Abstract
Resistance to apoptosis and uncontrolled proliferation are two hallmarks of cancer cells. p53 is crucial for apoptosis triggered by a broad range of stresses and a well-known gatekeeper for neoplastic transformation. Here we show that oncogenic IDH1 R132H/R132Q mutants robustly inhibit p53 expression and such an effect is attributed to 2-HG production. Mechanistically, 2-hydroxyglutarate (2-HG) stabilizes hypoxia-inducible factor-2α, which in turn activates the expression of miR-380-5p, a characterized microRNA against p53 expression. Rescue expression of p53 can inhibit the proliferation rate and impair the resistance of apoptosis induced by doxorubicin in IDH1 R132Q mouse embryonic fibroblast cells. Furthermore, p53 protein levels correlates negatively with IDH1 R132H levels in human glioma samples. Our results thus shed a new light on how p53 is down-regulated by 2-HG and suggests that impairment of p53-mediated apoptosis contributes to the tumorigenesis driven by IDH1 mutants.
Keywords: p53, hypoxia-inducible factor (HIF), brain tumor, microRNA (miRNA), cell metabolism, IDH1 mutant
Introduction
Isocitrate dehydrogenases (IDH)5 normally convert isocitrate to α-ketoglutarate (α-KG) in a NAD(P)+-dependent manner. Three isozymes with distinct subcellular distributions and co-factors exist: IDH1, IDH2, and IDH3. In the past few years, various somatic point mutations of either IDH1 or IDH2 have been frequently found in multiple cancers, such as glioma, acute myeloid leukemia, chondrosarcoma, cholangiocarcinoma, paraganglioma, colon cancer, prostate cancer, and lung cancer (1–13). Interestingly, these mutations including IDH1 R132H/Q/C/S/L/G/V/P, IDH2 R140Q/W/L, and R172K/M/G/T/S all confer upon IDHs an abnormal catalytic activity that converts α-KG to the oncometabolite 2-hydroxyglutarate (2-HG) (14–16).
2-HG and α-KG are structurally similar except that the hydroxyl group in 2-HG is replaced by the C2 carbonyl group in α-KG (17, 18). Accumulating lines of evidence ascribe the carcinogenicity of 2-HG to its competitive inhibition of dioxygenases with α-KG as a co-substrate due to their structural similarity. Elevated levels of 2-HG inhibits the methylcytosine dioxygenase TET2, leading to a hypermethylator phenotype in cells harboring various IDH1/2 mutations (16, 18, 20–22). In addition, α-KG-dependent histone demethylases are also inhibited by 2-HG (18, 23), which in turn results in hypermethylation of histone and the disruption of cell differentiation (23). Furthermore, several groups have reported that 2-HG could stabilize hypoxia-inducible factor-1α (HIF-1α) by inhibiting HIF prolyl hydroxylase, which is responsible for HIF-1α hydroxylation, a process required for subsequent ubiquitination and degradation of HIF-1α via proteosome pathway (18, 24).
Tumorigenesis is widely accepted as a multistep process resulting from abnormal activation of oncogenes and inactivation of tumor suppressor genes (25). p53 tumor suppressor is recognized as a gatekeeper for neoplastic transformation due to its critical role in triggering apoptotic cell death, cell cycle arrest, and senescence in response to diverse stressor including DNA damage, nutrient deprivation, and inappropriate mitogenic stimulation (26, 27). The notion that p53 function has to be disrupted for tumor progression is supported by previous studies showing that restoring p53 function is sufficient to cause regression of several types of tumors in mice (28, 29). The importance of p53 in preventing tumor initiation is also indicated by the presence of somatic mutations of p53 in ∼50% of all human cancers (30). We questioned whether p53 inactivation is also involved in tumorigenesis caused by IDH1 mutations.
In this study, we report that IDH1 mutations robustly inhibit p53 expression in mouse embryonic fibroblasts (MEF) and other cell types. Such inhibition results from 2-HG-mediated inhibition of prolyl hydroxylase and subsequent stabilization of HIF-2α. Increased HIF-2α transactivates the expression of miR-380-5p, which in turn down-regulates the p53 protein level. Consistently, p53 protein levels were decreased in human glioma samples with the IDH1 R132H mutation, implying that 2-HG-caused p53 deficiency may be a key component in tumorigenesis driven by IDH1 mutations.
Results
Oncogenic IDH1 Arg-132 mutant robustly down-regulates p53
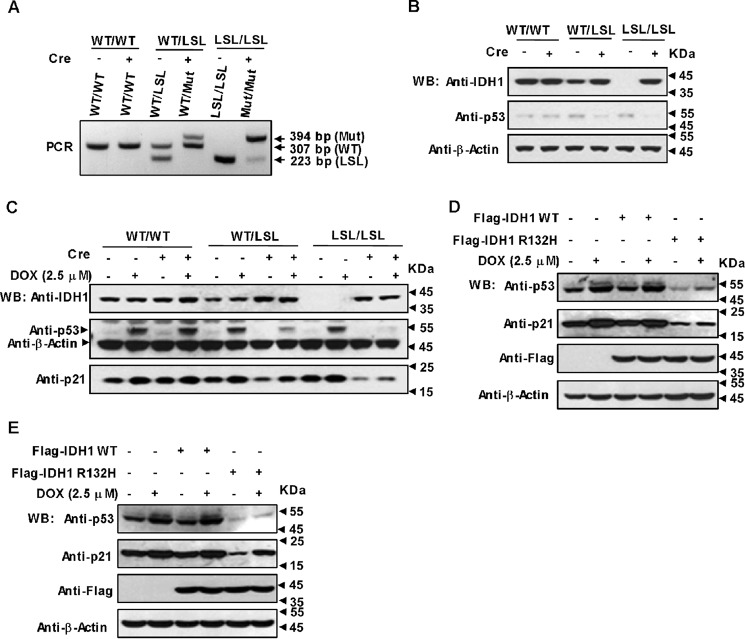
To find out whether the IDH1 mutation shows any inhibitory effect on p53, MEF cells with genotypes IDH1WT/WT, IDH1WT/LSL, and IDH1LSL/LSL were isolated from the embryos of conditional IDH1 R132Q knock-in mice (22, 31, 32), followed by excision of lox-stop-lox (LSL) cassette with Cre recombinase to generate cell lines with five different genotypes, IDH1WT/WT, IDH1WT/LSL, IDH1WT/Mut, IDH1LSL/LSL, and IDH1Mut/Mut (WT:WT; Mut:R132Q mutant). The genotypes and IDH1 protein levels of these cell lines were validated by polymerase chain reaction (PCR) and Western blotting (Fig. 1, A and B). Next we determined p53 expression in these cell lines. As shown in Fig. 1B, p53 expression was dramatically suppressed in IDH1WT/Mut and IDH1Mut/Mut MEFs, but not altered in IDH1WT/LSL and IDH1LSL/LSL MEFs with reduced or without WT IDH1 expression indicating that mutant IDH1 rather than WT IDH1 was responsible for the down-regulation of p53 expression. Interestingly, the IDH1 R132Q mutant could also significantly suppress p53 accumulation induced by doxorubicin (DOX) (Fig. 1C). Consistently, the expression of p21, one of p53 target genes, was down-regulated in IDH1 mutant cells to the same extent as p53 (Fig. 1C). To test if the IDH1 Arg-132 mutant also suppresses p53 expression in human cancer cells, we expressed WT IDH1 or its R132H mutant in HCT116 cells (Fig. 1D) and U2OS cells (Fig. 1E) and observed the same results as in MEFs. Taken together, the oncogenic IDH1 Arg-132 mutant is capable of down-regulating p53 dramatically.
Figure 1.
Oncogenic IDH1 Arg-132 mutant down-regulates p53. A, genotyping PCR of genomic DNA from IDH1WT/WT, IDH1WT/LSL, and IDH1LSL/LSL MEFs treated with or without Cre recombinase. After administration of Cre five different genotypes, IDH1WT/WT, IDH1WT/LSL, IDH1WT/Mut, IDH1LSL/LSL, and IDH1Mut/Mut were obtained. Bands associated with IDH1 R132Q mutant (Mut), wildtype (WT), and LSL alleles are indicated. B, p53 protein levels were dramatically decreased in IDH1WT/Mut and IDH1Mut/Mut MEFs. The same cell lines as displayed in A were detected for p53 and IDH1 expression with Western blotting (WB). C, the same cell lines as displayed in B were treated with or without 2.5 μm DOX for 4 h, followed by Western blotting with the antibodies indicated. D and E, IDH1 R132H mutant also inhibits p53 expression in cancerous cell lines U2OS and HCT116. U2OS cells (D) and HCT116 cells (E) were transfected with FLAG-tagged WT IDH1 or its R132H mutant. At 24 h post-transfection cells were treated with or without 2.5 μm DOX for 4 h, followed by detection of the indicated proteins.
p53 down-regulation by IDH1 R132H/R132Q depends on 2-HG production
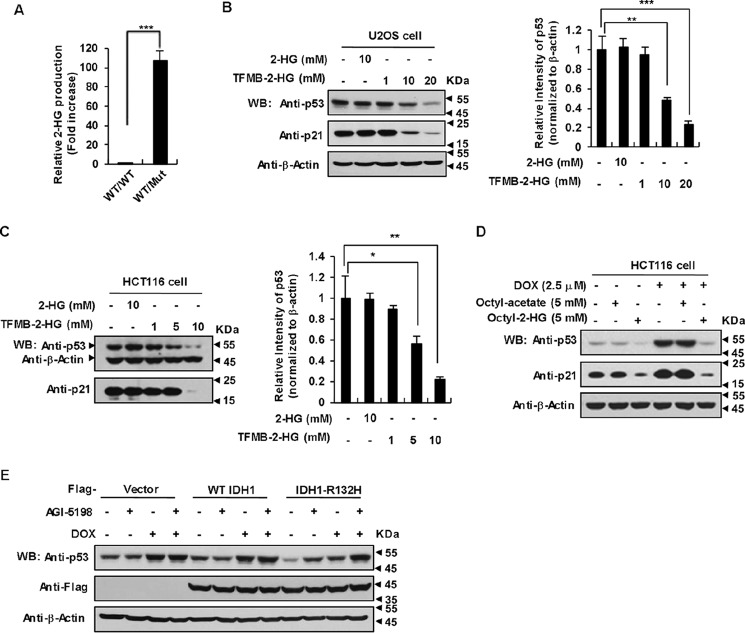
One of the most prominent feature of various IDH1 Arg-132 mutants is to produce extremely high levels of intracellular 2-HG, which disturbs a wide spectrum of biochemical reactions and thus leads to disorder of a broad range of cell biological functions (14, 15, 34). We spontaneously wanted to know whether IDH1 mutants induced down-regulation of p53 is the result of increased concentrations of 2-HG. First, we detected 2-HG by employing the LC-MS technique and found that dramatically high levels of 2-HG were produced in IDH1WT/Mut MEFs relative to IDH1WT/WT MEFs (Fig. 2A). It is important to point out that we used heterozygous, but not homozygous IDH1 R132Q knock-in (KI) MEF cells in all of the following experiments, because of the observation that all somatic gain-of-function mutations of IDH1 have been identified exclusively in one allele and heterozygous IDH1 mutation displayed the similar inhibitory effect on p53 as that of homozygous. Then we examined whether exogenous 2-HG could down-regulate p53. Treatment of U2OS cells with cell-permeable trifluoromethylbenzyl-(R)-2-HG (TFMB-2-HG) effectively increased the intracellular 2-HG level (Fig. S1A) and decreased the expression of p53 and p21 in a dose-dependent manner (Fig. 2B). Similar results were observed in the HCT116 cell line (Fig. 2C). To further bolster this conclusion, we treated HCT116 cells with another cell-permeable octyl-2-HG, whose permeability was confirmed by LC-MS (Fig. S1B). As expected, octyl-2-HG treatment markedly suppressed p53 expression in HCT116 cells even under exposure to 2.5 μm DOX (Fig. 2D). In addition, AGI-5198, a powerful and selective inhibitor of IDH1 R132H for 2-HG production (35), could efficiently suppress 2-HG production (Fig. S1C) and restored p53 expression in HCT116 cells harboring the IDH1 R132H mutant (Fig. 2E). These data demonstrate that 2-HG inhibits p53 expression in a broad range of mammalian cell types.
Figure 2.
p53 down-regulation by IDH1 R132H/R132Q depends on 2-HG production. A, IDH1 R132Q were produced at an extremely high level of 2-HG. The extracts of MEFs were subjected to LC-MS for analysis of relative 2-HG levels. Error bars show the standard deviations of three independent experiments (***, p < 0.001, unpaired Student's t test). B and C, TFMB-2-HG inhibits p53 expression in U2OS cells and HCT116 cells. U2OS (B) cells and HCT116 cells (C) were treated with the indicated amounts of TFMB-2-HG for 9 h, followed by Western blotting to detect p53 protein levels (left). Quantitation of signal intensities of Western blot bands of p53 (right) was performed by using ImageJ software. p53 levels were normalized to β-actin levels. Data are presented as the mean ± S.D. of three independent experiments (*, p < 0.05; **, p < 0.01; ***, p < 0.001, unpaired Student's t test). D, octyl-2-HG inhibit p53 accumulation induced by DOX in HCT116 cells. HCT116 cells were treated with DOX alone or in combination with octyl-2-HG (5 mm) for 8 h with octyl acetate as a control. Cell lysates were immunoblotted for p53 and p21. E, AGI-5198 restored p53 expression in HCT116 cells harboring IDH1 R132H mutant. HCT116 cells were infected with control lentivirus or lentivirus expressing FLAG-WT IDH1 or IDH1 R132H. At 48 h post-infection, cells were treated with or without 1.5 μm AGI-5198 for 2 days, followed by treating with 2.5 μm DOX (or not) for another 9 h in the presence of AGI-5198, as indicated.
2-HG down-regulates p53 at mRNA level
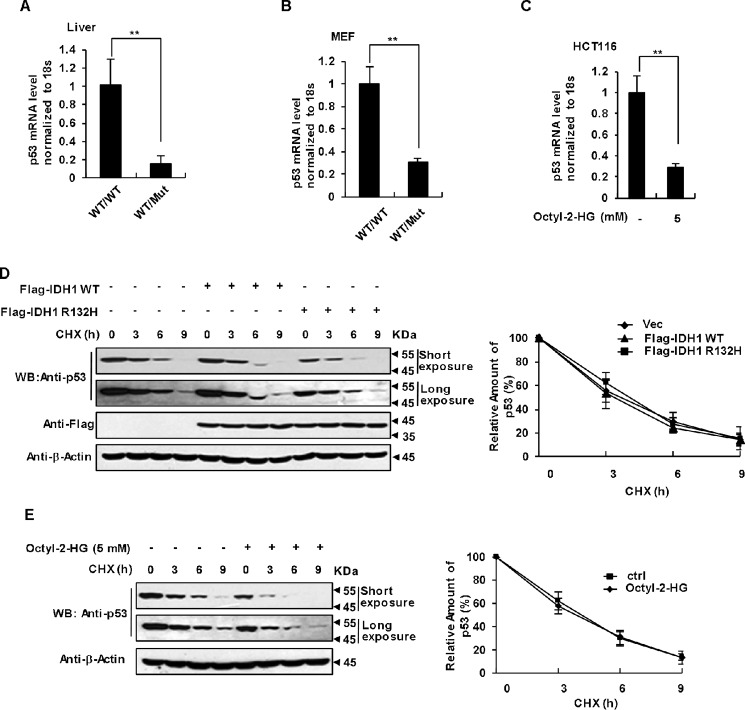
To dissect the mechanism underlying the p53 down-regulation mediated by the IDH1 R132 mutation, we examined whether this regulation occurs at mRNA or protein levels. We compared p53 mRNA levels in IDH1WT/WT and IDH1WT/Mut mice livers and observed that IDH1 R132Q KI mice livers had much less p53 mRNA than that in control mice livers (Fig. 3A). We also checked p53 mRNA levels in IDH1WT/WT and IDH1WT/Mut MEFs, and obtained exactly the same result (Fig. 3B). Furthermore, cell permeable octyl-2-HG treatment dramatically decreased the p53 mRNA levels in HCT116 cells (Fig. 3C). To our surprise, proteosome inhibitor MG132 and calpain inhibitor ALLN, but not lysosome inhibitors NH4Cl and chloroquine significantly blocked the inhibitory effect of 2-HG on p53 and restored its expression in HCT116 cells treated with octyl-2-HG (Fig. S2A) and MEF cells expressing the IDH1 R132Q mutant (Fig. S2B). One possibility is that 2-HG also down-regulated p53 by promoting its degradation in addition to the suppression of mRNA level. To validate this hypothesis, HCT116 cells were treated with cycloheximide (CHX), followed by detection of p53 protein levels and determination of p53 half-life. As shown in Fig. 3, D and E, both IDH1 R132H expression and 2-HG treatment failed to influence the half-life of p53. These results indicate that down-regulation of p53 expression by the IDH1 mutation appears to occur mainly at the mRNA level. A possible explanation to the observation that the proteosome inhibitor MG132 can partially antagonize 2-HG-induced p53 down-regulation is that 2-HG may also function to stimulate proteosome-mediated degradation of some positively transcriptional regulator of p53.
Figure 3.
2-HG down-regulates p53 at the transcriptional level. A, qRT-PCR analysis of relative p53 mRNA levels in IDH1WT/WT and IDH1WT/Mut mice livers. Data were normalized to 18S RNA and are presented relative to the levels in IDH1WT/WT mice. Mean ± S.D., n = 3 independent experiments, are shown (**, p < 0.01, unpaired Student's t test). B, qRT-PCR analysis of relative p53 mRNA levels in IDH1WT/WT and IDH1WT/Mut MEF. Data are presented as in A (**, p < 0.01, unpaired Student's t test). C, octyl-2-HG down-regulates p53 mRNA level in HCT116 cells. HCT116 cells were treated with the indicated amounts of octyl-2-HG for 9 h, followed by qRT-PCR analysis of relative p53 mRNA levels. Data are presented as mean ± S.D. of three independent experiments (**, p < 0.01, unpaired Student's t test). D, IDH1 Arg-132 mutant does not affect the turnover rate of p53 protein. HCT116 cells were transfected with the same amount of blank vector, FLAG-tagged IDH1, or IDH1 R132H. The levels of p53 at different time points after CHX treatment were determined by immunoblotting the total cell lysates (left panel) and quantified using ImageJ software (Bio-Rad) with β-actin as a loading control. Results plotted (right panel) are the amounts of p53 relative to that at time 0. Mean ± S.D., n = 3 independent experiments, are shown. E, 2-HG had no effect on the turnover rate of p53. HCT116 cells were treated with CHX alone or in combination with octyl-2-HG. Results were presented as in D.
IDH1 Arg-132 mutations down-regulate p53 via promoting miR-380-5p expression
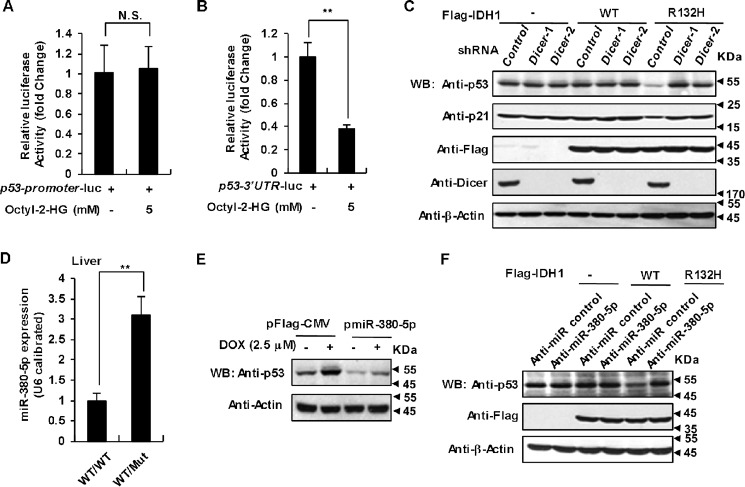
Next we asked whether 2-HG can diminish p53 transcription or p53 mRNA stability. To address this question, we constructed two chimeric luciferase reports containing p53-promoter and p53–3′ UTR because the 3′ UTR usually plays a very important role in the regulation of mRNA stability (36). 2-HG significantly suppressed the reporter activity of p53–3′ UTR-luc, but not p53-promoter-luc, implying that p53–3′ UTR may be required for 2-HG down-regulation of p53 mRNA (Fig. 4, A and B). miRNAs are 20–25-nucleotide short RNA molecules that function to down-regulate gene expression by targeting almost the 3′ UTR of mRNAs (37). To determine whether some miRNAs are involved in 2-HG caused p53 down-regulation, we utilized two independent short hairpin RNAs (shRNAs) to knockdown the expression of Dicer, one of the critical proteins in the maturation of miRNA (37). Loss of Dicer caused great up-regulation of p53 protein levels in HCT116 cells expressing mutated IDH1 (Fig. 4C), suggesting that p53 expression could be tightly regulated by miRNAs.
Figure 4.
2-HG down-regulates p53 through miR-380-5p. A, 2-HG fails to decrease p53 promoter activity. HEK-293T cells were transfected with the combinations of plasmids as indicated. After 8 h of transfection cells were treated with DMSO (solvent control) or 2-HG (5 mm) for another 36 h, followed by determination and quantification of relative luciferase activity. Data shown are mean ± S.D. of three independent experiments (unpaired Student's t test; N.S., not significant). B, 2-HG dramatically decreased p53-3′ UTR activities. HEK-293T cells were transfected with p53-3′ UTR-luc reporter, followed by 2-HG treatment and quantification of relative luciferase activity as in A. Mean ± S.D. (n = 3 independent experiments) are shown (**, p < 0.01, unpaired Student's t test). C, depletion of Dicer significantly up-regulates the p53 protein level in HCT116 cells expressing IDH1 R132H mutant. HCT116 cells transfected with WT IDH1 or its R132H mutant were infected with lentiviruses expressing two independent Dicer shRNAs or control shRNA. 96 h after infection, cells lysates were analyzed by Western blotting with the antibodies indicated. D, expression of miR-380-5p was significantly up-regulated in mice livers expressing IDH1 mutant. The expression of miR-380-5p in IDH1WT/WT and IDH1WT/Mut mice livers was analyzed with qRT-PCR and normalized to U6. Data shown are mean ± S.D. of three independent experiments (**, p < 0.01, unpaired Student's t test). E, miR-380-5p suppresses p53 expression. HCT116 cells transiently transfected with expression vectors for miR-380-5p and control miRNA were harvested after 48 h of transfection. Cell lysates were analyzed by Western blot. F, inhibition of miR-380-5p significantly increases p53 expression. HCT116 cells stably expressing WT IDH1 or its R132H mutant were transfected with chemically synthesized single-stranded anti-miR-380-5p oligonucleotides. 48 h after transfection, cells were analyzed by Western blot.
To find out which miRNA is responsible for the down-regulation of p53 by 2-HG, we performed a sequencing-based RNA profiling analysis using liver samples from IDH1WT/WT and IDH1WT/Mut mice. Several reports have revealed that a series of miRNAs, including miR-125b, miR-504, miR-25, miR-30d, miR-1285, and miR-380-5p directly target the 3′ UTR of p53 mRNA to down-regulate p53 protein levels (37–41). We thus analyzed the expression of these miRNAs in our sequencing results and found that expression of miR-380-5p was significantly increased in IDH1 mutant mice liver (Fig. S3A). This observation was confirmed by qRT-PCR data (Fig. 4D). Consistently, MEF cells (Fig. S3B) and HCT116 cells (Fig. S3C) expressing the IDH1 Arg-132 mutant also displayed much higher miR-380-5p levels. Overexpression of miR-380-5p dramatically down-regulated p53 in HCT116 cells (Fig. 4E). Moreover, inhibition of endogenous miR-380-5p by chemically synthesized single-stranded anti-miR-380-5p oligonucleotides in IDH1 mutant cells restored p53 expression (Fig. 4F). Taken together, our data demonstrate that IDH1 Arg-132 mutant down-regulates p53 by promoting miR-380-5p expression. There are two putative binding sites for miR-380-5p in the 3′ UTR of human p53 as shown in supporting Fig. S4A according to previous work (41). To determine whether these two putative sites are responsible for p53 down-regulation, we created p53-3′ UTR-luc vectors with either one or both of the sites deleted (Fig. S4B). The vectors were separately transfected into HCT116 cells together with the miR-380-5p expression plasmid or control pFlag-CMV vector. As expected, expression of miR-380-5p greatly decreased the luciferase activity of WT p53-3′ UTR-luc. However, deletion of both binding sites abolished the repressing effect of miR-380-5p on luciferase activity of p53-3′ UTR-luc (Fig. S4C). These data clearly demonstrate that miR-380-5p down-regulates p53 expression by binding to the two sites in the p53 3′ UTR.
IDH1 mutation down-regulation of p53 depends on HIF-2α
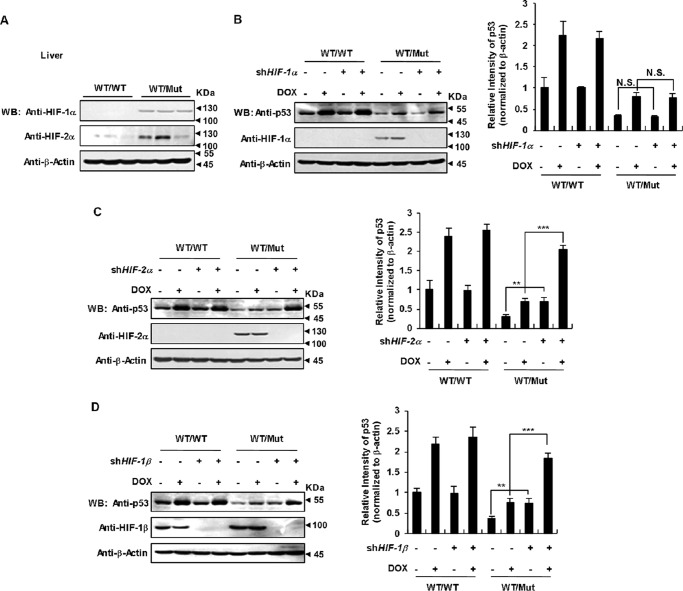
It was reported that 2-HG blocks prolyl hydroxylase and thereby stabilizes HIF-1α, leading to transcriptional activation of HIF-1α-dependent genes (18, 24). The regulation of HIF-2α protein stability is similar to that of the HIF-1α isoform and relies on α-KG-dependent hydroxylation and degradation (42). We detected HIF-1α and HIF-2α protein levels in IDH1 R132Q KI mice livers and found that both were increased (Fig. 5A). Similarly, both MEFs (Fig. S5A) and HCT116 cells (Fig. S5B) expressing IDH1 Arg-132 mutant also displayed much higher HIF-1α and HIF-2α protein levels under either normoxia or hypoxia conditions. We then used shRNAs to knock down HIF-1α and HIF-2α, respectively, in IDH1WT/Mut MEF to test whether HIFs were involved in p53 down-regulation. Loss of HIF-2α (Fig. 5C), but not HIF-1α (Fig. 5B), restored p53 expression in the mutant cells. In addition, shRNA-mediated silence of HIF-1β, which is necessary for HIFs transcriptional activity, restored p53 expression in IDH1 mutant cells (Fig. 5D). These data indicate that HIF-2α rather than HIF-1α is required for p53 down-regulation caused by IDH1 mutant.
Figure 5.
IDH1 Arg-132 mutant down-regulation of p53 depends on HIF-2α. A, IDH1 Arg-132 mutant increases HIF protein levels in transgenic mice livers. HIF protein levels in mice livers of the indicated genotypes were determined by Western blotting. B, knockdown of HIF-1α has no effect on p53 down-regulation caused by IDH1 Arg-132 mutant. IDH1WT/WT and IDH1WT/Mut MEFs were infected with lentiviruses expressing HIF-1α shRNA or control shRNA. 48 h after infection, cells were treated with or without 2.5 μm DOX, followed by Western blotting to detect the indicated proteins (left). Quantitation of signal intensities of Western blot (WB) bands of p53 (right) was performed by using ImageJ software. p53 levels were normalized to β-actin levels. Data are presented as the mean ± S.D. of three independent experiments (N.S. = not significant). C, IDH1 mutant down-regulation of p53 depends on HIF-2α. IDH1WT/WT and IDH1WT/Mut MEFs were infected with lentiviruses expressing HIF-2α shRNA or control shRNA. 48 h after infection, cells were treated with or without 2.5 μm DOX, followed by Western blot to detect the indicated proteins (left). Quantitation of signal intensities of Western blot bands was performed and presented as in A (right). D, HIF-1β knockdown reverses p53 suppression by the IDH1 Arg-132 mutant. IDH1WT/WT and IDH1Mut/Mut MEF were infected with lentiviruses expressing HIF-1β shRNA or control shRNA. 48 h after infection, cells were treated with or without 2.5 μm DOX, followed by detection of the proteins as indicated (left). Quantitation of signal intensities of Western blot bands was performed and presented as in A (**, p < 0.01; ***, p < 0.001, unpaired Student's t test).
HIF-2α promotes miR-380-5p transcription
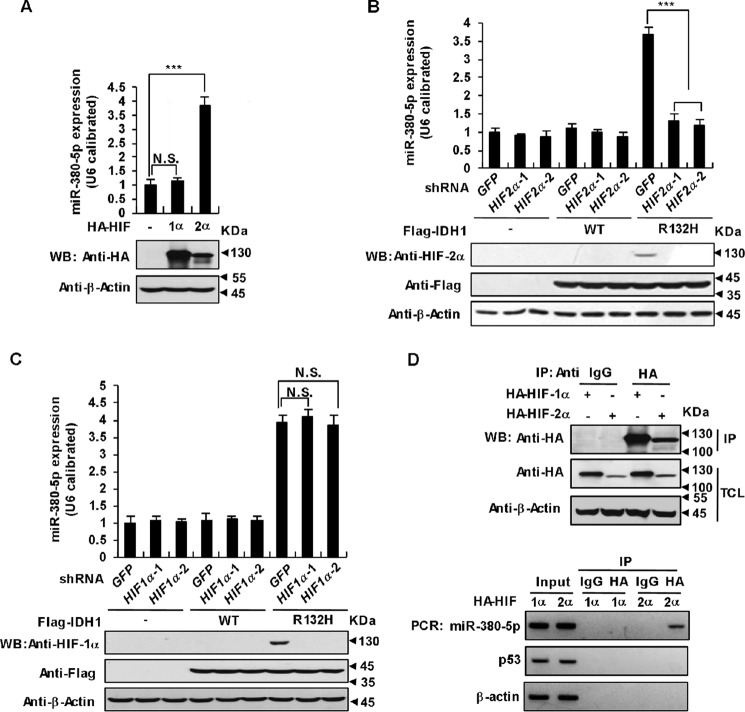
We next explored the regulatory relationship between HIF-2α and miR-380-5p in p53 down-regulation caused by IDH1 mutations. First, we overexpressed HIF-1α and HIF-2α in HCT116 cells and found that increased HIF-2α protein but not HIF-1α stimulated miR-380-5p expression as determined by qRT-PCR (Fig. 6A). Second, knockdown of HIF-2α in IDH1 mutant MEFs decreased miR-380-5p expression (Fig. 6B). As a control, shRNA-mediated silence of HIF-1α have no effect on miR-380-5p expression in IDH1 mutant MEFs (Fig. 6C). Third, we examined whether HIF-2α directly binds to the miR-380-5p promoter region and promotes its expression using chromatin immunoprecipitation (ChIP) assays given the fact that HIF-2α is a transcriptional factor. Indeed, DNA fragments derived from the miR-380-5p promoter region, but not from the actin or p53 promoter regions, were co-precipitated with HIF-2α (Fig. 6D). These data suggest that HIF-2α directly binds to the miR-380-5p promoter region and stimulates its transcription.
Figure 6.
HIF-2α promotes miR-380-5p transcription. A, overexpression of HIF-2α up-regulates miR-380-5p. HCT116 cells were transfected with HA-tagged HIF-1α or HIF-2α. 36 h after transfection the expression of miR-380-5p was analyzed with qRT-PCR and normalized to U6 (mean ± S.D., n = 3, ***, p < 0.001, unpaired Student's t test). B, knockdown of HIF-2α down-regulates miR-380-5p. HCT116 cells stably expressing WT IDH1 or its R132H mutant were infected with lentiviruses expressing two independent HIF-2α shRNAs or control shRNA, individually. 48 h after infection, the expression of miR-380-5p was analyzed and presented as in A (***, p < 0.001). C, knockdown of HIF-1α has no effect on miR-380-5p up-regulation caused by the IDH1 Arg-132 mutant. HCT116 cells stably expressing WT IDH1 or its R132H mutant were infected with lentiviruses expressing two independent HIF-1α shRNAs or control shRNA, individually. 48 h after infection, the expression of miR-380-5p was analyzed and presented as in A (N.S. = not significant). D, HIF-2α binds to the promoter regions of miR-380-5p. HCT116 cells were transfected with HA-tagged HIF-1α or HIF-2α. 36 h after transfection, ChIP assays were performed using IgG (control) or anti-HA antibodies. Proteins in precipitates and total cell lysates were determined by Western blot analysis (upper panel). Purified DNA was analyzed by standard PCR using primers targeting the indicated promoter regions (lower panel). Actin and p53 were used as negative controls.
p53 down-regulation is involved in IDH1 mutant-driven tumorigenesis
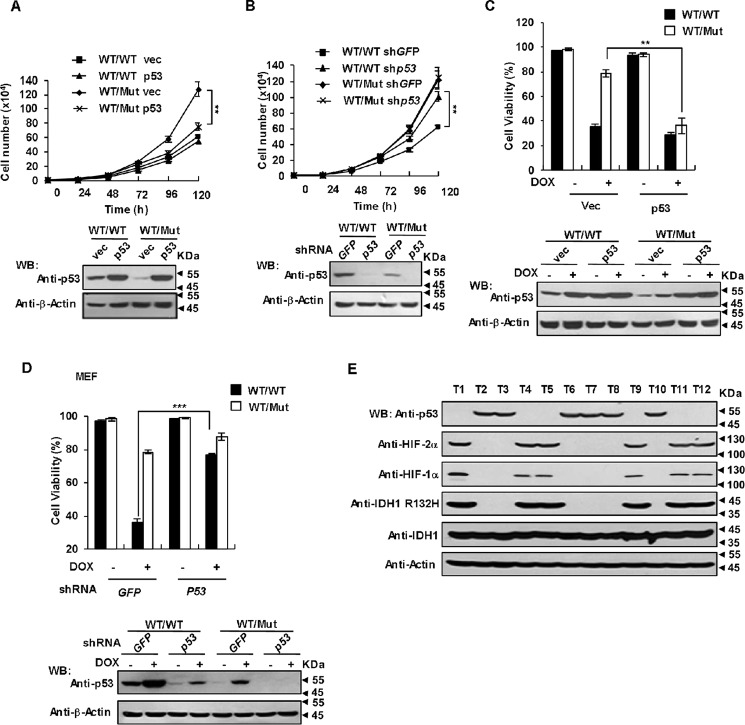
Because p53 plays a crucial role in triggering apoptotic cell death and cell cycle arrest in response to diverse stressors, and our data indicate that IDH1 R132H/R132Q mutations could dramatically suppress p53 expression, we examined whether such inhibition benefits cell proliferation and endow cells an ability to escape apoptosis induced by DNA damage agent. Rescue expression of p53 significantly attenuated the proliferation rate of IDH1WT/Mut cells but not IDH1WT/WT cells (Fig. 7A). Consistently, knockdown HIF-2α (Fig. S6A) or inhibition of endogenous miR-380-5p by anti-miR-380-5p oligonucleotides (Fig. S6B) in IDH1 mutant cells restored p53 expression and markedly suppressed the proliferation rate. On the contrary, 2-HG treatment (Fig. S6C) or knockdown of p53 (Fig. 7B) increased the proliferation rate of IDH1WT/WT cells but not IDH1WT/Mut cells. These findings suggest that inhibition of p53 by 2-HG confer IDH1-mutated cells a higher proliferating rate. Next we exposed IDH1WT/WT and IDH1WT/Mut MEFs to a lethal dose of DOX for 16 h and assessed their viability by Annexin V staining and flow cytometry. As shown in Fig. 7C, IDH1WT/Mut MEFs displayed substantial resistance to DOX-induced apoptosis compared with IDH1WT/WT cells. However, rescue expression of p53 rendered IDH1WT/Mut MEFs sensitive to DOX again. Consistent with this result, knockdown of HIF-2α (Fig. S6D) or anti-miR-380-5p oligonucleotides treatment (Fig. S6E) efficiently eliminated the resistance of IDH1WT/Mut MEFs to DOX-induced apoptosis. In addition, 2-HG treatment (Fig. S6F) or knockdown of p53 (Fig. 7D) desensitized IDH1WT/WT cells but not IDH1WT/Mut cells to DOX-induced apoptosis. Moreover, the p53 protein level was dramatically decreased in low grade glioma samples with the IDH1 R132H mutation compared with that expressing WT IDH1 (Fig. 7E), establishing a solid correlation between IDH1 R132H mutation and p53 down-regulation. We further determined the p53 genotype of human glioma samples by Sanger sequencing. As shown in Fig. S6G, samples 1, 3, 7, 8, 9, 10, 11, and 12 harbor p53 mutations. Thus, IDH1 R132 mutations down-regulate p53 expression, despite the status of p53. It is important to point out that all presently available evidence indicates that IDH1 mutations are co-present with the p53 mutation and occur before the acquisition of p53 mutations (43, 44). These evidences together with our data suggest that IDH1 R132 mutations induced down-regulation of WT p53 may play an important role in the early stage of gliomagenesis. Collectively, IDH1 R132 mutations can remarkably stimulate cell proliferation and protect cells from DNA damage-induced apoptosis by suppressing p53 expression, thereby favoring tumor formation.
Figure 7.
p53 down-regulation is involved in IDH1 mutant-driven tumorigenesis. A, rescue expression of p53 significantly attenuated the proliferation rate of IDH1 Arg-132 mutant cells. Both IDH1WT/WT and IDH1WT/Mut cells were infected with lentiviruses expressing p53. At 36 h after infection, proliferation rates were determined by growth curves (upper panel). Mean ± S.D., n = 3 independent experiments, are shown (**p < 0.01, unpaired Student's t test). Proteins in total cell lysates of the same cell lines were determined by Western blot analysis (WB) (lower panel). B, knockdown p53 increased the proliferation rate of IDH1WT/WT cells but not IDH1WT/Mut cells. IDH1WT/WT and IDH1WT/Mut MEFs were infected with lentiviruses expressing p53 shRNA or control shRNA. At 36 h after infection, proliferation rates (upper panel) were determined as in A (**, p < 0.01, unpaired Student's t test). Proteins in total cell lysates of the same cell lines were determined by Western blot analysis (lower panel). C, forced expression of p53 sensitize IDH1 Arg-132 mutant cells to DOX-induced apoptosis. IDH1WT/WT and IDH1WT/Mut MEF cells were infected with lentiviruses expressing p53. At 36 h after infection, cells were treated with or without 2.5 μm DOX for 16 h. The percentages of surviving cells (Annexin V negative) were determined by a flow cytometer. Data are presented as mean ± S.D. of three independent experiments (**, p < 0.01, unpaired Student's t test). D, knockdown of p53 desensitized of IDH1WT/WT cells rather than IDH1WT/Mut cells to DOX-induced apoptosis. IDH1WT/WT and IDH1WT/Mut MEF cells were infected with lentiviruses expressing p53 shRNA. At 36 h after infection, cells were treated with or without 2.5 μm DOX for 16 h. The percentages of surviving cells were determined and presented as in D (***, p < 0.001, unpaired Student's t test). E, IDH1 R132H mutant suppresses p53 expression in glioma. Clinical specimens of glioma were analyzed by Western blot with the antibodies as indicated.
In conclusion, our data demonstrates that the high level of 2-HG produced by cancer-associated IDH mutations can stabilize HIF-2α and consequently stimulate miR-380-5p expression, which in turn down-regulates p53. Impaired p53 expression confer cells a higher proliferation rate and resistance to apoptosis, which contributes to the oncogenicity of the IDH1 mutations.
Discussion
It is well-accepted that aberrant genes' expression caused by epigenetic alterations and HIF-1α accumulation in IDH1-mutated cells contribute to tumor progression. 2-HG is believed to competitively inhibit α-KG-dependent dioxygenases, including prolyl hydroxylase 2, which disrupts HIF-1α hydroxylation and leads to aberrant accumulation of HIF-1α (18, 24). Little is known about whether the HIF-2α protein level is also regulated by 2-HG and if so what role does HIF-2α play in tumorigenesis stimulated by 2-HG? We initially observed that the HIF-2α protein level was dramatically increased in cells harboring IDH1 mutations, similar to the case of HIF-1α. Next we tried to find out whether HIF-2α plays any role different from that HIF-1α does in tumorigenesis.
The p53 tumor suppressor has long been recognized as the gatekeeper for tumor formation. The notion that p53 function has to be disrupted for the progression of some tumors is well-accepted. Indeed, we observed that p53 protein levels were markedly decreased either in IDH1WT/Mut MEFs or HCT116 cells expressing IDH1 R132H or treated with permeable 2-HG. Moreover, IDH1WT/Mut MEFs took great advantage of suppression of p53 expression in proliferation and resistance to DOX-induced apoptosis. Importantly, the p53 protein level was abolished in low grade glioma samples with the IDH1 R132H mutation. Our data thus provided convincing evidence that p53 down-regulation is a key event in the IDH1 mutant that caused tumor formation. Our further observations indicated that down-regulation of p53 was at the mRNA level, and HIF-2α, but not HIF-1α, played a predominant a role in such regulation, raising an interesting question how was the p53 mRNA level reduced by HIF-2α.
In ∼50% of all human cancers, inactivation of p53 function is acquired by the presence of its somatic mutations (30). In addition, p53 is also inactivated through proteasome-mediated degradation caused by amplification of MDM2 or its homolog MDM4 in 10∼20% of total human cancer (45). In recent years, accumulating evidence has established that various miRNAs, including miR-125b, miR-504, miR-25, miR-30d, miR-1285, and miR-380-5p are involved in down-regulating p53 protein levels (37–41). These studies remind us that regulation of miRNAs against p53 by HIF-2α might be an intermediate event in 2-HG-induced down-regulation of p53. As expected, the expression of miR-380-5p was robustly activated by HIF-2α and required for down-regulation of p53 in IDH1 mutation cells. In summary, a high level of 2-HG produced by the IDH1 mutation stabilizes HIF-2α, which further activates transcription of miR-380-5p. Elevated miR-380-5p levels effectively mediate degradation of the p53 mRNA and eventually benefits cell proliferation and tumor formation. In summary, we find a novel mechanism underlying the tumorigenesis driven by 2-HG-producing IDH1 mutations. Our observation that HIF-2α rather than HIF-1α is involved in the suppression of p53 will help us to further understand the delicate difference of HIF-2α and HIF-1α in promoting tumor formation.
Experimental procedures
Cell culture and transfections
HEK293T cells, HCT116 cells, U2OS cells, and MEF were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 4 mm l-glutamine at 37 °C in a humidified incubator containing 5% CO2. HEK293T cells were transfected using polyethylenimine (catalog number 23966, Polysciences, Inc.) at a final concentration of 10 μm. Total DNA for each plate was equalized using relevant empty vectors. Transfected cells were harvested at 24 h post-transfection. Lentivirus for infection of MEF and HCT116 cells was packaged in HEK293T cells using Turbofect (catalog number R0532, Thermo Scientific) transfection reagent. At 36 h post-transfection, virus-containing culture supernatant was collected by centrifugation (13,000 × g, 5 min). After titration, an appropriate volume of the virus-containing supernatant was added to cells (1 × 105) in the presence of Polybrene (Sigma) at a final concentration of 10 ng/μl.
Generation of IDH1-KI MEF
IDH1WT/WT, IDH1WT/LSL, and IDH1LSL/LSL MEF were isolated from embryos of conditional IDH1 R132Q knock-in mice (22, 31, 32) at 13.5 days post-coitum. The LSL cassette flanking the IDH1 R132Q mutation (Mut) in IDH1WT/LSL and IDH1LSL/LSL MEF was excised by Cre recombinase-expressing adenovirus to generate IDH1WT/Mut and IDH1Mut/Mut MEF. IDH1WT/WT MEF were treated with adenovirus-Cre as a control. Primers for genotyping of Idh1 were 5′-ACCAGCACCTCCCAACTTGTAT-3′, 5′-AGGTTAGCTCTTGCCGATCCGT-3′, and 5′-CAGCAGCCTCTGTTCCACATAC-3′.
Reagents and antibodies
2-HG and CHX were obtained from Sigma. Octyl-2-HG was purchased from Cayman Chemical Co. TFMB-2-HG was synthesized, purified, and characterized as described previously (46). AGI-5198 and DOX were purchased from Selleck. Dual luciferase assay kits were purchased from Promega. Annexin V-FITC apoptosis detection kits were purchased from Sigma. Anti-FLAG M2 and anti-β-actin antibodies were purchased from Sigma. Anti-HIF-1α (number 36169) antibody was purchased from Cell Signaling Technology. Anti-HIF-2α (AF2997) antibody was purchased from R&D Systems. Anti-IDH1 (SC-49996), anti-p53 (SC-3243), and anti-HA (SC-7392) antibodies were purchased from Santa Cruz Biotechnology. Anti-IDH1 R132H (DIA H09) antibody was obtained from Dianova. Anti-p21 (10355-1-AP) antibody was purchased from Proteintech.
Constructs
WT IDH1 or its R132H mutant were cloned into BamHI and SmaI sites of the modified lentivirus vector FLAG-tagged pLV using the Exo III-assisted ligase-independent cloning method (33). For luciferase reporter assays, the human p53 promoter and p53 3′ UTR were cloned into the pGL2-Basic vector and pmir GLO vector, respectively (Promega). The primer sequences used for amplifying the p53 promoter were: 5′-GCGTGCTAGCTCGAGTCGGCGAGAATCCTG-3′ and 5′- CGGAATGCCAAGCTTTCTAGACTTTTGAGAAG-3′. The primer sequences used for amplifying the p53 3′ UTR were: 5′-TGTTTAAACGAGCTCCATTCTCCACTTCTTGTTCC-3′ and 5′-GACTCTAGACTCGAGGATGTTGACCCTTCCAGCTGG-3′. p53-3′ UTR-luc constructs with either one or both of two putative miR-380-5p-binding sites deleted were generated using a PCR-based, site-directed mutagenesis method employing Pfu polymerase. All plasmids were verified by DNA sequencing (sequences available upon request).
Immunoprecipitation and Western blotting
Cells were harvested in lysis buffer (20 mm Tris-HCl (pH 7.4), 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerolphosphate, 1 mm sodium orthovanadate, 1 μg/ml of leupeptin, 1 mm phenylmethylsulfonyl fluoride). For immunoprecipitations, lysates were incubated with antibodies indicated at 4 °C for 3 h. Immunoprecipitates were washed three times in lysis buffer and boiled in SDS-PAGE loading buffer. Proteins in total cell lysates or immunoprecipitates were fractionated by SDS-PAGE and transferred to PVDF membranes. Blots were blocked in 5% nonfat milk or BSA and incubated with the appropriate antibodies.
Luciferase reporter assay
HCT116 cells transfected with chimeric luciferase reporter plasmids were washed with PBS and lysed by lysis buffer. The supernatants were collected by centrifugation (13,000 × g, 5 min) and subjected to dual luciferase assays (Promega) by following the manufacturer's instructions. The transfection efficiency was normalized by co-expression of Renilla luciferase.
Flow cytometric cell death assay
Cells cultured in 6-well plates were left untreated or exposed to DOX for the times indicated in the figures. After treatment, suspended and trypsinized cells were centrifuged at 800 × g for 5 min, washed once with PBS, and stained with FITC-conjugated Annexin V for 10 min at 37 °C in the dark. Percentages of apoptotic cells were quantified by a fluorescence-activated cell using a flow cytometer.
Chromatin IP assay
The experiments were performed following the standard protocol of the SimpleChIP Enzymatic Chromation IP Kit (Cell Signaling, number 9002). Briefly, HCT116 cells transfected with HA-HIF-1α or HA-HIF-2α cultured in a 15-cm plate were cross-linked by 37% formaldehyde for 10 min at room temperature, and then glycine was added to stop the reaction. Cells were lysed, followed by nuclei preparation. 0.5 μl of micrococcal nuclease were added per each sample then incubated for 20 min at 37 °C with frequent mixing to digest DNA to ∼150–900 bp. After the nuclear membrane was broken by sonication, the same amount of chromatin was used for immunoprecipitation with anti-HA antibody or anti-IgG antibody (control). After the immunoprecipitation, chromatin was eluted from agarose beads with ChIP Elution Buffer and cross-links were reversed by incubating with NaCl and Proteinase K. DNA was purified using spin columns and quantification of DNA was determined by PCR or real-time quantitative PCR with primers targeting p53, actin, and the miR-380-5p TSS region. Primer sequences were: miR-380-5p forward: 5′-GTCAGTCATAGCACTAGTTCC-3′, miR-380-5p reverse: 5′-CTGAGGCCTGATGTAGTATTG-3′; p53 forward: 5′-CCTGACTCTGCACCCTCCTC-3′, p53 reverse: 5′-CGAGGCTCCTGGCACAAAGC-3′; actin forward: 5′-GAGCACAGAGCCTCGCCTTT-3′ and actin reverse: 5′-AGACAAAGACCCCGCCGGTT-3′.
RNA interference
The lentivirus-based vector pLKO.1 was used for shRNA expression. The sequences of the 19-nucleotide shRNAs used for gene knockdown were as follows (5′ to 3′): mouse HIF-1α shRNA: GGAAAGAACTAAACACACA; mouse HIF-2α shRNA: AGAATCAACTCTAGGGTTA; mouse HIF-1β shRNA: GGATAAACTTCGAGAGCAG; mouse p53 shRNA: GAATGAGGCCTTAGAGTTA; human Dicer shRNA-1: CACTGGTCAGGGAAGACAT; human Dicer shRNA-2: GCTCGAAATCTTACGCAAA; human HIF-2α shRNA-1: AACAGCATCTTTGATAGCA; human HIF-2α shRNA-2: CCCGGATAGACTTATTGCC; human HIF-1α shRNA-1: GTGATGAAAGAATTACCGAAT; human HIF-1α shRNA-2: TGCTCTTTGTGGTTGGATCTA.
Real-time RT-PCR analyses of mRNAs and miRNAs
For real-time RT-PCR mRNA quantification, total RNA was isolated using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. In all, 1.5 μg of total RNA was used to prepare cDNA using random primer mixtures. Real-time RT-PCR was performed by using the SYBR Green Real-time PCR Master Mix (TOYOBO, Shanghai). Primers for mp53 were 5′-GCCAGGAGACATTTTCAGGC-3′ and 5′-CTCCTCAACATCCTGGGGC-3′, for hp53 were 5′-GTTCCGAGAGCTGAATGAGG-3′ and 5′-TCTGAGTCAGGCCCTTCTGT-3′, for 18S RNA was 5′-CGACGACCCATTCGAACGTCT-3′.
For miRNAs, real-time RT-PCR was performed with the stem-loop primers as reported (19). U6 RNA served as an internal control. has-miR-380-5p primers were: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTGCGCAT-3′ (RT), 5′-GGGGTGGTTGACCATAGAAC-3′ (forward), and 5′-TGCGTGTCGTGGAGTC-3′ (reverse). mmu-miR-380-5p primers were: 5′-GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACTCGCATG-3′ (RT), 5′-GGGGATGGTTGACCATAGAAC-3′ (forward), and 5′-TGCGTGTCGTGGAGTC-3′ (reverse). U6 primers were: 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (RT), 5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward), and 5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse).
2-HG measurement by LC-MS
LC-MS analysis of 2-HG levels was performed as described (14). Briefly, cells were cultured to ∼80% confluence, washed in PBS, quenched in 1 ml of 80:20 methanol:water at −80 °C, and detached from the culture dish using a cell scraper. Quenched cells were centrifuged at 12,000 × g for 15 min at 4 °C, and 0.8 ml of supernatant was dried under nitrogen gas and dissolved in 100 ml of aqueous LC buffer. This mixture was centrifuged at 12,000 × g for 15 min and analyzed by LC-MS within 24 h. Total cell numbers and protein levels used for subsequent normalization of LC-MS signal intensities were determined from equivalently treated control plates. Sample separation and analysis were performed on a 50 × 2 mm, 4-mm Synergi Hydro-RP 80 A column, using a gradient of buffer A (10 mm tributylamine, 15 mm acetic acid, 3% (v/v) methanol, in water) and buffer B (methanol) using multiple reaction monitoring transitions. 2-HG levels were quantified by comparing peak areas with pure metabolite standards at known concentration.
Animal experiments and patient samples
All animal experimental protocols were approved by the Institutional Animal Care and Use Committee at Xiamen University. Glioma samples were obtained with the approval of the research ethics boards of Xiamen University and Huanhu Hospital and in accordance with the principles of the Declaration of Helsinki. Written informed consent was obtained from all patients.
Statistics
Two-tailed Student's t test was used to compare differences between treated and control groups. Differences with p values <0.05 were considered statistically significant: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Author contributions
B. J., W. Z., and Q. L. funding acquisition; B. J., W. Z., M. Shi, J. Z., A. C., H. M., M. Suleman, F. L., L. Z., J. W., Y. Z., M. L., S. W., C. O., H. W., X. H., H. Z., and Q. L. investigation; B. J. and W. Z. writing-original draft; B. J., W. Z., M. Shi, J. Z., A. C., H. W., X. H., H. Z., and Q. L. project administration; B. J., H. W., X. H., H. Z., and Q. L. writing-review and editing; W. Z. and M. Shi data curation.
Supplementary Material
Acknowledgments
We thank the University Health Network (UHN) and Tak W. Mak (Campbell Family Institute for Breast Cancer Research, Toronto, Canada) for kindly providing IDH1 R132Q mice.
This work was supported by the National Natural Science Foundation of China Grants 81372702, 81402285, and 31701252, National Science Foundation of China for Fostering Talents in Basic Research Grant J1310027, the Open Research Fund of State Key Laboratory of Cellular Stress Biology, Xiamen University Grant SKLCSB2018KF007, and Postdoctoral Science Foundation of China Grant 2017M622071. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S6.
- IDH
- isocitrate dehydrogenases
- α-KG
- α-ketoglutarate
- 2-HG
- 2-hydroxyglutarate
- HIF-1α
- hypoxia-inducible factor-1α
- MEF
- mouse embryonic fibroblast
- LSL
- lox-stop-lox
- DOX
- doxorubicin
- CHX
- cycloheximide
- TFMB-2-HG
- trifluoromethylbenzyl-(R)-2-HG
- miRNA
- microRNA
- qRT
- quantitative RT
- shRNA
- short hairpin RNA.
References
- 1. De Carli E., Wang X., and Puget S. (2009) IDH1 and IDH2 mutations in gliomas. New Engl. J. Med. 360, 2248; author reply 2249 [DOI] [PubMed] [Google Scholar]
- 2. Ducray F., Marie Y., and Sanson M. (2009) IDH1 and IDH2 mutations in gliomas. New Engl. J. Med. 360, 2248–2249; author reply 2249 10.1056/NEJMc090593 [DOI] [PubMed] [Google Scholar]
- 3. Parsons D. W., Jones S., Zhang X., Lin J. C., Leary R. J., Angenendt P., Mankoo P., Carter H., Siu I. M., Gallia G. L., Olivi A., McLendon R., Rasheed B. A., Keir S., Nikolskaya T., et al. (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321, 1807–1812 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Mardis E. R., Ding L., Dooling D. J., Larson D. E., McLellan M. D., Chen K., Koboldt D. C., Fulton R. S., Delehaunty K. D., McGrath S. D., Fulton L. A., Locke D. P., Magrini V. J., Abbott R. M., et al. (2009) Recurring mutations found by sequencing an acute myeloid leukemia genome. New Engl. J. Med. 361, 1058–1066 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Abbas S., Lugthart S., Kavelaars F. G., Schelen A., Koenders J. E., Zeilemaker A., van Putten W. J., Rijneveld A. W., Löwenberg B., and Valk P. J. (2010) Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood 116, 2122–2126 10.1182/blood-2009-11-250878 [DOI] [PubMed] [Google Scholar]
- 6. Amary M. F., Bacsi K., Maggiani F., Damato S., Halai D., Berisha F., Pollock R., O'Donnell P., Grigoriadis A., Diss T., Eskandarpour M., Presneau N., Hogendoorn P. C., Futreal A., Tirabosco R., and Flanagan A. M. (2011) IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 224, 334–343 10.1002/path.2913 [DOI] [PubMed] [Google Scholar]
- 7. Borger D. R., Tanabe K. K., Fan K. C., Lopez H. U., Fantin V. R., Straley K. S., Schenkein D. P., Hezel A. F., Ancukiewicz M., Liebman H. M., Kwak E. L., Clark J. W., Ryan D. P., Deshpande V., Dias-Santagata D., Ellisen L. W., Zhu A. X., and Iafrate A. J. (2012) Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist 17, 72–79 10.1634/theoncologist.2011-0386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pansuriya T. C., van Eijk R., d'Adamo P., van Ruler M. A., Kuijjer M. L., Oosting J., Cleton-Jansen A. M., van Oosterwijk J. G., Verbeke S. L., Meijer D., van Wezel T., Nord K. H., Sangiorgi L., Toker B., Liegl-Atzwanger B., et al. (2011) Somatic mosaic IDH1 and IDH2 mutations are associated with enchondroma and spindle cell hemangioma in Ollier disease and Maffucci syndrome. Nat. Genet. 43, 1256–1261 10.1038/ng.1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bleeker F. E., Lamba S., Leenstra S., Troost D., Hulsebos T., Vandertop W. P., Frattini M., Molinari F., Knowles M., Cerrato A., Rodolfo M., Scarpa A., Felicioni L., Buttitta F., Malatesta S., Marchetti A., and Bardelli A. (2009) IDH1 mutations at residue p.R132 (IDH1(R132)) occur frequently in high-grade gliomas but not in other solid tumors. Hum. Mutat. 30, 7–11 10.1002/humu.20937 [DOI] [PubMed] [Google Scholar]
- 10. Gaal J., Burnichon N., Korpershoek E., Roncelin I., Bertherat J., Plouin P. F., de Krijger R. R., Gimenez-Roqueplo A. P., and Dinjens W. N. (2010) Isocitrate dehydrogenase mutations are rare in pheochromocytomas and paragangliomas. J. Clin. Endocrinol. Metab. 95, 1274–1278 10.1210/jc.2009-2170 [DOI] [PubMed] [Google Scholar]
- 11. Kang M. R., Kim M. S., Oh J. E., Kim Y. R., Song S. Y., Seo S. I., Lee J. Y., Yoo N. J., and Lee S. H. (2009) Mutational analysis of IDH1 codon 132 in glioblastomas and other common cancers. Int. J. Cancer 125, 353–355 10.1002/ijc.24379 [DOI] [PubMed] [Google Scholar]
- 12. Sequist L. V., Heist R. S., Shaw A. T., Fidias P., Rosovsky R., Temel J. S., Lennes I. T., Digumarthy S., Waltman B. A., Bast E., Tammireddy S., Morrissey L., Muzikansky A., Goldberg S. B., Gainor J., et al. (2011) Implementing multiplexed genotyping of non-small-cell lung cancers into routine clinical practice. Ann. Oncol. 22, 2616–2624 10.1093/annonc/mdr489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sjoblöm T., Jones S., Wood L. D., Parsons D. W., Lin J., Barber T. D., Mandelker D., Leary R. J., Ptak J., Silliman N., Szabo S., Buckhaults P., Farrell C., Meeh P., Markowitz S. D., et al. (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314, 268–274 10.1126/science.1133427 [DOI] [PubMed] [Google Scholar]
- 14. Dang L., White D. W., Gross S., Bennett B. D., Bittinger M. A., Driggers E. M., Fantin V. R., Jang H. G., Jin S., Keenan M. C., Marks K. M., Prins R. M., Ward P. S., Yen K. E., Liau L. M., et al. (2009) Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 462, 739–744 10.1038/nature08617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ward P. S., Patel J., Wise D. R., Abdel-Wahab O., Bennett B. D., Coller H. A., Cross J. R., Fantin V. R., Hedvat C. V., Perl A. E., Rabinowitz J. D., Carroll M., Su S. M., Sharp K. A., Levine R. L., and Thompson C. B. (2010) The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting α-ketoglutarate to 2-hydroxyglutarate. Cancer Cell 17, 225–234 10.1016/j.ccr.2010.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Figueroa M. E., Abdel-Wahab O., Lu C., Ward P. S., Patel J., Shih A., Li Y., Bhagwat N., Vasanthakumar A., Fernandez H. F., Tallman M. S., Sun Z., Wolniak K., Peeters J. K., et al. (2010) Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell 18, 553–567 10.1016/j.ccr.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chowdhury R., Yeoh K. K., Tian Y. M., Hillringhaus L., Bagg E. A., Rose N. R., Leung I. K., Li X. S., Woon E. C., Yang M., McDonough M. A., King O. N., Clifton I. J., Klose R. J., et al. (2011) The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 12, 463–469 10.1038/embor.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu W., Yang H., Liu Y., Yang Y., Wang P., Kim S. H., Ito S., Yang C., Wang P., Xiao M. T., Liu L. X., Jiang W. Q., Liu J., Zhang J. Y., et al. (2011) Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 10.1016/j.ccr.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Raymond C. K., Roberts B. S., Garrett-Engele P., Lim L. P., and Johnson J. M. (2005) Simple, quantitative primer-extension PCR assay for direct monitoring of microRNAs and short-interfering RNAs. RNA 11, 1737–1744 10.1261/rna.2148705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noushmehr H., Weisenberger D. J., Diefes K., Phillips H. S., Pujara K., Berman B. P., Pan F., Pelloski C. E., Sulman E. P., Bhat K. P., Verhaak R. G., Hoadley K. A., Hayes D. N., Perou C. M., et al. (2010) Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell 17, 510–522 10.1016/j.ccr.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Turcan S., Rohle D., Goenka A., Walsh L. A., Fang F., Yilmaz E., Campos C., Fabius A. W., Lu C., Ward P. S., Thompson C. B., Kaufman A., Guryanova O., Levine R., Heguy A., et al. (2012) IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 483, 479–U137 10.1038/nature10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sasaki M., Knobbe C. B., Munger J. C., Lind E. F., Brenner D., Brüstle A., Harris I. S., Holmes R., Wakeham A., Haight J., You-Ten A., Li W. Y., Schalm S., Su S. M., Virtanen C., et al. (2012) IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature 488, 656–659 10.1038/nature11323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lu C., Ward P. S., Kapoor G. S., Rohle D., Turcan S., Abdel-Wahab O., Edwards C. R., Khanin R., Figueroa M. E., Melnick A., Wellen K. E., O'Rourke D. M., Berger S. L., Chan T. A., Levine R. L., et al. (2012) IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 483, 474–478 10.1038/nature10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhao S., Lin Y., Xu W., Jiang W., Zha Z., Wang P., Yu W., Li Z., Gong L., Peng Y., Ding J., Lei Q., Guan K. L., and Xiong Y. (2009) Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science 324, 261–265 10.1126/science.1170944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 26. Levine A. J., Hu W., and Feng Z. (2006) The P53 pathway: what questions remain to be explored? Cell Death Differ. 13, 1027–1036 10.1038/sj.cdd.4401910 [DOI] [PubMed] [Google Scholar]
- 27. Ko L. J., and Prives C. (1996) p53: puzzle and paradigm. Genes Dev. 10, 1054–1072 10.1101/gad.10.9.1054 [DOI] [PubMed] [Google Scholar]
- 28. Ventura A., Kirsch D. G., McLaughlin M. E., Tuveson D. A., Grimm J., Lintault L., Newman J., Reczek E. E., Weissleder R., and Jacks T. (2007) Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 10.1038/nature05541 [DOI] [PubMed] [Google Scholar]
- 29. Xue W., Zender L., Miething C., Dickins R. A., Hernando E., Krizhanovsky V., Cordon-Cardo C., and Lowe S. W. (2007) Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 10.1038/nature05529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hollstein M., Sidransky D., Vogelstein B., and Harris C. C. (1991) p53 mutations in human cancers. Science 253, 49–53 10.1126/science.1905840 [DOI] [PubMed] [Google Scholar]
- 31. Inoue S., Li W. Y., Tseng A., Beerman I., Elia A. J., Bendall S. C., Lemonnier F., Kron K. J., Cescon D. W., Hao Z., Lind E. F., Takayama N., Planello A. C., Shen S. Y., Shih A. H., et al. (2016) Mutant IDH1 downregulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell 30, 337–348 10.1016/j.ccell.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sasaki M., Knobbe C. B., Itsumi M., Elia A. J., Harris I. S., Chio I. I., Cairns R. A., McCracken S., Wakeham A., Haight J., Ten A. Y., Snow B., Ueda T., Inoue S., Yamamoto K., et al. (2012) d- 2-hydroxyglutarate produced by mutant IDH1 perturbs collagen maturation and basement membrane function. Genes Dev. 26, 2038–2049 10.1101/gad.198200.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li C., and Evans R. M. (1997) Ligation independent cloning irrespective of restriction site compatibility. Nucleic Acids Res. 25, 4165–4166 10.1093/nar/25.20.4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gross S., Cairns R. A., Minden M. D., Driggers E. M., Bittinger M. A., Jang H. G., Sasaki M., Jin S., Schenkein D. P., Su S. M., Dang L., Fantin V. R., and Mak T. W. (2010) Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J. Exp. Med. 207, 339–344 10.1084/jem.20092506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Rohle D., Popovici-Muller J., Palaskas N., Turcan S., Grommes C., Campos C., Tsoi J., Clark O., Oldrini B., Komisopoulou E., Kunii K., Pedraza A., Schalm S., Silverman L., Miller A., et al. (2013) An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science 340, 626–630 10.1126/science.1236062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mazumder B., Seshadri V., and Fox P. L. (2003) Translational control by the 3′-UTR: the ends specify the means. Trends Biochem. Sci. 28, 91–98 10.1016/S0968-0004(03)00002-1 [DOI] [PubMed] [Google Scholar]
- 37. Bartel D. P. (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 38. Kumar M., Lu Z., Takwi A. A., Chen W., Callander N. S., Ramos K. S., Young K. H., and Li Y. (2011) Negative regulation of the tumor suppressor p53 gene by microRNAs. Oncogene 30, 843–853 10.1038/onc.2010.457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Le M. T., Teh C., Shyh-Chang N., Xie H., Zhou B., Korzh V., Lodish H. F., and Lim B. (2009) MicroRNA-125b is a novel negative regulator of p53. Genes Dev. 23, 862–876 10.1101/gad.1767609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tian S., Huang S., Wu S., Guo W., Li J., and He X. (2010) MicroRNA-1285 inhibits the expression of p53 by directly targeting its 3′ untranslated region. Biochem. Biophys. Res. Commun. 396, 435–439 10.1016/j.bbrc.2010.04.112 [DOI] [PubMed] [Google Scholar]
- 41. Swarbrick A., Woods S. L., Shaw A., Balakrishnan A., Phua Y., Nguyen A., Chanthery Y., Lim L., Ashton L. J., Judson R. L., Huskey N., Blelloch R., Haber M., Norris M. D., Lengyel P., et al. (2010) miR-380-5p represses p53 to control cellular survival and is associated with poor outcome in MYCN-amplified neuroblastoma. Nat. Med. 16, 1134–1140 10.1038/nm.2227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., and Kaelin W. G. Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 10.1126/science.1059817 [DOI] [PubMed] [Google Scholar]
- 43. Dubbink H. J., Taal W., van Marion R., Kros J. M., van Heuvel I., Bromberg J. E., Zonnenberg B. A., Zonnenberg C. B., Postma T. J., Gijtenbeek J. M., Boogerd W., Groenendijk F. H., Smitt P. A., Dinjens W. N., and van den Bent M. J. (2009) IDH1 mutations in low-grade astrocytomas predict survival but not response to temozolomide. Neurology 73, 1792–1795 10.1212/WNL.0b013e3181c34ace [DOI] [PubMed] [Google Scholar]
- 44. Watanabe T., Nobusawa S., Kleihues P., and Ohgaki H. (2009) IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am. J. Pathol. 174, 1149–1153 10.2353/ajpath.2009.080958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Toledo F., and Wahl G. M. (2006) Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer 6, 909–923 10.1038/nrc2012 [DOI] [PubMed] [Google Scholar]
- 46. Losman J. A., Looper R. E., Koivunen P., Lee S., Schneider R. K., McMahon C., Cowley G. S., Root D. E., Ebert B. L., and Kaelin W. G. Jr. (2013) (R)-2-Hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science 339, 1621–1625 10.1126/science.1231677 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.