Abstract
Expression of placental growth factor (PGF) is closely associated with placental perfusion in early pregnancy. PGF is primarily expressed in placental trophoblasts, and its expression decreases in preeclampsia, associated with placental hypoxia. The transcription factors glial cells missing 1 (GCM1) and metal-regulatory transcription factor 1 (MTF1) have been implicated in the regulation of PGF gene expression through regulatory elements upstream and downstream of the PGF transcription start site, respectively. Here, we clarified the mechanism underlying placenta-specific PGF expression. We demonstrate that GCM1 up-regulates PGF expression through three downstream GCM1-binding sites (GBSs) but not a previously reported upstream GBS. Interestingly, we also found that these downstream GBSs also harbor metal-response elements for MTF1. Surprisingly, however, we observed that MTF1 is unlikely to regulate PGF expression in the placenta because knockdown or overexpression of GCM1, but not MTF1, dramatically decreased PGF expression or reversed the suppression of PGF expression under hypoxia, respectively. We also demonstrate that another transcription factor, Distal-less homeobox 3 (DLX3), interacts with the DNA-binding domain and the first transactivation domain of GCM1 and that this interaction inhibits GCM1-mediated PGF expression. Moreover, the GCM1–DLX3 interaction interfered with CREB-binding protein–mediated GCM1 acetylation and activation. In summary, we have identified several GBSs in the PGF promoter that are highly responsive to GCM1, have demonstrated that MTF1 does not significantly regulate PGF expression in placental cells, and provide evidence that DLX3 inhibits GCM1-mediated PGF expression. Our findings revise the mechanism for GCM1- and DLX3-mediated regulation of PGF gene expression.
Keywords: placenta, gene transcription, growth factor, transcription factor, acetylation, DLX3, GCM1, MTF1, PGF, trophoblast
Introduction
Vasculogenesis and angiogenesis are essential for placental development. These events are modulated by vascular endothelial growth factor (VEGF),2 placental growth factor (PGF), and their receptors kinase insert domain receptor (KDR) and Flt-1 (1). The human placenta is composed of villous tissues where fetal arterial and venous vessels are developed to convey nutrients and gases from the mother to the fetus (2, 3). Indeed, VEGF and PGF are preferentially responsible for branching and nonbranching angiogenesis in terminal villi, which results in a complex and branched capillary web and elongated capillary loops, respectively (3). A balance of branching and nonbranching angiogenesis is maintained for proper geometry of the villous vascular bed for healthy pregnancy outcomes.
PGF, which is a member of the VEGF family, is primarily expressed in placental trophoblasts, and its serum level increases throughout pregnancy and peaks between gestational weeks 28 and 32 (4, 5). In early pregnancy, a correlation of placental perfusion and PGF expression has been observed by measuring the systolic blood flow velocity at proximal and distal ends of the umbilical cord artery and maternal serum PGF levels (6). Clinically, a decreased circulating level of PGF has been detected as early as the first trimester and often leads to development of preeclampsia (PE) of which symptoms include hypertension and proteinuria (7). As PE is associated with poor uteroplacental circulation and placental hypoxia, PGF expression is also decreased under hypoxia (8, 9). Importantly, administration of recombinant human PGF has recently been shown to relieve the clinical symptoms of PE in rat and baboon disease models induced by uteroplacental ischemia (10, 11).
Previous studies have reported that glial cells missing 1 (GCM1), metal-regulatory transcription factor 1 (MTF1), and DLX3 are involved in the regulation of PGF gene expression in human placenta (12–14). GCM1 is a placental transcriptional factor responsible for trophoblast fusion, migration, and invasion and hormone production by transactivation of syncytin, HtrA4, and hCGβ genes (15–18). As cAMP signaling is crucial for trophoblast differentiation, GCM1 activity is up-regulated by the cAMP/PKA/CBP and cAMP/Epac1/Ca2+/calmodulin–dependent protein kinase I signaling cascades, respectively (19, 20). A GCM1-response element (GRE) harbors a consensus GCM1-binding sequence of 5′-ATGCGGG(T/C)-3′ (21). MTF1 was identified in the regulation of metallothionein gene expression in the presence of heavy metals such as copper, zinc, and cadmium (22). MTF1 binds to DNA sequence motifs known as metal-response elements (MREs) with a core consensus of 5′-TGC(A/G)C(G/A/T/C)C-3′ (23). Chang et al. (12) have demonstrated that GCM1 up-regulates PGF promoter activity through a GRE at position −818 to −810 relative to the transcription start site (TSS). Conversely, Depoix et al. (24) performed a DNA microarray analysis to demonstrate that PGF is induced in BeWo cells treated with the cAMP stimulant forskolin (FSK). The same group further demonstrated that mutation of the GRE identified by Chang et al. (12) fails to inhibit FSK-induced PGF expression and questioned the role of GCM1 in regulation of PGF gene expression. Interestingly, Nishimoto et al. (13) reported that MTF1 up-regulates PGF gene expression through triple repeats of MRE downstream of TSS. Nevertheless, it is hard to reconcile the placenta-specific gene expression pattern of PGF with MTF1, which is a ubiquitous transcription factor. The Distal-less homeobox 3 (DLX3) transcription factor harbors a conserved homeodomain with DNA-binding activity and regulates trophoblast differentiation (25, 26). Targeted gene disruption of Dlx3 in mice leads to embryonic lethality and placental vascularization defects in the labyrinth (27). DLX3 has recently been reported in the regulation of trophoblast differentiation and modulates PGF expression by inhibition of the transactivation activity of GCM1 in a homeodomain-dependent fashion (14, 28).
In search of GCM1 functions in placenta, we have recently conducted chromatin immunoprecipitation (ChIP)-chip analysis to identify PGF as a candidate target gene of GCM1 (16). In the present study, we investigated the functional role of GCM1 in regulation of PGF gene expression and provide a revised mechanism underlying the placenta-specific expression of PGF gene. We identify three novel tandem repeats of GRE downstream of the TSS of PGF gene, which also constitute the above described triple repeats of MRE. Importantly, the identified GREs/MREs are recognized by GCM1, but not MTF1, in vivo and in vitro by ChIP analysis and EMSA, respectively. We further demonstrate that the transcript level of GCM1, but not MTF1, correlates with that of PGF in response to hypoxia and FSK. GCM1 overexpression significantly reverses the suppression of PGF gene expression by hypoxia, which was not observed by MTF1 overexpression. We also show that DNA-binding domain (DBD) and the first transactivation domain (TAD) of GCM1 are involved in interaction with DLX3, leading to suppression of GCM1-mediated PGF expression. DLX3 very likely blocks the interaction between GCM1 and CBP to suppress CBP-mediated GCM1 acetylation and activation. Collectively, our results provide new insights into regulation of PGF gene expression by GCM1 and DLX3 in the human placenta.
Results
Identification of PGF as a GCM1 target gene
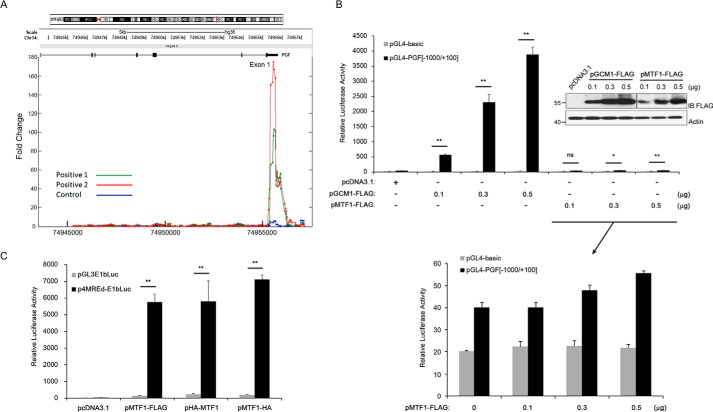
We recently performed ChIP-chip experiments in BeWo31 cells, which stably express GCM1 with an N-terminal HA tag (HA-GCM1) and identified PGF as a candidate GCM1 target gene (16). As shown in Fig. 1A, strong HA-GCM1 binding signals were detected and mapped to a genomic region downstream of TSS of PGF gene. Interestingly, MTF1 has been suggested to regulate PGF promoter activity through a genomic region harboring triple repeats of MRE positioned between nucleotides −511 and −468 relative to the translation initiation site (i.e. nucleotides +12 to +55 relative to TSS) (13), which also overlaps with the aforementioned HA-GCM1–binding region in ChIP-chip experiments. Because PGF is primarily expressed in placenta and MTF1 is a ubiquitous transcription factor, we wished to re-examine whether MTF1 or GCM1 plays a key role in regulation of PGF gene expression in placenta. We compared the effects of GCM1 and MTF1 on the pGL4-PGF(−1000/+100) luciferase reporter construct (note that nucleotides were numbered relative to TSS) in 293T cells in transient expression experiments. The luciferase activity directed by pGL4-PGF(−1000/+100) was significantly stimulated by GCM1-FLAG in a dose-dependent manner (Fig. 1B, upper). In contrast, transcriptional activation of the pGL4-PGF(−1000/+100) construct by MTF1-FLAG was marginally detected after a closer examination (Fig. 1B, lower). To rule out the possibility that the C-terminal FLAG tag may impair MTF1-FLAG activity, we compared the MTF1-FLAG expression plasmid with another expression plasmid encoding MTF1 with an N-terminal HA tag (HA-MTF1) or a C-terminal HA tag (MTF1-HA) in transient expression experiments using an established MTF1 reporter construct, p4MREd-E1bLuc. Indeed, significant stimulation of luciferase activity directed by p4MREd-E1bLuc was detected in the presence of MTF1-FLAG, HA-MTF1, or MTF1-HA (Fig. 1C), supporting that all three MTF1 expression constructs are functional in terms of transcriptional activity. Taken together, these results suggested that GCM1, but not MTF1, very likely plays an important role in regulation of PGF promoter activity.
Figure 1.
Identification of PGF as a GCM1 target gene. A, ChIP-chip isolation of genomic sites in the PGF gene occupied by GCM1. BeWo31 cells stably expressing HA-GCM1 were treated with 50 μm FSK for 24 h and then subjected to chemical cross-linking and immunoprecipitation using HA mAb (positive, green and red) or normal mouse serum (control, blue). The immunopurified genomic fragments were amplified and labeled for hybridization to the chips of Human Promoter 1.0R array (Affymetrix). Note that significant hybridization signals were detected in a genomic region downstream of the transcription start site of PGF gene. B, regulation of PGF promoter activity by GCM1 or MTF1. 293T cells were transfected with different combinations of pGL4-basic, pGL4-PGF(−1000/+100), pGCM1-FLAG, and increasing amounts of pMTF1-FLAG. Cells were harvested for luciferase assays at 48 h post-transfection. The levels of GCM1-FLAG and MTF1-FLAG proteins were analyzed by immunoblotting (IB) with FLAG mAb. For easier comparison, an enlargement of MTF-FLAG data is presented. C, analysis of MTF1 transactivation activity. 293T cells were transfected with pGL3E1bLuc, p4MREd-E1bLuc, and the indicated MTF1 expression plasmid. At 48 h post-transfection, cells were harvested for luciferase assays. Error bars in B and C represent S.D. of the mean of three independent experiments. *, p < 0.05; **, p < 0.01; ns, not significant.
Identification and characterization of GCM1 response elements in PGF gene
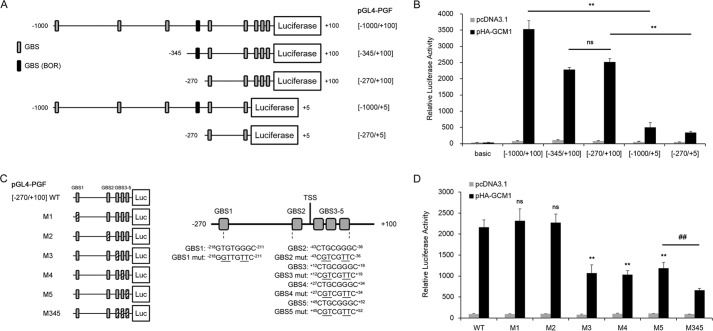
We characterized the genomic region −1000 to +100 (relative to TSS) of PGF gene for potential GREs using a series of luciferase constructs harboring different parts of this genomic region (Fig. 2A). 293T cells were transfected with pHA-GCM1 and the aforementioned PGF reporter plasmids in transient expression experiments. The luciferase activity directed by pGL4-PGF(−270/+100) was also enhanced by HA-GCM1, although the effect was lower compared with pGL4-PGF(−1000/+100) (Fig. 2B). Interestingly, deletion of nucleotides +6 to +100 dramatically diminished the enhancement effect of HA-GCM1 on the luciferase activity directed by pGL4-PGF(−1000/+5) and pGL4-PGF(−270/+5), suggesting that the deleted genomic region constitutes a major regulatory region responsive to GCM1. Chang et al. (12) have reported an upstream GRE (hereafter named as GBS-BOR) in the PGF promoter, i.e. nucleotides −818 to −810 relative to the translation initiation site or −295 to −288 relative to the TSS. Because the luciferase activity directed by pGL4-PGF(−345/+100) or pGL4-PGF(−270/+100) was not significantly different, it is very likely that GBS-BOR is not a functional GRE (Fig. 2B). Again, marginal effects of MTF-FLAG were observed on the tested PGF reporter constructs (Fig. S1). We then concentrated on the genomic region of −270 to +100 of PGF gene for identification of functional GREs. There are five potential GCM1-binding sites (i.e. GBS1–5) in the genomic region of −270 to +100 (Fig. 2, A and C). The GBSs were changed by site-directed mutagenesis individually (M1–5) or in combination (M345) in pGL4-PGF(−270/+100), and the resultant reporter constructs were cotransfected with pHA-GCM1 into 293T cells. As shown in Fig. 2D, mutagenesis of GBS3, -4, or -5 (the M3, -4, or -5 construct) alone significantly reduced the stimulatory effect of HA-GCM1 on the PGF promoter region of −270 to +100. Moreover, this effect was further decreased on the M345 construct with combined mutagenesis of GBS3, -4, and -5 (Fig. 2D). These results suggested that GBS3, -4, and -5 are functional GREs required for up-regulation of PGF gene expression by GCM1.
Figure 2.
Identification of GCM1 response elements in PGF gene. A, schematic representation of PGF promoter reporter constructs. Genomic fragments covering different parts of the region nucleotides −1000 to +100 were subcloned into pGL4-basic to generate a series of PGF reporter constructs harboring the indicated PGF promoter region. Potential GBSs scrutinized in this study and the previously reported GBS (BOR) in nucleotides −1000 to +100 are boxed. B, promoter analysis of PGF gene in response to GCM1. 293T cells were transfected with pHA-GCM1 and the indicated PGF reporter plasmid for 48 h and then harvested for luciferase assays. C, schematic representation of potential GBSs in the PGF promoter region responsive to GCM1. Positions of GBS1–5 relative to the TSS are indicated, and wildtype (WT) and mutant (underlined) sequences of GBS1–5 are listed. D, functional characterization of GCM1 response elements. 293T cells were transfected with pHA-GCM1 and WT or mutant pGL4-PGF(−270/+100) harboring individual (M1–5) or combined (M345) mutant GBS. At 48 h post-transfection, cells were harvested for luciferase assays. Error bars in B and D represent S.D. of the mean of three independent experiments. **, p < 0.01 compared with the WT in the presence of pHA-GCM1; ns, not significant compared with the WT in the presence of pHA-GCM1; ##, p < 0.01 compared with M5 in the presence of pHA-GCM1.
Association of GCM1, but not MTF1, with PGF promoter
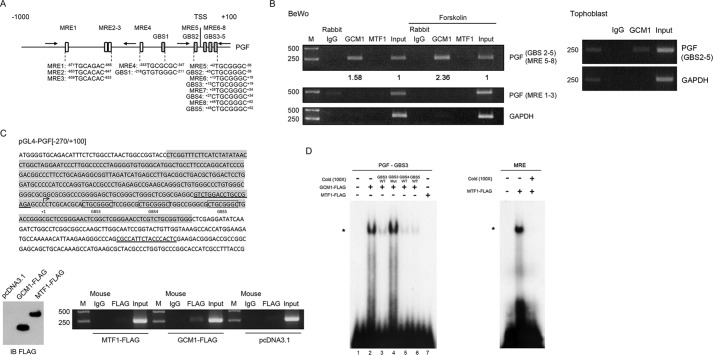
The identified GREs (GBS3–5) in PGF gene also harbor the previously reported triple repeats of MRE (here defined as MRE6–8) (13). Careful inspection of the PGF promoter region from nucleotides −1000 to +100 revealed additional potential MREs (MRE1–5; Fig. 3A). We then studied whether GCM1 or MTF1 is associated with PGF gene through GBS3–5/MRE6–8 and/or other MREs in placental BeWo cells by ChIP analysis. As shown in the left panel of Fig. 3B, GCM1, but not MTF1, was able to recognize the PGF promoter region harboring GBS3–5, which was enhanced in the presence of FSK. Association of GCM1 with GBS3–5 in the PGF promoter was also detected in primary human trophoblasts (Fig. 3B, right). We further performed ChIP analysis in 293T cells transfected with pGL4-PGF(−270/+100) and the expression plasmids encoding MTF1-FLAG and GCM1-FLAG, respectively. Specific interaction between GBS3–5 in pGL4-PGF(−270/+100) and GCM1-FLAG, but not MTF1-FLAG, was detected using a pGL4 vector–specific primer and a primer upstream of GBS3–5 (Fig. 3C). To study direct interaction between GCM1 and GBS3–5, we performed EMSA using a radiolabeled GBS3 oligonucleotide probe and recombinant GCM1-FLAG or MTF1-FLAG protein. Indeed, GCM1-FLAG specifically bound to the radiolabeled GBS3 probe, which was competed out by unlabeled GBS3, but not mutant GBS3, at a 100× molar ratio to the probe (Fig. 3D, left panel, lanes 2–4). Of note, MTF1-FLAG failed to bind to the GBS3 probe (Fig. 3D, left panel, lane 7), although it was able to bind to a radiolabeled MRE oligonucleotide probe of the consensus MTF1-binding sequence (Fig. 3D, right panel). Together with the observation that unlabeled GBS4 and GBS5 were capable of competing with the GBS3 probe for GCM1-FLAG (Fig. 3D, left panel, lanes 5and 6), these results strongly suggested that GCM1 is the bona fide transcription factor that recognizes GREs (GBS3–5) in the PGF promoter.
Figure 3.
Characterization of GCM1 binding to the PGF promoter. A, schematic representation of GBSs and MREs in PGF gene. Note that MRE5–8 overlap with GBS2–5. B and C, association of GCM1, but not MTF1, with the PGF promoter. ChIP analyses of GCM1-GBS or MTF1-MRE interaction were performed in BeWo cells treated with or without 50 μm FSK for 24 h using GCM1 or MTF1 Ab (B, left). ChIP analyses of GCM1-GBS interaction were also performed in primary human trophoblasts using GCM1 Ab (B, right). The numbers underneath indicate the band intensities normalized against input. In a separate experiment, 293T cells were transfected with pGL4-PGF(−270/+100), pGCM1-FLAG, and pMTF1-FLAG for 48 h. Cells were harvested for ChIP analysis using FLAG mAb. The sequence of nucleotide −270 to +100 in pGL4-PGF(−270/+100) is shaded in gray, primers for PCR are underlined, and GBS3–5 are boxed (C). The protein levels of GCM1-FLAG and MTF1-FLAG in the transfected 293T cells were analyzed by immunoblotting (IB). D, EMSA of GCM1 binding to the GBS3–5 (GREs) of PGF promoter. Recombinant GCM1-FLAG or MTF1-FLAG protein was incubated with radiolabeled PGF-GBS3 probe in the presence or absence of the indicated unlabeled oligonucleotide at a molar ratio of 100 over the probe followed by electrophoresis and autoradiography. In a separate experiment, recombinant MTF1-FLAG protein was incubated with radiolabeled MRE probe in the presence or absence of unlabeled MRE oligonucleotide in EMSA. The asterisk indicates the position of the protein–DNA complex.
Regulation of PGF gene expression by GCM1
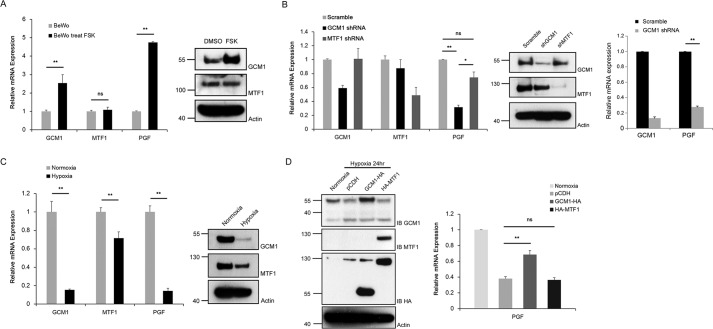
Trophoblast differentiation and GCM1 activity are stimulated by cAMP (20, 29–31). We tested the effect of FSK on the transcript and protein levels of PGF, GCM1, and MTF1 in BeWo cells. As shown in Fig. 4A, FSK up-regulated the transcript and protein levels of PGF and GCM1 but not MTF1. As a complementary approach, knocking down GCM1 in BeWo cells stably expressing GCM1 short-hairpin RNA (shRNA) significantly decreased the transcript level of PGF, whereas MTF1 knockdown failed to significantly affect PGF gene expression (Fig. 4B, left). Moreover, GCM1 knockdown also decreased the PGF transcript level in primary human trophoblasts (Fig. 4B, right). Because placental hypoxia and decreased PGF expression are associated with PE, we measured the GCM1, MTF1, and PGF transcript levels in BeWo cells under normoxic and hypoxic conditions. Compared with the MTF1 transcript level, the GCM1 and PGF transcript levels in BeWo cells were dramatically decreased by hypoxia (Fig. 4C, left panel). Correspondingly, the GCM1 protein level in BeWo cells was greatly reduced by hypoxia, whereas the MTF1 protein level was only modestly reduced (Fig. 4C, right panel). As hypoxia suppresses PGF expression, we introduced GCM1-HA or HA-MTF1 into hypoxic BeWo cells by a lentiviral expression system to test whether GCM1-HA or HA-MTF1 could reverse the PGF transcript level. As shown in Fig. 4D, GCM1-HA, but not HA-MTF1, significantly elevated the PGF transcript level in the BeWo cells under hypoxia. Taken together, these results suggested that GCM1 is a key factor controlling PGF gene expression in placenta in response to environmental cues such as FSK and hypoxia.
Figure 4.
Regulation of PGF gene expression by GCM1. A, stimulation of PGF gene expression by cAMP signaling. BeWo cells were treated with or without 50 μm FSK for 24 h. Cells were harvested for qRT-PCR and immunoblotting analyses of GCM1, MTF1, or PGF transcript and protein levels. B, GCM1 knockdown represses PGF gene expression. BeWo cells (left) or primary human trophoblasts (right) stably expressing scrambled, GCM1, or MTF1 shRNA were harvested for qRT-PCR and immunoblotting analyses for GCM1, MTF1, or PGF transcript and protein levels. C, down-regulation of PGF gene expression by hypoxia. BeWo cells were subjected to normoxic (1% O2) or hypoxic (21% O2) condition for 24 h and then harvested for qRT-PCR and immunoblotting analyses of GCM1, MTF1, or PGF transcript and protein levels. D, reversal of PGF gene expression by GCM1 under hypoxia. BeWo cells stably expressing empty vector (pCDH), GCM1-HA, or HA-MTF1 were subjected to normoxic or hypoxic condition for 24 h and then harvested for qRT-PCR and immunoblotting (IB) analyses of PGF transcript level and GCM1, HA-GCM1, MTF1, and HA-MTF1 protein levels. Error bars represent S.D. of the mean of three independent qRT-PCR experiments in A–D. *, p < 0.05; **, p < 0.01; ns, not significant.
Functional interaction between GCM1 and DLX3
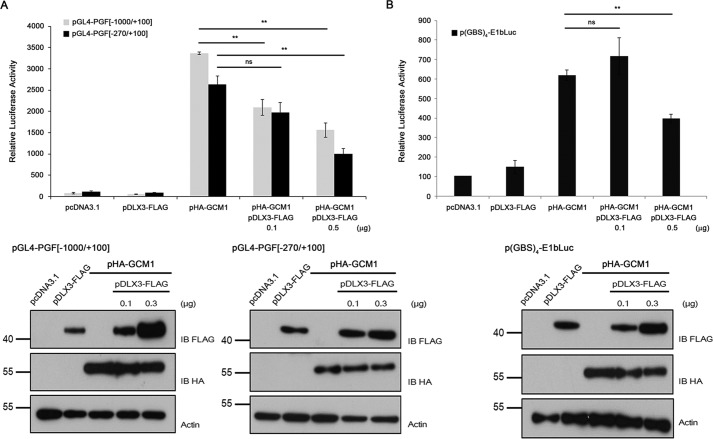
Li and Roberson (14) have recently reported that DLX3 and GCM1 modulate PGF promoter activity through the genomic region −369 to −320 (relative to TATA box, i.e. −622 to −592 relative to TSS. To test whether DLX3 affects GCM1-up-regulated PGF promoter activity, we transfected 293T cells with different combinations of pGL4-PGF(−1000/+100), pGL4-PGF(−270/+100), pHA-GCM1, and pDLX3-FLAG. DLX3-FLAG alone did not affect luciferase activity directed by pGL4-PGF(−1000/+100) or pGL4-PGF(−270/+100) (Fig. 5A). Instead, the stimulatory effect of HA-GCM1 on pGL4-PGF(−1000/+100) or pGL4-PGF(−270/+100) was significantly decreased when a higher amount of pDLX3-FLAG was cotransfected (Fig. 5A). A similar inhibitory effect of DLX3 on GCM1-mediated transcription was observed in 293T cells transfected with the GCM1 reporter construct p(GBS)4-E1bLuc, pHA-GCM1, and a higher amount of pDLX3-FLAG (Fig. 5B). These results suggest that DLX3 may suppress PGF expression through GCM1.
Figure 5.
Regulation of PGF promoter activity by DLX3. A, suppression of GCM1-mediated PGF promoter activation by DLX3. 293T cells were transfected with the pGL4-PGF(−1000/+100) or pGL4-PGF(−270/+100) reporter plasmid and pHA-GCM1 alone or pHA-GCM1 plus increasing amounts of pDLX3-FLAG for 48 h before being subjected to luciferase assays. B, DLX3 inhibits GCM1 transcriptional activity. 293T cells were transfected with the p(GBS)4-E1bLuc reporter plasmid and pHA-GCM1 alone or pHA-GCM1 plus increasing amounts of pDLX3-FLAG for 48 h before being subjected to luciferase assays. Representative immunoblots underneath show the protein levels of HA-GCM1 and DLX3-FLAG in the transfected cells. Error bars in A and B represent S.D. of the mean of three independent experiments. **, p < 0.01; ns, not significant.
Consequently, we examined the interaction between GCM1 and DLX3 by coimmunoprecipitation analysis of 293T cells transfected with pDLX3-FLAG and pHA-GCM1. Specific interaction between HA-GCM1 and DLX3-FLAG was detected and was further enhanced in the presence of the proteasome inhibitor MG132, which is known to stabilize GCM1 (Fig. 6A). To characterize endogenous GCM1–DLX3 interaction, we first performed immunofluorescence microscopy to detect colocalization of GCM1 and DLX3 in the nuclei of BeWo cells (Fig. 6B). We then performed coimmunoprecipitation analyses to demonstrate the interaction between endogenous DLX3 and GCM1 in BeWo cells (Fig. 6C). To map the interaction domain in GCM1 for DLX3, 293T cells were transfected with pHA-DLX3 and expression plasmids encoding Gal4-GCM1 fusion proteins harboring full-length or truncated GCM1 for coimmunoprecipitation analysis. As shown in Fig. 6C, HA-DLX3 interacts with the GCM1 polypeptide region, amino acids 1–167 or 167–349, corresponding to the N-terminal DBD or the first TAD of GCM1, respectively.
Figure 6.
Characterization of the interaction between GCM1 and DLX3. A, GCM1 specifically interacts with DLX3. 293T cells were transfected with the indicated combinations of pDLX3-FLAG, pHA-GCM1, and pGATA3-FLAG for 48 h and then harvested for coimmunoprecipitation analysis using FLAG and HA mAbs. Note that the interaction between DLX3-FLAG and HA-GCM1 was enhanced after 6 h of treatment with 10 μm proteasome inhibitor MG132. Specific interaction between GATA3-FLAG and HA-GCM1 was detected and used as a positive control. B, panels a–f, colocalization of GCM1 and DLX3. BeWo cells were stained with mouse anti-DLX3 and rabbit anti-GCM1 Abs. Cells were then incubated with secondary Abs and examined under a confocal microscope. Arrows indicate GCM1 and DLX3 nuclear colocalization. Bar in d, 20 μmeter; bar in f, 100 μmeter. C, endogenous interaction of GCM1 and DLX3. BeWo cells were harvested for immunoprecipitation (IP) with rabbit IgG or rabbit anti-DLX3 Ab followed by immunoblotting (IB) with rabbit anti-GCM1 Ab. D, mapping of the DLX3-interacting domains in GCM1. 293T cells were transfected with pHA-DLX3 and pGal4-FLAG or pGal4-GCM1-FLAG encoding for full-length or deleted GCM1 polypeptide. At 48 h post-transfection, cells were harvested for coimmunoprecipitation analysis using FLAG and HA mAbs.
DLX3 suppresses GCM1 activity by blockade of CBP-mediated GCM1 acetylation
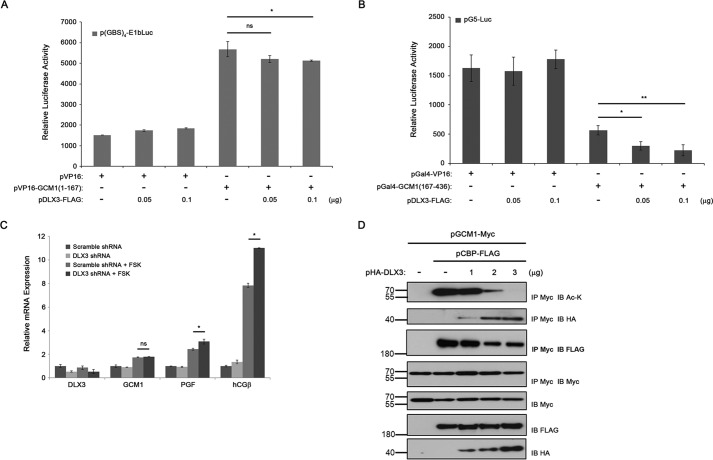
To explore the mechanism underlying the inhibitory effect of DLX3 on GCM1 activity, we studied whether DLX3 inhibits the DNA-binding or transactivation activity of GCM1 or both using a mammalian two-hybrid system. 293T cells were transfected with different combinations of p(GBS)4-E1bLuc, pVP16, pVP16-GCM1(1–167), and pDLX3-FLAG to measure the effect of DLX3-FLAG on the DNA-binding activity of GCM1. As shown in Fig. 7A, stimulation of the luciferase activity directed by p(GBS)4-E1bLuc was detected in the presence of VP16-GCM1(1–167), which is a fusion protein containing the TAD of VP16 and the DBD of GCM1. The transcriptional activity of VP16-GCM1(1–167) was only modestly inhibited by a higher amount of pDLX3-FLAG (Fig. 7A). To analyze the effect of DLX3 on the transcriptional activity of GCM1, 293T cells were transfected with different combinations of pG5-Luc, pVP16, pGal4-GCM1(167–436), and pDLX3-FLAG. Indeed, the luciferase activity directed by pG5-Luc was up-regulated by the Gal4-GCM1(167–436) fusion protein containing the DBD of GAL4 and the TAD of GCM1 (Fig. 7B). Interestingly, the transcriptional activity of Gal4-GCM1(167–436) was significantly decreased by DLX3-FLAG in a dose-dependent manner (Fig. 7B).
Figure 7.
DLX3 inhibits PGF expression through inhibition of CBP-mediated GCM1 acetylation and activation. A and B, DLX3 inhibits the TAD activity of GCM1. 293T cells were transfected with p(GBS)4-E1bLuc, pVP16, pVP16-GCM1(1–167), and increasing amounts of pDLX3-FLAG to assess the effect of DLX3 on the DBD activity of GCM1 (A). In a different experimental setting, 293T cells were transfected with pG5-Luc, pGal4-VP16, pGal4-GCM1(167–436), and increasing amounts of pDLX3-FLAG to assess the effect of DLX3 on the TAD activity of GCM1 (B). Cells were harvested for luciferase assays at 48 h post-transfection. C, DLX3 negatively regulates FSK-stimulated PGF gene expression. BeWo cells stably expressing scrambled or DLX3 shRNA were treated with or without 50 μm FSK for 24 h and then subjected to qRT-PCR analysis of DLX3, GCM1, PGF, and hCGβ transcript levels. D, suppression of CBP-mediated GCM1 acetylation by DLX3. 293T cells were transfected with pGCM1-Myc, pCBP-FLAG, and increasing amounts of pHA-DLX3. At 48 h post-transfection, cells were harvested for coimmunoprecipitation analysis with Myc, FLAG, HA, and Ac-Lys mAbs. Error bars in A–C represent S.D. of the mean of three independent experiments. IP, immunoprecipitation; IB, immunoblotting.
The biological function of DLX3 in regulation of PGF gene expression was studied by RNAi in BeWo cells treated with or without FSK. The PGF transcript level was not affected by DLX3 knockdown in the absence of FSK (Fig. 7C). As expected, the PGF transcript level was stimulated by FSK in the scrambled control cells. Interestingly, this FSK-stimulated PGF transcript expression was further elevated when DLX3 was knocked down (Fig. 7C). These observations suggest that DLX3 may interfere with the cAMP signaling pathway up-regulating GCM1-mediated PGF gene expression. We tested whether DLX3 jeopardizes the interaction between GCM1 and CBP, a transcriptional coactivator known to stimulate GCM1 acetylation and activity under cAMP (20). To this end, 293T cells were transfected with pGCM1-Myc, pCBP-FLAG, and pHA-DLX3 for coimmunoprecipitation analysis. As shown in Fig. 7D, acetylation of GCM1-Myc was detected in the presence of CBP-FLAG, which was decreased by increased interaction between GCM1-Myc and HA-DLX3. Correspondingly, the interaction between GCM1-Myc and CBP-FLAG was decreased (Fig. 7D). Therefore, DLX3 may suppress PGF gene expression by blocking the interaction between GCM1 and CBP and thereby inhibiting GCM1 acetylation and activation. The inhibitory effect of DLX3 on GCM1 activity was also effective on the expression of another GCM1 target gene, hCGβ, as DLX3 knockdown enhanced the stimulatory effect of FSK on hCGβ gene expression (Fig. 7C).
Discussion
It is generally believed that PGF is a critical factor regulating vasculogenesis under physiological and pathological conditions (32, 33). PGF expression is elevated during pregnancy and is crucial for normal pregnancy outcomes as decreased PGF expression is associated with preeclampsia featuring poor vasculogenesis and placental hypoxia (7). Indeed, administration of PGF was shown to reverse the clinical symptoms in animal models of preeclampsia, including hypertension, proteinuria, and endotheliosis (10, 11). Although GCM1 and MTF1 have been reported to regulate PGF gene expression in placenta (12–14), there are issues that need to be resolved before the physiological underpinnings of PGF expression can be truly revealed. In this regard, the regulatory elements for GCM1 in stimulation of PGF gene expression have not yet been clearly defined. Moreover, MTF1 is a ubiquitous transcription factor, which is difficult to reconcile with the primary expression of PGF gene in placenta.
In the present study, we reveal a new mechanism to revise the current knowledge about PGF gene expression in placenta. We redefined the key regulatory elements (GREs) in PGF gene to be GBS3–5 that are highly responsive to GCM1 and demonstrated that MTF1 barely regulates or does not recognize GBS3–5 in placental cells even though GBS3–5 also harbor previously reported MREs. This statement is supported by the following lines of evidence. First, ChIP-chip or ChIP analysis demonstrated the association of GCM1, but not MTF1, with GBS3–5. Second, transcriptional activation of PGF promoter by GCM1 was primarily dependent on GBS3–5. Third, direct binding of GCM1 to GBS3–5 was demonstrated by EMSA. Fourth, suppression of PGF gene expression by hypoxia was significantly reversed by GCM1, but not MTF1, in placental cells.
An upstream GRE (i.e. GBS-BOR) has been reported in PGF gene and localized to nucleotides −295 to −288 relative to TSS (12). Because pGL4-PGF(−345/+100) and pGL4-PGF(−270/+100) exhibit a similar response to GCM1 (Fig. 2B), i.e. deletion of nucleotides −345 to −269 did not affect GCM1-up-regulated PGF promoter activity, we think that the reported GBS-BOR is not a functional GRE. Conversely, deletion of nucleotides −1000 to −269 decreased the responsiveness of PGF promoter to GCM1 (Fig. 2B), suggesting potential upstream GREs within this region. This possibility was overshadowed by the fact that deletion of downstream GBS3–5 in pGL4-PGF(−1000/+5) and pGL4-PGF(−270/+5) further decreases the responsiveness of PGF promoter to a similar level (Fig. 2B). Therefore, we think that GBS3–5 constitute the essential GREs for GCM1 to regulate PGF gene expression in placenta. PGF expression has been reported in nonplacental cells, mostly tumor cells, in which PGF collaborates with VEGF family members to stimulate vessel formation for oxygen supply for tumor growth (34). Regulation of PGF gene expression in nonplacental cells has been attributed to MTF1, NF-κB, or FoxD1 (35–37). Therefore, MTF1 may be involved in the regulation of PGF gene expression in pathological conditions to support tumor growth in the absence of GCM1.
Li and Roberson (14, 28) have recently reported that DLX3 alone stimulates PGF gene expression in JEG-3 cells. Contradictorily, they also showed that DLX3 inhibits GCM1-up-regulated PGF gene expression. Our results in the present study did not support DLX3 positively stimulating PGF expression based upon two lines of evidence. First, DLX3 alone failed to activate the luciferase activity directed by pGL4-PGF(−1000/+100) that harbors the claimed DLX3 response element. Second, DLX3 knockdown did not affect the PGF transcript level in BeWo cells. Nevertheless, our results further indicated that DLX3 inhibits GCM1-transactivated PGF expression via interaction with the DBD and the first TAD of GCM1. We further ascribe this observation to the interference of CBP-mediated GCM1 acetylation and activation by DLX3, which was corroborated by the fact that DLX3 knockdown enhances GCM1 target gene expression (i.e. PGF and hCGβ) in response to the cAMP-stimulant FSK. Therefore, DLX3 may play a suppressor role to fine-tune GCM1-mediated trophoblast differentiation and PGF gene expression.
Experimental procedures
Plasmid constructs
HA- and FLAG-tagged GCM1 expression plasmids, pHA-GCM1 and pGCM1-FLAG, have been described previously (38). A DNA fragment encoding human MTF1 with an N-terminal HA tag or a C-terminal FLAG or HA tag was subcloned into the pcDNA3.1 expression vector (Invitrogen) to generate expression plasmids for HA-MTF1, MTF1-FLAG, or MTF1-HA. Likewise, DLX3 expression plasmids, DLX3-FLAG and HA-DLX3, were generated. The PGF promoter region from nucleotides −1000 to +100 (relative to TSS) was subcloned into pGL4-basic (Promega, Madison, WI) to generate the pGL4-PGF(−1000/+100) reporter plasmid. Derivatives of pGL4-PGF(−1000/+100) harboring different regions in the PGF promoter from nucleotides −1000 to +100 were also generated. The p4MREd-E1bLuc reporter plasmid was generated by subcloning four copies of the MREd in the mouse metallothionein-I gene into pGL3E1bLuc (20, 39). The p(GBS)4-E1bLuc reporter plasmid for GCM1 has been described previously (20). The pVP16, pGal4-VP16, and pG5-Luc plasmids were derived from the CheckMate mammalian two-hybrid system (Promega). The pVP16 plasmid harbors the herpes simplex virus VP16 transactivation domain, and the pGal4-VP16 plasmid encodes a fusion protein containing the yeast Gal4 DNA-binding domain and the VP16 transactivation domain.
Cell culture, transfection, and lentivirus transduction
293T and BeWo cells were obtained from the American Type Culture Collection (Manassas, VA) and maintained at 37 °C in minimal essential medium α medium (for 293T) or F-12K medium (for BeWo) supplemented with fetal bovine serum, streptomycin, and penicillin. Preparation of primary human trophoblasts from term placentas has been described previously (16, 17). For transient expression, cells were transfected with expression plasmids using Lipofectamine 2000 reagent (Invitrogen). For luciferase reporter assays, cells were harvested and analyzed with a commercial kit (Promega). Specific luciferase activities were normalized by protein concentration, which was measured using the BCA protein assay kit (Pierce). For generating stable knockdown cell lines, BeWo cells were infected by lentiviral pLKO.1-Puro shRNA expression plasmids harboring a scrambled sequence (5′-CCTAAGGTTAAGTCGCCCTCG-3′) or target sequences for GCM1 (5′-CCTCAGCAGAACTCACTAAAT-3′), MTF1 (5′-CAGAACTTACAATGGATATTA-3′), and DLX3 (5′-CCTTCTCTACTCCTCCAGTAA-3′) provided by the National RNAi Core Facility of Taiwan. The infected cells were subjected to antibiotic selection using 5 μg/ml puromycin, and the puromycin-resistant clones were pooled for studies.
ChIP assay
ChIP assays were performed to study in vivo association between PGF promoter and GCM1 or MTF1 in BeWo cells treated with or without 50 μm FSK for 24 h or primary human trophoblasts. In brief, the associated GCM1–genomic DNA or MTF1–genomic DNA complexes were immunoprecipitated by GCM1 or MTF1 (Santa Cruz Biotechnology, Dallas, TX) Ab and PCR-amplified for specific regions in the PGF promoter. Sequences of primers used for PCR are 5′-CAAACGCAGAGAGAGAGG-3′ and 5′-AGGAACTGGCAGAGACA-3′ for the genomic region harboring MRE1–3, 5′-GCTGGACTCCTGGATGC-3′ and 5′-CAGACGAGGTTCCCGAG-3′ for the genomic region harboring GBS2–5 (MRE5–8), and 5′-AAAAGCGGGGAGAAAGTAGG-3′ and 5′-CTAGCCTCCCGGGTTTCTCT-3′ for the GAPDH promoter. PCR products were analyzed on 2% agarose gels. Band intensities were quantitated by densitometric analysis using ImageJ software. In addition, 293T cells were transfected with pGL4-PGF(−270/+100), pMTF1-FLAG, and pGCM1-FLAG. At 48 h post-transfection, cells were harvested for ChIP assays to study in vivo association between the GBS3–5 (or MRE6–8) in pGL4-PGF(−270/+100) and GCM1-FLAG or MTF1-FLAG using FLAG mAb (Sigma-Aldrich). The immunoprecipitated complexes of genomic DNA and GCM1-FLAG or MTF1-FLAG were PCR-amplified for a specific region containing the GBS3–5 (or MRE6–8) in the PGF promoter construct. Sequences of primers used for PCR are 5′-GTCTGGACCTGCCGAGA-3′ and 5′-GAGTGGGTAGAATGGCG-3′.
Electrophoretic mobility shift assay (EMSA)
Recombinant GCM1-FLAG and MTF1-FLAG proteins were prepared from 293T cells transfected with pGCM1-FLAG and pMTF1-FLAG plasmids using anti-FLAG M2 affinity agarose gel for purification and FLAG peptide for elution. EMSA experiments using the PGF-GBS3 (5′-GCACGCACTGCGGGCTCCGGCG-3′) or MRE (5′-AGGGAGCTCTGCACTCCGCCCGAAAA-3′) oligonucleotide and the prepared GCM1-FLAG and MTF1-FLAG proteins were conducted under conditions as described previously (15).
Quantitative real-time PCR (qRT-PCR)
Cells were lysed using a RealTime ready cell lysis kit (Roche Applied Science) and reverse transcribed followed by analysis of transcripts of interest in a LightCycler® 480 real-time PCR instrument II (Roche Applied Science). The sequences of the primer sets for PCR analysis are as follows: 5′-CTGACAAGGCTTTTTTCTTCACA-3′ and 5′-CCAGACGGGACAGGTTT-3′ for GCM1, 5′-CACATACTGGTGAAAGACCC-3′ and 5′-GTGGACAGAAGGCTCAAG-3′ for MTF1, 5′-TCAGAGGTGGAAGTGGTACCCT-3′ and 5′-GCAGAGGCCGGCATTC-3′ for PGF, 5′-TACACCTACCACCACCAAT-3′ and 5′-CTCCTTCACCGACACTG-3′ for DLX3, 5′-GCTACTGCCCCACCATGACC-3′ and 5′-ATGGACTCGAAGCGCACATC-3′ for hGCβ, and 5′-AACTCCATCATGAAGTGTGACG-3′ and 5′-GATCCACATCTGCTGGAAGG-3′ for β-actin.
Coimmunoprecipitation analysis and immunofluorescence microscopy
To study the interaction between GCM1 and DLX3, 293T cells were transfected with pHA-GCM1 and pDLX3-FLAG. At 48 h post-transfection, cells were harvested in lysis buffer containing 50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 2 mm EDTA, 10% glycerol, 0.5% Nonidet P-40, 1 mm DTT, 5 mm NaF, 1 mm Na3VO4, 1 mm PMSF, and a protease inhibitor mixture (Sigma-Aldrich) followed by consecutive immunoprecipitation and immunoblotting with FLAG and HA (Sigma-Aldrich) mAbs. Interaction between endogenous GCM1 and DLX3 in BeWo cells and primary human trophoblasts was examined using rabbit anti-DLX3 (Proteintech, Rosemont, IL) and rabbit anti-GCM1 Abs. To map the GCM1 domain that interacts with DLX3, 293T cells were transfected with pDLX3-Myc and pGal4-FLAG or pGal4-GCM1-FLAG harboring full-length GCM1 or deletion mutants of GCM1. The proteins were immunoprecipitated with Myc mAb (Santa Cruz Biotechnology) followed by immunoblotting with FLAG mAb. To study CBP-mediated acetylation of GCM1, 293T cells were transfected with different combinations of pGCM1-Myc, pCBP-FLAG, and pHA-DLX3. At 48 h post-transfection, cells were harvested for immunoprecipitation using Myc mAb. The immune complexes were further analyzed by immunoblotting using Ac-Lys mAb (Cell Signaling Technology, Danvers, MA). Colocalization of GCM1 and DLX3 was performed in BeWo cells by costaining both factors with rabbit anti-GCM1 and mouse anti-DLX3 (Santa Cruz Biotechnology) Abs overnight and then coincubating with Alex Fluor 546–labeled secondary Ab for GCM1 and Alex Fluor 488–labeled secondary Ab (Invitrogen) for DLX3. Nuclei were stained by DAPI. Immunofluorescence was examined under a Leica TCS SP5 laser scanning confocal microscope (Wetzlar, Germany).
Statistical analysis
Differences were assessed by Student's t test. A p value of <0.05 was considered statistically significant (* or #, p < 0.05; ** or ##, p < 0.01).
Author contributions
Y.-H. C., M.-R. Y., L.-J. W., M.-H. C., and H. C. investigation; G.-D. C. methodology; H. C. conceptualization; H. C. supervision; H. C. funding acquisition; H. C. writing-original draft.
Supplementary Material
This work was supported by Ministry of Science and Technology Grants 103-2311-B-001-024 and 106-2311-B-001-013 and Academia Sinica, Taiwan (to H. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Fig. S1.
- VEGF
- vascular endothelial growth factor
- PGF
- placental growth factor
- GCM1
- glial cells missing 1
- MTF1
- metal-regulatory transcription factor 1
- GBS
- GCM1-binding site
- DLX3
- Distal-less homeobox 3
- CREB
- cAMP-response element–binding protein
- PE
- preeclampsia
- hCG
- human chorionic gonadotropin
- CBP
- CREB-binding protein
- GRE
- GCM1-response element
- MRE
- metal-response element
- TSS
- transcription start site
- FSK
- forskolin
- DBD
- DNA-binding domain
- TAD
- transactivation domain
- Ab
- antibody
- qRT-PCR
- quantitative real-time PCR
- Luc
- luciferase.
References
- 1. Cerdeira A. S., and Karumanchi S. A. (2012) Angiogenic factors in preeclampsia and related disorders. Cold Spring Harb. Perspect. Med. 2, a006585 10.1101/cshperspect.a006585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Benirschke K., and Kaufmann P. (2001) Early development of the human placenta, in Pathology of the Human Placenta, 4th Ed., pp. 42–49, Springer-Verlag, New York [Google Scholar]
- 3. Benirschke K., and Kaufmann P. (2001) Architecture of normal villous trees, in Pathology of the Human Placenta, 4th Ed., pp. 116–154, Springer-Verlag, New York [Google Scholar]
- 4. Khaliq A., Li X. F., Shams M., Sisi P., Acevedo C. A., Whittle M. J., Weich H., and Ahmed A. (1996) Localisation of placenta growth factor (PIGF) in human term placenta. Growth Factors 13, 243–250 [DOI] [PubMed] [Google Scholar]
- 5. Chau K., Hennessy A., and Makris A. (2017) Placental growth factor and pre-eclampsia. J. Hum. Hypertens. 31, 782–786 10.1038/jhh.2017.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Welch P. C., Amankwah K. S., Miller P., McAsey M. E., and Torry D. S. (2006) Correlations of placental perfusion and PlGF protein expression in early human pregnancy. Am. J. Obstet. Gynecol. 194, 1625–1631 10.1016/j.ajog.2006.01.012 [DOI] [PubMed] [Google Scholar]
- 7. Baumwell S., and Karumanchi S. A. (2007) Pre-eclampsia: clinical manifestations and molecular mechanisms. Nephron 106, c72–c81 10.1159/000101801 [DOI] [PubMed] [Google Scholar]
- 8. Soleymanlou N., Jurisica I., Nevo O., Ietta F., Zhang X., Zamudio S., Post M., and Caniggia I. (2005) Molecular evidence of placental hypoxia in preeclampsia. J. Clin. Endocrinol. Metab. 90, 4299–4308 10.1210/jc.2005-0078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gobble R. M., Groesch K. A., Chang M., Torry R. J., and Torry D. S. (2009) Differential regulation of human PlGF gene expression in trophoblast and nontrophoblast cells by oxygen tension. Placenta 30, 869–875 10.1016/j.placenta.2009.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Spradley F. T., Tan A. Y., Joo W. S., Daniels G., Kussie P., Karumanchi S. A., and Granger J. P. (2016) Placental growth factor administration abolishes placental ischemia-induced hypertension. Hypertension 67, 740–747 10.1161/HYPERTENSIONAHA.115.06783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Makris A., Yeung K. R., Lim S. M., Sunderland N., Heffernan S., Thompson J. F., Iliopoulos J., Killingsworth M. C., Yong J., Xu B., Ogle R. F., Thadhani R., Karumanchi S. A., and Hennessy A. (2016) Placental growth factor reduces blood pressure in a uteroplacental ischemia model of preeclampsia in nonhuman primates. Hypertension 67, 1263–1272 10.1161/HYPERTENSIONAHA.116.07286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chang M., Mukherjea D., Gobble R. M., Groesch K. A., Torry R. J., and Torry D. S. (2008) Glial cell missing 1 regulates placental growth factor (PGF) gene transcription in human trophoblast. Biol. Reprod. 78, 841–851 10.1095/biolreprod.107.065599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nishimoto F., Sakata M., Minekawa R., Okamoto Y., Miyake A., Isobe A., Yamamoto T., Takeda T., Ishida E., Sawada K., Morishige K., and Kimura T. (2009) Metal transcription factor-1 is involved in hypoxia-dependent regulation of placenta growth factor in trophoblast-derived cells. Endocrinology 150, 1801–1808 10.1210/en.2008-0949 [DOI] [PubMed] [Google Scholar]
- 14. Li S., and Roberson M. S. (2017) Dlx3 and GCM-1 functionally coordinate the regulation of placental growth factor in human trophoblast-derived cells. J. Cell. Physiol. 232, 2900–2914 10.1002/jcp.25752 [DOI] [PubMed] [Google Scholar]
- 15. Yu C., Shen K., Lin M., Chen P., Lin C., Chang G. D., and Chen H. (2002) GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 277, 50062–50068 10.1074/jbc.M209316200 [DOI] [PubMed] [Google Scholar]
- 16. Wang L. J., Cheong M. L., Lee Y. S., Lee M. T., and Chen H. (2012) High-temperature requirement protein A4 (HtrA4) suppresses the fusogenic activity of syncytin-1 and promotes trophoblast invasion. Mol. Cell. Biol. 32, 3707–3717 10.1128/MCB.00223-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheong M. L., Wang L. J., Chuang P. Y., Chang C. W., Lee Y. S., Lo H. F., Tsai M. S., and Chen H. (2016) A positive feedback loop between glial cells missing 1 and human chorionic gonadotropin (hCG) regulates placental hCGβ expression and cell differentiation. Mol. Cell. Biol. 36, 197–209 10.1128/MCB.00655-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baczyk D., Drewlo S., Proctor L., Dunk C., Lye S., and Kingdom J. (2009) Glial cell missing-1 transcription factor is required for the differentiation of the human trophoblast. Cell Death Differ. 16, 719–727 10.1038/cdd.2009.1 [DOI] [PubMed] [Google Scholar]
- 19. Chang C. W., Chang G. D., and Chen H. (2011) A novel cyclic AMP/Epac1/CaMKI signaling cascade promotes GCM1 desumoylation and placental cell fusion. Mol. Cell. Biol. 31, 3820–3831 10.1128/MCB.05582-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chang C. W., Chuang H. C., Yu C., Yao T. P., and Chen H. (2005) Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol. Cell. Biol. 25, 8401–8414 10.1128/MCB.25.19.8401-8414.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schreiber J., Enderich J., and Wegner M. (1998) Structural requirements for DNA binding of GCM proteins. Nucleic Acids Res. 26, 2337–2343 10.1093/nar/26.10.2337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Laity J. H., and Andrews G. K. (2007) Understanding the mechanisms of zinc-sensing by metal-response element binding transcription factor-1 (MTF-1). Arch. Biochem. Biophys. 463, 201–210 10.1016/j.abb.2007.03.019 [DOI] [PubMed] [Google Scholar]
- 23. Günther V., Lindert U., and Schaffner W. (2012) The taste of heavy metals: gene regulation by MTF-1. Biochim. Biophys. Acta 1823, 1416–1425 10.1016/j.bbamcr.2012.01.005 [DOI] [PubMed] [Google Scholar]
- 24. Depoix C., Tee M. K., and Taylor R. N. (2011) Molecular regulation of human placental growth factor (PlGF) gene expression in placental villi and trophoblast cells is mediated via the protein kinase a pathway. Reprod. Sci. 18, 219–228 10.1177/1933719110389337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gehring W. J., Müller M., Affolter M., Percival-Smith A., Billeter M., Qian Y. Q., Otting G., and Wüthrich K. (1990) The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329 10.1016/0168-9525(90)90253-3 [DOI] [PubMed] [Google Scholar]
- 26. Chui A., Tay C., Cocquebert M., Sheehan P., Pathirage N. A., Donath S., Fournier T., Badet J., Evain-Brion D., Brennecke S. P., Kalionis B., and Murthi P. (2012) Homeobox gene Distal-less 3 is a regulator of villous cytotrophoblast differentiation and its expression is increased in human idiopathic foetal growth restriction. J. Mol. Med. 90, 273–284 10.1007/s00109-011-0836-1 [DOI] [PubMed] [Google Scholar]
- 27. Morasso M. I., Grinberg A., Robinson G., Sargent T. D., and Mahon K. A. (1999) Placental failure in mice lacking the homeobox gene Dlx3. Proc. Natl. Acad. Sci. U.S.A. 96, 162–167 10.1073/pnas.96.1.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li S., and Roberson M. S. (2017) DLX3 interacts with GCM1 and inhibits its transactivation-stimulating activity in a homeodomain-dependent manner in human trophoblast-derived cells. Sci. Rep. 7, 2009 10.1038/s41598-017-02120-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Knerr I., Schubert S. W., Wich C., Amann K., Aigner T., Vogler T., Jung R., Dötsch J., Rascher W., and Hashemolhosseini S. (2005) Stimulation of GCMa and syncytin via cAMP mediated PKA signaling in human trophoblastic cells under normoxic and hypoxic conditions. FEBS Lett. 579, 3991–3998 10.1016/j.febslet.2005.06.029 [DOI] [PubMed] [Google Scholar]
- 30. Keryer G., Alsat E., Tasken K., and Evain-Brion D. (1998) Cyclic AMP-dependent protein kinases and human trophoblast cell differentiation in vitro. J. Cell Sci. 111, 995–1004 [DOI] [PubMed] [Google Scholar]
- 31. Wice B., Menton D., Geuze H., and Schwartz A. L. (1990) Modulators of cyclic AMP metabolism induce syncytiotrophoblast formation in vitro. Exp. Cell Res. 186, 306–316 10.1016/0014-4827(90)90310-7 [DOI] [PubMed] [Google Scholar]
- 32. Dewerchin M., and Carmeliet P. (2014) Placental growth factor in cancer. Expert Opin. Ther. Targets 18, 1339–1354 10.1517/14728222.2014.948420 [DOI] [PubMed] [Google Scholar]
- 33. De Falco S. (2012) The discovery of placenta growth factor and its biological activity. Exp. Mol. Med. 44, 1–9 10.3858/emm.2012.44.1.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carmeliet P., Moons L., Luttun A., Vincenti V., Compernolle V., De Mol M., Wu Y., Bono F., Devy L., Beck H., Scholz D., Acker T., DiPalma T., Dewerchin M., Noel A., et al. (2001) Synergism between vascular endothelial growth factor and placental growth factor contributes to angiogenesis and plasma extravasation in pathological conditions. Nat. Med. 7, 575–583 10.1038/87904 [DOI] [PubMed] [Google Scholar]
- 35. Zhang H., Palmer R., Gao X., Kreidberg J., Gerald W., Hsiao L., Jensen R. V., Gullans S. R., and Haber D. A. (2003) Transcriptional activation of placental growth factor by the forkhead/winged helix transcription factor FoxD1. Curr. Biol. 13, 1625–1629 10.1016/j.cub.2003.08.054 [DOI] [PubMed] [Google Scholar]
- 36. Cramer M., Nagy I., Murphy B. J., Gassmann M., Hottiger M. O., Georgiev O., and Schaffner W. (2005) NF-κB contributes to transcription of placenta growth factor and interacts with metal responsive transcription factor-1 in hypoxic human cells. Biol. Chem. 386, 865–872 10.1515/BC.2005.101 [DOI] [PubMed] [Google Scholar]
- 37. Green C. J., Lichtlen P., Huynh N. T., Yanovsky M., Laderoute K. R., Schaffner W., and Murphy B. J. (2001) Placenta growth factor gene expression is induced by hypoxia in fibroblasts: a central role for metal transcription factor-1. Cancer Res. 61, 2696–2703 [PubMed] [Google Scholar]
- 38. Yang C. S., Yu C., Chuang H. C., Chang C. W., Chang G. D., Yao T. P., and Chen H. (2005) FBW2 targets GCMa to the ubiquitin-proteasome degradation system. J. Biol. Chem. 280, 10083–10090 10.1074/jbc.M413986200 [DOI] [PubMed] [Google Scholar]
- 39. Dubé A., Harrisson J. F., Saint-Gelais G., and Séguin C. (2011) Hypoxia acts through multiple signaling pathways to induce metallothionein transactivation by the metal-responsive transcription factor-1 (MTF-1). Biochem. Cell Biol. 89, 562–577 10.1139/o11-063 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.