Abstract
Purpose
Mounting evidence highlights the essential role of TRIM44 in tumor initiation and malignant progression in several cancers; however, the function of TRIM44 in osteosarcoma (OS) remains unknown. In this study, we aim to investigate the role of TRIM44 and reveal its regulation by deregulated miRNAs in OS.
Materials and methods
The expression profiles of TRIM44 were examined by immunohistochemistry, Western blotting, and qRT-PCR. The biological functions of TRIM44 were investigated through siRNA-mediated knockdown experiments. The regulation of TRIM44 by miR-410 was confirmed by Western blotting, dual luciferase reporter assays, and rescue experiments.
Results
TRIM44 was upregulated in OS tissues and cell lines, and its overexpression was positively correlated with TNM stage, metastasis, and recurrence. Knockdown of TRIM44 in OS cells suppressed cell proliferation, migration, invasion, and epithelial–mesenchymal transition. In addition, we identified TRIM44 as a novel target gene of miR-410 and miR-410 was remarkably downregulated in OS. Moreover, overexpression of miR-410 suppressed proliferation, migration, invasion, and epithelial–mesenchymal transition of OS cells by directly targeting TRIM44 expression. Furthermore, reintroduction of TRIM44 partially reversed miR-410-induced inhibitory effects on OS cells.
Conclusion
Collectively, our findings indicate that the miR-410/TRIM44 link is critical in the control of OS progression.
Keywords: TRIM44, miR-410, osteosarcoma, proliferation, invasion
Introduction
Osteosarcoma (OS) is the most common malignant bone cancer encountered in children and adolescents, with high incidence, rapid progression, and great metastatic potential.1,2 It is derived from primitive transformed cells that display osteoblastic differentiation and generate malignant osteoid tissue.3 Despite the numerous improvements in surgery and preoperative and postoperative chemotherapy, the prognosis of OS patients remains poor.4 Therefore, it is imperative to investigate the molecular mechanism underlying OS tumorigenesis, to provide more clues for clinical instructions, to identify novel therapeutic targets, and to improve patient prognosis.
TRIM44 is a member of the TRIM protein family characterized by a RING finger domain, two B-box zinc-fingers, and a coiled-coil region.5 Proteins in this large family have been implicated in a range of biological functions and consequently, their abnormal expression leads to diverse pathological conditions including inflammation, viral infection, neurodegenerative diseases, and tumorigenesis.6–10 Recently, several reports have uncovered the oncogenic potential of TRIM44 in cancer development and progression, such as non-small cell lung cancer, gastric cancer, testicular germ cell tumor, and esophageal squamous cell carcinoma.10–13 However, the expression and functions of TRIM44 in OS, as well as the mechanism underlying its dysregulation in tumorigenesis remain uncharacterized.
It has been well established that miRNAs, a class of small non-coding RNAs, could modulate the expression of numerous downstream target genes, thereby being involved in a wide range of biological processes, such as proliferation, apoptosis, migration, invasion, and metabolism.14–16 To date, several miRNAs have been implicated in the development and progression of OS, such as miR-155, miR-216a, miR-489, and miR-410 of which miR-410 was identified to display tumor-suppressive properties.17–20 However, the important role of miR-410 in OS still needs to be further elucidated.
In this study, we investigated the expression profile and biological functions of TRIM44 in an integrative investigation system including clinical OS tissues and cellular models, aiming to define the expression status and downstream signaling pathway of TRIM44 in OS. Moreover, we also identified TRIM44 as a direct functional target of miR-410, and miR-410/TRIM44 axis’ importance in the control of OS.
Materials and methods
Tissue specimens
A total of 90 primary OS tissue samples and their paired 42 adjacent tissues with available clinical information were obtained between 2013 and 2017 from Shanghai Cancer Center, Fudan University. Simultaneously, eight fresh OS tissues and their corresponding adjacent tissues were obtained from the Department of Surgery, Shanghai Cancer Center of Fudan University. Detailed clinicopathological information was confirmed by two pathologists independently. All these OS patients were treatment-naïve before surgery and this study was approved by the Ethics Committee of the Shanghai Cancer Center of Fudan University. Written informed consent was obtained from every patient.
Immunohistochemistry
Immunohistochemistry was performed as previously described.21 Briefly, slides went through antibody incubation overnight at 4°C with TRIM44 antibody (11511-1-AP; Proteintech, Chicago, IL, USA). The staining scores were then evaluated by two independent pathologists. TRIM44 expression was scored according to four grades by staining intensity (1+, 2+, 3+) and the percentages of TRIM44-positive cells: 1 (<10%), 2 (10%–33%), 3 (34%–66%), and 4 (>67%). A final staining score was generated by multiplying the scores of these two numbers ranging from 1 to 12. For further assessment, the negative and weak staining TRIM44 cases were defined as negative expression with a score index ≤6, and TRIM44-positive OS cases with a score index >6 as high expression.
Western blotting
Whole cell lysates or tissues were obtained with lysis buffer consisting of 50 mM Tris/HCl (pH 7.5), 0.5% Nonidet P-40 (NP-40), 1 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.2 mM PMSF, 10 μM pepstatin A, and 1 mM leupeptin. Equal amounts of clear cell lysates (20–50 μg) were used for western blotting analysis as described previously.14 All the membranes were blocked with 5% milk solution and then incubated with the following primary antibodies overnight: TRIM44 (ab153383; Abcam, Cambridge, MA, USA), E-cadherin (ab76055; Abcam), N-cadherin (ab98952; Abcam), fibronectin (ab2413; Abcam), and GAPDH (ab9484; Abcam). The next day, the membranes were washed with TBST and then incubated with secondary antibodies for 1 h at room temperature. GAPDH was used as an internal control and protein bands were analyzed using ImageJ software.
RNA isolation and RT-PCR assays
RNA isolation and qRT-PCR were performed according to the manufacturer’s instructions as previously described.22 Trizol reagent (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) was used for total RNA extraction. Total RNAs of 0.5 μg were used as template for reverse transcription using poly-(T)20 primers and M-MLV reverse transcriptase (Promega Corporation, Fitchburg, WI, USA). RT-PCR was performed using SYBR Green Mix following the manufacturer’s protocol (Bio-Rad Laboratories Inc., Hercules, CA, USA). The primers for TRIM44, GAPDH, miR-410, and U6 are listed in Table 1. GAPDH or U6 was used as the endogenous control and all experiments were repeated more than twice. The 2−ΔΔCT equation was used to assess the relative expression levels.
Table 1.
The sequences of primers for qRT-PCR
| Gene | Primers |
|---|---|
| TRIM44 | Forward: GTGGACATCCAAGAGGCAAT |
| Reverse: AGCAAGCCTTCATGTGTCCT | |
| miR-410 | Forward: CCGCACGATATAACACAGATG |
| Reverse: GTGCAGGGTCCGAGGTATTC | |
| U6 | Forward: CTCGCTTCGGCAGCACA |
| Reverse: AACGCTTCACGAATTTGCGT | |
| GAPDH | Forward: GATTCCACCCATGGCAAATTC |
| Reverse: AGCATCGCCCCACTTGATT |
Cell lines and transfection
The human OS cell lines MG-63, U2OS, HOS, Saos-2 and the osteoblastic cell line NHOst were purchased from American Type Culture Collection ([ATCC], Manassas, VA, USA) or the Chinese Academy of Medical Sciences (Beijing, People’s Republic of China). All the cells were cultured in RPMI 1640 medium with 10% FBS. All mentioned cells were maintained at 37°C with 5% CO2. Cells were transiently transfected with negative control siRNA, TRIM44 siRNA, negative control, miR-410 mimics and TRIM44-no UTR using Turbofect Reagent (Thermo Fisher Scientific) following the manufacturer’s instructions. The sequences for TRIM44 siRNA or negative control siRNA were TRIM44 siRNA, 5′-GUACCGCACGUCAUUCGUAUC-3′; TRIM44 siRNA, 5′-CCGCUAUGAUCGAAUUGGUGG-3′. The negative control and miR-410 mimics were purchased from the GenePharma company (Shanghai, People’s Republic of China) and the sequences were as follows: negative control, 5′-UUCUCCGAACGUGUCACGUACGUGACACGUUCGGAGAA-3′; miR-410 mimics 3′-AAUAUAACACAGAUGGCCUGUACAGGCCAUCUGUGUUAUAUU-5′. TRIM44 expression vector without 3′UTR was constructed by inserting its CDS region into the psiCHECK2 vector and purchased from GenePharma. Transfection efficiency was confirmed 48 h after transfection by qRT-PCR or Western blotting.
Cell viability assay
To assess cell survival, the Cell Counting Kit-8 (CCK-8) (Dojindo Molecular Technologies, Rockville, MD, USA) was used according to the manufacturer’s instructions. Cells were seeded at 1.5× 103 cells per well in 96-well culture plates 24 h post-transfection. Cell viability was determined by adding WST-8 at a final concentration of 10% to each well, and the absorbance of these samples was measured at 450 nm using a Microplate Reader (SpecrtraMax M5e; Molecular Devices LLC, Sunnyvale, CA, USA) every 24 h for 5 days. The experiments were performed more than three times.
Wound healing assay
To evaluate cell motility, cells were seeded on 6-well plates with appropriate medium containing 10% FBS. Until the cells achieved full confluency, we made a cell-free gap by scratching the bottom of the plate with a sterilized 200 μL pipette tip, and the cells were further incubated for 48 h. The images of the wound scratch were obtained by using an inverted Olympus IX50 microscope at 0 h and 48 h post-scratching. The percentage of wound closure was calculated as the wound area at a given time compared with the initial wound surface. The experiments were performed no less than three times.
Transwell assay
Transwell assay was carried out as described previously.22 Cells were suspended in 200 μL serum-free medium and seeded into the upper chamber pre-coated with Matrigel. An amount of 500 μL medium containing 15% FBS was added to the lower chamber as a chemoattractive factor. At 48 h post-transfection, cells that invaded the lower chamber were fixed and stained with 10% crystal violet. The experiments were performed in triplicate.
Bioinformatics prediction and luciferase reporter assay
Bioinformatic analysis was used to predict the potential miRNAs that could target TRIM44 using TargetScan (http://www.targetscan.org/) and miRanda (http://microrna.org/). Luciferase reporter assay was carried out as described previously. Briefly, OS cells were transiently transfected with the luciferase reporter vector containing wild-type (WT) or mutant (MUT) 3′UTR of TRIM44 and miR-410 mimics or negative control using TurboFect Reagent (Thermo Fisher Scientific). The full length 3′UTR of the TRIM44 gene and the MUT variant was amplified by PCR and was separately cloned into the luciferase reporter psiCHECK2 vector. The WT plasmid was constructed containing the full length 3′UTR of TRIM44 with miR-410 complementary sequence. The MUT variant plasmid with a degenerate 3′UTR was created by replacing “CAAUAUA” with “GUUAUAU” in the miR-410 complementary sequence. At 24 h post-transfection, cells were harvested and analyzed for Firefly and Renilla luciferase activities using the Dual-Luciferase Reporter Assay System (E1910; Promega). Activities were normalized to Renilla luciferase. This experiment was performed more than three times.
Statistical analysis
All these bio-functional experiments were performed in biological triplicate. The relationship between TRIM44 expression and clinicopathological characteristics was analyzed by the chi-square test. The Student’s two-tailed t-test was used to determine mean difference among groups. P<0.05 was considered statistically significant. All the data were presented as mean ± SEM.
Results
TRIM44 is upregulated in OS tissues
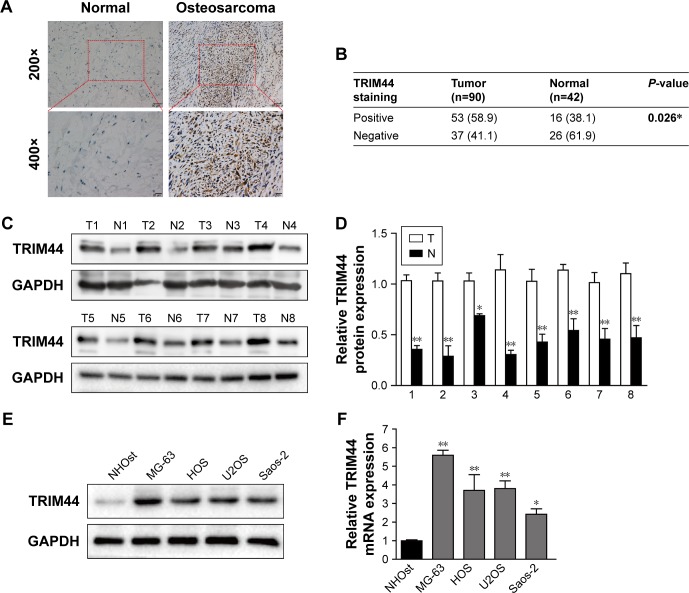
To determine the clinical relevance of TRIM44 in OS, TRIM44 expression was detected in 90 paraffin-embedded archived OS tissues and their paired 42 adjacent non-cancerous tissues by immunohistochemistry assay. As shown in Figure 1A, TRIM44 was predominantly localized in the nucleus of tumor cells and TRIM44 expression in OS samples was significantly higher than its expression in their paired normal tissues. The increased expression of TRIM44 was found in approximately 58.9% (53/90) of the clinical OS tissue samples, while it was 38.1% (16/42) in paired adjacent normal tissues (Figure 1B, P=0.026). To further confirm this finding, we examined the expression level of TRIM44 by Western blotting in eight pairs of OS tissues and matched adjacent non-cancerous tissues. The protein expression of TRIM44 in eight pairs of OS tissues was upregulated compared with their paired normal tissues (Figure 1C and D). Consistently, compared with normal osteoblastic cell line NHOst, the protein and mRNA expression of TRIM44 was also higher in all these four OS cell lines (Figure 1E and F).
Figure 1.
Upregulation of TRIM44 in human osteosarcoma tissues and cell lines.
Notes: (A) Expression of TRIM44 was detected by immunohistochemistry in osteosarcoma tissues and their paired adjacent non-cancerous tissues. The samples were stained with an anti-TRIM44 antibody. (B) Statistical analysis of TRIM44 expression in osteosarcoma tissues and adjacent normal tissues. Data in bold indicates statistical difference (P<0.05). (C, D) Western blotting analysis of TRIM44 in eight paired osteosarcoma tissues and their corresponding non-cancerous tissues. GAPDH was used as a loading control, data shown were mean ± SEM values (n=3). (E, F) Expression of TRIM44 was upregulated in all four osteosarcoma cell lines compared with normal osteoblastic cell line NHOst, confirmed by Western blotting and RT-PCR. The average TRIM44 mRNA expression was normalized to the endogenous control GAPDH. Error bars represent the mean ± SEM values from three independent experiments, *P<0.05, **P<0.01 by two-tailed Student’s t-test.
The association between TRIM44 expression and its clinicopathological factors was further analyzed in the OS cohort (n=90). As shown in Table 2, TRIM44 expression was dramatically correlated with TNM stage (P<0.001), metastasis (P=0.016), and recurrence (P=0.024). However, no statistically significant association was detected between TRIM44 expression and age, sex, and tumor size. Collectively, these data indicated that TRIM44 was upregulated and correlated with progression in OS.
Table 2.
Correlation between TRIM44 expression and clinicopathological factors of osteosarcoma patient
| Clinicopathological parameters | N | TRIM44 expression, n (%)
|
P-value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Total number | 90 | 37 (41.1) | 53 (58.9) | |
| Age (years) | 0.188 | |||
| <18 | 56 | 26 (46.4) | 30 (53.6) | |
| ≥18 | 34 | 11 (32.4) | 23 (67.6) | |
| Sex | 0.875 | |||
| Male | 44 | 20 (45.5) | 24 (54.5) | |
| Female | 46 | 17 (37.0) | 29 (63.0) | |
| Tumor size (cm) | 0.190 | |||
| <7 | 39 | 13 (33.3) | 26 (66.7) | |
| ≥7 | 51 | 24 (47.1) | 27 (52.9) | |
| TNM stage | <0.001* | |||
| I | 43 | 26 (60.5) | 17 (39.5) | |
| II–III | 47 | 11 (23.4) | 36 (76.6) | |
| Metastasis | 0.016* | |||
| Lung | 34 | 8 (23.5) | 26 (76.5) | |
| Other | 11 | 4 (36.4) | 7 (63.6) | |
| No | 45 | 25 (55.6) | 20 (44.4) | |
| Recurrence | 0.024* | |||
| Yes | 42 | 12 (28.6) | 30 (71.4) | |
| No | 48 | 25 (52.1) | 23 (47.9) | |
Notes:
Analyzed by the chi-square test. Data in bold indicates statistical difference (P<0.05).
Silencing TRIM44 inhibits cell proliferation, invasion, and epithelial–mesenchymal transition (EMT)
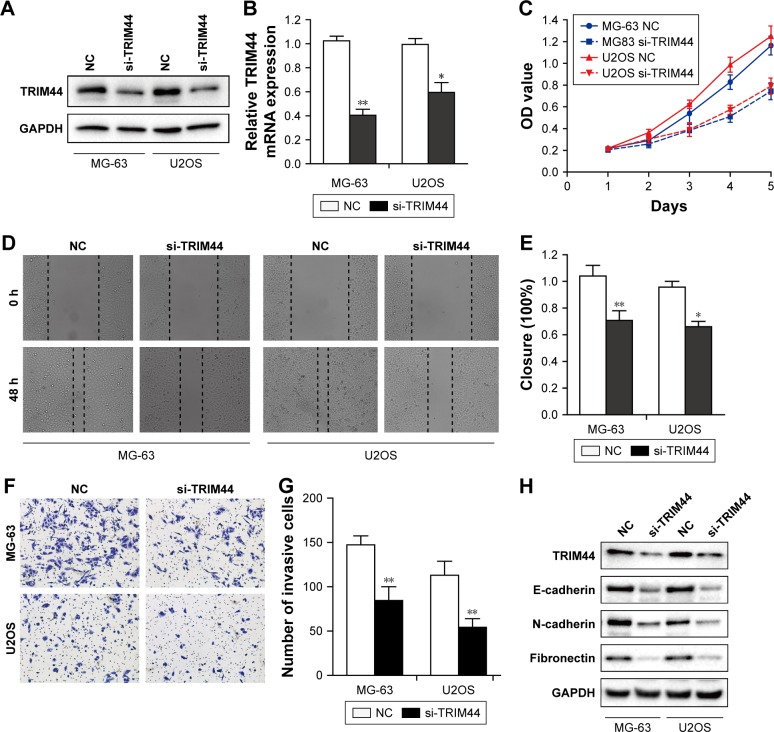
As TRIM44 was upregulated in OS cell lines, especially in MG-63 and U2OS cells, small interfering RNA-mediated TRIM44 knockdown was performed in these two cell lines. Both the protein and mRNA expression of TRIM44 were markedly decreased upon knockdown by specific siRNAs in OS cells (Figure 2A and B). The CCK-8 assay showed that the TRIM44 knockdown displayed a marked decrease in cell viability of MG-63 and U2OS cells (P<0.01, Figure 2C). Next, we investigated the effects of TRIM44 expression on migration and invasion of OS cells by wound healing and Transwell invasion assay. The results of wound healing assay showed that it took much longer for scratch wounds to close in TRIM44-downregulated cells compared to controls (P<0.05, Figure 2D and E). Moreover, TRIM44 knockdown led to significant suppression of cell invasive abilities in MG-63 and U2OS cells (P<0.01, Figure 2F and G). Furthermore, we examined the EMT markers in these two cell lines upon TRIM44 knockdown, surprisingly, E-cadherin was increased, while N-cadherin and fibronectin were reduced upon TRIM44 knockdown (Figure 2H). These data indicate a critical role of TRIM44 in cell proliferation, invasion, and EMT in OS cells.
Figure 2.
TRIM44 knockdown inhibits cell proliferation, invasion, and EMT.
Notes: (A) Western blotting was used to detect the protein expression of TRIM44 in MG-63 and U2OS cells upon siRNA transfection. (B) RT-PCR was performed to examine the mRNA expression of TRIM44 in MG-63/U2OS cells after transfection. (C) CCK8 assay revealed that knockdown of TRIM44 inhibited cell viability. (D, E) The wound healing assay showed that TRIM44 knockdown decreased the cell migration ability. (F, G) Representative images and quantification of invasive cells. (H) Western blotting analysis of EMT markers after transfection with TRIM44 siRNA. Error bars represent the mean ± SEM values of three independent experiments (*P<0.05, **P<0.01 by two-tailed Student’s t-test).
Abbreviations: EMT, epithelial–mesenchymal transition; CCK8, Cell Counting Kit-8; OD, optical density; NC, negative control.
MiR-410 represses TRIM44 expression by directly targeting its 3′UTR
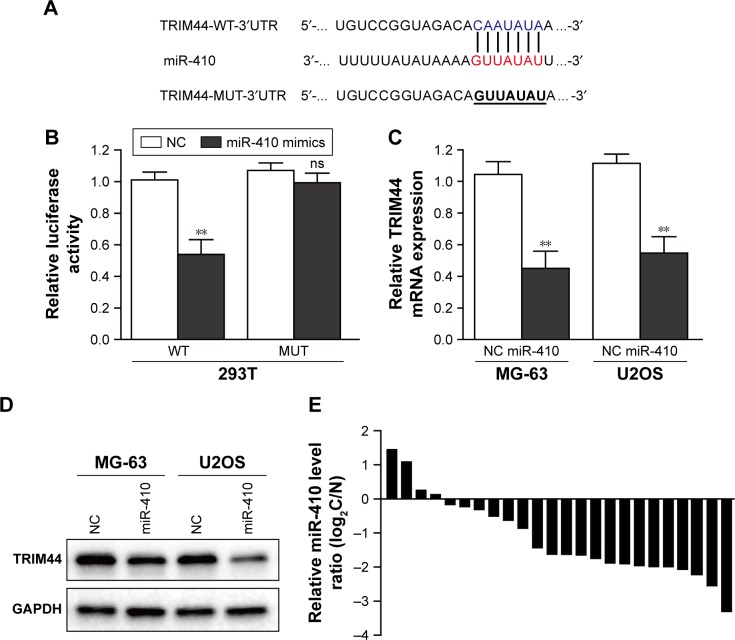
To investigate the underlying mechanism of TRIM44 upregulation, two web-based target analysis (TargetScan and miRanda) tools were used to identify miRNAs that potentially target TRIM44. The list of potential miRNAs which were predicted to target TRIM44 by two algorithms were shown in Figure S1. Among them, we focused on miR-410, which was reported to be involved in tumor progression, including OS. To test the specificity of the interaction between miR-410 and TRIM44, we constructed WT and MUT TRIM44 3′UTR luciferase reporter gene (Figure 3A). The luciferase reporter assay showed that when 293T cells were co-transfected with the WT luciferase reporter plasmids and miR-410 mimics, the luciferase activity diminished greatly as compared with negative control. However, the difference between miR-410 mimics and negative control would disappear when the binding site was mutated (Figure 3B). Moreover, we detected the mRNA level and protein level of TRIM44 in miR-410 mimics transfected OS cells. As clearly illustrated in Figure 3C and D, overexpression of miR-410 significantly repressed TRIM44 expression in MG-63 and U2OS cells. Furthermore, miR-410 expression was downregulated in OS tissues compared with adjacent normal tissues (80%, 16/20, Figure 3E). Collectively, these results suggest that miR-410 represses TRIM44 expression by directly targeting the 3′UTR.
Figure 3.
miR-410 directly targets TRIM44, and miR-410 is downregulated in osteosarcoma tissues.
Notes: (A) The putative miR-410-binding sequence in the 3′UTR of TRIM44. (B) 293T cells were co-transfected with the WT or MUT luciferase reporter plasmids and miR-410 mimics or negative control. The luciferase activity was evaluated 24 h post-transfection. (C, D) Western blot and qRT-PCR analysis of TRIM44 expression in osteosarcoma cells transfected with miR-410 mimics or negative control. (E) Detection of miR-410 expression in 20 paired osteosarcoma tissues and its corresponding noncancerous tissues. All data are presented as the mean ± SEM of experiments performed in triplicate (**P<0.01).
Abbreviations: WT, wild-type; MUT, mutant; NC, negative control; ns, non-significant.
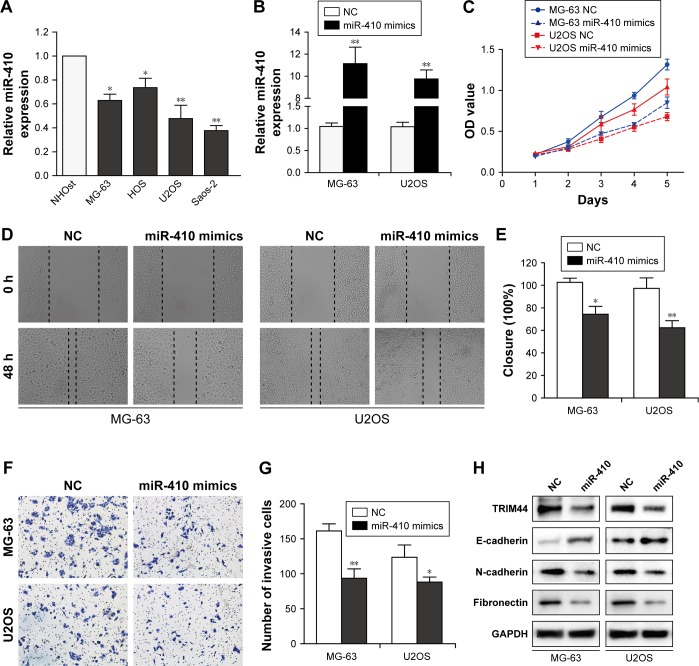
MiR-410 suppresses cell proliferation, invasion, and EMT in OS cells
To study the biological role of miR-410 in OS cells, we first examined the expression of miR-410 in four OS cells (MG-63, HOS, Saos2, and U2OS) and normal osteoblastic cell line NHOst. As shown in Figure 4A, miR-410 expression was significantly downregulated in OS cells compared with NHOst. Next, we overexpressed miR-410 in MG-63 and U2OS cells by transfection with miR-410 mimics or negative control (Figure 4B). Mimicking the effects of TRIM44 knockdown, miR-410 overexpression markedly inhibited cell growth, migration, and invasion (Figure 4C–G). Moreover, ectopic expression of miR-410 increased E-cadherin and reduced N-cadherin and fibronectin (Figure 4H). These results demonstrate that miR-410 suppresses cell proliferation, invasion, and EMT in OS cells.
Figure 4.
miR-410 suppresses cell proliferation, invasion, and EMT of osteosarcoma cells.
Notes: (A) The expression of miR-410 was detected by qRT-PCR in osteosarcoma cells. (B) qRT-PCR analysis of miR-410 expression in MG-63 and U2OS cells after transfection with miR-410 mimics or negative control. Effects of ectopic miR-410 on the proliferation (C), migration (D and E), and invasion (F and G) of MG-63 and U2OS cells were validated by CCK-8, wound healing, and Transwell assays, respectively. (H) Western blotting analysis of EMT markers after transfection with miR-410 overexpression. All experiments were repeated three times and data are shown as mean ± SEM (*P<0.05, **P<0.01).
Abbreviations: EMT, epithelial–mesenchymal transition; CCK8, Cell Counting Kit-8; OD, optical density; NC, negative control.
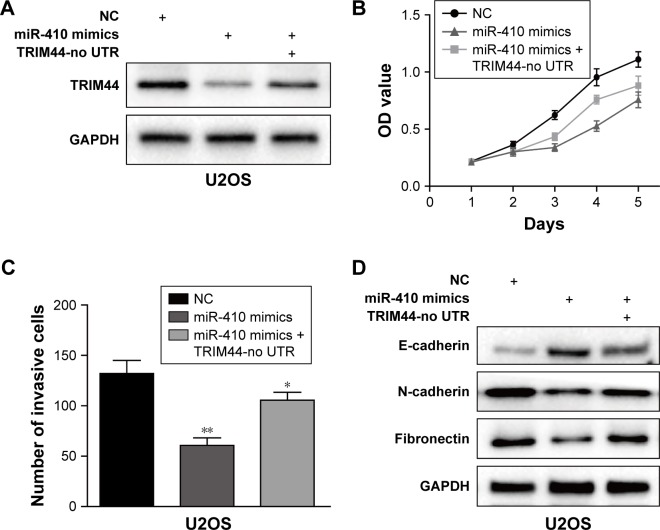
Reintroduction of TRIM44 attenuates miR-410-induced inhibition of cell proliferation and invasion
To confirm whether TRIM44 is a functional target of miR-410, we performed gain-of-function assays by synthesizing a TRIM44 overexpression plasmid without 3′UTR (TRIM44-no UTR) in U2OS cells. As shown in Figure 5A, TRIM44-no UTR transfection could restore the TRIM44 expression, which was suppressed by miR-410 in U2OS cells. Moreover, the functional assays showed that overexpression of TRIM44 partially reversed the inhibitory effects of miR-410 on U2OS cells, which was supported by CCK-8 and Transwell invasion assays (Figure 5B and C). In addition, reintroduction of TRIM44 could also partially abrogate the effects of miR-410 on EMT process in U2OS cells (Figure 5D). These data provide further support that TRIM44 is a critical functional target of miR-410.
Figure 5.
Ectopic expression of TRIM44 partially rescues the miR-410-induced effects on osteosarcoma cells.
Notes: (A) Western blot analysis of TRIM44 expression after transfection with TRIM44 no-UTR and miR-410 mimics or negative control. CCK-8 and Transwell assays were performed to examine the cell viability (B) and invasive ability (C), respectively. (D) Western blot analysis of the EMT markers’ expression after transfection with TRIM44 no-UTR and miR-410 mimics or negative control. All data are presented as the mean ± SEM of experiments performed in triplicate (*P<0.05, **P<0.01).
Abbreviations: EMT, epithelial–mesenchymal transition; CCK8, Cell Counting Kit-8; OD, optical density; NC, negative control.
Discussion
OS is one of the most common cancers in children and adolescents, and always has an aggressive clinical outcome. Nevertheless, the molecular mechanism underlying OS tumorigenesis and progression remains poorly investigated. Recently, several studies indicated that TRIM44, a member of the TRIM family, was upregulated and correlated with cancer progression in multiple human cancers. In this study, we explored the clinical relevance, biological functions, as well as the regulatory mechanism of TRIM44 in OS.
A series of cancers have recently been demonstrated to overexpress TRIM44 proteins. Kawabata et al showed that TRIM44 is a poor prognostic factor for breast cancer and increases aggressiveness by modulating NF-κB pathway.23 Xing et al showed that TRIM44 is upregulated and significantly correlated with poor behaviors in lung cancer, and knockdown of TRIM44 inhibits cell proliferation, migration, invasion, and EMT via mTOR signaling pathway.24 Our study, for the first time, has shown that TRIM44 was upregulated in OS tissues when compared with matched adjacent normal tissues, and TRIM44 overexpression was positively correlated with TNM stage, metastasis, and recurrence, implying its cancer-promoting role in the initiation and progression of OS. Silencing TRIM44 expression by specific siRNAs inhibits cell survival in OS cells. Furthermore, our results provide new evidence that TRIM44 promotes cell migration and invasion in OS, which may be mediated through activation of the EMT process. Our data suggest that TRIM44 is potentially an oncogenic gene.
It has been well established that MiRNAs are abnormally expressed in carcinogenesis. A bunch of miRNAs have been demonstrated to regulate the progression of OS proliferation, invasion, and metastasis through repressing their specific target genes. For these reasons, we suspected there could be some miRNAs involved in TRIM44 upregulation in OS. The TRIM44 transcript was predicted to be a direct target of miR-410 by bioinformatics tools, and was further confirmed by luciferase reporter assay, qRT-PCR, and Western blotting. There is a growing body of literature indicating the suppressive role of miR-410 in human cancers, including pancreatic cancer, gastric cancer, glioma, and breast cancer.25–28 Xiong et al have shown that miR-410 attenuates gemcitabine resistance by suppressing HMGB1-mediated autophagy in pancreatic ductal adenocarcinoma.29 In addition, miR-410 exerts a tumor-suppressing role in breast cancer progression by downregulating oncogenic Snail.27 Surprisingly, miR-410 also served as an oncogenic miRNA in some other types of cancer.30–32 Therefore, we believe that miR-410 may play a contradictory role in tumorigenesis depending on the type of cancer. Here, we identified that miR-410 was frequently downregulated in OS tissues and cell lines, indicating that miR-410 may be implicated in OS carcinogenesis. Consistent with previous studies, we also found overexpression of miR-410 inhibited cell proliferation, migration, invasion, and EMT, which mimics the cellular outcomes of TRIM44 knockdown. More intriguingly, ectopic expression of TRIM44 reversed the inhibitory effects of miR-410 on OS cells. To conclude, our data indicate that TRIM44 is another main functional target gene of miR-410, and miR-410 functions as a crucial tumor suppressor in OS.
Conclusion
Our findings showed that TRIM44 may be a critical driver in OS tumorigenesis, and downregulated miR-410 is responsible for the upregulation of TRIM44. MiR-410 has an important role in cell proliferation and invasion by targeting TRIM44 in OS. This newly characterized miR-410/TRIM44 axis provides new insights into the mechanisms underlying OS development, with opportunities for intervention for patients in the future.
Supplementary material
Several miRNAs were computationally predicted by using two independent miRNAs databases.
Acknowledgments
This study was supported by grants from the Natural Science Foundation of China (nos 81772295 and 81660408), the Science Foundation of Jiangxi Province (no 20142BAB205046), and the Graduate Student Innovation Foundation of Jiangxi Province (no YC2015-B010).
Footnotes
Author contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
Reference
- 1.Geller DS, Gorlick R. Osteosarcoma: a review of diagnosis, management, and treatment strategies. Clin Adv Hematol Oncol. 2010;8(10):705–718. [PubMed] [Google Scholar]
- 2.Parkin DM, Stiller CA, Draper GJ, Bieber CA. The international incidence of childhood cancer. Int J Cancer. 1988;42(4):511–520. doi: 10.1002/ijc.2910420408. [DOI] [PubMed] [Google Scholar]
- 3.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008;466(9):2114–2130. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: data from the Surveillance, Epidemiology, and End Results Program. Cancer. 2009;115(7):1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi: 10.1038/nrc3139. [DOI] [PubMed] [Google Scholar]
- 6.Yang B, Wang J, Wang Y, et al. Novel function of Trim44 promotes an antiviral response by stabilizing VISA. J Immunol. 2013;190(7):3613–3619. doi: 10.4049/jimmunol.1202507. [DOI] [PubMed] [Google Scholar]
- 7.Urano T, Usui T, Takeda S, et al. TRIM44 interacts with and stabilizes terf, a TRIM ubiquitin E3 ligase. Biochem Biophys Res Commun. 2009;383(2):263–268. doi: 10.1016/j.bbrc.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Ong CA, Shannon NB, Ross-Innes CS, et al. Amplification of TRIM44: pairing a prognostic target with potential therapeutic strategy. J Natl Cancer Inst. 2014;106(5):dju050. doi: 10.1093/jnci/dju050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kashevarova AA, Nazarenko LP, Skryabin NA, et al. Array CGH analysis of a cohort of Russian patients with intellectual disability. Gene. 2014;536(1):145–150. doi: 10.1016/j.gene.2013.11.029. [DOI] [PubMed] [Google Scholar]
- 10.Luo Q, Lin H, Ye X, et al. Trim44 facilitates the migration and invasion of human lung cancer cells via the NF-kappaB signaling pathway. Int J Clin Oncol. 2015;20(3):508–517. doi: 10.1007/s10147-014-0752-9. [DOI] [PubMed] [Google Scholar]
- 11.Yamada Y, Takayama KI, Fujimura T, et al. A novel prognostic factor TRIM44 promotes cell proliferation and migration, and inhibits apoptosis in testicular germ cell tumor. Cancer Sci. 2017;108(1):32–41. doi: 10.1111/cas.13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kawaguchi T, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 is related to invasive potential and malignant outcomes in esophageal squamous cell carcinoma. Tumour Biol. 2017;39(6) doi: 10.1177/1010428317700409. 1010428317700409. [DOI] [PubMed] [Google Scholar]
- 13.Kashimoto K, Komatsu S, Ichikawa D, et al. Overexpression of TRIM44 contributes to malignant outcome in gastric carcinoma. Cancer Sci. 2012;103(11):2021–2026. doi: 10.1111/j.1349-7006.2012.02407.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fang Z, Zhang L, Liao Q, et al. Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer. J Exp Clin Cancer Res. 2017;36(1):17. doi: 10.1186/s13046-017-0489-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xia JT, Chen LZ, Jian WH, et al. MicroRNA-362 induces cell proliferation and apoptosis resistance in gastric cancer by activation of NF-kappaB signaling. J Transl Med. 2014;12:33. doi: 10.1186/1479-5876-12-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen S, Huang K, Wu Y, et al. A miR-135b-TAZ positive feedback loop promotes epithelial-mesenchymal transition (EMT) and tumorigenesis in osteosarcoma. Cancer Lett. 2017;407:32–44. doi: 10.1016/j.canlet.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 17.Bhattacharya S, Chalk AM, Ng AJ, et al. Increased miR-155-5p and reduced miR-148a-3p contribute to the suppression of osteosarcoma cell death. Oncogene. 2016;35(40):5282–5294. doi: 10.1038/onc.2016.68. [DOI] [PubMed] [Google Scholar]
- 18.Ji Q, Xu X, Li L, et al. miR-216a inhibits osteosarcoma cell proliferation, invasion and metastasis by targeting CDK14. Cell Death Dis. 2017;8(10):e3103. doi: 10.1038/cddis.2017.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Yang G, Qian Y. Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol Carcinog. 2017;56(4):1312–1321. doi: 10.1002/mc.22593. [DOI] [PubMed] [Google Scholar]
- 20.Zhao D, Jia P, Wang W, Zhang G. VEGF-mediated suppression of cell proliferation and invasion by miR-410 in osteosarcoma. Mol Cell Biochem. 2015;400(1–2):87–95. doi: 10.1007/s11010-014-2265-2. [DOI] [PubMed] [Google Scholar]
- 21.Fang Z, Deng J, Zhang L, et al. TRIM24 promotes the aggression of gastric cancer via the Wnt/beta-catenin signaling pathway. Oncol Lett. 2017;13(3):1797–1806. doi: 10.3892/ol.2017.5604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han X, Fang Z, Wang H, et al. CUL4A functions as an oncogene in ovarian cancer and is directly regulated by miR-494. Biochem Biophys Res Commun. 2016;480(4):675–681. doi: 10.1016/j.bbrc.2016.10.114. [DOI] [PubMed] [Google Scholar]
- 23.Kawabata H, Azuma K, Ikeda K, et al. TRIM44 is a poor prognostic factor for breast cancer patients as a modulator of NF-kappaB signaling. Int J Mol Sci. 2017;18(9):1931. doi: 10.3390/ijms18091931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xing Y, Meng Q, Chen X, et al. TRIM44 promotes proliferation and metastasis in nonsmall cell lung cancer via mTOR signaling pathway. Oncotarget. 2016;7(21):30479–30491. doi: 10.18632/oncotarget.8586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo R, Gu J, Zhang Z, Wang Y, Gu C. MicroRNA-410 functions as a tumor suppressor by targeting angiotensin II type 1 receptor in pancreatic cancer. IUBMB Life. 2015;67(1):42–53. doi: 10.1002/iub.1342. [DOI] [PubMed] [Google Scholar]
- 26.Shen J, Niu W, Zhou M, et al. MicroRNA-410 suppresses migration and invasion by targeting MDM2 in gastric cancer. PLoS One. 2014;9(8):e104510. doi: 10.1371/journal.pone.0104510. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Zhang YF, Yu Y, Song WZ, et al. miR-410-3p suppresses breast cancer progression by targeting Snail. Oncol Rep. 2016;36(1):480–486. doi: 10.3892/or.2016.4828. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Zhang J, Feng Y, et al. MiR-410 regulates MET to influence the proliferation and invasion of glioma. Int J Biochem Cell Biol. 2012;44(11):1711–1717. doi: 10.1016/j.biocel.2012.06.027. [DOI] [PubMed] [Google Scholar]
- 29.Xiong J, Wang D, Wei A, et al. MicroRNA-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting HMGB1-mediated autophagy. Oncotarget. 2017;8(64):107500–107512. doi: 10.18632/oncotarget.22494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Ke X, Pu Q, et al. MicroRNA-410 acts as oncogene in NSCLC through downregulating SLC34A2 via activating Wnt/beta-catenin pathway. Oncotarget. 2016;7(12):14569–14585. doi: 10.18632/oncotarget.7538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li D, Yang Y, Zhu G, et al. MicroRNA-410 promotes cell proliferation by targeting BRD7 in non-small cell lung cancer. FEBS Lett. 2015;589(17):2218–2223. doi: 10.1016/j.febslet.2015.06.031. [DOI] [PubMed] [Google Scholar]
- 32.Liu C, Zhang A, Cheng L, Gao Y. miR410 regulates apoptosis by targeting Bak1 in human colorectal cancer cells. Mol Med Rep. 2016;14(1):467–473. doi: 10.3892/mmr.2016.5271. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Several miRNAs were computationally predicted by using two independent miRNAs databases.