Abstract
Lipids display large structural complexity, with ∼40,000 different lipids identified to date, ∼4000 of which are sphingolipids. A critical factor determining the biological activities of the sphingolipid, ceramide, and of more complex sphingolipids is their N-acyl chain length, which in mammals is determined by a family of six ceramide synthases (CerS). Little information is available about the CerS regions that determine specificity toward different acyl-CoA substrates. We previously demonstrated that substrate specificity resides in a region of ∼150 residues in the Tram-Lag-CLN8 domain. Using site-directed mutagenesis and biochemical analyses, we now narrow specificity down to an 11-residue sequence in a loop located between the last two putative transmembrane domains (TMDs) of the CerS. The specificity of a chimeric protein, CerS5(299–309→CerS2), based on the backbone of CerS5 (which generates C16-ceramide), but containing 11 residues from CerS2 (which generates C22–C24-ceramides), was altered such that it generated C22–C24 and other ceramides. Moreover, a chimeric protein, CerS4(291–301→CerS2), based on CerS4 (which normally generates C18–C22 ceramides) displayed significant activity toward C24:1-CoA. Additional data supported the notion that substitutions of these 11 residues alter the specificities of the CerS toward their cognate acyl-CoAs. Our findings may suggest that this short loop may restrict adjacent TMDs, leading to a more open conformation in the membrane, and that the CerS acting on shorter acyl-CoAs may have a longer, more flexible loop, permitting TMD flexibility. In summary, we have identified an 11-residue region that determines the acyl-CoA specificity of CerS.
Keywords: ceramide, ceramide synthase, sphingolipid, membrane, lipid
Introduction
Recent advances in analytical techniques, such as MS, have revealed an unexpected complexity in lipid structures (1), with ∼40,000 lipids identified to date (http://www.lipidmaps.org).5 Of these, ∼4000 individual sphingolipid (SL)6 structures have been identified (2). SLs differ in the headgroups added at the C1 position, the length and degree of saturation of the sphingoid long-chain base, and of the acyl chain that is attached to the sphingoid backbone via an amide bond at C2 (3, 4).
The acyl chain composition of ceramide, the simplest SL, is determined by the activity of a family of ceramide synthases (CerS), of which six are found in mammals (5, 6), with each generating ceramides of a different acyl chain length (7). Since the identification of the first mammalian CerS in 2002 (8), some effort has been invested in attempting to determine the basis for the acyl chain specificity of the mammalian CerS; thus, it is known that the amino acids involved in acyl chain specificity are located in the Tram-Lag-CLN8 (TLC) domain (9), a region of ∼200 residues also found in other proteins (10). The active site was suggested to reside within a region known as the Lag1p motif, containing two conserved histidine residues (11, 12), which is within the TLC domain (13, 14). Other regions of the CerS include the homeobox (Hox)-like domain (14) that is found in CerS2–S6 (6) and contains residues that are critical for activity (15), but they do not appear to play a role in determining acyl-CoA specificity (9).
CerS are located in the endoplasmic reticulum (ER) membrane (6), although the exact number of transmembrane domains (TMDs) and their topology have not been resolved experimentally (16). Although most prediction programs give similar suggestions for the topology of the N and C termini, there is a significant disagreement about almost every other feature of CerS topology (3). That being said, recent experimental data support an odd number of TMDs (17).
As stated above, we previously determined that the domain responsible for acyl chain specificity resides in a minimal region of 150 residues in the TLC domain (9). This was shown by generation of chimeric proteins consisting of portions of CerS2 (which generates C22:0–C24:1-ceramides) and CerS5 (which generates C16:0-ceramide) and consequently examining which ceramide species were generated by the chimeric proteins. We now provide data in which we considerably narrow down the region involved in determining acyl chain specificity, and we identify a region of 11 residues that appears to be critically involved in determining the acyl chain specificity, not only of CerS5 and CerS2 but also of CerS4.
Results
N terminus of CerS faces the lumen of the ER
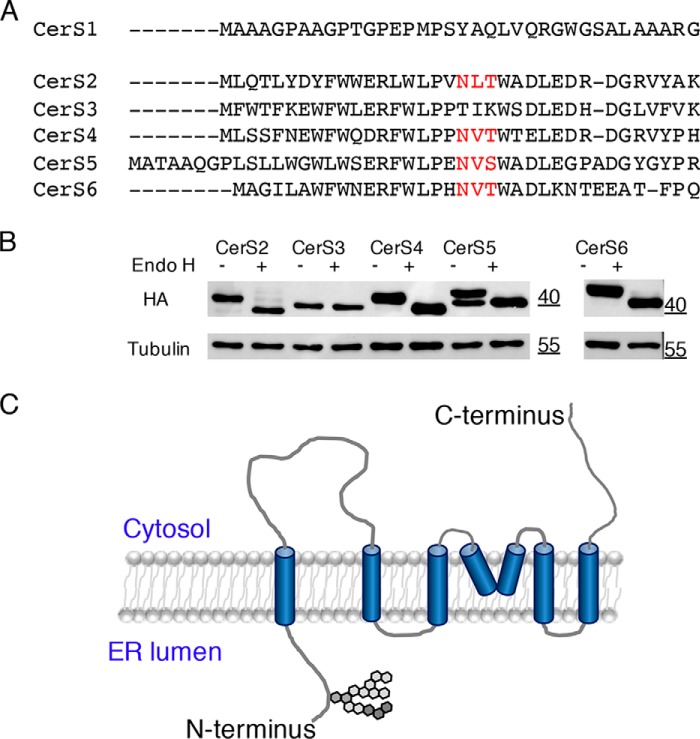
To further analyze CerS topology in the ER (16–18), we took advantage of a conserved N-glycosylation motif (NX(T/S)) found in CerS2 and CerS4–CerS6. This sequence is not found in CerS3, and CerS1 does not align with the other CerS at the N terminus (Fig. 1A). Consequently, upon treatment of CerS2–CerS6 with endoH, a decrease in molecular weight consistent with deglycosylation was observed for all CerS except CerS3 (Fig. 1B). Interestingly, CerS5 activity increased upon endoH treatment (1189 ± 63 pmol/min/mg before endoH treatment and 1768 ± 204 pmol/min/mg after endoH treatment; n = 3). Mutation of the key asparagine residue within the N-glycosylation motif in CerS5 (CerS5(N26Q)) resulted in a similar reduction in molecular weight (data not shown). However, a small decrease in CerS5 activity (1887 ± 325 pmol/min/mg for CerS5 and 1353 ± 327 pmol/min/mg for CerS5(N26Q)) was observed, perhaps suggesting an important role for the N-glycan in protein folding. Irrespective of the effect of deglycosylation on CerS activity, these data unambiguously demonstrate that the N terminus of CerS faces the lumen of the ER (Fig. 1C), where the first step in N-glycosylation takes place (20), as suggested in some but not all previous topological predictions (9, 17, 18). This is of relevance to the studies below where specific sequences and motifs are modified, based to some extent on the putative topology of the CerS.
Figure 1.
N terminus of CerS is in the ER lumen. A, multiple alignments of human CerS1–CerS6. The N-glycosylation consensus sequence in CerS2 and CerS4–6 is in red. B, cell lysates (2 mg of protein/ml) from HEK cells overexpressing the indicated CerS-HA were incubated with or without endoH prior to Western blotting using an anti-HA antibody. Molecular weight markers are indicated. Tubulin was used as a loading control. This experiment was repeated three times with similar results. C, revised putative topology of the CerS. Unlike previous models (9), we now suggest that the N terminus of the CerS faces the ER lumen and that the fourth TMD does not fully penetrate the ER membrane.
Acyl-CoA specificity is associated with one of two frameshifts in CerS2 and CerS5 sequences
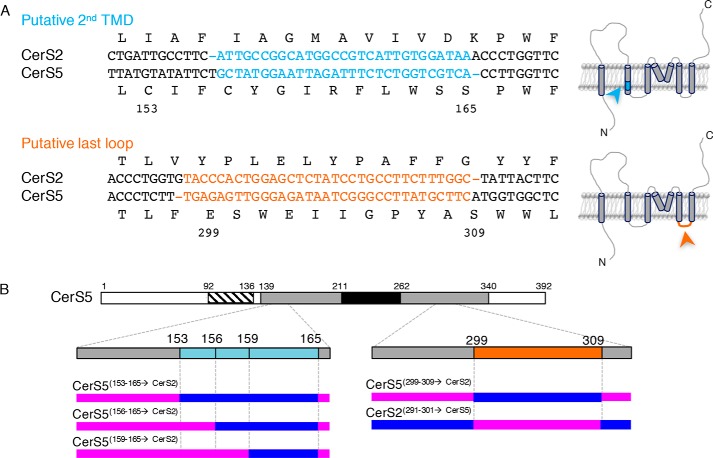
Pairwise alignment of human CerS2 and CerS5 revealed two frameshifts in regions of low sequence similarity. The first is in the second putative TMD, between residues 156 and 165 of CerS5, and the other frameshift is in the putative last loop between residues 299 and 309 of CerS5 (Fig. 2A). CerS2 and CerS5 belong to two different branches of the CerS phylogenetic tree (9), which can essentially be defined based on these frameshifts.
Figure 2.
Alignment of CerS2 and CerS5. A, multiple alignments of human CerS2 and CerS5 reveal two frameshifts between the sequences in the putative second TMD and in the last lumenal loop. Amino acid residues are shown with the residue numbers corresponding to CerS5. The right-hand panel indicates the location of each frameshift based on the putative topology designated in Fig. 1, and color-coded accordingly. B, schematic representation of mammalian CerS5 and its three main domains: Hox-like domain (stripes); TLC domain (gray); Lag1p motif (black). The five chimeras generated based on the frameshifts are indicated. Regions derived from CerS5 are in pink and from CerS2 in blue.
Two sets of chimeric proteins were generated (Fig. 2B) to determine whether the frameshifts play a role in defining the acyl-CoA specificities of Cer2 and CerS5. Thus, in CerS5(153–165→CerS2), all of the sequence is derived from CerS5 with the exception of residues 153–165, which are from CerS2. To analyze acyl chain specificity, each chimeric protein was assayed under optimal kinetic conditions for the parent CerS using the relevant acyl-CoA. Thus, CerS2 activity was assayed using very-long–chain acyl-CoAs and a reaction time of 20 min with 40 μg of protein, whereas CerS5 activity was assayed with C16-acyl-CoA using 1 μg of protein for 5 min (21, 22).
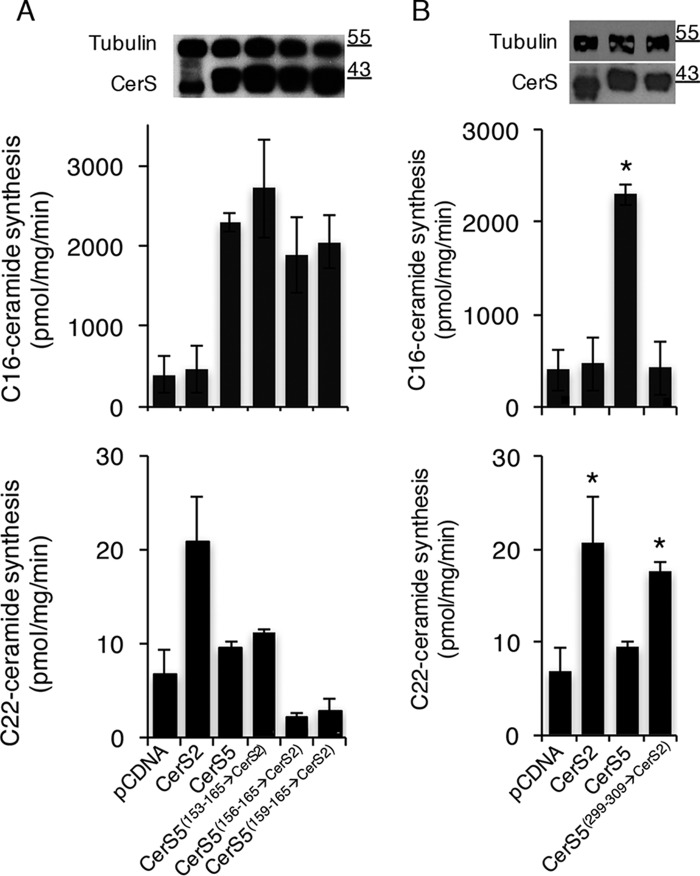
We first examined the role of the second putative TMD domain (Fig. 2A), because CerS5 contains more polar and bulky residues than CerS2 in this region. Three constructs were prepared, where the corresponding residues of CerS5 were replaced with 13, 10, or 6 residues of CerS2 (Fig. 2B). However, CerS5(153–165→CerS2), CerS5(156–165→CerS2), and CerS5(159–165→CerS2) all displayed the same specificity toward C16-CoA as CerS5, and no activity was detected using C22-CoA (Fig. 3A).
Figure 3.
Role of the last putative loop of CerS5 in acyl chain specificity. Homogenates (40 μg of protein for assays using C22-CoA (A) and 1 μg for assays using C16-CoA (B)) were prepared from cells overexpressing chimeric proteins. CerS5 activity was assayed using C16-CoA for 5 min (upper panel), and CerS2 activity was assayed using C22-CoA for 20 min (lower panel). Results are means ± S.D. for a typical experiment repeated 2–4 times in duplicate with similar results. *, p < 0.05. Levels of protein expression, ascertained by Western blotting using an anti-HA antibody and anti-tubulin as a loading control, are shown. Molecular weight markers are indicated.
We then examined the role of the putative last loop of CerS5 (Fig. 2A). Intriguingly, CerS5(299–309→CerS2) was able to utilize very-long–chain acyl-CoAs, generating C22:0-ceramide (Fig. 3B) and C24:1-ceramide (data not shown) under optimal conditions for CerS2 but demonstrated no activity when using C16-CoA (Fig. 3B) under optimal conditions for CerS5. These data indicate that these 11 residues might be involved in determining acyl-CoA specificity. The corresponding chimeric protein, based on CerS2, i.e. CerS2(291–301→CerS5), displayed no activity with either C16-CoA or C24:1-CoA (data not shown).
Critical role for the putative last loop of CerS in determining acyl-CoA specificity
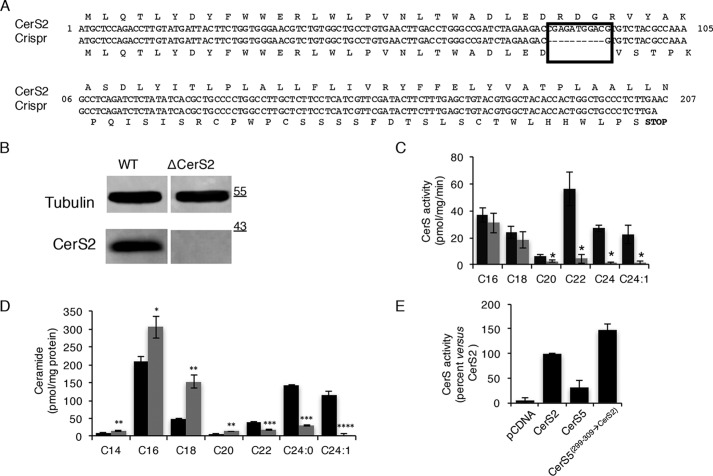
We generated CerS2−/− HEK293T cells using CRISPR/Cas9 technology. The main motivation for doing this was to refute the possibility that apparent changes in acyl-CoA specificity could be due to more efficient dimerization (17) of CerS5(299–309→CerS2) with endogenous CerS2, leading to an increase in CerS2 activity rather than to altered acyl-CoA specificity of CerS5(299–309→CerS2). A HEK293T clone, which contained a deletion of 10 nucleotides in both alleles of the genomic DNA sequence, was obtained, resulting in a premature stop codon after 63 residues (Fig. 4A). As a consequence, no CerS2 protein was generated (Fig. 4B), and no very-long acyl chain ceramides were synthesized (Fig. 4C) or detected in the cells (Fig. 4D). The SL profile of the CerS2−/− HEK293T cells was similar to that of the CerS2 null mouse (23, 24), with decreased levels of very-long acyl chain ceramides and increased levels of long acyl chain ceramides (Fig. 4D). Overexpression of CerS5(299–309→CerS2) in HEK293T/CerS2−/− cells resulted in a similar increase of very-long acyl chain ceramide synthesis (Fig. 4E) as with overexpression in WT HEK cells, excluding the possibility that the activity of CerS5(299–309→CerS2) toward very-long acyl chain CoAs (Fig. 3B) was related to heterodimerization of CerS5(299–309→CerS2) with endogenous CerS2.
Figure 4.
Generation and characterization of HEK/CerS2−/− cells. A, alignment of CerS2 genomic DNA and protein sequence in WT HEK cells (top) and HEK/CerS2−/− cells (bottom). Rectangle marks amino acids that were deleted in HEK/CerS2−/− cells. B, CerS2 expression ascertained by Western blotting using an anti-CerS2 antibody. An anti-tubulin antibody was used as loading control. Molecular weight markers are indicated. C, homogenates (30 μg of protein) from WT HEK cells (black) or HEK/CerS2−/− cells (gray) were assayed using the indicated acyl-CoA for 30 min. Results are means ± S.D. n = 4. D, fatty acid composition of ceramides in wild type (WT) HEK cells (black) and HEK/CerS2−/− cells (gray). Results are means ± S.D. n = 4. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001. E, homogenates (40 μg of protein) were prepared from HEK/CerS2−/−cells overexpressing CerS5(299–309→CerS2), CerS2, or CerS5 and assayed using C24:1-CoA for 20 min. Results are means ± S.D. n = 4.
We next characterized the activity of CerS5(299–309→CerS2) toward various CoAs. An obstacle in using an in vitro assay for a chimeric protein composed of different regions of different CerS is that each of the parent CerS exhibits different kinetic parameters with respect to reaction time and amount of protein (21, 22). To attempt to overcome this complication, we assayed CerS5(299–309→CerS2) activity with increasing amounts of protein while utilizing different acyl-CoAs (Fig. 5A). CerS5(299–309→CerS2) generated C22:0-ceramide and C16:0-ceramide to a similar extent7 but also generated C18:0-, C20:0-, and C24:1-ceramides (Fig. 5A), suggesting that CerS5(299–309→CerS2) has lost its specificity toward C16-CoA and shows specificity toward all of the CoAs tested. Importantly, upon increasing amounts of C22-CoA, CerS5(299–309→CerS2) generated more C22-ceramide than CerS2 itself (Fig. 5B), indicating that under assay conditions optimized for CerS2, CerS5(299–309→CerS2) generates bona fide amounts of very-long–chain ceramides. However, the SL profile of HEK cells upon overexpression of CerS5(299–309→CerS2) did not change significantly compared with controls (Fig. 5C) with the exception of a small increase in C14- and C18-ceramide; however, levels of C16-ceramide, which is generated by CerS5, were unaltered (25).
Figure 5.
Acyl chain specificity of CerS5(299–309→CerS2). A, homogenates were prepared from WT HEK cells overexpressing CerS5(299–309→CerS2), and activity was assayed using the indicated acyl-CoAs with increasing amounts of protein for 20 min. Results are means ± S.E. of two individual experiments performed in duplicate. B, homogenates (40 μg of protein) were prepared from WT HEK cells overexpressing CerS2 (triangles), CerS5 (circles), or CerS5(299–309→CerS2) (squares), and activity was assayed using C22-CoA for 30 min. Results are from a typical experiment in which CerS activity was assayed in duplicate and repeated four times. Levels of expression of all CerS constructs were similar as determined by Western blotting. C, fatty acid composition of ceramide was determined in WT HEK cells overexpressing pcDNA (black) or CerS5(299–309→CerS2) (gray). n = 6. *, p < 0.05; **, p < 0.01.
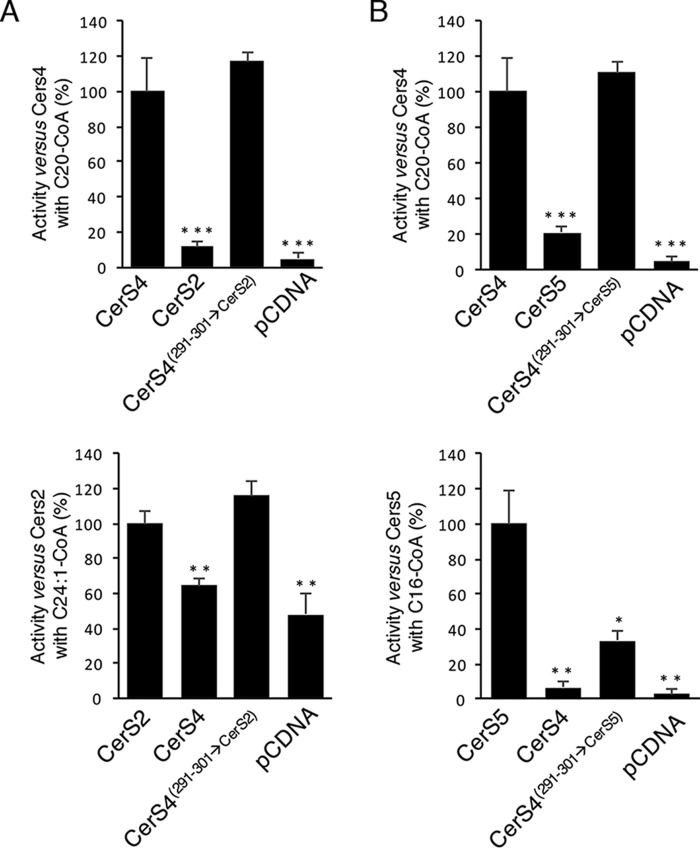
To determine whether the putative last loop determines the acyl-CoA specificity of other CerS, we generated two additional chimeras in which the corresponding 11 residues in CerS4 were altered to the corresponding sequence in CerS2 or CerS5. Similar to CerS5(299–309→CerS2), CerS4(291–301→CerS2) displayed significant activity using C24:1-CoA (Fig. 6A) and gained specificity toward other acyl-CoAs (data not shown). CerS4(291–301→CerS5) generated C16-ceramide to a higher extent than CerS4 but significantly less than the WT CerS5 (Fig. 6B). Both CerS4(291–301→CerS2) and CerS4(291–301→CerS5) displayed activity using C20-CoA (Fig. 6, A and B).
Figure 6.
Acyl chain specificity of CerS4(291–301→CerS2) and CerS4(291–301→CerS5). Homogenates were prepared from cells overexpressing the indicated constructs, and CerS activity was measured using 20 μg of protein for 20 min. A, CerS4(291–301→CerS2) activity was examined with C20-CoA (upper panel) and C24:1-CoA (lower panel). B, CerS4(291–301→CerS5) activity was examined with C20-CoA (upper panel) and C16-CoA (lower panel). Results are means ± S.E. n = 3. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Discussion
Studies performed ∼25 years ago indicated that ceramide is generated in the ER (26, 27), but it was only when CerS was identified that their localization in the ER membrane could be unambiguously determined (8, 17, 18, 29). However, solubilization and reconstitution of the CerS have proved extremely difficult (17, 31), and therefore structure–function studies are few and far between.
In this study, we extend our earlier work and suggest a new putative topology in which the N terminus is lumenal, and the fourth TMD is unlikely to completely cross the ER membrane (Fig. 1C). This revised topology, along with the discovery of an unexpected frameshift when aligning CerS2 and CerS5, led us to focus on the loop between the 5th and 6th TMDs and to the principal finding of this study, namely that CerS specificity can be altered by modifying the sequence of only 11 amino acid residues within this lumenal loop. How the two CerS substrates gain access to this loop is currently unknown, but it should be noted that acyl-CoAs are generated in the cytosol, whereas sphingosine is presumably generated within the membrane.
Whether changes in this loop cause loss of specificity or gain of activity is unclear because in vitro assays using CerS5(299–309→CerS2) indicate that this chimeric protein is able to generate ceramides with a variety of different acyl chain lengths, albeit under conditions optimized for the activity of CerS2, which requires a longer reaction time and more protein to obtain similar levels of enzymatic activity as CerS5, which uses shorter acyl chains. Under these conditions, C22:0-ceramide is the main ceramide generated; moreover, when using increasing amounts of C22:0 acyl-CoA in the reaction mixture, CerS5(299–309→CerS2) is more effective in generating C22:0-ceramide than the cognate CerS2. Thus, altering 11 residues in the CerS5 sequence unequivocally allows it to generate C22:0-ceramide, although CerS5(299–309→CerS2) is also able to generate other ceramides. The reason that C22:0-ceramide levels were not elevated in cells upon overexpression may be related to the availability of acyl-CoAs in cells or to a currently unknown level of regulation of chimeric CerS upon their overexpression.
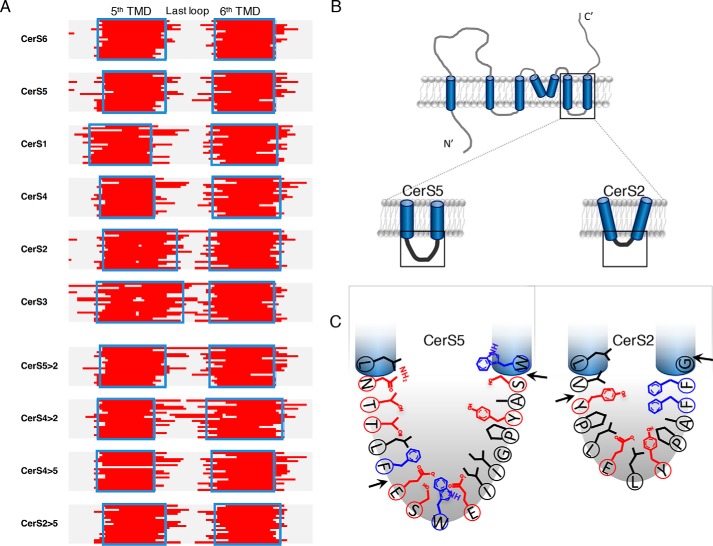
Because the structure of the CerS is not known, any discussion of how altering these residues affects specificity is by definition somewhat speculative. All topology predictions are consistent with this region forming a loop (Fig. 7A and Fig. S7A), but we are unable to determine experimentally at this stage how the loop folds to determine acyl-CoA specificity. An obvious proposition would be that the very-long–chain acyl-CoAs, such as those required by CerS2, might fit in a more extended loop, whereas the long-chain acyl-CoAs, such as those required by CerS5, might fit in a more compact loop. However, the opposite is true, as the CerS5 loop contains more residues than the CerS2 loop and thus the adjacent TMDs might be shorter (Fig. 7). Moreover, examination of the TMDs adjacent to the loop indicate that the loop in WT CerS5 and CerS6 consists of 15 and 16 residues, CerS1 and CerS4, 21 and 20 residues, respectively, and CerS2 and CerS3, only 11 and 9 residues (Fig. 7A). This indicates that the CerS that utilize the longest acyl-CoAs have the shortest number of residues in this loop. We suggest that the short loop may restrict the positioning of the adjacent TMDs such that they have a more open conformation in the membrane, whereas the CerS that use shorter acyl-CoAs may have a longer and more flexible loop affecting the flexibility of the TMDs and thus permitting use of shorter length acyl-CoAs (Fig. 7B). TMD prediction of the chimeric proteins (Fig. 7A) is consistent with this suggestion as the TMDs are intermediate in length. Importantly, the CerS2(291–301→CerS5) chimera has a longer loop (18 versus 11 residues) consistent with its ability to accommodate C16-CoA. The suggestions of a relationship between the number of residues in the loop and acyl-CoA specificity remain speculative but are nevertheless consistent with the altered specificities of the chimeric proteins.
Figure 7.
Residues in the loop between TMD5 and -6 in CerS2 and CerS5. A, predictions of the fifth and sixth TMDs (red) by 19 different programs in the six human CerS (top) and chimeric proteins (bottom). Blue rectangles indicate residues where the majority of the programs (>10/19) predict a TMD. See also Fig. S7A. B, conformation of TMDs 5 and 6 might be restricted according to the loop length. C, residues of CerS5 (left) and CerS2 (right) within the last putative loop. The arrows indicate the region that was altered between CerS5 and CerS2. Polar residues are indicated in red and bulky residues in blue.
In summary, we have identified a critical region that appears to determine the acyl-CoA specificity of the CerS. The precise mechanistic details of how this lumenal region gains accessibility to acyl-CoAs, which are generated in the cytosol, and the relationship to the CerS active site, await insight into the three-dimensional structure of the CerS.
Experimental procedures
Materials
NBD-Sphinganine (NBD-Sph) and fatty acyl-CoAs were from Avanti Polar Lipids (Alabaster, AL). Defatted BSA, a protease inhibitor mixture, anti-hemagglutinin (HA), anti-tubulin, anti-CerS2 antibodies, and polyethyleneimine were from Sigma. Horseradish peroxidase was from The Jackson Laboratory (Bar Harbor, ME). An ECL detection system was from Thermo Fisher Scientific (Waltham, MA). EndoH was from New England Biolabs Inc. (Ipswich, MA). Silica gel 60 TLC plates were from Merck (Billerica, MA). All solvents were of analytical grade and were purchased from Bio-Lab (Jerusalem, Israel).
Bioinformatics
Alignments of CerS were performed using ClustalW (32).
CerS constructs
CerS5(N26Q) and the chimeras CerS5(153–165→CerS2), CerS5(156–165→CerS2), CerS5(159–165→CerS2), CerS5(299–309→CerS2), CerS5(298–309→CerS2), CerS2(291–301→CerS5), CerS4(291–301→CerS2), and CerS4(291–301→CerS5) were subcloned from CerS5, CerS2, or CerS4 in a pcDNA3.1 vector carrying a C-terminal HA tag, using restriction-free cloning (33). Primers are given in Table 1. The terminology used for each construct is given in Fig. 2B. All sequences were confirmed prior to use.
Table 1.
Primers used in this study
F means forward; R means reverse.
| Construct | Primers | |
|---|---|---|
| CerS5(N26Q) | F: | GGCTACCCGAGCAAGTGAGCTGGGCTGATC |
| R: | GATCAGCCCAGCTCACTTGCTCGGGTAGCC | |
| CerS5(153–165→ CerS2) | F: | GCATGTGGAGATTCACATTTTATTTAATTGCCTTCATTGCCGGCATGGCCGTCATTGTGGATAAACCTTGGTTCTGGGACATCCGACAG |
| R: | CTGTCGGATGTCCCAGAACCAAGGTTTATCCACAATGACGGCCATGCCGGCAATGAAGGCAATTAAATAAAATGTGAATCTCCACATGC | |
| CerS5(156–165→ CerS2) | F: | GATTCACATTTTATTTATGTATATTCATTGCCGGCATGGCCGTCATTGTGGATAAACCTTGGTTCTGGGACATCCG |
| R: | CGGATGTCCCAGAACCAAGGTTTATCCACAATGACGGCCATGCCGGCAATGAATATACATAAATAAAATGTGAATC | |
| CerS5 (159–165→CerS2) | F: | CATTTTATTTATGTATATTCTGCTATGGAATGGCCGTCATTGTGGATAAACCTTGGTTCTGGGACATCCGACAG |
| R: | CTGTCGGATGTCCCAGAACCAAGGTTTATCCACAATGACGGCCATTCCATAGCAGAATATACATAAATAAAATG | |
| CerS5(299–309→CerS2) | F: | GATTCTGAACACGACCCTCTTTTACCCACTGGAGCTCTATCCTGCCTTCTTTGGCTGGTGGCTCCTCAATGGCC |
| R: | GGCCATTGAGGAGCCACCAGCCAAAGAAGGCAGGATAGAGCTCCAGTGGGTAAAAGAGGGTCGTGTTCAGAATC | |
| CerS2(291–301→CerS5) | F: | CTGGATCCTGCATTGCACCCTGTTTGAGAGTTGGGAGATAATCGGGCCTTATGCTTCATATTACTTCTTCAATTCCATGATGGG |
| R: | CCCATCATGGAATTGAAGAAGTAATATGAAGCATAAGGCCCGATTATCTCCCAACTCTCAAACAGGGTGCAATGCAGGATCCAG | |
| CerS4(291–301→CerS2) | F: | GATCCTCTACACCACATACTACTACCCACTGGAGCTCTATCCTGCCTTCTTTGGCTACTACTTCTTCAACGGGCTTC |
| R: | GAAGCCCGTTGAAGAAGTAGTAGCCAAAGAAGGCAGGATAGAGCTCCAGTGGGTAGTAGTATGTGGTGTAGAGGATC | |
| CerS4(291–301→CerS5) | F: | GATCCTCTACACCACATACTACGAGAGTTGGGAGATAATCGGGCCTTATGCTTCATACTACTTCTTCAACGGGCTTC |
| R: | GAAGCCCGTTGAAGAAGTAGTATGAAGCATAAGGCCCGATTATCTCCCAACTCTCGTAGTATGTGGTGTAGAGGATC |
Cell culture and transfection
HEK293T cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 100 IU/ml penicillin, 100 μg/ml streptomycin, and 110 μg/ml sodium pyruvate. Transfections were performed with the polyethyleneimine reagent using 8 μg of plasmid per 10-cm culture dish. 36–48 h after transfection, cells were removed from culture dishes and washed twice with PBS. Cell homogenates were prepared in 20 mm Hepes-KOH, pH 7.2, 25 mm KCl, 250 mm sucrose, and 2 mm MgCl2 containing a protease inhibitor mixture. Protein was determined using the Bradford reagent (Bio-Rad).
EndoH treatment
HEK cell lysates (2 mg protein/ml) were incubated with 0.5% SDS and 40 mm DTT at 100 °C for 10 min, subsequent to treatment with 50 mm sodium acetate, with or without 25 units of endoH/1 μg of protein for 1 h at 37 °C. For assay of CerS activity, samples were not denatured, and 50 units of endoH/1 μg of protein were used for 2 h at 37 °C.
Western blotting
Proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. HA-tagged constructs were identified using a mouse anti-HA antibody (1:10,000) and goat anti-mouse horseradish peroxidase (1:10,000) as the secondary antibody. Equal loading was confirmed using a mouse anti-tubulin antibody. Detection was performed using the ECL detection system.
Ceramide synthase assays
Cell homogenates were incubated with 15 μm NBD-Sph, 20 μm defatted BSA, and 50 μm fatty acyl-CoA in a 20-μl reaction volume at 37 °C for various times and using various amounts of tissue homogenate as indicated in the figure legends. Reactions were terminated by addition of chloroform/methanol (1:2, v/v) and lipids extracted (34). Lipids were dried under N2, resuspended in chloroform/methanol (9:1, v/v), and separated by TLC using chloroform/methanol, 2 m NH4OH (40:10:1, v/v/v) as the developing solvent. NBD-labeled lipids were visualized using a Typhoon 9410 variable mode imager and quantified by ImageQuantTL (GE Healthcare, Chalfont St Giles, UK).
Generation of Crispr/Cas9 mediated CerS2−/− HEK cells
CerS2−/− HEK cells were generated using Crispr/Cas9 (35). The guide sequence (GCCGATCTAGAAGACCGAGA) was designed using the chopchop tool (http://chopchop.cbu.uib.no),5 cloned into a pSpCas9(BB)-2A-GFP vector, and transfected into HEK293T cells using polyethyleneimine. Single cells were sorted by GFP 24 h after transfection and grown for 1–2 weeks. Proliferating cell colonies were collected, and CerS2 activity was assayed. The DNA of colonies with low CerS2 activity was sequenced using primers AGCTACTCCCTCTTGATGCC (forward) and TCAGGAAGACAATGGTGCCT (reverse), and verification of CerS2 knockout was determined by Western blotting and MS (LC-ESI-MS/MS).
Liquid chromatography electrospray ionization tandem MS
SL analysis by LC-ESI-MS/MS using an ABI 4000 quadrupole-linear ion trap mass spectrometer was performed as described previously (36).
Statistics
Statistical significance was assessed using an unpaired one-tailed Student's t test, with a p value under 0.05 (*), 0.01 (**), 0.001 (***), or 0.0001 (****) considered statistically significant.
Prediction of transmembrane domains
Transmembrane domain prediction was performed with the following programs using default parameters: DAS TMfilter (37); HMMTOP (38); MEMSAT (39); MEMSAT-SVM (40); oreinTM (pred-tmr2) (41); PHD (42); Philius (43); Phobius (44); SOSUI (45); Split 4.0 (46); SVMTM (47); TMHMM (19); Tmpred (28), and TOPCONS (30). TOPCONS component programs (Octopus, Polyphobius, SCAMPI, and SPOCTOPUS) were also assessed separately. An amino acid was considered part of a TMD if at least 10 of the 19 prediction programs identified it as such.
Author contributions
R. T., I. D. Z., and G. V. investigation; R. T. and I. D. Z. writing-original draft; I. D. Z., S. K., and A. H. F. methodology; S. B.-D., A. H. M., and A. H. F. conceptualization; S. B.-D. validation; S. K. data curation; A. H. M. funding acquisition; A. H. M. and A. H. F. writing-review and editing.
Supplementary Material
This work was supported in part by National Institutes of Health Grant GM076217 and Israel Science Foundation Grant 1728/15. The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article was selected as one of our Editors' Picks.
This article contains supporting data for Fig. 7A.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
The apparent lack of activity of CerS5(299–309→CerS2) toward C16-CoA in Fig. 3B was due to short assay times and the low amounts of protein that were used, corresponding to optimal assay conditions of 1 μg of protein for 5-min reaction time that is normally used for CerS5, whereas activity towards C16-CoA in Fig. 5A was assayed using more protein (5–60 μg) and for a longer reaction time (20 min), during which time more C16-CoA is formed.
- SL
- sphingolipid
- CerS
- ceramide synthase
- CoA
- coenzyme A
- endoH
- endoglycosidase H
- ER
- endoplasmic reticulum
- LC-ESI-MS/MS
- liquid chromatography-electrospray ionization-tandem mass spectrometry
- HEK
- human embryonic kidney
- NBD-Sph
- NBD-sphinganine
- TLC
- Tram-Lag-CLN8
- TMD
- transmembrane domain.
References
- 1. Ekroos K. (ed) (2012) in Lipidomics Perspective: From Molecular Lipidomics to Validated Clinical Diagnostics, pp. 1–19, Wiley-VCH Verlag GmbH & Co; KGaA, Weinheim, Germany [Google Scholar]
- 2. Futerman A. H. (2015) in Biochemistry of Lipids, Lipoproteins and Membranes (Ridgway N. D., and McLeod R. S., eds) Sixth Ed., pp. 297–326, Elsevier, Boston [Google Scholar]
- 3. Tidhar R., and Futerman A. H. (2013) The complexity of sphingolipid biosynthesis in the endoplasmic reticulum. Biochim. Biophys. Acta 1833, 2511–2518 10.1016/j.bbamcr.2013.04.010 [DOI] [PubMed] [Google Scholar]
- 4. Merrill A. H. (2011) Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 111, 6387–6422 10.1021/cr2002917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mullen T. D., Hannun Y. A., and Obeid L. M. (2012) Ceramide synthases at the centre of sphingolipid metabolism and biology. Biochem. J. 441, 789–802 10.1042/BJ20111626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pewzner-Jung Y., Ben-Dor S., and Futerman A. H. (2006) When do lasses (longevity assurance genes) become CerS (ceramide synthases)? Insights into the regulation of ceramide synthesis. J. Biol. Chem. 281, 25001–25005 10.1074/jbc.R600010200 [DOI] [PubMed] [Google Scholar]
- 7. Levy M., and Futerman A. H. (2010) Mammalian ceramide synthases. IUBMB Life 62, 347–356 10.1002/iub.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Venkataraman K., Riebeling C., Bodennec J., Riezman H., Allegood J. C., Sullards M. C., Merrill A. H. Jr., and Futerman A. H. (2002) Upstream of growth and differentiation factor 1 (uog1), a mammalian homolog of the yeast longevity assurance gene 1 (LAG1), regulates N-stearoyl-sphinganine (C18-(dihydro)ceramide) synthesis in a Fumonisin B1-independent manner in mammalian cells. J. Biol. Chem. 277, 35642–35649 10.1074/jbc.M205211200 [DOI] [PubMed] [Google Scholar]
- 9. Tidhar R., Ben-Dor S., Wang E., Kelly S., Merrill A. H. Jr., and Futerman A. H. (2012) Acyl chain specificity of ceramide synthases is determined within a region of 150 residues in the Tram-Lag-CLN8 (TLC) domain. J. Biol. Chem. 287, 3197–3206 10.1074/jbc.M111.280271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Winter E., and Ponting C. P. (2002) TRAM, LAG1 and CLN8: members of a novel family of lipid-sensing domains? Trends Biochem. Sci. 27, 381–383 10.1016/S0968-0004(02)02154-0 [DOI] [PubMed] [Google Scholar]
- 11. Spassieva S., Seo J.-G., Jiang J. C., Bielawski J., Alvarez-Vasquez F., Jazwinski S. M., Hannun Y. A., and Obeid L. M. (2006) Necessary role for the Lag1p motif in (dihydro)ceramide synthase activity. J. Biol. Chem. 281, 33931–33938 10.1074/jbc.M608092200 [DOI] [PubMed] [Google Scholar]
- 12. Kageyama-Yahara N., and Riezman H. (2006) Transmembrane topology of ceramide synthase in yeast. Biochem. J. 398, 585–593 10.1042/BJ20060697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jiang J. C., Kirchman P. A., Zagulski M., Hunt J., and Jazwinski S. M. (1998) Homologs of the yeast longevity gene LAG1 in Caenorhabditis elegans and human. Genome Res. 8, 1259–1272 10.1101/gr.8.12.1259 [DOI] [PubMed] [Google Scholar]
- 14. Venkataraman K., and Futerman A. H. (2002) Do longevity assurance genes containing Hox domains regulate cell development via ceramide synthesis? FEBS Lett. 528, 3–4 10.1016/S0014-5793(02)03248-9 [DOI] [PubMed] [Google Scholar]
- 15. Mesika A., Ben-Dor S., Laviad E. L., and Futerman A. H. (2007) A new functional motif in hox domain-containing ceramide synthases: identification of a novel region flanking the Hox and TLC domains essential for activity. J. Biol. Chem. 282, 27366–27373 10.1074/jbc.M703487200 [DOI] [PubMed] [Google Scholar]
- 16. Futerman A. H., and Riezman H. (2005) The ins and outs of sphingolipid synthesis. Trends Cell Biol. 15, 312–318 10.1016/j.tcb.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 17. Laviad E. L., Kelly S., Merrill A. H. Jr., and Futerman A. H. (2012) Modulation of ceramide synthase activity via dimerization. J. Biol. Chem. 287, 21025–21033 10.1074/jbc.M112.363580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mizutani Y., Kihara A., and Igarashi Y. (2005) Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem. J. 390, 263–271 10.1042/BJ20050291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krogh A., Larsson B., von Heijne G, Sonnhammer E. L. (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580 10.1006/jmbi.2000.4315 [DOI] [PubMed] [Google Scholar]
- 20. Breitling J., and Aebi M. (2013) N-Linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb. Perspect. Biol. 5, a013359 10.1101/cshperspect.a013359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lahiri S., Lee H., Mesicek J., Fuks Z., Haimovitz-Friedman A., Kolesnick R. N., and Futerman A. H. (2007) Kinetic characterization of mammalian ceramide synthases: determination of K(m) values towards sphinganine. FEBS Lett. 581, 5289–5294 10.1016/j.febslet.2007.10.018 [DOI] [PubMed] [Google Scholar]
- 22. Tidhar R., Sims K., Rosenfeld-Gur E., Shaw W., and Futerman A. H. (2015) A rapid ceramide synthase activity using NBD-sphinganine and solid phase extraction. J. Lipid Res. 56, 193–199 10.1194/jlr.D052001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pewzner-Jung Y., Park H., Laviad E. L., Silva L. C., Lahiri S., Stiban J., Erez-Roman R., Brügger B., Sachsenheimer T., Wieland F., Prieto M., Merrill A. H. Jr., and Futerman A. H. (2010) A critical role for ceramide synthase 2 in liver homeostasis: I. alterations in lipid metabolic pathways. J. Biol. Chem. 285, 10902–10910 10.1074/jbc.M109.077594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Volpert G., Ben-Dor S., Tarcic O., Duan J., Saada A., Merrill A. H. Jr., Pewzner-Jung Y., and Futerman A. H. (2017) Oxidative stress elicited by modifying the ceramide acyl chain length reduces the rate of clathrin-mediated endocytosis. J. Cell Sci. 130, 1486–1493 10.1242/jcs.199968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Laviad E. L., Albee L., Pankova-Kholmyansky I., Epstein S., Park H., Merrill A. H. Jr., and Futerman A. H. (2008) Characterization of ceramide synthase 2: tissue distribution, substrate specificity, and inhibition by sphingosine 1-phosphate. J. Biol. Chem. 283, 5677–5684 10.1074/jbc.M707386200 [DOI] [PubMed] [Google Scholar]
- 26. Mandon E. C., Ehses I., Rother J., van Echten G., and Sandhoff K. (1992) Subcellular localization and membrane topology of serine palmitoyltransferase, 3-dehydrosphinganine reductase, and sphinganine N-acyltransferase in mouse liver. J. Biol. Chem. 267, 11144–11148 [PubMed] [Google Scholar]
- 27. Hirschberg K., Rodger J., and Futerman A. H. (1993) The long-chain sphingoid base of sphingolipids is acylated at the cytosolic surface of the endoplasmic reticulum in rat liver. Biochem. J. 290, 751–757 10.1042/bj2900751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ikeda M., Arai M., Okuno T., and Shimizu T. (2003) TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res. 31, 406–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Riebeling C., Allegood J. C., Wang E., Merrill A. H. Jr., and Futerman A. H. (2003) Two mammalian longevity assurance gene (LAG1) family members, trh1 and trh4, regulate dihydroceramide synthesis using different fatty acyl-CoA donors. J. Biol. Chem. 278, 43452–43459 10.1074/jbc.M307104200 [DOI] [PubMed] [Google Scholar]
- 30. Tsirigos K. D., Peters C., Shu N., Käll L., and Elofsson A. (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res. 43, W401–W407 10.1093/nar/gkv485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lahiri S., and Futerman A. H. (2005) LASS5 is a bona fide dihydroceramide synthase that selectively utilizes palmitoyl-CoA as acyl donor. J. Biol. Chem. 280, 33735–33738 10.1074/jbc.M506485200 [DOI] [PubMed] [Google Scholar]
- 32. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 33. van den Ent F., and Löwe J. (2006) RF cloning: a restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys. Methods 67, 67–74 10.1016/j.jbbm.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 34. Bligh E. G., and Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 10.1139/o59-099 [DOI] [PubMed] [Google Scholar]
- 35. Ran F. A., Hsu P. D., Wright J., Agarwala V., Scott D. A., and Zhang F. (2013) Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8, 2281–2308 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Merrill A. H. Jr., Sullards M. C., Allegood J. C., Kelly S., and Wang E. (2005) Sphingolipidomics: high-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods 36, 207–224 10.1016/j.ymeth.2005.01.009 [DOI] [PubMed] [Google Scholar]
- 37. Cserzö M., Eisenhaber F., Eisenhaber B., and Simon I. (2002) On filtering false positive transmembrane protein predictions. Protein Eng. 15, 745–752 10.1093/protein/15.9.745 [DOI] [PubMed] [Google Scholar]
- 38. Tusnády G. E., and Simon I. (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17, 849–850 10.1093/bioinformatics/17.9.849 [DOI] [PubMed] [Google Scholar]
- 39. Jones D. T. (2007) Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 23, 538–544 10.1093/bioinformatics/btl677 [DOI] [PubMed] [Google Scholar]
- 40. Nugent T., and Jones D. T. (2009) Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 10, 159 10.1186/1471-2105-10-159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Pasquier C., and Hamodrakas S. J. (1999) An hierarchical artificial neural network system for the classification of transmembrane proteins. Protein Eng. 12, 631–634 10.1093/protein/12.8.631 [DOI] [PubMed] [Google Scholar]
- 42. Rost B. (1996) PHD: predicting one-dimensional protein structure by profile-based neural networks. Methods Enzymol. 266, 525–539 10.1016/S0076-6879(96)66033-9 [DOI] [PubMed] [Google Scholar]
- 43. Reynolds S. M., Käll L., Riffle M. E., Bilmes J. A., and Noble W. S. (2008) Transmembrane topology and signal peptide prediction using dynamic bayesian networks. PLoS Comput. Biol. 4, e1000213 10.1371/journal.pcbi.1000213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Käll L., Krogh A., and Sonnhammer E. L. (2007) Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35, W429–W432 10.1093/nar/gkm256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hirokawa T., Boon-Chieng S., and Mitaku S. (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379 10.1093/bioinformatics/14.4.378 [DOI] [PubMed] [Google Scholar]
- 46. Juretić D., Zoranić L., and Zucić D. (2002) Basic charge clusters and predictions of membrane protein topology. J. Chem. Inf. Comput. Sci. 42, 620–632 10.1021/ci010263s [DOI] [PubMed] [Google Scholar]
- 47. Yuan Z., Mattick J. S., and Teasdale R. D. (2004) SVMtm: support vector machines to predict transmembrane segments. J. Comput. Chem. 25, 632–636 10.1002/jcc.10411 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.