Abstract
BACKGROUND
Early during human development, the trophoblast lineage differentiates to commence placentation. Where the placenta contacts the uterine decidua, extravillous trophoblast (EVT) cells differentiate and invade maternal tissues. EVT cells, identified by expression of HLA-G, invade into uterine blood vessels (endovascular EVT), as well as glands (endoglandular EVT), and open such luminal structures towards the intervillous space of the placenta. Endoglandular invasion diverts the contents of uterine glands to the intervillous space, while glands near the margin of the placenta that also contain endoglandular EVT cells open into the reproductive tract. Cells of the trophoblast lineage have thus been recovered from the uterine cavity and endocervical canal. An emerging non-invasive technology [trophoblast retrieval and isolation from the cervix (TRIC)] isolates and examines EVT cells residing in the cervix to explore their origin, biology and relationship to pregnancy and fetal status.
OBJECTIVE AND RATIONALE
This review explores the origins and possible uses of trophoblast cells obtained during ongoing pregnancies (weeks 5–20) by TRIC. We hypothesize that endoglandular EVT cells at the margins of the expanding placenta enter the uterine cavity and are carried together with uterine secretion products to the cervix where they can be retrieved from a Papanicolaou (Pap) smear. The advantages of TRIC for investigation of human placentation and prenatal testing will be considered. Evidence from the literature, and from archived in utero placental histological sections, is presented to support these hypotheses.
SEARCH METHODS
We used 52 out of 80 publications that appeared between 1966 and 2017 and were found by searching the PubMed and Google Scholar databases. The studies described trophoblast invasion of uterine vessels and glands, as well as trophoblast cells residing in the reproductive tract. This was supplemented with literature on human placental health and disease.
OUTCOMES
The literature describes a variety of invasive routes taken by EVT cells at the fetal–maternal interface that could displace them into the reproductive tract. Since the 1970s, investigators have attempted to recover trophoblast cells from the uterus or cervix for prenatal diagnostics. Trophoblast cells from Pap smears obtained at 5–20 weeks of gestation have been purified (>95% β-hCG positive) by immunomagnetic isolation with nanoparticles linked to anti-HLA-G (TRIC). The isolated cells contain the fetal genome, and have an EVT-like expression profile. Similar EVT-like cells appear in the lumen of uterine glands and can be observed entering the uterine cavity along the margins of the placenta, suggesting that they are the primary source of cervical trophoblast cells. Cells isolated by TRIC can be used to accurately genotype the embryo/fetus by targeted next-generation sequencing. Biomarker protein expression quantified in cervical trophoblast cells after TRIC correlates with subsequent pregnancy loss, pre-eclampsia and fetal growth restriction. A key remaining question is the degree to which EVT cells in the cervix might differ from those in the basal plate and placental bed.
WIDER IMPLICATIONS
TRIC could one day provide a method of risk assessment for maternal and fetal disease, and reveal molecular pathways disrupted during the first trimester in EVT cells associated with placental maldevelopment. As perinatal interventions emerge for pregnancy disorders and inherited congenital disorders, TRIC could provide a key diagnostic tool for personalized precision medicine in obstetrics.
Keywords: trophoblast, pregnancy, prenatal diagnosis, female reproductive tract, genetic diagnosis, endometrium, immunohistochemistry, placenta, obstetric disorders
Introduction
Pregnancy in humans, while most often producing a favorable outcome, is subject to significant morbidity and mortality to the mother, fetus and resulting child characterized by multiple congenital anomalies and clinical syndromes (Di Renzo, 2009; Romero, 2009). Because the fetus and placenta are nearly inaccessible during pregnancy, predicting adverse outcomes is challenging without the use of invasive approaches such as amniocentesis or chorionic villous sampling (CVS). Biomarkers in the maternal circulation have been explored for screening in the first trimester, but so far are only adequately reliable to be considered diagnostic for a small proportion of pregnancy pathologies. Identification of at-risk pregnancies early in gestation offers the best opportunities to discover etiologies and develop corrective interventions. A position paper by senior staff of the National Institute of Child Health and Human Development in the USA points out that the knowledge barrier to understanding the mechanisms associated with feto-placental disorders ‘is primarily due to limitations in current non-invasive “interrogation” of the placenta’ (Guttmacher et al., 2014).
For years, circulating protein biomarkers combined with ultrasonic monitoring have been used to detect neural tube defects and chromosomal abnormalities, as well as for determining risk for pregnancy disorders such as early-onset pre-eclampsia and other placental maldevelopment syndromes (SMFM, 2016). Various combinations of molecules accumulate in maternal blood that are indicative of placental maldevelopment-associated pathologies, including pregnancy-associated plasma protein A (PAPP-A), placental protein 13 (LGALS13 or PP13), inhibin, hCG, placental growth factor (PlGF or PGF) and soluble fms-like tyrosine kinase-1 (FLT1) (Wright et al., 2017). While these biomarkers have been used in combination with sonographic and Doppler velocimetry methods in the first and second trimesters, there are significant limitations in terms of their clinical utility based on their low sensitivity and positive predictive values for detection of small for gestational age newborns (ACOG, 2015; McCowan et al., 2017; Parry et al., 2017).
A promising approach for interrogating the placenta that is the subject of this review utilizes trophoblast cells that migrate from the placenta into the reproductive tract (Imudia et al., 2010). It is possible to recover hundreds of placental cells safely and non-invasively from the endocervical canal using a Papanicolaou (Pap) procedure (Canestrini and Testa, 1978; Fejgin et al., 2001; Cioni et al., 2005; Imudia et al., 2009), and then separate them from maternal cells for downstream analysis of fetal DNA, RNA, proteins and other informative molecules (Drewlo and Armant, 2017).
Methods
The PubMed and Google Scholar databases were comprehensively searched for relevant publications describing the migration of trophoblast cells into endometrial glands and the uterine lumen, as well as investigations of trophoblast cells residing in the reproductive tract and cervix. The publications identified between 1966 and 2017 included those published on cervical trophoblast from the time fetal cells were first found in Pap smears, as well as those exploring subsets of trophoblast in endometrial arterioles, venules and glands. Publications were identified by searching with combinations of the keywords, human, cervix, endocervical canal, cervical mucus, trophoblast, fetal cells, pregnancy, transcervical sampling, retrieval, isolation, DNA, genome, extravillous, invasion, glands, arteries, veins. Eighty publications were reviewed and 52 were selected that reported significant findings on the topic of trophoblast cells residing in the uterine cavity and cervix. An additional 69 references with keywords related to human placental health and disease were integrated into the review to expand on the topic and provide pertinent background information.
Trophoblast cells in maternal blood and the reproductive tract
The prospect of prenatal diagnosis with less risk than amniocentesis or CVS has been sought since it came to light that fetal cells are present in accessible locations, including maternal blood and the reproductive tract. The presence of Y chromosome-containing fetal cells deported into the maternal circulation has been recognized since the early 1980s (Iverson et al., 1981), and in the reproductive tract since the 1970s (Shettles, 1971).
Non-invasive prenatal testing using maternal blood samples
While successes were reported with isolation of fetal trophoblast and hematopoietic cells from the maternal circulation, their numbers were challenging (on the order of 1 per million maternal cells), and the field shifted toward the exploitation of cell-free fetal DNA (Norwitz and Levy, 2013; Wou et al., 2015). Non-invasive prenatal testing (NIPT), using cell-free fetal DNA in maternal blood, can screen for the most common chromosome number disorders (13, 18, 21, X, Y) beginning around 10 weeks of pregnancy (Bianchi et al., 2014). Cell-free fetal DNA is a slight misnomer, as it is derived from turnover of tissue in the placenta, rather than the fetus. The fetal, or placental, fraction must exceed 4% of the total cell-free DNA in maternal plasma to be reliably sequenced and interpreted (Wong and Lo, 2016). Although detection of trisomies 13, 18 and 21 has been impressive (98–99%) with false positive rates of ~0.13% (Gil et al., 2017), NIPT has not been accepted for detection of single gene mutations. Moreover, trisomies observed through NIPT screening should be confirmed by invasive diagnostic testing (Benn et al., 2015; Gregg et al., 2016). It has been reported that NIPT has high sensitivity and specificity for detection of paternally contributed genetic traits, such as sex and Rhesus D antigen in D-negative women (Devaney et al., 2011; Johnson et al., 2017), and the possibility exists for its eventual use as a diagnostic tool (Drury et al., 2016).
Trophoblast cells in the female reproductive tract and cervical canal
Subsequent to the initial observation of Shettles (1971), a number of investigators attempted to replicate and extend the transcervical approach for obtaining intact placental cells, not only in cervical smears or aspirated mucus, but also through more invasive endocervical and intrauterine lavage (Adinolfi and Sherlock, 1997, 2001; Bischoff and Simpson, 2006; Evans and Kilpatrick, 2010; Imudia et al., 2010; Wou et al., 2015). Early attempts during the 1970s to repeat Shettles’ findings were met with both success (Rhine et al., 1975) and failure (Bobrow and Lewis, 1971; Manuel et al., 1974; Amankwah and Bond, 1978; Goldberg et al., 1980). The reliability of Y-body fluorescence produced with quinacrine mustard to detect male cells, as used by Shettles, was the principal impediment to acceptance of the approach and further advancement at that time.
Attention turned away from exploitation of trophoblast cells in the reproductive tract for a decade, but then resumed to produce some important advances that are outlined in Table I. With the development of fluorescence in situ hybridization (FISH) and PCR, and the realization that adequate numbers of fetal cells for prenatal testing were unlikely to be available in maternal blood, interest returned during the 1990s to trophoblast cells residing in the uterus and endocervical canal (Griffith-Jones et al., 1992; Pertl et al., 1994; Ville et al., 1994; Briggs et al., 1995; Adinolfi et al., 1995c; Fung et al., 1995; Ishai et al., 1995; Kingdom et al., 1995; Rodeck et al., 1995). Using FISH and PCR, success in determining fetal sex from transcervical specimens was variable, but encouraging. An approach using X22 heterozygosity to distinguish the maternal genome from a female fetal genome was reported (Adinolfi and Cirigliano, 2000). Trophoblast protein markers and morphological assessment also identified trophoblast cell populations within specimens collected transcervically (Chaouat et al., 1994; Bahado-Singh et al., 1995; Bulmer et al., 1995). Early successes demonstrated the potential of transcervical sampling for clinical diagnosis including the detection of fetal trisomy 18 by FISH (Adinolfi et al., 1993), fetal rhesus blood group D antigen gene in rhesus negative patients (Adinolfi et al., 1995a), fetal chromosome 21 using both FISH and chromosome-specific short tandem repeat (STR) amplification (Adinolfi et al., 1995b), fetal chromosome number variants by FISH and PCR (Massari et al., 1996; Sherlock et al., 1997; Xi Zhao et al., 2003; Bussani et al., 2007), fetal hemoglobin genotypes responsible for sickle cell disease and thalassemia (Adinolfi et al., 1997; Cirigliano et al., 1999), and fetal triple X (Xi Zhao et al., 2003).
Table I.
Key findings, from 1971 to 2016, using transcervical retrieval of trophoblast cells.
| Advancements (chronological) | Retrieval method | Trophoblast detection method | References |
|---|---|---|---|
| Trophoblast cells found in cervix | Cervical swab smeared on slide | Y-body stain with quinacrine mustard | Shettles (1971) |
| Confirmation of cervical trophoblast cells | Cervical swabs smeared on slide | PCR for Y-derived sequences | Griffith-Jones et al. (1992) |
| Identification of syncytial trophoblast fragments and protein markers | Uterine lavage | Histology and immunocytochemistry | Griffith-Jones et al. (1992); Chaouat et al. (1994) |
| Detection of fetal trisomy | Uterine lavage | FISH for chromosomes 18 and Y | Adinolfi et al. (1993) |
| Fetal RHD detected in Rh negative women | Cervical mucus aspiration and uterine lavage | PCR | Adinolfi et al. (1995a) |
| Paternal STR allele detection | Endocervical lavage and mucus aspiration | PCR-seq to detect STRs | Adinolfi et al. (1995b); Kingdom et al. (1995) |
| Trisomy 21 detected by STR analysis | Uterine lavage, mucus aspiration and cytobrush | PCR-seq for STRs | Adinolfi et al. (1995c) |
| Detection of fetal SMA and myotonic dystrophy | Endocervical lavage | PCR-seq for STRs | Massari et al. (1996) |
| Trophoblast enrichment by morphology | Endocervical collection and micromanipulation based on morphology | FISH and PCR for Y chromosome | Sherlock et al. (1997) |
| Detection of fetal hemoglobinopathies | Cervical mucus aspiration and micromanipulation based on morphology | PCR-seq for STRs and hemoglobin mutations | Adinolfi et al. (1997); Cirigliano et al. (1999) |
| Identification of female trophoblast cells | Cervical mucus aspiration | PCR-seq for X22 | Adinolfi and Cirigliano (2000) |
| Reliable (97%) cervical trophoblasts recovery | Endocervical collection by cytobrush | FISH | Fejgin et al. (2001) |
| LCM isolation of trophoblasts | Uterine lavage, labeled with anti-HLA-G for LCM | PCR-seq for X, Y and paternal STRs | Bulmer et al. (2003) |
| Trophoblast enrichment with marker antigens | Immunofluorescence labeling and isolation by micromanipulation | PCR-seq for STRs | Katz-Jaffe et al. (2005); Mantzaris et al. (2005) |
| Enrichment of trophoblast based on morphology (152/181) | Uterine lavage and micromanipulation | PCR-seq for STRs | Bussani et al. (2007) |
| Reduced numbers of cervical trophoblast in pathological pregnancies | Endocervical collection by cytobrush | Immunocytochemistry for HLA-G | Imudia et al. (2009) |
| Isolation of >900 EVT cells to >95% purity from Pap specimens by TRIC | Endocervical collection by cytobrush | Immunomagnetic isolation of trophoblast (EVT) with anti-HLA-G | Bolnick et al. (2014) |
| EVT isolation by TRIC is unaltered by gestational age at 5–20 weeks or BMI | Endocervical collection by cytobrush and TRIC | Immunocytochemistry for HLA-G | Fritz et al. (2015b) |
| Trophoblast isolation by size and LCM | External cervical collection by cytobrush and ISET/LCM | Single cell STR genotyping | Pfeifer et al. (2016) |
| EVT protein profiles reflect pregnancy outcomes | Endocervical collection by cytobrush and TRIC | Immunocytochemistry for biomarkers of placental insufficiency | Fritz et al. (2015a); Bolnick et al. (2016b) |
| DNA profiling of EVT by NGS | Endocervical collection by cytobrush and TRIC | Fetal DNA isolation and targeted NGS for SNPs and STRs | Jain et al. (2016) |
STR, short tandem repeats; PCR-seq, Sanger sequencing of fluorescence-labeled PCR products; SMA, spinal muscular atrophy; LCM, laser capture microdissection; EVT, extravillous trophoblast; TRIC, trophoblast retrieval and isolation from the cervix; ISET, isolation by size of epithelial tumor; NGS, next-generation sequencing; SNP, single nucleotide polymorphism; RHD, rhesus blood group D antigen.
Genetic interrogation of trophoblast cells obtained transcervically relied principally upon morphological identification and micromanipulation or the use of trophoblast-specific markers (Adinolfi and Sherlock, 2001), limiting its usefulness in a clinical setting. Efforts to develop antibodies or antibody panels to distinguish trophoblast cells in transcervical specimens were undertaken during this period (Chaouat et al., 1994; Bulmer et al., 1995). Laser capture microdissection after identification of trophoblast cells by immunostaining with antibodies to selective lineage markers successfully provided fetal DNA for PCR genotyping (Bulmer et al., 2003; Mantzaris and Cram, 2015). Some success in DNA allelic profiling by single-cell approaches has been reported (Katz-Jaffe et al., 2005; Pfeifer et al., 2016), but these advances have not led to a consistent and efficient procedure that could be translated into clinical practice. Success rates for obtaining trophoblast cells remained variable and largely unacceptable for prenatal testing regardless of the invasiveness of the method used to obtain specimens (ErgIn et al., 2001; Fejgin et al., 2001; Cioni et al., 2003; Mantzaris et al., 2005; Bussani et al., 2007; Mantzaris and Cram, 2015). However, it was also clear from these investigations that during the first trimester trophoblast cells indeed reside freely in the maternal reproductive tract despite its structural separation from the chorionic villi within the chorion laeve and decidua capsularis.
Routes for trophoblast migration into the reproductive tract
If it is hypothesized that trophoblast cells can be retrieved from the reproductive tract during early pregnancy, a rationale is needed for their displacement from the placenta. At the tips of anchoring villi, cytotrophoblast cell columns attach to the maternal decidua and differentiating EVT cells invade the maternal tissues. Thereafter, they follow the interstitial route of invasion. Further routes of EVT invasion based on interstitial trophoblast cells include the uterine blood vessels (endovascular EVT subdivided into endoarterial EVT in uterine arteries and endovenous EVT in uterine veins) and uterine glands (endoglandular EVT) (Moser and Huppertz, 2017). The endovascular and endoglandular EVT activities are responsible for the tissue remodeling that establishes the hemotrophic and histiotrophic nutrition of the fetus and embryo, respectively (Benirschke et al., 2012a; Moser et al., 2015). Within the trophoblast cell column, cells express HLA-G as they differentiate from villous to extravillous, which parallels the physiological change from proliferative (HLA-G negative) to invasive (HLA-G positive) properties, and correlates with a switch in the expression of integrin subunits that mobilizes the cells (Merviel et al., 2001; Benirschke et al., 2012b). HLA-G expressing EVT cells can be visualized in the decidua basalis within the stroma, within vessels and within uterine glands, and occasionally in the uterine cavity (Fig. 1). HLA-G is a reliable phenotypic marker of differentiated EVT (McMaster et al., 1995; Apps et al., 2008; Moser et al., 2011), and is also expressed by trophoblast cells residing in the endocervical canal (Imudia et al., 2009).
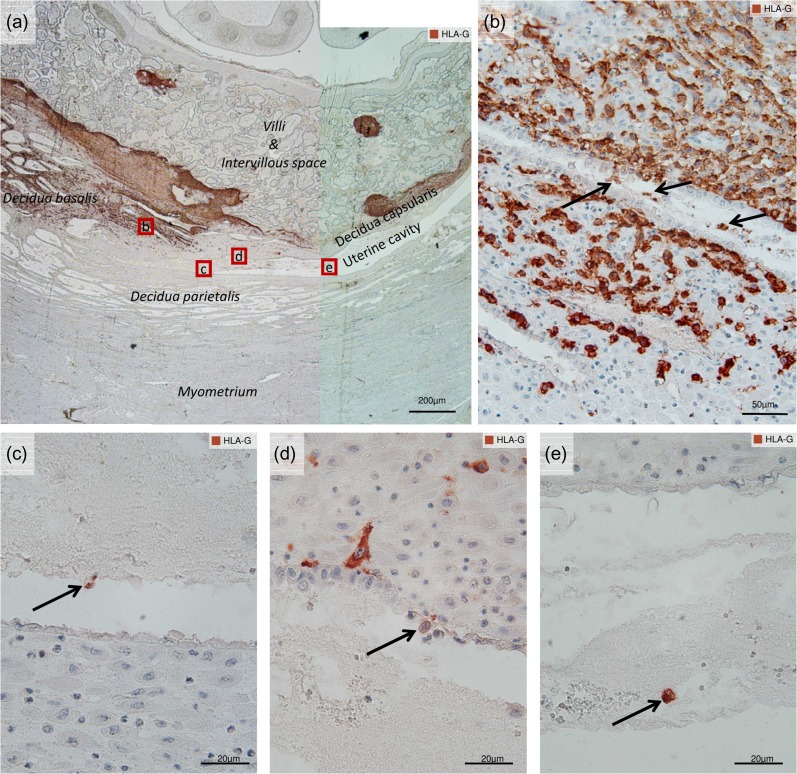
Figure 1.
Extravillous trophoblast cells in the human uterine cavity. Immunocytochemical staining in utero with an antibody against HLA-G (and Hemalaun nuclear counterstain) in paraffin sections of an archived placenta (most likely early first trimester). The dark brown labeling of HLA-G serves as a marker for extravillous trophoblast (EVT) cells in the invasive zone between fetal and maternal regions. (a) An overview at the margin of the placenta showing villi and intervillous space, decidua basalis, decidua parietalis, decidua capsularis and the uterine cavity, as labeled. Details of the red insets in (a) follow: (b) demonstrates endoglandular EVTs (arrows) in the lumen of a gland near the edge of the placenta. (c) Shows an HLA-G positive EVT cell (arrow) located in the uterine cavity. (d) Shows an EVT cell (arrow) that has replaced the uterine epithelium, while others nearby approach the epithelium. (e) Shows another EVT cell located in the uterine cavity, possibly surrounded by glandular secretions.
During placental development, there are two possible routes that EVT cells could take to reach the uterine cavity and, subsequently, the cervix. First, interstitial EVT cells expressing HLA-G penetrate the uterine epithelium near the margin of the placenta, replace the uterine epithelium from the basal side, and enter the uterine cavity (Fig. 2). Second, at the edge of the developing placenta, in the transitional zone of decidua basalis and decidua parietalis, endoglandular EVT cells invade uterine glands from the basal side, replace the glandular epithelium, and occupy the glandular lumen (Moser et al., 2010, 2015). Eroded uterine glands can be visualized (Fig. 3a and b) especially at the margin of the developing placenta (Moser and Huppertz, 2017). The developing placenta rapidly expands laterally (Craven et al., 2000; Nanaev et al., 2000), continuously exposing new glands to EVT invasion at the margin of the placenta. These invaded glands can secrete further laterally into the uterine cavity (Fig. 2), and thus could expel EVT cells together with the glandular secretion products into the uterine cavity. After they reach the uterine cavity, EVT cells could be carried together with the secretion products toward the cervix, as illustrated in Fig. 4.
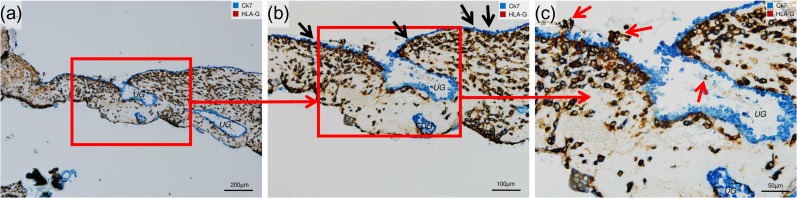
Figure 2.
EVT cells replace the uterine epithelium. Immunocytochemical double staining of invaded decidua (7 weeks gestational age) for cytokeratin 7 (Ck7, blue, serves as marker for glandular and uterine epithelium) and HLA-G (dark brown, serves as marker for EVT). No nuclear counterstain. (a) Overview shows the transitional zone between decidua capsularis (to the left) and decidua basalis (to the right) with uterine cavity above and intervillous space below. The decidua basalis includes prominent uterine glands (UG) with blue-labeled epithelia. (b) Inset shown in (a). Black arrows indicate the uterine epithelium. (c) Inset shown in (b). Higher magnification shows EVT cells (red arrows) breaking through the uterine epithelium at the opening of the UG, and an endoglandular EVT cell in the lumen of the gland.
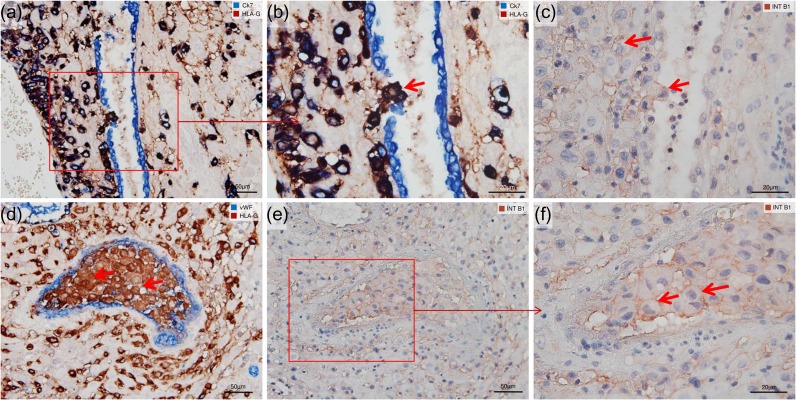
Figure 3.
Endoglandular and endovascular trophoblast cells express integrin ITGB1. Immunocytochemical single and double staining of serial sections of invaded decidua (11 weeks gestational age) for integrin subunit ITGB1 (INT B1, brown), CK7 (blue, serves as marker for glandular and uterine epithelium), HLA-G (dark brown, serves as marker for EVT), and von Willebrand Factor (vWF, blue, serves as marker for vascular endothelium). Colors of the labels are indicated in each panel. No nuclear counterstain in (a, b, d). Nuclei were counterstained with Hemalaun in (c, e, f). (a) Overview showing a UG partly surrounded by EVT, while in (b) a higher magnification of inset shown in (a) clearly demonstrates endoglandular EVT cells (red arrow) breaking through the glandular epithelium towards the glandular lumen. (c) Adjacent section to (b) labeled for ITGB1. Like the interstitial EVT, the endoglandular EVT (red arrows) express ITGB1 on their cell surface. (d) Shows endovascular EVT cells within a trophoblast plug (red arrows) of a uterine blood vessel lined with vWF-positive endothelia. (e) Shows the same trophoblast plug from an adjacent serial section stained for ITGB1. A higher magnification of the inset, shown in (f), reveals that endovascular EVT cells (red arrows) express ITGB1 on their cell surfaces.
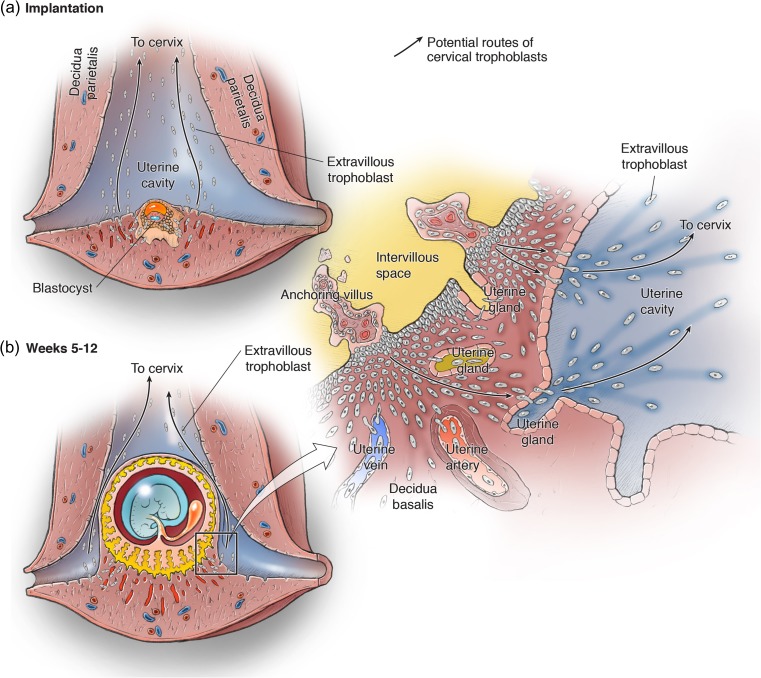
Figure 4.
Origin of cervical trophoblast cells during placental development. The developing conceptus is shown within the uterus at the implantation site (a) and later during the placentation period of weeks 5–12 of gestation (b). EVT cells originate from trophoblast cell columns at the base of the anchoring villi, and follow the interstitial, endovascular, and endoglandular invasion routes. The inset in (b) is expanded to the right, showing the transitional zone of the decidua basalis at the margin of the placenta. As demonstrated in Figs 1 and 2, interstitial EVT cells can invade and replace the uterine epithelium from the basal side, and then enter the uterine cavity. In the decidua basalis, endoglandular EVT cells invade and reach the lumen of UGs. We speculate that, at the lateral margin of the placenta, they are transported together with the glandular secretions into the uterine cavity. Once EVT cells have reached the uterine cavity, they could migrate towards the cervix, or be carried there by the uterine secretion products (arrows).
Interstitial, as well as endovascular and endoglandular, EVT cells react with antibodies against the pro-invasive proteins matrix metalloproteinase 1 (MMP1) (Weiss et al., 2016) and integrin β1 (ITGB1) (Kemp et al., 2002), which is shown in Fig. 3c–f. The expression of these molecules points to the invasive capacity, and thus to a common (extravillous) origin of the cells. The migratory potential of the EVT was established decades ago (Billington, 1966), supporting the hypothesis that EVT cells migrate from the uterine cavity toward the cervix. The developing placenta expands rapidly, hence, there could be a continuous supply of EVT cells entering the endocervical canal.
As the placenta expands to fill the uterine cavity, it would be expected that migration of EVT cells into the cervix would cease. Accordingly, it is observed that endocervical EVT cells are recovered in Pap specimens only until 20 weeks of gestational age (GA), after which cell recoveries rapidly plummet in the authors’ (D.R.A. and S.D.) experience (Fritz et al., 2015b). Whether trophoblast cells not expressing HLA-G remain longer in the cervix is unclear.
As indicated by the literature cited in Table I, there are numerous reports of syncytial trophoblast, and even villous fragments, recovered through transcervical collections. It has been suggested that ulceration of the decidua capsularis might permit release of these elements into the uterine cavity (Adinolfi and Sherlock, 2001). Direct evidence for this route is limited, and the inconsistencies reported for retrieval of villous trophoblast elements after uterine or endocervical lavage (Bahado-Singh et al., 1995; Miller et al., 1999; Fang et al., 2005; Bussani et al., 2007) suggest that it might not be a natural route. Further investigation is needed to resolve this issue.
Trophoblast Retrieval and Isolation From the Cervix
Technical issues of TRIC
Although endocervical specimens from pregnant women could provide useful information to clinicians, the approach initially did not prove robust, due fully or in large part to the excess of maternal cells from which trophoblast cells must be distinguished for efficient and accurate analysis (Imudia et al., 2010). Trophoblast retrieval and isolation from the cervix (TRIC) was developed as a solution to the technical challenge of analyzing placental cells in real time, leveraging the unique expression of HLA-G on the surface of EVT cells, exclusive of adult tissues in the reproductive tract (Loke et al., 1997; McMaster et al., 1998), to obtain a homogeneous population of fetal cells with an EVT-like phenotype (Bolnick et al., 2014). The TRIC procedure separates trophoblast cells from maternal cells using immunomagnetic nanoparticles, to provide placental cells for downstream molecular analyses. The information that TRIC provides could complement data obtained using current NIPT procedures that are based on biophysical and biochemical measurements.
Retrieval of trophoblast cells from the endometrial canal using a cytobrush can be considered minimally invasive. The office procedure is essentially a Pap smear, and can be performed successfully between 5 and 20 weeks GA (Fritz et al., 2015b). Pap smears are recommended during pregnancy, and several studies that surveyed approximately 1900 pregnant women found the cytobrush procedure to be safe and associated with no serious adverse outcomes (Orr et al., 1992; Rivlin et al., 1993; Paraiso et al., 1994; Holt et al., 2005). Our (S.D. and D.R.A.) experience with over 1000 endocervical samples collected from ongoing pregnancies has revealed no increase above baseline frequencies in pregnancy loss, excessive bleeding or infection (manuscript in preparation).
Analytical issues of TRIC
The isolation of EVT cells by TRIC is enhanced with inclusion of adequate quality controls. Ideally, endocervical specimens will contain 1 000 000 or more cells, determined in a simple cell count, although specimens with as few as 50 000 cells can provide highly purified trophoblast cells. However, yield will be affected by the initial cell number, since there are approximately 2000 maternal cells for every trophoblast cell (Imudia et al., 2009). An aliquot containing 5–10% of the endocervical specimen should be examined by immunocytochemical labeling with anti-HLA-G to provide an estimate of the number of EVT cells that can be expected in the final isolate (Bolnick et al., 2014). Trophoblast cells are also distinct from the maternal cells in their expression of β-hCG, which can be used to determine the purity of the final isolate. Typically, 95–99% of the cells isolated by TRIC are β-hCG positive, but some samples range lower, down to 75–85% (Bolnick et al., 2014; Fritz et al., 2015b). Trophoblast cells obtained by TRIC, when examined by FISH or single-cell PCR for the X and Y chromosomes, consistently demonstrate either XY or XX signals, in agreement with the sex of the babies delivered. Moreover, cells isolated from male specimens are seldom XX, verifying the fetal origin and purity of cells obtained by TRIC (Bolnick et al., 2014). Conversely, the cells excluded during immunomagnetic isolation are uniformly female and β-hCG negative, as expected of the maternal cells in the cervix.
Specificity of TRIC
Cells obtained by TRIC have been profiled by immunocytochemistry to characterize their lineage and phenotype. The trophoblast marker proteins cytokeratin 7 and placental lactogen (CSH1) are uniformly expressed in the isolated cell fraction, in addition to β-hCG (Bolnick et al., 2014). Because TRIC employs affinity to HLA-G, a protein associated with EVT differentiation, selective isolation of that trophoblast population would be expected. As expected, the isolated cells express several EVT markers (integrin subunit α1 [ITGA1], cadherin 5, platelet and endothelial cell adhesion molecule 1, MMP9, HLA-G), and lack villous trophoblast markers (ITGA6, cadherin 1, pregnancy specific beta-1-glycoprotein 1). These protein expression profiles are consistent with EVT cells present in the basal plate and lumen of the uterus during pregnancy (Moser et al., 2010, 2015), as well as observations during EVT differentiation in vitro and in vivo (Damsky et al., 1994; Zhou et al., 1997). The use of anti-HLA-G appears to narrowly target TRIC for capture of cells with an EVT phenotype. Therefore, it remains feasible that other placental cells with a different lineage or phenotype also populate endocervical specimens, but are not captured by TRIC.
Pregnancy pathologies and TRIC
For TRIC to be a truly robust approach to investigate fetal genetics, human placentation and associated pathologies, it is critical that adequate numbers of cells are available from all pregnancies regardless of their GA or the presence of pathology. It has been suggested that placental HLA-G expression is reduced by pre-eclampsia (Yie et al., 2004; Zhu et al., 2012), which could compromise the recovery of EVT cells by TRIC: these studies found reduced expression with GA, and examined the effect of pre-eclampsia at term. HLA-G protein levels were significantly reduced in placentas of women with pre-eclampsia, but maintained about half the expression level of healthy pregnancies (Zhu et al., 2012). Because HLA-G is expressed with the onset of EVT differentiation (McMaster et al., 1995), fewer EVT cells are present in fetal membrane or decidua of pre-eclamptic than normal pregnancies, but equivalent amounts of HLA-G mRNA are found when normalized to trophoblast content (Colbern et al., 1994). Cytotrophoblast cells isolated from second trimester placentas that are induced to differentiate to EVT on Matrigel upregulate HLA-G mRNA and protein, but levels are significantly reduced in cultures of cells from pre-eclamptic pregnancies (Lim et al., 1997). While disease might reduce HLA-G expression by EVT cells, there could remain sufficient amounts for immunomagnetic isolation by TRIC. A significant 4–5-fold reduction in the relative number of HLA-G positive cells present in endocervical specimens was observed in women with an ectopic or blighted ovum pregnancy (Imudia et al., 2009). However, a survey of 224 ongoing pregnancies demonstrated that the number of cells obtained by TRIC is reduced by ~50% (P < 0.05) prior to an early pregnancy loss, but provides sufficient numbers (hundreds) of EVT cells for most downstream analyses (Fritz et al., 2015b). The Fritz et al. (2015b) study further revealed no significant reduction of EVT cells isolated by TRIC in women who later developed pre-eclampsia or fetal growth restriction (FGR), and regression analysis showed no change in the number of EVT cells in specimens from women with an elevated BMI. TRIC is available 4–5 weeks earlier than other modalities for prenatal testing, at 5 weeks GA, and cell yield is unaffected by GA until 20 weeks, when it abruptly declines (Fritz et al., 2015b). Thus, TRIC can be useful for investigation of early placentation and prenatal testing during a period of development when there is currently a significant gap in knowledge.
Utility of TRIC for Placental Research
The availability of EVT cells during ongoing pregnancies as a result of the TRIC methodology is unique and offers a powerful research tool that not only enables the use of cellular and molecular evaluation in the first trimester, but also provides information within the context of subsequent pregnancy outcomes. The literature provides strong support for the hypothesis that EVT cells are required for a normal term pregnancy by facilitating key physiological changes during placentation.
Trophoblast development and routes of invasion
The trophoblast lineage initially emerges during preimplantation development, and produces the trophectoderm of the blastocyst (Wiley, 1988; Collins and Fleming, 1995). As the first epithelium to form during human development, trophoblast cells retain expression of cytokeratin 7 even after differentiation to a non-epithelial EVT phenotype (Gauster et al., 2013). Trophoblast cells in the blastocyst initially differentiate by fusion to produce a syncytiotrophoblast during implantation, which can remove the uterine epithelium and invade the stroma (Fig. 4a). Later, the chorionic villi form (Fig. 4b) and the villous trophoblast generate a stratified epithelium composed of mononuclear cytotrophoblast underlying a syncytiotrophoblast that directly contacts maternal fluids in the intervillous space. At the basal plate (Fig. 4b, expanded view of inset), proliferating cytotrophoblast cells at the bases of anchoring villi generate columns where they contact the decidua. Distally, the cytotrophoblast cells differentiate into EVT cells that invade the decidua and advance into uterine blood vessels and glands. Endovascular EVT cells invade and open the uterine veins (He et al., 2017; Moser and Huppertz, 2017; Moser et al., 2017; Windsperger et al., 2017), as well as remodel the spiral arteries by disrupting the smooth muscle and tunica media, replacing the endothelia, and expressing endothelial proteins (Zhou et al., 1997). The remodeled arteries form wide, low-resistance conduits of maternal blood that can more efficiently perfuse the chorionic villi to support the growing conceptus.
Trophoblast invasion in pregnancy pathologies
Incomplete or failed remodeling of the spiral arteries is hypothesized to be associated with dysregulated placental perfusion, in which perfusion of the placenta might be irregular, too fast or inadequate (Chaddha et al., 2004; Burton et al., 2009). When severe, dysregulated placental perfusion may even cause an early pregnancy loss. Pregnancies that endure could have an inadequately perfused placenta, and the associated pathology of FGR, or the hypertensive disorder pre-eclampsia (Burton and Jauniaux, 2004). There is also evidence that preterm labor can be associated with dysregulated placental perfusion (Papageorghiou et al., 2004; Hossain and Paidas, 2007). The incidence of early pregnancy loss approaches 15% (Larsen et al., 2013). FGR occurs in 4–8% of all pregnancies in developed countries, often defined as below the fifth percentile of newborn weight (Savchev et al., 2014). Pre-eclampsia has a frequency of ~5–7%, while the early onset form occurs at 0.5–1%. Pre-eclampsia is clinically defined by the presence of maternal hypertension after 20 weeks of pregnancy, occurring in a previously normotensive patient, and is mostly accompanied by proteinuria (Davey and MacGillivray, 1988; Tranquilli, 2013). Maternal death or serious long-term morbidity can occur, while abruption, preterm delivery and FGR contribute to perinatal fetal death and infant morbidity, with the associated costs of extensive neonatal hospitalization, and lifelong disabilities (Roberts et al., 1998). Until recently, pre-eclampsia could be alleviated only by removal of the placenta (Roberts et al., 1998), requiring delivery of the fetus and accounting for 15% of premature deliveries and the associated infant morbidity and mortality (Meis et al., 1998). Intervention prior to 16 weeks GA with regimens of low-dose aspirin (Bujold et al., 2014) or Pravastatin (Ramma and Ahmed, 2014) have shown promise in reducing the rate of pre-eclampsia in high-risk patients, providing a course of action when anticipated early.
Screening based on TRIC and other techniques
Evidence has been assembled using TRIC to demonstrate that while displaced from the environment of the placenta, EVT cells residing in the endocervical canal nevertheless provide information about pregnancy status prior to clinical diagnosis of malplacentation syndromes. In two studies (Fritz et al., 2015a; Bolnick et al., 2016b) a set of proteins (PP13, galectin 14, PAPP-A, PGF, alpha fetoprotein, FLT1, endoglin) associated with dysregulated placental perfusion were measured in EVT cells obtained by TRIC between 5 and 19 weeks. The results demonstrated significant differences in expression levels for all but one (PP13) of the seven proteins, as assessed by relative protein quantification, when comparing samples from control pregnancies to those that either ended with an early pregnancy loss (Fritz et al., 2015a), or later developed pre-eclampsia and/or FGR (Bolnick et al., 2016b). These relatively small pilot studies require expansion to screen a larger patient pool, and further suggest that EVT cells obtained by TRIC offer a valuable resource for exploratory studies using ‘omics’ approaches to identify new, more valuable biomarkers of obstetrical pathology, and for investigating the etiology of these diseases through pathway analysis.
TRIC, in providing access to EVT cells in ongoing pregnancies, is the first minimally invasive procedure that can potentially screen patients early in the first trimester to assess risk for dysregulated placental development and perfusion. If EVT molecular profiles prove to be capable of reliably identifying at-risk patients, novel interventions to prevent adverse pregnancy outcomes could more efficiently scrutinized by focusing on high-risk patients identified using TRIC; thereby reducing the number of subjects required to obtain definitive results. For example, a meta-analysis of 32 217 women participating in randomized trials to test the effect of various antiplatelet agents on pre-eclampsia showed a lack of benefit (Askie et al., 2007). A limitation of such large studies is the infrequency of the adverse outcomes under study, thus, significantly reducing statistical power.
Recently, an algorithm was employed using first trimester biophysical and biochemical test results, as well as patient medical history, to identify women with a risk >1:100 for preterm pre-eclampsia in a double-blind, placebo-controlled trial (Rolnik et al., 2017). Using this approach, 1620 women were selected for inclusion in the study after screening 26 941, and a significant reduction of preterm pre-eclampsia was obtained in the aspirin treatment group, compared to placebo. The predictive algorithm used in the Rolnik et al. (2017) study is not recommended currently by the US Preventive Services Task Force because screening based on clinical history appears to prevent pre-eclampsia in a reasonable fraction of cases without the additional costs of biophysical and biochemical testing (Roberts and Himes, 2017). In addition, other studies show that combining maternal characteristics, serum biomarkers and uterine artery pulsatility index between 11 and 13 + 6 weeks identifies only 60% of preterm pre-eclampsia, with a fixed false positive rate of 10% (Sonek et al., 2017). Prediction of FGR (i.e. small for GA) is suboptimal as well (McCowan et al., 2017). Because TRIC offers the ability to obtain information earlier in gestation that reflects embryonic/fetal status, it might be possible to use this approach to develop better intervention strategies.
Utility of TRIC for prenatal testing
Prediction of chromosomal and genetic disorders early in pregnancy provides vital information, allowing parents and caregivers to be proactive and prepare for the best possible outcome if congenital or inherited disorders are present. Down syndrome (trisomy 21) and other aneuploidies occur in over 0.2% of births worldwide, and are on the rise due to advanced women’s age at the time of pregnancy. Single-gene mutations account for over 6000 diseases, 10% of all pediatric hospital admissions, and 20% of infant deaths. Currently, definitive diagnosis of fetal genetic traits requires invasive procedures; either CVS beginning at 9–10 weeks GA, or amniocentesis beginning at 12–14 weeks GA (Eisenberg and Wapner, 2002). Comprehensive interrogation of the fetal genome before 9 weeks is now unavailable. However, NIPT is useful as an early screen for common aneuploidies (Bianchi et al., 2014). By 10 weeks GA, fetal DNA fragments comprise 4–10% of total cell-free DNA in maternal plasma, which can be distinguished from the maternal fraction by massively parallel sequencing and computational comparisons of single nucleotide polymorphisms (SNPs) or STRs (Wong and Lo, 2016). However, it is the opinion of professional medical organizations that a diagnostic test, which would require invasive procedures, should be offered to patients who have a positive cell-free DNA test result (ACOG/SMFM, 2015; Benn et al., 2015; Gregg et al., 2016).
TRIC is capable of providing highly purified EVT cells containing the intact and complete fetal genome for analysis, with minimal maternal DNA present that must be distinguished. As a result, fetal DNA analysis becomes more straightforward, simple and inexpensive than NIPT. While similar advantages have motivated investigators pursuing fetal cells in maternal blood, the cervix offers one or two orders of magnitude more cells and fetal DNA, making it a more practical approach for development of prenatal genetic tests. Early investigations demonstrated that fetal cells in the reproductive tract could provide clinically relevant genetic information (Canestrini and Testa, 1978), although the overwhelming background of maternal cells present made it impractical.
TRIC has been used successfully with FISH and PCR to unequivocally determine fetal sex (Bolnick et al., 2014, 2016a). DNA fingerprinting by targeted next-generation sequencing of STRs and SNPs has now been successfully conducted with EVT cells obtained by TRIC (Jain et al., 2016). There was a 100% correspondence between the SNP and STR haplotypes of fetal DNA obtained by TRIC and corresponding placental DNA in every specimen. These findings demonstrate single nucleotide resolution of fetal DNA, establishing the feasibility of safely analyzing single gene disorders as early as 5 weeks GA, earlier than other non-invasive or invasive methods. Future studies will test the limitations of TRIC for genetic testing and its potential suitability as a diagnostic method. Sequencing of loci amplified by multiplex PCR can be extremely robust, whereas whole exome and whole genome sequencing will require whole genome amplification, which can introduce bias and sequencing errors. However, as the methodologies improve, more downstream analytical approaches will become available with the limited amounts of DNA available through TRIC.
Conclusions
The existence of trophoblasts in the uterine cavity and endocervical canal has been known for many years; however, the technology is only now becoming available to access the information within those cells to probe the status of the placenta and pregnancy in real time. The TRIC procedure, in skilled hands, can isolate hundreds of fetal cells as early as 5 weeks of pregnancy from a simple pap smear. While single cell technology still lacks clinical precision, TRIC, as an intermediate solution, offers unique insights into human pregnancy and fetal development. It has advantages over cell-free fetal DNA in maternal serum because the isolated EVT cells contain the entire fetal genome for genotyping, while the problem of confined placental mosaicism limits the certainty of aneuploidy detection, as in NIPT and CVS. Although we do not yet understand the exact biological mechanisms that release EVT cells into the reproductive tract and the route that trophoblast cells take to the cervix, recent advances suggest that a new era of prenatal diagnosis lies ahead. There remains a paucity of information about differences that might exist between EVT cells in the placenta and those residing in the cervix that might color their utility as a barometer on pregnancy status and health, which needs to be resolved in future studies. Currently, measurements of disease biomarkers in maternal blood (e.g. PAPP-A, PP13, inhibin, hCG, PGF, FLT1) are under development to assess pregnancy. EVT cells accessible in the reproductive tract could potentially serve in that role at an earlier GA and with greater precision, since deficiencies of the EVT likely contribute to placental maldevelopment. With the use of TRIC for early prenatal testing now feasible, the future could hold opportunities to screen fetuses and mothers at risk in preparation for corrective interventions to reduce morbidity and mortality.
Acknowledgements
We thank Dr Steven J. Korzeniewski of Wayne State University for critical reading of the article, and for his helpful comments. We would also like to thank Monika Sundl of Medical University of Graz for her expert technical assistance.
Authors’ role
All authors contributed to the study design, literature analysis and drafting of the article, as well as critical reading and editing.
Funding
Grants from the National Institutes of Health (HD071408, HL128628, HD092205), a March of Dimes Basil O'Connor Starter Scholar Research Award (S.D.), PerkinElmer Health Sciences, Inc., and the W.K. Kellogg Foundation.
Conflict of interest
D.R.A. and S.D. have pending patents, receive payment for intellectual property that has been licensed by Wayne State University to PerkinElmer, Inc., and are principals in Advanced Reproductive Testing, LLC.
References
- ACOG Committee Opinion No. 638: first-trimester risk assessment for early-onset preeclampsia. Obstet Gynecol 2015;126:e25–e27. [DOI] [PubMed] [Google Scholar]
- ACOG/SMFM Committee Opinion No. 640: cell-free DNA screening for fetal aneuploidy. Obstet Gynecol 2015;126:e31–e37. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Cirigliano V.. Detection of fetal cells in transcervical samples using X22 marker. J Med Genet 2000;37:E1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adinolfi M, Davies A, Sharif S, Soothill P, Rodeck C.. Detection of trisomy 18 and Y-derived sequences in fetal nucleated cells obtained by transcervical flushing. Lancet 1993;342:403–404. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, el-Hashemite N, Sherlock J, Ward RH, Petrou M, Rodeck C.. Prenatal detection of Hb mutations using transcervical cells. Prenat Diagn 1997;17:539–543. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Sherlock J.. First trimester prenatal diagnosis using transcervical cells: an evaluation. Hum Reprod Update 1997;3:383–392. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Sherlock J.. Fetal cells in transcervical samples at an early stage of gestation. J Hum Genet 2001;46:99–104. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Sherlock J, Kemp T, Carritt B, Soothill P, Kingdom J, Rodeck C.. Prenatal detection of fetal RhD DNA sequences in transcervical samples. Lancet 1995. a;345:318–319. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Sherlock J, Soothill P, Rodeck C.. Molecular evidence of fetal-derived chromosome 21 markers (STRs) in transcervical samples. Prenat Diagn 1995. b;15:35–39. [DOI] [PubMed] [Google Scholar]
- Adinolfi M, Sherlock J, Tutschek B, Halder A, Delhanty J, Rodeck C.. Detection of fetal cells in transcervical samples and prenatal diagnosis of chromosomal abnormalities. Prenat Diagn 1995. c;15:943–949. [DOI] [PubMed] [Google Scholar]
- Amankwah KS, Bond EC.. Unreliability of prenatal determination of fetal sex with the use of Y-body fluorescence in midcervical smears. Am J Obstet Gynecol 1978;130:300–301. [PubMed] [Google Scholar]
- Apps R, Gardner L, Moffett A.. A critical look at HLA-G. Trends Immunol 2008;29:313–321. [DOI] [PubMed] [Google Scholar]
- Askie LM, Duley L, Henderson-Smart DJ, Stewart LA, Group PC.. Antiplatelet agents for prevention of pre-eclampsia: a meta-analysis of individual patient data. Lancet 2007;369:1791–1798. [DOI] [PubMed] [Google Scholar]
- Bahado-Singh RO, Kliman H, Feng TY, Hobbins J, Copel JA, Mahoney MJ.. First-trimester endocervical irrigation: feasibility of obtaining trophoblast cells for prenatal diagnosis. Obstet Gynecol 1995;85:461–464. [DOI] [PubMed] [Google Scholar]
- Benirschke K, Burton G, Baergen RN.. Early Developement of the Human Placenta. Pathology of the Human Placenta. Berlin Heidelberg: Springer, 2012. a;41–53. [Google Scholar]
- Benirschke K, Burton G, Baergen RN.. Nonvillous Parts and Trophoblast Invasion. Pathology of the Human Placenta. Berlin Heidelberg: Springer, 2012. b;157–240. [Google Scholar]
- Benn P, Borrell A, Chiu RW, Cuckle H, Dugoff L, Faas B, Gross S, Huang T, Johnson J, Maymon R et al. Position statement from the chromosome abnormality screening committee on behalf of the board of the international society for prenatal diagnosis. Prenat Diagn 2015;35:725–734. [DOI] [PubMed] [Google Scholar]
- Bianchi DW, Parker RL, Wentworth J, Madankumar R, Saffer C, Das AF, Craig JA, Chudova DI, Devers PL, Jones KW et al. DNA sequencing versus standard prenatal aneuploidy screening. N Engl J Med 2014;370:799–808. [DOI] [PubMed] [Google Scholar]
- Billington WD. Vascular migration of transplanted trophoblast in the golden hamster. Nature 1966;211:988–989. [DOI] [PubMed] [Google Scholar]
- Bischoff FZ, Simpson JL.. Endocervical fetal trophoblast for prenatal genetic diagnosis. Curr Opin Obstet Gynecol 2006;18:216–220. [DOI] [PubMed] [Google Scholar]
- Bobrow M, Lewis BV.. Unreliability of fetal sexing using cervical material. Lancet 1971;2:486. [DOI] [PubMed] [Google Scholar]
- Bolnick AD, Fritz R, Jain C, Kadam L, Bolnick JM, Kilburn BA, Singh M, Diamond MP, Drewlo S, Armant DR.. Trophoblast retrieval and isolation from the cervix for noninvasive, first trimester, fetal gender determination in a carrier of congenital adrenal hyperplasia. Reprod Sci 2016. a;23:717–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick JM, Kilburn BA, Bajpayee S, Reddy N, Jeelani R, Crone B, Simmerman N, Singh M, Diamond MP, Armant DR.. Trophoblast retrieval and isolation from the cervix (TRIC) for noninvasive prenatal screening at 5 to 20 weeks of gestation. Fertil Steril 2014;102:135–142 e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick JM, Kohan-Ghadr HR, Fritz R, Bolnick AD, Kilburn BA, Diamond MP, Armant DR, Drewlo S.. Altered biomarkers in trophoblast cells obtained noninvasively prior to clinical manifestation of perinatal disease. Sci Rep 2016. b;6:32382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs J, Miller D, Bulmer JN, Griffith-Jones M, Rame V, Lilford R.. Non-syncytial sources of fetal DNA in transcervically recovered cell populations. Hum Reprod 1995;10:749–754. [DOI] [PubMed] [Google Scholar]
- Bujold E, Roberge S, Nicolaides KH.. Low-dose aspirin for prevention of adverse outcomes related to abnormal placentation. Prenat Diagn 2014;34:642–648. [DOI] [PubMed] [Google Scholar]
- Bulmer JN, Cioni R, Bussani C, Cirigliano V, Sole F, Costa C, Garcia P, Adinolfi M.. HLA-G positive trophoblastic cells in transcervical samples and their isolation and analysis by laser microdissection and QF-PCR. Prenat Diagn 2003;23:34–39. [DOI] [PubMed] [Google Scholar]
- Bulmer JN, Rodeck C, Adinolfi M.. Immunohistochemical characterization of cells retrieved by transcervical sampling in early pregnancy. Prenat Diagn 1995;15:1143–1153. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Jauniaux E.. Placental oxidative stress: from miscarriage to preeclampsia. J Soc Gynecol Investig 2004;11:342–352. [DOI] [PubMed] [Google Scholar]
- Burton GJ, Woods AW, Jauniaux E, Kingdom JC.. Rheological and physiological consequences of conversion of the maternal spiral arteries for uteroplacental blood flow during human pregnancy. Placenta 2009;30:473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussani C, Cioni R, Mattei A, Fambrini M, Marchionni M, Scarselli G.. Prenatal diagnosis of common aneuploidies in transcervical samples using quantitative fluorescent-PCR analysis. Mol Diagn Ther 2007;11:117–121. [DOI] [PubMed] [Google Scholar]
- Canestrini C, Testa R.. Platelet antiaggregating activity of alclofenac: a preliminary pharmacodynamic and kinetic study in rabbit. Pharmacol Res Commun 1978;10:173–183. [DOI] [PubMed] [Google Scholar]
- Chaddha V, Viero S, Huppertz B, Kingdom J.. Developmental biology of the placenta and the origins of placental insufficiency. Semin Fetal Neonatal Med 2004;9:357–369. [DOI] [PubMed] [Google Scholar]
- Chaouat G, Lochu P, Ville Y, Rhali H, Bedossa P, Selva J, Bergere M, D’Auriol L, Bellet D, Vidault M et al. Transcervical sampling: a preliminary prospective study. Ann N Y Acad Sci 1994;731:197–200. [DOI] [PubMed] [Google Scholar]
- Cioni R, Bussani C, Bucciantini S, Scarselli G.. Fetal cells in a transcervical cell sample collected at 5 weeks of gestation. J Matern Fetal Neonatal Med 2005;18:271–273. [DOI] [PubMed] [Google Scholar]
- Cioni R, Bussani C, Scarselli B, Bucciantini S, Barciulli F, Scarselli G.. Fetal cells in cervical mucus in the first trimester of pregnancy. Prenat Diagn 2003;23:168–171. [DOI] [PubMed] [Google Scholar]
- Cirigliano V, Sherlock J, Petrou M, Ward RH, Rodeck C, Adinolfi M.. Transcervical cells and the prenatal diagnosis of haemoglobin (Hb) mutations. Clin Genet 1999;56:357–361. [DOI] [PubMed] [Google Scholar]
- Colbern GT, Chiang MH, Main EK.. Expression of the nonclassic histocompatibility antigen HLA-G by preeclamptic placenta. Am J Obstet Gynecol 1994;170:1244–1250. [DOI] [PubMed] [Google Scholar]
- Collins JE, Fleming TP.. Epithelial differentiation in the mouse preimplantation embryo: making adhesive cell contacts for the first time. Trends Biochem Sci 1995;20:307–312. [DOI] [PubMed] [Google Scholar]
- Craven CM, Zhao L, Ward K.. Lateral placental growth occurs by trophoblast cell invasion of decidual veins. Placenta 2000;21:160–169. [DOI] [PubMed] [Google Scholar]
- Damsky CH, Librach C, Lim KH, Fitzgerald ML, McMaster MT, Janatpour M, Zhou Y, Logan SK, Fisher SJ.. Integrin switching regulates normal trophoblast invasion. Development 1994;120:3657–3666. [DOI] [PubMed] [Google Scholar]
- Davey DA, MacGillivray I.. The classification and definition of the hypertensive disorders of pregnancy. Am J Obstet Gynecol 1988;158:892–898. [DOI] [PubMed] [Google Scholar]
- Devaney SA, Palomaki GE, Scott JA, Bianchi DW.. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. J Am Med Assoc 2011;306:627–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Renzo GC. The great obstetrical syndromes. J Matern Fetal Neonatal Med 2009;22:633–635. [DOI] [PubMed] [Google Scholar]
- Drewlo S, Armant DR.. Quo vadis, trophoblast? Exploring the new ways of an old cell lineage. Placenta 2017;60:S27–S31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drury S, Mason S, McKay F, Lo K, Boustred C, Jenkins L, Chitty LS.. Implementing non-invasive prenatal diagnosis (NIPD) in a National Health Service laboratory; from dominant to recessive disorders. Adv Exp Med Biol 2016;924:71–75. [DOI] [PubMed] [Google Scholar]
- Eisenberg B, Wapner RJ.. Clinical procedures in prenatal diagnosis. Best Pract Res Clin Obstet Gynaecol 2002;16:611–627. [DOI] [PubMed] [Google Scholar]
- ErgIn T, Baltaci V, Zeyneloglu HB, Duran EH, Ergenel IM, Batioglu S.. Non-invasive early prenatal diagnosis using fluorescent in situ hybridization on transcervical cells: comparison of two different methods for retrieval. Eur J Obstet Gynecol Reprod Biol 2001;95:37–41. [DOI] [PubMed] [Google Scholar]
- Evans MI, Kilpatrick M.. Noninvasive prenatal diagnosis: 2010. Clin Lab Med 2010;30:655–665. [DOI] [PubMed] [Google Scholar]
- Fang CN, Kan YY, Hsiao CC.. Detection of fetal cells from transcervical mucus plug before first-trimester termination of pregnancy by cytokeratin-7 immunohistochemistry. J Obstet Gynaecol Res 2005;31:500–507. [DOI] [PubMed] [Google Scholar]
- Fejgin MD, Diukman R, Cotton Y, Weinstein G, Amiel A.. Fetal cells in the uterine cervix: a source for early non-invasive prenatal diagnosis. Prenat Diagn 2001;21:619–621. [DOI] [PubMed] [Google Scholar]
- Fritz R, Kohan-Ghadr HR, Bolnick JM, Bolnick AD, Kilburn BA, Diamond MP, Drewlo S, Armant DR.. Noninvasive detection of trophoblast protein signatures linked to early pregnancy loss using trophoblast retrieval and isolation from the cervix (TRIC). Fertil Steril 2015. a;104:339–346 e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz R, Kohan-Ghadr HR, Sacher A, Bolnick AD, Kilburn BA, Bolnick JM, Diamond MP, Drewlo S, Armant DR.. Trophoblast retrieval and isolation from the cervix (TRIC) is unaffected by early gestational age or maternal obesity. Prenat Diagn 2015. b;35:1218–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung HY, Wong YF, Haines CJ.. Detection of fetal Y-specific DNA from the cervix. Acta Obstet Gynecol Scand 1995;74:248–250. [DOI] [PubMed] [Google Scholar]
- Gauster M, Blaschitz A, Siwetz M, Huppertz B.. Keratins in the human trophoblast. Histol Histopathol 2013;28:817–825. [DOI] [PubMed] [Google Scholar]
- Gil MM, Accurti V, Santacruz B, Plana MN, Nicolaides KH.. Analysis of cell-free DNA in maternal blood in screening for aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2017;50:302–314. [DOI] [PubMed] [Google Scholar]
- Goldberg MF, Chen AT, Ahn YW, Reidy JA.. First-trimester fetal chromosomal diagnosis using endocervical lavage: a negative evaluation. Am J Obstet Gynecol 1980;138:436–440. [DOI] [PubMed] [Google Scholar]
- Gregg AR, Skotko BG, Benkendorf JL, Monaghan KG, Bajaj K, Best RG, Klugman S, Watson MS.. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med 2016;18:1056–1065. [DOI] [PubMed] [Google Scholar]
- Griffith-Jones MD, Miller D, Lilford RJ, Scott J, Bulmer J.. Detection of fetal DNA in trans-cervical swabs from first trimester pregnancies by gene amplification: a new route to prenatal diagnosis? Br J Obstet Gynaecol 1992;99:508–511. [DOI] [PubMed] [Google Scholar]
- Guttmacher AE, Maddox YT, Spong CY.. The Human Placenta Project: placental structure, development, and function in real time. Placenta 2014;35:303–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He N, van Iperen L, de Jong D, Szuhai K, Helmerhorst FM, van der Westerlaken LA, Chuva de Sousa Lopes SM. Human extravillous trophoblasts penetrate decidual veins and lymphatics before remodeling spiral arteries during early pregnancy. PLoS One. 2017;12:e0169849. doi: 10.1371/journal.pone.0169849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J, Stiltner L, Jamieson B, Fashner J.. Clinical inquiries. Should a nylon brush be used for Pap smears from pregnant women? J Fam Pract 2005;54:463–464. [PubMed] [Google Scholar]
- Hossain N, Paidas MJ.. Adverse pregnancy outcome, the uteroplacental interface, and preventive strategies. Semin Perinatol 2007;31:208–212. [DOI] [PubMed] [Google Scholar]
- Imudia AN, Kumar S, Diamond MP, Decherney AH, Armant DR.. Transcervical retrieval of fetal cells in the practice of modern medicine: a review of the current literature and future direction. Fertil Steril 2010;93:1725–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imudia AN, Suzuki Y, Kilburn BA, Yelian FD, Diamond MP, Romero R, Armant DR.. Retrieval of trophoblast cells from the cervical canal for prediction of abnormal pregnancy: a pilot study. Hum Reprod 2009;24:2086–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishai D, Amiel A, Diukman R, Cogan O, Lichtenstein Z, Abramovici H, Fejgin MD.. Uterine cavity lavage: adding FISH to conventional cytogenetics for embryonic sexing and diagnosing common chromosomal aberrations. Prenat Diagn 1995;15:961–965. [DOI] [PubMed] [Google Scholar]
- Iverson GM, Bianchi DW, Cann HM, Herzenberg LA.. Detection and isolation of fetal cells from maternal blood using the flourescence-activated cell sorter (FACS). Prenat Diagn 1981;1:61–73. [DOI] [PubMed] [Google Scholar]
- Jain CV, Kadam L, van Dijk M, Kohan-Ghadr HR, Kilburn BA, Hartman C, Mazzorana V, Visser A, Hertz M, Bolnick AD et al. Fetal genome profiling at 5 weeks of gestation after noninvasive isolation of trophoblast cells from the endocervical canal. Sci Transl Med 2016;8:363re364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JA, MacDonald K, Clarke G, Skoll A.. No. 343-Routine non-invasive prenatal prediction of fetal RHD genotype in Canada: the time is here. J Obstet Gynaecol Can 2017;39:366–373. [DOI] [PubMed] [Google Scholar]
- Katz-Jaffe MG, Mantzaris D, Cram DS.. DNA identification of fetal cells isolated from cervical mucus: potential for early non-invasive prenatal diagnosis. BJOG 2005;112:595–600. [DOI] [PubMed] [Google Scholar]
- Kemp B, Kertschanska S, Kadyrov M, Rath W, Kaufmann P, Huppertz B.. Invasive depth of extravillous trophoblast correlates with cellular phenotype: a comparison of intra- and extrauterine implantation sites. Histochem Cell Biol 2002;117:401–414. [DOI] [PubMed] [Google Scholar]
- Kingdom J, Sherlock J, Rodeck C, Adinolfi M.. Detection of trophoblast cells in transcervical samples collected by lavage or cytobrush. Obstet Gynecol 1995;86:283–288. [DOI] [PubMed] [Google Scholar]
- Larsen EC, Christiansen OB, Kolte AM, Macklon N.. New insights into mechanisms behind miscarriage. BMC Med 2013;11:154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KH, Zhou Y, Janatpour M, McMaster M, Bass K, Chun SH, Fisher SJ.. Human cytotrophoblast differentiation/invasion is abnormal in pre-eclampsia. Am J Pathol 1997;151:1809–1818. [PMC free article] [PubMed] [Google Scholar]
- Loke YW, King A, Burrows T, Gardner L, Bowen M, Hiby S, Howlett S, Holmes N, Jacobs D.. Evaluation of trophoblast HLA-G antigen with a specific monoclonal antibody. Tissue Antigens 1997;50:135–146. [DOI] [PubMed] [Google Scholar]
- Mantzaris D, Cram DS.. Potential of syncytiotrophoblasts isolated from the cervical mucus for early non-invasive prenatal diagnosis: evidence of a vanishing twin. Clin Chim Acta 2015;438:309–315. [DOI] [PubMed] [Google Scholar]
- Mantzaris D, Cram D, Healy C, Howlett D, Kovacs G.. Preliminary report: correct diagnosis of sex in fetal cells isolated from cervical mucus during early pregnancy. Aust N Z J Obstet Gynaecol 2005;45:529–532. [DOI] [PubMed] [Google Scholar]
- Manuel M, Park IJ, Jones HW Jr. Prenatal sex determination by fluorescent staining of cells for the presence of Y chromatin. Am J Obstet Gynecol 1974;119:853–854. [DOI] [PubMed] [Google Scholar]
- Massari A, Novelli G, Colosimo A, Sangiuolo F, Palka G, Calabrese G, Camurri L, Ghirardini G, Milani G, Giorlandino C et al. Non-invasive early prenatal molecular diagnosis using retrieved transcervical trophoblast cells. Hum Genet 1996;97:150–155. [DOI] [PubMed] [Google Scholar]
- McCowan LM, Thompson JM, Taylor RS, Baker PN, North RA, Poston L, Roberts CT, Simpson NA, Walker JJ, Myers J et al. Prediction of small for gestational age infants in healthy nulliparous women using clinical and ultrasound risk factors combined with early pregnancy biomarkers. PLoS One 2017;12:e0169311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster MT, Librach CL, Zhou Y, Lim KH, Janatpour MJ, DeMars R, Kovats S, Damsky C, Fisher SJ.. Human placental HLA-G expression is restricted to differentiated cytotrophoblasts. J Immunol 1995;154:3771–3778. [PubMed] [Google Scholar]
- McMaster M, Zhou Y, Shorter S, Kapasi K, Geraghty D, Lim KH, Fisher S.. HLA-G isoforms produced by placental cytotrophoblasts and found in amniotic fluid are due to unusual glycosylation. J Immunol 1998;160:5922–5928. [PubMed] [Google Scholar]
- Meis PJ, Goldenberg RL, Mercer BM, Iams JD, Moawad AH, Miodovnik M, Menard MK, Caritis SN, Thurnau GR, Bottoms SF et al. The preterm prediction study: risk factors for indicated preterm births. Maternal-Fetal Medicine Units Network of the National Institute of Child Health and Human Development. Am J Obstet Gynecol 1998;178:562–567. [DOI] [PubMed] [Google Scholar]
- Merviel P, Challier JC, Carbillon L, Foidart JM, Uzan S.. The role of integrins in human embryo implantation. Fetal DiagnTher 2001;16:364–371. [DOI] [PubMed] [Google Scholar]
- Miller D, Briggs J, Rahman MS, Griffith-Jones M, Rane V, Everett M, Lilford RJ, Bulmer JN.. Transcervical recovery of fetal cells from the lower uterine pole: reliability of recovery and histological/immunocytochemical analysis of recovered cell populations. Hum Reprod 1999;14:521–531. [DOI] [PubMed] [Google Scholar]
- Moser G, Gauster M, Orendi K, Glasner A, Theuerkauf R, Huppertz B.. Endoglandular trophoblast, an alternative route of trophoblast invasion? Analysis with novel confrontation co-culture models. Hum Reprod 2010;25:1127–1136. [DOI] [PubMed] [Google Scholar]
- Moser G, Huppertz B.. Implantation and extravillous trophoblast invasion: from rare archival specimens to modern biobanking. Placenta 2017;56:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser G, Orendi K, Gauster M, Siwetz M, Helige C, Huppertz B.. The art of identification of extravillous trophoblast. Placenta 2011;32:197–199. [DOI] [PubMed] [Google Scholar]
- Moser G, Weiss G, Gauster M, Sundl M, Huppertz B.. Evidence from the very beginning: endoglandular trophoblasts penetrate and replace uterine glands in situ and in vitro. Hum Reprod 2015;30:2747–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser G, Weiss G, Sundl M, Gauster M, Siwetz M, Lang-Olip I, Huppertz B.. Extravillous trophoblasts invade more than uterine arteries: evidence for the invasion of uterine veins. Histochem Cell Biol 2017;147:353–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanaev AK, Kosanke G, Kemp B, Frank HG, Huppertz B, Kaufmann P.. The human placenta is encircled by a ring of smooth muscle cells. Placenta 2000;21:122–125. [DOI] [PubMed] [Google Scholar]
- Norwitz ER, Levy B.. Noninvasive prenatal testing: the future is now. Rev Obstet Gynecol 2013;6:48–62. [PMC free article] [PubMed] [Google Scholar]
- Orr JW Jr, Barrett JM, Orr PF, Holloway RW, Holimon JL.. The efficacy and safety of the cytobrush during pregnancy. Gynecol Oncol 1992;44:260–262. [DOI] [PubMed] [Google Scholar]
- Papageorghiou AT, Yu CK, Nicolaides KH.. The role of uterine artery Doppler in predicting adverse pregnancy outcome. Best Pract Res Clin Obstet Gynaecol 2004;18:383–396. [DOI] [PubMed] [Google Scholar]
- Paraiso MF, Brady K, Helmchen R, Roat TW.. Evaluation of the endocervical cytobrush and cervex-brush in pregnant women. Obstet Gynecol 1994;84:539–543. [PubMed] [Google Scholar]
- Parry S, Sciscione A, Haas DM, Grobman WA, Iams JD, Mercer BM, Silver RM, Simhan HN, Wapner RJ, Wing DA et al. Role of early second-trimester uterine artery Doppler screening to predict small-for-gestational-age babies in nulliparous women. Am J Obstet Gynecol 2017;217:594 e591–594 e510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertl B, Davies A, Soothill P, Rodeck C, Adinolfi M.. Detection of fetal cells in endocervical samples. Ann N Y Acad Sci 1994;731:186–192. [DOI] [PubMed] [Google Scholar]
- Pfeifer I, Benachi A, Saker A, Bonnefont JP, Mouawia H, Broncy L, Frydman R, Brival ML, Lacour B, Dachez R et al. Cervical trophoblasts for non-invasive single-cell genotyping and prenatal diagnosis. Placenta 2016;37:56–60. [DOI] [PubMed] [Google Scholar]
- Ramma W, Ahmed A.. Therapeutic potential of statins and the induction of heme oxygenase-1 in preeclampsia. J Reprod Immunol 2014;101–102:153–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhine SA, Cain JL, Cleary RE, Palmer CG, Thompson JF.. Prenatal sex detection with endocervical smears: successful results utilizing Y-bodyfluoroscence. Am J Obstet Gynecol 1975;122:155–160. [DOI] [PubMed] [Google Scholar]
- Rivlin ME, Woodliff JM, Bowlin RB, Moore JL Jr, Martin RW, Grossman JH III, Morrison JC. Comparison of cytobrush and cotton swab for Papanicolaou smears in pregnancy. J Reprod Med 1993;38:147–150. [PubMed] [Google Scholar]
- Roberts JM, Creasy RK, Resnik R.. Pregnancy Related Hypertension. Maternal Fetal Medicine. Philadelphia: W.B. Saunders, 1998;833–872. [Google Scholar]
- Roberts JM, Himes KP.. Pre-eclampsia: screening and aspirin therapy for prevention of pre-eclampsia. Nat Rev Nephrol 2017;13:602–604. [DOI] [PubMed] [Google Scholar]
- Rodeck C, Tutschek B, Sherlock J, Kingdom J.. Methods for the transcervical collection of fetal cells during the first trimester of pregnancy. Prenat Diagn 1995;15:933–942. [DOI] [PubMed] [Google Scholar]
- Rolnik DL, Wright D, Poon LC, O’Gorman N, Syngelaki A, de Paco Matallana C, Akolekar R, Cicero S, Janga D, Singh M et al. Aspirin versus placebo in pregnancies at high risk for preterm preeclampsia. N Engl J Med 2017;377:613–622. [DOI] [PubMed] [Google Scholar]
- Romero R. Prenatal medicine: the child is the father of the man. 1996. J Matern Fetal Neonatal Med 2009;22:636–639. [DOI] [PubMed] [Google Scholar]
- Savchev S, Figueras F, Sanz-Cortes M, Cruz-Lemini M, Triunfo S, Botet F, Gratacos E.. Evaluation of an optimal gestational age cut-off for the definition of early- and late-onset fetal growth restriction. Fetal Diagn Ther 2014;36:99–105. [DOI] [PubMed] [Google Scholar]
- Sherlock J, Halder A, Tutschek B, Delhanty J, Rodeck C, Adinolfi M.. Prenatal detection of fetal aneuploidies using transcervical cell samples. J Med Genet 1997;34:302–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shettles LB. Use of the Y chromosome in prenatal sex determination. Nature 1971;230:52–53. [DOI] [PubMed] [Google Scholar]
- SMFM Practice Bulletin No. 163: screening for fetal aneuploidy. Obstet Gynecol 2016;127:e123–e137. [DOI] [PubMed] [Google Scholar]
- Sonek J, Krantz D, Carmichael J, Downing C, Jessup K, Haidar Z, Ho S, Hallahan T, Kliman HJ, McKenna D.. First-trimester screening for early and late preeclampsia using maternal characteristics, biomarkers, and estimated placental volume. Am J Obstet Gynecol 2017;218:126-e1. [DOI] [PubMed] [Google Scholar]
- Tranquilli AL. Introduction to ISSHP new classification of preeclampsia. Pregnancy Hypertens 2013;3:58–59. [DOI] [PubMed] [Google Scholar]
- Ville Y, Lochu P, Rhali H, D’Auriol L, Bedossa P, Bergere M, Baud M, Selva J, Chaouat G, Nicolaides K et al. [Are desquamated trophoblastic cells retrieved from the cervix suitable for a prenatal diagnosis?]. Contracept Fertil Sex 1994;22:475–477. [PubMed] [Google Scholar]
- Weiss G, Sundl M, Glasner A, Huppertz B, Moser G.. The trophoblast plug during early pregnancy: a deeper insight. Histochem Cell Biol 2016;146:749–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley LM. Trophectoderm: the first epithelium to develop in the mammalian embryo. Scanning Microsc 1988;2:417–426. [PubMed] [Google Scholar]
- Windsperger K, Dekan S, Pils S, Golletz C, Kunihs V, Fiala C, Kristiansen G, Knofler M, Pollheimer J.. Extravillous trophoblast invasion of venous as well as lymphatic vessels is altered in idiopathic, recurrent, spontaneous abortions. Hum Reprod 2017;32:1208–1217. [DOI] [PubMed] [Google Scholar]
- Wong FC, Lo YM.. Prenatal diagnosis innovation: genome sequencing of maternal plasma. Annu Rev Med 2016;67:419–432. [DOI] [PubMed] [Google Scholar]
- Wou K, Feinberg JL, Wapner RJ, Simpson JL., Cell-free DNA. versus intact fetal cells for prenatal genetic diagnostics: what does the future hold? Expert Rev Mol Diagn 2015;15:989–998. [DOI] [PubMed] [Google Scholar]
- Wright E, Audette MC, Ye XY, Keating S, Hoffman B, Lye SJ, Shah PS, Kingdom JC.. Maternal vascular malperfusion and adverse perinatal outcomes in low-risk nulliparous women. Obstet Gynecol 2017;130:1112–1120. [DOI] [PubMed] [Google Scholar]
- Xi Zhao X, Suzumori K, Sato T.. Prenatal diagnosis of triple X using fetal cells obtained by endocervical lavage. Prenat Diagn 2003;23:549–551. [DOI] [PubMed] [Google Scholar]
- Yie SM, Li LH, Li YM, Librach C.. HLA-G protein concentrations in maternal serum and placental tissue are decreased in preeclampsia. Am J Obstet Gynecol 2004;191:525–529. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Fisher SJ, Janatpour M, Genbacev O, Dejana E, Wheelock M, Damsky CH.. Human cytotrophoblasts adopt a vascular phenotype as they differentiate. A strategy for successful endovascular invasion? J Clin Invest 1997;99:2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Han T, Yin G, Wang X, Yao Y.. Expression of human leukocyte antigen-G during normal placentation and in preeclamptic pregnancies. Hypertens Pregnancy 2012;31:252–260. [DOI] [PubMed] [Google Scholar]