Abstract
Despite the potential importance of retrieval-based targeting, few Golgi cisternae-localized proteins have been demonstrated to be targeted by retrieval, and the putative retrieval signals remain unknown. Golgi phosphoprotein of 130 kDa (GPP130) is a cis-Golgi protein that allows assay of retrieval-based targeting because it redistributes to endosomes upon treatment with agents that disrupt lumenal pH, and it undergoes endosome-to-Golgi retrieval upon drug removal. Analysis of chimeric molecules containing domains from GPP130 and the plasma membrane protein dipeptidylpeptidase IV indicated that GPP130 targeting information is contained entirely within its lumenal domain. Dissection of the lumenal domain indicated that a predicted coiled-coil stem domain adjacent to the transmembrane domain was both required and sufficient for pH-sensitive Golgi localization and endosome-to-Golgi retrieval. Further dissection of this stem domain revealed two noncontiguous stretches that each conferred Golgi localization separated by a stretch that conferred endosomal targeting. Importantly, in the absence of the endosomal determinant the Golgi targeting of constructs containing either or both of the Golgi determinants became insensitive to pH disruption by monensin. Because monensin blocks endosome-to-Golgi transport, the finding that the endosomal determinant confers monensin sensitivity suggests that the endosomal determinant causes GPP130 to traffic to endosomes from which it is normally retrieved. Thus, our observations identify Golgi and endosomal targeting determinants within a lumenal predicted coiled-coil domain that appear to act coordinately to mediate retrieval-based targeting of GPP130.
INTRODUCTION
The distinct protein compositions that define organelles in the secretory pathway are maintained by retention of resident proteins in their home compartment and retrieval of escaped residents from distal compartments. With regard to targeting to the stacked portion of the mammalian Golgi apparatus, the relative importance of retention (Weisz et al., 1993; Nilsson et al., 1994) versus retrieval (Johnston et al., 1994; Hoe et al., 1995; Harris and Waters, 1996; Linstedt et al., 1997) depends on which of two conflicting views of transport through the Golgi is considered. The stable compartments model (Orci et al., 2000) predicts an important role for retention to ensure that Golgi residents are excluded from transport vesicles that carry cargo between cisternae in a forward direction. The cisternal progression model (Glick et al., 1997; Bonfanti et al., 1998) predicts an important role for retrieval to ensure that, as cisternae mature, Golgi residents are included in backward-trafficking retrieval vesicles. As evidence continues to emerge in support of aspects of each model, it will probably also follow that the sorting of Golgi residents in stacked cisternae involves mechanisms that result in both de-enrichment in forward-progressing vesicle carriers as well as enrichment in retrieval carriers.
Although the extent to which retrieval-based targeting underlies Golgi stack organization is unknown, if it occurs, it is likely to be mediated by incorporation of Golgi proteins into COPI-coated vesicles (Love et al., 1998; Lanoix et al., 1999; but for another view, see Orci et al., 2000). Curiously, the integral membrane enzymes targeted to the stacked Golgi contain little or no targeting information in their cytoplasmic domains, whereas sorting of integral membrane proteins into COPI vesicles is typically mediated by cytoplasmically oriented sorting signals. In fact, for many of the studied cases, the key determinant for targeting to the Golgi stack appears to be in the transmembrane domain where it acts independently or in conjunction with determinants in the membrane-adjacent portion of the lumenal domain (reviewed by Munro, 1998). The transmembrane domain signal appears to depend on short length rather than any specific targeting sequence (Bretscher and Munro, 1993; Masibay et al., 1993). How transmembrane domain length might lead to COPI vesicle incorporation is a matter of speculation. One possibility is that the signal acts independently of a specific receptor; for example, the membrane composition at the site of retrieval vesicle formation could favor incorporation of short transmembrane domains. Another possibility is that a targeting determinant, whether it be a short transmembrane domain or a sequence stretch positioned outside the membrane, interacts with a retrieval receptor bearing a cytoplasmic COPI-coat binding sequence.
Isolation of Golgi retrieval receptors, if they exist, may prove difficult if the key sorting interaction takes place within a bilayer. Also, retrieval within the Golgi stack is difficult to demonstrate, in part, because it takes place over short distances between adjacent cisternae in a stack. The Golgi phosphoprotein of 130 kDa (GPP130) possesses targeting attributes that may bypass these concerns. Similar to other Golgi stack proteins, GPP130 has a type II membrane topology, but unlike these other proteins, which on average have transmembrane domains with a stretch of 15 strongly hydrophobic residues (Bretscher and Munro, 1993), GPP130 has a stretch of 20 hydrophobic amino acids (Linstedt et al., 1997). Most importantly, unlike other stack residents, GPP130 exhibits pH-sensitive targeting (Linstedt et al., 1997). After treatment of cells with agents, such as monensin, that block acidification of lumenal compartments, GPP130 moves out of the cis-Golgi to endosomes. On drug removal, endosome-localized GPP130 returns to the Golgi. These observations suggest that GPP130 contains sequence determinants that mediate endosomal targeting and endosome-to-Golgi retrieval. Furthermore, because the neutralizing agents that cause GPP130 redistribution are known to block endosome-to-Golgi traffic (Brown et al., 1986; Chapman and Munro, 1994; Clague et al., 1994), and because both human GPP130 and its rat counterpart Golgi integral membrane protein-cis acquire late Golgi carbohydrate modifications (Yuan et al., 1987), these observations suggest that GPP130 is targeted to the early Golgi by retrieval from distal compartments, including endosomes. Given that GPP130 retrieval is readily assayed after drug washout, our goal was to identify GPP130 Golgi-targeting determinants and test their role in retrieval.
Here we report that GPP130's targeting information resides in its lumenal domain within a 210 amino acid sequence stretch predicted to form a coiled-coil stem structure adjacent to the Golgi membrane. Surprisingly, two noncontiguous regions within this stretch acted independently to confer monensin-insensitive Golgi targeting. These were separated by a stretch that, in isolation, conferred endosomal targeting, and, when present together with one or both of the Golgi determinants, conferred monensin-sensitive Golgi targeting. These observations identify a novel type of Golgi targeting signal and suggest that it is comprised of separate determinants acting together to mediate GPP130 retrieval to the Golgi.
MATERIALS AND METHODS
Reagents and Antibodies
Cycloheximide was dissolved directly in normal medium at 100 μg/ml just before use. Monensin (Sigma-Aldrich, St. Louis, MO) was dissolved in isopropanol just before use to make a 10 mM stock, and the resulting stock was diluted 1000-fold into normal medium. The cDNA encoding dipeptidyl peptidase IV (DPPIV) was provided by O. Weisz (University of Pittsburgh, Pittsburgh, PA), and the monoclonal antibody (mAb) against DPPIV was provided by A. Hubbard (Johns Hopkins University, Baltimore, MD). The GPP130 mAb A1/118, used at 1:400, as well as the giantin polyclonal antibody, used at 1:500, have been described previously (Linstedt et al., 1997). The anti-hemagglutinin (HA) mAb 12CA5, used at 1:200, was provided by J. Woolford (Carnegie Mellon University, Pittsburgh, PA). Fixable fluorescein isothiocyanate (FITC)-dextran (Molecular Probes, Eugene, OR) was dissolved directly in normal medium at 1 mg/ml just before use.
Expression Constructs
All constructs were in the mammalian expression vector pECE, included an N-terminal HA epitope tag where indicated, and were generated as follows. 1–38: A fragment corresponding to the GPP130 cytoplasmic and transmembrane domain (TMD) (amino acid residues [aa] 1–38) was amplified by polymerase chain reaction (PCR) and cloned in-frame into a NotI site present in DPPIV at the beginning of the sequence that encodes its lumenal domain. The resulting fusion of the GPP130 cytoplasmic and transmembrane sequence with the DPPIV lumenal sequence (aa 47-Stop) was excised and cloned into pECE with the use of BamHI and KpnI. 40–696: An excised PvuII fragment encoding the membrane proximal stretch of the GPP130 lumenal domain was cloned in-frame into the NotI site of DPPIV. A fragment containing the N-terminal DPPIV sequence (aa 1–34) fused to GPP130 was then excised with KpnI and AccI and used to replace the corresponding N-terminal region in pECE-GPP130, producing contiguous GPP130 C-terminal sequence (aa 40–696). HADPPIV: A fragment encoding rat DPPIV was excised with the use of HindIII and XhoI and inserted in-frame with the use of a HindIII site at the 3′ end of sequences encoding an N-terminal HA epitope (in pRD54 Gal I:HA). The resulting HADPPIV was then cloned into pECE with the use of BamHI and XhoI sites. HAGPP130: An EcoRI site at the 3′ end of the HA epitope sequence was joined in frame to an EcoRI site nine nucleotides upstream of the starting ATG in GPP130. 38–248: The GPP130 region corresponding to the predicted coiled-coil (aa 38–248) was PCR amplified with NotI and XbaI containing primers and cloned in 3′ of the DPPIV cytoplasmic and TMD (aa 1–34) with the use of the NotI site and XbaI site. 248–294: A BstBI-XmnI fragment was excised from GPP130 (aa 248–294), made blunt, and used to replace a NotI-XbaI fragment in DPPIV, thus removing all but the cytoplasmic and TMD of DPPIV (aa 1–34). 295–696: HA-DPPIV was cut with XmnI and DraI to release the HADD fragment (aa 1–46), which was then inserted in frame into pECEGPP130 that had been subjected to a partial digest with XmnI to remove all GPP130 sequence except that corresponding to the long acidic C terminus (aa 295–696). Δ40–247: After the BstB1 in HAGPP130 was cut and made blunt, a partial digest with PvuII was performed followed by religation to generate an otherwise intact and in frame clone lacking the coil-encoding sequence. 38–107: The GPP130 region corresponding to an N-terminal part of the coil (aa 38–107) was PCR amplified with NotI and XbaI containing primers and cloned in 3′ of the DPPIV cytoplasmic and TMD (aa 1–34) with the use of the NotI site and XbaI site. 89–294: First, an XmnI-DraI fragment was excised from DPPIV containing the cytoplasmic and TMD sequences (aa 1–46) and used to replace an XmnI-XmnI fragment containing the corresponding GPP130 domains that was removed by partial digestion. This, HADDG(89-Stop), was then cut with XbaI, made blunt, partially digested with XmnI, and then religated leaving the desired segment (aa 89–294) in frame with the DPPIV. 89–175: The construct 89–294 was cut with AccI and BstBI, made blunt, and religated leaving the desired GPP130 segment (aa 89–175) in frame with the DPPIV, as confirmed by sequencing. 176–248: First, a BamHI-DraI fragment was excised from DPPIV containing the cytoplasmic and TMD sequences (aa 1–46) and used to replace an BglII-AccI (blunted) fragment containing the corresponding GPP130 domains. This was then cut with BstBI and XbaI, made blunt, and then religated leaving the desired segment (aa 176–248) in frame with the DPPIV. Δ89–175: After the AccI site in HAGPP130 was cut and made blunt, a partial digest with XmnI was performed followed by religation to generate an otherwise intact and in frame clone lacking the coil-B encoding sequence (aa 89–175). 38–175: The coil construct (38–248) was digested with AccI and XbaI, made blunt, and religated to leave the desired segment (aa 38–175) followed by a stop codon. 89–248: HADDG(89-Stop) was cut with BstBI and XbaI, made blunt, and then religated, leaving only the desired segment (aa 89–248) in frame with the DPPIV.
Transfection and Immunofluorescence Microscopy
HeLa cells were transfected with the use of the Ca3(PO4)2 method as described (Ausubel et al., 1995), trypsinized, and plated the following day onto 12-mm glass coverslips. On day 3, the cells were treated as indicated in the figure legends and analyzed by indirect immunofluorescence as described previously (Linstedt et al., 1997). Treatments included immediate fixation, fixation after a 4-h incubation in media containing 100 μg/ml cycloheximide, fixation after a 1-h monensin treatment (incubation in media containing 10 μM monensin and 100 μg/ml cycloheximide), and fixation after a 3-h washout (after the 1-h monensin treatment the cells were washed 4 × 1.5 ml with media containing cycloheximide and recultured for 3 h in media containing cycloheximide). Transfection of Chinese hamster ovary (CHO) and COS-7 cells was as described (Linstedt et al., 1997).
Coimmunoprecipitation Experiments
On day 2 posttransfection, each 10-cm plate of COS-7 cells was washed in phosphate-buffered saline and lysed in 1 ml of 10 mM HEPES, pH 7.2, 100 mM KCl, 0.5% Triton X-100, plus 10 μg/ml pepstatin and leupeptin, plus 50 mM phenylmethylsulfonyl fluoride. Lysates were passed through a 25-gauge needle and incubated at 4°C for 15 min. Insoluble material was removed by centrifugation and the resulting lysate was precleared by incubation with protein A-Sepharose for 30 min at 4°C. The precleared lysate was incubated with A1/118 antibodies covalently attached to protein A-Sepharose for 2 h at 4°C. The unbound material was precipitated with trichloroacetic acid. The beads were washed five times with 1 ml of 10 mM HEPES, pH 7.2, 100 mM KCl, 0.5% Triton X-100 and boiled in reducing sample buffer. Bound and unbound samples were analyzed by immunoblotting as described (Linstedt and Hauri, 1993) with the use of anti-GPP130 (1:100) or anti-HA (1:100) monoclonal antibodies.
RESULTS
GPP130 pH-sensitive Targeting and Endosome-to-Golgi Retrieval Are Mediated by Its Predicted Coiled-Coil Domain
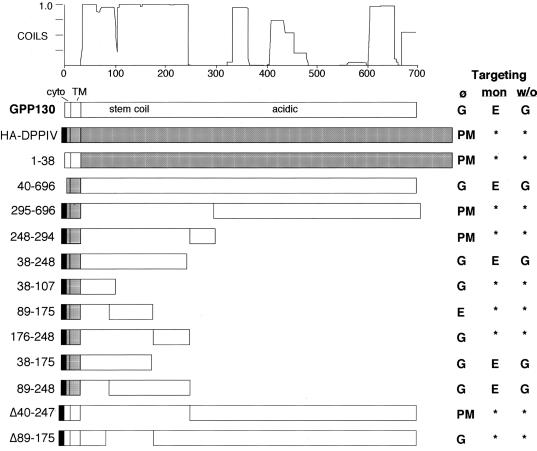
GPP130 has cytoplasmic, transmembrane, and lumenal domains of 12, 20, and 664 amino acid residues, respectively (Linstedt et al., 1997). To determine the potential role of each domain in the localization of GPP130, we generated chimeric molecules containing topologically equivalent domains from DPPIV, an integral plasma membrane protein with a type II topology similar to that of GPP130. Figure 1 depicts all constructs reported in this study. As indicated, most constructs contained an N-terminal HA epitope tag. Construct names refer to the GPP130 sequence present by amino acid number. Cells transfected with these constructs were treated with cycloheximide for at least 3 h before fixation to exclude any Golgi staining simply due to newly synthesized protein transiting the secretory pathway. They were then costained for the chimera and for the Golgi marker giantin.
Figure 1.
Schematic representation of GPP130/DPPIV chimeras. GPP130 sequences are represented by open boxes, DPPIV sequences are shaded, and the HA epitope is filled. Names of constructs are based on GPP130 amino acid numbering. Note that all constructs have cytoplasmic (cyto) and transmembrane (TM) domains. The localization of each chimera after no treatment (Ø), monensin treatment (mon), and monensin washout (w/o) is indicated to the right as either Golgi (G), endosome (E), plasma membrane (PM), or unchanged (∗). At the top is a plot of the predicted GPP130 coiled-coil domains derived by the COILS program (Lupas et al., 1991) with a window size of 28.
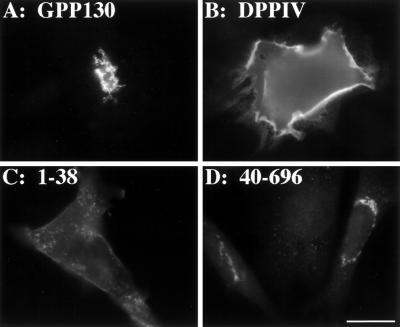
As expected, transfected full-length GPP130 was Golgi localized (Figure 2A) and full-length DPPIV accumulated on the plasma membrane (Figure 2B). A chimera containing the cytoplasmic and transmembrane domains of GPP130 (amino acids 1–38) fused to the lumenal domain of DPPIV, exhibited a plasma membrane pattern (Figure 2C). This indicated that the lumenal domain of GPP130 is required for proper Golgi targeting. Conversely, cells transfected with a chimera containing the lumenal domain of GPP130 (amino acids 40–696) fused to the cytoplasmic and transmembrane domains of DPPIV exhibited a normal Golgi pattern (Figure 2D). This pattern was indistinguishable from the Golgi marker giantin stained in the same cells. Taken together, these results indicated that the lumenal domain of GPP130 was required for Golgi targeting, and that, when fused to the cytoplasmic and transmembrane domains of DPPIV, the lumenal domain was also sufficient for Golgi targeting.
Figure 2.
GPP130 lumenal domain is required and sufficient for Golgi targeting. CHO cells transfected with constructs for full-length GPP130 (A), full-length DPPIV (B), chimera 1–38 containing GPP130 cytoplasmic and transmembrane domains (C), and chimera 40–696 containing the GPP130 lumenal domain (D) were treated with cycloheximide, fixed, and stained with the use of antibodies against GPP130 (A and D) or DPPIV (B and C). Untransfected CHO cells did not stain with either antibody. Bar, 10 μm.
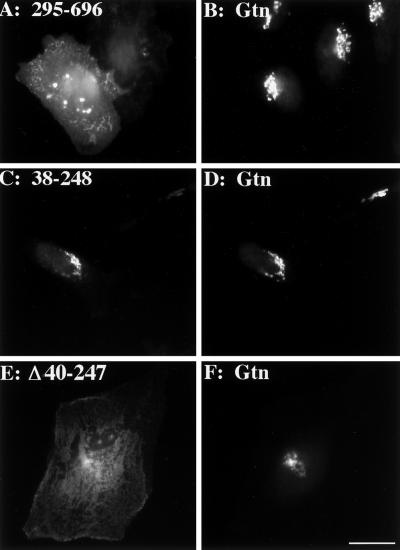
Conceptually, the GPP130 lumenal domain can be divided into two parts. The first part, encompassing membrane proximal residues 35–244, can be considered a stem region because it is membrane adjacent and strongly predicted to form a coiled-coil structure (Linstedt et al., 1997). Hereafter, this region will be referred to as the coil domain. The remaining part, encompassing residues 245 to the C terminus at 696, is uncommonly rich in aspartic and glutamic acid residues. To test these parts for Golgi targeting information, we generated chimeric proteins consisting of nonoverlapping segments of the lumenal domain of GPP130, fused to the cytoplasmic and transmembrane domains of HA epitope-tagged DPPIV (Figure 1). The large acidic C terminus of GPP130 lacked apparent targeting information because chimeras such as 295–696 containing this GPP130 region clearly exhibited surface staining (Figure 3A) distinct from giantin staining in the same cells (Figure 3B). In contrast, the coil-containing chimera, 38–248, yielded a Golgi pattern (Figure 3C) that precisely colocalized with giantin staining in the same cells (Figure 3D).
Figure 3.
Predicted coiled-coil stem domain (aa 38–248) within the GPP130 lumen is necessary and sufficient for Golgi localization. HeLa cells transfected with chimera 295–696 containing the acidic region (A and B); chimera 38–248 containing the coil domain (C and D); and Δ40–247, a version of GPP130 lacking the coil domain (E and F), were treated with cycloheximide for 4 h then fixed and double stained with the use of anti-HA (A, C, and E) and anti-giantin (Gtn; B, D, and F). Bar, 10 μm.
Because the Golgi targeting information in the GPP130 lumenal domain mapped to the coil domain, we next tested whether the predicted coil domain is also required. A deletion construct of the full-length GPP130 cDNA was generated with this region excised. The corresponding protein, Δ40–247, was plasma membrane localized (Figure 3E), and this was confirmed by surface staining of nonpermeabilized cells (our unpublished observations). Therefore, the coil was both necessary and sufficient for GPP130 Golgi targeting.
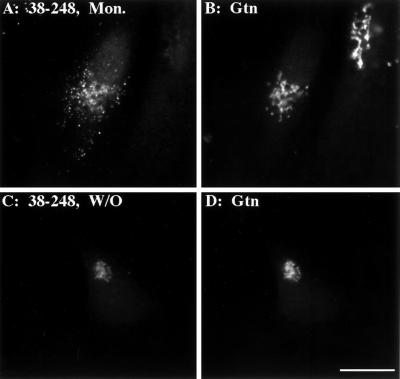
Because endogenous GPP130 exhibits pH-sensitive Golgi targeting and endosome-to-Golgi retrieval (Linstedt et al., 1997), we next tested whether the GPP130 coil contained the necessary determinants to mediate this behavior. Interestingly, the coil-containing chimera (38–248) retained the ability to reversibly redistribute to endosomes. The chimera, which was initially coincident with giantin in untreated cells (Figure 3C), accumulated, upon monensin treatment, in peripheral structures (Figure 4A) that lacked giantin staining (Figure 4B). On drug washout, the coil chimera was able to traffic from endosomes back to the Golgi yielding a Golgi pattern (Figure 4C) coincident with giantin staining (Figure 4D). Because both the monensin treatment and washout incubations included cycloheximide, we could exclude any contribution made by newly synthesized protein. Therefore, GPP130's pH-sensitive targeting and endosome-to-Golgi retrieval are mediated by its predicted coiled-coil domain.
Figure 4.
Predicted GPP130 coiled-coil stem mediates reversible redistribution to endosomes. HeLa cells transfected with the coil chimera 38–248 were incubated in 10 μM monensin in normal medium for 1 h (A and B) or 10 μM monensin for 1 h followed by washing and incubation in normal medium for 3 h (C and D). All incubations contained 100 μg/ml cycloheximide to prevent new protein synthesis. The cells were then fixed and double stained with anti-HA (A and C) and anti-giantin (B and D). Bar, 10 μm.
Isolation of Independently Acting Golgi and Endosomal Targeting Determinants in GPP130 Coiled-Coil Domain
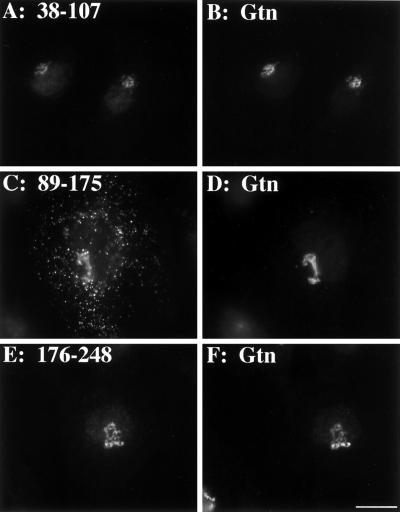
The fact that GPP130 undergoes reversible redistribution to endosomes upon treatment with monensin suggests that it contains, in addition to Golgi-targeting determinants, determinants required for endosomal targeting and endosome-to-Golgi retrieval (Linstedt et al., 1997). In an attempt to test whether these determinants could be separated, we subdivided the coil domain into three segments and tested each for targeting when placed on the DPPIV cytoplasmic and transmembrane domains (Figure 1; 38–107, 89–175, 176–248). We also made a number of further subdivisions but the proteins produced were unstable. Surprisingly, the nonoverlapping 38–107 and 176–248 chimeras each yielded Golgi localization patterns (Figure 5, A and E) that were coincident with giantin staining in the same cells (Figure 5, B and F, respectively). Therefore, Golgi-targeting information in the GPP130 lumenal domain mapped to two noncontiguous stretches of the GPP130 stem region. Except for the heptad repeat motif that underlies the predicted coiled-coil structure in this region, we did not find any obvious sequence similarities shared by these two sequence stretches.
Figure 5.
Separation of endosomal and Golgi-targeting determinants. HeLa cells transfected with chimera 38–107 (A and B), chimera 89–175 (C and D), or chimera 176–248 (E and F) were cycloheximide treated, fixed, and double stained with the use of anti-HA (A, C, and E) and anti-giantin (B, D, and F). Bar, 10 μm.
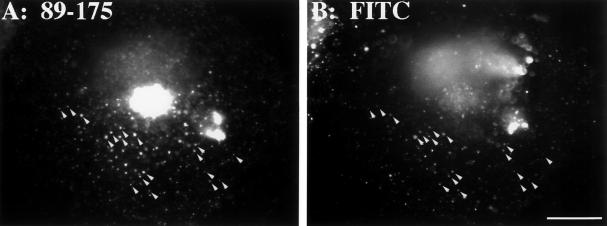
In contrast to the 38–107 and 176–248 constructs, the 89–175 chimera produced a mostly punctate peripheral pattern (Figure 5C) that was distinct from the giantin pattern in the same cells (Figure 5D). This suggested that this region might contain the hypothesized endosomal determinant. To test whether the punctate peripheral structures were endosomal we performed a 30-min FITC dextran uptake on cells transfected with the coil-B chimera. As expected, many of the structures that contained coil-B (Figure 6A) became labeled with the internalized dextran (Figure 6B). Together, the experiments testing the localization of the three segments of the predicted coiled-coil domain indicate that this domain contains two Golgi determinants separated by an endosomal determinant.
Figure 6.
Internalized FITC-dextran partially colocalizes with the endosomal determinant-containing chimera. COS-7 cells transfected with chimera 89–175 were incubated for 30 min with 1 mg/ml fixable FITC-dextran. After fixation the anti-HA staining pattern (A) was compared with the FITC pattern (B). Arrowheads mark a subset of the many puncta that were costained. These cells were not cycloheximide treated. Bar, 10 μm.
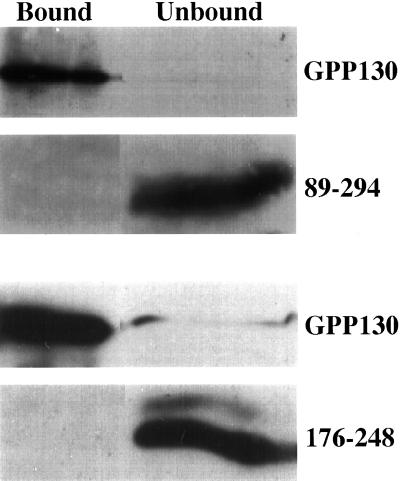
To ensure that the Golgi targeting we observed for DPPIV/GPP130 chimeras was not due to association with endogenous GPP130, we immunoprecipitated GPP130 under native conditions and tested for coprecipitation of two Golgi-localized chimeras. Because these chimeras did not contain the monoclonal anti-GPP130 antibody epitope, any coprecipitation would indicate association with GPP130. Bound and unbound fractions were subsequently probed with anti-GPP130 antibodies to determine the efficiency of the immunoprecipitation and with anti-HA antibodies to determine the extent of association. The GPP130 antibody beads quantitatively bound the endogenous GPP130 in cells transfected with two independent Golgi-localized chimeras; yet, although each HADPPIV/GPP130 chimera was abundantly expressed, neither was detected on the GPP130 antibody beads (Figure 7). This result strongly suggests that the targeting of the DPPIV/GPP130 chimeras occurs independently of endogenous GPP130.
Figure 7.
Golgi-localized chimeras do not stably interact with endogenous GPP130. Nonionic detergent lysates of cells transfected with either the 89–294 chimera (top panels) or the 176–248 chimera (bottom panels) were incubated with immobilized anti-GPP130. Bound and unbound material was collected and analyzed by immunoblotting to detect GPP130 and the chimeric proteins as indicated. Note that there was no detectable chimeric protein associated with immunoprecipitated GPP130, even though GPP130 was essentially depleted from the lysates.
To investigate the roles of the mapped Golgi and endosomal determinants in the GPP130 redistribution response to pH disruption, we first determined the localization of the chimeras containing Golgi determinants (38–107 and 176–248) in cells that were treated with monensin for 60 min. Significantly, in monensin-treated cells both 38–107 (Figure 8A) and 176–248 (Figure 8C) remained Golgi-localized as indicated by their coincidence with giantin staining in the same cells (Figure 8, B and D, respectively). This was in marked contrast to the accumulation in peripheral punctate structures observed for both the construct containing the entire coil (Figure 4A) and for endogenous GPP130 in parallel processed cells (our unpublished observations). One interpretation of this result is that the endosomal determinant, present in 89–175, somehow confers monensin sensitivity to GPP130 targeting. To test this idea, we next determined the targeting of a deletion construct in which the endosomal determinant was excised from the GPP130 cDNA. The resulting protein, Δ89–175, when expressed in HeLa cells, was targeted to the Golgi and it remained Golgi-localized upon monensin treatment (Figure 8E) as indicated by its coincidence with giantin (Figure 8F). Therefore, although the endosomal determinant is not required for GPP130 Golgi targeting it is required for the monensin-induced redistribution of GPP130 to endosomes.
Figure 8.
GPP130 endosomal determinant is required for pH-sensitive Golgi targeting. HeLa cells transfected with chimera 38–107 containing a Golgi determinant (A and B); chimera 176–248 containing a Golgi determinant (C and D); or Δ89–175, a version of GPP130 lacking the endosomal determinant (E and F), were treated with 10 μM monensin for 1 h in the presence of cycloheximide. They were then fixed and double stained with the use of anti-HA (A, C, and E) and anti-giantin (B, D, and F). Unlike wild type, chimeras with either of the isolated Golgi determinants remained Golgi localized after monensin treatment. This was also the case for GPP130 lacking the endosomal determinant. Bar, 10 μm.
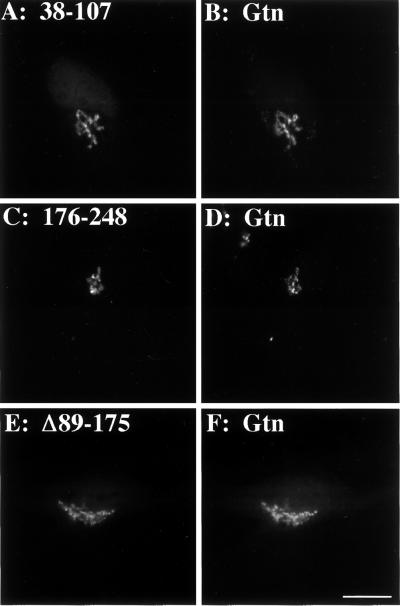
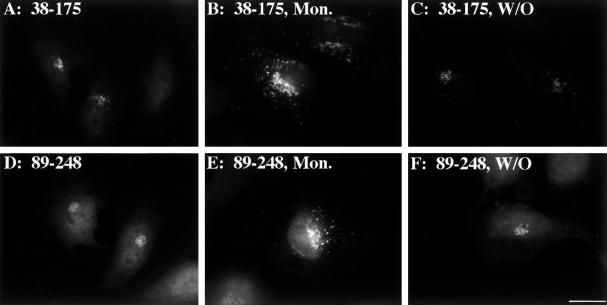
We next sought to test whether the mapped endosomal determinant is also sufficient to bestow monensin-induced endosomal redistribution upon the isolated Golgi determinants, and, if so, whether either of these Golgi determinants would then contain the necessary information for endosome-to-Golgi retrieval upon monensin washout. To this end, constructs were generated in which the endosomal determinant (89–175) was present together with either of the isolated monensin-insensitive Golgi determinants (38–107 or 176–248). When expressed in HeLa cells, the corresponding proteins, 38–175 (Figure 9A) and 89–248 (Figure 9D), were Golgi localized as expected. Interestingly, even without monensin treatment, a small amount of 38–175 staining was apparent in peripheral structures, suggesting that the presence of the endosomal determinant was causing some displacement out of the Golgi at steady state. On monensin treatment, both 38–175 (Figure 9B) and 89–248 (Figure 9E) redistributed to peripheral structures that did not colocalize with giantin. Thus, not only is the endosomal determinant required for GPP130 monensin sensitivity, but the presence of the endosomal determinant is also sufficient to cause monensin-sensitive Golgi targeting of the otherwise monensin-insensitive isolated Golgi determinants. Furthermore, upon monensin washout, both 38–175 (Figure 9C) and 89–248 (Figure 9F) mostly returned to the Golgi. This strongly suggests that endosome-to-Golgi retrieval information is present in each of the isolated Golgi determinants. In summary, GPP130's pH-sensitive Golgi targeting and endosome-to-Golgi retrieval depend on separate Golgi and endosome determinants present in its predicted coiled-coil stem domain. As described below, these findings are consistent with a model in which the endosomal determinant mediates cycling of GPP130 to endosomes and one or both of the Golgi determinants mediate its retrieval back to the early Golgi.
Figure 9.
Endosomal determinant confers pH sensitivity to the isolated GPP130 Golgi determinants. HeLa cells transfected with chimera 38–175 (A–C) or chimera 89–248 (D–F) were incubated in normal medium 4 h (A and D), incubated in 10 μM monensin in normal medium for 1 h (B and E), or incubated in 10 μM monensin for 1 h followed by washing and incubation in normal medium for 3 h (C and F). All incubations contained 100 μg/ml cycloheximide to prevent new protein synthesis. The cells were then fixed and double stained. Anti-HA staining is shown. The presence of the endosomal determinant restored the monensin-induced reversible redistribution to endosomes. Bar, 10 μm.
DISCUSSION
The sequence stretches in GPP130 that mediate Golgi targeting were mapped and tested for their role in endosome-to-Golgi retrieval. A stretch of 210 aa (residues 38–248) in the lumenal domain of GPP130 conferred all three targeting characteristics of GPP130: steady-state Golgi localization, endosomal targeting upon monensin treatment, and Golgi retrieval upon monensin washout, when attached to the cytoplasmic and transmembrane domains of a plasma membrane protein. Chimeras containing stretches of GPP130 outside of the 210-aa region, including those containing GPP130's cytoplasmic and transmembrane domains, were plasma membrane localized. Although the GPP130 lumenal domain is glycosylated, this has no apparent role in targeting because the glycosylation sites are not present in the targeting domain (Linstedt et al., 1997). Within the targeting domain, which is strongly predicted to form a coiled-coil structure, each of two segments, 38–107 and 176–248, appeared capable of mediating both Golgi targeting and endosome-to-Golgi retrieval, whereas a third segment, 89–175, appeared to be responsible for endosomal targeting. In addition, 89–175 mediated the monensin-sensitivity of GPP130 Golgi targeting.
These observations suggest a model in which the steady-state localization of GPP130 in the early Golgi is maintained by rapid retrieval from distal compartments, including the trans-Golgi network (TGN) and endosomes. As a consequence, when endosome-to-Golgi retrieval is blocked by pH disruption, GPP130 accumulates in endosomes. Because the endosomal determinant is required for monensin sensitivity, this determinant may actively divert a fraction of cycling GPP130 from the TGN to endosomes. Retrieval may not normally involve significant traffic to the cell surface because externally added anti-GPP130 antibodies are not specifically internalized in otherwise untreated cells (Linstedt et al., 1997). Because both the Golgi retrieval and endosomal determinants are lumenal, it is possible that they interact with Golgi-retrieval and endosomal-targeting receptors, respectively. Competition in the TGN between the Golgi retrieval and endosomal receptors may determine the extent to which GPP130 normally cycles to endosomes. Further experiments are required to validate this and other aspects of the model.
The findings reported here suggest that GPP130 is unusual in that its complex targeting characteristics are conferred solely through its lumenal domain, specifically through sequence elements present in its coiled-coil stem domain. GPP130 is recovered from cells as a homodimer lacking any stably associated partner protein (Puri and Linstedt, unpublished data). This suggests that it forms a parallel coiled-coil structure with itself, and that if its targeting domains interact with other proteins, such as the hypothetical receptors mentioned above, they probably do so not through coiled-coil formation but rather through transient interactions mediated by amino acid side chains on the exterior of the coiled-coil structure. Coiled-coil structure in this position is not sufficient for Golgi targeting as suggested by our unpublished observation that placement of another strongly predicted coiled-coil domain after the cytoplasmic and transmembrane domains of DPPIV produced a protein localized to the plasma membrane not the Golgi.
The lumenal location of GPP130's coiled-coil targeting domain is unusual, but other lumenal Golgi-targeting determinants have been identified (Nilsson et al., 1996; reviewed by Munro, 1998), notably in yeast (Graham and Krasnov, 1995; Vowels and Payne, 1998). The lumenal location of the endosomal determinant in GPP130 is also noteworthy. Although further work is necessary to characterize the endosomes that contain the 89–175 chimera, as well as those that contain GPP130 after monensin treatment, they are unlikely to be part of the early endosomal system because they do not costain with transferrin receptor (our unpublished observations). On the other hand, GPP130 after redistribution in response to chloroquine shows considerable overlap with the late endosomal protein mannose-6-phosphate receptor (Linstedt et al., 1997), suggesting that the lumenal endosomal determinant mediates targeting to a late endosomal compartment. Currently, we can only speculate as to why proteins that reside in the early Golgi contain endosomal targeting information. Perhaps the function of GPP130, which remains unknown, is required in endosomes under certain conditions. Analysis of more differentiated cell types may provide further insight into the regulation of GPP130 Golgi and endosomal targeting.
In summary, we have identified what appears to be a new type of Golgi-targeting determinant that appears to mediate retrieval. As stated above, GPP130 retrieval may involve interaction with a receptor that concentrates it in retrieval vesicles that originate from the late Golgi and endosomes. Intriguingly, COPI-coated vesicles, which have been implicated in retrograde transport within the Golgi, could also mediate GPP130 retrieval from endosomes. Subunits of the COPI coat have been detected on endosomes, and furthermore, at least one of these, βCOP, is required for endosomal carrier vesicle formation in vitro (Aniento et al., 1996). Significantly, both association of βCOP with endosomes and carrier vesicle formation require an acidic lumenal pH. An analogous pH-sensitive COPI coat recruitment could operate in TGN-to-early Golgi transport as well as endosome-to-Golgi transport. By isolating the domains that mediate retrieval of GPP130, our experiments not only define a new type of targeting signal but also generate the necessary reagents to identify and characterize the entire GPP130 retrieval sorting complex.
ACKNOWLEDGMENTS
We thank S. Puri, M. Puthenveedu, and A. Mehta for helpful suggestions; G. Apodaca and O. Weisz for critical reading of the manuscript; and A. Hubbard, J. Woolford, and O. Weisz for generous contribution of essential reagents. This work was supported by a National Institutes of Health grant GM-56779–02 to A.D.L.
Abbreviations used:
- aa
amino acid residues
- DPPIV
dipeptidyl peptidase IV
- GPP130
Golgi phosphoprotein of 130 kDa
- HA
hemagglutinin
- TMD
transmembrane domain
REFERENCES
- Aniento F, Gu F, Parton RG, Gruenberg J. An endosomal βCOP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol. 1996;133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York, NY: John Wiley & Sons; 1995. [Google Scholar]
- Bonfanti L, Mironov AA, Jr, Martinez-Menarguez JA, Martella O, Fusella A, Baldassarre M, Buccione R, Geuze HJ, Mironov AA, Luini A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell. 1998;95:993–1003. doi: 10.1016/s0092-8674(00)81723-7. [DOI] [PubMed] [Google Scholar]
- Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brown WJ, Goodhouse J, Farquhar MG. Mannose-6-phosphate receptors for lysosomal enzymes cycle between the Golgi complex and endosomes. J Cell Biol. 1986;103:1235–1247. doi: 10.1083/jcb.103.4.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman RE, Munro S. Retrieval of TGN proteins from the cell surface requires endosomal acidification. EMBO J. 1994;13:2305–2312. doi: 10.1002/j.1460-2075.1994.tb06514.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clague MJ, Urbe S, Aniento F, Gruenberg J. Vacuolar ATPase activity is required for endosomal carrier vesicle formation. J Biol Chem. 1994;269:21–24. [PubMed] [Google Scholar]
- Glick BS, Elston T, Oster G. A cisternal maturation mechanism can explain the asymmetry of the Golgi stack. FEBS Lett. 1997;414:177–181. doi: 10.1016/s0014-5793(97)00984-8. [DOI] [PubMed] [Google Scholar]
- Graham TR, Krasnov VA. Sorting of yeast alpha 1,3 mannosyltransferase is mediated by a lumenal domain interaction, and a transmembrane domain signal that can confer clathrin-dependent Golgi localization to a secreted protein. Mol Biol Cell. 1995;6:809–824. doi: 10.1091/mbc.6.7.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SL, Waters MG. Localization of a yeast early Golgi mannosyltransferase, Och1p, involves retrograde transport. J Cell Biol. 1996;132:985–998. doi: 10.1083/jcb.132.6.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe MH, Slusarewicz P, Misteli T, Watson R, Warren G. Evidence for recycling of the resident medial/trans Golgi enzyme, N-acetylglucosaminyltransferase I, in ldlD cells. J Biol Chem. 1995;270:25057–25063. doi: 10.1074/jbc.270.42.25057. [DOI] [PubMed] [Google Scholar]
- Johnston PA, Stieber A, Gonatas NK. A hypothesis on the traffic of MG160, a medial Golgi sialoglycoprotein, from the trans-Golgi network to the Golgi cisternae. J Cell Sci. 1994;107:529–537. doi: 10.1242/jcs.107.3.529. [DOI] [PubMed] [Google Scholar]
- Lanoix J, Ouwendijk J, Lin CC, Stark A, Love HD, Ostermann J, Nilsson T. GTP hydrolysis by arf-1 mediates sorting and concentration of Golgi resident enzymes into functional COP I vesicles. EMBO J. 1999;18:4935–4948. doi: 10.1093/emboj/18.18.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri H-P. Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kD. Mol Biol Cell. 1993;4:679–693. doi: 10.1091/mbc.4.7.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Mehta A, Suhan J, Reggio H, Hauri H-P. Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol Biol Cell. 1997;8:1073–1087. doi: 10.1091/mbc.8.6.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love HD, Lin CC, Short CS, Ostermann J. Isolation of functional Golgi-derived vesicles with a possible role in retrograde transport. J Cell Biol. 1998;140:541–51. doi: 10.1083/jcb.140.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Masibay AS, Balaji PV, Boeggeman EE, Quasba PK. Mutational analysis of the Golgi retention signal of bovine beta-1,4-galactosyltransferase. J Biol Chem. 1993;268:9908–9916. [PubMed] [Google Scholar]
- Munro S. Localization of proteins to the Golgi apparatus. Trends Cell Biol. 1998;8:11–15. doi: 10.1016/S0962-8924(97)01197-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Hoe MH, Slusarewicz P, Rabouille C, Watson R, Hunte F, Watzele G, Berger EG, Warren G. Kin recognition between medial Golgi enzymes in HeLa cells. EMBO J. 1994;13:562–574. doi: 10.1002/j.1460-2075.1994.tb06294.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Rabouille C, Hui N, Watson R, Warren G. The role of the membrane-spanning domain and stalk region of N-acetylglucosaminyltransferase I in retention, kin recognition and structural maintenance of the Golgi apparatus in HeLa cells. J Cell Sci. 1996;109:1975–1989. doi: 10.1242/jcs.109.7.1975. [DOI] [PubMed] [Google Scholar]
- Orci L, Amherdt M, Ravazzola M, Perrelet A, Rothman JE. Exclusion of Golgi residents from transport vesicles budding from Golgi cisternae in intact cells. J Cell Biol. 2000;150:1263–1270. doi: 10.1083/jcb.150.6.1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vowels JJ, Payne GS. A role for the lumenal domain in Golgi localization of the Saccharomyces cerevisiae guanosine diphosphatase. Mol Biol Cell. 1998;9:1351–1365. doi: 10.1091/mbc.9.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz OA, Swift AM, Machamer CE. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Barriocanal JG, Bonifacino JS, Sandoval IV. Two integral membrane proteins located in the cis-middle and trans-part of the Golgi system acquire sialylated N-linked carbohydrates and display different turnovers and sensitivity to cAMP-dependent phosphorylation. J Cell Biol. 1987;105:215–227. doi: 10.1083/jcb.105.1.215. [DOI] [PMC free article] [PubMed] [Google Scholar]