Abstract
4-1BBL is a member of the tumor necrosis factor (TNF) superfamily and is the ligand for the TNFR superfamily receptor, 4-1BB. 4-1BB plays an immunomodulatory role in T cells and NK cells, and agonists of this receptor have garnered strong attention as potential immunotherapy agents. Broadly speaking, the structural features of TNF superfamily members, their receptors, and ligand-receptor complexes are similar. However, a published crystal structure of human 4-1BBL suggests that it may be unique in this regard, exhibiting a three-bladed propeller-like trimer assembly that is distinctly different from that observed in other family members. This unusual structure also suggests that the human 4-1BB/4-1BBL complex may be structurally unique within the TNF/TNFR superfamily, but to date no structural data have been reported. Here we report the crystal structure of the human 4-1BB/4-1BBL complex at 2.4-Å resolution. In this structure, 4-1BBL does not adopt the unusual trimer assembly previously reported, but instead forms a canonical bell-shaped trimer typical of other TNF superfamily members. The structure of 4-1BB is also largely canonical as is the 4-1BB/4-1BBL complex. Mutational data support the 4-1BBL structure reported here as being biologically relevant, suggesting that the previously reported structure is not. Together, the data presented here offer insight into structure/function relationships in the 4-1BB/4-1BBL system and improve our structural understanding of the TNF/TNFR superfamily more broadly.
Keywords: structural biology, structure-function, T-cell biology, crystal structure, tumor necrosis factor (TNF), protein-protein interaction, protein structure
Introduction
Tumor necrosis factor receptor (TNFR)3 superfamily receptors are critical regulators of diverse biological processes and play a particularly important role in the regulation of immune cell biology. Recently, it has been shown that agonism of various co-stimulatory members of the TNFR superfamily can help to activate useful immune cell subsets and elicit anti-tumor immune responses (1, 2). 4-1BB (also known as CD137 and TNFRSF9) is one family member that has drawn strong attention in this regard (3–5). Structural data revealing how TNFR superfamily receptors engage their ligands has shed light on the basic mechanism of these important signaling systems and led to a general paradigm for signaling in this receptor family (6, 7). Cell surface-bound trimeric ligands engage receptor molecules on the surface of another cell in 3:3 stoichiometry bringing large numbers of receptor chains into close proximity. This receptor clustering facilitates recruitment and cooperative assembly of downstream signaling molecules that associate with the receptor cytoplasmic domains and each other, to transduce and amplify the output signal (8). 4-1BB would be expected to exhibit a similar signaling mechanism. However, despite a reported structure of human 4-1BB ligand (4-1BBL, also known as CD137L and TNFSF9) (9) and murine 4-1BB (10) to date, no structural data have been reported for the 4-1BB/4-1BBL complex.
Broadly speaking, most TNFR superfamily ligands are structurally similar. They are type II transmembrane proteins with a single conserved extracellular domain known as the TNF homology domain (THD). THDs all adopt a jellyroll β-sandwich fold consisting of two β-sheets comprised of strands A′AHCF and B′BGDE lettered alphabetically from the N to C terminus. These THDs trimerize through packing of the A′AHCF (inner) sheet of one monomer with the inner sheet of the neighboring monomer as well as the edge E-strand of the outer B′BGDE sheet in a closed loop arrangement. The overall structure of these trimers is usually a condensed bell shape (Fig. 1A), although in some family members (e.g. OX40, GITRL), monomer units are shorter and more splayed resembling a blooming flower (Fig. 1B) (6, 11, 12). A published crystal structure of human 4-1BBL suggests that it is a notable exception to this architecture (9). In the reported structure, an altered β-sheet structure is observed in which sheets are comprised by strands A′HCF and ABGDE instead of the typical arrangement described above and trimer assembly is driven by interaction of the A′HCF sheet with a linear stretch of amino acids at the C terminus of the neighboring monomer that lie outside the THD. The result is a highly divergent three-bladed propeller structure (Fig. 1C). TNFR superfamily members canonically bind their ligands at the interface between ligand monomer units (6). The unusual trimer assembly of 4-1BBL in the reported crystal structure results in monomer–monomer interfaces that are radically different from that in other TNFR superfamily ligands. Thus, the structure of the 4-1BB/4-1BBL complex could also be quite different from what has been observed for other family members.
Figure 1.
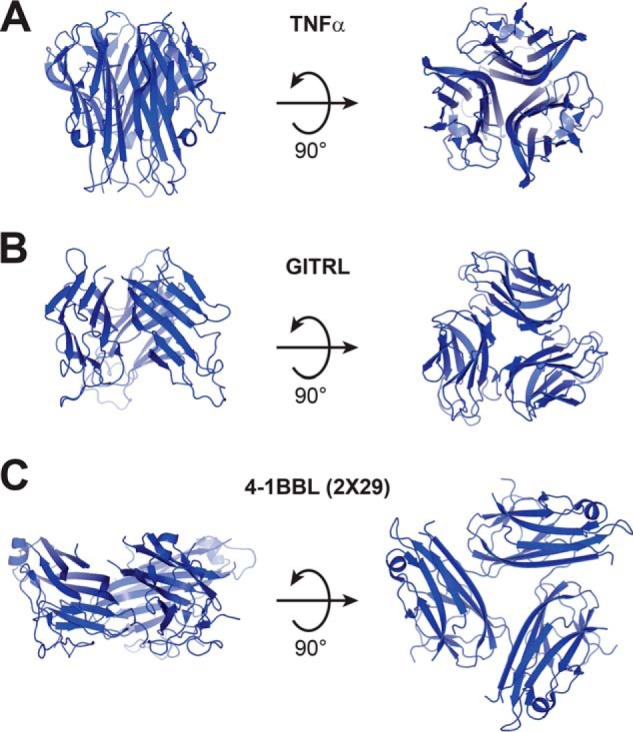
Structural features of TNFR superfamily ligands. A, TNFα is representative of the canonical bell-shaped trimer structure observed for most family members (PDB code 3ALQ). B, GITRL is representative of an atypical subgroup of ligands with generally similar structure, but a shorter THD and a more open blooming flower-like trimer conformation (PDB code 2Q1M). C, a published crystal structure of 4-1BBL (e4-1BBL as described by Won et al. (9), PDB ID 2X29) showing a highly unusual three-bladed propeller structure, which is distinct from all other TNFR superfamily ligands.
To better understand the 4-1BB/4-1BBL interaction and gain further insight into the signaling mechanism of this receptor system, we set out to solve the crystal structure of the human 4-1BB/4-1BBL complex. Here, we report this structure, solved at 2.4-Å resolution. Notably, we were able to crystallize the complex using a 4-1BBL construct containing only the THD. This construct excludes a significant number of residues that appeared to be involved in trimer formation and/or monomer folding in the previously reported 4-1BBL structure. Despite deletion of these residues, we found that the THD-only 4-1BBL retained a stable trimer structure in solution, bound to 4-1BB, and retained 4-1BB agonist activity. In the crystal structure of the 4-1BB/4-1BBL complex, we did not observe the unusual three-bladed propeller structure previously reported for 4-1BBL, but instead observed a trimer structure that closely resembles that of other bell-shaped TNF superfamily members with receptor interactions that are also largely canonical. Published mutagenesis data as well as data of our own support this structure as being biologically relevant. The findings presented here contribute to our mechanistic understanding of 4-1BB signaling and provide insight for the design of molecules to stimulate or antagonize this signaling for therapeutic or academic purposes.
Results
Residues flanking the THD of 4-1BBL can be deleted/mutated without functional consequence
A previously described attempt to crystallize the human 4-1BB/4-1BBL complex was unsuccessful (9). One possible reason for this failure was the inclusion of a substantial number of residues at the N and C termini of the 4-1BBL extracellular domain that are not part of the THD and are disordered in the reported crystal structure of 4-1BBL (9). These extra residues in the Won et al. (9) construct, termed “e4-1BBL” and consisting of residues 58–254 of the full-length protein, may disrupt crystal packing of the 4-1BBL/4-1BB complex. Minimizing the number of these residues may improve the likelihood of crystallization. Constructs consisting of only the THD have been used successfully in crystallizing other TNF superfamily ligands and ligand/receptor complexes. However, some residues outside the THD of 4-1BBL (comprised by residues 91–240 (6)) appear to contribute to the folded monomer structure and trimer formation in the previously reported e4-1BBL structure, specifically, residues 80–90 and 241–246.
Despite the apparent contributions of extra-THD residues to trimer formation and monomer folding in the e4-1BBL structure, we considered the possibility that these residues may not, in fact, be required for proper folding and trimer assembly. A cross-species 4-1BBL sequence comparison shows low sequence conservation across residues 80–90 and 241–246 compared with regions within the THD, even at positions that are heavily buried in the e4-1BBL trimer interface or contributing to the e4-1BBL monomer hydrophobic core (Fig. S1). Such low conservation is unexpected at positions that are structurally or functionally important. Furthermore, a structure of the closest sequence homolog of 4-1BBL, LIGHT, published after the e4-1BBL structure, shows a canonical bell-shaped trimer structure (13). Thus, we questioned whether 4-1BBL might be capable of forming a canonical trimer structure in which case the previously reported structure could be inaccurate.
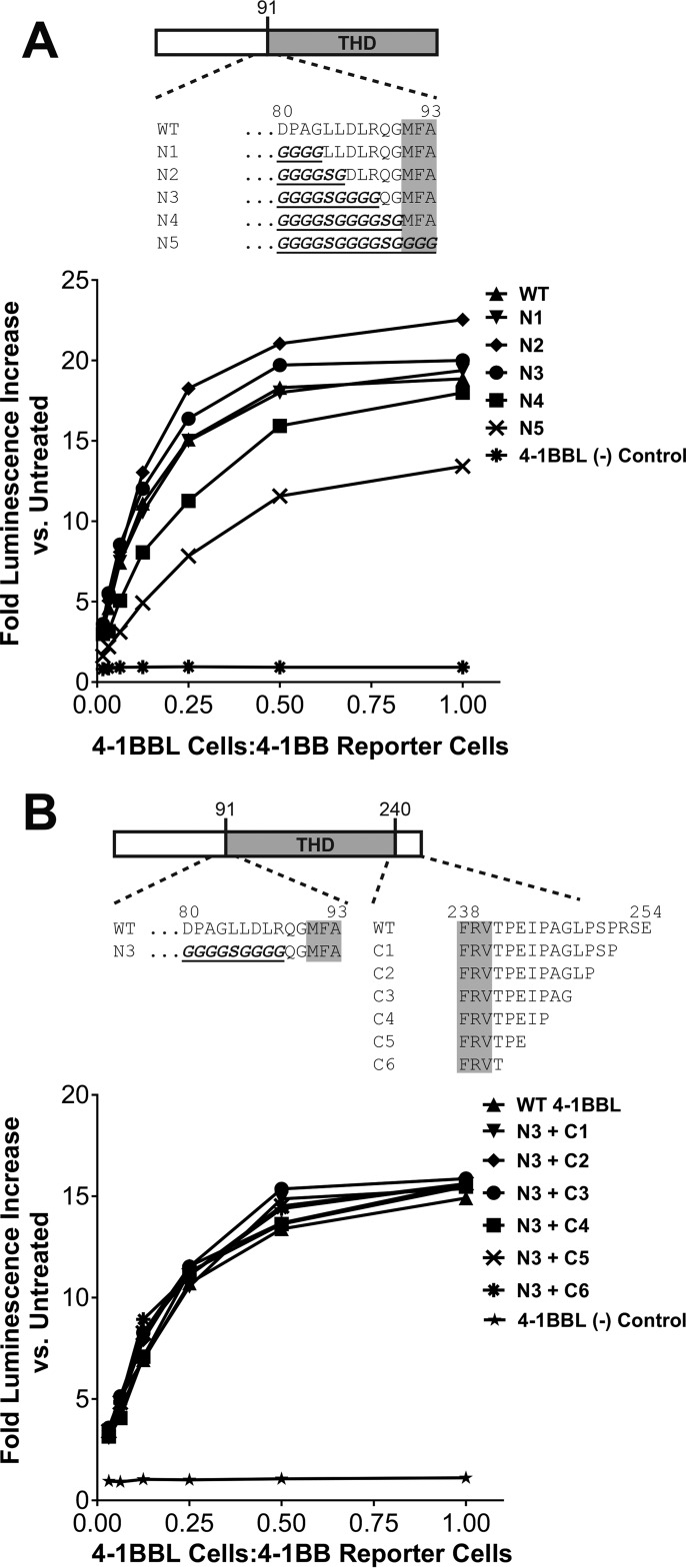
To further explore our hypothesis, we deleted/mutated the residues flanking the 4-1BBL THD and assessed the structural and functional consequences. We initially explored this question in the context of native cell-surface 4-1BBL using a cell line expressing full-length WT 4-1BBL, or a series of variants in which the residues immediately preceding or following the THD were mutated or deleted, respectively. We co-cultured these 4-1BBL expressing cells with a 4-1BB-dependent NF-κB luciferase reporter cell line to assess any functional impact of the mutations on 4-1BB signaling. We found that most residues upstream of the THD's N-terminal boundary that were included in e4-1BBL could be mutated to Gly/Ser without impact on the ligand's ability to drive 4-1BB signaling (Fig. 2A). Mutation of residues 80–88 had essentially no impact on function versus WT. However, mutation of residues 89–93 appeared to adversely affect function in a progressive manner. This result approximated the N-terminal functional edge of the THD as residue 89. Notably, residues 80–88 appeared to be dispensable in this experiment despite their contribution to the folded monomer structure of e4-1BBL.
Figure 2.
Residues outside the THD of 4-1BBL are not required for function. A, HEK293 cells expressing full-length WT 4-1BBL or variants in which an increasing number of residues upstream of the N-terminal boundary of the TNF homology domain were mutated to Gly/Ser (labeled N1–N5) were incubated with 4-1BB-dependent NF-κB luciferase reporter cells to measure the 4-1BB agonist activity of the ligand constructs. Sequence details for each construct are shown in the schematic. B, same as in A except that 4-1BBL residues 80–88 were fixed as Gly4SerGly4 (N3 variant from A) and an increasing number of residues extending beyond the C-terminal boundary of the THD were deleted in constructs labeled C1–C6. Sequence details for each construct are shown in the schematic. In both experiments 293 cells not expressing 4-1BBL were included as a negative control.
With residues 80–88 fixed as Gly4SerGly4, we subsequently found that essentially all residues beyond the C-terminal end of the 4-1BBL THD (residues 242–254) could be deleted without functional consequence in the 4-1BB reporter assay (Fig. 2B). Notably, many of these residues appear to contribute substantially to the trimer interface in the e4-1BBL structure. Although inconsistent with the previously reported structure, the collective results from these mutagenesis studies indicated that residues outside the THD of 4-1BBL are not required for productive engagement with 4-1BB. This suggested that a “THD-only” construct might be useful for crystallizing the 4-1BBL/4-1BB complex.
A THD-only 4-1BBL construct forms a stable trimer in solution and maintains receptor-binding and agonist functions
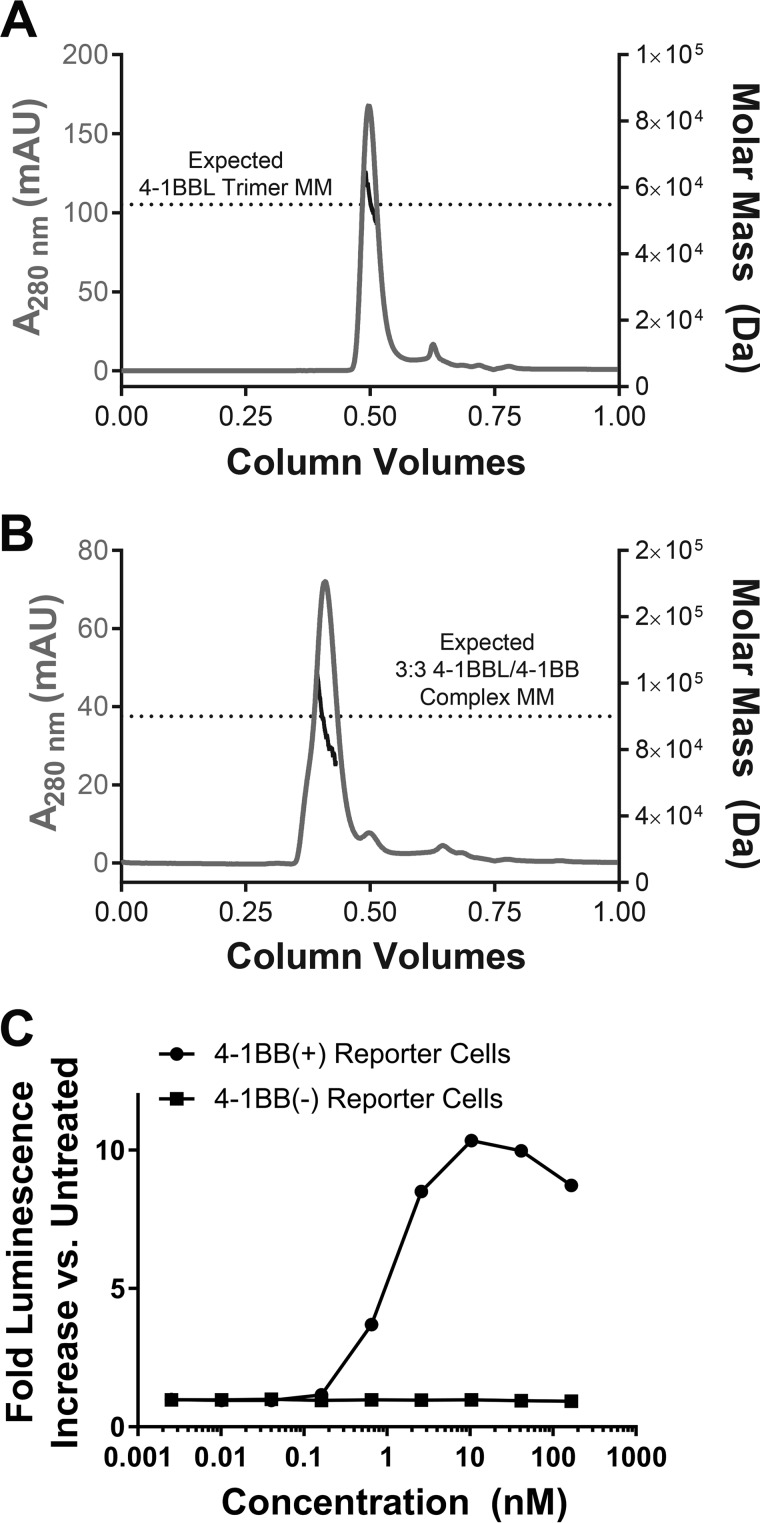
Having observed that residues outside the 4-1BBL THD did not appear important for function in native cell-surface 4-1BBL, we next sought to explore whether a soluble THD-only 4-1BBL construct would maintain the trimeric structure and function in solution. We successfully expressed and purified THD-only 4-1BBL (residues 89–242) as a soluble protein from Escherichia coli. Size exclusion chromatography revealed that THD-only 4-1BBL was monodisperse in solution and static light-scattering data indicated an averaged molar mass of 55,300 Da, consistent with a trimeric assembly of the recombinant protein (expected molar mass of 54,897 Da) (Fig. 3A). We also sought to confirm the ability of the THD-only 4-1BBL trimer to form a complex with the extracellular domain of 4-1BB. We expressed the 4-1BB extracellular region for this purpose (residues 24-160), where this construct contained three mutations to eliminate an unpaired cysteine residue and potential N-linked glycosylation sites that could otherwise introduce heterogeneity unfavorable for subsequent crystallization (see “Materials and methods”). We mixed THD-only 4-1BBL with the 4-1BB construct described at an ∼1:1 molar ratio and examined the mixture by size exclusion chromatography (SEC) and light scattering. The mixture eluted from the SEC column in a single peak at a shorter retention time versus 4-1BBL alone and light-scattering data gave an increased averaged molar mass of 90,600 Da, consistent with complex formation (Fig. 3B). This value is slightly less than the expected molar mass of 100,196 Da for the 3:3 4-1BBL/4-1BB complex. However, the difference (∼9600 Da) is less than the molar mass of one 4-1BB monomer (15,100 Da) and was likely due to inaccuracy in the mixing stoichiometry, and/or some degree of dissociation during the chromatographic separation.
Figure 3.
THD-only 4-1BBL is a biologically active stable trimer in solution. A, SEC chromatogram and overlaid plot of static light-scattering derived molar mass data for THD-only 4-1BBL. The expected trimer molar mass is indicated with a dotted line. B, SEC chromatogram and overlaid plot of static light-scattering derived molar mass data for the THD-only 4-1BBL/4-1BB complex with components mixed at ∼1:1 molar ratio. The expected molar mass for the 3:3 4-1BB/4-1BBL complex is indicated with a dotted line. C, 4-1BB NF-κB luciferase reporter assay activity for THD-only 4-1BBL. Assay is as described in the legend to Fig. 2 and under “Materials and methods.” Data are shown using reporter cells that are either positive or negative for expression of 4-1BB.
We sought to further confirm that THD-only 4-1BBL adopted a functionally relevant confirmation by testing its activity in the 4-1BB NF-κB reporter cell assay. In this assay, THD-only 4-1BBL successfully drove 4-1BB NF-κB signaling with an EC50 of ∼2 nm, suggesting high-affinity of the 4-1BBL trimer for the cellular 4-1BB receptor (Fig. 3C). Together, SEC/light-scattering data and reporter assay results show that THD-only 4-1BBL forms a stable trimer in solution, maintains its ability to bind 4-1BB, and adopts a functionally relevant conformation.
The structure of THD-only 4-1BBL
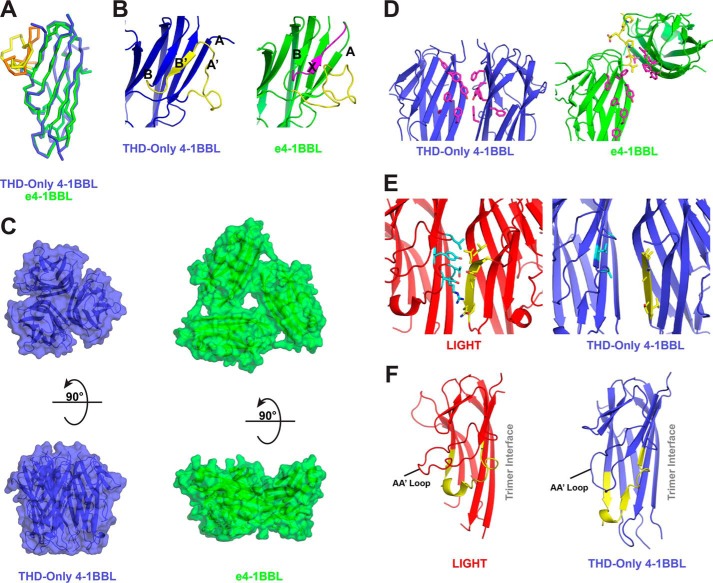
Having confirmed the structural and functional integrity of THD-only 4-1BBL, we crystallized its complex with the 4-1BB extracellular domain and determined its structure at 2.4-Å resolution (Table 1). The monomer structure of THD-only 4-1BBL shows that it is globally quite similar (r.m.s. deviation of 0.7 Å for β-sheet Cα atoms) to the e4-1BBL structure previously reported by Won et al. (9) and exhibits the jellyroll β-sandwich architecture typical of all TNF superfamily members (6, 7) (Fig. 4A). Locally, however, there is one notable difference. In the THD-only structure, residues 111–130 adopt a conformation similar to that observed in most other TNF superfamily members. Together, these residues form β-strands A′ and B′ with a short loop connecting them. These A′ and B′ strands dock in antiparallel fashion against strands A and B, respectively and cap the edge of the β-sandwich (Fig. 4B). In contrast, in the e4-1BBL structure, equivalent strands to A′ and B′ are absent. Instead, N-terminal residues outside the THD (residues 80–88) form a unique strand (here termed strand X) that docks onto the edge of strand B in parallel fashion where the antiparallel B′ strand would ordinarily be found in TNF superfamily members (Fig. 4B). This displaces the residues that would typically comprise strands A′ and B′ and results in an extended loop that protrudes off the side of the 4-1BBL monomer in e4-1BBL. This feature is not observed in THD-only 4-1BBL.
Table 1.
Crystallographic data refinement and statistics
| Data statistics | |
| Space group | I213 |
| Cell parameters (Å, °) | a = b = c = 161.57, α = β = γ = 90 |
| Mosaicity (°) | 0.08 |
| Resolution limits (Å)a | 38.00–2.40 (2.46–2.40)b |
| Wavelength (Å) | 0. 97946 |
| Rmerge | 0.165 (1.449) |
| Rmeas | 0.173 (1.523) |
| Rpim | 0.053 (0.467) |
| I/σ(I) | 12.4 (2.5) |
| CC1/2 | 0.99 (0.80) |
| Completeness (%) | 100 (100) |
| Redundancy | 20.6 (20.6) |
| Refinement statistics | |
| Rwork/Rfree | 20.3/24.7 (32.7/34.5) |
| No. of non-hydrogen atoms | 4200 |
| B factor (Wilson/mean, Å2) | 71/64.8 |
| Bond lengths (Å) | 0.013 |
| Bond angles (°) | 1.564 |
| Ramachandran (additional allowed, %) | 0.6 |
a Resolution limit estimates are based on CC1/2 > 0.30; Mn(I/σ)>2.00.
b Values in parentheses are for highest resolution shell.
Figure 4.
The structure of THD-only 4-1BBL. A, overlay of THD-only 4-1BBL monomer (blue) with e4-1BBL monomer from PDB code 2X29 (green). The THD-only 4-1BBL construct is comprised of residues 89–242 of the full-length protein, whereas e4-1BBL is comprised of residues 58–254. The 111–130 residue region, which differs significantly between the two structures, is highlighted for THD-only 4-1BBL (orange) and e4-1BBL (yellow). B, structural comparison of the 111–130 residue region (yellow) in THD-only 4-1BBL (blue, left) and e4-1BBL (green, right). The “X” strand that displaces the A′ and B′ strands in e4-1BBL is highlighted (magenta). C, comparison of the trimer structures of THD-only 4-1BBL (blue) and e4-1BBL (green). D, detailed view of the trimer interface of THD-only 4-1BBL (blue, left) and e4-1BBL (green, right). Tiled aromatic residues that typically pack together to form the trimer core in TNFR superfamily ligands (magenta sticks) are shown as is a linear stretch of C-terminal residues that participates in the e4-1BBL trimer interface (yellow sticks). For each illustration, the monomer on the left-hand side is shown in the same orientation. E, comparison of E-strand contacts in the trimer interfaces of LIGHT (left, red) and THD-only 4-1BBL (right, blue). E-strand residues (yellow) and E-strand contacting residues (cyan) are shown as sticks. F, comparison of GH loop (yellow) conformations in LIGHT (left, red) and THD-only 4-1BBL (right, blue). The AA′ loop, which changes conformation, and the GH loop are indicated as is the side of the monomer involved in the trimer interface.
Another striking difference between THD-only 4-1BBL and e4-1BBL is in the architecture of the trimer assembly. THD-only 4-1BBL forms a symmetric, bell-shaped homotrimer as observed for other TNF superfamily members (Fig. 4C) (6, 14). This is in sharp contrast to the atypical three-bladed propeller trimer assembly observed in e4-1BBL (Fig. 4C). The trimer interface in the THD-only 4-1BBL structure is typical for the TNF superfamily with tiled aromatic residues from the “inner” A′AHCF sheets of neighboring monomers packing together to help form the core of the trimer assembly (12, 15) (Fig. 4D). In e4-1BBL, instead of interacting with one another, these same inner sheet aromatics interact with a linear stretch of residues that extend beyond the C-terminal boundary of the THD (residues 240–246) to form an unusual trimer interface (Fig. 4D). Notably, most of these C-terminal residues are absent in THD-only 4-1BBL.
Although THD-only 4-1BBL forms a largely canonical trimer structure, at ∼660 Å2, the size of the THD-only 4-1BBL trimer interface is somewhat smaller than for most other TNF superfamily members (typically 900–1000 Å2). LIGHT is 4-1BBL's closest homolog by sequence. PDBeFold also identifies LIGHT as the closest structural homolog by monomer structure and PISA identifies the LIGHT trimer as having one of the most similar trimer interfaces to THD-only 4-1BBL (16, 17). A comparison with LIGHT provides some insight into the reduced trimer interface size in 4-1BBL. In the LIGHT trimer and the trimers of most other canonical TNF superfamily members, inner sheet residues of neighboring monomers pack closely with each other and also with the edge of the neighboring β-sandwich. In LIGHT, E-strand residues at the β-sheet edge bury 370 Å2 of surface out of a total of 915 A2 in the monomer–monomer interface. In contrast in the THD-only 4-1BBL structure, the equivalent E-strand residues bury just 92 Å2. This difference largely accounts for the reduced trimer interface in 4-1BBL as compared with LIGHT. The reduction in E-strand interactions in the 4-1BBL trimer is attributable to a large number of side chain truncating substitutions versus LIGHT at E-strand positions (E178A, L179A, V181A, Q183T, and Q184V using LIGHT numbering) and at positions on the neighboring monomer that normally interact with these E-strand residues (L120G, R232A, and Y234V using LIGHT numbering) (Fig. 4E). Furthermore, two additional residues that contribute to E-strand contacts in LIGHT (Asp-229 and Thr-231) have no structural equivalent in 4-1BBL due to differences in GH loop and H-strand conformations.
In both the THD-only 4-1BBL and previously reported e4-1BBL structures, the G- and H-strands of 4-1BBL are longer than in other TNFR superfamily ligands. As a result, the GH loop is positioned adjacent to the CD loop at the bottom of the 4-1BBL monomer. In contrast, for other family members the GH loop is positioned higher and tucks underneath the AA′ loop causing it to protrude out further than in 4-1BBL (Fig. 4F). The end result is a 4-1BBL monomer that is wider at the bottom and narrower at the middle/top than other family members. This in turn gives rise to a trimer that is more box-like than the typical bell-shape observed for the family (e.g. TNFα, Fig. 1A). Furthermore, the change in GH loop conformation pulls GH loop residues away from the trimer interface and, as a result, these residues contribute fewer monomer–monomer contacts than in LIGHT or other family members (Fig. 4F). Loss of these contacts coupled with the side chain truncations described above lead to the reduced trimer interface size observed and a distinct groove between 4-1BBL monomers that is plainly visible when viewing the trimer along its symmetry axis (Fig. 4C).
The structure of 4-1BB
Globally, human 4-1BB is similar to other TNFR superfamily members with an elongated structure comprised by small repeating cysteine-rich motifs constrained by disulfide bonds (Fig. 5A). Naismith and Sprang (18) established that these repeating motifs could be classified into A- and B-types according to their sequence and structural characteristics. A- and B-type modules typically pack together in pairs to yield the canonical cysteine-rich domain (CRD) units characteristic of TNFR superfamily members (18). Using the Naismith/Sprang nomenclature, 4-1BB is comprised from the N to C terminus by modules of types B2, A1, B2, A2, A1, A1, B1, consistent with predictions based on primary sequence. With the exception of the first B2 module, which forms a truncated CRDI, these modules combine in pairs to give CRDs II–IV.
Figure 5.
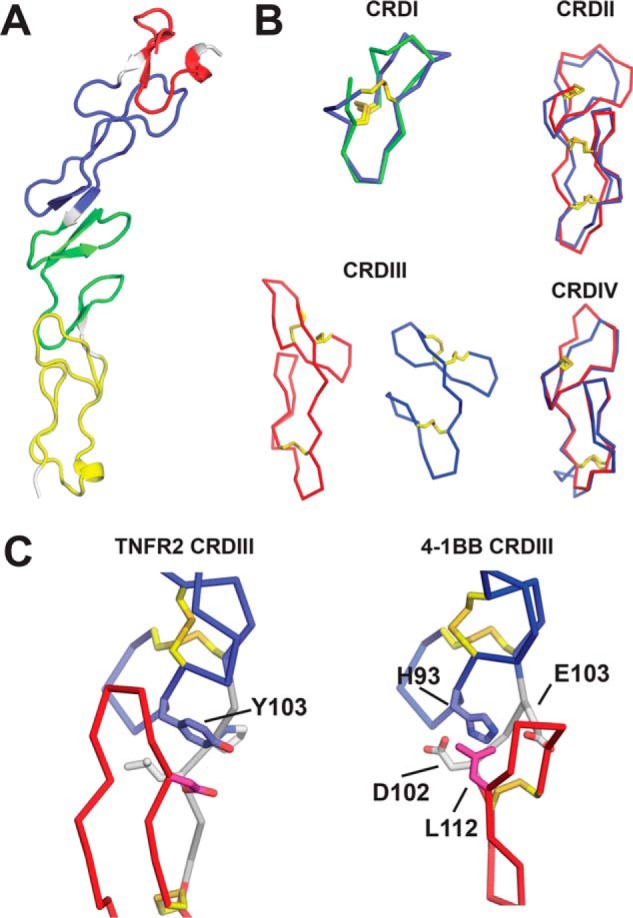
The structure of 4-1BB. A, cartoon of the 4-1BB structure with cysteine-rich domain I (CRDI) (red), CRDII (blue), CRDIII (green), and CRDIV (yellow) indicated. Linker regions connecting the CRDs are colored gray. B, comparison of 4-1BB CRD structures (blue) with those of TNFR2 (red) and TRAILR2 (green). Disulfide bonds (yellow sticks) are also shown. C, comparison of the A2/B1 interface in TNFR2 CRDIII to the A2/A1 interface in 4-1BB CRDIII. In both structures, the N-terminal A2 modules (blue) and downstream B1 or A1 modules (red) are indicated. Linker regions connecting the modules (gray) are also shown as are disulfide bonds (yellow sticks). Key residues involved in the A2/B1 and A2/A1 interfaces are shown and labeled (sticks) and colored to match the module in which they are located.
PDBeFold identifies TNFR2 as the closest sequence/structural homolog of 4-1BB (17). CRDII and -IV in 4-1BB are canonical in structure and very similar to CRDII and -IV of TNFR2 (Cα r.m.s. deviation = 1.51 Å for 36/39 residues in CRDI, Cα r.m.s. deviation = 1.20 Å for 31/34 residues in CRDIV) (Fig. 5B). This close resemblance to TNFR2 CRDs also places CRDII and -IV of 4-1BB in the “L-2” and “L-4” structural families identified by Magis et al. (14) which, respectively, contain the CRDII and CRDIV structures from many TNFR superfamily members. CRDI and -III, however, are more atypical. CRDI is truncated, containing only a single unusually compact B2 module with a short helical element in place of what would typically be strand 1 of the unit. Notably, this N-terminal B2 domain quite closely resembles the “N1” domain found at the N terminus of TRAILR2 (Cα r.m.s. deviation = 0.548 Å for 11/14 residues in TRAILR2), which also lacks a preceding A-module and mimics a B2 domain, but contains only one disulfide (Fig. 5B). CRDIII is also atypical containing an A2 and A1 module instead of a typical A/B pair. Interestingly, despite this unusual arrangement, 4-1BB CRDIII mimics the overall shape of the TNFR2 CRDIII, but is more compact (Fig. 5B). Furthermore, the interface between these A2 and A1 modules in 4-1BB mimics that usually seen for an A/B pair. In an A/B CRD, a conserved aromatic residue in the A-module typically folds down and packs tightly against the loop of the B-module, which extends upwards. This can be seen for Tyr-103 in the canonical CRDIII of TNFR2 (Fig. 5C). In 4-1BB CRDIII, His-93 assumes this role and, with neighboring Glu-103 and Asp-105 residues charge compensating/hydrogen bonding, it folds and packs closely against Leu-112 of the A1 module below (Fig. 5C). In this way, the downstream A1 module in 4-1BBL CRDIII mimics a B-module. Notably this A1 module lacks the conserved aromatic residue that would normally be used to interact with a downstream B module and would pull the A1 loop downward. The absence of this residue and a downstream B module seem to allow this A1 module to pucker upward in the B-module-mimicking fashion described.
The structure of the 4-1BBL/4-1BB complex
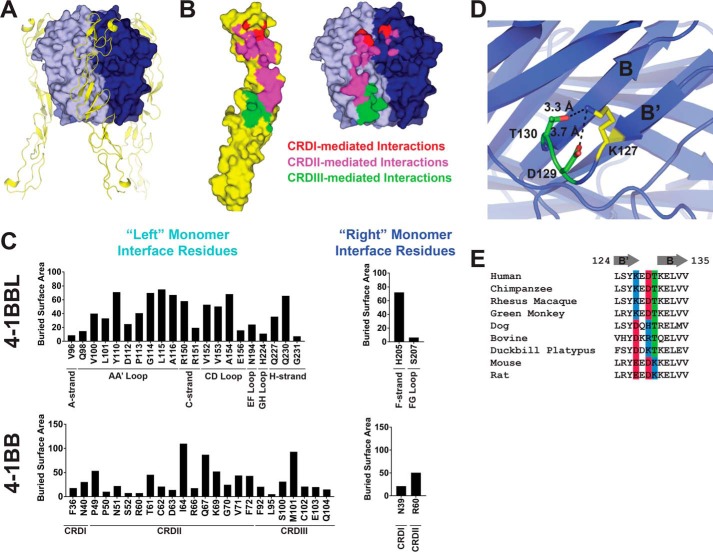
In examining the 4-1BB/4-1BBL complex, we observed that 4-1BB binds to 4-1BBL in a similar manner as all other TNFR superfamily receptors bind to their ligands, at the interface between ligand monomers with contacts made predominantly through CRDII (69% of 4-1BB buried surface area) and III (23% of 4-1BB buried surface area) and minor contacts made by CRDI (8% of 4-1BB buried surface area) (Fig. 6, A–C) (6). This is consistent with reported mutational studies that identified CRDII and -III as the major contributing regions to ligand binding with CRDI being largely dispensable (19, 20). In most TNF superfamily ligand/receptor complexes, the upper portion of the receptor CRDII bridges two adjacent ligand monomers and contacts the AA′ loop of the “left” monomer and the DE loop of the “right” monomer (6). In the 4-1BBL/4-1BB complex, the AA′ contact is maintained, but the DE loop contact is absent and most DE loop residues in 4-1BBL (168–175) are disordered. CRDII does make limited contact with the right ligand monomer, however, interacting with F-strand and FG loop residues as depicted in Fig. 6, B and C. CRDI also bridges the ligand monomer–monomer interface making minor contacts. The lower portion of CRDII and the upper portion of CRDIII typically form contacts with the CD and EF loops of the left and right ligand monomers, respectively, in other TNFR superfamily members. However, in the 4-1BB/4-1BBL complex, these regions contact only the left 4-1BBL monomer, interacting with the CD loop as well as GH loop and H-strand residues (Fig. 6, B and C). Notably, these GH loop and H-strand residues are part of the downwardly extended GH loop structure described above (Fig. 4F). The absence of receptor contacts with the EF and DE loops of the right 4-1BBL monomer result in a 4-1BB/4-1BBL interface that is highly skewed compared with other TNF superfamily members with 92% of the buried surface area contributed by interaction with a single (left) monomer (Fig. 6, B and C). The interfaces of other TNF superfamily ligand/receptor complexes are much less skewed with buried surface being much more evenly distributed across the two neighboring monomers. An average of 54 ± 6% of buried surface area is contributed by the left monomer and 46 ± 6% by the right monomer (Table S1). Despite this skewedness, the total size of the 4-1BBL/4-1BB interface is 916 Å2, which is just slightly below the average of 1113 ± 116 Å2 for other TNF superfamily ligand/receptor complexes (Table S1).
Figure 6.
The structure of the 4-1BB/4-1BBL complex. A, structure of the 4-1BB/4-1BBL complex. 4-1BBL is shown in surface representation with neighboring monomer units indicated (blue and cyan). 4-1BB (yellow) is shown as a cartoon. B, open book representation of the 4-1BB/4-1BBL interface (4-1BB has been rotated 180° and translated with respect to 4-1BBL). 4-1BB (left) and 4-1BBL (right) are shown in surface representation with interface residues highlighted. 4-1BB CRDI residues and their contacting partners (red), CRDII residues and their contacting partners (magenta), and CRDIII residues and their contacting partners (green) are indicated. C, buried surface area contributions of individual residues in 4-1BBL (top) and 4-1BB (bottom). Relevant subdomains and secondary structure regions are indicated. Residues involved in the interface with the left and right 4-1BBL monomers as depicted in A and B are segregated in the left and right columns. D, structural detail of the intrachain interactions formed by Lys-127 in 4-1BBL. The B- and B′-strands are labeled. E, sequence alignment of the B′- and B-strand regions of 4-1BBL from diverse species. Positions 127, 129, and 130, which appear to play a role in shaping the local structure of the B′B-strand region in the human 4-1BBL structure reported here, are highlighted and colored according to their side chain charge properties: positive (blue), negative (red), or polar (green).
A residue level dissection of the 4-1BB/4-1BBL interface shows that on the 4-1BBL side of the interface, 23 residues participate in the interaction with receptor with buried surface area contributions spread fairly evenly across the interface (Fig. 6C). Hydrophobic and aromatic residues contribute ∼62% of the buried surface area with polar/charged residues making up the remainder. On the 4-1BB side of the interface, 25 residues contact 4-1BBL and buried surface area contributions are distributed less evenly. Ile-64 and Met-101 contribute disproportionately comprising 12 and 11% of the total buried surface area, respectively. 52% of the 4-1BB buried surface is contributed by hydrophobic and aromatic residues and 48% by polar and charged residues. This distribution is similar to 4-1BBL, but skewed slightly more toward polar and charged residues.
Mutagenesis data probing the 4-1BB/4-1BBL interaction have been previously reported by Won et al. (9) and can be interpreted in the context of the structure reported here. L115A and Q227A mutations were observed to have significant effects on 4-1BBL/4-1BB binding. This result is consistent with our observation that both of these residues contribute significantly to the receptor/ligand interface in the 4-1BBL/4-1BB structure (Fig. 6C). 4-1BBL DE loop mutations R171G and S172G were found to have minimal effect on receptor binding by Won et al. (9). Consistent with this result, these residues are disordered in the structure here and do not contribute to the receptor interface. Interestingly, Won et al. (9) also reported that the K127A mutation significantly impacted binding, but we do not observe any direct participation of Lys-127 in the 4-1BB/4-1BBL interface. Instead, Lys-127 appears to play a role in the structure of the B′- and B-strands by forming hydrogen bond/electrostatic interactions with nearby Asp-129 and Thr-130 (Fig. 6D). The K127A mutation may disrupt these interactions, leading to structural changes in the B′-strand and nearby AA′ loop, which is extensively involved in receptor contacts. Notably, although Lys-127 is not strictly conserved across species, position 127 is nearly always a charged residue (Lys, Glu, or Asp) and when the charge of position 127 is reversed to negative, there is a distinct tendency for the charge at positions 129 or 130 to reverse to positive in a complementary fashion (Fig. 6E). This observation is consistent with the notion that the intrachain electrostatic/hydrogen bonding interactions observed for Lys-127 may be structurally and functionally important for 4-1BBL.
Discussion
Here, we report the crystal structure of the human 4-1BBL/4-1BB complex. As indicated, one of the most surprising features of this structure is the striking difference in the 4-1BBL trimer assembly compared with a previously reported structure of 4-1BBL alone (e4-1BBL) (9). In light of both existing data and new data generated here, we believe that the 4-1BBL structure described in this work represents the biologically relevant assembly for the molecule. Although the previously reported 4-1BBL structure was supported by mutational data, we find that the same mutational data are consistent with the structure reported here and that the effects (or lack thereof) of some mutations are more easily explained in the context of our structure. For example, Won et al. (9) found that individual Q89A, L115A, H205A, V240A, P242K, P242E, and P242D mutations had no significant effect on 4-1BBL trimerization although, based on their reported structure, they would have been expected to have significant effects. If interpreted in the context of the structure here, it is not surprising that these mutations had little effect on trimer assembly as none of them are observed to play major roles in trimer assembly and are located either away from or at the periphery of the trimer interface. In contrast, the effects of F199A, V140A, and Y142A mutations, all of which were found by Won et al. (9) to have significant impact on trimer formation or biophysical behavior, can be rationalized based on the structure here, given they participate heavily in the trimer interface. Furthermore, our own mutational data support the notion that the 4-1BBL trimer assembly reported here represents the biologically relevant state. We found that all residues outside the THD of 4-1BBL (defined as residues 89–242 in the construct examined here) could be deleted without a significant effect on trimer assembly, receptor binding, or biological activity. This finding is inconsistent with the previously reported structure of 4-1BBL in which residues outside the THD contributed 37% of the buried surface at the trimer interface.
The reason why an alternative trimer assembly was observed by Won et al. (9) seems likely to be traceable to the alternative β-strand configuration present in the e4-1BBL structure. As outlined above, in the e4-1BBL structure, a stretch of residues outside the THD (residues 80–88) appears to displace the B′ strand typically present in TNFR superfamily ligands (Fig. 4B). This displacement results in marked rearrangement of the A′- and B′-strands as well as the AA′ loop when compared with the structure reported here. There is also a distinct shift in the upper portion of A-strand in the e4-1BBL monomer. When superimposed with the 4-1BBL trimer structure described here, the structural changes in e4-1BBL cause steric clashes and close contacts between neighboring 4-1BBL monomers (Fig. S2). These clashes would likely prohibit formation of the trimer assembly that we observed here, perhaps facilitating formation of the alternative assembly observed. Although the cause of the structural rearrangements in the e4-1BBL structure is not immediately clear, we speculate that they may have resulted from different expression/purification procedures, or possibly have been driven by crystallization conditions.
Generally, human 4-1BB's structure is quite similar to that of other TNFR superfamily members with the exception of a truncated CRDI and unusual CRDIII comprised by two A-type modules rather than the typical A/B pair. As noted above, CRDI of 4-1BB also resembles the truncated CRDI of TRAILR2, which is known to be involved in ligand-independent pre-association of receptor monomers. It is possible that CRDI could be involved in similar pre-assembly of 4-1BB on the cell surface, although, to our knowledge, no clear evidence for such pre-assembly has been presented for 4-1BB in the literature. Despite the unusual pairing of two A-type modules in CRDIII of 4-1BB, we observed that the domain structurally mimics a more conventional A/B-type CRD. It is possible that similar atypical CRDs could occur in other TNFR superfamily members, although sequence analysis does not clearly identify any candidates, suggesting it may be a unique feature of 4-1BB.
During preparation of this manuscript, a crystal structure of murine 4-1BB was published (10). A comparison of the human and mouse receptors reveals that the two are very similar in overall structure with 99 of 109 Cα atoms in the mouse receptor aligning with the human receptor with an r.m.s. deviation of 1.03 Å. However, CRDI and a portion of the A1 module from CRDII are disordered in the murine structure precluding analysis. Notably, the close matching of the two structures is maintained even in the unusual A2/A1 modules comprising the 4-1BB CRDIII. The aforementioned His-93, Glu-103, Asp-105, and Leu-112 residues that facilitate structural mimicry of a conventional A/B CRD in CRDIII are conserved in murine 4-1BB and adopt the same structural configuration.
With the mouse and human 4-1BB receptors being largely similar in overall structure, it is perhaps surprising that Yi et al. (20) reported that mouse and human 4-1BB differentially relied on CRDs II and III for binding to 4-1BBL with human relying more heavily on CRDIII and mouse relying more heavily on CRDII. Indeed, in addition to their structural similarity, mouse and human 4-1BB show 65% sequence identity in their extracellular domains. However, sequence conservation in mouse and human 4-1BBL is lower (40% identity in the THD region) and conservation at positions that contact receptor in the human structure is extremely low at just 4% identity. These data suggest that mouse 4-1BB may bind to mouse 4-1BBL in a manner distinct from human 4-1BB/4-1BBL.
The structure of the human 4-1BB/4-1BBL complex described here gives insight into the signaling mechanism of this ligand/receptor pair. We observe that like all other TNFR superfamily receptor/ligand pairs, 4-1BB and 4-1BBL associate in a 3:3 with 4-1BB binding at the interface between 4-1BBL monomers. Interestingly, although 4-1BB binds to 4-1BBL at the monomer–monomer interface, 4-1BB disproportionately interacts with one monomer of the pair (Fig. 6, B and C). This “skewedness” is unusual among TNFR superfamily ligand/receptor interactions in which the receptor typically interacts with both ligand monomers approximately equally. What functional implications, if any, arise from this skewedness are not clear. However, with 92% of buried surface area contributed by interaction with a single monomer, an interesting possibility is that human 4-1BB binding to 4-1BBL may not require ligand trimer formation. It is not clear under what circumstances 4-1BBL might exist in a nontrimer state. The THD-only 4-1BBL construct used here formed a stable monodisperse trimer in solution. Nevertheless, 1:1 4-1BB/4-1BBL interactions remain a notable possibility.
Finally, because the 4-1BBL/4-1BB complex structure clearly defines both surfaces involved in ligand/receptor interactions and the geometry of presumably productive receptor engagement, it provides valuable information for the design of both antagonist and agonist agents. The receptor relies most heavily on CRDII and -III for interaction with the ligand, so targeting these domains for antagonist agents would most likely prove fruitful. Of particular interest could be epitopes centering around residues Ile-64 and Met-101, which contribute disproportionately to the interface-buried surface area. On the ligand side, residues of the AA′ loop provide the largest portion of receptor-binding surface and thus could make an attractive target antagonist target (Fig. 6C). Because TNFR superfamily receptor signaling is driven largely by clustering of large numbers of receptors, there may not be particular epitopes of interest for 4-1BB agonist design. However, one could use the model of the 4-1BB/4-1BBL structure described here as a guide in the design of agonists presuming that a good candidate should either facilitate or at least allow receptor clustering with a similar geometry.
In summary, we report here the structure of the human 4-1BB/4-1BBL complex. This structure reveals a 4-1BBL trimer assembly that is distinctly different from that previously observed in a crystal structure of human 4-1BBL alone. Our own mutagenesis and functional data as well as previously published mutational data support the structure reported here as the biologically relevant conformation of both the ligand and the ligand/receptor complex. The details that this structure reveals with regard to ligand/receptor interactions should be valuable for both mechanistic understanding of signaling in this important receptor system as well as the design of agents for its manipulation.
Materials and methods
Preparation of HEK293F cells expressing WT or variants of human 4-1BBL
Genes encoding full-length WT human 4-1BBL or variants in which residues flanking the THD were deleted or mutated were cloned into an Orip/EBNA-1-based episomal mammalian expression vector (pOE, MedImmune). This vector has been described previously (21). The expressed amino acid sequence for WT 4-1BBL was MEYASDASLDPEAPWPPAPRARACRVLPWALVAGLLLLLLLAAACAVFLACPWAVSGARASPGSAASPRLREGPELSPDDPAGLLDLRQGMFAQLVAQNVLLIDGPLSWYSDPGLAGVSLTGGLSYKEDTKELVVAKAGVYYVFFQLELRRVVAGEGSGSVSLALHLQPLRSAAGAAALALTVDLPPASSEARNSAFGFQGRLLHLSAGQRLGVHLHTEARARHAWQLTQGATVLGLFRVTPEIPAGLPSPRSE. 4-1BBL variant constructs were made with mutations/deletions in the 80–93 and 242–254 residue regions as described above (Fig. 2). 30 ml of HEK293F cells (Invitrogen) at a density of 1.1 × 106 cells/ml were transfected with the expression vectors using the 293Fectin reagent (Invitrogen) (30 μg of DNA, 50 μl of 293Fectin) and cells were cultured in 293 FreeStyle medium (Invitrogen) at 37 °C and 8% CO2 and fed using a proprietary feed solution (MedImmune LLC). At 3 days post-transfection, transgene expression was confirmed by Western blotting using a biotinylated polyclonal anti-human 4-1BBL antibody (R&D Systems) and a streptavidin horseradish peroxidase conjugate (Pierce) (data not shown) and cells were used for co-culture reporter cell assays.
Co-culture reporter cell assay of 4-1BBL and 4-1BBL variants expressed on HEK293F cells
A human 4-1BB-dependent NF-κB reporter cell line was generated according to a previously described procedure (22), where Jurkat E6-1 cells were stably transduced to constitutively express full-length human 4-1BB, and a firefly luciferase reporter gene under control of an NF-κB response element. A control Jurkat E6-1 cell line containing only the luciferase reporter gene was also generated. Reporter cells were cultured in RPMI media (Invitrogen), 10% heat-inactivated FBS, 1× GlutaMAX (Invitrogen), and 1× Anti-anti (Invitrogen) and maintained at 37 °C and 5% CO2. For assays, reporter cells were resuspended in phenol red-free RPMI + 5% FBS and 50,000 cells were transferred to the wells of a 96-well plate in a volume of 50 μl. HEK293F cells expressing WT or variant 4-1BBL constructs were prepared as outlined above and resuspended in phenol red RPMI + 5% heat-inactivated FBS. Serial dilutions of these HEK293F cells in a volume of 50 μl were added to the plate containing the Jurkat reporter cells and the cell mixture was incubated for 4 h at 37 °C and 5% CO2. Following incubation, 100 μl of reconstituted Bright-Glo luciferase substrate (Promega) was added to the wells of the plate, mixed by gentle pipetting and incubated at room temperature for 2 min. Luminescence was then read using an EnVision plate reader (PerkinElmer Life Sciences) equipped with ultra-sensitive luminescence detection.
Expression and purification of THD-only 4-1BBL
A gene encoding the THD of human 4-1BBL (residues 89–242) was cloned into a pET15b-based vector (Novagen). A His6 tag and a TEV protease cleavage site were appended to the N terminus of the 4-1BBL THD. The expressed amino acid sequence was MKHHHHHHENLYFQGSQGMFAQLVAQNVLLIDGPLSWYSDPGLAGVSLTGGLSYKEDTKELVVAKAGVYYVFFQLELRRVVAGEGSGSVSLALHLQPLRSAAGAAALALTVDLPPASSEARNSAFGFQGRLLHLSAGQRLGVHLHTEARARHAWQLTQGATVLGLFRVTP. Chemically competent BL21(DE3)pLysS cells (Novagen) were transformed with this vector and cells were grown in M9ZB media (Teknova) supplemented with 2% glucose at 37 °C until reaching A600 = 1. Protein expression was induced by addition of isopropyl 1-thio-β-d-galactopyranoside at a final concentration of 1 mm and expression was allowed to proceed for 4 h at 37 °C. Cells were then harvested by centrifugation, resuspended in 100 ml of 50 mm HEPES, pH 7.5, 300 mm NaCl, 5 mm MgCl2, and 20 μg/ml of DNase I and lysed using a microfluidizer. The lysate was clarified by centrifugation and the cleared lysate was loaded onto a 5-ml HisTrap column (GE Healthcare) for purification. After loading, the column was rinsed with running buffer (50 mm HEPES, pH 7.5, 300 mm NaCl, 10 mm imidazole) until the absorbance at 280 nm (A280) reached baseline level and the protein was eluted using a 0–100% gradient of elution buffer (50 mm HEPES, pH 7.5, 300 mm NaCl, 500 mm imidazole) over 20 column volumes. Purity of the recovered protein was confirmed by SDS-PAGE, glycerol was added to the sample at a final concentration of 5% and the sample was exhaustively dialyzed against 10 mm HEPES, pH 7.5, 20 mm NaCl, 5% glycerol.
To remove the N-terminal His tag, purified THD-only 4-1BBL (final concentration of 0.3 mg/ml) was mixed with His-tagged TEV protease (expressed and purified in-house from ATCC glycerol stock catalog number MBA-145) at a 1:10 TEV:4-1BBL ratio (w/w) in 50 mm HEPES, pH 7.5, 20 mm NaCl, and 2 mm DTT and the mixture was incubated overnight at room temperature with gentle shaking. After this incubation, the mixture was passed through a 0.2-μm filter and loaded onto a 5-ml HisTrap column (GE Healthcare) pre-equilibrated with running buffer (50 mm, Tris, pH 7.5, 20 mm NaCl) to remove the His-tagged TEV protease and any uncleaved 4-1BBL protein. The column was rinsed until A280 reached baseline level. TEV-cleaved 4-1BBL was eluted from the column with a linear 0–100% gradient of elution buffer (50 mm Tris, pH 7.5, 20 mm NaCl, 500 mm imidazole) over 30 column volumes. TEV-cleaved 4-1BBL eluted at a low concentration of imidazole (∼50 mm). SDS-PAGE confirmed that His tag TEV, uncleaved 4-1BBL, and cleaved His tag were retained on the column at this concentration and that the recovered cleaved 4-1BBL was of high purity. The purified TEV-cleaved 4-1BBL was dialyzed into 50 mm Tris, pH 7.5, with 50 mm NaCl and protein concentration was calculated by A280 using a Nanodrop (Thermo). The expected intact mass of the cleaved 4-1BBL monomer subunit was confirmed by Q-TOF LC/MS analysis (expected mass 16,422 Da; measured mass 16,423 Da).
Expression and purification of human 4-1BB extracellular region
A gene encoding the extracellular region of human 4-1BB (residues 24-160) with C121S, N138D, and N149Q mutations was cloned as an N-terminal fusion to a hingeless human IgG1 Fc as previously described (23). 4-1BB and other members of the TNFR superfamily are comprised by cysteine-rich repeats containing large numbers of disulfide bonds. However, 4-1BB has an odd number of cysteines in its extracellular region indicating at least one must be unpaired. A sequence alignment with murine 4-1BB showed that Cys-121 is not conserved (Ser-121 in mouse) and is also not located in a consensus position for cysteine as seen in the cysteine-rich domains of other TNFR superfamily members (18). Thus, to avoid unwanted disulfide bond formation, we introduced the C121S mutation. The N138D and N149Q mutations were introduced to remove potential N-linked glycosylation sites which, if occupied, could introduce heterogeneity and steric bulk unfavorable for subsequent crystallization. The expressed amino acid sequence was MPLL LLLPLLWAGALALQDPCSNCPAGTFCDNNRNQICSPCPPNSFSSAGGQRTCDICRQCKGVFRTRKECSSTSNAECDCTPGFHCLGAGCSMCEQDCKQGQELTKKGCKDCSFGTFNDQKRGICRPWTDCSLDGKSVLVQGTKERDVVCGPGSLGGPSVFLFPPKPKDTLMISRTPEVTCVVVDVSHEDPEVKFNWYVDGVEVHNAKTKPREEQYNSTYRVVSVLTVLHQDWLNGKEYKCKVSNKALPAPIEKTISKAKGQPREPQVYTLPPSREEMTKNQVSLTCLVKGFYPSDIAVEWESNGQPENNYKTTPPVLDSDGSFFLYSKLTVDKSRWQQGNVFSCSVMHEALHNHYTQKSLSLSPGK. This sequence also includes a CD33 secretion signal. The sequence corresponding to 4-1BB is in bold underlined italics. 150 ml of HEK293F cells (Invitrogen) at a density of 1.1 × 106 cells/ml were transfected with the expression vector using the 293Fectin reagent (Invitrogen) (100 μg of DNA, 150 μl of 293Fectin) and cells were cultured in 293 FreeStyle medium (Invitrogen) at 37 °C and 8% CO2. Cells were periodically fed using a proprietary feed solution (MedImmune LLC) and culture volume was doubled on days 3 and 6 using 293 FreeStyle medium (Invitrogen). At day 10, cells were harvested and the culture supernatant containing the secreted 4-1BB-Fc fusion protein was recovered by centrifugation. This supernatant was loaded onto a 5-ml HiTrap rProteinA column (GE Healthcare) pre-equilibrated with running buffer (10 mm phosphate, pH 7.2, 150 mm NaCl). The column was rinsed with running buffer until A280 reached baseline levels. The 4-1BB-Fc fusion protein was eluted using Pierce Gentle Ag/Ab Elution Buffer, pH 6.6 (Thermo), and the recovered protein was dialyzed into 10 mm phosphate, pH 7.2, 150 mm NaCl.
The Fc portion of the 4-1BB-Fc molecule was cleaved using IdeS protease in a manner similar to that previously described (23). The purified 4-1BB-Fc was mixed at a 1:15 ratio (IdeS:4-1BB-Fc, w/w) in 10 mm phosphate, pH 7.2, 150 mm NaCl and the mixture was incubated overnight at room temperature with gentle mixing. The final 4-1BB concentration was 1.2 mg/ml. SDS-PAGE was used to confirm successful cleavage. The mixture was then 0.2-μm filtered and applied a 5-ml HiTrap rProtein A column (GE Healthcare) in-line prior to a 5-ml HisTrap HP column. The protein A column was used to capture cleaved Fc and uncleaved fusion protein and the HisTrap column was used to capture His-tagged IdeS. 4-1BB ECD was collected in the unbound fraction. Recovered protein was quantitated by A280 using a Nanodrop instrument (Thermo). Purity was confirmed by SDS-PAGE. Expected intact mass of the cleaved 4-1BB ECD was confirmed by Q-TOF LC/MS analysis (expected mass 15,358 Da; observed: 15,358 Da).
Size exclusion chromatography and static light-scattering analysis of THD-only 4-1BBL and the 4-1BBL/4-1BB complex
For THD-only 4-1BBL analysis, 20 μl of purified THD-only 4-1BBL (with N-terminal His tag and TEV protease site intact) at a concentration of 2.3 mg/ml was injected onto a Superdex 75 10/300 GL SEC column (GE Healthcare) and analyzed at a flow rate of 0.75 ml/min using 10 mm phosphate, pH 7.2, 150 mm NaCl as the running buffer. For analysis of the 4-1BBL/4-1BB complex, THD-only 4-1BBL and 4-1BB (Fc portion removed by IdeS cleavage) were mixed in an ∼1:1 molar ratio (total protein concentration of 2.1 mg/ml), incubated at room temperature for 30 min, then injected onto the size exclusion column and analyzed in the same manner as for uncomplexed 4-1BBL. For both samples, the eluate from the SEC column was analyzed by static light scattering using a Dawn HELIOS II instrument (Wyatt Technologies) connected in-line immediately after the SEC column. Scattering data were measured at angles spanning 64° to 134° (scattering was isotropic) and analysis was carried out using software provided by the manufacturer. Reported weight averaged molar mass values (Mw) are mean values calculated from measurements taken throughout the eluting peak as shown in Fig. 3.
Reporter cell assay of soluble THD-only 4-1BBL
Human 4-1BB-NF-κB Jurkat E6-1 reporter cells were resuspended in phenol red-free RPMI + 5% FBS and 50,000 cells were transferred to the wells of a 96-well plate in a volume of 50 μl. Serial dilutions of THD-only 4-1BBL were prepared in 50 μl of the same media and added to the plate containing the reporter cells. The cell mixture was incubated for 4 h at 37 °C and 5% CO2. Following incubation, 100 μl of reconstituted Bright-Glo luciferase substrate (Promega) was added to the wells of the plate, mixed by gentle pipetting and incubated at room temperature for 2 min. Luminescence was then read using an EnVision plate reader (PerkinElmer Life Sciences) equipped with ultra-sensitive luminescence detection.
Crystallization, data collection, structure solution, and refinement
Approximately equimolar amounts of purified human 4-1BB ECD and THD-only 4-1BBL, purified as described above, were mixed together (with 5% excess of 4-1BB ECD), concentrated to 5 mg/ml of total protein and the 4-1BB/4-1BBL complex purified by size exclusion chromatography using a Superdex 75 10/300 GL SEC column equilibrated in 25 mm HEPES, pH 7.5, 100 mm NaCl. The running buffer was the same composition. The recovered complex was concentrated to 10 mg/ml and subjected to screening for crystallization conditions using a Phoenix robot (Art Robbins Instruments), 96-well 2-drop Intelli plates (Art Robbins Instruments), and commercial screens. Screening was carried out using the sitting drop vapor diffusion method. Droplets were prepared by mixing 300 nl of protein sample with an equal volume of reservoir solution. Crystallization optimization in hanging drop format was carried out using greased Linbro plates (Hampton Research) and varying volumes of protein and reservoir. Diffraction quality crystals were harvested from optimized hanging drop conditions consisting of 2 μl of protein sample mixed with 1 μl of 0.16 m potassium citrate tribasic monohydrate and 16% (w/v) PEG 3350 solution. The same buffer was used as the reservoir solution. Harvested crystals were cryopreserved in reservoir solution supplemented with 20% glycerol and flash cooled in liquid nitrogen. Data were collected from a single crystal on beamline BL9-2 of Stanford Synchrotron Radiation Lightsource equipped with Pilatus 6M PAD detector (Paul Scherer Institute, Switzerland). The cubic crystal diffracted to 2.4-Å resolution and belonged to I213 space group. Diffraction data were collected over an oscillation range of 180° an increment of 0.5°, and 0.8-s exposure per image. Processing of diffraction data were performed using XDS (24). The asymmetric unit contained two 4-1BB/4-1BBL complexes. Structure solution was carried out using molecular replacement as implemented in MOLREP (25) in the CCP4 program suite (26). The search model for 4-1BBL was a previously reported 4-1BBL structure (PDB code 2X29) (9) and the search model for 4-1BB was the structure of the TNFR superfamily homolog CD40 (chain R, PDB code 3QD6) (27). The CHAINSAW program (28) from the CCP4 suite was used to prepare models for molecular replacement runs. Alternating cycles of manual rebuilding with “O” (29) and macromolecular refinement with Refmac5 (30) were used to build the model for the 4-1BBL/4-1BB complex. The final electron density map covers all cloned amino acids of 4-1BBL with the following exceptions: Gly-Ser upstream of residue Gln-89 (these two amino acids are a residual artifact of the TEV protease cleavage site), residues 168 to 175 and 189 to 191. These regions show no traceable electron density. For 4-1BB, all cloned residues with the exception of 24, 25, and 157–160 are visible in the electron density map. Also missing in the 4-1BB map are Gly-Ser-Leu-Gly residues that follow residue 160 and are a residual sequence that results from the cloning and the IdeS cleavage strategy. Model and experimental structure factors have been deposited with the PDB under accession number 6BWV.
Author contributions
R. N. G. and M. B. conceptualization; R. N. G., V. Y. O., H. A., and S. N. formal analysis; R. N. G. and M. B. supervision; R. N. G., V. Y. O., H. A., S. N., L. G., and A. B. investigation; R. N. G., V. Y. O., S. N., L. G., A. B., and M. B. methodology; R. N. G., V. Y. O., S. N., and A. B. writing-original draft; R. N. G. and M. B. project administration; R. N. G., V. Y. O., S. N., A. B., and M. B. writing-review and editing.
Supplementary Material
Acknowledgments
Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by United States Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, NIGMS (including Grant P41GM103393).
All authors are or have been employees of and may hold financial interests in MedImmune LLC. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Table S1 and Figs. S1 and S2.
This work is dedicated to the memory of our friend and colleague, Luba Grinberg.
The atomic coordinates and structure factors (code 6BWV) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TNFR
- tumor necrosis factor receptor
- THD
- TNF homology domain
- SEC
- size exclusion chromatography
- r.m.s. deviation
- root mean square deviation
- CRD
- cysteine-rich domain
- TEV
- tobacco etch virus
- PDB
- Protein Data Bank.
References
- 1. Sanmamed M. F., Pastor F., Rodriguez A., Perez-Gracia J. L., Rodriguez-Ruiz M. E., Jure-Kunkel M., and Melero I. (2015) Agonists of co-stimulation in cancer immunotherapy directed against CD137, OX40, GITR, CD27, CD28, and ICOS. Semin. Oncol. 42, 640–655 10.1053/j.seminoncol.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 2. Schaer D. A., Hirschhorn-Cymerman D., and Wolchok J. D. (2014) Targeting tumor-necrosis factor receptor pathways for tumor immunotherapy. J. Immunother. Cancer 2, 7 10.1186/2051-1426-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chester C., Sanmamed M. F., Wang J., and Melero I. (2018) Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood 131, 49–57 [DOI] [PubMed] [Google Scholar]
- 4. Bartkowiak T., and Curran M. A. (2015) 4-1BB agonists: multi-potent potentiators of tumor immunity. Front. Oncol. 5, 117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vinay D. S., and Kwon B. S. (2012) Immunotherapy of cancer with 4-1BB. Mol. Cancer Ther. 11, 1062–1070 10.1158/1535-7163.MCT-11-0677 [DOI] [PubMed] [Google Scholar]
- 6. Bodmer J. L., Schneider P., and Tschopp J. (2002) The molecular architecture of the TNF superfamily. Trends Biochem. Sci. 27, 19–26 10.1016/S0968-0004(01)01995-8 [DOI] [PubMed] [Google Scholar]
- 7. Vanamee E. S., and Faustman D. L. (2018) Structural principles of tumor necrosis factor superfamily signaling. Sci. Signal. 11, 4910 10.1016/scisignal.aao4910 [DOI] [PubMed] [Google Scholar]
- 8. Li J., Yin Q., and Wu H. (2013) Structural basis of signal transduction in the TNF receptor superfamily. Adv. Immunol. 119, 135–153 10.1016/B978-0-12-407707-2.00005-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Won E. Y., Cha K., Byun J. S., Kim D. U., Shin S., Ahn B., Kim Y. H., Rice A. J., Walz T., Kwon B. S., and Cho H. S. (2010) The structure of the trimer of human 4-1BB ligand is unique among members of the tumor necrosis factor superfamily. J. Biol. Chem. 285, 9202–9210 10.1074/jbc.M109.084442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bitra A., Doukov T., Wang J., Picarda G., Benedict C. A., Croft M., and Zajonc D. M. (2018) Crystal structure of murine 4-1BB and its interaction with 4-1BBL support a role for galectin-9 in 4-1BB signaling. J. Biol. Chem. 293, 1317–1329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chattopadhyay K., Ramagopal U. A., Mukhopadhaya A., Malashkevich V. N., Dilorenzo T. P., Brenowitz M., Nathenson S. G., and Almo S. C. (2007) Assembly and structural properties of glucocorticoid-induced TNF receptor ligand: implications for function. Proc. Natl. Acad. Sci. U.S.A. 104, 19452–19457 10.1073/pnas.0709264104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Compaan D. M., and Hymowitz S. G. (2006) The crystal structure of the costimulatory OX40-OX40L complex. Structure 14, 1321–1330 10.1016/j.str.2006.06.015 [DOI] [PubMed] [Google Scholar]
- 13. Liu W., Zhan C., Cheng H., Kumar P. R., Bonanno J. B., Nathenson S. G., and Almo S. C. (2014) Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly. Structure 22, 1252–1262 10.1016/j.str.2014.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Magis C., van der Sloot A. M., Serrano L., and Notredame C. (2012) An improved understanding of TNFL/TNFR interactions using structure-based classifications. Trends Biochem. Sci. 37, 353–363 10.1016/j.tibs.2012.06.002 [DOI] [PubMed] [Google Scholar]
- 15. Eck M. J., and Sprang S. R. (1989) The structure of tumor necrosis factor-α at 2.6-Å resolution: implications for receptor binding. J. Biol. Chem. 264, 17595–17605 [DOI] [PubMed] [Google Scholar]
- 16. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 17. Krissinel E., and Henrick K. (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr. 60, 2256–2268 10.1107/S0907444904026460 [DOI] [PubMed] [Google Scholar]
- 18. Naismith J. H., and Sprang S. R. (1998) Modularity in the TNF-receptor family. Trends Biochem. Sci. 23, 74–79 10.1016/S0968-0004(97)01164-X [DOI] [PubMed] [Google Scholar]
- 19. Yi L., Yan Z., Jia H., Wang X., Zhao Y., and Zhang H. (2017) CD137-CRDI is not necessary in the role of contacting its natural ligand. Immunol. Cell Biol. 95, 24–32 10.1038/icb.2016.64 [DOI] [PubMed] [Google Scholar]
- 20. Yi L., Zhao Y., Wang X., Dai M., Hellström K. E., Hellström I., and Zhang H. (2014) Human and mouse CD137 have predominantly different binding CRDs to their respective ligands. PLoS ONE 9, e86337 10.1371/journal.pone.0086337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dimasi N., Gao C., Fleming R., Woods R. M., Yao X. T., Shirinian L., Kiener P. A., and Wu H. (2009) The design and characterization of oligospecific antibodies for simultaneous targeting of multiple disease mediators. J. Mol. Biol. 393, 672–692 10.1016/j.jmb.2009.08.032 [DOI] [PubMed] [Google Scholar]
- 22. Leyland R., Watkins A., Mulgrew K. A., Holoweckyj N., Bamber L., Tigue N. J., Offer E., Andrews J., Yan L., Mullins S., Oberst M. D., Coates Ulrichsen J., Leinster D. A., McGlinchey K., Young L., et al. (2017) A novel murine GITR ligand fusion protein induces antitumor activity as a monotherapy that is further enhanced in combination with an OX40 agonist. Clin. Cancer Res. 23, 3416–3427 10.1158/1078-0432.CCR-16-2000 [DOI] [PubMed] [Google Scholar]
- 23. Novarra S., Grinberg L., Rickert K. W., Barnes A., Wilson S., and Baca M. (2016) A hingeless Fc fusion system for site-specific cleavage by IdeS. MAbs 8, 1118–1125 10.1080/19420862.2016.1186321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Vagin A., and Teplyakov A. (2010) Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 26. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., et al. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 10.1107/S0907444910045749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. An H. J., Kim Y. J., Song D. H., Park B. S., Kim H. M., Lee J. D., Paik S. G., Lee J. O., and Lee H. (2011) Crystallographic and mutational analysis of the CD40-CD154 complex and its implications for receptor activation. J. Biol. Chem. 286, 11226–11235 10.1074/jbc.M110.208215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stein N. (2008) CHAINSAW: a program for mutating pdb files used as templates in molecular replacement. J. Appl. Cryst. 41, 641–643 10.1107/S0021889808006985 [DOI] [Google Scholar]
- 29. Jones T. A., and Kjeldgaard B. M. M. (1990) O: a macromolecule modeling environment. in Crystallographic and Modeling Methods in Molecular Design (Bugg C. E., and Ealick S. E., eds) Springer, New York [Google Scholar]
- 30. Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., and Vagin A. A. (2011) REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 67, 355–367 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.