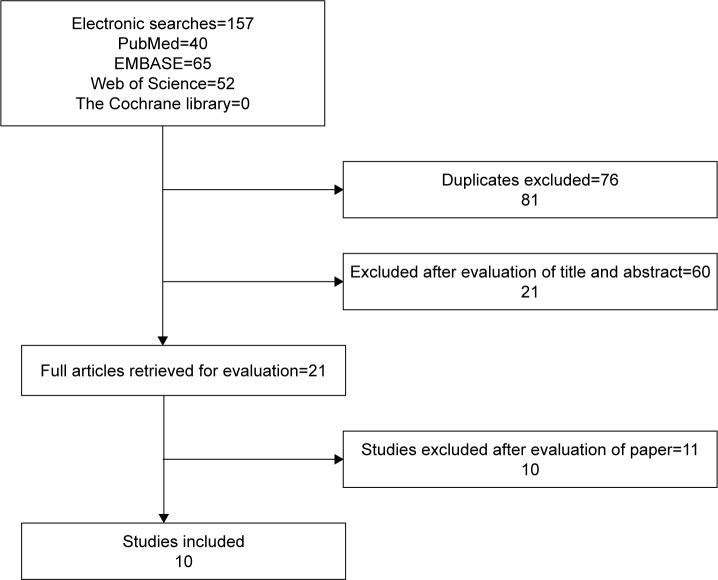
According to the inclusion and exclusion criteria, we selected a total of ten articles with data from approximately 1,135 people for meta-analysis, and the detailed selection steps are shown in Figure 1.
Figure 1.
Study flow chart showing the process for selecting eligible publications.
We summarize the major features of the selected studies in Table 1. Among these ten studies, nine measured miR-17-5p expression using polymerase chain reaction (PCR), while one study performed PCR and in situ hybridization (ISH). Across these the ten studies, patients were diagnosed with CRC HCC, colon cancer (CC), gastric cancer (GC), childhood infratentorial ependymoma (CIE), pancreatic cancer (PC), osteosarcoma (OSA), nasopharyngeal carcinoma (NPC), and LC. Eight studies directly provided HRs and 95% CIs, one study provided only a survival curve, and one study provided only the risk ratios (RRs); thus, we merged RRs and HRs.
Table 1.
Characteristics of the included studies
| Tumor type | Reference | Region | Sample size | Specimen source | Test method | Definition of high miR-17-5p expression | Rate of high miR-17-5p expression | Tumor stage | Follow-up time (months) | Survival | HR |
|---|---|---|---|---|---|---|---|---|---|---|---|
| HCC | Chen et al10 | China | 120 | Tissue | PCR | Median | 56.7 | Edmondson–Steiner grade (I–IV) | 2–46 | OSMA DFSMA |
RR |
| CIE | Zakrzewska et al15 | Poland | 53 | Tissue | PCR | Median | NR | WHO (I–III) | Up to 120 | OSMA EFSMA |
R |
| GC | Wang et al17 | China | 65 | Plasma | PCR | Median | 50.8 | TNM (I–IV) | Up to 40 | OSUA | R |
| CC | Diaz et al18 | Spain | 62 | Tissue | PCR | Tertile | 33.9 | TNM (I–IV) | 68–99 | OSUA DFSUA |
R |
| HCC | Zheng et al11 | China | 96 | Serum | PCR | Median (2.705) | 50 | TNM (I–IV) | From resection to death or May 1, 2012 | OSMA | R |
| PC | Yu et al19 | Japan | 80 | Tissue | PCR | Cutoff value (5.69) | 37.5 | UICC (I–IV) | Up to 100 | OSMA | R |
| CRC | Fang et al13 | China | 295 | Tissue | PCR ISH | Staining score (8–12) | 30.2 | TNM (I–IV) | Up to 96 | OSMA | R |
| NPC | Chen et al16 | China | 81 | Tissue | PCR | Staining score (tai) | 75.3 | TNM (I–IV) | Up to 100 | OSUA | SC |
| LC | Chen et al12 | China | 221 | Serum | PCR | Median (33 months) | 45.7 | TNM (I–IV) | Up to 50 | OSUA | R |
| OSA | Zhu et al14 | China | 62 | Plasma | PCR | Mean | NR | TNM (I–III) | 10–70 (OS) 10–60 (PFS) |
OSMA PFSMA |
R |
Abbreviations: HR, hazard ratio; HCC, hepatocellular carcinoma; CIE, childhood infratentorial ependymoma; GC, gastric cancer; CC, colon cancer; PC, pancreatic cancer; CRC, colorectal cancer; NPC, nasopharyngeal carcinoma; LC, lung cancer; OSA, osteosarcoma; PCR, polymerase chain reaction; ISH, in situ hybridization; NR, not reported; WHO, World Health Organization; TNM, tumour node metastases; UICC, Union for International Cancer Control; OS, overall survival; PFS, progress-free survival; DFS, disease-free survival; MA, multivariate analysis; UA, univariate analysis; RR, risk ratio; R, reported; SC, survival curve.
miR-17-5p expression and OS
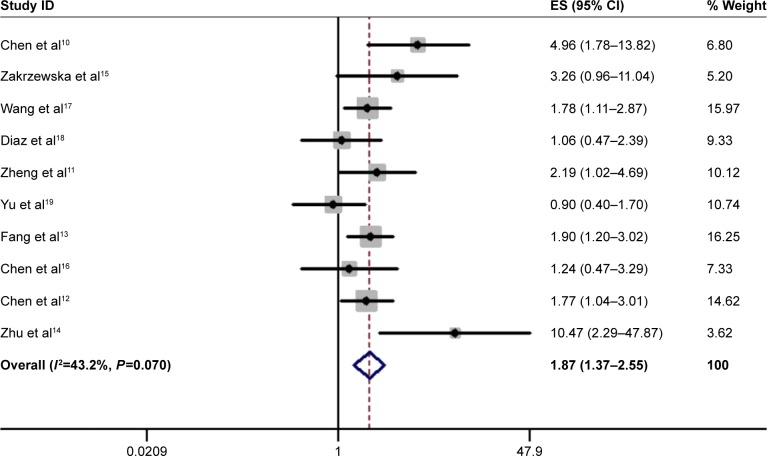
We included ten studies of OS in our analysis. Considering that potential heterogeneity may have been introduced in this study, we used a random effect model to conservatively estimate the effect of combined HRs and 95% CIs. We found that patients with high expression of miR-17-5p had a worse prognosis than those with low expression of miR-17-5p (HR=1.87; 95% CI, 1.37–2.55; P=0.000) (Figure 2).
Figure 2.
Forest plot of the overall survival analysis.
Note: Weights are from random-effects analysis.
Abbreviations: CI, confidence interval; ES, effect size.
To lessen the impact of heterogeneity, subgroup analyses were performed for sample size, ethnicity, sources of detected sample source, definition method, tumor types, and statistical mode (Table 2). First, we found that the expression level of miR-17-5p had a statistically significant effect on OS in subgroups with a sample size greater than or equal to 100 (HR=2.15; 95% CI, 1.37–3.37; P=0.001) as well as subgroups with a sample size of less than 100 (HR=1.72; 95% CI, 1.11–2.68; P=0.016). Second, we observed that miR-17-5p had a statistically significant effect on OS in the Asians patient subgroup (HR=1.92; 95% CI, 1.37–2.71; P=0.000), but no statistical significance was found in the European patient subgroup. Third, when we conducted a subgroup analysis for the sample source, we found that miR-17-5p was more strongly associated with the plasma/serum subgroup (HR=2.13; 95% CI, 1.36–3.31; P=0.001) than the tissue subgroup (HR=1.69; 95% CI, 1.05–2.71; P=0.031) using a random effects model. Fourth, miR-17-5p has a stronger correlation with median definition than other definition methods (HR=2.08; 95% CI, 1.54–2.81; P=0.000). Fifth, in a subgroup analysis stratified by the cancer type, all three of the subgroups showed a strong correlation between miR-17-5p and OS with a random effects model, indicating that miR-17-5p has a predictive effect for different kinds of tumors. Finally, considering that the combination of univariate analysis and multivariate analysis is unreasonable, we performed a subgroup analysis for statistical mode, and the results were consistent with the overall combined effect. Therefore, the conclusion is stable and credible.
Table 2.
The pooled HRs, 95% CIs, heterogeneity, and P-values of overall survival stratified by sample size, ethnicity, sample sources, definition method, tumor type, and statistical mode
| Stratified analysis | Number of studies | Number of patients | Pooled HR (95% CI) | P-value | Heterogeneity
|
Test of interaction
|
||
|---|---|---|---|---|---|---|---|---|
| I2 (%) | p-value | Model | P-value | |||||
| Sample size | 0.49 | |||||||
| ≥100 | 3 | 636 | 2.15 (1.37–3.37) | 0.001 | 38.6 | 0.196 | Random | |
| <100 | 7 | 499 | 1.72 (1.11–2.68) | 0.016 | 48.6 | 0.070 | Random | |
| Dominant ethnicity | 0.82 | |||||||
| Asian | 8 | 1,020 | 1.92 (1.37–2.71) | 0.000 | 47.2 | 0.066 | Random | |
| European | 2 | 115 | 1.69 (0.57–5.00) | 0.344 | 55.6 | 0.133 | Random | |
| Source | 0.48 | |||||||
| Tissue | 6 | 691 | 1.69 (1.05–2.71) | 0.031 | 50.7 | 0.071 | Random | |
| Serum/plasma | 4 | 444 | 2.13 (1.36–3.31) | 0.001 | 40.5 | 0.169 | Random | |
| Definition | 0.80 | |||||||
| Median | 5 | 555 | 2.08 (1.54–2.81) | 0.000 | 1.5 | 0.398 | Random | |
| Stain score | 2 | 376 | 1.77 (0.57–5.50) | 0.327 | 76.4 | 0.015 | Random | |
| Others | 3 | 204 | 1.76 (1.15–2.67) | 0.008 | 0 | 0.438 | Random | |
| Tumor types | 0.15 | |||||||
| Digestive system | 6 | 718 | 1.73 (1.20–2.49) | 0.003 | 45.2 | 0.104 | Random | |
| Respiratory system | 2 | 302 | 1.63 (1.02–2.60) | 0.040 | 0 | 0.531 | Random | |
| Others | 2 | 115 | 5.33 (1.72–16.50) | 0.004 | 27.3 | 0.241 | Random | |
| Statistical mode | 0.19 | |||||||
| Univariate analysis | 4 | 429 | 1.59 (1.17–2.17) | 0.003 | 0 | 0.663 | Random | |
| Multivariate analysis | 6 | 706 | 2.43 (1.39–4.26) | 0.002 | 61.4 | 0.024 | Random | |
Note: Data in bold P<0.05.
Abbreviations: CI, confidence interval; HR, hazard ratio.
miR-17-5p expression and DFS
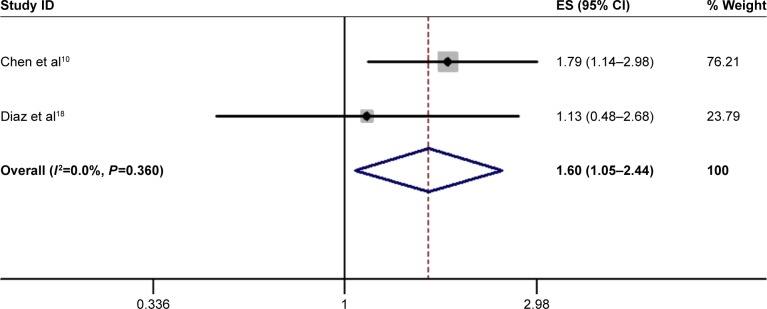
Two studies focused on DFS analysis. Heterogeneity results showed a small amount of heterogeneity (I2=0; P=0.360), so we use a fixed effect model to compute the combined HRs and 95% CIs for DFS. The results showed that the DFS of patients with elevated miR-17-5p expression was more affected than that of patients with decreased miR-17-5p expression (HR=1.60; 95% CI, 1.05–2.44; P=0.027) (Figure 3). Because only two studies reported DFS, the data are too limited to perform subgroup analysis for DFS.
Figure 3.
Forest plot of disease free survival analysis.
Abbreviations: CI, confidence interval; ES, effect size.
miR-17-5p expression and clinicopathological characteristics
Meta-analysis indicated that elevated miR-17-5p expression was associated with lymph node invasion (OR=1.28; 95% CI, 1.05–1.56; P=0.016) and venous invasion (OR=1.92; 95% CI, 1.40–2.63; P=0.000) (Table 3). No significant correlations were found for age (P=0.24), gender (P=0.212), tumor invasion depth (P=0.400), distant metastasis (P=0.499), tumor stage (P=0.804), differentiation grade (P=0.077), tumor size (P=0.291), or hepatitis history (P=0.550). Due to insufficient data, we failed to detect any correlations between elevated miR-17-5p expression and other clinicopathological characteristics.
Table 3.
Meta-analysis results of the correlation between elevated miR-17-5p expression and multiple clinicopathological characteristics
| Stratified analysis | Number of studies | Number of patients | Pooled OR (95% CI) | P-value | Heterogeneity
|
||
|---|---|---|---|---|---|---|---|
| I2 (%) | P-value | Model | |||||
| Age (old vs young) | 6 | 663 | 1.08 (0.95–1.24) | 0.240 | 0 | 0.578 | Fixed |
| Gender (male vs female) | 6 | 663 | 0.92 (0.80–1.05) | 0.212 | 0 | 0.809 | Fixed |
| Invasion depth (T3+T4 vs T1+T2)* | 2 | 160 | 1.45 (0.61–3.45) | 0.400 | 87.7 | 0.004 | Random |
| Lymph node invasion (≥2 vs <2) | 2 | 201 | 1.28 (1.05–1.56) | 0.016 | 0 | 0.404 | Fixed |
| Distant metastasis (yes vs no) | 3 | 242 | 1.24 (0.66–2.32) | 0.499 | 83.4 | 0.002 | Random |
| Tumor stage (III+IV vs I+II) | 4 | 463 | 1.09 (0.56–2.10) | 0.804 | 88.8 | 0.000 | Random |
| Venous invasion (yes vs no) | 2 | 200 | 1.92 (1.40–2.63) | 0.000 | 0 | 0.776 | Fixed |
| Differentiation grade (poorly vs well and moderately) | 2 | 161 | 1.33 (0.97–1.84) | 0.077 | 0 | 0.361 | Fixed |
| Tumor size (>5 vs <5) (cm) | 3 | 281 | 0.90 (0.62–1.30) | 0.291 | 53.4 | 0.117 | Random |
| Hepatitis history (yes vs no) | 3 | 216 | 1.20 (0.66–2.16) | 0.550 | 58.9 | 0.119 | Random |
Notes: According to the size of the tumor and/or the extent of the primary tumor are defined as different T stages. Data in bold P<0.05.
Abbreviations: CI, confidence interval; OR, odds ratio.
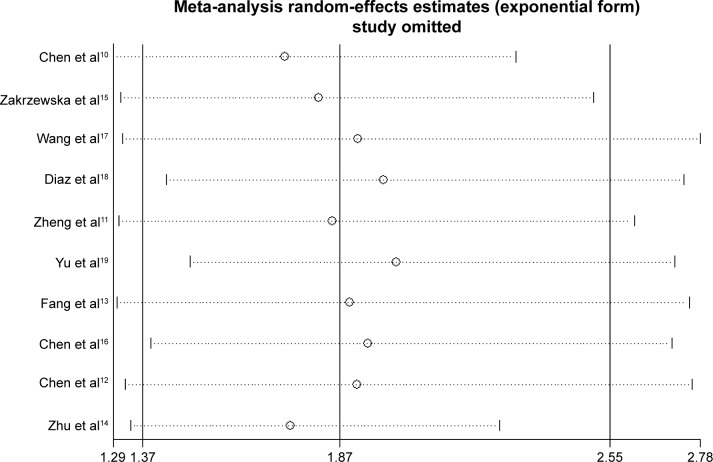
Sensitivity analysis
Sensitivity analysis was implemented to assess the influence of each individual study on the integrated results of OS. We excluded each study, re-merged the meta-analysis results and then compared the results of the unexcluded meta-analysis to investigate the stability of the results. This analysis showed that the combined HR for the effect of miR-17-5p expression on OS was not remarkably influenced after eliminating any of the individual studies (Figure 4).
Figure 4.
Sensitivity analysis of overall survival in this meta-analysis.
Publication bias
We use Egger’s tests and funnel plots to detect the publication bias. The results of the funnel plot for OS are shown in Figure 5. Since the Egger’s test has a P-value greater than 0.05, no publication bias was found. Due to the small number of studies that reported DFS, we were not able to determine the publication bias for DFS.
Figure 5.
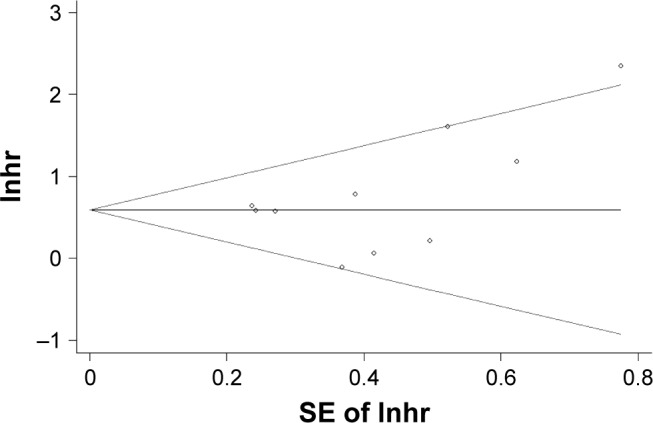
Begg’s funnel plot with pseudo 95% CI of the publication bias for overall survival.
Abbreviations: lnhr, logarithm of the hazard ratio; SE, standard error.
Discussion
In recent years, miRNAs have been considered to be closely related to carcinogenic processes such as angiogenesis, proliferation, differentiation, invasion, apoptosis, and metastasis.9 It is presumed that more than one third of human genes are regulated by miRNAs,22 which serve as negative regulatory molecules of gene expression at the posttranscriptional translation phase and are widely involved in the development and progression of human diseases, particularly human cancer.23 Since the publication of reviews about miRNA-17-5p in multiple journals, miR-17-5p has gradually attracted the attention of many researchers as a molecule of interest. miR-17-5p has also been found to be closely associated with the development of tumors.9,24–26 However, as no quantitative systematic reviews about miR-17-5p have been published so far, we conducted this meta-analysis.
In this study, we strictly adhered to the MOOSE guidelines to perform the meta-analysis and to assess the quality of the included studies using the scales recommended by the Cochrane Collaboration.21 The results indicated that high expression of miR-17-5p plays an unfavorable role in OS and DFS. Considering the potential heterogeneity of the included studies, we used a random effects model to combine the data and then performed subgroup analysis, sensitivity analysis, and publication bias detection; the findings of these analyses show that our results are stable and credible.
Furthermore, we carried out subgroup analyses to explore the prognostic significance of miR-17-5p in OS, and we have successfully drawn some valuable conclusions for clinical practice. To rule out the impact of different races on prognosis, we divided the ten studies into patient groups of Asian and European ethnicities. We observed that the elevated expression of miR-17-5p predicts significantly worse OS in Asian patients (pooled HR=1.92; 95% CI, 1.37–2.71; P=0.000), but no statistical significance was found in European patients (combined HR=1.69; 95% CI, 0.57–5.00, P=0.344) (Table 2). This difference may be due to the different genetic backgrounds, geographical locations, climates etc., of the different races. Therefore, we concluded that miR-17-5p might be a more valuable predictor of cancer prognosis in Asian populations. To illustrate the relationship between the expression level of miR-17-5p and prognosis using different sample sources, we divided the patients into two subgroups, serum/plasma and tissue, and we observed that the serum/plasma group had a worse prognosis (combined HR=2.13; 95% CI, 1.36–3.31; P=0.001) than tissue-derived specimens (combined HR=1.69; 95% CI, 1.05–2.71; P=0.031) (Table 2). Although the detection of miR-17-5p in tumor specimens has been widely used to study tumor prognosis, plasma/serum specimen detection is more convenient and effective, and it can efficiently assess the risk of hospital stay and postoperative recurrence. The expression of miR-17-5p in the blood is more predictive than in tissue, which can be applied in clinical research. Therefore, the detection of miR-17-5p in plasma/serum during postoperative follow-up periods might be an effective way to closely monitor disease-progression and treatment in cancer patients.
Moreover, we carried out meta-analyses with respect to pathological characteristics. We found that elevated miR-17-5p expression is predominantly correlated with lymph node invasion (combined OR=1.28; 95% CI, 1.05–1.56; P=0.016) and venous invasion (combined OR=1.92; 95% CI, 1.40–2.63; P=0.000). No statistically significant correlations were found for age, gender, tumor invasion depth, distant metastasis, tumor stage, differentiation grade, or hepatitis history. Therefore, we concluded that miR-17-5p may be involved in the progression of lymphatic metastasis and vein invasion in cancer.
To the best of our knowledge, miR-17-5p has been found to be closely associated with cancers through complicated regulatory mechanisms.9 For instance, Yang et al27 observed that miR-17-5p can facilitate the migration and proliferation of human liver cancer cells through the p38-MAPK-HSP27 pathway. Li et al28 discovered that miR-17-5p can boost the invasion and migration of human breast tumor cells by inhibiting HMG box-containing protein 1 (HBP1), a tumor suppressor. Qu et al29 observed that miR-17-5p can negatively regulate transforming growth factor-beta receptor 2 (TGFBR2), which serves as an original regulator of the TGF-beta signaling pathway, and reduction or loss of its expression can lead to uncontrolled cell proliferation and invasion, thus promoting cell growth and migration. Wang et al30 noticed that miR-17-5p can promote the proliferation of OSA in a breast cancer cell 2 (BRCC2)-dependent mechanism, which plays a key role in suppressing tumorigenesis. To some extent, these mechanisms explain why patients with elevated miR-17-5p expression have multiple lymphatic metastases and venous violations, but the specific mechanism still requires further investigation.
These studies had several limitations. First, the definition of high miR-17-5p expression was not the same across studies; thus, it was difficult to define miR-17-5p overexpression in various cancers. Second, the study included a total of only ten articles; therefore, the data are not sufficient. Third, there was considerable heterogeneity among the different studies in terms of cancer type, specimen source, detection method and definition of high miR-17-5p expression. Fourth, eligible studies only included positive results, and there may be publication bias. Fifth, combining univariate and multivariate analysis is somewhat unreasonable. Finally, part of the HR value was calculated using a survival curve, which may lead to some error.
Conclusion and recommendations
Our meta-analysis has explored the predictive value of miR-17-5p in different types of carcinomas. miR-17-5p may serve as a new tumor marker for predicting prognosis and for evaluating the clinicopathological characteristics of different kinds of carcinomas. Ultimately, considering the limitations of this study, these conclusions require additional large-sample and high-quality research to further clarify the relationship between miRNA-17-5p and prognosis.
Acknowledgments
The authors acknowledge their research group fund: Guangzhou Science and Technology Plan Project (201707010112).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Kleemann M, Bereuther J, Fischer S, et al. Investigation on tissue specific effects of pro-apoptotic micro RNAs revealed miR-147b as a potential biomarker in ovarian cancer prognosis. Oncotarget. 2017;8(12):18773–18791. doi: 10.18632/oncotarget.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svoronos AA, Engelman DM, Slack FJ. OncomiR or tumor suppressor? The duplicity of microRNAs in cancer. Cancer Res. 2016;76(13):3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmadi S, Sharifi M, Salehi R. Locked nucleic acid inhibits miR-92a-3p in human colorectal cancer, induces apoptosis and inhibits cell proliferation. Cancer Gen Ther. 2016;23(7):199–205. doi: 10.1038/cgt.2016.10. [DOI] [PubMed] [Google Scholar]
- 5.Zhao X, He W, Li J, et al. MiRNA-125b inhibits proliferation and migration by targeting SphK1 in bladder cancer. Am J Transl Res. 2015;7(11):2346–2354. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang JX, Xu Y, Gao Y, et al. Decreased expression of miR-939 contributes to chemoresistance and metastasis of gastric cancer via dysregulation of SLC34A2 and Raf/MEK/ERK pathway. Mol Cancer. 2017;16(1):18. doi: 10.1186/s12943-017-0586-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Zhang J, Sun Q, Zhang Z, Ge S, Han ZG, Chen WT. Loss of microRNA-143/145 disturbs cellular growth and apoptosis of human epithelial cancers by impairing the MDM2-p53 feedback loop. Oncogene. 2013;32(1):61–69. doi: 10.1038/onc.2012.28. [DOI] [PubMed] [Google Scholar]
- 8.Liang B, Chen Y, Yuan W, et al. Down-regulation of miRNA-451a and miRNA-486-5p involved in benzene-induced inhibition on erythroid cell differentiation in vitro and in vivo. Arch Toxicol. 2018;92(1):252–272. doi: 10.1007/s00204-017-2033-7. [DOI] [PubMed] [Google Scholar]
- 9.Fuziwara CS, Kimura ET. Insights into regulation of the miR-17-92 cluster of miRNAs in cancer. Front Med (Lausanne) 2015;2:64. doi: 10.3389/fmed.2015.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen L, Jiang M, Yuan W, Tang H. miR-17-5p as a novel prognostic marker for hepatocellular carcinoma. J Invest Surg. 2012;25(3):156–161. doi: 10.3109/08941939.2011.618523. [DOI] [PubMed] [Google Scholar]
- 11.Zheng J, Dong P, Gao S, Wang N, Yu F. High expression of serum miR-17-5p associated with poor prognosis in patients with hepatocellular carcinoma. Hepatogastroenterology. 2013;60(123):549–552. doi: 10.5754/hge12754. [DOI] [PubMed] [Google Scholar]
- 12.Chen Q, Si Q, Xiao S, et al. Prognostic significance of serum miR-17-5p in lung cancer. Med Oncol. 2013;30(1):353. doi: 10.1007/s12032-012-0353-2. [DOI] [PubMed] [Google Scholar]
- 13.Fang L, Li H, Wang L, et al. MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget. 2014;5(10):2974–2987. doi: 10.18632/oncotarget.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu HX, Liao QD, Yin K, Da Zhong D, Wang CG. Circulating miR-17-5p as a potential biomarker for diagnosis and prognosis in osteosarcoma. Int J Clin Exp Pathol. 2017;10(3):3409–3416. [Google Scholar]
- 15.Zakrzewska M, Fendler W, Zakrzewski K, et al. Altered microRNA expression is associated with tumor grade, molecular background and outcome in childhood infratentorial ependymoma. PLoS One. 2016;11(7):e0158464. doi: 10.1371/journal.pone.0158464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen C, Lu Z, Yang J, et al. MiR-17-5p promotes cancer cell proliferation and tumorigenesis in nasopharyngeal carcinoma by targeting p21. Cancer Med. 2016;5(12):3489–3499. doi: 10.1002/cam4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang M, Gu H, Wang S, et al. Circulating miR-17-5p and miR-20a: molecular markers for gastric cancer. Mol Med Rep. 2012;5(6):1514–1520. doi: 10.3892/mmr.2012.828. [DOI] [PubMed] [Google Scholar]
- 18.Diaz R, Silva J, Garcia JM, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47(9):794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 19.Yu J, Ohuchida K, Mizumoto K, Fujita H, Nakata K, Tanaka M. MicroRNA miR-17-5p is overexpressed in pancreatic cancer, associated with a poor prognosis, and involved in cancer cell proliferation and invasion. Cancer Biol Ther. 2010;10(8):748–757. doi: 10.4161/cbt.10.8.13083. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Dong L, Liu Y, et al. A c-Myc/miR-17-5p feedback loop regulates metastasis and invasion of hepatocellular carcinoma. Tumour Biol. 2016;37(4):5039–5047. doi: 10.1007/s13277-015-4355-5. [DOI] [PubMed] [Google Scholar]
- 21.Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA. 2000;283(15):2008–2012. doi: 10.1001/jama.283.15.2008. [DOI] [PubMed] [Google Scholar]
- 22.Lin SB, Gregory RI. MicroRNA biogenesis pathways in cancer. Nature Rev Cancer. 2015;15(6):321–333. doi: 10.1038/nrc3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shen J, Hung MC. Signaling-mediated regulation of microRNA processing. Cancer Res. 2015;75(5):783–791. doi: 10.1158/0008-5472.CAN-14-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dellago H, Bobbili MR, Grillari J. MicroRNA-17-5p: at the crossroads of cancer and aging – a mini-review. Gerontology. 2017;63(1):20–28. doi: 10.1159/000447773. [DOI] [PubMed] [Google Scholar]
- 25.Ventura A, Young AG, Winslow MM, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133(2):217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang F, Yin Y, Wang F, et al. miR-17-5p Promotes migration of human hepatocellular carcinoma cells through the p38 mitogen-activated protein kinase-heat shock protein 27 pathway. Hepatology. 2010;51(5):1614–1623. doi: 10.1002/hep.23566. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Bian C, Liao L, Li J, Zhao RC. miR-17-5p promotes human breast cancer cell migration and invasion through suppression of HBP1. Breast Cancer Res Treat. 2011;126(3):565–575. doi: 10.1007/s10549-010-0954-4. [DOI] [PubMed] [Google Scholar]
- 29.Qu Y, Zhang H, Duan J, et al. MiR-17-5p regulates cell proliferation and migration by targeting transforming growth factor-β receptor 2 in gastric cancer. Oncotarget. 2016;7(22):33286–33296. doi: 10.18632/oncotarget.8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W, Zhang L, Zheng K, Zhang X. miR-17-5p promotes the growth of osteosarcoma in a BRCC2-dependent mechanism. Oncol Rep. 2016;35(3):1473–1482. doi: 10.3892/or.2016.4542. [DOI] [PubMed] [Google Scholar]