Abstract
Clinical pharmacogenomics (PGx) has the potential to make pharmacotherapy safer and more effective by utilizing genetic patient data for drug dosing and selection. However, widespread adoption of PGx depends on its successful integration into routine clinical care through clinical decision support tools, which is often hampered by insufficient or fragmented infrastructures. This paper describes the setup and implementation of a unique multimodal, multilingual clinical decision support intervention consisting of digital, paper-, and mobile-based tools that are deployed across implementation sites in seven European countries participating in the Ubiquitous PGx (U-PGx) project.
Keywords: clinical decision support systems, pharmacogenomics
INTRODUCTION
Pharmacogenomics (PGx), i.e., using genetic data to guide drug-dosing and selection, emerged as a promising strategy for making pharmacotherapy safer and more effective.1–4 A successful implementation of PGx into clinical practice strongly depends on the availability of clinical decision support (CDS) tools that translate raw genetic test results into concise and clinically actionable therapeutic recommendations, and make those results available to healthcare providers at the point of care.5
Several projects utilizing different variants of PGx testing and CDS have been launched; these projects are described and compared in detail in Supplementary Material S1.6–23
A common factor of these successful PGx implementation projects is the delivery of CDS via the electronic health record (EHR), either as an interruptive alert at the time of prescribing, or as part of the patie'nt’s digital record. Although the availability of EHRs in hospital settings significantly increased within the past decade and reached adoption rates of >50% in most developed countries, nationwide availability still cannot be expected in most regions.24,25 Moreover, a lack of interoperability between different existing EHR systems as well as their fragmented availability beyond hospital settings still constitute substantial barriers to the efficient and secure sharing of PGx data.25–28
In this paper, we describe the development and implementation of a unique, multi-modal, and multi-lingual PGx CDS strategy across 7 European countries in the context of the Ubiquitous PGx (U-PGx) project that enables the delivery of PGx CDS in the presence of diverse and fragmented healthcare infrastructures.
IMPLEMENTING CDS IN THE U-PGx PROJECT
Project setting
Widespread adoption of PGx-guided prescribing in routine care will heavily depend on the availability of robust data from large-scale clinical studies that demonstrate improved clinical outcomes and cost-effectiveness of PGx testing when applied to broad patient populations. The Ubiquitous PGx (U-PGx) project was initiated to address this need by implementing PGx panel testing and CDS across 7 European countries and measuring patient outcomes and cost-effectiveness.29 The project started in January 2016 with a total duration of 5 years and a budget of 15 million Euros from the Horizon 2020 EU research program.
The project includes a clinical study which was initiated in early 2017. It is designed as a prospective, block-randomized, controlled study, and a total of 8100 patients are planned to be enrolled over the course of three years (Clinicaltrials.gov NCT03093818). Patient recruitment takes place at one or more healthcare institutions in each of the 7 participating countries.
As part of the intervention, patients who receive a first prescription of a drug, for which a pharmacogenomic guideline is available, are tested for a panel of more than 48 clinically relevant PGx variants across 13 important pharmacogenes relevant for dose optimization of 41 drugs.29 PGx testing is deployed in a mixed “reactive-preemptive” approach, meaning that a full PGx panel is ordered at the time of first prescription of a drug and dosage can be optimized based on PGx guidelines. PGx panel results are then readily available for optimizing prescription of other drugs prescribed in future interactions with the healthcare system.
Special requirements for the implementation of CDS in the U-PGx project
The international setup of the U-PGx project entailed a special set of demands that are unique among projects utilizing CDS for PGx.
While having to deal with heterogeneity (e.g., different types of EHRs) is not uncommon in larger CDS implementation projects,30 this challenge is aggravated within the U-PGx project, where technical framework conditions range from complete absence of any information technology (IT) infrastructure at some implementation sites to sophisticated IT systems with the ability to provide active CDS via automated alerts at others (see Table 1). Furthermore, differences do not only exist between different participating countries but also between participating sites within the same country. Ensuring a standardized intervention while still making optimal use of existing technical capabilities and meeting essential country-specific requirements is therefore a key requirement in the U-PGx project.
Table 1.
Characteristics of Existing IT Infrastructures at the U-PGx Implementation Sites
| Infrastructure characteristics | NL | GB | IT | ES | AT | SI | GR |
|---|---|---|---|---|---|---|---|
| EHR inpatient setting | Yes | Yes | Partially | Yes | Partially | Partially | No |
| EHR outpatient setting | Yes | Partially | Partially | Yes | No | No | No |
| Text reports | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Active CDS | Yes (for PGx, DDIs, contraindications, and drug dose) | Yes (for allergies and DDI) | No | Yes (for allergies) | No | No | No |
| Passive CDS | No | Yes | Yes | Yes | Yes | No | No |
| LIMS | Yes | Yes | Yes | Yes | Yes | Yes | No |
| Structured laboratory results | Yes | Yes | Yes | Yes | Yes | No | No |
Abbreviations: AT, Austria; DDI, Drug-drug interactions; EHR, Electronic health record; ES, Spain; GB, Great Britain; GR, Greece; IT, Italy; LIMS, Laboratory information management system; NL, The Netherlands; PGx, Pharmacogenomics; SI, Slovenia.
Participating healthcare institutions as of August 2017: NL: A network of primary care physicians and pharmacies established by the department of Clinical Pharmacy & Toxicology of the Leiden University Medical Center (LUMC); Department of Neurology, LUMC; Outpatient Pharmacy, LUMC. GB: The Royal Liverpool University Hospital. IT: Medical Oncology and Radiotherapy Oncology Unit of the National Cancer Institute in Aviano and Treviso, Medical Oncology and Radiotherapy Oncology Unit of the San Filippo Neri Hospital in Rome. ES: Departments of Pharmacy and Cardiology of the San Cecilio University Hospital in Granada. AT: Department of Nephrology and Dialysis of the Vienna General Hospital. SI: Kidney transplant center, Nephrology Clinic, University Clinical Center Ljubljana; Health Care Center Ljubljana, Health Care Center Kocevje, Health Care Center Litija; University Psychiatry Clinic Ljubljana.GR: Department of Pharmacy, University of Patras; Psychiatric Clinic, Cardiology Clinic and Oncology Clinic of the General University Hospital in Patras.
Another fundamental challenge lies in maximizing the accessibility of PGx results within and between different healthcare settings. Fragmented health IT infrastructures within countries and lacking interoperability, as encountered in this project, entail a significant risk of experiencing silo effects, meaning that information is trapped in one system or institution and sharing between different systems or institutions is impeded. Preemptive PGx testing is based on the rationale that testing patients for an entire panel of the most important PGx variants at once may be cost-effective, because they will likely profit several times from their PGx results in future interactions with the healthcare system.31 Avoiding silo effects by ensuring the accessibility and sharing of PGx results within and between different healthcare settings, e.g., in- and outpatient settings, and healthcare providers is therefore vital for a successful implementation of a preemptive PGx strategy.
Besides these technical requirements, the international setting of the project also poses unique challenges caused by the diversity of languages and regulatory frameworks.
Devising a multi-modal CDS implementation strategy
To overcome challenges associated with implementing PGx CDS on an international level and across multiple clinical sites with largely diverging IT infrastructures, the U-PGx consortium has devised a unique implementation strategy drawing from a spectrum of CDS delivery modes deployed inside and outside of EHRs that complement each other while still ensuring a standardized intervention by using a centralized knowledge base (see Figure 1).
Figure 1.
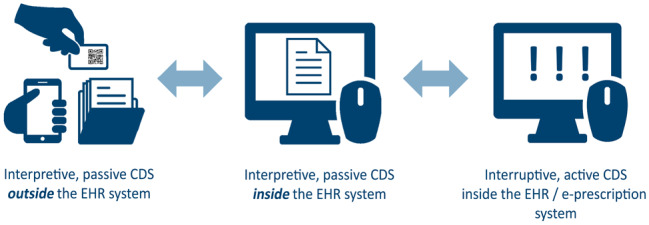
Decision support solutions in U-PGx. The U-PGx CDS strategy combines several complementary modes of delivering patient-specific PGx therapeutic recommendations to healthcare providers at the point of care, with or without integration into local EHRs. Active, interruptive CDS alerts clinicians of relevant gene-drug interactions via a pop-up message in the EHR or e-prescription system at the time of prescribing. Passive CDS is delivered either inside the EHR system as a digital report, or outside the EHR system via mobile- and paper-based solutions. The different decision support solutions deployed in the U-PGx project including the underlying knowledgebase are described in detail below.
U-PGx knowledge base
Technical realization
For U-PGx, the curation of the knowledge base and the automated translation of genetic data to associated phenotypes and recommendations are handled by the Genetic Information Management Suite (GIMS), a Drupal-based content management system developed and operated by the U-PGx partner bio.logis Genetic Information Management GmbH.32,33 Using a web-based content management system for knowledge base maintenance offers several advantages compared to using local or static solutions, such as a central workflow for editing, translating, reviewing, and validating content and a transparent change history across all participating sites (see Supplementary Material S2).
Content curation
The Dutch Pharmacogenetics Working Group (DPWG) is an ongoing effort to develop concise and clinically actionable recommendations for risk-phenotypes based on systematic literature review, and is formally associated with U-PGx. Up to the time of this writing, the DPWG has authored guidelines for 92 gene-drug pairs across 17 genes, all of which are incorporated into the G-Standaard, a comprehensive Dutch drug database, and regularly updated.29,34 This subset of the G-Standaard, containing the PGx-based therapeutic recommendations, including the data structure that links genotype-predicted phenotypes with active ingredients and therapeutic recommendations was adopted unchanged for the U-PGx knowledge base and is also available to interested parties via an open-source license (see Supplementary Material S3).
Chemical substances and active ingredients are identified by their Chemical Abstracts Service (CAS) number and the Anatomical Therapeutic Chemical (ATC) Classification System code within the knowledge base, respectively; a comprehensive systematic vocabulary such as Systematized Nomenclature of Medicine – Clinical Terms (SNOMED CT) is currently not utilized.35–37 An overview and further description of the knowledge base data model is provided in Supplementary Material S4.
Based on the genotype variants included in the U-PGx panel, rules for translating from genotypes to haplotypes and phenotypes were curated by PGx experts in the project.
DPWG guidelines for clinically actionable phenotype-drug pairs covered by the U-PGx genotyping panel were translated from Dutch to the local languages of each participating country (English, German, Greek, Slovenian, Spanish, and Italian) by certified translators and validated by consortium members. Furthermore, representatives from clinical implementation sites curated a list of the most common local trade names for all drugs covered by the project. Phenotype designations (e.g., “CYP2D6 ultrarapid metabolizer”) remained in English to preserve a standardized designation across all implementation sites. For the initial phase of the project, the phenotype and genotype terminology of the G-Standaard was adopted unchanged (see Supplementary Material S3); efforts to standardize and harmonize existing PGx terminologies and therapeutic recommendations developed by different working groups are currently underway.38,39
Decision support tools
U-PGx GIMS does not only act as a centralized knowledge base in the project but also serves as the main portal for the upload of genetic data obtained from the U-PGx genotyping platform, and the retrieval of patient-specific PGx reports in various formats. For this purpose, GIMS offers a wide range of secure data transfer capabilities, ranging from simple file (.csv) imports and uploads to modern web-based application programming interface technologies, including Hyper Text Transfer Protocol Secure (HTTPS)-based representation state transfer (RESTful) services and common standards like Health Level Seven International (HL7) and Fast Healthcare Interoperability Resources (FHIR).40,41
As illustrated in Figure 2, U-PGx GIMS enables PGx CDS by the following means: (1) the secure transfer of PGx test results and patient-specific dosing recommendations in a structured format for incorporation into local EHRs for use in passive or active CDS; (2) the generation of a PGx report in Portable Document Format or Open Document (ODT) format that can be filed either in the patient’s digital or paper-based health record; and (3) the generation of a “Safety-Code” card that enables mobile-based PGx CDS independent of existing IT infrastructures (see Supplementary Material S2).
Figure 2.
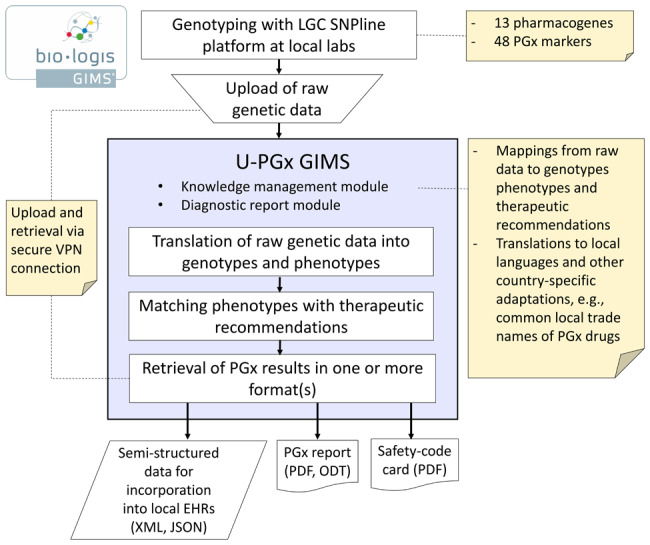
Retrieval of PGx results and dosing recommendations for patients in the U-PGx project.
To conform to privacy and data security regulations, all data that are exchanged between the implementation site and the centralized GIMS (i.e., PGx test results, PGx reports) are done with pseudonyms. Matching of PGx reports with identifying patient information occurs locally at each implementation site.
PGx report
Delivering PGx CDS for an entire panel of PGx variants in a paper-based form requires a careful report design to avoid overwhelming clinicians. The U-PGx report was therefore structured to provide information most relevant at the point of care – such as for which drugs a dosage adjustment is recommended for the respective patient – right at the beginning of the report, whereas additional information – such as the patients detailed PGx results – are provided on the following pages (see Supplementary Material S2).
The report is generated by the GIMS Diagnostic Report Module, which is certified as a medical device and holds the Conformité Européene (CE) mark in accordance with European legislation (EEC 93/42, EC 2007/47).
Safety-code card
To complement paper-based CDS solutions at clinical sites that lack an EHR infrastructure and to maximize the accessibility and sharing of PGx results within and between different healthcare settings and healthcare professionals, the “Safety-Code” card system is deployed at all participating institutions.42,43
This card is part of a mobile clinical decision support (CDSS) called the Medication Safety Code system which enables quick retrieval of patient-relevant PGx drug dosing guidelines even in the absence of a local EHR infrastructure.
Designed as a credit card-sized plastic card, the “Safety-Code” card contains a quick response (QR) code that can be decoded and interpreted by common smartphones and other devices (Figure 3). After scanning the QR code, the medical professional is led to a website that provides drug dosing recommendations customized to the PGx profile of the patient.
Figure 3.

Front and back side of an exemplary Safety-Code card for a fictional patient recruited in the U-PGx project
Furthermore, the “Safety-Code” card contains an overview of the patients’ most important PGx test results including a list of drugs for which PGx-based dosing adjustments are recommended. Patients participating in the PREemptive Pharmacogenomic testing for prevention of Adverse drug Reactions (PREPARE) study are asked to carry their “Safety-Code” cards with them and display them to medical professionals when pharmacotherapy is initiated or altered, which has the additional benefit of promoting patient engagement. Card contents are generated through GIMS (see Supplementary Material S2), and physical cards are printed locally at implementation sites with dedicated card printers.
The basic architecture of the Medication Safety Code system is publicly available via an open-source license (see Supplementary Material S3).
CDS implementation at clinical sites
Depending on their existing IT infrastructure and associated technical capabilities, each of the participating countries uses at least 2 complementary CDS methods for providing health care providers with patient-specific PGx-based therapeutic recommendations (Table 2). While main methods differ per country, all sites deploy the “Safety-Code” card as an adjunct method to optimize information diffusion within and between different healthcare providers.
Table 2.
Utilization of CDS tools in Each Participating Country
| Planned CDS intervention | NL | UK | I | E | A | SLO | GR |
|---|---|---|---|---|---|---|---|
| Automatic alerts via her | xx | ||||||
| Paper-based PGx reports | x | xx | xx | xx | |||
| Digital PGx reports via EHR | xx | xx | xx | x | |||
| Safety-Code card | x | x | x | x | x | x | x |
Each participating country uses one of the listed CDS interventions as their main method for providing physicians and pharmacists with patient-specific drug dosing recommendations, depending on their existing infrastructure and IT capabilities. Furthermore, each country deploys additional methods to facilitate the transfer of results between different healthcare settings (e.g., inpatient and outpatient setting). Main methods are marked with xx, adjunct methods are marked with x.
Lessons learned
While a final reflection and assessment of our implementation efforts can only be conducted after completion of the project, we nevertheless want to share the most important experiences collected over the course of this initial project phase.
Firstly, sufficient time should be ensured for the curation of the knowledge base, and in particular, the curation of the mapping between the raw data output of the genotyping platform. In addition, it is advisable to establish a workflow for dealing with rare variants that may not be covered by the knowledge base. For U-PGx, such variants are reported to a dedicated mailing list by implementation site representatives, reviewed by experts in the consortium, and added to the knowledge base.
Furthermore, the ability to quickly interpret and return partial genotyping results should be considered. At early implementation stages, some sites had problems with assays for 1 or 2 genes in the panel, and requested the generation of PGx reports based solely on the results of the remaining genes. This was not anticipated in the initial design of the reporting software for pre-emptive PGx.
Finally, in case an integration into local EHR infrastructure is envisaged, sufficient lead time should be scheduled for establishing communication and collaboration with the local IT department and dealing with often encountered bureaucratic obstacles, such as obtaining necessary permits. This observation resonates with the experiences reported earlier by Herr et al.30
DISCUSSION AND CONCLUSION
While automated alerts displayed via the EHR at the time of prescribing are commonly viewed as the gold standard for delivering CDS, their implementation is tied to the availability of an adequate technical infrastructure which is currently still insufficient in many healthcare settings.44,45 By developing and implementing a multi-modal CDS concept, we demonstrate the feasibility of implementing PGx CDS at multiple clinical sites across 7 countries in the presence of largely fragmented and diverse health care infrastructures. We use several complementary methods, including digital, paper-based, and mobile CDS solutions that allow each participating institution to choose their preferred combination of CDS tools that best fit their institutional preferences and technical requirements.
As of June 2017, all CDS tools had been finalized and rolled out in the countries that were randomized to start with the study arm, i.e., Greece, Slovenia, and Spain. Roll-out of CDS at sites that are currently recruiting patients for the control arm (i.e., UK, the Netherlands, Austria, and Italy) will be commenced in summer 2018. The adoption and usability of the different CDS tools deployed in the U-PGx project will be evaluated at several time points throughout the study period; intermediary results will be used for continuous improvement of the tools.
We hope that the project will successfully address the remaining barriers to widespread adoption of PGx-guided prescribing and preemptive testing strategies.
COMPETING INTERESTS
DS has developed concepts of genetic information management (GIM) that were realized by the company founded as, the bio.logis Genetic Information Management GmbH. For this company, she is the CEO. She is also medical director of bio.logis Center of Human Genetics, a diagnostic institution that is applying and testing the GIM-systems in the context of medical care. Enrico Just is the CEO of the bio.logis Genetic Information Management GmbH. RK is an employee of bio.logis Genetic Information Management GmbH. The other authors have no competing interests to declare.
CONTRIBUTORS
KB wrote major parts of the manuscript. JJS, HJG, and MS were involved in the initial conception of the U-PGx systems. KB, RK, HX, and MS were directly involved in software engineering of components of the U-PGx systems. All authors were involved in shaping some aspects of the U-PGx systems. All authors assisted in revising the manuscript and gave final approval of the version to be published.
FUNDING
This work was supported by European Community’s Horizon 2020 Programme grant number 668353 (U-PGx).
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
Supplementary Material
REFERENCES
- 1. Pirmohamed M, Burnside G, Eriksson N et al. . A randomized trial of genotype-guided dosing of warfarin. New Engl J Med 2013;369:2294–2303. [DOI] [PubMed] [Google Scholar]
- 2. Verhoef TI, Ragia G, de Boer A et al. . A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med 2013;369:2304–2312. [DOI] [PubMed] [Google Scholar]
- 3. Mallal S, Phillips E, Carosi G et al. . HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med 2008;358:568–579. [DOI] [PubMed] [Google Scholar]
- 4. Coenen MJH, de Jong DJ, van Marrewijk CJ et al. . Identification of patients with variants in TPMT and dose reduction reduces hematologic events during thiopurine treatment of inflammatory bowel disease. Gastroenterology 2015;149:907–17.e7. [DOI] [PubMed] [Google Scholar]
- 5. Swen JJ, Huizinga TW, Gelderblom H et al. . Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med 2007;4:e209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. O’Donnell PH, Bush A, Spitz J et al. . The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther 2012;92:446–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O’Donnell PH, Danahey K, Jacobs M et al. . Adoption of a clinical pharmacogenomics implementation program during outpatient care--initial results of the University of Chicago “1,200 Patients Project.” Am J Med Genet C Semin Med Genet 2014;166C:68–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gottesman O, Scott SA, Ellis SB et al. . The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clin Pharmacol Ther 2013;94:214–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Scott SA, Owusu Obeng A, Botton MR et al. . Institutional profile: translational pharmacogenomics at the Icahn School of Medicine at Mount Sinai. Pharmacogenomics 2017;18:1381–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Johnson JA, Elsey AR, Clare-Salzler MJ et al. . Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics 2013;14:723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weitzel KW, Alexander M, Bernhardt BA et al. . The IGNITE network: a model for genomic medicine implementation and research. BMC Med Genomics 2016;9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nelson DR, Conlon M, Baralt C et al. . University of Florida Clinical and Translational Science Institute: transformation and translation in personalized medicine. Clin Transl Sci 2011;4:400–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Weitzel KW, Elsey AR, Langaee TY et al. . Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 2014;166C:56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hicks JK, Stowe D, Willner MA et al. . Implementation of clinical pharmacogenomics within a large health system: from electronic health record decision support to consultation services. Pharmacotherapy 2016;36:940–948. [DOI] [PubMed] [Google Scholar]
- 15. Teng K, DiPiero J, Meese T et al. . Cleveland Clinic’s Center for personalized healthcare: setting the stage for value-based care. Pharmacogenomics 2014;15:587–591. [DOI] [PubMed] [Google Scholar]
- 16. Hoffman JM, Haidar CE, Wilkinson MR et al. . PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet C Semin Med Genet 2014;166C:45–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bell GC, Crews KR, Wilkinson MR et al. . Development and use of active clinical decision support for preemptive pharmacogenomics. J Am Med Inform Assoc 2013;21(e1):e93–e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pulley JM, Denny JC, Peterson JF et al. . Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT Project. Clin Pharmacol Ther 2012;92:87–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Peterson JF, Bowton E, Field JR et al. . Electronic health record design and implementation for pharmacogenomics: a local perspective. Genet Med 2013;15:833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lunenburg CATC, Henricks LM, Guchelaar H-J et al. . Prospective DPYD genotyping to reduce the risk of fluoropyrimidine-induced severe toxicity: ready for prime time. Eur J Cancer 2016;54:40–48. [DOI] [PubMed] [Google Scholar]
- 21. Lunenburg CA, van Staveren MC, Gelderblom H et al. . Evaluation of clinical implementation of prospective DPYD genotyping in 5-fluorouracil- or capecitabine-treated patients. Pharmacogenomics 2016;17:721–729. [DOI] [PubMed] [Google Scholar]
- 22. Bielinski SJ, Olson JE, Pathak J et al. . Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014;89:25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caraballo PJ, Hodge LS, Bielinski SJ et al. . Multidisciplinary model to implement pharmacogenomics at the point of care. Genet Med Published Online First: September 22, 2016, doi:10.1038/gim.2016.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Office of the National Coordinator for Health Information Technology. “Non-federal Acute Care Hospital Electronic Health Record Adoption,” Health IT Quick-Stat #47. /quickstats/pages/FIG-Hospital-EHR-Adoption.php. Accessed November 7, 2017. [Google Scholar]
- 25. Zelmer J, Ronchi E, Hyppönen H et al. . International health IT benchmarking: learning from cross-country comparisons. J Am Med Inform Assoc 2017;24:371–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kierkegaard P. eHealth in Denmark: a case study. J Med Syst 2013;37:9991. [DOI] [PubMed] [Google Scholar]
- 27. Kierkegaard P. Interoperability after deployment: persistent challenges and regional strategies in Denmark. Int J Qual Health Care 2015;27:147–153. [DOI] [PubMed] [Google Scholar]
- 28. Rathert C, Porter TH, Mittler JN et al. . Seven years after Meaningful Use: Physicians’ and nurses’ experiences with electronic health records. Health Care Manage Rev Published Online First: June 13, 2017, doi:10.1097/HMR.0000000000000168 [DOI] [PubMed] [Google Scholar]
- 29. van der Wouden CH, Cambon-Thomsen A, Cecchin E et al. . Implementing pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther 2017;101:341–358. [DOI] [PubMed] [Google Scholar]
- 30. Herr TM, Bielinski SJ, Bottinger E, et al. Practical considerations in genomic decision support: The eMERGE experience. J Pathol Inform. 2015;6:50. [DOI] [PMC free article] [PubMed]
- 31. Samwald M, Xu H, Blagec K et al. . Incidence of exposure of patients in the United States to multiple drugs for which pharmacogenomic guidelines are available. PLoS One 2016;11:e0164972.27764192 [Google Scholar]
- 32.bio.logis Genetic Information Management GmbH |. https://www.biologis.com/. Accessed July 31, 2017.
- 33.Drupal - Open Source CMS. Drupal.org USA: Drupal Association. 2016. https://www.drupal.org/. Accessed November 8, 2017.
- 34. G-Standaard — Z-Index https://www.z-index.nl/english. The Netherlands: Z-Index BV. Accessed April 19, 2017.
- 35. WHO Collaborating Centre for Drug Statistics Methodology, ATC Classification Index with DDDs Norway: WHO Collaborating Centre for Drug Statistics Methodology https://www.whocc.no/atc_ddd_index/. Accessed November 8, 2017.
- 36. Chemical Substances - CAS REGISTRY USA: Chemical Abstracts Service. http://www.cas.org/content/chemical-substances. Accessed November 8, 2017.
- 37. Heymans S, McKennirey M, Phillips J. Semantic validation of the use of SNOMED CT in HL7 clinical documents. J Biomed Semantics 2011;2:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Caudle KE, Dunnenberger HM, Freimuth RR et al. . Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med Published Online First: July 21, 2016, doi:10.1038/gim.2016.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bank P, Caudle K, Swen J et al. . Comparison of the Guidelines of the Clinical Pharmacogenetics Implementation Consortium and the Dutch Pharmacogenetics Working Group. Clin Pharmacol Ther Published Online First: October 10, 2017, doi:10.1002/cpt.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fielding RT, Taylor RN. Architectural Styles and the Design of Network-based Software Architectures. Doctoral dissertation: University of California, Irvine; 2000. [Google Scholar]
- 41. HL7 - Health Level Seven International http://www.hl7.org/. Accessed November 15, 2017.
- 42. Blagec K, Romagnoli KM, Boyce RD et al. . Examining perceptions of the usefulness and usability of a mobile-based system for pharmacogenomics clinical decision support: a mixed methods study. PeerJ 2016;4:e1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Medication Safety Code Initiative | Personalized Medicine in Your Pocket http://safety-code.org/. Accessed August 7, 2017.
- 44. Jha AK, DesRoches CM, Campbell EG et al. . Use of Electronic Health Records in U.S. Hospitals. New Engl J Med 2009;360:1628–1638. [DOI] [PubMed] [Google Scholar]
- 45. Kawamoto K. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. BMJ 2005;330:765 See https://www.ncbi.nlm.nih.gov/pmc/articles/PMC555881/. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


