Abstract
The human malaria parasite Plasmodium falciparum proliferates in red blood cells following repeated cycles of invasion, multiplication, and egress. P. falciparum serine repeat antigen 5 (PfSERA5), a putative serine protease, plays an important role in merozoite egress. However, regulation of its activity leading to merozoite egress is poorly understood. In this study, we show that PfSERA5 undergoes phosphorylation prior to merozoite egress. Immunoprecipitation of parasite lysates using anti-PfSERA5 serum followed by MS analysis identified calcium-dependent protein kinase 1 (PfCDPK1) as an interacting kinase. Association of PfSERA5 with PfCDPK1 was corroborated by co-sedimentation, co-immunoprecipitation, and co-immunolocalization analyses. Interestingly, PfCDPK1 phosphorylated PfSERA5 in vitro in the presence of Ca2+ and enhanced its proteolytic activity. A PfCDPK1 inhibitor, purfalcamine, blocked the phosphorylation and activation of PfSERA5 both in vitroas well as in schizonts, which, in turn, blocked merozoite egress. Together, these results suggest that phosphorylation of PfSERA5 by PfCDPK1 following a rise in cytosolic Ca2+ levels activates its proteolytic activity to trigger merozoite egress.
Keywords: phosphoproteomics, plasmodium, posttranslational modification (PTM), protein phosphorylation, malaria, CDPK1, merozoites egress, Plasmodium falciparum, SERA5
Introduction
Despite continued efforts to control malaria, it still remains a major public health problem in the tropical world. The development and spread of artemisinin-resistant parasites in recent years has raised serious concerns regarding the availability of effective tools to eliminate malaria (1, 2). Sustained efforts are thus required to understand parasite biology, identify novel drug targets, and develop new therapeutic interventions against malaria. The clinical manifestations of malaria are largely attributed to its asexual stages, during which merozoites repeatedly invade, multiply within, and exit red blood cells for their propagation. The exit of invasive merozoites from infected erythrocytes, a process referred to as parasite egress, is a critical step in blood-stage parasite growth. A cascade of proteolytic activities has been implicated in the release of merozoites from mature schizonts. However, an understanding of the regulation of the activities of such proteases leading to egress still remains elusive (3–7).
A number of putative Plasmodium proteases, such as cysteine proteases (falcipain-2, SERA9 proteins, dipeptidyl aminopeptidase 3 (DPAP 3)), aspartic protease (plasmepsin II) and serine protease (subtilisin-like protease 1 (SUB1)) are involved in merozoite egress from P. falciparum schizonts (8–12). Of particular interest are members of the SERA family of putative proteases that possess a central papain-like protease domain (13). P. falciparum encodes nine SERA proteins, SERA 1–9 (14). Among these, SERA 1–5 and SERA 9 have a serine residue in place of the conventional cysteine residue at the active site, whereas three SERA proteins, SERA 6–8, have cysteine at the key catalytic position (15, 16). Such substitutions at the catalytic sites of enzyme homologs are well known in protozoan and metazoan proteases, some of which play key regulatory roles (17). SERAs are proteins ∼100–130 kDa in size and are mainly localized in the parasitophorous vacuole (13). Many members of the PfSERA family are proteolytically processed by PfSUB1 (11). Of the nine members of the PfSERA family, PfSERA5 and PfSERA6 are refractory to genetic deletion, suggesting that they are essential for parasite growth and viability (18, 19). PfSERA5 is the best studied PfSERA family member and is a promising vaccine candidate as well as drug target (20, 21). Inhibitors of PfSUB1, an enzyme involved in PfSERA5 processing, stalls merozoite egress and parasite maturation, suggesting that PfSERA5 and its processing enzyme PfSUB1 are important drug targets (11, 12). Although PfSERA5 is highly expressed at the late schizont stage, its proteolytic activity has been an issue of debate because of low proteolytic activity displayed by recombinant PfSERA5 expressed in Escherichia coli (13, 22). A few recent studies have suggested that PfSERA5 serves as a pseudoprotease that regulates the kinetics/efficiency of P. falciparum merozoite egress from mature schizonts (23, 24).
Posttranslational modifications (PTMs) of proteins, such as phosphorylation/dephosphorylation, palmitoylation, ubiquitination, and sumoylation, are fast emerging as important mechanisms for the regulation of functional activity in various organisms. Cross-talk between these PTMs may also play a role in regulating parasite development (25). However, little is known about the parasite proteases that undergo PTMs and about the role of PTMs in regulating their protease activities. In most organisms, including malaria parasites, of the many possible PTMs, the role of phosphorylation in regulation of the cell cycle has been studied most extensively (26–28). Given that PfSERA5 is essential for parasite survival, we explored the potential role of posttranslational modification, especially the role of phosphorylation of PfSERA5, in the regulation of its protease and merozoite egress activities. Immunoprecipitation using anti-phosphoserine antibodies followed by MS analysis showed that PfSERA5 is phosphorylated at the schizont stage. We identified PfCDPK1 as an interacting kinase that phosphorylates PfSERA5. Purfalcamine, a known chemical inhibitor of PfCDPK1 (29), inhibited the native protease activity at the schizont stage and also blocked merozoite egress. These results provide insight into the regulation of PfSERA5 protease activity and its link with merozoite egress.
Results
PfSERA5 exists in the phosphorylated form at schizont stages of P. falciparum
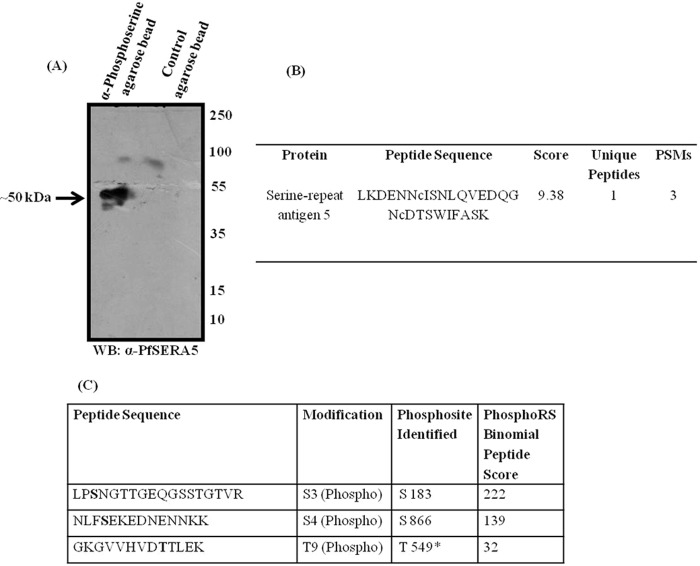
Protein phosphorylation, in most eukaryotes, including Plasmodium spp., is one of the major regulatory processes that controls the functional activities of diverse proteins to modulate cellular processes (30). PfSERA5 expression in P. falciparum is tightly regulated so that it is primarily expressed at the schizont stage. To determine whether phosphorylation plays a role in the regulation of PfSERA5 activity, we investigated the phosphorylation status of PfSERA5 in P. falciparum schizonts by immunoprecipitation using anti-phosphoserine antibodies coupled to agarose. When eluates were probed using mouse antiserum raised against PfSERA5 by Western blotting, an ∼50-kDa band corresponding to the processed form of PfSERA5 was detected (Fig. 1A). When immunoprecipitates generated from P. falciparum schizont lysates with anti-phosphoserine antibodies were digested with trypsin and analyzed by tandem MS (LC-MS/MS), peptides corresponding to a number of parasite proteins, including PfSERA5, were detected (Table S1). A peptide corresponding to the PfSERA5 sequence was identified with high confidence (Fig. 1B). To identify the phosphorylation sites in PfSERA5, antibodies raised against PfSERA5 were used for immunoprecipitation from P. falciparum schizonts lysates, and the resulting immunoprecipitates were subjected to phosphopeptide analysis by LC-MS/MS. As shown in Fig. 1C, three phosphopeptides corresponding to PfSERA5 were detected with phosphorylation on Ser-183, Thr-549, and Ser-866 (the spectra for these peptides are shown in Fig. S1). Together, these results suggest that PfSERA5 exists in the phosphorylated state at the schizont stage.
Figure 1.
PfSERA5 is phosphorylated at the schizont stage in P. falciparum. A, representative immunoblot showing a band of ∼50 kDa in eluates of the affinity pulldown from P. falciparum schizont extracts using anti-phosphoserine antibody when probed with anti-PfSERA5 mouse serum. WB, Western blot. B, identification of PfSERA5 from the LC-MS/MS analysis of the above mentioned immunoprecipitates. Other proteins identified by this pulldown are summarized in Table S1. PSMs, peptide spectrum matches. C, the sites of phosphorylation in PfSERA5 as identified by LC-MS/MS analysis after immunoprecipitation using anti-PfSERA5 serum from P. falciparum schizont extracts.
PfCDPK1 associates with PfSERA5 in the parasitophorous vacuole of P. falciparum schizonts
Given that PfSERA5 is phosphorylated in late schizonts, we next attempted to identify the kinase(s) that may associate with and phosphorylate PfSERA5. LC-MS/MS analysis of immunoprecipitates from P. falciparum schizont extract using anti-PfSERA5 serum revealed several kinases associated with PfSERA5 (Table S2A). Among these kinases, PfCDPK1 is localized to the parasitophorous vacuole (PV) of P. falciparum-infected erythrocytes, similar to the localization observed for PfSERA5. Co-localization studies using immunofluorescence assays confirmed co-localization of PfSERA5 and PfCDPK1 in the PV of schizonts (Fig. 2A). Significant co-localization with a Pearson's correlation coefficient of >0.85 was observed with PfCDPK1 and PfSERA5 (Fig. 2A).
Figure 2.

PfSERA5 and PfCDPK1 co-localize in the PV of P. falciparum schizonts. A, confocal microscopy analysis of P. falciparum schizonts demonstrates that PfSERA5 and PfCDPK1 co-localize in the PV. DAPI, 4′,6-diamidino-2-phenylindole. DIC, differential interference contrast. Scale bar, 2 μm. B, Western blotting (WB) shows that PfSERA5 and PfCDPK1 are concentrated in fractions 6–8 when the protein extract of schizont-stage parasites was subjected to ultracentrifugation on a glycerol density gradient. This indicates that both of these proteins may be part of a complex within the parasite.
Sedimentation analysis of P. falciparum schizont lysates using glycerol density gradient centrifugation was performed to ascertain whether PfSERA5 and PfCDPK1 are present together in the same fraction(s). Western blotting using antibodies raised against PfSERA5 and PfCDPK1 for 16 fractions and LC-MS/MS analysis showed that both proteins co-sediment together in fractions 5 to 8 (Fig. 2B and Table S2B). Together, these results indicate a possible association of PfSERA5 and PfCDPK1.
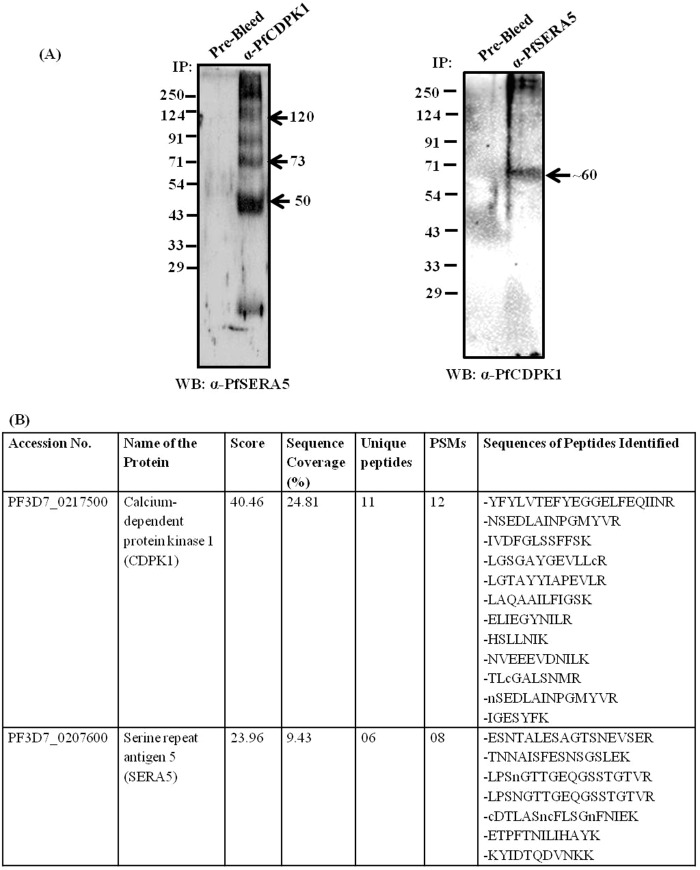
To confirm the interaction between PfSERA5 and PfCDPK1, immunoprecipitation analysis was performed with P. falciparum schizont lysates using either anti-PfSERA5 mouse serum or anti-PfCDPK1 rat serum. Western blotting of immunoprecipitates of parasite lysate generated with anti-PfSERA5 mouse serum showed the presence of PfCDPK1 (Fig. 3A). Similarly, immunoprecipitates generated with anti-PfCDPK1 rat serum from P. falciparum schizont lysates showed the presence of full-length and proteolytically processed fragments of PfSERA5, as detected by Western blotting (Fig. 3A). Analysis by MS further confirmed the presence of PfCDPK1 and PfSERA5 in the immunoprecipitates of P. falciparum lysate generated using anti-PfCDPK1 serum (Fig. 3B). The list of unique peptides of both proteins identified by LC-MS/MS analysis is summarized in Fig. 3B. A detailed list of all proteins identified in the immunoprecipitates is shown in Table S3. These results unequivocally suggest that PfCDPK1 interacts with PfSERA5 at the late schizont stage. Further, the recombinant PfCDPK1 and recombinant protease domain of PfSERA5 (PfSERA550) were found to interact with each other in an ELISA-based binding assay. We found that indeed both recombinant proteins interact with each other in vitro (Fig. S2).
Figure 3.
PfSERA5 is associated with PfCDPK1 at the schizont stage of P. falciparum. A, representative immunoblots show an interaction between PfSERA5 and PfCDPK1 in the parasite. Protein extracts of mature schizonts were independently used for immunoprecipitation (IP) using anti-PfSERA5 serum and anti-PfCDPK1 serum, and eluates were separated on SDS-PAGE and used for Western blotting (WB) with anti-CDPK1 and anti-PfSERA5 serum, respectively. B, the same eluates were trypsin-digested and analyzed by LC-MS/MS. The table shows the sequences of the unique peptides corresponding to PfCDPK1 and PfSERA5 identified by mass spectrometric analysis of the immunoprecipitate using anti-PfCDPK1 serum. A detailed list of all the proteins identified in this analysis is summarized in Table S3. PSMs, peptide spectrum matches.
PfCDPK1 phosphorylates PfSERA5 and enhances its protease activity
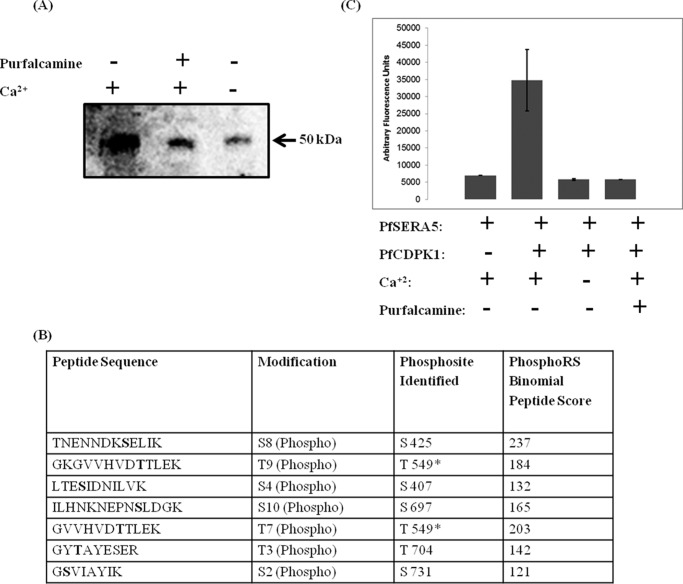
Given that PfSERA5 and PfCDPK1 co-localize in the PV, we tested whether PfCDPK1 can phosphorylate PfSERA5 in vitro. Recombinant PfCDPK1 was incubated with the recombinant protease domain of PfSERA5 (PfSERA550) in the presence of [γ-32P]ATP with or without Ca2+. Recombinant PfCDPK1 phosphorylated PfSERA550 in the presence of Ca2+ (Fig. 4A). Phosphorylation of PfSERA550 by PfCDPK1 was inhibited by purfalcamine, a known inhibitor of PfCDPK1 (Fig. 4A). Subsequently, the phosphorylation site(s) were mapped on in vitro phosphorylated PfSERA550 by MS. This analysis identified seven phosphorylated peptides corresponding to PfSERA5, each containing a single phosphorylated Ser/Thr residue (Fig. 4B). Among these phosphorylation sites, phosphorylation at Thr-549 was also identified in PfSERA5 immunoprecipitated from parasite extract using anti PfSERA5 sera, as shown in Fig. 1C.
Figure 4.
PfCDPK1 phosphorylates and enhances the enzymatic activity of recombinant PfSERA550. Recombinant PfCDPK1 phosphorylates PfSERA550 in vitro. A, an autoradiograph showing the incorporation of the radiolabeled phosphate group in recombinant PfSERA550. B, the sites of phosphorylation in PfSERA550 as identified by LC-MS/MS analysis. The phosphorylation of Thr-549 is the common site, which was also detected in LC-MS/MS analysis following immunoprecipitation with anti-PfSERA5 serum from schizont-stage protein extracts (Fig. 1C). The asterisk denotes the common phosphorylation site(s) identified in in vitro phosphorylation studies as well as in schizont-stage immunoprecipitation experiments. C, preincubation of recombinant PfCDPK1 with recombinant PfSERA550 significantly amplifies its protease activity, as measured by the hydrolysis of the fluorogenic synthetic peptide substrate LLVY-AMC. The fluorescence signal was completely abrogated by addition of inhibitors of PfCDPK1, purfalcamine or EGTA.
Next we evaluated the possible role of phosphorylation in the modulation of protease activity of PfSERA5. Recombinant PfSERA550 has been shown to cleave LLVY-AMC, a synthetic fluorescent peptide substrate (13, 22). Using LLVY-AMC as a substrate, we analyzed the activity of recombinant PfSERA550 after incubation with recombinant PfCDPK1 in the presence or absence of Ca2+. As shown in Fig. 4C, phosphorylated PfSERA550 showed a considerable increase in protease activity following incubation with PfCDPK1 in the presence of Ca2+ compared with nonphosphorylated PfSERA550. Addition of purfalcamine or EGTA inhibited the activation of PfSERA5 protease activity (Fig. 4C), suggesting that PfCDPK1 phosphorylates PfSERA5 and modulates its activity.
PfCDPK1 activity regulates PfSERA5 function in P. falciparum schizonts to trigger egress
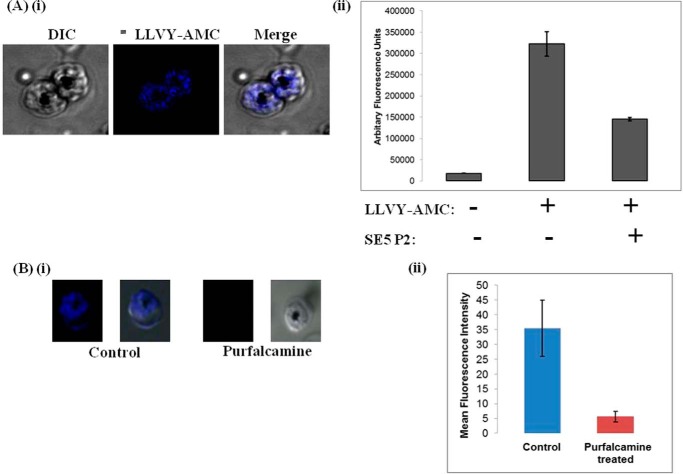
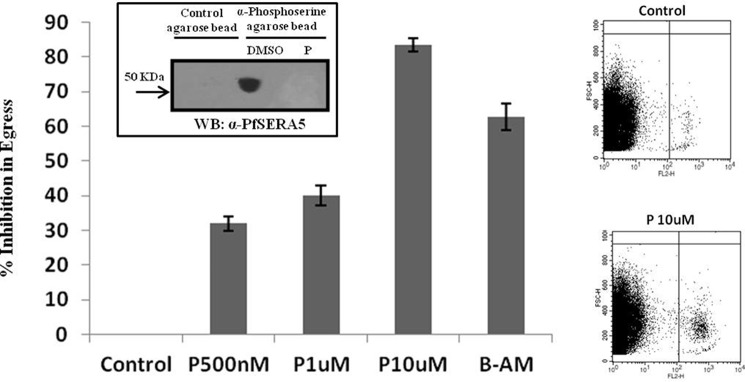
Purfalcamine, a PfCDPK1 inhibitor, or a specific PfSERA5 inhibitory peptide, SE5 P2 (22), were examined next to probe the possible role of PfCDPK1 and PfSERA5 in merozoites egress. We have shown previously that SE5 P2, a peptide from the C-terminal catalytic domain of PfSERA5, inhibits its activity (22). To analyze the activity of PfSERAs, especially PfSERA5, P. falciparum schizonts were incubated with the fluorogenic substrate LLVY-AMC. These assays were performed in the presence or absence of either purfalcamine or SE5 P2 peptide. In the past, we have successfully used similar assay to analyze the activity of cysteine protease(s) in intact parasitized erythrocytes using a fluorogenic small peptide substrate, Z-Phe-Arg-AMC (Z-FR-AMC) (31). As shown in Fig. 5A, blue fluorescence resulting from the cleavage of LLVY-AMC was detected in the control schizonts. Purfalcamine- or SE5 P2-treated parasites showed little or no fluorescence signal, indicating a block in serine protease(s)–mediated proteolytic activity (Fig. 5B). Next, the biological effect of purfalcamine on parasite egress was evaluated. P. falciparum schizonts treated with different concentrations of the PfCDPK1 inhibitor purfalcamine were assayed for egress ∼20 h after treatment using flow cytometry. A dose-dependent block in merozoite egress was observed after purfalcamine treatment (Fig. 6). A similar block was also observed after BAPTA-AM treatment (Fig. 6). To find out whether this block in merozoite egress is due to inhibition of phosphorylation of PfSERA5, we tested the phosphorylation status of PfSERA5 in purfalcamine-treated parasite extracts. As shown in Fig. 6, inset, purfalcamine significantly blocked the phosphorylation of PfSERA5. Together, these results show that inhibition of PfCDPK1 activity or PfSERA5 phosphorylation produces a block in merozoite egress, suggesting that these two proteins are critical players for merozoite egress.
Figure 5.
Purfalcamine and SE5 P2 peptide block protease activity. A, i, a synthetic fluorescent peptide substrate, LLVY-AMC, was added to P. falciparum cultures at early schizont stage, and its hydrolysis was visualized by blue fluorescence. DIC, differential interference contrast. ii, the fluorescence signal was inhibited by addition of the PfSERA5 inhibitory peptide SE5 P2 (500 μm), and this change was quantified spectrophotometrically. B, i and ii, hydrolysis of the synthetic substrate peptide was also inhibited by purfalcamine, a known inhibitor of PfCDPK1. This inhibition was captured and quantified using confocal microscopy. Together, these results indicate possible regulation of PfSERA5 activity by phosphorylation.
Figure 6.
Effect of the PfCDPK1 inhibitor purfalcamine on merozoites egress. Schizont-stage parasites (40–42 hpi) were treated with DMSO control, purfalcamine (500 nm, 1 μm, and 10 μm), or BAPTA-AM at 50 μm in duplicates and assayed for egress by flow cytometer. Data from two independent experiments (mean ± S.E.) showed a dose-dependent effect of purfalcamine on parasite egress. P, purfalcamine; B-AM, BAPTA-AM. Egress in the DMSO control was considered 100% for all experiments. The representative FACS raw data file shows loss of schizonts in the DMSO control, whereas the presence of stalled schizonts is seen in the presence of purfalcamine at 10 μm concentration. Inset, purfalcamine treatment abrogated the phosphorylation of PfSERA5 in schizont-stage parasites. Western blotting (WB) using anti-PfSERA5 serum shows a band at ∼50 kDa in DMSO-treated schizont extracts after pulldown using anti-phosphoserine–agarose, whereas no such band was visible in purfalcamine-treated parasite extracts.
Discussion
A role for the members of PfSERA family of putative proteases in merozoite egress from infected erythrocytes and sporozoite egress from oocysts has been suggested (11, 32). However, relatively weak in vitro catalytic activity and differences in the crystal structure of PfSERA5 compared with papain-like cysteine proteases has raised questions about the role of PfSERA5 as an actual protease during the egress process (13, 23). A couple of genetic studies have recently described PfSERA5 as a pseudoprotease that is essential for merozoites egress from human erythrocytes, although its functional role is unclear (23, 24). However, the role of posttranslational modifications in regulating the protease activity of SERA5 has never been addressed. It is well known that phosphorylation by protein kinases can regulate protease activities; a number of human metalloproteases, cysteine proteases, and threonine proteases have been shown to be activated/deactivated by kinases (33). To find out whether posttranslational modifications of proteases or kinases in apicomplexan parasites play a role in regulating biological processes, we investigated the phosphorylation status of PfSERA5 and its role in Plasmodium merozoite egress from infected erythrocytes.
To determine whether PfSERA5 is phosphorylated, P. falciparum schizont extracts were immunoprecipitated using anti-phosphoserine antibodies. Western blotting was performed using anti-PfSERA5 serum, which identified a specific band of PfSERA5 in immunoprecipitates. Mass spectrometry analysis of these immunoprecipitates and phosphopeptide analysis of immunoprecipitates made from P. falciparum extracts using anti-PfSERA5 sera identified phosphorylation sites within PfSERA5, confirming the phosphorylation status of PfSERA5 at the schizont stage. These results are in line with previous reports where seven phosphorylation sites were described within PfSERA5 at asexual blood stages (26, 28). We next looked for the kinase(s) that interact with and phosphorylate PfSERA5. Because most of the SERAs are localized in the PV, we focused on PfCDPK1, a kinase identified in the co-immunoprecipitates that is localized in the PV. A number of experimental strategies, including density gradient sedimentation, co-localization by immunofluorescence assays, and co-immunoprecipitation studies with P. falciparum schizont extracts unequivocally suggested an interaction between PfSERA5 and PfCDPK1. The ability of PfCDPK1 to phosphorylate PfSERA5 was tested in vitro in the presence of Ca2+. One of the phosphorylation sites on PfSERA5 following in vitro phosphorylation by PfCDPK1 was common to the phosphorylation sites found on PfSERA5 in the parasite at late schizont stage. Purfalcamine, a known PfCDPK1 inhibitor (29), blocked the phosphorylation of recombinant PfSERA5 by recombinant PfCDPK1 in vitro as well as phosphorylation of PfSERA5 in vivo in schizonts.
Having demonstrated that PfSERA5 exists in phosphorylated form at the late schizont stage and that PfCDPK1 is possibly involved in PfSERA5 phosphorylation, we next examined the significance of PfSERA5 phosphorylation and the consequences of blocking this phosphorylation using a PfCDPK1 inhibitor. Phosphorylation of PfSERA5 by PfCDPK1 in vitro greatly enhanced the proteolytic activity of PfSERA5, as measured using the fluorescent peptide substrate LLVY-AMC (Fig. 4). Purfalcamine blocked phosphorylation of PfSERA5 by PfCDPK1 and inhibited the rise in proteolytic activity of PfSERA5 (Fig. 4). We also examined these activities in vivo in the presence of purfalcamine and a PfSERA5-specific peptide that has been shown previously to inhibit PfSERA5 activity in vitro as well as in cultured parasites (22). Fluorescence measurements of parasites incubated with LLVY-AMC substrate and treated with purfalcamine or the PfSERA5 inhibitory peptide SE5 P2 showed a considerable drop in fluorescence signal from AMC release compared with untreated parasites. These results thus demonstrated that purfalcamine or SE5 P2 peptide blocked the PfSERA5 activity in vivo, confirming the need for PfSERA5 and its phosphorylation by PfCDPK1 for activation of its protease activity. We further analyzed the effect of purfalcamine on parasite development, especially in merozoite egress, as a role for PfSERAs, in particularly PfSERA5, has been suggested earlier in merozoite egress (11, 12, 19). Incubation of schizont-stage parasites with purfalcamine blocked merozite egress, demonstrating that both PfCDPK1 and PfSERA5 activities are required for merozoite egress from infected erythrocytes. Further, we found that phosphorylation of PfSERA5 was blocked in purfalcamine-treated schizont extracts (Fig. 6, inset). Overall, these results demonstrate that PfCDPK1 phosphorylates PfSERA5 following a rise in Ca2+ in mature schizonts and that this phosphorylation step activates the proteolytic activity of PfSERA5 to trigger merozoite egress. Because Collins et al. (24) have proposed a role of PfSERA5 in timely rupture of invasive merozoites, we speculate that its phosphorylation is a key regulator in the later stages of egress, which needs to be explored further. The process of egress is a complicated and multiproteolytic cascade of events; multiple phosphorylation events are expected to take place, involving multiple kinases and their substrates. A role for PfCDPK1 and PfCDPK5 in merozoite egress has been suggested earlier (29, 34, 35). PfCDPK1 has also been shown to be essential for merozoite invasion and plays a role in the phosphorylation of PfMTIP and PfGAP45, components of the glideosome complex, which provides gliding motility to the merozoite during invasion (36–38). These datasets suggest that multiple kinases and proteases participate in the processes of egress and invasion during blood-stage growth of malaria parasites.
In conclusion, the results of this study demonstrate that PfSERA5 is phosphorylated in P. falciparum schizonts by PfCDPK1 following a rise in intracellular Ca2+, which has been shown previously to trigger egress. Phosphorylation enhances the protease activity of PfSERA5. Although this is one of the first reports to show that the activity of a parasite protease can be regulated by a kinase, many human proteases have been shown to be regulated by different kinases (33). The evidence discussed above thus clearly provides data about a link between PfSERA5 protease activity and its phosphorylation status, which is essential for the timely egress of merozoites from infected erythrocytes. Understanding the kinase–protease cross-talk may lead to the introduction of combined therapies targeting specific components of both families as part of the new generation of anti-malarial therapies.
Experimental procedures
In vitro parasite culture
P. falciparum 3D7 was cultured in 4% hematocrit with RPMI 1640 with l-glutamine (Invitrogen) supplemented with 0.5% Albumax I (Invitrogen), 2.1 mg ml−1 sodium bicarbonate (Sigma-Aldrich), and 25 μg ml−1 gentamycin reagent solution (Invitrogen) according to a protocol described previously (39). Parasite cultures were maintained in a mixed-gas environment (5% O2, 5% CO2, and 90% N2) at 37 °C. Parasites were synchronized by two consecutive sorbitol treatments 4 h apart at early ring stage and allowed to mature into schizonts (44–46 h post-invasion (hpi)) for further experiments.
Cloning, expression, and purification of PfSERA5 and generation of PfSERA5 antiserum
The plasmid expressing the catalytically active region of PfSERA550 (∼50-kDa domain) was obtained from Anthony N. Hodder (Walter and Eliza Hall Institute, Australia). Protein was expressed and purified by a protocol described previously (13, 22). Polyclonal antibodies against recombinant PfSERA550 were generated in mice and used for further experiments.
Glycerol density gradient centrifugation
Late schizonts (44–46 hpi) from a P. falciparum 3D7 culture were lysed in 0.5% NP-40 in HEPES-buffered saline (10 mm HEPES, 2 mm MgCl2, 10 mm KCl, 0.5 mm EDTA, and 150 mm NaCl (pH 7.0)) containing protease and phosphatase inhibitor mixtures (Roche Diagnostics Corp.) for 15–20 min at 4 °C. Lysates were cleared by centrifugation at 13,000 rpm for 10 min, and ∼500 μl of lysate was layered on top of an 11-ml ultracentrifuge tube containing a 45–5% glycerol density gradient. Tubes were centrifuged at 38,000 rpm for 19 h at 4 °C in an SW41 rotor (Beckman Coulter Life Sciences). The molecular mass standards BSA (66 kDa), aldolase, catalase (220 kDa), ferritin (440 kDa), and thyroglobulin (660 kDa) were also prepared in the same buffer and run in parallel. Twenty fractions of 500 μl were collected from each gradient, and equal volumes of each fraction were mixed with Laemmli buffer and analyzed by Western blotting with antibodies against PfSERA5 and PfCDPK1.
Indirect immunofluorescence assay
Synchronized P. falciparum 3D7 cultures at late schizont stage (44–46 hpi) were smeared on glass slides, air-dried, and fixed in chilled absolute methanol for 30 min. Nonspecific binding sites on the cells were blocked in 3% BSA prepared in PBS. Smears were probed with anti-PfSERA5 mouse serum and anti-PfCDPK1 rat serum at dilutions of 1:50 and 1:100, respectively, for 1 h at 37 °C. The slides were then washed once with PBST (PBS + 0.02% Tween 20) and three times with PBS. Smears were probed with Alexa Fluor 594–conjugated goat anti-mouse IgG antibody (Invitrogen) and Alexa Fluor 488–conjugated donkey anti-rat IgG antibody (Invitrogen) at dilutions of 1:500 and 1:200, respectively, for 1 h at 37 °C. All antibodies were diluted in 1% BSA prepared in PBS. Slides were washed, mounted, and sealed. Nuclear DNA was visualized with 4′,6-diamidino-2-phenylindole (Invitrogen). Images were acquired using a Nikon A1R confocal microscope using the NIS Elements software.
Co-immunoprecipitation
Immunoprecipitation experiments were performed using the Pierce Cross-link Immunoprecipitation Kit (product no. 26147; Pierce Biotechnology Inc., Rockford, IL) according to the instructions provided by the manufacturer. Briefly, erythrocytes infected with synchronized P. falciparum 3D7 late schizonts (44–46 hpi) were pelleted at 1000 × g for 5 min, treated with 0.15% saponin prepared in PBS for 10 min at 4 °C, and centrifuged at 2500 × g for 20 min. The parasite pellet obtained was washed two to three times with PBS by centrifugation at 15,000 × g for 1 min. The parasites were subsequently lysed using lysis buffer (250 mm Tris, 150 mm NaCl, 1 mm EDTA, 1% NP-40, and 5% glycerol (pH 7.4)) containing protease and phosphatase inhibitor cocktails (Roche Diagnostics) for 15–20 min at 4 °C with intermittent mixing. Lysates were clarified by centrifugation at 15,000 × g for 30 min. The protein concentration of the supernatants was determined by the BCA protein estimation assay kit (Pierce Biotechnology Inc., Rockford, IL). Approximately 1 mg of total protein was incubated overnight at 4 °C with about 10 μg of anti-PfSERA5 and/or anti-PfCDPK1 serum cross-linked to 10 μl of protein A/G–Sepharose resin using disuccinimidyl suberate as a cross-linker with constant mixing. An equal amount of protein was allowed to bind to the resin conjugated to normal mouse or rat serum as a control. Following binding, resins were washed with wash buffer and bound proteins were eluted using the elution buffer (Tris-glycine (pH 2.8)). The eluates were separated on SDS-PAGE and analyzed by Western blotting using antisera against PfSERA5 and PfCDPK1.
Immunoprecipitation assay for the identification of phosphorylated PfSERA5 in schizont extracts
For the detection of native phosphorylated PfSERA5, cell lysates prepared from late schizonts (44–46 hpi) of the P. falciparum 3D7 strain (as mentioned earlier) were incubated with mouse monoclonal anti-phosphoserine antibodies coupled to agarose (product no. A8076, Sigma-Aldrich) for 12–15 h (or overnight) at 4 °C to allow binding. About 2 mg of total cellular protein was used for binding to either anti-phosphoserine agarose or control agarose beads. The bound phosphoproteins were eluted with Tris-glycine buffer (pH 2.8) and analyzed by Western blotting using anti-PfSERA5 mouse serum. The eluates were also digested with trypsin for identification of proteins by MS as described below.
Immunoprecipitation using anti-phosphoserine–agarose was also performed using schizont (44–46 hpi) lysates treated with DMSO or purfalcamine. The eluates were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membranes. The membranes were probed with anti-PfSERA5 serum.
Protein digestion and identification by LC-MS/MS
Proteins in the immunoprecipitated samples were proteolysed by in-solution trypsin digestion. Samples were brought to a final volume of 100 μl in 50 mm ammonium bicarbonate buffer to adjust the pH to 8.0, reduced with 10 mm DTT (final concentration) for 1 h at room temperature, and alkylated with 40 mm iodoacetamide (Sigma-Aldrich) for 1 h at room temperature in the dark. Proteins were digested by the addition of trypsin-Gold (Promega) at a ratio of 1:50 (w/w) of trypsin:protein. For complete digestion, samples were placed in a water bath at 37 °C for 18 h. The digestion was stopped by acidification using formic acid (0.1% final concentration). The peptide mixtures were concentrated in a SpeedVac and analyzed by LC-MS/MS.
Enrichment of phosphopeptides
For phosphorylation studies of PfSERA5 by LC-MS/MS (with both recombinant PfSERA5 and PfSERA5 from parasite lysates), the trypsin-digested peptide mixtures were first enriched for phosphopeptides before mass spectrometric analysis. Phosphopeptides from the digested peptide pool were enriched using immobilized metal affinity chromatography (IMAC) as described by Villén and Gygi (40). Briefly, vacuum-dried peptides were resuspended in 120 μl of IMAC binding buffer (40% acetonitrile and 25 mm formic acid) and allowed to bind to 10 μl of pre-equilibrated iron–nitrilotriacetic acid IMAC beads (Sigma-Aldrich). The peptides were incubated for 1 h at room temperature with constant mixing. Following the incubation, the flow-through containing nonphosphorylated peptides was collected separately. The resin was washed three times with 120 μl of IMAC binding buffer. The bound phosphopeptides were eluted three times with 40 μl of IMAC elution buffer (50 mm K2HPO4 in NH4OH (pH 10.0)). The eluates were pooled, and the pH was neutralized with 40 μl of 10% formic acid. The flow-through and eluates were vacuum-dried, desalted, and analyzed by LC-MS/MS.
Mass spectrometry analysis was performed on an Orbitrap Velos Pro MS coupled to Easy n-LC 1000 (Thermo Fisher Scientific). Tryptically digested peptide mixtures were loaded on to a reverse phase C-18 precolumn (Acclaim PepMap, 75 μm × 2 cm, 3 μm, 100 Å, Thermo Fisher Scientific) that was in line with an analytical column (Acclaim PepMap, 50 μm × 15 cm, 2 μm, 100 Å). The peptides were separated using a gradient of 5% solvent B (0.1% formic acid in 95/5 acetonitrile/water) to 50% solvent B over 60 min. For recombinant proteins, a relatively smaller gradient was used. The eluted peptides were injected into the Orbitrap Velos Pro MS, and MS1 data were acquired using a mass range from 350–2000 Da in full scan mode at 60,000 resolution. The top 20 precursors were allowed to fragment using collision-induced dissociation in an ion trap with a collision energy of 35. For phosphopeptide analysis, precursors were fragmented using high-energy collision dissociation and detected in an Orbitrap at a resolution of 7500. The raw data were analyzed using Proteome Discoverer 1.4 (Thermo Fisher Scientific) with the SEQUEST algorithm against the P. falciparum database downloaded from PlasmoDB and UniProt. The identification of peptides was validated using Percolator. The sites of phosphorylation were scored using PhosphoRS 3.0.
In vitro kinase assay
Phosphorylation assays were performed as described previously (38). Briefly, recombinant PfCDPK1 (40 nm) was allowed to phosphorylate 1 μg of recombinant PfSERA5 in kinase assay buffer (50 mm MgCl2, 50 mm Tris (pH 8.0), 20 mm sodium orthovanadate, 20 mm sodium fluoride, 1 mm DTT, and 2.5 mm CaCl2) for 1 h at 30 °C using radioactive [γ-33P]ATP as the source of the phosphate group. Syntide-2 was used as a control substrate for PfCDPK1. The reaction was stopped by adding 4× SDS-PAGE Laemmli buffer. The reaction mixture was separated on a polyacrylamide gel, and phosphate incorporation was detected using Storage Phosphor Screen (GE Healthcare). Purfalcamine, a known inhibitor of PfCDPK1 (29), was used to confirm the role of PfCDPK1 in phosphorylating PfSERA5.
Protease activity assay with recombinant PfSERA5 following phosphorylation by recombinant PfCDPK1
To evaluate the effect of PfCDPK1-mediated phosphorylation on the proteolytic activity of recombinant PfSERA5, 1 μg of PfSERA5 was incubated with recombinant PfCDPK1 (40 nm) in a kinase assay buffer in a 50-μl reaction volume as described previously (38). Here, the radioactive [γ-33P]ATP was replaced with nonradioactive ATP as the source of the phosphate group. The reaction was allowed to proceed for 1 h at 30 °C. The reaction mixture was diluted to 400 μl in PfSERA5 activity assay buffer (0.1 m NaHCO3 (pH 7.5), 5 mm CaCl2, and 0.1% Tween 20) and incubated with 20 μm 7-amino-4-methyl coumarin (LLVY-AMC) (Sigma-Aldrich) at room temperature. Released AMC was detected and measured using a LS50B PerkinElmer Life Sciences fluorimeter (excitation 355 nm, emission 460 nm) over a period of 7–8 h, with readings taken every 30 min. The values at which the fluorescence plateaued were considered for representation.
In situ protease activity assay
Erythrocytes infected with P. falciparum 3D7 schizonts (44–46 hpi) were separated from uninfected erythrocytes by centrifugation on 65% Percoll solution to achieve 85–90% purity (as described previously). Purified schizonts were added to individual wells of a 96-well plate to 1% hematocrit in a 200-μl medium volume. The culture was treated with 1 μm purfalcamine for 2 h at 37 °C. Appropriate control wells were set up simultaneously. After the treatment, the wells were loaded with a fluorogenic serine protease substrate (20 μm LLVY-AMC, Sigma-Aldrich) for 20–30 min. Cells were scraped from each well and mounted on glass slides, and AMC fluorescence was observed using the 405 filter of a Nikon A1 confocal microscope. Image analysis was performed using the ImagePro analyzer (v6.2). An alternative method of quantifying the released AMC was employed while studying the effect of SE5 P2 peptide (final concentration of 500 μm) on serine protease activity in the schizonts. Here, the release of AMC was monitored using a LS50B PerkinElmer Life Sciences fluorimeter (excitation 355 nm, emission 460 nm) after incubation of SE5 P2 with mature schizonts for 2 h at 37 °C.
Merozoite egress inhibition assay
To determine the effect of inhibition of PfSERA5 phosphorylation by inhibiting PfCDPK1 on P. falciparum merozoite egress, egress inhibition assays were performed as described previously (41). Briefly, late-stage schizonts (42–44 hpi) were treated with different concentrations of purfalcamine (0.5–10 μm), 50 μm BAPTA-AM, and appropriate solvent controls for 8–10 h in duplicate wells. Following incubation, parasites from treated and control wells were collected, washed, and stained with ethidium bromide (10 mg/ml) and ethidium bromide–positive cells were scored by flow cytometry on a FACSCalibur (BD Biosciences) using CellQuest software. The fluorescence signal (FL-2) was detected with a 90-nm band pass filter using an excitation laser of 488 nm collecting typically 100,000 cells per sample. Post-acquisition data were analyzed using FlowJo software (Tree Star, Inc.) by determining the proportion of FL-2–positive cells representing schizonts. Inhibition of egress of merozoites was calculated as follows: % inhibition of merozoite egress = 100 ((T-C)/(I-C)), where I is the frequency of schizonts (percent) at the start of the assay, T is the frequency of schizonts (percent) after treatment, and C is the frequency of schizonts (percent) in the control.
Sequences of SE5 P2 and P3 peptides
The sequences of SE5 P2 and P3 peptides were as follows: SE5 P2, Ac-KASPEFYHNLYFKNF-NH2; SE5 P3, Ac-NVGKKNLFSEKEDN-NH2.
Author contributions
G. R. I., I. K., C. E. C., and P. M. conceptualization; G. R. I., A. B., G. K., I. K., and P. M. resources; G. R. I., S. S., I. K., S. A., M. A. S., A. B., E. S., and A. M. data curation; G. R. I., S. S., I. K., S. A., C. E. C., and P. M. formal analysis; C. E. C., I. K., and P. M. supervision; C. E. C. and P. M. funding acquisition; G. R. I., S. S., I. K., S. A., M. A. S., A. B., E. S., G. P., and A. M. validation; G. R. I., S. S., I. K., S. A., M. A. S., E. S., and A. M. investigation; G. R. I., I. K., S. A., and P. M. visualization; G. R. I., S. S., I. K., S. A., M. A. S., A. B., E. S., G. P., and A. M. methodology; G. R. I., I. K., S. A., C. E. C., and P. M. writing-original draft; G. R. I., I. K., C. E. C., and P. M. project administration; P. M., G. R. I., I. K., S. A., E. S., A. M., and C. E. C. writing-review and editing.
Supplementary Material
Acknowledgments
We thank Dr. Brendan Crabb and Dr. Anthony N. Hodder for kindly providing the PfSERA5P50 expression clone. We also thank the Rotary Blood Bank for providing human red blood cells and Surbhi Dabral for help with confocal microscopy.
This work was supported by the Department of Biotechnology, Government of India (BT/01/CEIB/11/V/01). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3 and Tables S1–S3.
- SERA
- serine repeat antigen
- Pf
- Plasmodium falciparum
- PTM
- posttranslational modification
- PV
- parasitophorous vacuole
- hpi
- hour(s) post-invasion
- IMAC
- immobilized metal affinity chromatography
- LLVY-AMC
- N-succinyl-Leu-Leu-Val-Tyr-AMC
- AMC
- 7-amino-4-methylcoumarin
- Z
- benzyloxycarbonyl
- BAPTA-AM
- 1,2-bis(2-aminophenoxy) ethane-N,N,N′,N′-tetraacetic acid tetrakis (acetoxymethyl ester).
References
- 1. Mbengue A., Bhattacharjee S., Pandharkar T., Liu H., Estiu G., Stahelin R. V., Rizk S. S., Njimoh D. L., Ryan Y., Chotivanich K., Nguon C., Ghorbal M., Lopez-Rubio J. J., Pfrender M., Emrich S., et al. (2015) A molecular mechanism of artemisinin resistance in Plasmodium falciparum malaria. Nature 520, 683 10.1038/nature14412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ashley E. A., Dhorda M., Fairhurst R. M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J. M., Mao S., Sam B., Sopha C., Chuor C. M., Nguon C., Sovannaroth S., Pukrittayakamee S., et al. (2014) Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 371, 411–423 10.1056/NEJMoa1314981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hadley T., Aikawa M., and Miller L. H. (1983) Plasmodium knowlesi: studies on invasion of rhesus erythrocytes by merozoites in the presence of protease inhibitors. Exp. Parasitol. 55, 306–311 10.1016/0014-4894(83)90027-9 [DOI] [PubMed] [Google Scholar]
- 4. Lyon J. A., and Haynes J. D. (1986) Plasmodium falciparum antigens synthesized by schizonts and stabilized at the merozoite surface when schizonts mature in the presence of protease inhibitors. J. Immunol. 136, 2245–2251 [PubMed] [Google Scholar]
- 5. Soni S., Dhawan S., Rosen K. M., Chafel M., Chishti A. H., and Hanspal M. (2005) Characterization of events preceding the release of malaria parasite from the host red blood cell. Blood Cells Mol. Dis. 35, 201–211 10.1016/j.bcmd.2005.05.006 [DOI] [PubMed] [Google Scholar]
- 6. Salmon B. L., Oksman A., and Goldberg D. E. (2001) Malaria parasite exit from the host erythrocyte: a two-step process requiring extraerythrocytic proteolysis. Proc. Natl. Acad. Sci. 98, 271–276 10.1073/pnas.98.1.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wickham M. E., Culvenor J. G., and Cowman A. F. (2003) Selective inhibition of a two-step egress of malaria parasites from the host erythrocyte. J. Biol. Chem. 278, 37658–37663 10.1074/jbc.M305252200 [DOI] [PubMed] [Google Scholar]
- 8. Le Bonniec S., Deregnaucourt C., Redeker V., Banerjee R., Grellier P., Goldberg D. E., and Schrével J. (1999) Plasmepsin ii, an acidic hemoglobinase from the Plasmodium falciparum food vacuole, is active at neutral pH on the host erythrocyte membrane skeleton. J. Biol. Chem. 274, 14218–14223 10.1074/jbc.274.20.14218 [DOI] [PubMed] [Google Scholar]
- 9. Hanspal M., Dua M., Takakuwa Y., Chishti A. H., and Mizuno A. (2002) Plasmodium falciparum cysteine protease falcipain-2 cleaves erythrocyte membrane skeletal proteins at late stages of parasite development: presented in part in abstract form at the 43rd annual meeting of the American Society of Hematology, Orlando, FL, 2001. Blood 100, 1048–1054 10.1182/blood-2002-01-0101 [DOI] [PubMed] [Google Scholar]
- 10. Dua M., Raphael P., Sijwali P. S., Rosenthal P. J., and Hanspal M. (2001) Recombinant falcipain-2 cleaves erythrocyte membrane ankyrin and protein 4.1. Mol. Biochem. Parasitol. 116, 95–99 10.1016/S0166-6851(01)00306-1 [DOI] [PubMed] [Google Scholar]
- 11. Yeoh S., O'Donnell R. A., Koussis K., Dluzewski A. R., Ansell K. H., Osborne S. A., Hackett F., Withers-Martinez C., Mitchell G. H., Bannister L. H., Bryans J. S., Kettleborough C. A., and Blackman M. J. (2007) Subcellular discharge of a serine protease mediates release of invasive malaria parasites from host erythrocytes. Cell 131, 1072–1083 10.1016/j.cell.2007.10.049 [DOI] [PubMed] [Google Scholar]
- 12. Arastu-Kapur S., Ponder E. L., Fonović U. P., Yeoh S., Yuan F., Fonović M., Grainger M., Phillips C. I., Powers J. C., and Bogyo M. (2008) Identification of proteases that regulate erythrocyte rupture by the malaria parasite Plasmodium falciparum. Nat. Chem. Biol. 4, 203–213 10.1038/nchembio.70 [DOI] [PubMed] [Google Scholar]
- 13. Hodder A. N., Malby R. L., Clarke O. B., Fairlie W. D., Colman P. M., Crabb B. S., and Smith B. J. (2009) Structural insights into the protease-like antigen Plasmodium falciparum SERA5 and its noncanonical active-site serine. J. Mol. Biol. 392, 154–165 10.1016/j.jmb.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 14. Gardner M. J., Tettelin H., Carucci D. J., Cummings L. M., Aravind L., Koonin E. V., Shallom S., Mason T., Yu K., Fujii C., Pederson J., Shen K., Jing J., Aston C., Lai Z., et al. (1998) Chromosome 2 sequence of the human malaria parasite Plasmodium falciparum. Science 282, 1126–1132 10.1126/science.282.5391.1126 [DOI] [PubMed] [Google Scholar]
- 15. Bzik D. J., Li W. B., Horii T., and Inselburg J. (1988) Amino acid sequence of the serine-repeat antigen (sera) of Plasmodium falciparum determined from cloned cDNA. Mol. Biochem. Parasitol. 30, 279–288 10.1016/0166-6851(88)90097-7 [DOI] [PubMed] [Google Scholar]
- 16. Kiefer M. C., Crawford K. A., Boley L. J., Landsberg K. E., Gibson H. L., Kaslow D. C., and Barr P. J. (1996) Identification and cloning of a locus of serine repeat antigen (sera)-related genes from Plasmodium vivax. Mol. Biochem. Parasitol. 78, 55–65 10.1016/S0166-6851(96)02607-2 [DOI] [PubMed] [Google Scholar]
- 17. Pils B., and Schultz J. (2004) Inactive enzyme-homologues find new function in regulatory processes. J. Mol. Biol. 340, 399–404 10.1016/j.jmb.2004.04.063 [DOI] [PubMed] [Google Scholar]
- 18. Putrianti E. D., Schmidt-Christensen A., Arnold I., Heussler V. T., Matuschewski K., and Silvie O. (2010) The Plasmodium serine-type sera proteases display distinct expression patterns and non-essential in vivo roles during life cycle progression of the malaria parasite. Cell. Microbiol. 12, 725–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McCoubrie J. E., Miller S. K., Sargeant T., Good R. T., Hodder A. N., Speed T. P., de Koning-Ward T. F., and Crabb B. S. (2007) Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect. Immun. 75, 5565–5574 10.1128/IAI.00405-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Aoki S., Li J., Itagaki S., Okech B. A., Egwang T. G., Matsuoka H., Palacpac N. M., Mitamura T., and Horii T. (2002) Serine repeat antigen (sera5) is predominantly expressed among the sera multigene family of plasmodium falciparum, and the acquired antibody titers correlate with serum inhibition of the parasite growth. J. Biol. Chem. 277, 47533–47540 10.1074/jbc.M207145200 [DOI] [PubMed] [Google Scholar]
- 21. Palacpac N. M., Arisue N., Tougan T., Ishii K. J., and Horii T. (2011) Plasmodium falciparum serine repeat antigen 5 (se36) as a malaria vaccine candidate. Vaccine 29, 5837–5845 10.1016/j.vaccine.2011.06.052 [DOI] [PubMed] [Google Scholar]
- 22. Kanodia S., Kumar G., Rizzi L., Pedretti A., Hodder A. N., Romeo S., and Malhotra P. (2014) Synthetic peptides derived from the C-terminal 6 kDa region of Plasmodium falciparum sera5 inhibit the enzyme activity and malaria parasite development. Biochim. Biophys. Acta 1840, 2765–2775 10.1016/j.bbagen.2014.04.013 [DOI] [PubMed] [Google Scholar]
- 23. Stallmach R., Kavishwar M., Withers-Martinez C., Hackett F., Collins C. R., Howell S. A., Yeoh S., Knuepfer E., Atid A. J., Holder A. A., and Blackman M. J. (2015) Plasmodium falciparum sera5 plays a non-enzymatic role in the malarial asexual blood-stage lifecycle. Mol. Microbiol. 96, 368–387 10.1111/mmi.12941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Collins C. R., Hackett F., Atid J., Tan M. S. Y., and Blackman M. J. (2017) The Plasmodium falciparum pseudoprotease sera5 regulates the kinetics and efficiency of malaria parasite egress from host erythrocytes. PLoS Pathog. 13, e1006453 10.1371/journal.ppat.1006453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaur I., Zeeshan M., Saini E., Kaushik A., Mohmmed A., Gupta D., and Malhotra P. (2016) Widespread occurrence of lysine methylation in Plasmodium falciparum proteins at asexual blood stages. Sci. Rep. 6, 35432 10.1038/srep35432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lasonder E., Green J. L., Camarda G., Talabani H., Holder A. A., Langsley G., and Alano P. (2012) The Plasmodium falciparum schizont phosphoproteome reveals extensive phosphatidylinositol and cAMP-protein kinase a signaling. J. Proteome Res. 11, 5323–5337 10.1021/pr300557m [DOI] [PubMed] [Google Scholar]
- 27. Solyakov L., Halbert J., Alam M. M., Semblat J.-P., Dorin-Semblat D., Reininger L., Bottrill A. R., Mistry S., Abdi A., Fennell C., Holland Z., Demarta C., Bouza Y., Sicard A., Nivez M. P., et al. (2011) Global kinomic and phospho-proteomic analyses of the human malaria parasite Plasmodium falciparum. Nat. Commun. 2, 565 10.1038/ncomms1558 [DOI] [PubMed] [Google Scholar]
- 28. Treeck M., Sanders J. L., Elias J. E., and Boothroyd J. C. (2011) The phosphoproteomes of Plasmodium falciparum and Toxoplasma gondii reveal unusual adaptations within and beyond the parasites' boundaries. Cell Host Microbe 10, 410–419 10.1016/j.chom.2011.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kato N., Sakata T., Breton G., Le Roch K. G., Nagle A., Andersen C., Bursulaya B., Henson K., Johnson J., Kumar K. A., Marr F., Mason D., McNamara C., Plouffe D., Ramachandran V., et al. (2008) Gene expression signatures and small-molecule compounds link a protein kinase to Plasmodium falciparum motility. Nat. Chem. Biol. 4, 347–356 10.1038/nchembio.87 [DOI] [PubMed] [Google Scholar]
- 30. Stephens B. J., Han H., Gokhale V., and Von Hoff D. D. (2005) Prl phosphatases as potential molecular targets in cancer. Mol. Cancer Ther. 4, 1653–1661 10.1158/1535-7163.MCT-05-0248 [DOI] [PubMed] [Google Scholar]
- 31. Korde R., Bhardwaj A., Singh R., Srivastava A., Chauhan V. S., Bhatnagar R. K., and Malhotra P. (2008) A prodomain peptide of Plasmodium falciparum cysteine protease (falcipain-2) inhibits malaria parasite development. J. Med. Chem. 51, 3116–3123 10.1021/jm070735f [DOI] [PubMed] [Google Scholar]
- 32. Aly A. S., and Matuschewski K. (2005) A malarial cysteine protease is necessary for Plasmodium sporozoite egress from oocysts. J. Exp. Med. 202, 225–230 10.1084/jem.20050545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. López-Otín C., and Hunter T. (2010) The regulatory crosstalk between kinases and proteases in cancer. Nat. Rev. Cancer 10, 278–292 10.1038/nrc2823 [DOI] [PubMed] [Google Scholar]
- 34. Azevedo M. F., Sanders P. R., Krejany E., Nie C. Q., Fu P., Bach L. A., Wunderlich G., Crabb B. S., and Gilson P. R. (2013) Inhibition of Plasmodium falciparum cdpk1 by conditional expression of its j-domain demonstrates a key role in schizont development. Biochem. J. 452, 433–441 10.1042/BJ20130124 [DOI] [PubMed] [Google Scholar]
- 35. Dvorin J. D., Martyn D. C., Patel S. D., Grimley J. S., Collins C. R., Hopp C. S., Bright A. T., Westenberger S., Winzeler E., Blackman M. J., Baker D. A., Wandless T. J., and Duraisingh M. T. (2010) A plant-like kinase in Plasmodium falciparum regulates parasite egress from erythrocytes. Science 328, 910–912 10.1126/science.1188191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ridzuan M. A., Moon R. W., Knuepfer E., Black S., Holder A. A., and Green J. L. (2012) Subcellular location, phosphorylation and assembly into the motor complex of gap45 during Plasmodium falciparum schizont development. PLoS ONE 7, e33845 10.1371/journal.pone.0033845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Green J. L., Rees-Channer R. R., Howell S. A., Martin S. R., Knuepfer E., Taylor H. M., Grainger M., and Holder A. A. (2008) The motor complex of Plasmodium falciparum phosphorylation by a calcium-dependent protein kinase. J. Biol. Chem. 283, 30980–30989 10.1074/jbc.M803129200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bansal A., Singh S., More K. R., Hans D., Nangalia K., Yogavel M., Sharma A., and Chitnis C. E. (2013) Characterization of Plasmodium falciparum calcium-dependent protein kinase 1 (pfcdpk1) and its role in microneme secretion during erythrocyte invasion. J. Biol. Chem. 288, 1590–1602 10.1074/jbc.M112.411934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Trager W., and Jensen J. B. (1976) Human malaria parasites in continuous culture. Science 193, 673–675 10.1126/science.781840 [DOI] [PubMed] [Google Scholar]
- 40. Villén J., and Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 10.1038/nprot.2008.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Agarwal S., Singh M. K., Garg S., Chitnis C. E., and Singh S. (2013) Ca2+-mediated exocytosis of subtilisin-like protease 1: a key step in egress of Plasmodium falciparum merozoites. Cell. Microbiol. 15, 910–921 10.1111/cmi.12086 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.