Abstract
Purpose of Review
Atopic dermatitis (AD) is a chronic, relapsing inflammatory skin disorder that is a major public health burden worldwide. AD lesions are often colonized by Staphylococcus aureus and Staphylococcus epidermidis. An important aspect of Staphylococcus spp. is their propensity to form biofilms, adhesive surface-attached colonies that become highly resistant to antibiotics and immune responses, and recent studies have found that clinical isolates colonizing AD skin are often biofilm-positive. Biofilm formation results in complex bacterial communities that have unique effects on keratinocytes and host immunity. This review will summarize recent studies exploring the role of staphyloccocal biofilms in atopic dermatitis and the implications for treatment.
Recent Findings
Recent studies suggest an important role for biofilms in the pathogenesis of numerous dermatologic diseases including AD. S. aureus biofilms have been found to colonize the eccrine ducts of AD skin, and these biofilms influence secretion of keratinocyte cytokines and trigger differentiation and apoptosis of keratinocytes. These activities may act to disrupt barrier function and promote disease pathogenesis as well as allergen sensitization.
Summary
Formation of biofilm is a successful strategy that protects the bacteria from environmental danger, antibiotics, and phagocytosis, enabling chronic persistence in the host. An increasing number of S. aureus skin isolates are resistant to conventional antibiotics, and staphylococcal biofilm communities are prevalent on the skin of individuals with AD. Staphylococcal colonization of the skin impacts skin barrier function and plays multiple important roles in AD pathogenesis.
Keywords: Atopic dermatitis, Biofilm, Staphylococci, Microbiome, Barrier function, Epidermis
Introduction
Atopic dermatitis (AD) is a chronic skin condition characterized by eczematous lesions and intense itching [1]. AD presents in about 10% of children and 7% of adults in the United States [2]. Industrialized countries have a higher prevalence of AD, although an increasing number of cases are observed in developing countries [3]. AD presents most commonly in early childhood, with up to 60% of patients developing symptoms within the first year of life [3]. Lesions commonly present on the face in infancy and progress to other sites of the body, particularly skin surfaces that are subject to flexure [1]. In adulthood, these lesions often undergo lichenification [1]. AD affects many facets of daily life for patients, families, and caretakers as itching, scratching, and loss of sleep significantly impact quality of life.
While dry, itchy skin can cause significant discomfort, this disorder is also characterized by a compromised barrier in the skin and possibly other epithelial surfaces, facilitating IgE sensitization to environmental allergens. Penetration of these allergens may contribute to the eventual development of allergic rhinitis and asthma later in childhood in many AD patients, a process known as the atopic march [4]. There are many contributors to the pathogenesis of AD including genetic susceptibilities and dysbiosis of the skin microbiota [3, 5–9]. Genetic predisposition to the development of AD involves genes expressing proteins that contribute to skin barrier function. The most common example is filaggrin, a structural protein that is incorporated into the cornified envelope, a highly crosslinked mixture of structural proteins that surrounds a network of keratin filaments in the outer layer of the epidermis [10]. Mature filaggrin is proteolytically processed from the profilaggrin precursor, which also releases amino acid degradation products that play a vital role in retaining moisture in the skin. However, in addition to the genetic predisposition to AD, there is accumulating evidence demonstrating the central role that the skin microbiome plays in the pathogenesis of AD. Ninety percent of patients with AD are colonized with S. aureus while only 5–20% of healthy individuals are typically colonized [1, 11]. A major challenge with S. aureus is its propensity to form biofilm, which contributes to increasing severity in many diseases [12, 13]. Importantly, staphylococcal biofilms were found to be nearly ubiquitous in AD lesional skin [14••]. Those biofilms are complex microbial communities that provide an advantage to the bacteria in terms of evading the host immune response and resisting antibiotic action. The objective of this review is to summarize what is currently known about staphylococcal biofilms and how they impact skin keratinocytes and influence host immune responses. We will review the basics of biofilms and biofilm formation, how skin-colonizing organisms interact with each other in biofilms, how biofilms may trigger and exacerbate AD, and how these recent developments influence potential directions for clinical management.
Epidemiology of Atopic Dermatitis and the Hygiene Hypothesis
The most valuable AD prevalence and trend data were collected by the International Study of Asthma and Allergies on Childhood (ISAAC), the only global study to use uniformly validated methodology to allow comparisons of populations worldwide [15]. The prevalence of eczema differs between developing and industrialized nations [16], with rates in industrialized nations increasing to as much as 15–30% of children and 2–10% of adults [17]. The ISAAC data was initially collected during 1994–1995 and re-collected 5–10 years later in 56 countries [18]. These data revealed that 58% of participating centers reported an increase in eczema prevalence among older children (13–14 years) [18, 19] and 84% reported increased prevalence of eczema among younger children (6–7 years), with the highest increases seen in Western Europe, Canada, South America, Australasia, and the Far East [18]. While these substantial differences argue that environmental factors and genetic predisposition are key players for eczema development worldwide [18], this also raises the possibility that that skin microbial fluctuations modulate the gene-environment interactions at the skin surface [20].
Although there is a general agreement that microorganisms are potential components of many skin disorders, there is limited literature about how they relate to the genetic and environmental variation that also contributes to the disease [20]. The association of AD with factors that are linked to microbial exposure, such as daycare attendance, living on a farm environment, household pets, endotoxin exposure, and early antibiotic use supports the microbial component of AD [15]. The “revised” hygiene hypothesis theorizes that a decrease in early childhood exposures to infection, and by extension microbial exposure, increases the susceptibility to allergic disease [15], suggesting that diversity in the early microbiota might be important in allergy development and prevention [21]. Other changes in lifestyle in industrialized countries, such as increased skin washing, not only leads to removal of harmful pathogens but also removes antimicrobial peptides produced by the keratinocytes to protect the skin barrier [22]. This concept is timely because it is predicted that two-thirds of the human population will be living in urban areas by 2050, resulting in declining contact with the natural environment [23, 24].
Barrier Function in the Skin
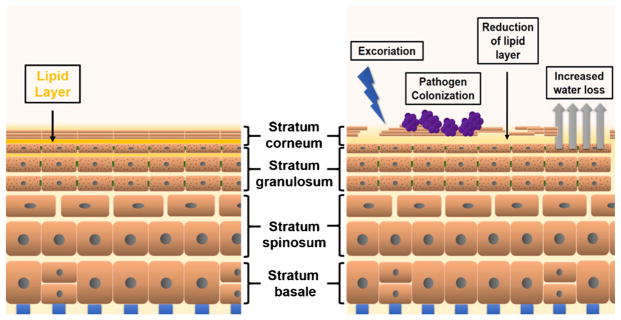
The skin acts as a barrier to environmental insults or allergens and is also a key in preventing water loss from the tissue. The epidermis, the upper layer of the skin, consists primarily of keratinocytes that are regenerated regularly. The skin can be subdivided into several layers with functional specialization (Fig. 1): the outermost stratum corneum, the underlying stratum granulosum, and the lower stratum spinosum and stratum basale [10]. Throughout life, skin is continually being regenerated through the migration of keratinocytes from inner layers outward to the stratum corneum. As keratinocytes migrate upward from the stratum basale through the epidermis, they differentiate and proliferate, lose their organelles, undergo cornification, and slough from the skin [4, 25]. Specifically, the stratum basale contains proliferating, undifferentiated keratinocytes; these migrate up into the stratum spinosum, where they cease proliferating and begin to differentiate. Keratinocytes in the stratum granulosum still contain organelles and are characterized by the presence of granules containing keratohyalin. Finally, the outermost stratum corneum is comprised of dead, flattened keratinocytes that are crosslinked together by corneodesmosomes to form a waxy, dense barrier, a process called cornification [10].
Fig. 1.
Barrier function in healthy and AD skin. Keratinocytes proliferate in the stratum basale and migrate to the stratum granulosum where lipids are secreted into the stratum corneum. The stratum corneum houses keratinocytes that have lost organelles, flatten, and eventually slough off. In AD, increased water loss is a result of a loss of the lipid layer surrounding corneocytes in the inner stratum corneum that acts as a barrier to water-soluble substances. With excoriation, pathogens such as S. aureus are able to colonize the skin more readily
AD can be characterized by defects in the skin barrier that predominate in the stratum corneum. Filaggrin (encoded by the gene FLG) is a structural protein that is crucial to the formation of an intact normal skin barrier; it condenses keratin fibers from the cytoskeleton into tight clusters [25] and is crucial to maintaining the physical strength of the stratum corneum. Mature filaggrin is the result of proteolytic processing of the profilaggrin precursor, which also results in the release of peptides that are essential for skin homeostasis, e.g., natural moisturizing factor (NMF), and for maintenance of an acidic pH [26].
The FLG gene is the most widely studied in AD; loss-of-function mutations, truncations, and null mutations have been identified as contributors to atopy [25]. Patients with filaggrin mutations exhibit perturbed barrier function as a result of the loss of structural integrity in the cornified envelope, which normally functions to minimize water loss and to protect the lower layers of the skin from exposure to external antigens or environmental factors. Although FLG defects predispose individuals to atopic conditions, sensitization is also necessary as shown in mice exposed to external antigens [27–29]. Both lesional and normal-appearing skin of individuals with AD have been shown to have abnormal barrier function, as demonstrated by elevated transepidermal water loss (TEWL) compared to healthy controls. These findings support the idea that AD is a disease of barrier dysfunction [30].
The epidermis is rich in various proteases with multiple targets that are under strict regulation. These proteases act to degrade superfluous proteins, activate downstream pathways that impact terminal differentiation, and cleave precursors of structural proteins or other proteases into mature, processed versions [31]. Given the tight regulation of skin barrier function by the balance between critical proteases such as the kallikreins and protease inhibitors such as SPINK5, it is no surprise that changes in protease levels or activity in the skin contribute to defects in barrier function seen in AD [32–35]. Recently, filaggrin knockdown keratinocytes were shown to have increased endogenous cysteine protease activity, suggesting that the epidermal phenotype observed in FLG deficiency may be due in part to unleashed cysteine protease activity. Indeed, when these cysteine proteases were inhibited, keratin and tight junction proteins were significantly rescued [36]. In addition to endogenous control of protease levels, exogenous factors can also modulate protease secretion in the epidermis. For example, S. aureus induces increased secretion of kallikreins by keratinocytes [37]. These serine proteases can act to degrade filaggrin as well as desmoglein-1, an important component of desmosomes [32, 37]. Cleavage of desmoglein-1 is a normal aspect of desquamation, but dysregulation of this process can lead to impaired barrier function. Different strains of S. aureus and S. epidermidis show varying effects on protease activity of keratinocytes, suggesting that a detailed understanding of staphylococcal colonization at the strain level will be important for understanding the impact on keratinocytes in AD patients.
Skin Microbiome
A crucial factor impacting skin barrier function and both local and systemic immune responses in AD is the skin-specific microbiome. Microbes can influence human health through interactions at host epithelial surfaces, including skin and oral, respiratory, and urogenital mucosae. The interaction of microbiota with the skin surfaces is extensive: including microbial colonization of skin follicles, the total surface area involved is estimated to be 30 square meters [38]. The skin microbiome is less well characterized than the gut microbiome; however, there is increasing interest given recent studies demonstrating that skin commensals influence host immunity [39–41]. The distribution of microbial communities changes with gender, age, and fluctuations in immune status [41] and is also sensitive to changes in humidity or seasonal weather [42]. In contrast, birthing method and feeding method have little effect on the skin microbiome [43]. The composition of the skin microbiome varies widely between different skin sites, dependent in part on whether the skin site is dry, oily, or moist [40, 44••].
The skin microbiome can fluctuate in various states of disease. Dysbiosis is observed in AD, with loss of microbial diversity and over-abundance of certain microbial species. S. aureus prevalence greatly increases during AD flares and decreases after the lesion resolves [44••]. To assess the evolution of dysbiosis in the skin microbiome, a study was conducted to characterize the skin microbiome within the first 6 months of life. Colonization at the antecubital fossa with commensal staphylococcal species (e.g., S. epidermidis and S. cohnii) at 2 months of age was associated with decreased incidence of AD at 1 year [43]. Children who had developed AD at 1 year of age showed a decrease in skin commensals, suggesting that these species are protective. Notably, S. aureus was not observed in any samples collected from infants before the onset of AD symptoms [43]. More studies are needed to assess when S. aureus colonization most commonly occurs and the implications it carries for inflammatory responses.
Recent developments in sequencing technology allow for assessment of microbial communities, including hard-to-culture organisms. 16S rRNA sequencing has been used in many studies to assess the taxa present in a microbial community of the skin. Although 16S rRNA sequencing is relatively affordable, this method typically resolves taxa down to the genus or species level and does not provide strain-level resolution of the microbiome [45, 46••]. Recent studies have emphasized that specific strains of S. aureus or S. epidermidis can differ in the expression of critical virulence or protective factors (e.g., proteases or antimicrobial peptides) that may play important roles in pathogenesis of AD. Thus, in order to fully understand strain-specific effects on atopy, shotgun metagenomic sequencing is necessary, since it can resolve species- and strain-level variation. Both 16S rRNA and shotgun metagenomic sequencing were recently used to verify that the relative abundance of S. aureus rises to a striking degree during AD flares and decreases after treatment [44••, 46••]. S. epidermidis abundance was also observed to increase during AD flares, but to a lesser degree. Byrd et al. showed that S. aureus strains varied between individuals, however, each individual typically was colonized by a single strain of S. aureus. S. epidermidis populations were shown to be more heterogeneous and could vary at different skin sites from the same individual [46••]; such variation is likely due to differences in the skin microenvironment [40, 47••]. mice were colonized with S. aureus strains isolated from healthy controls and S. epidermidis from AD patients, non-inflammatory responses were elicited. However, severe inflammation was noted when mice were colonized with S. aureus strains isolated from AD patients [46••]. The same S. aureus strains also induced an influx of CD4+ T cells and increased secretion of IL-13, demonstrating the ability of specific strains to elicit different inflammatory responses.
S. aureus in AD
S. aureus is known to initiate and aggravate inflammation in AD lesions by secreting a number of factors that modulate host immunity or compromise barrier function in the skin. Staphylococcal alpha toxin, a cytolytic secreted factor, induces cell death in keratinocytes, which is further potentiated in the presence of Th2 cytokines [48]. Furthermore, decreased expression of filaggrin increases the susceptibility of keratinocytes to cytolysis by alpha toxin, due to concomitant decrease in sphingomyelinase levels [49]. In addition to the production of toxins, S. aureus secretes a large number of proteases that are important virulence factors. Among these, the V8 protease and exfoliative toxins A and B have each been demonstrated to cleave desmoglein-1, a critical structural protein within the corneodesmosomes that anchor differentiated keratinocytes to one another [50–52]. Such proteases therefore degrade the barrier function in the skin, increasing water loss and allowing greater exposure to external antigens.
Greater than 80% of S. aureus isolated from patients with AD also secrete superantigens, such as Staphylococcal enterotoxin B (SEB) and Toxic Shock Syndrome Toxin-1 (TSST-1). These toxins crosslink MHC-II and T cell receptors leading to the hyperactivation of T cells. These superantigens lead to significant inflammation in AD and contribute to atopy, as specific IgE against these molecules is often observed [6]. Toxin-producing S. aureus also induces corticosteroid resistance in peripheral blood mononuclear cells (PBMCs) in vitro, as PBMCs stimulated with superantigen were resistant to dexamethasone [53]. In patients with AD, S. aureus isolates from patients that demonstrated corticosteroid resistance exhibited a greater ability to produce superantigens than isolates from corticosteroid-responsive AD or the general population [54].
Staphylococcal Biofilms in AD Skin
Epithelial surfaces are constitutively colonized by bacteria, which commonly exist in the form of biofilm communities. For example, S. epidermidis forms biofilms between squamous epithelial cells in normal skin that vary in thickness depending on the type of skin site (e.g., dry vs. moist), and they colonize sebaceous glands and hair follicles [55]. Furthermore, positive Congo red staining of the epidermis of patients with AD revealed that S. aureus biofilms exist in the eccrine ducts [14••]. Congo red typically stains amyloid proteins, which in the skin are normally found in the dermis as macular amyloid, but the matrix of staphylococcal biofilms contains amyloid and thus stains with Congo red as well. Among S. aureus and S. epidermidis isolates from AD patients, 85% were strong biofilm producers. Interestingly, while staphylococci were found across the body regardless of lesion site, biofilms were only observed in AD lesions [14••]. While the characterization of S. aureus biofilms in the skin is at an early stage, the implications of these findings are intriguing, given that biofilms are associated with refractory, recurrent infections that resist immune responses and antibiotic treatment.
Basics of Biofilm Formation
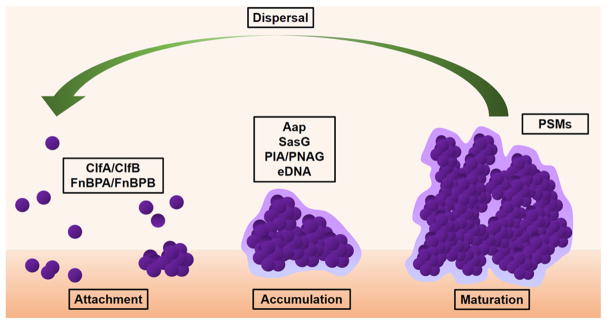
Biofilms are a growth adaptation to environmental stressors; the biofilm growth mode confers resistance to immune defenses and antibiotics [56]. Biofilms are surface-attached microbial communities typically surrounded by extracellular polymeric substances (EPS). EPS is a composite of extracellular DNA, exopolysaccharides, and proteins unique to bacterial biofilms [57]. Staphylococcal biofilm formation begins by adherence of bacteria to a primary surface followed by accumulation of cells via intracellular adhesion mechanisms, and finally the formation of a mature biofilm [58, 59] (Fig. 2). Staphylococcal biofilms can attach to a variety of surfaces, including abiotic material and human tissues, including the skin [60]. The ability of S. epidermidis in particular to adhere to abiotic surfaces and form biofilms is the reason it is the species responsible for the largest number of device-related infections [61]. Both S. aureus and S. epidermidis express a large number of MSCRAMM (microbial surface component recognizing adhesive matrix molecules) adhesion proteins that mediate adherence to host extracellular matrix proteins [62–70]. Several of these staphylococcal MSCRAMM-matrix interactions are relevant in AD. For example, the stratum corneum of AD skin has increased fibronectin relative to healthy control skin, and S. aureus fibronectin-binding protein (FnBP) A and B can interact with fibronectin in human skin [71]. Likewise, the S. aureus MSCRAMM clumping factor B (ClfB) that binds to fibrinogen and several other extracellular matrix proteins was shown to be important in biofilm formation under calcium-depleted conditions [72]. ClfB was recently implicated in facilitating attachment of S. aureus to the stratum corneum [73]. While the binding activity of ClfB varies among S. aureus strains assessed, these studies provide a molecular basis for how S. aureus may initiate colonization on AD skin.
Fig. 2.
Stages of biofilm formation. Bacterial biofilms begin with attachment to a biotic or abiotic surface. Attachment to host tissue typically occurs via MSCRAMM adhesins such as clumping factors (ClfA or ClfB) or fibronectin-binding proteins (FnBPs). Through either polysaccharide (PIA/PNAG) or protein (Aap/SasG) interactions mediating cell-to-cell adhesion, cells begin to accumulate. Remodeling of the biofilm and dispersal of planktonic bacteria is dependent on phenol-soluble modulins (PSMs) under the control of the Agr quorum sensing system
Following attachment, the nascent biofilm forms upon accumulation of bacterial cells via intercellular adhesion events, which occur via two primary mechanisms: polysaccharide-and protein-dependent. The biofilm polysaccharide, poly-N-acetylglucosamine (PNAG, also called polysaccharide intercellular adhesin, PIA), is produced by the biosynthetic enzymes of the icaADBC operon [74–77]. The PNAG polysaccharide has been shown to contribute to staphylococcal biofilm strength under conditions of high shear stress [78]. The alternate mechanism involves bacterial surface proteins, which directly engage one another to allow staphylococcal cells to adhere together in the biofilm. The accumulation-associated protein (Aap) of S. epidermidis is the prototypic member of this protein family; Aap contains an N-terminal A domain upstream of multiple tandem B repeats followed by an extended proline/glycine-rich stalk region [79] that terminates in an LPXTG sortase motif that is covalently attached to the cell wall peptidoglycan [80]. The S. aureus ortholog SasG adopts nearly the same domain arrangement. Proteolytic processing of Aap or SasG removes the A domain, unmasking the B-repeat region, which then allows formation of a protein-dependent biofilm, even in the absence of PNAG [81–83]. The B-repeat superdomain of Aap self-assembles in the presence of Zn2+ ions to form twisted, rope-like filaments between staphylococcal cells [84–88]; similar Zn2+-dependent assembly has been demonstrated for SasG B repeats [89, 90•]. S. epidermidis strains isolated from AD skin contained both the ica operon and aap gene [14••]. More work will be needed to assess the relative importance of PNAG-dependent versus Aap/SasG-dependent biofilm formation among AD-isolated staphylococcal strains.
Mixed Biofilms
Multi-species biofilms are also prevalent; in fact, the majority of microbes in nature likely exist as members of polymicrobial communities [91]. Such mixed-species biofilms represent an interwoven community of organisms with even more complex interactions [92–94]. There are many examples in which bacterial or fungal species synergize by forming cooperative multi-species biofilms, such as the interactions of Candida albicans with Streptococcus gordonii in the oral cavity, or C. albicans with S. aureus in denture stomatitis infections [91]. In some cases, specific interactions between heterologous macromolecules are known to facilitate the inter-species cooperation, as in the case of C. albicans protein Als3 directly binding to Streptococcus gordonii surface protein SspB [95]. Likewise, the staphylococcal biofilm adhesion proteins Aap and SasG have been shown to form heterophilic assemblies, suggesting that these two staphylococcal species might be able to form mixed biofilms [90•]. Indeed, a recent paper demonstrated that S. aureus and S. epidermidis can form mixed biofilms in vitro [96], and at least one example has been published of an infected prosthetic joint that was colonized by a mixed S. aureus and S. epidermidis biofilm [97]. Given the prevalence of both S. aureus and S. epidermidis in AD skin, it is interesting to speculate that such mixed staphylococcal biofilms may play an important role in the pathogenesis of AD.
Control of Staphylococcal Colonization of Skin by Antimicrobial Peptides
Antimicrobial peptides (AMPs) play a key role in cutaneous immunity [25, 98–100] and are secreted by keratinocytes. AMPs can be constitutively active, while others are induced by infection to combat microbes. Key inducible keratinocyte AMPs are human β-defensin 2, β-defensin 3, and cathelicidin (LL-37), which exert their antimicrobial effect by disrupting bacterial cell membranes. These three AMPs have anti-staphylococcal activity and are strongly induced in psoriasis, an inflammatory skin condition. The levels of these AMPs are much lower in AD due to the presence of Th2 cytokines that downregulate AMP expression [99, 101, 102]. AMPs are also important in modulating innate and adaptive immunity, as they can recruit and activate innate and adaptive immune cells [98]. S. epidermidis and other coagulase-negative staphylococci (CoNS) of the skin microbiota can also modulate antimicrobial responses in the skin both directly and indirectly. Certain strains of commensal CoNS provide protection from S. aureus colonization through the direct production of staphylococcal AMPs [7], secretion of lipopeptides that stimulate the release of β-defensin from keratinocytes [8], and the induction of immune cell recruitment via IL-1 and IL-17 secreted from macrophages [40]. The commensal staphylococcal AMPs target S. aureus to inhibit colonization and can act synergistically with keratinocyte-expressed AMPs [98, 103••]. Interestingly, the CoNS strains that expressed AMPs were found to frequently colonize normal skin but were rarely detected on AD lesional skin [103••]. Furthermore, AD skin also exhibits decreased levels of keratinocyte-secreted AMPs [26, 104], which is correlated with increased S. aureus colonization [103••]. Recently, it was shown that the human AMP LL-37 when combined with antimicrobial peptides produced by the commensal Staphylococcus hominis can inhibit S. aureus survival more effectively than human or bacterial AMPs individually. Furthermore, restoring strains of S. epidermidis or S. hominis that inhibited S. aureus growth to the skin of two AD subjects led to significant decreases in S. aureus colonization compared to vehicle alone. These findings suggest that interactions between microbial communities in the skin play a central role in the pathogenesis of AD and that restoration of antimicrobial commensal strains can be an effective way to control S. aureus colonization [103••].
The Impact of Staphylococcal Biofilms on Immune Responses and Keratinocyte Function
A number of studies have begun to assess the specific roles that staphylococcal biofilms play in the pathogenesis of AD. As mentioned, by virtue of growing the biofilm, staphylococci become much more resistant to antibiotic action and immune responses such as phagocytosis. Phagocytosis effectively kills planktonic bacteria and sets the stage for adaptive immune responses [57, 105•]. The formation of a biofilm provides protection to the bacteria within by shielding them from innate immune cells, especially macrophages and neutrophils. Studies have shown that neutrophils are inhibited by S. aureus via neutrophilic lysins such as alpha toxin, which is upregulated upon S. aureus biofilm formation following neutrophil exposure [106]. Macrophages can either be classically activated in order to present antigen and defend against intracellular pathogens, or these cells can undergo “alternate” activation which is crucial in wound healing and contributes to bacterial persistence [57, 107]. The alternate activation of macrophages contributes to chronicity of these infections, which could be important in disease processes such as AD [106]. Biofilms offer protection from macrophage phagocytosis through several mechanisms. The sheer size of biofilms and the density of the extracellular biofilm matrix have been suggested to render them resistant to engulfment—referred to as “frustrated phagocytosis” [105•, 108]. In addition, macrophage phagocytosis is inhibited by specific proteins secreted from S. aureus biofilms, later identified to be alpha toxin, LukA, and LukB. Increased macrophage cytotoxicity was also observed in the presence of S. aureus biofilms. S. epidermidis biofilms containing increased levels of dormant bacteria led to decreased activation of murine macrophages and less secretion of inflammatory cytokines, suggesting that biofilms aid in immune evasion [109].
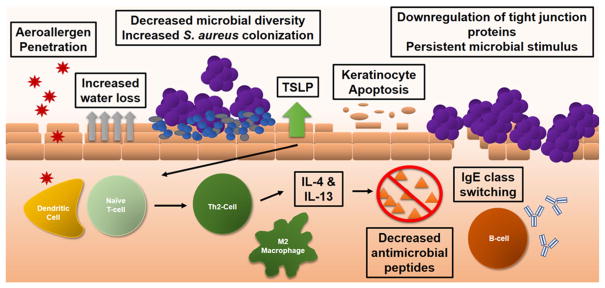
In addition to the immune evasion properties mediated by biofilms that lead to recurrent, hard-to-treat infections, staphylococcal biofilms exert direct effects on keratinocytes. For example, a potentially significant impact of S. aureus in AD patients is its ability to trigger apoptosis in keratinocytes. Keratinocytes exposed to S. aureus biofilms were shown to lose viability and undergo apoptosis after only 3 h of exposure, while those exposed to planktonic culture at 3 h were not statistically different from the control group of keratinocytes alone. Cell morphology was also consistent with keratinocyte apoptosis [110•], although the mechanism for apoptosis was not investigated. This is of importance as damage to epithelial cells releases dsRNA, initiating TLR-3-mediated secretion of thymic stromal lymphopoietin (TSLP) [111]. TSLP secretion results in a strong itch response [112] that can exacerbate excoriation of the skin. Furthermore, TSLP induces dermal dendritic cell activation and recruitment of Th2 cells that secrete IL-4 and IL-13, which have a suppressive effect on AMPs [26], further limiting protection from pathogens. It was also recently shown that extracts of S. aureus biofilms inhibited the terminal differentiation of keratinocytes [113•]. The biofilm extracts induced secretion of IL-6 from the keratinocytes, leading to a decrease in expression of the important differentiation markers keratin 1 and 10, as well as filaggrin (Fig. 3). Furthermore, this block of terminal differentiation renders the keratinocytes more susceptible to the cytotoxic effects of staphylococcal alpha toxin, which was shown to be secreted by S. aureus biofilms grown on reconstructed human epidermal tissue [114].
Fig. 3.
Immune dysfunction in atopic dermatitis. Genetic predispositions, such as FLG mutations, can weaken the physical strength of the epidermal barrier. In addition, colonization by S. aureus also causes inflammation and excoriation, worsening barrier function. With loss of barrier function, aeroallergens are able to interact with dendritic cells, and via antigen presentation, activate naïve T cells. The presence of TSLP enables naïve T cells to differentiate and expand as Th2 cells, which secrete IL-4 and IL-13. These cytokines are important in class switching of B cells to secrete IgE as well as their ability to diminish the secretion of antimicrobial peptides
Clinical Implications
Treatment of AD focuses on preventing or reducing bacterial colonization in lesions and controlling inflammation using moisturizers and topical corticosteroids [1]. Bleach baths are also used to reduce bacterial load present on the skin, and recently, these have been shown to inhibit S. aureus biofilm formation and reverse pre-formed S. aureus biofilms [115]. However, when experiments were repeated on skin biopsies from patients with AD, a 0.16% sodium hypochlorite solution was needed to eradicate 90% of the bacteria present on the biopsy, whereas only 0.005–0.01% sodium hypochlorite solutions were tested on keratinocytes for toxicity [115]. Further studies will be needed to assess the effects of higher amounts of sodium hypochlorite on keratinocytes and to explore the possibility of recurrence of bacterial colonization.
As described earlier, AMPs are key to preventing S. aureus colonization [98–100, 116, 117]. The isolation of commensal strains that produce protective AMPs has been used to assess the effects of AMP replacement on S. aureus colonization. In AD patients, investigators observed a significant difference in S. aureus colonization in skin treated with the commensal bacteria versus vehicle control [103••]. Recent studies have also explored the therapeutic use of human keratinocyte AMPs in treating biofilms; LL-37 was able to eradicate pre-existing MRSA biofilms in a wounded skin model without compromising keratinocyte function [56, 118]. However, additional in vivo studies are needed to determine if replenishing AMPs is an effective treatment option.
As dysbiosis is a driving force in AD, restoration of balance in microbial communities is a target of upcoming treatments. Recently, commensal skin bacteria from healthy human individuals have been transplanted to the skin of mice with induced AD to re-establish a balanced microbiome [119]. To counterbalance the observed decrease in gram-negative (GN) species in patients with AD, culturable GN strains from AD patients and healthy volunteers were tested for their effects on S. aureus. Roseomonas mucosa isolated from healthy volunteers, but not from AD patients, was shown to inhibit S. aureus growth in vitro. These findings suggest that specific GN strains can exert a bacteriostatic effect on S. aureus. When R. mucosa strains from healthy volunteers were transplanted onto the skin of mice, the authors observed decreased colonization of S. aureus, along with improved transepithelial water loss measurements and decreased redness and swelling in the ears [119]. In future studies, it will be interesting to assess the overall contribution of transplanted individual species or even particular strains on AD outcomes. These methods can also be applied to other organisms such as S. epidermidis or other commensal staphylococcal species.
Conclusion
The severity of AD is significantly influenced by the colonization of S. aureus and S. epidermidis, which colonize the skin via microbial communities known as biofilms. Recent studies have reported staphylococcal biofilms colonizing eccrine ducts adjacent to lesional skin in patients with AD, and a number of studies have demonstrated significant impacts of staphylococcal biofilms on the differentiation, apoptosis, or cytokine secretion by keratinocytes. These studies highlight the importance of staphylococcal biofilms in the pathogenesis of AD and highlight the importance of studying host-microbial interactions and their implications for host immunity in AD and allergic disease. Further understanding of S. aureus biofilms in the context of AD will allow for development of better treatments to reduce skin colonization, reduce flares, and dampen the rampant Th2 response that likely contributes to the development of additional co-morbidities.
Acknowledgments
Research by the authors on atopic dermatitis, host epithelial responses, and staphylococcal biofilms has been supported by NIH grants U19 AI070235 (to GKH, JBM, and ABH) and R01 GM094363 (to ABH). We gratefully acknowledge the editorial assistance of Angela Sadler.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Dr. Herr reports the following disclosures: Advisory board member for Hoth Therapeutics, Inc.; Owns equity in Chelexa BioSciences, LLC; Co-inventor on patent EP23106821 licensed to Chelexa BioSciences, LLC; and Co-inventor on patent application US 20140308326 A1. Ms. Gonzalez, Dr. Biagini Myers, and Dr. Khurana Hershey declare no conflicts of interest relevant to this manuscript.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
•• Of major importance
- 1.Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016;387(10023):1109–22. doi: 10.1016/S0140-6736(15)00149-X. [DOI] [PubMed] [Google Scholar]
- 2.Drucker AM, Wang AR, Li WQ, Sevetson E, Block JK, Qureshi AA. The burden of atopic dermatitis: summary of a report for the National Eczema Association. J Invest Dermatol. 2017;137(1):26–30. doi: 10.1016/j.jid.2016.07.012. [DOI] [PubMed] [Google Scholar]
- 3.Biagini Myers JM, Khurana Hershey GK. Eczema in early life: genetics, the skin barrier, and lessons learned from birth cohort studies. J Pediatr. 2010;157(5):704–14. doi: 10.1016/j.jpeds.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng T, Yu J, Oh MH, Zhu Z. The atopic march: progression from atopic dermatitis to allergic rhinitis and asthma. Allergy Asthma Immunol Res. 2011;3(2):67–73. doi: 10.4168/aair.2011.3.2.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brandt EB, Sivaprasad U. Th2 cytokines and atopic dermatitis. J Clin Cell Immunol. 2011;2(3):110. doi: 10.4172/2155-9899.1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Travers JB. Toxic interaction between Th2 cytokines and Staphylococcus aureus in atopic dermatitis. J Invest Dermatol. 2014;134(8):2069–71. doi: 10.1038/jid.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kobayashi T, Glatz M, Horiuchi K, Kawasaki H, Akiyama H, Kaplan DH, et al. Dysbiosis and Staphylococcus aureus colonization drives inflammation in atopic dermatitis. Immunity. 2015;42(4):756–66. doi: 10.1016/j.immuni.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salava A, Lauerma A. Role of the skin microbiome in atopic dermatitis. Clin Transl Allergy. 2014;4:33. doi: 10.1186/2045-7022-4-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Williams MR, Gallo RL. The role of the skin microbiome in atopic dermatitis. Curr Allergy Asthma Rep. 2015;15(11):65. doi: 10.1007/s11882-015-0567-4. [DOI] [PubMed] [Google Scholar]
- 10.Ovaere P, Lippens S, Vandenabeele P, Declercq W. The emerging roles of serine protease cascades in the epidermis. Trends Biochem Sci. 2009;34(9):453–63. doi: 10.1016/j.tibs.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Otto M. Staphylococcus colonization of the skin and antimicrobial peptides. Expert Rev Dermatol. 2010;5(2):183–95. doi: 10.1586/edm.10.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. Staphylococcus aureus biofilms: Properties, regulation and roles in human disease. Virulence. 2011;2(5):445–59. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dasgupta MK. Biofilms and infection in dialysis patients. Semin Dial. 2002;15(5):338–46. doi: 10.1046/j.1525-139x.2002.00084.x. [DOI] [PubMed] [Google Scholar]
- 14••.Allen HB, Vaze ND, Choi C, Hailu T, Tulbert BH, Cusack CA, et al. The presence and impact of biofilm-producing staphylococci in atopic dermatitis. JAMA Dermatol. 2014;150(3):260–5. doi: 10.1001/jamadermatol.2013.8627. The initial study reporting near-ubiquitous S. aureus biofilms in AD lesional skin and showing the activation of TLR2 adjacent to the sweat ducts. [DOI] [PubMed] [Google Scholar]
- 15.Nutten S. Atopic dermatitis: global epidemiology and risk factors. Ann Nutr Metab. 2015;66(Suppl 1):8–16. doi: 10.1159/000370220. [DOI] [PubMed] [Google Scholar]
- 16.Williams H, Flohr C. How epidemiology has challenged 3 prevailing concepts about atopic dermatitis. J Allergy Clin Immunol. 2006;118(1):209–13. doi: 10.1016/j.jaci.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 17.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358(14):1483–94. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 18.Williams H, Stewart A, von Mutius E, Cookson W, Anderson HR. Is eczema really on the increase worldwide? J Allergy Clin Immunol. 2008;121(4):947–54. e15. doi: 10.1016/j.jaci.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 19.Strachan D, Sibbald B, Weiland S, Ait-Khaled N, Anabwani G, Anderson HR, et al. Worldwide variations in prevalence of symptoms of allergic rhinoconjunctivitis in children: the International Study of Asthma and Allergies in Childhood (ISAAC) Pediatr Allergy Immunol. 1997;8(4):161–76. doi: 10.1111/j.1399-3038.1997.tb00156.x. [DOI] [PubMed] [Google Scholar]
- 20.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–53. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang M, Karlsson C, Olsson C, Adlerberth I, Wold AE, Strachan DP, et al. Reduced diversity in the early fecal microbiota of infants with atopic eczema. J Allergy Clin Immunol. 2008;121(1):129–34. doi: 10.1016/j.jaci.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 22.Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010;160(1):1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.United Nations. World Urbanization Prospects; the 2007 revision. United Nations Department of Economic and Social Affairs, Population Division; New York: 2008. [Google Scholar]
- 24.Bendiks M, Kopp MV. The relationship between advances in understanding the microbiome and the maturing hygiene hypothesis. Curr Allergy Asthma Rep. 2013;13(5):487–94. doi: 10.1007/s11882-013-0382-8. [DOI] [PubMed] [Google Scholar]
- 25.Agrawal R, Woodfolk JA. Skin barrier defects in atopic dermatitis. Curr Allergy Asthma Rep. 2014;14(5):433. doi: 10.1007/s11882-014-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ong PY, Leung DY. Bacterial and viral infections in atopic dermatitis: a comprehensive review. Clin Rev Allergy Immunol. 2016;51(3):329–37. doi: 10.1007/s12016-016-8548-5. [DOI] [PubMed] [Google Scholar]
- 27.Lakatos G, Soproni K, Doka A, Miklosi A. A comparative approach to dogs’ (Canis familiaris) and human infants’ comprehension of various forms of pointing gestures. Anim Cogn. 2009;12(4):621–31. doi: 10.1007/s10071-009-0221-4. [DOI] [PubMed] [Google Scholar]
- 28.Kawasaki H, Nagao K, Kubo A, Hata T, Shimizu A, Mizuno H, et al. Altered stratum corneum barrier and enhanced percutaneous immune responses in filaggrin-null mice. J Allergy Clin Immunol. 2012;129(6):1538–46. e6. doi: 10.1016/j.jaci.2012.01.068. [DOI] [PubMed] [Google Scholar]
- 29.Oyoshi MK, Murphy GF, Geha RS. Filaggrin-deficient mice exhibit TH17-dominated skin inflammation and permissiveness to epicutaneous sensitization with protein antigen. J Allergy Clin Immunol. 2009;124(3):485–93. 93 e1. doi: 10.1016/j.jaci.2009.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gupta J, Grube E, Ericksen MB, Stevenson MD, Lucky AW, Sheth AP, et al. Intrinsically defective skin barrier function in children with atopic dermatitis correlates with disease severity. J Allergy Clin Immunol. 2008;121(3):725–30. e2. doi: 10.1016/j.jaci.2007.12.1161. [DOI] [PubMed] [Google Scholar]
- 31.de Veer SJ, Furio L, Harris JM, Hovnanian A. Proteases: common culprits in human skin disorders. Trends Mol Med. 2014;20(3):166–78. doi: 10.1016/j.molmed.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Fischer J, Meyer-Hoffert U. Regulation of kallikrein-related peptidases in the skin—from physiology to diseases to therapeutic options. Thromb Haemost. 2013;110(3):442–9. doi: 10.1160/TH12-11-0836. [DOI] [PubMed] [Google Scholar]
- 33.Deraison C, Bonnart C, Lopez F, Besson C, Robinson R, Jayakumar A, et al. LEKTI fragments specifically inhibit KLK5, KLK7, and KLK14 and control desquamation through a pH-dependent interaction. Mol Biol Cell. 2007;18(9):3607–19. doi: 10.1091/mbc.E07-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weidinger S, Baurecht H, Wagenpfeil S, Henderson J, Novak N, Sandilands A, et al. Analysis of the individual and aggregate genetic contributions of previously identified serine peptidase inhibitor Kazal type 5 (SPINK5), kallikrein-related peptidase 7 (KLK7), and filaggrin (FLG) polymorphisms to eczema risk. J Allergy Clin Immunol. 2008;122(3):560–8. e4. doi: 10.1016/j.jaci.2008.05.050. [DOI] [PubMed] [Google Scholar]
- 35.Walley AJ, Chavanas S, Moffatt MF, Esnouf RM, Ubhi B, Lawrence R, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001;29(2):175–8. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 36.Wang XW, Wang JJ, Gutowska-Owsiak D, Salimi M, Selvakumar TA, Gwela A, et al. Deficiency of filaggrin regulates endogenous cysteine protease activity, leading to impaired skin barrier function. Clin Exp Dermatol. 2017;42(6):622–31. doi: 10.1111/ced.13113. [DOI] [PubMed] [Google Scholar]
- 37.Williams MR, Nakatsuji T, Sanford JA, Vrbanac AF, Gallo RL. Staphylococcus aureus induces increased serine protease activity in keratinocytes. J Invest Dermatol. 2017;137(2):377–84. doi: 10.1016/j.jid.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gallo RL. Human skin is the largest epithelial surface for interaction with microbes. J Invest Dermatol. 2017;137(6):1213–4. doi: 10.1016/j.jid.2016.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dybboe R, Bandier J, Skov L, Engstrand L, Johansen JD. The role of the skin microbiome in atopic dermatitis: a systematic review. Br J Dermatol. 2017 doi: 10.1111/bjd.15390. https://doi.org/10.1111/bjd.15390. [DOI] [PubMed]
- 40.Belkaid Y, Segre JA. Dialogue between skin microbiota and immunity. Science (New York, NY) 2014;346(6212):954–9. doi: 10.1126/science.1260144. [DOI] [PubMed] [Google Scholar]
- 41.SanMiguel A, Grice EA. Interactions between host factors and the skin microbiome. Cell Mol Life Sci. 2015;72(8):1499–515. doi: 10.1007/s00018-014-1812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kong HH. Skin microbiome: genomics-based insights into the diversity and role of skin microbes. Trends Mol Med. 2011;17(6):320–8. doi: 10.1016/j.molmed.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennedy EA, Connolly J, Hourihane JO, Fallon PG, McLean WH, Murray D, et al. Skin microbiome before development of atopic dermatitis: early colonization with commensal staphylococci at 2 months is associated with a lower risk of atopic dermatitis at 1 year. J Allergy Clin Immunol. 2017;139(1):166–72. doi: 10.1016/j.jaci.2016.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44••.Kong HH, Oh J, Deming C, Conlan S, Grice EA, Beatson MA, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–9. doi: 10.1101/gr.131029.111. A study describing dysbiosis in active AD lesions using 16S rRNA sequencing, showing increased prevalence of both S. aureus and S. epidermidis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT. Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics. PLoS One. 2014;9(4):e93827. doi: 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46••.Byrd AL, Deming C, Cassidy SKB, Harrison OJ, Ng WI, Conlan S, et al. Staphylococcus aureus and Staphylococcus epidermidis strain diversity underlying pediatric atopic dermatitis. Sci Transl Med. 2017;9(397):eaal4651. doi: 10.1126/scitranslmed.aal4651. A study using metagenomic shotgun sequencing to identify strain-level differences in S. aureus and S. epidermidis colonization in pediatric AD patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47••.Oh J, Byrd AL, Deming C, Conlan S, Program NCS, Kong HH, et al. Biogeography and individuality shape function in the human skin metagenome. Nature. 2014;514(7520):59–64. doi: 10.1038/nature13786. The first metagenomic survey of different healthy human skin sites, which lays the foundation for studies to assess changes of the skin microbiome in disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brauweiler AM, Goleva E, Leung DY. Th2 cytokines increase Staphylococcus aureus alpha toxin-induced keratinocyte death through the signal transducer and activator of transcription 6 (STAT6) J Invest Dermatol. 2014;134(8):2114–21. doi: 10.1038/jid.2014.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brauweiler AM, Bin L, Kim BE, Oyoshi MK, Geha RS, Goleva E, et al. Filaggrin-dependent secretion of sphingomyelinase protects against staphylococcal alpha-toxin-induced keratinocyte death. J Allergy Clin Immunol. 2013;131(2):421–7. e1–2. doi: 10.1016/j.jaci.2012.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat Med. 2000;6(11):1275–7. doi: 10.1038/81385. [DOI] [PubMed] [Google Scholar]
- 51.Hanakawa Y, Schechter NM, Lin C, Nishifuji K, Amagai M, Stanley JR. Enzymatic and molecular characteristics of the efficiency and specificity of exfoliative toxin cleavage of desmoglein 1. J Biol Chem. 2004;279(7):5268–77. doi: 10.1074/jbc.M311087200. [DOI] [PubMed] [Google Scholar]
- 52.Amagai M, Yamaguchi T, Hanakawa Y, Nishifuji K, Sugai M, Stanley JR. Staphylococcal exfoliative toxin B specifically cleaves desmoglein 1. J Invest Dermat. 2002;118(5):845–50. doi: 10.1046/j.1523-1747.2002.01751.x. [DOI] [PubMed] [Google Scholar]
- 53.Hauk PJ, Hamid QA, Chrousos GP, Leung DY. Induction of corticosteroid insensitivity in human PBMCs by microbial superantigens. J Allergy Clin Immunol. 2000;105(4):782–7. doi: 10.1067/mai.2000.105807. [DOI] [PubMed] [Google Scholar]
- 54.Schlievert PM, Case LC, Strandberg KL, Abrams BB, Leung DY. Superantigen profile of Staphylococcus aureus isolates from patients with steroid-resistant atopic dermatitis. Clin Infect Dis. 2008;46(10):1562–7. doi: 10.1086/586746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costerton W, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest. 2003;112(10):1466–77. doi: 10.1172/JCI20365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pletzer D, Hancock RE. Antibiofilm peptides: potential as broad-spectrum agents. J Bacteriol. 2016;198(19):2572–8. doi: 10.1128/JB.00017-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Watters C, Fleming D, Bishop D, Rumbaugh KP. Host responses to biofilm. Prog Mol Biol Transl Sci. 2016;142:193–239. doi: 10.1016/bs.pmbts.2016.05.007. [DOI] [PubMed] [Google Scholar]
- 58.Otto M. Staphylococcal infections: mechanisms of biofilm maturation and detachment as critical determinants of pathogenicity. Annu Rev Med. 2013;64:175–88. doi: 10.1146/annurev-med-042711-140023. [DOI] [PubMed] [Google Scholar]
- 59.Otto M. Staphylococcal biofilms. Curr Top Microbiol Immunol. 2008;322:207–28. doi: 10.1007/978-3-540-75418-3_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vlassova N, Han A, Zenilman JM, James G, Lazarus GS. New horizons for cutaneous microbiology: the role of biofilms in dermatological disease. Br J Dermatol. 2011;165(4):751–9. doi: 10.1111/j.1365-2133.2011.10458.x. [DOI] [PubMed] [Google Scholar]
- 61.Otto M. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat Rev Microbiol. 2009;7(8):555–67. doi: 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moormeier DE, Bose JL, Horswill AR, Bayles KW. Temporal and stochastic control of Staphylococcus aureus biofilm development. MBio. 2014;5(5):e01341–14. doi: 10.1128/mBio.01341-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharp JA, Echague CG, Hair PS, Ward MD, Nyalwidhe JO, Geoghegan JA, et al. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS One. 2012;7(5):e38407. doi: 10.1371/journal.pone.0038407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Foster TJ, Hook M. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol. 1998;6(12):484–8. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 65.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, et al. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115(2):217–28. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 66.Zhang X, Wu M, Zhuo W, Gu J, Zhang S, Ge J, et al. Crystal structures of Bbp from Staphylococcus aureus reveal the ligand binding mechanism with Fibrinogen alpha. Protein Cell. 2015;6(10):757–66. doi: 10.1007/s13238-015-0205-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, et al. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008;4(11):e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiang H, Feng Y, Wang J, Liu B, Chen Y, Liu L, et al. Crystal structures reveal the multi-ligand binding mechanism of Staphylococcus aureus ClfB. PLoS Pathog. 2012;8(6):e1002751. doi: 10.1371/journal.ppat.1002751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Askarian F, Ajayi C, Hanssen AM, van Sorge NM, Pettersen I, Diep DB, et al. The interaction between Staphylococcus aureus SdrD and desmoglein 1 is important for adhesion to host cells. Sci Rep. 2016;6:22134. doi: 10.1038/srep22134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbu EM, Ganesh VK, Gurusiddappa S, Mackenzie RC, Foster TJ, Sudhof TC, et al. Beta-neurexin is a ligand for the Staphylococcus aureus MSCRAMM SdrC. PLoS Pathog. 2010;6(1):e1000726. doi: 10.1371/journal.ppat.1000726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cho SH, Strickland I, Boguniewicz M, Leung DY. Fibronectin and fibrinogen contribute to the enhanced binding of Staphylococcus aureus to atopic skin. J Allergy Clin Immunol. 2001;108(2):269–74. doi: 10.1067/mai.2001.117455. [DOI] [PubMed] [Google Scholar]
- 72.Abraham NM, Jefferson KK. Staphylococcus aureus clumping factor B mediates biofilm formation in the absence of calcium. Microbiology. 2012;158(Pt 6):1504–12. doi: 10.1099/mic.0.057018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fleury OM, McAleer MA, Feuillie C, Formosa-Dague C, Sansevere E, Bennett DE, et al. Clumping Factor B promotes adherence of Staphylococcus aureus to corneocytes in atopic dermatitis. Infect Immun. 2017;85(6):e00994. doi: 10.1128/IAI.00994-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vuong C, Voyich JM, Fischer ER, Braughton KR, Whitney AR, DeLeo FR, et al. Polysaccharide intercellular adhesin (PIA) protects Staphylococcus epidermidis against major components of the human innate immune system. Cell Microbiol. 2004;6(3):269–75. doi: 10.1046/j.1462-5822.2004.00367.x. [DOI] [PubMed] [Google Scholar]
- 75.Formosa-Dague C, Feuillie C, Beaussart A, Derclaye S, Kucharikova S, Lasa I, et al. Sticky matrix: adhesion mechanism of the Staphylococcal polysaccharide intercellular adhesin. ACS Nano. 2016;10(3):3443–52. doi: 10.1021/acsnano.5b07515. [DOI] [PubMed] [Google Scholar]
- 76.Cramton SE, Gerke C, Schnell NF, Nichols WW, Gotz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67(10):5427–33. doi: 10.1128/iai.67.10.5427-5433.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cue D, Lei MG, Lee CY. Genetic regulation of the intercellular adhesion locus in Staphylococci. Front Cell Infect Microbiol. 2012;2:38. doi: 10.3389/fcimb.2012.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaeffer CR, Hoang TN, Sudbeck CM, Alawi M, Tolo IE, Robinson DA, et al. Versatility of biofilm matrix molecules in Staphylococcus epidermidis clinical isolates and importance of polysaccharide intercellular adhesin expression during high shear stress. mSphere. 2016;1(5):e00165. doi: 10.1128/mSphere.00165-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yarawsky AE, English LR, Whitten ST, Herr AB. The proline/glycine-rich region of the biofilm adhesion protein Aap forms an extended stalk that resists compaction. J Mol Biol. 2017;429(2):261–79. doi: 10.1016/j.jmb.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hussain M, Herrmann M, von Eiff C, Perdreau-Remington F, Peters G. A 140-kilodalton extracellular protein is essential for the accumulation of Staphylococcus epidermidis strains on surfaces. Infect Immun. 1997;65(2):519–24. doi: 10.1128/iai.65.2.519-524.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paharik AE, Kotasinska M, Both A, Hoang TN, Buttner H, Roy P, et al. The metalloprotease SepA governs processing of accumulation-associated protein and shapes intercellular adhesive surface properties in Staphylococcus epidermidis. Mol Microbiol. 2017;103(5):860–74. doi: 10.1111/mmi.13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rohde H, Burdelski C, Bartscht K, Hussain M, Buck F, Horstkotte MA, et al. Induction of Staphylococcus epidermidis biofilm formation via proteolytic processing of the accumulation-associated protein by staphylococcal and host proteases. Mol Microbiol. 2005;55(6):1883–95. doi: 10.1111/j.1365-2958.2005.04515.x. [DOI] [PubMed] [Google Scholar]
- 83.Corrigan RM, Rigby D, Handley P, Foster TJ. The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology. 2007;153(Pt 8):2435–46. doi: 10.1099/mic.0.2007/006676-0. [DOI] [PubMed] [Google Scholar]
- 84.Conrady DG, Brescia CC, Horii K, Weiss AA, Hassett DJ, Herr AB. A zinc-dependent adhesion module is responsible for intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A. 2008;105(49):19456–61. doi: 10.1073/pnas.0807717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Herr AB, Conrady DG. Thermodynamic analysis of metal ion-induced protein assembly. Methods Enzymol. 2011;488:101–21. doi: 10.1016/B978-0-12-381268-1.00005-7. [DOI] [PubMed] [Google Scholar]
- 86.Conrady DG, Wilson JJ, Herr AB. Structural basis for Zn2+-dependent intercellular adhesion in staphylococcal biofilms. Proc Natl Acad Sci U S A. 2013;110(3):E202–11. doi: 10.1073/pnas.1208134110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shelton CL, Conrady DG, Herr AB. Functional consequences of B-repeat sequence variation in the staphylococcal biofilm protein Aap: deciphering the assembly code. Biochem J. 2017;474(3):427–43. doi: 10.1042/BCJ20160675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chaton CT, Herr AB. Defining the metal specificity of a multi-functional biofilm adhesion protein. Protein Sci. 2017;26:1964–73. doi: 10.1002/pro.3232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Geoghegan JA, Corrigan RM, Gruszka DT, Speziale P, O’Gara JP, Potts JR, et al. Role of surface protein SasG in biofilm formation by Staphylococcus aureus. J Bacteriol. 2010;192(21):5663–73. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90•.Formosa-Dague C, Speziale P, Foster TJ, Geoghegan JA, Dufrene YF. Zinc-dependent mechanical properties of Staphylococcus aureus biofilm-forming surface protein SasG. Proc Natl Acad Sci U S A. 2016;113(2):410–5. doi: 10.1073/pnas.1519265113. A study demonstrating Zn2+-dependent intercellular adhesion between S. aureus cells mediated by SasG, and heterophilic adhesion between S. aureus and S. epidermidis mediated by SasG/Aap, using single-cell force microscopy. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peters BM, Jabra-Rizk MA, O’May GA, Costerton JW, Shirtliff ME. Polymicrobial interactions: impact on pathogenesis and human disease. Clin Microbiol Rev. 2012;25(1):193–213. doi: 10.1128/CMR.00013-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14(2):93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wolcott R, Costerton JW, Raoult D, Cutler SJ. The polymicrobial nature of biofilm infection. Clin Microbiol Infect. 2013;19(2):107–12. doi: 10.1111/j.1469-0691.2012.04001.x. [DOI] [PubMed] [Google Scholar]
- 94.Gabrilska RA, Rumbaugh KP. Biofilm models of polymicrobial infection. Future Microbiol. 2015;10(12):1997–2015. doi: 10.2217/fmb.15.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Silverman RJ, Nobbs AH, Vickerman MM, Barbour ME, Jenkinson HF. Interaction of Candida albicans cell wall Als3 protein with Streptococcus gordonii SspB adhesin promotes development of mixed-species communities. Infect Immun. 2010;78(11):4644–52. doi: 10.1128/IAI.00685-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stewart EJ, Payne DE, Ma TM, VanEpps JS, Boles BR, Younger JG, et al. Effect of antimicrobial and physical treatments on growth of multispecies Staphylococcal biofilms. Appl Environ Microbiol. 2017;83(12):e03483–16. doi: 10.1128/AEM.03483-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stoodley P, Conti SF, DeMeo PJ, Nistico L, Melton-Kreft R, Johnson S, et al. Characterization of a mixed MRSA/MRSE biofilm in an explanted total ankle arthroplasty. FEMS Immunol Med Microbiol. 2011;62(1):66–74. doi: 10.1111/j.1574-695X.2011.00793.x. [DOI] [PubMed] [Google Scholar]
- 98.Zhang LJ, Gallo RL. Antimicrobial peptides. Curr Biol. 2016;26(1):R14–9. doi: 10.1016/j.cub.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 99.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347(15):1151–60. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 100.Powers CE, McShane DB, Gilligan PH, Burkhart CN, Morrell DS. Microbiome and pediatric atopic dermatitis. J Dermatol. 2015;42(12):1137–42. doi: 10.1111/1346-8138.13072. [DOI] [PubMed] [Google Scholar]
- 101.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171(6):3262–9. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 102.Hata TR, Gallo RL. Antimicrobial peptides, skin infections, and atopic dermatitis. Semin Cutan Med Surg. 2008;27(2):144–50. doi: 10.1016/j.sder.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103••.Nakatsuji T, Chen TH, Narala S, Chun KA, Two AM, Yun T, et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci Transl Med. 2017;9(378):eaah4680. doi: 10.1126/scitranslmed.aah4680. A study describing AMPs that inhibit S. aureus from strains of coagulase-negative staphylococci common on the skin of healthy individuals but rare in AD patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.de Koning HD, Kamsteeg M, Rodijk-Olthuis D, van Vlijmen-Willems IM, van Erp PE, Schalkwijk J, et al. Epidermal expression of host response genes upon skin barrier disruption in normal skin and uninvolved skin of psoriasis and atopic dermatitis patients. J Invest Dermatol. 2011;131(1):263–6. doi: 10.1038/jid.2010.278. [DOI] [PubMed] [Google Scholar]
- 105•.Scherr TD, Hanke ML, Huang O, James DB, Horswill AR, Bayles KW, et al. Staphylococcus aureus biofilms induce macrophage dysfunction through leukocidin AB and alpha-toxin. MBio. 2015;6(4):e01021. doi: 10.1128/mBio.01021-15. A paper identifying protein factors secreted from S. aureus biofilms that inhibit macrophage phagocytosis, illustrating how S. aureus biofilms can evade host defense. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Scherr TD, Heim CE, Morrison JM, Kielian T. Hiding in plain sight: interplay between Staphylococcal biofilms and host immunity. Front Immunol. 2014;5:37. doi: 10.3389/fimmu.2014.00037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Paharik AE, Horswill AR. The staphylococcal biofilm: adhesins, regulation, and host response. Microbiol Spectr. 2016;4(2) doi: 10.1128/microbiolspec.VMBF-0022-2015. VMBF-0022-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Thurlow LR, Hanke ML, Fritz T, Angle A, Aldrich A, Williams SH, et al. Staphylococcus aureus biofilms prevent macrophage phagocytosis and attenuate inflammation in vivo. J Immunol. 2011;186(11):6585–96. doi: 10.4049/jimmunol.1002794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Cerca F, Andrade F, Franca A, Andrade EB, Ribeiro A, Almeida AA, et al. Staphylococcus epidermidis biofilms with higher proportions of dormant bacteria induce a lower activation of murine macrophages. J Med Microbiol. 2011;60(Pt 12):1717–24. doi: 10.1099/jmm.0.031922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110•.Tankersley A, Frank MB, Bebak M, Brennan R. Early effects of Staphylococcus aureus biofilm secreted products on inflammatory responses of human epithelial keratinocytes. J Inflamm (Lond) 2014;11:17. doi: 10.1186/1476-9255-11-17. A paper demonstrating that S. aureus biofilm conditioned media induces significantly stronger inflammatory responses in human keratinocytes compared to planktonic conditioned media. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Takai T. TSLP expression: cellular sources, triggers, and regulatory mechanisms. Allergol Int. 2012;61(1):3–17. doi: 10.2332/allergolint.11-RAI-0395. [DOI] [PubMed] [Google Scholar]
- 112.Wilson SR, The L, Batia LM, Beattie K, Katibah GE, McClain SP, et al. The epithelial cell-derived atopic dermatitis cytokine TSLP activates neurons to induce itch. Cell. 2013;155(2):285–95. doi: 10.1016/j.cell.2013.08.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113•.Son ED, Kim HJ, Park T, Shin K, Bae IH, Lim KM, et al. Staphylococcus aureus inhibits terminal differentiation of normal human keratinocytes by stimulating interleukin-6 secretion. J Dermatol Sci. 2014;74(1):64–71. doi: 10.1016/j.jdermsci.2013.12.004. A study exploring the impact of S. aureus on keratinocyte differentiation, describing the increase in IL-6 and decrease in filaggrin and other differentiation markers upon exposure to S. aureus. [DOI] [PubMed] [Google Scholar]
- 114.den Reijer PM, Haisma EM, Lemmens-den Toom NA, Willemse J, Koning RI, Demmers JA, et al. Detection of alpha-toxin and other virulence factors in biofilms of Staphylococcus aureus on polystyrene and a human epidermal model. PLoS One. 2016;11(1):e0145722. doi: 10.1371/journal.pone.0145722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Eriksson S, van der Plas MJA, Morgelin M, Sonesson A. Antibacterial and antibiofilm effects of sodium hypochlorite against Staphylococcus aureus isolates derived from patients with atopic dermatitis. Br J Dermatol. 2017;177(2):513–21. doi: 10.1111/bjd.15410. [DOI] [PubMed] [Google Scholar]
- 116.Batoni G, Maisetta G, Esin S. Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta. 2016;1858(5):1044–60. doi: 10.1016/j.bbamem.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 117.Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. 2011;242(1):233–46. doi: 10.1111/j.1600-065X.2011.01027.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haisma EM, de Breij A, Chan H, van Dissel JT, Drijfhout JW, Hiemstra PS, et al. LL-37-derived peptides eradicate multidrug-resistant Staphylococcus aureus from thermally wounded human skin equivalents. Antimicrob Agents Chemother. 2014;58(8):4411–9. doi: 10.1128/AAC.02554-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Myles IA, Williams KW, Reckhow JD, Jammeh ML, Pincus NB, Sastalla I, et al. Transplantation of human skin microbiota in models of atopic dermatitis. JCI Insight. 2016;1(10):e86955. doi: 10.1172/jci.insight.86955. [DOI] [PMC free article] [PubMed] [Google Scholar]