Abstract
Congenital heart defects (CHD) are structural malformations found at birth with a prevalence of 1%. The clinical trajectory of CHD is highly variable and thus in need of robust diagnostics and therapeutics. Major surgical interventions are often required for most CHDs. In Africa, despite advances in life sciences infrastructure and improving education of medical scholars, the limited clinical data suggest that CHD detection and correction are still not at par with the rest of the world. But the toll and genetics of CHDs in Africa has seldom been systematically investigated. We present an expert review on CHD with lessons learned on Africa. We found variable CHD phenotype prevalence in Africa across countries and populations. There are important gaps and paucity in genomic studies of CHD in African populations. Among the available genomic studies, the key findings in Africa were variants in GATA4 (P193H), MTHFR 677TT, and MTHFR 1298CC that were associated with atrial septal defect, ventricular septal defect (VSD), Tetralogy of Fallot (TOF), and patent ductus arteriosus phenotypes and 22q.11 deletion, which is associated with TOF. There were no data on epigenomic association of CHD in Africa, however, other studies have shown an altered expression of miR-421 and miR-1233-3p to be associated with TOF and hypermethylation of CpG islands in the promoter of SCO2 gene also been associated with TOF and VSD in children with non-syndromic CHD. These findings signal the urgent need to develop and implement genetic and genomic research on CHD to identify the hereditary and genome–environment interactions contributing to CHD. These projected studies would also offer comparisons on CHD pathophysiology between African and other populations worldwide. Genomic research on CHD in Africa should be developed in parallel with next generation technology policy research and responsible innovation frameworks that examine the social and political factors that shape the emergence and societal embedding of new technologies.
Keywords: : congenital heart defects, genomics, epigenomics, global health, responsible innovation
Introduction
Congenital heart defects (CHD) are among the most common human congenital anomalies, occurring in an accepted approximation of 6 to 8 out of 1000 live-births (Hoffman and Kaplan, 2002; van der Linde et al., 2011) and accounting for nearly one-third of all major congenital anomalies. CHD are a group of structural heart and great vessel disorders present at birth.
This expert review examines the genes and genomic loci that are reportedly associated with the CHD phenotypic distribution in different populations. Based on these observations and literature analysis, we propose a case for a closer look at the African continent in regard to CHD biomarkers and CHD genomics, especially for neglected or understudied populations. In addition, we present an original enrichment analysis as part of our literature analyses so as to evaluate possible interactions between known genes that have been implicated in non-syndromic CHD showing the complexity of CHD.
Clinical Context for CHD
Errors in septation, proper partitioning of the great vessels, and valve formation are the most common aberrations in cardiac development. Frequently identified risk factors predisposing newborns to developing CHD include genetics factors such as consanguinity, advanced maternal age that is associated with aneuploidies, environmental factors such as a vitamin A (Botto et al., 2001) and folic acid-deficient diet during pregnancy, maternal drug/teratogen exposure, and previous obstetric history of abortions and still births (Abqari et al., 2016; Fung et al., 2013; Ul Haq et al., 2011).
The epidemiology of CHD varies across different populations with a recent study in China indicating approximately 11.1 per 1000 live births (Qu et al., 2016) are affected by CHD, which was higher than previously reported. The Metropolitan Atlanta Congenital Defects Program (MACDP) surveillance system in Atlanta, reported an overall prevalence of 81.4/10,000 births (Reller et al., 2008). A study in Northern England from 1985 to 2003 indicated a CHD livebirth prevalence of 79.7 per 10,000 livebirths (Dadvand et al., 2009).
The European surveillance of congenital anomalies, a network of population-based registers of congenital anomalies in Europe showed that CHD are the most common non-chromosomal anomaly with a prevalence of 5.8–6.5/1000 births (Bourdial et al., 2012; Dolk et al., 2010). In Asia, a study conducted in Iran, utilizing data over a 1-year period (2007–2008) showed a prevalence of 8.6 per 1000 live births, with an upward trend during this period (Nikyar et al., 2011). A nationwide study of birth prevalence of CHD in Norway over a 15-year period (1994–2009) showed a prevalence of 133.2 per 10,000 live births, which is on the high side compared to the average prevalence of 8/1000 live births (Leirgul et al., 2014).
Data on clinical epidemiology on CHD in Africa vary greatly due to differences in diagnostic methods and expertise. In the Niger Delta region of Nigeria, a 4-year prospective echocardiographic data for frequency and pattern of CHD showed a prevalence of 14.4 per 1000 children (Otaigbe and Tabansi, 2014). Recently, echocardiography data from a cardiac referral hospital in Cameroon showed that 27.2% of suspected heart diseases cases reported involved definite structural CHD (Nkoke et al., 2017). CHDs are usually found in the pediatric age group (0–15 years), however, it is not rare to see adults with corrected or uncorrected CHDs, especially in Africa. Some patients may not manifest any symptomatic characteristic of CHD and thus, the condition might not be noticed until complications arise later.
The variable CHD prevalence presented in the various studies across both developed and developing countries shows the complexity in clinical CHD epidemiology. Even in high resource countries, CHD can be missed at birth resulting in infant mortality (Ng and Hokanson, 2010; Wren et al., 2008). In developing nations, missed diagnoses has the potential to be much worse secondary to limited resources (Ekure and Adeyemo, 2015). Fortunately, with the advancement in prenatal cardiovascular diagnostics and corrective strategies such as cardiothoracic surgery, the number of infantile deaths due to CHD has declined, and more than 75% of children with CHD survive into adulthood, including those with complex abnormalities (Gilboa et al., 2010), in high resource countries; unfortunately, many countries in Africa do not share these success stories despite the improvement in health infrastructure secondary to financial and policy limitations.
CHD can be linked to recognizable genetic syndromes that are due to aneuploidy such as Down syndrome, copy-number variations such as 22q11.2 deletion syndrome and single gene disorders such as Noonan syndrome (Fahed et al., 2013). Isolated or sporadic CHD (also referred to as non-syndromic CHDs) make up most CHDs. Advances are being made to elucidate the etiology of these conditions due to progress in genomic technology and animal models; however, most non-syndromic CHD remains without a known etiology. In addition to genetic causes, environmental exposures and nutritional deficiencies of pregnant mothers during pregnancy has long been suspected as some of the plausible risk for non-syndromic CHD.
In addition to DNA coding genetic changes and environmental and nutritional associations with CHD, epigenetic mechanisms are a new area of investigation. Here, “gene silencing” in the parent are activated in the offspring leading to NS-CHD (Fahed and Nemer, 2012). Epigenetic changes resulting in gene enrichment may in part explain the incomplete penetrance and low recurrence rates in familial NS-CHD (Sifrim et al., 2016).
Syndromic CHDs such as Noonan or Alagille syndrome have a monogenic mode of inheritance (Prendiville et al., 2014). In contrast, most of the observed non-syndromic CHD (NS-CHD) occur sporadically and families with clear monogenic inheritance of non-syndromic CHDs are scarce (Garg et al., 2003; Kirk et al., 2007). Incomplete penetrance and genetic heterogeneity are thought to contribute to apparent non-Mendelian inheritance in affected families. Non-syndromic CHD are usually explained by a multifactorial inheritance model that comprises a multitude of susceptibility genes with low-penetrance mutations (common variants) or intermediate-penetrance mutations (rare variants) superposed on adverse environmental factors (Burn et al., 1998; Oyen et al., 2010; Roos-Hesselink et al., 2005).
Non-syndromic CHD phenotypes worldwide have also been linked to parental consanguinity (Stavsky et al., 2017) and recessive etiology (McGregor et al., 2010). Genes that have been implicated in CHD can be categorized into ligand receptors, transcriptional factors, and structural/contractile proteins (Wessels and Willems, 2010) (Table 1).
Table 1.
Genes Involved in Heart Development Associated with Congenital Heart Defects
| Gene | Cytogenic location | Function | Mim number | Chd associated phenotype | Reference |
|---|---|---|---|---|---|
| Transcription genes | |||||
| CITED2 | 6q24.1 | Transcriptional modulator/co-activator | 602937 | ASD, VSD | MacDonald et al. (2013) |
| FOXH1 | 8q24.3 | Forkhead activin signal transducer-1 | 603621 | VSD, TGA | Barnes and Black (2016) |
| FOXP1 | 3p13 | Forkhead box P1 protein | 605515 | Bekheirnia et al. (2017) | |
| GATA4 | 8p23.1 | Zinc-finger transcription factor | 600576 | ASD, TOF, PDA, AVSD | Ang et al. (2016) |
| GATA6 | 18q11.2 | Zinc-finger transcription factor | 601656 | ASD, TOF, PDA, AVSD | Li et al. (2014a) |
| IRX4 | 5p15.33 | Iroquois homeobox 4 transcription factor | 606199 | VSD | Cheng et al. (2011) |
| MED13 L | 12q24.21 | Mediator complex transcriptional co-activator | 608771 | d-TGA | Asadollahi et al. (2013) |
| NKX2-5 | 5q35.1 | Homeobox-containing transcription factor | 600584 | ASD, AVSD, TOF, VSD | Xu et al. (2017) |
| NKX2-6 | 8p21.2 | Homeobox-containing transcription factor | 611770 | TA | Wang et al. (2015) |
| TBX5 | 12q24.21 | T-box genes transcription factors | 601620 | VSD | Chen et al. (2017) |
| TBX20 | 7p14.2 | T-box genes transcription factors | 606061 | ASD | Hu et al. (2013) |
| ZFPM2/FOG2 | 8q23.1 | Zinc-finger transcription factor/coactivator | 603693 | TOF | Zhang et al. (2014) |
| ZIC3 | Xq26.3 | Zinc-finger transcription factor | 300265 | Congenital heart defects | Cowan et al. (2014) |
| Structural proteins | |||||
| ACTC1 | 15q14 | Cardiac smooth muscle actin | 102540 | ASD | Greenway et al. (2014) |
| MYH6 | 14q11.2 | Cardiac muscle myosin Heavy chain | 160710 | ASD | Posch et al. (2011) |
| MYH7 | 14q11.2 | Cardiac muscle myosin Heavy chain | 160760 | Left ventricular noncompaction | Khodyuchenko et al. (2015) |
| MYH11 | 16p13.11 | Smooth muscle myosin heavy chain | 160745 | PDA | Harakalova et al. (2013) |
| NOTCH1 | 9q34.3 | Transmembrane protein | 109730 | AVD, LVOTO | Theodoris et al. (2015); Preuss et al. (2016) |
| Other | |||||
| CRELD1 | 3p25.3 | Cell adhesion | 607170 | AVSD | Guo et al. (2014) |
ASD, atrial septal defect; VSD, ventricular septal defects; TGA, transposition of the great arteries; TOF, tetralogy of fallot; AVSD, atrioventricular septal defects; PDA, patent ductus arteriosus; TA, tricuspid atresia; AVD, aortic valve disease; LVOTO, left-ventricular outflow tract obstructions.
These genes (Table 1) are expressed in different stages of fetal heart development and perturbations at different times in different genes could potentially affect formation of the heart structure (Cao et al., 2016; Sanchez-Castro et al., 2016; Xiong et al., 2013; Yoshida et al., 2016). Sequencing of genes encoding transcription factors such as GATA4, NKX2-5, and TBX5 in NS-CHD has identified variants that affect heart development. We now know that the molecules in these pathways affect both spatial and temporal gene expression patterns (Munshi, 2012; von Gise and Pu, 2012), and that disrupting these pathways results in abnormal heart development. However, there are variable phenotypes associated with any of these causal genes due to other influencing factors.
The complexity of CHD may be linked to embryonic cardiac development that involves multiple pathways with extensive cross-talking and promiscuous ligand–receptor interactions, secondary signal transduction pathways, and a network of transcription factors that determines the expression of cardio-specific effector genes (Fahed and Nemer, 2012). Of special interest to developing countries, dietary issues such as deficient periconceptional folic acid supplementation (which is critical for neural tube formation) may also contribute to the complexity of CHD. Maternal polymorphisms of “pharmacogenes” of folic acid metabolism especially methylene tetrahydrofolate reductase (MTHFR) such as MTHFR (677C>T) has been classified as a risk factor for CHD (Pishva et al., 2013; Van Beynum et al., 2006).
CHD continue to remain a critical health issue in Africa as the continent works to achieve the sustainable development goals (SDG) of reducing child mortality. A few decades ago, CHD diagnoses were not common in Africa. With improvements in health service delivery and healthcare professional training (Budzee et al., 2010), CHD diagnosis and treatment at an early age is allowing for children with CHD to live (Ejim et al., 2013; Methlouthi et al., 2016; Nkoke et al., 2017; Sadoh et al., 2016; Sani et al., 2007).
Despite the variation in different phenotypes of CHD diagnosed in the health facilities across Africa, the genetic basis of CHD in African populations remains a great challenge.
Literature Search
Article sources and databases employed
A comprehensive search method of published articles up to February 2018 was completed using PubMed (National Library of Medicine)/Medline and Google Scholar. Search terms included “genetics,” “congenital heart defects,” “Africa,” “epidemiology,” “diagnosis,” “CHD,” “burden,” “non-syndromic,” “transcriptional factors,” and “congenital heart malformations.” Authors that are known to have published in the field were specifically searched for and relevant articles included. Our search enabled us to discuss in detail the burden of CHD in Africa and understand the importance of genetic architecture in African populations.
Selection criteria
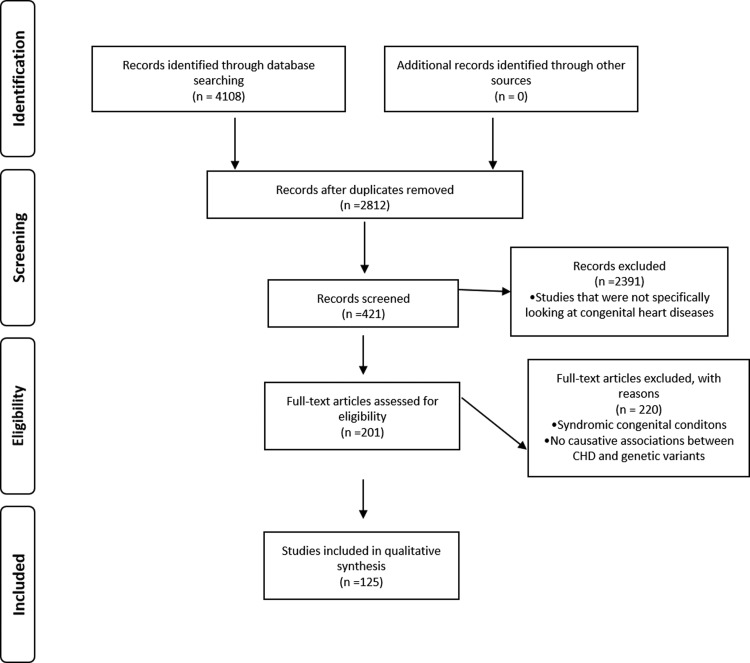
Abstract reviews were performed for relevant published articles related to CHD and relevant full articles were retrieved in this review (Fig. 1).
FIG. 1.
Summary of search and selection of the literature review used in the review.
A total of 4108 articles were consulted after search; exclusion criteria were performed based on article relevance to the expert review and articles that were not written in English. Subsequently, 125 articles that gave required information were included in this review. The inclusion criteria focused on academic, research, clinical case studies, and reports describing prevalence; clinical phenotypes; and genetics related to non-syndromic CHD. We must state, there were articles that fit these criteria except authors could not get the full articles or abstracts not in English and so were excluded. Articles for genes involved in non-syndromic CHD were used. Studies that found no causative associations between CHD and genetic variants were excluded.
Data collection
A summarized version of the different studies is presented in different sections of this review to elaborate the importance of such a review to, especially, populations in health transition in Africa. The following information is used for the summary, country of study, city, cohort, study type, encountered CHD phenotypes, and genes.
Key Findings from the Literature Search
The CHD odyssey in Africa
The last decade has seen the publication of several clinical epidemiological studies showing the differential prevalence of various subtype of CHD across different African populations, which is summarized in Table 2.
Table 2.
Spectrum of Congenital Heart Defects Phenotypes in African Countries
| Country | City | Study cohort | Study | Chd phenotypes and clinical prevalence (%) | Genetic study results | References |
|---|---|---|---|---|---|---|
| Angola | Luanda | Children Adolescents |
Clinical | CoA (2.1), TGA (2.1) PDA (35), PS (16.8), TOF (1.4), TA (1.4), VSD (49), DORV (1.4) | ND | Manuel et al. (2014) |
| Luanda | Children | Clinical | ASD (6.2), VSD (39.5), AVSD, AVCDD, TGA, PDA (33.8) | ND | Nunes et al. (2017) | |
| Burkina Faso | Bobo-Dioulasso | Children | Clinical | ASD, AVSD, DORV, TGA, TO, VSD | ND | Tougouma et al. (2016) |
| Cameroon | Children | Genetic | TOF, TA | 22q11.2 deletion | Wonkam et al., (2017) | |
| Yaoundé | Children Adult |
Clinical | i-VSD (38.8), TOF (26.1), i-PS (2.6), DORV (2.1), i-ASD (2.8), AVSD (7.3) | ND | Tantchou Tchoumi et al. (2011); Tantchou Tchoumi and Butera (2013) | |
| Yaoundé | Adolescent | Clinical | Shone's anomaly | ND | Nkoke et al. (2014) | |
| Yaoundé | Children Adolescent |
Clinical | VSD (37.2), PS (15.0), PDA (13.7), ASD (11.1), TOF (8.2), AVCD (5.3), TA (1.5), TGA + ASD (0.6) | ND | Chelo et al. (2016) | |
| Yaoundé | Children Adolescent |
Clinical | ASD (17.0), VSD (30.0), PDA (18.9), AV-Canal (2.3), TOF (5.3), PS (29.1) TGA (<1.0) | ND | Nkoke et al. (2017) | |
| Egypt | Children Adults (Mothers) |
Genetic | ASD (28.7), VSD (32.5), PDA (17.5) | MTHFR 677TT, 1298CC | Zidan et al. (2013) | |
| Cairo | Neonates infants | Clinical | PAVSD (6.2), VSD (50.8), TOF (4.6), AVSD (6.2), DORV (1.5) | ND | Udink ten Cate et al. (2013) | |
| Cairo | Adults | Clinical | ASD, VSD, TOF | ND | Farouk et al. (2015) | |
| Children??? | Genetic | ASD, VSD, AVSD, AVCDD | GATA4 gene variants | Al-Azzouny et al. (2016) | ||
| Genetic | VSD (47.3), ASD (16.4), TOF (7.3), PS (3.6) | GATA4 gene exon 1 (P193H) | Shaker et al. (2017) | |||
| Ghana | Accra | Adults | Clinical | ASD (26.0), VSD (34.0), PDA (12.0), TOF (12.0) | ND | Edwin et al. (2017) |
| Accra | Children | Clinical | TOF, PA-VSD, DORV | ND | Edwin et al. (2016) | |
| Libya | Tripoli | Children Adolescent |
Clinical | ASD (23.0), AVSD (19.0), VSD (14.0) | ND | Elmagrpy et al. (2011) |
| Malawi | Blantyre | Pediatric | Clinical | ASD (2.0), AVSD (5.2), DORV/TGA (4.4), TOF (10.0), VSD (24.0), PDA (7.2), PS (1.2) | ND | Kennedy and Miller (2013) |
| Morocco | Casablanca | Pediatric Adolescents |
Clinical | AVSD (29.0), ASD (19.9), TOF (5.4), VSD (21.5), PDA (16.7) | ND | Benhaourech et al. (2016) |
| Nigeria | Abidjan | Children Adolescents Adults |
Clinical | ASD (13.8), AVSD (7.7), CoA (2.3), TGA (3.8), PDA (7.7), PS (8.1), TOF (8.8), VSD (38.6), | ND | Métras et al. (1979) |
| Ibadan | Pediatric | Clinical | VSD (35.0), PDA (22.0), TOF (10.0), ASD (7.5), PS (9.0), CoA (2.0) | ND | Jaiyesimi and Antia (1981) | |
| Kano | Children | Clinical | i-VSD | ND | Asani et al. (2005) | |
| Ilesa | Pediatric Children |
Clinical | TOF | ND | Okeniyi and Kuti (2008) | |
| Lagos | Children | Clinical | TOF (16.9), PDA with ASD (8.3) | ND | Animasahun et al. (2015); Animasahun et al.(2016) | |
| Enugu | Children Adults |
Clinical | ASD, VSD, TOF, | ND | Ejim et al. (2013) | |
| Lagos Abuja |
Children | Clinical | ASD (8.7), AVSD (8.2), DORV (0.6), PDA (12.1), TGA (1.5), TOF (7.8), VSD (46.6), TA (1.2), Tricuspid atresia (2.1) | ND | Sadoh et al. (2013) | |
| Enugu | Children | Clinical | i-VSD (21.5), TOF (9.2), AVSD (4.2), TA (4.2), i-ASD (2.8), DORV (2.8), PDA (2.8) | ND | Chinawa et al. (2013) | |
| Port Harcourt | Children | Clinical | ASD (2.5), AVSD (2.4), DORV (0.9), PDA (14.5), TGA (3.6), TOF (8.6), TA (0.9), VSD (27.1), Tricuspid atresia (0.9) | ND | Otaigbe and Tabansi (2014) | |
| Benin City | Children | Clinical | VSD (50.0), PDA (14.29), TA (21.43), Tricuspid atresia (7.17), | ND | Sadoh and Osarogiagbon (2013) | |
| Lagos | Children | Clinical | Single ventricle physiology hearts (15.7), ASD/VSD/PDA (13.9), TGA (13.9), TA (12.6), PS (12.2), VSD/PS (4.8), CoA 94.8) | ND | Ekure et al. (2017) | |
| Rwanda | Kigali | Children | Clinical | PDA (53.3), ASD (3.0), PV (23.3) | ND | Senga et al. (2013) |
| Kirehe, Southern Kayonza districts |
Children Adults |
Clinical | ND | ND | Kwan et al. (2013) | |
| Kigali | Children Adolescents |
Clinical Genetic | ASD, AVSD, CoA, Dextrocardia, MVP, PDA, PS, TOF, TA, VSD | 7q11.23, deletion, 22q11.2 deletion, 13qter deletion | Teteli et al. (2014) | |
| Senegal | Clinical | VSD (25.0), TOF (13.0) | ND | Diop et al. (1996) | ||
| South Africa | Soweto | Children | Clinical | VSD (2.07), PS (0.17), ASD (0.42) | ND | McLaren et al. (1979) |
| Cape Town | Children | Genetics | 22q11.2 deletion (4.8) | De Decker et al. (2016) | ||
| Bloemfontein | Adults | Clinical | Septal defects (ASD, VSD, AVSD)- (41.2), TOF (1.3), CoA (9.0), PDA (3.9) | ND | Long et al. (2016) | |
| Sudan | Khartoum | Pediatric adolescent |
Clinical | AVSD (48.0), VSD (23.0), TOF (6.0), PDA (7.0) | ND | Ali (2009) |
| Tunisia | Consanguineous Family | Clinical Genetic | ASD, AVCD | PMM D5S394 & D5S2069 overlapping NKX2-5 gene | Nouira et al. (2008) | |
| Sfax | Children | Clinical | ASD (12.9), VSD (31.0), TOF (6.2), TGA (2.7), CoA (4.3), | ND | Abid et al. (2014) | |
| Togo | Lomé | Pediatric Adolescents |
Clinical | VSD (24.0), PDA (21.0), AVSD (9.0), ASD (18.0), TOF (19.5) | ND | Kokou et al. (1996) |
| Uganda | Kampala | Pediatric Adolescents |
Clinical | VSD (36.3), PDA (27.2), TGA (4.5), TOF (2.27), PS (4.5) | ND | Caddell and Connor (1966) |
| Kampala | Children | Clinical | ASD (20.0), VSD (15.0), PDA (6.6), TOF (3.3), CoA (1.6) | ND | Wood et al. (1969) | |
| Kampala | Children | Clinical | VSD, PDA, TOF, ASD | ND | Freers et al. (1996) | |
| Children | Clinical | VSD, PDA, TOF, ASD | ND | Ellis et al. (2007) | ||
| Kampala | Pediatric Adolescents |
Clinical | Cyanotic (28.9), Acyanotic (71.1) | ND | Batte et al. (2017) | |
| Kampala | Adults | Clinical | VSD (23), AVSD (13), TOF (13) | ND | Grimaldi et al. (2014) |
22qDS, 22q11.2 deletion syndrome; PMM, polymorphic microsatellite markers; AVCD, atrioventricular conduction defect; NS-CHD, non-syndromic congenital heart defects; NR, not reported; MTHFR, methylene tetrahydrofolate reductase; AVCDD, A-V canal disturbance defect; DCMP, dilated cardiomyopathy; EMF, endomyocardial fibrosis; HTN HD, hypertensive heart disease; RHD, rheumatic heart disease; IHD, ischemic heart disease; CoA, coarctation of the aorta; DORV, double-outlet right ventricle; PFO, patent foramen ovale; PA, pulmonary atresia; HDC, hypokinetic dilated cardiomyopathy; PT, pericardial tamponade; PAVSD, pulmonary atresia with ventricular septum defect; ND, not determined; RAA, right aortic arch; MD, mirror dextrocardia; AS, aortic stenosis; PVS, pulmonary valve stenosis.
A search for studies conducted in African populations found no study on birth incidence or population prevalence. Very rarely, genetic causes were investigated with the 22q11 deletion syndrome being the most investigated, in three independent studies in Cameroon, Rwanda, and South Africa (Table 2).
The trends and patterns of CHD in Nigerian children over a 51-year period (1964–2015) were recently published showing the most prevalent forms of CHD as ventricular septal defect (VSD) accounting for 40.6% of all CHDs, 11.3% as atrial septal defect (ASD), 11.8%, as Tetralogy of Fallot (TOF), and 18.4% as patent ductus arteriosus (PDA) (Abdulkadir and Abdulkadir, 2016). This study also reported decline in the occurrence of pulmonary stenosis (PS) and a 6% increase in the burden of VSD over 5 decades. These recent findings agree with a previous study on mortality related to CHD in Nigerian children, which indicated that VSD, PDA, and TOF were the most common causes of mortality (Akang et al., 1993).
A hospital-based necropsy study on CHD over an 8-year period in Nigeria demonstrated, the most common conditions were VSDs, ASDs, TOF, PDA and transposition of great arteries (TGA) with a combined overall prevalence being 22.2% at age 2 months (Thomas et al., 2013). TOF, the most common cyanotic CHD, has a prevalence of 4.9/1000 among children presenting at the Lagos State University Teaching Hospital from 2007 to 2015 (Animasahun et al., 201611). Mozambique, a sub-Saharan African country with a life expectancy at birth of 50 years is also faced with CHD challenges requiring the involvement of nongovernmental organizations to make arrangements for free surgery, reported a 9.65% CHD-related mortality at a median age of 23 months over a 10-year period (2001–2011) (Mirabel et al., 2017).
Arthur (1995) reported over two decades ago that sudden deaths due to cardiac heart diseases in children in Accra, Ghana were quite common. A 2-year study conducted in Ghana on conotruncal heart defect (TOF, pulmonary atresia with ventricular septal defect (PA-VSD), double-outlet right ventricle (DORV), common arterial trunk, and transposition of the great arteries) repair and outcomes after treatment reported that 20% of the 6168 heart defects were of conotruncal anomalies (Edwin et al., 2016). This indicates that Ghana is gradually being able to diagnose CHD conditions due to trained staff and improved infrastructure. At the same time, Ghana is experiencing an increased CHD burden as seen by the increase in new cases diagnosed within a recent 2-year period.
A 1200 patient cohort in Cameroon presenting with abnormal echocardiogram during a 2-year period showed that approximately 52% of children under 10 years had CHD mostly relating to PDA, TOF, and septal defects (Jingi et al., 2013). In the East African country of Kenya, a 5-year retrospective study at the Kenyatta National Hospital showed that the most common CHDs for children (average age 1.5 years) were VSDs, ASDs, PDAs, TOF, and TGA (Awori and Ogendo, 2013).
A prospective cross-sectional study conducted at the pediatric cardiology and inpatient clinic in El Fayoum University hospital showed approximately ASDs and VSDs made up 30% of cases among children less than 12 years with a mortality of 4.8% among the same cohort (Atwa and Safar, 2014). Prevalence of CHD in Tunisia is 6.8 per 1000 live births with common CHD diagnosed conditions been VSD, ostium secundum ASD, pulmonary valve abnormalities, coarctation of the aorta, TOF, and TGA (Abid et al., 2014) with a 23% mortality rate over a 1-year period before access to surgery.
Despite the clinical epidemiological spectrum of CHD in Africa displaying variable prevalence of CHD phenotypes with the common occurrence being VSD, ASD, and TOF not much has been done in terms of genetics/genomic studies, which can influence our understanding of epidemiological data obtained.
Genetic basis of non-syndromic CHD: global populations
Variations in transcriptional factors and CHD
With the understanding that CHD development is a multifactorial condition involving both genetic predisposition and environmental influences, it is becoming important that the genetic factors predisposing newborns to heart malformations be investigated. There has been a tremendous progress in understanding the molecular mechanisms involved in cardiac development that has enabled researchers and the clinical community to make prediction on plausible genetic markers for CHD.
Previous studies have shown an evolutionary conserved programme of cardiogenesis initiated by complex interactions of transcription factors with their regulators, receptors, ligands, signaling pathways, and protein networks (Fahed and Nemer, 2012). Some of these factors involved in the process including CRELD1, GATA4, NKX2.5, TBX5, TBX1, JAG1, TFAP2B, PTPN11, NOTCH1, and HAND1/2 (Clark et al., 2006; McCulley and Black, 2012) have been implicated in sporadic CHDs. The correct formation of the heart is dependent on the correct differential expression, co-expression profile, and interaction of each of these genes involved in cardiogenesis at the right stage of the heart formation process. We performed an enrichment analysis using version 10.5 of STRING (Jensen et al., 2009; Szklarczyk et al., 2017) querying genes involved in fetal heart development to assess co-expression (Fig. 2).
FIG. 2.
Genes (Ligand receptors, transcriptional, structural) involved in cardiogenesis and their co-expression profiles in Homo sapiens and other organisms. Using the genes implicated in cardiogenesis as query, co-expression profiles were generated using version 10.5 of STRING.
Genes associated with CHD in humans have been supported by animal studies. Gene regulatory networks in zebrafish are increasing our understanding of heart development. Our enrichment analysis conducted is supported by data that elucidated that altered gene expression of transcription factors TBX5 and NKX2-5 in zebrafish can lead to sub-functionalization (Hill et al., 2017; Rodius et al., 2016) with a dynamic interactome analysis. There is time-dependent expression of cardiac ontogeny and regulation in heart formation with any alteration leading to significant consequences for heart formation (Li et al., 2014). Differential expression of the transcription factors TBX5 and TBX20 has been demonstrated to affect cardiac development given evidence to the critical effects of cardiac gene expression and combinatorial regulatory interactions on the correct formation of the heart (Plageman and Yutzey, 2004).
Collectively, these data demonstrate clear differences in both the expression and function of transcription factors and suggest that the modulation of cardiac gene expression potentially plays a role in CHD. With this in mind, single nucleotide polymorphisms (SNPs) that may affect gene and protein profiles of cardiogenesis-associated genes may be detrimental to fetal heart development. Several studies conducted in other populations mostly of Caucasians and Asian decent have shown several SNPs in genes involved in cardiogenesis as potential etiologies to non- syndromic CHD (Table 3).
Table 3.
Selected Genetic Polymorphisms Identified in Genes Linked with Heart Development and Associated with Non-Syndromic Congenital Heart Defects
| Gene | Nucleotide/Protein change | Effect | Phenotype | Population | Reference |
|---|---|---|---|---|---|
| IRX1 | p.Gln240Glu p.Ser298Asn p.Ala381Glu |
Non-synonymous | TrA, TGA | Asian (Chinese) | Guo et al. (2017) |
| PITX2 | p.R91Q p.T129S |
Loss-of-function mutation | TGA, VSD | Asian (Chinese) | Panepistēmio tēs Krētēs. et al. (2014) |
| HAND2 | p.L47P | Loss-of-function mutation | TOF | Asian (Chinese) | Lu et al. (2016) |
| GATA4 | c.431C4T p.Ala144Val | Possibly damaging | PA, ASD | Asian (Japanese) | Yoshida et al. (2016) |
| c.1064C > G, T355S c.1129A > G, S377G c.1138G > A, V380 M c.1180C > A, P394T c.1273G > A, D425 N |
Missense | ASD, VSD, PDA, DORV, TOF, PS | Asians (Indians) | Bose et al. (2017) | |
| c.788 C > G | Missense | VSD | Asian (Chinese) | Xiong et al. (2013) | |
| GATA6 | p.D404Y p.E460X |
Decreased transactivation activity |
TOF, VSD | Asian (Chinese) | Wang et al. (2012) |
| c.151G>A/(E51K), c.551G>A/(S184 N) c.733G>C/(G245R), | Reduction in the transactivation capacity of downstream genes | TOF, TrA | Asian (Chinese) | Lin et al. (2010); Wang et al. (2014) | |
| NR2F2 | c.220_222dup/p.Gln75dup c.1022C>A/p.Ser341Tyr c.614A>T/p.Asn205Ile c.753G>C/p.Glu251Asp c.1234G>T/p.Ala412Ser c.509A>T/p.Asp170Val c.970þ1G>A (14;15)(q23;q26.3) |
Reductions in transcriptional activity | TOF, AVSD, CoA, | Caucasians | Al Turki et al. (2014) |
| PRDM6 | c.1646G>A/p.Arg549Gln c.788G>C/p.Cys263Ser c.1385A>G/p.Gln462Arg |
Epigenetic regulation of ductus remodeling | PDA | Caucasians | Li et al. (2016a) |
| FOXC1 | Copy number variation | CoA | Caucasians | Sanchez-Castro et al. (2016) | |
| TFAP2B | c.601 + 5G>A c.435_438delCCGG | PDA | Asian (Chinese) | Chen et al. (2011) | |
| c.1006 G > A | TOF, PA, PDA | Asian (Chinese) | Xiong et al. (2013) | ||
| NKX2-5 | 1433A>G | Affects septation during cardiac morphogenesis and maturation and maintenance of the atrioventricular node |
TOF PLSVC, BAV | Asian (Chinese) Caucasians | Cao et al. (2015); Cao et al. (2016); Dargis et al. (2016) |
| c.608A>G/p.E203G c.646C>T/p.R216C c.852G>A/p.N226D 1212G>T (3′UTR) |
VSD | Asian (Indians) | Dinesh et al. (2010) | ||
| p.K192X | No transcriptional activity | BAV | Asian (Chinese) | Qu et al. (2014) | |
| NKX2.6 | c.454A>C/p.K152Q | Reduced its transcriptional activating function | VSD | Asian (Chinese) | Li et al. (2016b) |
| TBX5 | c.322C>A/p.Pro108Thr c.791G>A/p.Arg264Lys |
Affect protein function | TrA VSD, PA |
Asian (Japanese) | Yoshida et al. (2016) |
| TBX20 | c.991A>G p.Thr331Ala | Possibly damaging | VSD | Asian (Japanese) | Yoshida et al. (2016) |
| CITED2 | c.550G > A, Gly184Ser c.574A > G, Ser192Gly c.573-578del6, Ser192fs |
Non-synonymous | MD, RAA, TOF AS AS, PVS, VSD, ASD |
Asian (Chinese) | Xu et al. (2014) |
| c.428 G > C p.Gly143Ala | Non-synonymous | PDA | Asian (Chinese Tibetan) | Liu et al. (2017) | |
| c.508_534del27 p.Ser170_Gly178del c.534_535ins27 p.Gly178_Ser179ins9 c.592_597delAGCGGC p.Ser198_Gly199del |
Non-synonymous | VSD, ASD | Caucasians | Sperling et al. (2005) | |
| c.C418T, p. P140S c.C548T p. S183 L c.A586G p. S196G c.481–483delAGC p.Ser161delAGC c.574–579delAGCGGC p.Ser192_Gly193delAGCGGC |
Non-synonymous | ASD, VSD | Asian (Chinese) | Liu et al. (2014) | |
| FOXP1 | c.1702C>T, p.Pro568Ser | Non-synonymous | AVSD | Caucasian | Chang et al. (2013) |
| MYH6 | E501Stop, A230P IVS37-2A.G, R1116S, A1443D, R1865Q H252Q V700 M A1366D |
Non-sense mutation, Splice site mutation, Missense mutations | TrA ASD TGA PFO AS |
Caucasians | Granados-Riveron et al. (2010) |
| c.50G>A, R17H c.1615T>C, C539R c.1628A>G, K543R |
missense mutations | ASD | Caucasians |
Posch et al. (2011) | |
| ZFPM2/FOG2 | p.V339I, p.A426 p.M703 L, p.T843 M |
Non-synonymous | DORV TOF |
Asian (Chinese) | Zhang et al., 2014); Qian et al. (2017) |
| c. 972C>G, H324Q c. 3442G>A, E1148 K |
Non-synonymous | TOF TOF/PFO |
Asian (Chinese) | ||
| ACTC1 | c.251T>C, p.Met84Thr p.Glu101Lys p.Met125Val |
Non-synonymous | ASD, VSD, PS, | Lebanese | Augiere et al. (2015) |
The causal genes have mostly been found to be factors that interact with each other during the formation of the heart and muscles. Some of these transcription factors like NKX2-5 has been found to influence septation in heart differentiation and maturation and are responsible for maintenance of the atrioventricular node throughout one's lifetime. In most of these studies, it is observed that CHD phenotypic outcome is not singularly linked to a single SNP in a gene. Though SNPs may potentially affect heart morphogenesis, the genes involved in cardiogenesis may interact with each other and have various cardio-morphogenesis functions.
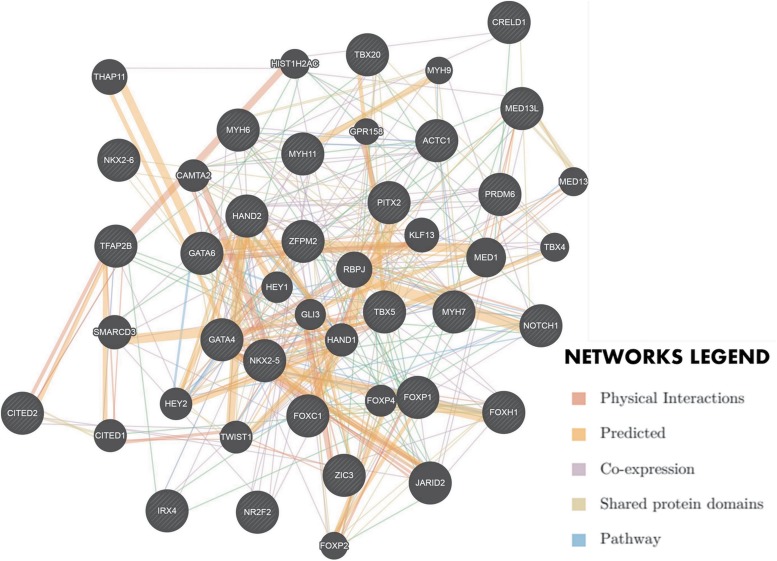
The complex network of gene–gene and protein–protein interactions involved in cardiogenesis could be a reason why there is so much phenotypic variation in CHD. An enrichment analysis was performed to show this complex interaction network using geneMANIA cytoscape plugin by determining the interactions of wild-type genes (Montojo et al., 2010) (Fig. 3).
FIG. 3.
Complex interactions of genes involved in cardiogenesis. (Network created using GeneMANIA Cytoscape plugin). Genes implicated in congenital heart defects were provided as query (shaded gray nodes) to see whether they have any direct interaction and a number of additional cardiogenesis genes were predicted to be related (unshaded gray nodes) in an enrichment analysis.
From the interaction network, it can be deduced that genetic variants share common pathways. This may explain the genetic heterogeneity of CHD (Reamon-Buettner and Borlak, 2004). Some previous studies have shown that the interaction of transcription factors GATA4 and NKX2-5 was critical during heart formation (Kinnunen et al., 2015; Slagle and Conlon, 2016). There is a lack of information on the genetic architecture (cardiogenesis genes) of CHD on the African continent even in countries with good economic standing. A case–control study in an Egyptian cohort showed a novel nonsynonymous sequence variation in GATA4 (P193H) detected in 9.1% of the study population with septal defects (Shaker et al., 2017).
Epigenetics: a game changer in CHD?
During cell differentiation and embryonic morphogenesis, epigenetic modifications can arise leading to effects that play a critical role in gene regulation and expression affecting genomic functions. Epigenetics involves heritable changes in gene activity, regulation, and expression that can occur as a result of several factors without modification in the DNA sequence. Epigenetic mechanisms include DNA methylation, chromatin modification, and microRNAs (Saetrom et al., 2007). DNA methylation, one of the first epigenetic mechanisms to be studied is involved in a variety of biological processes such embryonic development, X-chromosome inactivation, and genomic imprinting (Bird, 2002).
DNA methylation changes in CHD have recently been found in myocardial biopsies obtained from patients with TOF and VSD. Two mechanisms were identified to be involved in TOF and VSD namely hypermethylation of distinct CpG islands in the promoter of SCO2 leading to reduced gene expression and differential methylation influencing alteration and formation of alternate splicing that affect protein function in genes like TNN1, MYL7, PDZ, PDLIM3, and TNNT2 (Grunert et al., 2016).
Another case–control study involving pediatric patients with sporadic or isolated TOF and healthy controls showed that in addition to two identified SNPs (rs11582932T>G, rs11265385T>G), two methylation changes in the promoter regions (B1-1 and B3) significantly decreased the VANGL2 mRNA and protein levels in TOF patients (Yuan et al., 2014). VANGL2 is a critical gene that regulates cell polarity and polarized cell movements (Wansleeben et al., 2010) and studies suggest that it plays a significant role in heart development (Ramsbottom et al., 2014; Yu et al., 2012).
DNA methylation is known to be catalyzed by DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B) and methyl-CpG-binding domain protein 2 (MBD2). The expression profile of these methyltransferases and accompanying polymorphisms in the binding protein domains have a potential influence of biological process and corresponding clinical outcomes. A case–control study looking at the expression profile of DNMTs, MBD2, and long interspersed nuclear elements (LINE-1) methylation status in infants with TOF showed that lower levels of LINE-1 methylation significantly caused aberrant MBD2 mRNA levels with lower DNMT1 and DNMT3B likely playing a role in TOF pathogenesis in a process involving hypomethylation of genes, which is critical cardiogenesis (Sheng et al., 2013).
Chromatin modification that involves remodeling and histone modification have significant roles in activating or silencing gene expression. The SWI/SNF complex is a family of ATP-dependent chromatin remodeling proteins that regulates transcription in the heart in tandem with co-activator and repressor complexes. This complex is made of Brahma (Brm) or Brg1 ATPase, which is critical to chromatin remodeling machinery for vertebrate heart formation (Lei et al., 2012). Interactions between Brg1 ATPase and DNA-binding protein Parp1 with chromatin was shown to be involved in a prototypical shift in cardiac MHC isoform expression in pathological cardia hypertrophy.
It is also shown that α-MHC and Serca2a genes are highly expressed in healthy hearts whiles their expression is suppressed in heart hypertrophy. These suppression events are regulated by SWI/SNF containing Brm/Brg1-associated HDAC co-repressor complexes (Chang et al., 2011; El-Osta, 2011; Hang et al., 2010). These chromatin modeling complexes may play a critical regulatory role in CHD acquisition as their activities will affect expression or silencing of genes involved in heart formation, growth, and contractility throughout life.
The discovery of small non-coding RNAs known as microRNAs (miRNAs) of 20–22 nucleotides has evidentially shown to be crucial in post-translational mechanisms. miRNAs are classified as epigenetic markers with the specific biological processes of most miRNAs remaining unknown. However, several studies have shown how they control various physiological processes that can eventually have pathogenic roles in several diseases. Studies have shown that miRNAs operate by binding with specific sequences in the 3′UTR of target genes involved in development and physiology.
A functional analysis from a recent study in two Chinese Hans populations found out that miR-9 and miR-30a downregulated expression of TBX5, a vital transcription factor involved in cardiac development in a dosage-dependent manner at the transcriptional and translational levels, respectively (Wang et al., 2017). These two miRNAs hybridized to the 3′UTR of the TBX5 because of an SNP (rs6489956) that caused a change in binding affinity. Another recent study has shown that miR-199a-5p, ATF6, and GRP78 3′-UTR binding sites interact with the downregulation of miR-199a-5p having a favorable effect on myocardial unfolded protein response against hypoxia-induced ER stress in CHD leading to myocardial protection (Zhou et al., 2017).
It has recently been shown that altered expression levels of circulating miRNAs, miR-421, miR-1233-3p, and miR-625-5p in TOF patients with symptomatic right heart failure may play a critical role in diseases progression (Abu-Halima et al., 2017). The phenotypic manifestation of CHD can potentially be influenced by epigenetic mechanisms aside SNPs occurring in genes involved in cardiogenesis.
Copy number variation and CHD genetic paradox
Studies using chromosomal microarray have added many candidate genes to the list of potential etiologies for CHD (Soemedi et al., 2012). As previously mentioned, CHD does not follow classical Mendelian inheritance and the etiology of this complex disease remains unsolved for most cases. The phenomenon that results in sections of the genome being deleted or duplicated is known as copy number variation (CNV) and has become a key contributor to CHD (Lander and Ware, 2014). The role of CNVs in CHD occurs in genomic regions responsible for cardiogenesis that affect heart formation.
Recently, Kim et al. (2016), highlighted the pathogenicity of CNVs of >300kb in size in non-syndromic CHD to be significantly associated with decreased transplant-free survival after surgery. Rare and or large CNVs present a greater burden in isolated CHD with an increased risk of death suggesting that CNV burden is an important modifier to CHD acquisition, phenotype, and survival (Kim et al., 2016). Different phenotypes including CoA (15q 11.2 dup), DORV (20p12.2 dup, 22q11 del), TOF (8p23.2 dup, 10p12.31 dup), TOF (22q11.22del), and TGA (Dup2 2q11) were associated with CNVs of children born with CHD who exhibited microdeletions and duplications (Campos et al., 2015; Mercer-Rosa et al., 2013).
CNVs potentially play a significant role in CHD influencing the phenotypic outcome in both simple and complex conditions. CNVs affect large chromosomal segments that involve millions of DNA base pairs and multiple contiguous genes that make the identification of a single causal gene for CHD and expressive phenotypes a major challenge. Also, finding CNVs across different segments of the chromosome and associating it to different phenotypes remains challenged due to expertise to analyze data that are generated.
Literature Analysis and Discussion
Innovative policy and next generation technology research: focusing on African populations
The global burden of congenital anomalies, of which CHDs are the most common, indicates that 5% of deaths in children under five are due to CHDs (Hoffman, 2013). Despite the first heart transplant occurring on the African continent many decades ago, we discuss the fact that Africa still has a long way to go in the following dialog.
The first challenge is related to obtaining proper clinical data on the spectrum of commonly occurring CHD in Africa. Birth incidence, population prevalence, and qualitative CHD descriptions have not been well characterized in Africa. Prevalence of CHD in most African countries have generally been extrapolated by using data from previous studies from other jurisdiction (mostly developed countries), which may indicate that the figure in the public domain may not be a true reflection. This extrapolation is mostly emphasized by certain factors such as limited diagnostic proficiencies, lack of effective health-related statistics, a comprehensive database on newborns including any abnormalities observed and diagnosed. Also, one of the most common sources of determining prevalence is using hospital-based information, which does not necessarily give accurate statistics compared to using population-based studies.
Most African countries appear to be having high prevalence of CHD due to supposed risk factors associated with CHD such as age of mother, exposure to different teratogens, maternal malnutrition, and infections exposed to during pregnancy. Despite these challenges, there has been several studies in some African countries that addresses the burden and phenotypic spectra of CHD in these countries (Table 2), thus representing a reasonable estimation or burden of CHD in these countries.
Second, clinical epidemiology in African populations present different dynamics as the available data indicate that many African countries are under resourced to deal with the burden of congenital anomalies in general and, and specifically CHD, with regards to available surgical options and postsurgery management; This calls for an urgent implementation of a database of commonly and rare occurring CHD across the African continent in the relevant health facilities that attend to such newborns with CHD, to plan health resources adequately. This will be beneficial in monitoring the SDG on child mortality and most importantly, allow for effective and quality research into CHD in Africa, leading to effective planning, monitoring, and management of children with CHD in Africa.
Third, the few available clinical reports indicate that mortality and morbidity associated CHD in Africa remains a huge challenge even as health services and infrastructure improve. However, cases of mortality related to CHD appears to be under reported due to inadequacies of available professional for proper diagnosis, and the health information system, which affect data acquisition to properly monitor the burden of CHD remarkable. The African continent is generally lagging compared to the remarkable improvement in CHD management globally over the last few decades. Investments in infrastructure and specialist training can improve the diagnosis and amenable treatment of so-called complicated lesions.
Nonetheless, CHD go beyond the clinical, phenotypic, and even the surgical corrections involved. Understanding the causes or triggers of CHD in African populations and improving diagnostic accuracy are paramount to using preventive approaches if any to address the epidemic. One of the most common tools for assessing chromosomal deletions or rearrangements is karyotyping and fluorescent in situ hybridization analysis, which remains one of the starting points for genetic assessment of CHD patients (Gohring et al., 2011). These tests continue to remain relevant for the diagnosis of syndromic conditions such as trisomy 21, trisomy 13, trisomy 18, and known microdeletions (22q11.2 deletion and 7q11 deletion), as evidenced by the few available studies on the African continent, from South Africa and Cameroon, notably.
For isolated CHDs without any associated syndromes, not much has been done on the African continent and it is increasingly becoming relevant to use techniques that can help identify single gene variants, epigenetic markers, and possible CNVs in genes involved in cardiogenesis.
Understanding the genetic architecture of CHD patients of African descent will help better understand some of the triggers that lead to especially sporadic or isolated incidents that will help build a reliable register for public health education especially at ante- and postnatal clinics. Genetic investigations of CHD have the potential to improve prognosis from valuable information generated and may serve as a tool for predicting recurrent risk, assess familial inheritance pattern, therapeutic options for CHD patients who have undergone successful surgery, and appraise the need for further family screening.
A candidate gene approach using Sanger sequencing can be performed on known genes to evaluate disease causing mutations and their frequencies. However, considering the complexity of CHD and its genetic heterogeneity, the use of advanced genomic platforms will provide more insights. Next generation technologies that can be used to sequence whole genomes (WGS) and exomes (WES), perform CNV analysis, transcriptome analysis, and homozygosity mapping will be useful for broader understanding of the genetic architecture of CHD in African populations to influence policy formulation. The use of Next generation sequencing (NGS) employing targeted panel sequencing, whole exome and genome sequencing allows for evaluation of massive gene numbers in one run is urgently needed for clinical use and research in African population.
RNASeq/Ampliseq that uses the NGS platform can be used to sequence cardiac transcripts from CHD patients from African populations who have undergone successful surgery to evaluate the transcriptomics of genes in cardiogenesis. ATAC-Seq (assay for transposase-accessible chromatin) and THS-seq (transposome hypersensitive sites sequencing) an innovative NGS platform can be used as a rapid and sensitive method for integrative epigenomic analysis (Buenrostro et al., 2013, 2015; Sos et al., 2016).
The use of chromosomal microarray analysis validated with NGS can be useful in identifying CNVs associated with NS-CHDs (Helm and Freeze, 2016; Zhu et al., 2016). With challenges in CNV data analysis, algorithms that use whole exome sequencing data such as CoNVex can be used to identify CNVs associated with non-syndromic congenital heart diseases (Sifrim et al., 2016). Molecular analysis coupled with multiplex ligation-dependent probe amplification (MLPA) technique is also useful in determining CNV in non-syndromic CHD (Campos et al., 2015).
With the goal of the Human Heredity and Health in Africa (H3Africa) Initiative to develop array chips with genes from African populations, biomarkers can be developed using sequencing and expression profile data of CHD patients in Africa, which will serve as a diagnostic and prognostic platform for management. This will translate to the incorporation of validated genetic data into clinical practice of CHD management including genetic counseling and pre/postnatal screening. The clinical utility of including genetic data in clinical practice can improve health outcomes of conditions through the use of genetic therapeutic dosing algorithms, diagnosis, and public health discourse (Dotson et al., 2016; McCormick and Calzone, 2016).
With the knowledge and understanding of epigenetics and environmental exposure including teratogens, which pregnant mothers are exposed to in certain parts of Africa during pregnancy, public health policies in African countries especially resource limited settings where getting access to surgical intervention is limited can be formulated as preventive measures to avert potential non-syndromic CHDs. Industrialization, which has been linked to epigenetic changes and pre- and postnatal care involving multivitamin supplementation should be linked with responsible innovative public health policies that seek to insulate pregnant women from exposures that may affect fetal development.
Outlook and key lessons for Africa
Research focusing on the genomics and epigenomics of non-syndromic CHD in African populations using next generation technologies can lead to biomarker development with improved diagnosis and prognosis. “–Omics” research is significant to the discovery and development of novel diagnostics for CHD in African populations. The current article has highlighted the lack of genomic and epigenomic research in Africa, which could be key to providing answers to unanswered questions on most especially non-syndromic CHD development and phenotyping. This therefore calls for much needed research to understand the genetic architecture of CHD in African populations to improve management of CHD.
Perspective on CHD epigenomics in Africa
Africa been touted as the cradle of mankind (Ozdemir et al., 2014) with a wide genetic diversity. Yet, the epigenetic association of CHD in African populations seems to be overlooked by both researchers and policy makers. Over the past decades, most African countries have observed significant developments with corresponding industrialization. Industrialization affects biological circadian rhythms involving regulatory pathways resulting in rhythmic epigenetic modifications and the formation of epigenomes (Salavaty, 2015). DNA methylation and histone modifications serve to integrate environmental signals, nutrition, and xenobiotics for the cells to modulate the functional output of their genome. Most pregnant mothers in African countries are exposed to numerous environmental epigenetic risk factors that may affect their epigenome with corresponding effects on cardiogenes putting them at risk of having babies with CHDs.
Smoking, alcohol, consumption of herbal medicines, and exposure to environmental pollutants such as smokes from burning fire, car exhaust fumes, and inappropriate nutrition (malnutrition and eating of junk food) are becoming prevalent in Africa increasing incidence of noncommunicable diseases including CHDs (Hofman, 2014; Jenkins, 2003). Pregnant women are normally prescribed nutritional supplements like folate and iron to help with fetal development and prevent them from being anemic.
However, it is observed that at times these women do not adhere to their supplements as recommended, a common phenomenon in some African countries (Arega Sadore et al., 2015; Gebreamlak et al., 2017; Shewasinad and Negash, 2017) leading to anemia, which is not healthy for the growing foetus potentially influencing heart malformation (Feng et al., 2015; Linask and Huhta, 2010; Linask, 2013). Research evidence have shown how the epigenome can influence malformation of the heart and lead to CHD phenotypes. An audit and research into the epigenetic role of CHD being observed in African settings using available next generation technologies (“-omics” research) and algorithms should be spearheaded especially looking at the influential risk industrialization and urbanization poses to the epigenome.
Key lessons: Responsible innovative policies
Despite the need and effort to refine the real toll of CHD, the few available data indicate that CHD are a burden in Africa with significant populations having no access to proper diagnosis, medical care, and interventions such as corrective surgery and family support. Though mostly diagnosed in infants, there are asymptomatic adults living with CHD. One of the challenges to the early detection and management of CHD in Africa is the lack of policy direction for monitoring and screening newborns.
Basic screening such as pulse oximetry is available in most African health facilities, however, mostly physical examination has become the norm for determining the wellness of newborns. In addition, for most African populations, there is not much data on the genetic predisposition to CHD, despite, the identifiable preventable causes that can be translated into prevention strategies. Apart from screening strategies, “-omics” research such as genomics and epigenomics is critical in African research and diagnostics as it will produce knowledge-based innovation that remains fundamental to understanding the precipitants, pathophysiology, prevention, and management of CHD. This calls for a holistic public health approach looking at risks assessment and diagnostic strategies (clinical and genomics) being put into effect.
In other jurisdictions like Canada, United States of America, United Kingdom, and New Zealand, innovative public health strategies such as cardiovascular genetic research and newborn screening strategies for critical CHD have been implemented to improve the clinical management of CHD (Cloete et al., 2017; Lancet, 2012; National Collaborating Centre for and Children's, 2008; Oster et al., 2013; Riehle‐Colarusso et al., 2016). Policy innovations such as the following for African countries can lead to effective tracking, monitoring, and management of CHD incidences and epidemiology.
(1) Public health strategies and education should be intensified on CHD as it is observed that aside the common communicable and noncommunicable diseases that occur in most African populations, little is known of CHD to which parents and at times physicians miss the signs of, which lead to an increase in CHD-related mortality. Pulse oximetry screening in addition to physical examination should be made a routine in screening of newborns for CHD. Early detection will increase the survival rate and proper management of CHD patients.
(2) Interventional strategies for preventable risk factors associated with CHD such as nutritional deficiency in pregnant mothers, diabetes, and smoking should be implemented. CHD screening and interventional measures should be incorporated into health insurance coverage policies to reduce the financial limitations of CHD management.
(3) Investigating possible environmental exposures, medications, herbal concoctions, and consistent antenatal monitoring will help identify CHD babies early to effectively strategize intervention measures.
(4) Enhancement of birth surveillance, postnatal visits, and improved tracking of research to identify potential causes and long-term outcomes. As part of the policy framework, a centralized database is required to link access to health records across health facilities in African countries to allow continuity of management and follow-up.
(5) Support for public health, clinical and genetic research to outline the precipitants of CHD of diverse African populations will inform effective genetic counseling and future decisions for parents with CHD-children and CHD-adults.
(6) Responsible next generation technology policy research that will lay innovation frameworks for biomarker development for improved CHD diagnosis and prognosis. As part of responsible next generation technology policy research, biobanks and related infrastructure, where samples of CHD patients are stored for research purposes, should be established with proper management structures to ensure confidentiality and protection of patients' identity and samples.
Abbreviations Used
- 22qDS
22q11.2 deletion syndrome
- AS
aortic stenosis
- ASD
atrial septal defect
- AVCD
atrioventricular conduction defect
- AVCDD
A-V canal disturbance defect
- AVD
aortic valve disease
- AVSD
atrioventricular septal defects
- CHD
congenital heart defect
- CNV
copy number variation
- CoA
coarctation of the aorta
- DCMP
dilated cardiomyopathy
- DORV
double-outlet right ventricle
- EMF
endomyocardial fibrosis
- HDC
hypokinetic dilated cardiomyopathy
- HTN HD
hypertensive heart disease
- IHD
ischemic heart disease
- LVOTO
left-ventricular outflow tract obstructions
- MD
mirror dextrocardia
- MTHFR
methylene tetrahydrofolate reductase
- ND
not determined
- NGS
next generation sequencing
- NR
not reported
- NS-CHD
non-syndromic congenital heart defects
- PA
pulmonary atresia
- PAVSD
pulmonary atresia with ventricular septum defect
- PDA
patent ductus arteriosus
- PFO
patent foramen ovale
- PMM
polymorphic microsatellite markers
- PT
pericardial tamponade
- PVS
pulmonary valve stenosis
- RAA
right aortic arch
- RHD
rheumatic heart disease
- SDG
sustainable development goals
- SNP
single nucleotide polymorphism
- TA
tricuspid atresia
- TGA
transposition of the great arteries
- TOF
tetralogy of fallot
- TrA
truncus arteriosus
- VSD
ventricular septal defect
- WES
whole exome sequencing
- WGS
whole genome sequencing
Acknowledgment
Nicholas Ekow Thomford (PhD) is supported by the Faculty of Health Sciences, University of Cape Town, Prof. Ambroise Wonkam and Prof. Collet Dandara of the Division of Human Genetics University of Cape Town.
Author Disclosure Statement
The authors declare there are no conflicts of interest.
References
- Abdulkadir M, and Abdulkadir Z. (2016). A systematic review of trends and patterns of congenital heart disease in children in Nigeria from 1964–2015. Afr Health Sci 16, 367–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abid D, Elloumi A, Abid L, et al. (2014). Congenital heart disease in 37,294 births in Tunisia: Birth prevalence and mortality rate. Cardiol Young 24, 866–871 [DOI] [PubMed] [Google Scholar]
- Abqari S, Gupta A, Shahab T, et al. (2016). Profile and risk factors for congenital heart defects: A study in a tertiary care hospital. Ann Pediatr Cardiol 9, 216–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Halima M, Meese E, Keller A, Abdul-Khaliq H, and Radle-Hurst T. (2017). Analysis of circulating microRNAs in patients with repaired Tetralogy of Fallot with and without heart failure. J Transl Med 15, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akang EE, Osinusi KO, Pindiga HU, Okpala JU, and Aghadiuno PU. (1993). Congenital malformations: A review of 672 autopsies in Ibadan, Nigeria. Pediatr Pathol 13, 659–670 [DOI] [PubMed] [Google Scholar]
- Al-Azzouny MA, El Ruby MO, Issa HA, et al. (2016). Detection and putative effect of GATA4 gene variants in patients with congenital cardiac septal defects. Cell Mol Biol (Noisy-le-Grand, France) 62, 10–14 [PubMed] [Google Scholar]
- Al Turki S, Manickaraj AK, Mercer CL, et al. (2014). Rare variants in NR2F2 cause congenital heart defects in humans. Am J Hum Genet 94, 574–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali SK. (2009). Cardiac abnormalities of Sudanese patients with Down's syndrome and their short-term outcome. Cardiovasc J Afr 20, 112–115 [PMC free article] [PubMed] [Google Scholar]
- Animasahun BA, Madise-Wobo AD, Falase BA, and Omokhodion SI. (2016). The burden of Fallot's tetralogy among Nigerian children. Cardiovasc Diagnosis Ther 6, 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Animasahun BA, Madise-Wobo AD, Omokhodion SI, and Njokanma OF. (2015). Children With Tetralogy of Fallot in an Urban Centre in Africa. J Cardiovasc Thoracic Res 7, 168–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arega Sadore A, Abebe Gebretsadik L, and Aman Hussen M. (2015). Compliance with Iron-Folate Supplement and Associated Factors among Antenatal Care Attendant Mothers in Misha District, South Ethiopia: Community Based Cross-Sectional Study. J Environ Public Health 2015, 1–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur JT. (1995). Sudden deaths: Cardiac and non-cardiac in children in Accra. West Afr J Med 14, 108–111 [PubMed] [Google Scholar]
- Asadollahi R, Oneda B, Sheth F, et al. (2013). Dosage changes of MED13L further delineate its role in congenital heart defects and intellectual disability. Eur J Hum Genet EJHG 21, 1100–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asani MO, Sani MU, Karaye KM, Adeleke SI, and Baba U. (2005). Structural heart diseases in Nigerian children. Niger J Med 14, 374–377 [DOI] [PubMed] [Google Scholar]
- Atwa ZT. and Safar HH. (2014). Outcome of congenital heart diseases in Egyptian children: Is there gender disparity? Egypt Pediatric Assoc Gaz 62, 35–40 [Google Scholar]
- Augiere C, Megy S, El Malti R, et al. (2015). A novel alpha cardiac actin (ACTC1) mutation mapping to a domain in close contact with myosin heavy chain leads to a variety of congenital heart defects, arrhythmia and possibly midline defects. PLoS One 10, e0127903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awori M, and Ogendo S. (2013). The spectrum of paediatric congenital heart disease at the Kenyatta National Hospital: Implications for surgical care. Ann Afr Surg 10, 8–10 [Google Scholar]
- Barnes RM. and Black BL. (2016). Nodal Signaling and Congenital Heart Defects. In: Etiology and Morphogenesis of Congenital Heart Disease Nakanishi T., Markwald R., Baldwin H., Keller B., Srivastava D., (Eds). Springer, Tokyo, pp. 183–192 [PubMed] [Google Scholar]
- Batte A, Lwabi P, Lubega S, et al. (2017). Wasting, underweight and stunting among children with congenital heart disease presenting at Mulago hospital, Uganda. BMC Pediatr 17, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bekheirnia MR, Bekheirnia N, Bainbridge MN, et al. (2017). Whole-exome sequencing in the molecular diagnosis of individuals with congenital anomalies of the kidney and urinary tract and identification of a new causative gene. Genet Med 19, 412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhaourech S, Drighil A, and Hammiri A. (2016). Congenital heart disease and Down syndrome: Various aspects of a confirmed association. Cardiovasc J Afr 27, 287–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev 16, 6–21 [DOI] [PubMed] [Google Scholar]
- Bose DDV, Shetty M JK, and Kutty AVM. (2017). Identification of intronic-splice site mutations in GATA4 gene in Indian patients with congenital heart disease. Mutat Res 803–805, 26–34 [DOI] [PubMed] [Google Scholar]
- Botto LD, Loffredo C, Scanlon KS, et al. (2001). Vitamin A and cardiac outflow tract defects. Epidemiology 12, 491–496 [DOI] [PubMed] [Google Scholar]
- Bourdial H, Jamal-Bey K, Edmar A, et al. (2012). Congenital heart defects in La Réunion Island: A 6-year survey within a EUROCAT-affiliated congenital anomalies registry. Cardiol Young 22, 547–557 [DOI] [PubMed] [Google Scholar]
- Budzee A, Tantchou Tchoumi JC, Ambassa JC, et al. (2010). The Cardiac Center of Shisong Hospital: The first cardio-surgical center in West and Central Africa is inaugurated in Cameroon. Pan Afr Med J 4, 4. [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ. (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10, 1213–1218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Wu B, Chang HY, and Greenleaf WJ. (2015). ATAC-seq: A Method for Assaying Chromatin Accessibility Genome-Wide. Curr Protoc Mol Biol 109, 21 29 21–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn J, Brennan P, Little J, et al. (1998). Recurrence risks in offspring of adults with major heart defects: Results from first cohort of British collaborative study. Lancet 351, 311–316 [DOI] [PubMed] [Google Scholar]
- Caddell JL, and Connor DH. (1966). Congenital heart disease in Ugandan children. Br Heart J 28, 766–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos CM, Zanardo EA, Dutra RL, Kulikowski LD, and Kim CA. (2015). Investigation of copy number variation in children with conotruncal heart defects. Arq Bras Cardiol 104, 24–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Lan W, Li Y, et al. (2015). Single nucleotide polymorphism of NKX2-5 gene with sporadic congenital heart disease in Chinese Bai population. Int J Clin Exp Pathol 8, 14917–14924 [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Wang J, Wei C, et al. (2016). Genetic variations of NKX2-5 in sporadic atrial septal defect and ventricular septal defect in Chinese Yunnan population. Gene 575, 29–33 [DOI] [PubMed] [Google Scholar]
- Chang L, Kiriazis H, Gao X-M, Du X-J, and El-Osta A. (2011). Cardiac genes show contextual SWI/SNF interactions with distinguishable gene activities. Epigenetics 6, 760–768 [DOI] [PubMed] [Google Scholar]
- Chang SW, Mislankar M, Misra C, et al. (2013). Genetic abnormalities in FOXP1 are associated with congenital heart defects. Hum Mutat 34, 1226–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelo D, Nguefack F, Menanga AP, et al. (2016). Spectrum of heart diseases in children: An echocardiographic study of 1,666 subjects in a pediatric hospital, Yaounde, Cameroon. Cardiovasc Diagn Ther 6, 10–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HX, Zhang X, Hou HT, et al. (2017). Identification of a novel and functional mutation in the TBX5 gene in a patient by screening from 354 patients with isolated ventricular septal defect. Eur J Med Genet 60, 385–390 [DOI] [PubMed] [Google Scholar]
- Chen Y-W, Zhao W, Zhang Z-F, et al. (2011). Familial nonsyndromic patent ductus arteriosus caused by mutations in TFAP2B. Pediatr Cardiol 32, 958–965 [DOI] [PubMed] [Google Scholar]
- Cheng Z, Wang J, Su D, et al. (2011). Two novel mutations of the IRX4 gene in patients with congenital heart disease. Hum Genet 130, 657–662 [DOI] [PubMed] [Google Scholar]
- Chinawa JM, Eze JC, Obi I, et al. (2013). Synopsis of congenital cardiac disease among children attending University of Nigeria Teaching Hospital Ituku Ozalla, Enugu. BMC Res Notes 6, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KL, Yutzey KE, and Benson DW. (2006). Transcription factors and congenital heart defects. Annu Rev Physiol 68, 97–121 [DOI] [PubMed] [Google Scholar]
- Cloete E, Gentles T, Alsweiler J, et al. (2017). Should New Zealand introduce nationwide pulse oximetry screening for the detection of critical congenital heart disease in newborn infants. N Z Med J 130, 64. [PubMed] [Google Scholar]
- Cowan J, Tariq M, and Ware SM. (2014). Genetic and functional analyses of ZIC3 variants in congenital heart disease. Hum Mutat 35, 66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadvand P, Rankin J, Shirley MDF, Rushton S, and Pless-Mulloli T. (2009). Descriptive epidemiology of congenital heart disease in Northern England. Paediatr Perinat Epidemiol 23, 58–65 [DOI] [PubMed] [Google Scholar]
- Dargis N, Lamontagne M, Gaudreault N, et al. (2016). Identification of gender-specific genetic variants in patients with bicuspid aortic valve. Am J Cardiol 117, 420–426 [DOI] [PubMed] [Google Scholar]
- De Decker R, Bruwer Z, Hendricks L, et al. (2016). Predicted v. real prevalence of the 22q11.2 deletion syndrome in children with congenital heart disease presenting to Red Cross War Memorial Children's Hospital, South Africa: A prospective study. S Afr Med J 106, S82–S86 [DOI] [PubMed] [Google Scholar]
- Dinesh SM, Kusuma L, Smitha R, et al. (2010). Single-nucleotide polymorphisms of NKX2.5 found in congenital heart disease patients of Mysore, South India. Genet Test Mol Biomarkers 14, 873–879 [DOI] [PubMed] [Google Scholar]
- Diop IB, Ndiaye M, Ba SA, et al. (1996). [Congenital heart disease surgery in Senegal. Indications, evaluation and perspectives]. Dakar Med 41, 85–90 [PubMed] [Google Scholar]
- Dolk H, Loane M, and Garne E. 2010. The Prevalence of Congenital Anomalies in Europe. Adv Exp Med Biol 686:349–364 [DOI] [PubMed] [Google Scholar]
- Dotson WD, Bowen MS, Kolor K, and Khoury MJ. (2016). Clinical utility of genetic and genomic services: Context matters. Genet Med 18, 672–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwin F, Entsua-Mensah K, Sereboe LA, et al. (2016). Conotruncal Heart Defect Repair in Sub-Saharan Africa. World J Pediatr Congenit Heart Surg 7, 592–599 [DOI] [PubMed] [Google Scholar]
- Edwin F, Zühlke L, Farouk H, et al. (2017). Status and challenges of care in Africa for adults with congenital heart defects. World J Pediatr Congenit Heart Surg 8, 495–501 [DOI] [PubMed] [Google Scholar]
- Ejim E, Ubani-Ukoma C, Nwaneli U, and Onwubere B. (2013). Common echocardiographic abnormalities in Nigerians of different age groups. Niger J Clin Pract 16, 360. [DOI] [PubMed] [Google Scholar]
- Ekure EN. and Adeyemo AA. (2015). Clinical epidemiology and management of congenital heart defects in a developing country. In: Congenital Heart Disease Molecular Genetics, Principles of Diagnosis and Treatment. Muenke M, Kruszka PS, Sable CA, Belmont JW, eds. Basel: Karger, 46–56 [Google Scholar]
- Ekure EN, Bode-Thomas F, Sadoh WE, et al. (2017). Congenital Heart Defects in Nigerian Children: Preliminary Data From the National Pediatric Cardiac Registry. World J Pediatr Congenit Heart Surg 8, 699–706 [DOI] [PubMed] [Google Scholar]
- El-Osta A. (2011). Remodeling is at the heart of chromatin: The heartaches of chromatin. Epigenetics 6, 884–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis J, Martin R, Wilde P, et al. (2007). Echocardiographic, chest X-ray and electrocardiogram findings in children presenting with heart failure to a Ugandan paediatric ward. Trop Doct 37, 149–150 [DOI] [PubMed] [Google Scholar]
- Elmagrpy Z, Rayani A, Shah A, Habas E, and Aburawi EH. (2011). Down syndrome and congenital heart disease: Why the regional difference as observed in the Libyan experience? Cardiovasc J Afr 22, 306–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahed AC, Gelb BD, Seidman JG, and Seidman CE. (2013). Genetics of congenital heart disease: The glass half empty. Circ Res 112, 707–720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahed CA, and Nemer MG. 2012. Genetic causes of syndromic and non-syndromic congenital heart disease. Mutat Hum Genet Dis. InTech. Doi: 10.5772/48477 [DOI] [Google Scholar]
- Farouk H, Shaker A, El-Faramawy A, et al. (2015). Adult Congenital Heart Disease Registry at Cairo University. World J Pediatr Congenit Heart Surg 6, 53–58 [DOI] [PubMed] [Google Scholar]
- Feng Y, Wang S, Chen R, et al. (2015). Maternal folic acid supplementation and the risk of congenital heart defects in offspring: A meta-analysis of epidemiological observational studies. Sci Rep 5, 8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freers J, Mayanja-Kizza H, Ziegler JL, and Rutakingirwa M. (1996). Echocardiographic diagnosis of heart disease in Uganda. Trop Doct 26, 125–128 [DOI] [PubMed] [Google Scholar]
- Fung A, Manlhiot C, Naik S, et al. (2013). Impact of prenatal risk factors on congenital heart disease in the current era. J Am Heart Assoc 2, e000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg V, Kathiriya IS, Barnes R, et al. (2003). GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 [DOI] [PubMed] [Google Scholar]
- Gebreamlak B, Dadi AF, and Atnafu A. (2017). High Adherence to Iron/Folic Acid Supplementation during Pregnancy Time among Antenatal and Postnatal Care Attendant Mothers in Governmental Health Centers in Akaki Kality Sub City, Addis Ababa, Ethiopia: Hierarchical Negative Binomial Poisson Regression. PLoS One 12, e0169415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilboa SM, Salemi JL, Nembhard WN, Fixler DE, and Correa A. (2010). Mortality Resulting From Congenital Heart Disease Among Children and Adults in the United States, 1999 to 2006. Circulation 122, 2254–2263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohring G, Giagounidis A, Busche G, et al. (2011). Cytogenetic follow-up by karyotyping and fluorescence in situ hybridization: Implications for monitoring patients with myelodysplastic syndrome and deletion 5q treated with lenalidomide. Haematologica 96, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granados-Riveron JT, Ghosh TK, Pope M, et al. (2010). Alpha-cardiac myosin heavy chain (MYH6) mutations affecting myofibril formation are associated with congenital heart defects. Hum Mol Genet 19, 4007–4016 [DOI] [PubMed] [Google Scholar]
- Greenway SC, Mcleod R, Hume S, et al. (2014). Exome sequencing identifies a novel variant in ACTC1 associated with familial atrial septal defect. Can J Cardiol 30, 181–187 [DOI] [PubMed] [Google Scholar]
- Grimaldi A, Ammirati E, Vermi AC, et al. (2014). Cardiac surgery for patients with heart failure due to structural heart disease in Uganda: Access to surgery and outcomes. Cardiovasc J Afr 25, 204–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunert M, Dorn C, Cui H, et al. (2016). Comparative DNA methylation and gene expression analysis identifies novel genes for structural congenital heart diseases. Cardiovas Res. 112, 464–477 [DOI] [PubMed] [Google Scholar]
- Guo Y, Shen J, Li F, et al. (2014). Potential role of CRELD1 gene in the pathogenesis of atrioventricular septal defect. Chinese J Med Genet 31, 263–267 [DOI] [PubMed] [Google Scholar]
- Guo C, Wang Q, Wang Y, et al. (2017). Exome sequencing reveals novel IRXI mutation in congenital heart disease. Mol Med Rep 15, 3193–3197 [DOI] [PubMed] [Google Scholar]
- Hang CT, Yang J, Han P, et al. (2010). Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 466, 62–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harakalova M, Van Der Smagt J, De Kovel CG, et al. (2013). Incomplete segregation of MYH11 variants with thoracic aortic aneurysms and dissections and patent ductus arteriosus. Eur J Hum Genet 21, 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm BM. and Freeze SL. (2016). Genetic Evaluation and use of chromosome microarray in patients with isolated heart defects: Benefits and challenges of a new model in cardiovascular care. Front Cardiovasc Med 3, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JT, Demarest B, Gorsi B, Smith M, and Yost HJ. (2017). Heart morphogenesis gene regulatory networks revealed by temporal expression analysis. Development 144, 3487–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JI. (2013). The global burden of congenital heart disease. Cardiovasc J Afr 24, 141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JIE, and Kaplan S. (2002). The incidence of congenital heart disease. J Am Coll Cardiol 39, 1890–1900 [DOI] [PubMed] [Google Scholar]
- Hofman K. (2014). Non-communicable diseases in South Africa: A challenge to economic development. S Afr Med J 104, 647. [DOI] [PubMed] [Google Scholar]
- Hu Z, Shi Y, Mo X, et al. (2013). A genome-wide association study identifies two risk loci for congenital heart malformations in Han Chinese populations. Nat Genet 45, 818–821 [DOI] [PubMed] [Google Scholar]
- Jaiyesimi F, and Antia AU. (1981). Congenital heart disease in Nigeria: A ten-year experience at UCH, Ibadan. Ann Trop Paediatr 1, 77–85 [DOI] [PubMed] [Google Scholar]
- Jenkins CD. (2003). Building Better Health: A Handbook of Behavioral Change. Washington: Pan American Health Organization [Google Scholar]
- Jensen LJ, Kuhn M, Stark M, et al. , (2009). STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Research 37, D412–D416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingi AM, Jacques J, Noubiap N, et al. (2013). The spectrum of cardiac disease in the West Region of Cameroon: A hospital-based cross-sectional study. Int Arch Med 6, 1–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy N, and Miller P. (2013). The spectrum of paediatric cardiac disease presenting to an outpatient clinic in Malawi. BMC Res Notes 6, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodyuchenko T, Zlotina A, Pervunina T, et al. (2015). Congenital heart defects are rarely caused by mutations in cardiac and smooth muscle actin genes. Biomed Res Int 2015, 127807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DS, Kim JH, Burt AA, et al. (2016). Burden of potentially pathologic copy number variants is higher in children with isolated congenital heart disease and significantly impairs covariate-adjusted transplant-free survival. J Thorac Cardiovasc Surg 151, 1147–1151e1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen S, Valimaki M, Tolli M, et al. (2015). Nuclear Receptor-Like Structure and Interaction of Congenital Heart Disease-Associated Factors GATA4 and NKX2-5. PLoS One 10, e0144145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk EP, Sunde M, Costa MW, et al. (2007). Mutations in Cardiac T-Box factor gene TBX20 are associated with diverse cardiac pathologies, including defects of septation and valvulogenesis and cardiomyopathy. Am J Hum Genet 81, 280–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokou O, Agbèrè AR, Balaka B, et al. (1996). The Use of Doppler Echocardiography in the Diagnosis of Congenital Heart Disease in the Pediatric Department of CHU-Tokoin, at Lomé (Togo). Sante 6, 161–164 [PubMed] [Google Scholar]
- Kwan GF, Bukhman AK, Miller AC, et al. (2013). A Simplified Echocardiographic Strategy for Heart Failure Diagnosis and Management Within an Integrated Noncommunicable Disease Clinic at District Hospital Level for Sub-Saharan Africa. JACC Heart Fail 1, 230–236 [DOI] [PubMed] [Google Scholar]
- Lancet T. (2012). A new milestone in the history of congenital heart disease. Lancet 30;379(9835):2401. [DOI] [PubMed] [Google Scholar]
- Lander J, and Ware SM. (2014). Copy number variation in congenital heart defects. Curr Genet Med Rep 2, 168–178 [Google Scholar]
- Lei I, Sham MH, and Wang Z. (2012). ATP-dependent chromatin remodeling complex SWI/SNF in cardiogenesis and cardiac progenitor cell development. Front Biol 7, 202–211 [Google Scholar]
- Leirgul E, Fomina T, Brodwall K, et al. (2014). Birth prevalence of congenital heart defects in Norway 1994–2009—A nationwide study. Am Heart J 168, 956–964 [DOI] [PubMed] [Google Scholar]
- Li N, Subrahmanyan L, Smith E, et al. (2016a). Mutations in the Histone Modifier PRDM6 Are Associated with Isolated Nonsyndromic Patent Ductus Arteriosus. Am J Hum Genet 98, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu C, Xu Y, et al. (2016b). Identification of candidate genes for congenital heart defects on proximal chromosome 8p. Sci Rep 6, 36133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Martinez-Fernandez A, Hartjes KA, et al. (2014). Transcriptional atlas of cardiogenesis maps congenital heart disease interactome. Physiol Genomics 46, 482–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Huo Z, Liu X, et al. (2010). A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet 55, 662–667 [DOI] [PubMed] [Google Scholar]
- Linask KK, and Huhta J. (2010). Folate protection from congenital heart defects linked with canonical Wnt signaling and epigenetics. Curr Opin Pediatr 22, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linask KK. (2013). The heart-placenta axis in the first month of pregnancy: Induction and prevention of cardiovascular birth defects. J Pregnancy 2013, 320413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Su Z, Tan S, et al. (2017). Functional analyses of a novel CITED2 nonsynonymous mutation in chinese tibetan patients with congenital heart disease. Pediatr Cardiol 38, 1226–1231 [DOI] [PubMed] [Google Scholar]
- Liu Y, Wang F, Wu Y, et al. (2014). Variations of CITED2 are associated with congenital heart disease (CHD) in Chinese population. PLoS One 9, e98157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long MA, Smit FE, and Brown SC. (2016). Twenty years of adult congenital heart surgery. World J Pediatr Congenit Heart Surg 7, 619–625 [DOI] [PubMed] [Google Scholar]
- Lu CX, Gong HR, Liu XY, et al. (2016). A novel HAND2 loss-of-function mutation responsible for tetralogy of Fallot. Int J Mol Med 37, 445–451 [DOI] [PubMed] [Google Scholar]
- Macdonald ST, Bamforth SD, Braganca J, et al. (2013). A cell-autonomous role of Cited2 in controlling myocardial and coronary vascular development. Eur Heart J 34, 2557–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel V, Morais H, Manuel A, David B, and Gamboa S. (2014). Ventricular septal defect in children and adolescents in Angola: Experience of a tertiary center. Rev Port Cardiol 33, 637–640 [DOI] [PubMed] [Google Scholar]
- Mccormick KA, and Calzone KA. (2016). The impact of genomics on health outcomes, quality, and safety. Nurs Manage 47, 23–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcculley DJ, and Black BL. (2012). Transcription factor pathways and congenital heart disease. Curr Top Dev Biol 100, 253–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcgregor TL, Misri A, Bartlett J, et al. (2010). Consanguinity mapping of congenital heart disease in a South Indian population. PLoS One 5, e10286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclaren MJ, Lachman AS, and Barlow JB. (1979). Prevalence of congenital heart disease in black schoolchildren of Soweto, Johannesburg. Br Heart J 41, 554–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer-Rosa L, Pinto N, Yang W, Tanel R, and Goldmuntz E. (2013). 22q11.2 Deletion syndrome is associated with perioperative outcome in tetralogy of Fallot. J Thorac Cardiovasc Surg 146, 868–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Methlouthi J, Mahdhaoui N, Bellaleh M, et al. (2016). Incidence of congenital heart disease in newborns after pulse oximetry screening introduction. Tunis Med 94, 231–234 [PubMed] [Google Scholar]
- Métras D, Turquin H, Coulibaly AO, and Ouattara K. (1979). [Congenital cardiopathies in a tropical environment. Study of 259 cases seen at Abidjan from 1969–1976]. Arch Mal Coeur Vaiss 72, 305–310 [PubMed] [Google Scholar]
- Mirabel M, Lachaud M, Offredo L, et al. (2017). Cardiac surgery in low-income settings: 10 years of experience from two countries. Arch Cardiovasc Dis 110, 82–90 [DOI] [PubMed] [Google Scholar]
- Montojo J, Zuberi K, Rodriguez H, et al. (2010). GeneMANIA Cytoscape plugin: Fast gene function predictions on the desktop. Bioinformatics 26, 2927–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munshi NV. (2012). Gene regulatory networks in cardiac conduction system development. Circ Res 110, 1525–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Collaborating Centre For WS and Children's H. (2008). National Institute for Health and Clinical Excellence: Guidance. Antenatal Care: Routine Care for the Healthy Pregnant Woman. London: RCOG Press. National Collaborating Centre for Women's and Children's Health [Google Scholar]
- Ng B, and Hokanson J. (2010). Missed congenital heart disease in neonates. Congenit Heart Dis 5, 292–296 [DOI] [PubMed] [Google Scholar]
- Nikyar B, Sedehi M, Mirfazeli A, Qorbani M and Golalipour M-J. (2011). Prevalence and Pattern of Congenital Heart Disease among Neonates in Gorgan, Northern Iran (2007–2008). Iranian J Pediatr 21, 307–312 [PMC free article] [PubMed] [Google Scholar]
- Nkoke C, Balti E, Menanga A, et al. (2017). Trends in pediatric echocardiography and the yield for congenital heart disease in a major cardiac referral hospital in Cameroon. Transl Pediatr 6, 40–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nkoke C, Lekoubou A, Yonta EW, Dzudie A, and Kengne AP. (2014). Shone's anomaly: A report of one case in sub-Saharan Africa. Cardiovasc Diagn Ther 4, 495–498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouira S, Kamoun I, Ouragini H, et al. (2008). Clinical and genetic investigation of atrial septal defect with atrioventricular conduction defect in a large consanguineous Tunisian Family. Arch Med Res 39, 429–433 [DOI] [PubMed] [Google Scholar]
- Nunes MaS, Magalhães MP, Uva MS, et al. (2017). A multinational and multidisciplinary approach to treat CHD in paediatric age in Angola: Initial experience of a medical-surgical centre for children with heart disease in Angola. Cardiol Young 27, 1755–1763 [DOI] [PubMed] [Google Scholar]
- Okeniyi J, and Kuti B. (2008). Cerebral malaria in children with cyanotic heart diseases: The need for a closer look. Congenit Heart Dis 3, 73–76 [DOI] [PubMed] [Google Scholar]
- Oster ME, Riehle-Colarusso T, Simeone RM, et al. (2013). Public health science agenda for congenital heart defects: Report from a centers for disease control and prevention experts meeting. J Am Heart Assoc 2, e000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaigbe BE, and Tabansi PN. (2014). Congenital heart disease in the Niger Delta region of Nigeria: A four-year prospective echocardiographic analysis: Cardiovascular topic. Cardiovasc J Afr 25, 265–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen N, Poulsen G, Wohlfahrt J, et al. (2010). Recurrence of discordant congenital heart defects in families. Circ Cardiovasc Genet 3, 122–128 [DOI] [PubMed] [Google Scholar]
- Ozdemir V, Downey RA, Lin B, et al. (2014). Special issue “OMICS IN AFRICA”: Power to the people—moving 21st century integrative biology from lab to village to innovation ecosystems. OMICS 18, 399–401 [DOI] [PubMed] [Google Scholar]
- PanepistēMio TēS KrēTēS. D, Wei D, Gong X-H, et al. (2014). Novel PITX2c loss-of-function mutations associated with complex congenital heart disease. Int J Mol Med 33, 1201–1208 [DOI] [PubMed] [Google Scholar]
- Pishva SR, Vasudevan R, Etemad A, et al. (2013). Analysis of MTHFR and MTRR gene polymorphisms in Iranian ventricular septal defect subjects. Int J Mol Sci 14, 2739–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plageman TF, Jr. and Yutzey KE. (2004). Differential expression and function of Tbx5 and Tbx20 in cardiac development. J Biol Chem 279, 19026–19034 [DOI] [PubMed] [Google Scholar]
- Posch MG, Waldmuller S, Muller M, et al. (2011). Cardiac alpha-myosin (MYH6) is the predominant sarcomeric disease gene for familial atrial septal defects. PLoS One 6, e28872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendiville T, Jay PY, and Pu WT. (2014). Insights into the genetic structure of congenital heart disease from human and murine studies on monogenic disorders. Cold Spring Harb Perspect Med 4, pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preuss C, Capredon M, Wunnemann F, et al. , (2016). Family Based Whole Exome Sequencing Reveals the Multifaceted Role of Notch Signaling in Congenital Heart Disease. PLoS Genet 12, e1006335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Xiao D, Guo X, et al. (2017). Multiple gene variations contributed to congenital heart disease via GATA family transcriptional regulation. J Transl Med 15, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu XK, Qiu XB, Yuan F, et al. (2014). A novel NKX2.5 loss-of-function mutation associated with congenital bicuspid aortic valve. Am J Cardiol 114, 1891–1895 [DOI] [PubMed] [Google Scholar]
- Qu Y, Liu X, Zhuang J, et al. (2016). Incidence of congenital heart disease: The 9-year experience of the guangdong registry of congenital heart disease, China. PLoS One 11, e0159257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsbottom SA, Sharma V, Rhee HJ, et al. (2014). Vangl2-regulated polarisation of second heart field-derived cells is required for outflow tract lengthening during cardiac development. PLoS Genet 10, e1004871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buettner SM, and Borlak J. (2004). Genetic analysis of cardiac-specific transcription factors reveals insights into congenital heart disease. Monatsschrift Kinderheilkunde 152, 1183–1188 [DOI] [PubMed] [Google Scholar]
- Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT. and Correa A. (2008). Prevalence of Congenital Heart Defects in Metropolitan Atlanta, 1998–2005. J Pediatr 153, 807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riehle‐Colarusso TJ, Bergersen L, Broberg CS, et al. (2016). Databases for congenital heart defect public health studies across the lifespan. J Am Heart Assoc 5, e004148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodius S, Androsova G, Götz L, et al. (2016). Analysis of the dynamic co-expression network of heart regeneration in the zebrafish. Sci Rep 6, 26822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos-Hesselink JW, Kerstjens-Frederikse WS, Meijboom FJ, and Pieper PG. (2005). Inheritance of congenital heart disease. Neth Heart J 13, 88–91 [PMC free article] [PubMed] [Google Scholar]
- Sadoh W, Ikhurionan P, and Imarengiaye C. (2016). Pre-anesthetic echocardiographic findings in children undergoing non-cardiac surgery at the University of Benin Teaching Hospital, Nigeria. Cardiovasc J Afr 27, 276–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoh W, and Osarogiagbon W. (2013). Underlying congenital heart disease in Nigerian children with pneumonia. Afr Health Sci 13, 607–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadoh WE, Uzodimma CC, and Daniels Q. (2013). Congenital heart disease in Nigerian Children. World J Pediatr Congenit Heart Surg 4, 172–176 [DOI] [PubMed] [Google Scholar]
- Saetrom P, Snove O, Jr. and Rossi JJ. (2007). Epigenetics and microRNAs. Pediatr Res 61, 17R-23R [DOI] [PubMed] [Google Scholar]
- Salavaty A. (2015). Carcinogenic effects of circadian disruption: An epigenetic viewpoint. Chin J Cancer 34, 375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Castro M, Eldjouzi H, Charpentier E, et al. (2016). Search for rare copy-number variants in congenital heart defects identifies novel candidate genes and a potential role for FOXC1 in patients with coarctation of the aortaclinical perspective. Circ Cardiovasc Genet 9, 86–94 [DOI] [PubMed] [Google Scholar]
- Sani MU, Mukhtar-Yola M, and Karaye KM. (2007). Spectrum of congenital heart disease in a tropical environment: An echocardiography study. J Natl Med Assoc 99, 665–669 [PMC free article] [PubMed] [Google Scholar]
- Senga J, Rusingiza E, Mucumbitsi J, et al. (2013). Catheter interventions in congenital heart disease without regular catheterization laboratory equipment: The chain of hope experience in Rwanda. Pediatr Cardiol 34, 39–45 [DOI] [PubMed] [Google Scholar]
- Shaker O, Omran S, Sharaf E, et al. (2017). A novel mutation in exon 1 of GATA4 in Egyptian patients with congenital heart disease. Turk J Med Sci 47, 217–221 [DOI] [PubMed] [Google Scholar]
- Sheng W, Qian Y, Wang H, et al. (2013). Association between mRNA levels of DNMT1, DNMT3A, DNMT3B, MBD2 and LINE-1 methylation status in infants with tetralogy of Fallot. Int J Mol Med 32, 694–702 [DOI] [PubMed] [Google Scholar]
- Shewasinad S, and Negash S. (2017). Adherence and Associated Factors of Prenatal Iron Folic Acid Supplementation among Pregnant Women Who Attend Ante Natal Care in Health Facility at Mizan-Aman Town, Bench Maji Zone, Ethiopia, 2015. J Pregnancy Child Health. Doi: 10.4172/2376-127X.1000335 [DOI] [Google Scholar]
- Sifrim A, Hitz MP, Wilsdon A, et al. (2016). Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nat Genet 48, 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slagle CE, and Conlon FL. (2016). Emerging field of cardiomics: High-throughput investigations into transcriptional regulation of cardiovascular development and disease. Trends Genet 32, 707–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Wilson Ian j, Bentham J, et al. (2012). Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. Am J Hum Genet 91, 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sos BC, Fung HL, Gao DR, et al. (2016). Characterization of chromatin accessibility with a transposome hypersensitive sites sequencing (THS-seq) assay. Genome Biol 17, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperling S, Grimm CH, Dunkel I, et al. (2005). Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum Mutat 26, 575–582 [DOI] [PubMed] [Google Scholar]
- Stavsky M, Robinson R, Sade MY, et al. (2017). Elevated birth prevalence of conotruncal heart defects in a population with high consanguinity rate. Cardiol Young 27, 109–116 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Morris JH, Cook H, et al. (2017). The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45, D362–D368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantchou Tchoumi JC, and Butera G. (2013). Profile of cardiac disease in Cameroon and impact on health care services. Cardiovasc Diagn Ther 3, 236–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantchou Tchoumi JC, Butera G, Giamberti A, Ambassa JC, and Sadeu JC. (2011). Occurrence and pattern of congenital heart diseases in a rural area of sub-Saharan Africa. Cardiovasc J Afr 22, 63–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teteli R, Uwineza A, Butera Y, et al. (2014). Pattern of congenital heart diseases in Rwandan children with genetic defects. Pan Afr Med J 19, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theodoris CV, Li M, White MP, et al. , (2015). Human disease modeling reveals integrated transcriptional and epigenetic mechanisms of NOTCH1 haploinsufficiency. Cell 160, 1072–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M, Olusoji O, and Awolola N. (2013). Spectrum of congenital heart diseases in an African population: A necropsy study. World J Cardiovasc Dis 3, 34–39 [Google Scholar]
- Tougouma SJ, Kissou SA, Yameogo AA, et al. (2016). [Cardiopathies in children hospitalized at the University hospital Souro Sanou, Bobo-Dioulasso: Echocardiographic and therapeutic aspects]. Pan Afr Med J 25, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udink Ten Cate FEA, Sreeram N, Hamza H, et al. (2013). Stenting the arterial duct in neonates and infants with congenital heart disease and duct-dependent pulmonary blood flow: A multicenter experience of an evolving therapy over 18 years. Cathet Cardiovasc Interv 82, E233–E243 [DOI] [PubMed] [Google Scholar]
- Ul Haq F, Jalil F, Hashmi S, et al. (2011). Risk factors predisposing to congenital heart defects. Ann Pediatr Cardiol 4, 117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Beynum IM, Kapusta L, Den Heijer M, et al. (2006). Maternal MTHFR 677C>T is a risk factor for congenital heart defects: Effect modification by periconceptional folate supplementation. Eur Heart J 27, 981–987 [DOI] [PubMed] [Google Scholar]
- Van Der Linde D, Konings EE, Slager MA, et al. (2011). Birth prevalence of congenital heart disease worldwide: A systematic review and meta-analysis. J Am Coll Cardiol 58, 2241–2247 [DOI] [PubMed] [Google Scholar]
- Von Gise A, and Pu WT. (2012). Endocardial and epicardial epithelial to mesenchymal transitions in heart development and disease. Circ Res 110, 1628–1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Liu D, Zhang RR, et al. (2017). A TBX5 3′UTR variant increases the risk of congenital heart disease in the Han Chinese population. Cell Discov 3, 17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Luo XJ, Xin YF, et al. (2012). Novel GATA6 mutations associated with congenital ventricular septal defect or tetralogy of fallot. DNA Cell Biol 31, 1610–1617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Mao JH, Ding KK, et al. (2015). A novel NKX2.6 mutation associated with congenital ventricular septal defect. Pediatr. Cardiol 36, 646–656 [DOI] [PubMed] [Google Scholar]
- Wang X, Ji W, Wang J, et al. (2014). Identification of two novel GATA6 mutations in patients with nonsyndromic conotruncal heart defects. Mol Med Rep 10, 743–748 [DOI] [PubMed] [Google Scholar]
- Wansleeben C, Feitsma H, Montcouquiol M, et al. (2010). Planar cell polarity defects and defective Vangl2 trafficking in mutants for the COPII gene Sec24b. Development 137, 1067–1073 [DOI] [PubMed] [Google Scholar]
- Wessels M, and Willems P. (2010). Genetic factors in non-syndromic congenital heart malformations. Clin Genet 78, 103–123 [DOI] [PubMed] [Google Scholar]
- Wonkam A, Toko R, Chelo D, et al. (2017). The 22q11.2 deletion syndrome in congenital heart defects. Global Heart 12, 115–120 [DOI] [PubMed] [Google Scholar]
- Wood JB, Serumaga J, and Lewis MG. (1969). Congenital heart disease at necropsy in Uganda. A 16-years survey at Mulago Hospital, Kampala. Br Heart J 31, 76–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wren C, Reinhardt Z, and Khawaja K. (2008). Twenty-year trends in diagnosis of life-threatening neonatal cardiovascular malformations. Arch Dis Child Fetal Neonatal Ed 93, F33–F35 [DOI] [PubMed] [Google Scholar]
- Xiong F, Li Q, Zhang C, et al. (2013). Analyses of GATA4, NKX2.5, and TFAP2B genes in subjects from southern China with sporadic congenital heart disease. Cardiovasc Pathol 22, 141–145 [DOI] [PubMed] [Google Scholar]
- Xu M, Wu X, Li Y, et al. (2014). CITED2 Mutation and methylation in children with congenital heart disease. J Biomed Sci 21, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y-J, Qiu X-B, Yuan F, et al. (2017). Prevalence and spectrum of NKX2.5 mutations in patients with congenital atrial septal defect and atrioventricular block. Mol Med Report 15, 2247–2254 [DOI] [PubMed] [Google Scholar]
- Yoshida A, Morisaki H, Nakaji M, et al. (2016). Genetic mutation analysis in Japanese patients with non-syndromic congenital heart disease. J Hum Genet 61, 157–162 [DOI] [PubMed] [Google Scholar]
- Yu H, Ye X, Guo N, and Nathans J. (2012). Frizzled 2 and frizzled 7 function redundantly in convergent extension and closure of the ventricular septum and palate: Evidence for a network of interacting genes. Development 139, 4383–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Gao Y, Wang H, et al. (2014). Promoter methylation and expression of the VANGL2 gene in the myocardium of pediatric patients with Tetralogy of Fallot. Birth Defects Res A Clin Mol Teratol 100, 973–984 [DOI] [PubMed] [Google Scholar]
- Zhang W, Shen L, Deng Z, et al. (2014). Novel missense variants of ZFPM2/FOG2 identified in conotruncal heart defect patients do not impair interaction with GATA4. PLoS One 9, e102379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Jia WK, Jian Z, et al. (2017). Downregulation of microRNA-199a-5p protects cardiomyocytes in cyanotic congenital heart disease by attenuating endoplasmic reticulum stress. Mol Med Rep 16, 2992–3000 [DOI] [PubMed] [Google Scholar]
- Zhu X, Li J, Ru T, et al. (2016). Identification of copy number variations associated with congenital heart disease by chromosomal microarray analysis and next-generation sequencing. Prenat Diagn 36, 321–327 [DOI] [PubMed] [Google Scholar]
- Zidan HE, Rezk NA, and Mohammed D. (2013). MTHFR C677T and A1298C gene polymorphisms and their relation to homocysteine level in Egyptian children with congenital heart diseases. Gene 529, 119–124 [DOI] [PubMed] [Google Scholar]