Abstract
Aims: Thalassemia is a dangerous hematolytic genetic disease. In south China, ∼24% Chinese carry alpha-thalassemia or beta-thalassemia gene mutations. Given the fact that the invasive sampling procedures can only be performed by professionals in experienced centers, it may increase the risk of miscarriage or infection. Thus, most people are worried about the invasive operation. As such, a noninvasive and accurate prenatal diagnosis is needed for appropriate genetic counseling for families with high risks. Here we sought to develop capture probes and their companion analysis methods for the noninvasive prenatal detection of deletional and nondeletional thalassemia.
Materials and Methods: Two families diagnosed as carriers of either beta-thalassemia gene or Southeast Asian deletional alpha-thalassemia gene mutation were recruited. The maternal plasma and amniotic fluid were collected for prenatal diagnosis. Probes targeting exons of the genes of interest and the highly heterozygous SNPs within the 1Mb flanking region were designed. The target capture sequencing was performed with plasma DNA from the pregnant woman and genomic DNA from the couples and their children. Then the parental haplotype was constructed by the trios-based strategy. The fetal haplotype was deduced from the parental haplotype with a hidden Markov model-based algorithm.
Results: The fetal genotypes were successfully deduced in both families noninvasively. The noninvasively constructed haplotypes of both fetuses were identical to the invasive prenatal diagnosis results with an accuracy rate of 100% in the target region.
Conclusion: Our study demonstrates that the effective noninvasive prenatal diagnosis of alpha-thalassemia and beta-thalassemia can be achieved with the targeted capture sequencing and the haplotype-assisted analysis method.
Keywords: : thalassemia, noninvasive prenatal test, target region capture, massively parallel sequencing, SNP
Introduction
Thalassemia is one of the most common genetic blood disorders. In Guangxi, China, about 24% of people carry alpha-thalassemia or beta-thalassemia gene mutations (Xiong et al., 2010). Individuals affected by beta-thalassemia mainly receive regular transfusion program and chelation therapy to prolong their life expectancy. More seriously, babies suffering from the severe Hb Bart's alpha-thalassemia, usually die after birth due to fetal hydrops. To provide appropriate genetic counseling for families with high risk, invasive prenatal diagnosis has been widely used since 1972 (Kan et al., 1974; Orlandi et al., 1988; Ko et al., 1989). They are still used either for diagnosis or confirmation nowadays and they give very reliable results. Although rare, this kind of invasive operation may increase the risk of miscarriage or infection and it can only be performed by professionals in experienced centers. Therefore, noninvasive and accurate prenatal diagnosis is needed for appropriate genetic counseling for families with high risk.
The discovery of cell-free fetal DNA in the maternal plasma (Lo et al., 1997) makes it possible to detect fetal genetic disease noninvasively (Bianchi et al., 2012; Lim et al., 2013; Jin et al., 2014; Lv et al., 2015; Yoo et al., 2015) Methods based on real-time polymerase chain reaction (PCR) (Chiuet al., 2002; Tungwiwat et al., 2006), mass spectrometry (Tsang et al., 2007), digital PCR (Lun et al., 2008), and coamplification at lower denaturation temperature PCR (Galbiati et al., 2011) have been used in noninvasive prenatal detection of paternal inheritance-specific thalassemia mutations. Later on, the noninvasive detection of maternal inherited mutations has been realized by the combinational application of massively parallel sequencing technology and the haplotype-assisted linkage analysis (Lo et al., 2010; Lam et al., 2012; Meng et al., 2014; New et al., 2014; You et al., 2014). However, in most of these studies, the whole genome or SNPs distributed all over the entire chromosome were sequenced and analyzed, making the cost too high to be endured in clinical applications.
Targeted sequencing has been reported in noninvasive prenatal diagnosis of fetal beta-thalassemia (Lam et al., 2012), and in the selection of highly heterozygous SNPs distributed across beta-globin clusters suitable for the development of noninvasive detection methods (Papasavva et al., 2013). However, these studies only reported the prenatal detection of either single nucleotide variant (SNV) or small insertion or deletion (INDEL). Haplotyping-assisted methods have not been applied in the detection of large deletions, such as the Southeast Asian (SEA)-type alpha-thalassemia deletion. SEA-type deletion is common in South China, and the high morbidity of SEA deletion thalassemia results in a high risk of giving birth to a fetus affected by Hb Bart's syndrome. Some studies reported that homozygous SEA deletion can be excluded by assessing paternal-specific SNP (Yan et al., 2011) or microsatellite markers located within the breakpoints (Ho et al., 2007). However, the exact genotype could not be determined by these methods. Although the SEA deletion can be detected by the analysis of the copy number variation in the plasma sample (Ge et al., 2013), it was only performed in samples with high fetal DNA fraction. The haplotyping-assisted method has been proved to successfully work with low fetal DNA fraction samples (Xu et al., 2015). The purpose of this study is to develop the target capture sequencing-based method for prenatal detection of deletional and nondeletional thalassemia.
To achieve the capture sequencing-based noninvasive prenatal detection of fetal alpha-thalassemia and beta-thalassemia, a set of probes that target the highly heterozygous SNP flanking of the genes of interest (HBA1, HBA2, and HBB) was developed, whose size of target region for alpha-thalassemia and beta-thalassemia analysis is only about 154 and 287 kb, respectively. The results in this study demonstrated that the fetal genotypes in alpha-thalassemia and beta-thalassemia mutations were noninvasively constructed from a small set of informative SNPs of one beta-thalassemia-affected family and one SEA deletion-type alpha-thalassemia-affected family with a sequential application of hidden Markov model (HMM) and Viterbi algorithm. The results also indicated that this method could be applied in the detection of SEA deletion where all of the family members used for haplotype construction were carriers of SEA-type deletion.
Materials and Methods
Sample collection
Two families affected by thalassemia were recruited at the Prepotency Division of Affiliated Hospital of Guilin Medical University with informed consent and ethical approval from the Ethics Committee of the Affiliated Hospital of Guilin Medical University. Genetic counseling was provided to the families. Peripheral blood of the parents and their first child was collected into EDTA-containing tubes. Plasma of pregnant woman was separated within 8 h. Eight milliliters of amniotic fluids collected by amniocentesis from the two pregnant women were used to validate the results of noninvasive detection. DNA of the blood and amniotic fluids was extracted with magnetic bead adsorption/automatic nucleic acid extraction method (Zhishan Biotechnology). All samples were shipped in dry ice to the BGI Clinical Laboratory for noninvasive prenatal assays. The plasma cell-free DNA was extracted with the QIAamp Circulating Nucleic Acid Kit (Qiagen). The carrier status of recruited families was determined by a thalassemia gene detection kit (Yaneng Biotechnology Company) (Jin et al., 2014) and Gap-PCR and PCR-reverse dot blot for detecting six Chinese common alpha-thalassemia mutations.
Target capture sequencing
One microgram of genomic DNA from whole blood or the amniotic fluid was used for library preparation. Briefly, the genomic DNA was sonicated to yield fragments with an average size ranging from 200 to 250 bp. After end repair, “A”-overhanging, and adapter ligation, four cycles of PCR were performed with index primers. Cell-free DNA libraries were prepared using the KAPA library preparation kit (KAPA Biosystems) according to the manufacturer's protocol. After the ligation of adapter, eight cycles of PCR were performed with index primers. Customized NimbleGen SeqCap EZ probes were used to enrich the target region. After hybridization, 14-cycle PCR was used to amplify the library. Then, paired-end 101 bp sequencing was performed on the Illumina HiSeq™ 2500.
The customized probes covered 3.7 Mb of the target region, including the regions used for deduction of fetal genotypes in the alpha-thalassemia and beta-thalassemia mutations, which comprises the HBA1, HBA2, or HBB gene, the selected highly heterozygous SNPs distributed within the 1Mb flanking region of the gene of interest, and regions used for other research purposes. To increase the number of informative SNPs to be identified in different families, SNPs with the minor allele frequency between 0.3 and 0.5 in 1000 Genome project, dbSNP, and HapMap database were selected. Also, SNPs located in tandem repeat or segmental duplication regions (UCSC) were excluded. A total of 1140 SNPs in the alpha-thalassemia mutation region and 2001 SNPs in the beta-thalassemia mutation region were selected for probe design. The size of target region for alpha-thalassemia and beta-thalassemia analysis is about 154 and 287 kb, respectively.
Sequencing data process
The paired-end reads were mapped against the reference genome with the BWA software (0.7.12), and the low-quality reads (mapping quality <13, base quality <13) were discarded. The SNP calling for the family members was performed with GATK software and optimized to detect alleles with frequency larger than 1%, as described by Frampton et al. (2013).
Estimation of fetal DNA fraction in the maternal plasma
Fetal DNA fraction was calculated according to the formula: fetal DNA fraction = 2dfetus/(dmother+dfetus), where dmother stands for the counts of allele shared by the mother and the fetus, and dfetus stands for the counts of allele specifically detected in plasma samples.
Noninvasive deduction of fetal genotype
The genotype information of the parents and their first child was used to construct the parental haplotype and its linkage to the pathogenic allele following Mendel' laws as described by Meng et al. (2014). Haplotypes transmitted from the parents to the first child were called haplotype 0 (hap0), and those not inherited by the first child called haplotype 1 (hap1). Then, a HMM was constructed to deduce the maternal or paternal transmitted haplotype with the parental haplotype information and the base distribution information in the plasma, respectively. The paternal transmitted haplotype was deduced with SNPs homozygous in mother and heterozygous in father, while the maternal transmitted haplotype was deduced with SNPs heterozygous in mother and homozygous in father. The initial state distribution was set at {0.5,0.5}, and the transition probability matrix was set by the recombination rate between two adjacent sites calculated by the genetic distance data in HapMap. The observation probability matrix was calculated by allele counts at each phased site (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/gtmb). Then, the Viterbi algorithm was used to determine the most likely path with the observed data and to deduce the inherited maternal haplotype or paternal haplotype in the fetus (Fig. 1 and Supplementary Fig. S1) (Details can be seen in Supplementary Data).
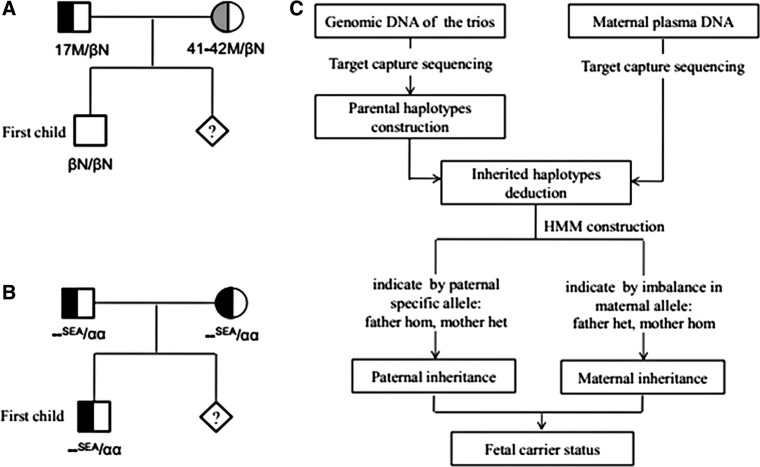
FIG. 1.
(A) The pedigree of HBB mutation in family A. (B) The pedigree of alpha-thalassemia mutation in family B. (C) The flowchart of the noninvasive prenatal diagnosis assays. HMM, hidden Markov model; SEA, Southeast Asian deletion.
Estimation of numbers of SNPs and sequence depth required for an accurate fetal genotype deduction
We analyzed the correlation between the accuracy of maternal haplotype deduction and the available numbers of informative SNPs. Briefly, the zygosity in each SNP locus of the proband and the fetal haplotype was randomly simulated. A binomial distribution model of the allele frequency constructed from the real plasma data of family A was used to simulate the read distribution in each locus of the plasma data. The inherited haplotype was deduced from the simulated data, and its accuracy was determined by comparing it with the simulated status. For status of SNP numbers ranging from 0 to 100, 1e7 times simulation experiments were performed in each status. Then, the accuracy was calculated as the percentage of the times that the fetal haplotype was correctly determined. To estimate the sequence depth for an accurate fetal haplotype deduction, 10%, 20%, 30%, 40%, 50%, 60%, and 70% of the raw data were randomly extracted for further analysis, and the locus genotype, which was incorrectly compared to that of amniotic fluid sample, was calculated then.
Results
Hematological parameters and genotypes of two recruited families with thalassemia
Both recruited families were from Guilin, Guangxi province in China. In family A, the parents are microcytic and hypochromic with increased levels of HbA2. The molecular diagnosis showed that the mother was a carrier of CD17 (A > T) mutation at HBB gene, the father was a carrier of CD41-42(-TTCT) mutation at HBB gene, and their first child did not carry any HBB gene mutation (Fig. 1A and Supplementary Table S2). In family B, the hematological parameters showed that the parents and their first child were all microcytic and hypochromic, but with normal hemoglobin electrophoresis results. Their molecular diagnosis results indicated that they were all carriers of the Southeast Asia-type alpha-thalassemia mutation (–SEA/N) (Fig. 1B and Supplementary Table S2). The maternal plasma of these two pregnant women was collected at 23 and 22 gestation weeks for noninvasive prenatal diagnosis assays. Their amniotic fluid was also collected for direct molecular diagnosis.
Noninvasive prenatal diagnosis of fetal genotype
The workflow is illustrated in Figure 1C. The basic statistics of sequence data is summarized in Table 1. The mean depth of genomic DNA sample is about 160× , ranging from 92× to 230× in the target region. The sequencing depth of plasma sample is about 747× in family A and 230.6× in family B.
Table 1.
Statistics of Target Region Sequencing Data
| Whole targeted region | Region used for fetal genotype deduction | SNP supported the transmitted haplotype in plasma | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case | Genotype | Mapped reads (Mb) | SNP marker | Mapped reads (Mb) | Dedup mean depth | depth ≥20 × | SNP marker | Phased SNP | M-hap0 | M-hap1 | F-hap0 | F-hap1 |
| Family A | ||||||||||||
| Mother | 17 M/βN | 18.77 | 12424 | 0.78 | 180.20 | 99.68% | 857 | 503 | 0 | 280 | 0 | 223 |
| Father | 41–42 M/βN | 21.66 | 12576 | 0.94 | 206.99 | 98.94% | 937 | |||||
| First child | βN/βN | 15.66 | 13014 | 0.53 | 133.26 | 99.44% | 834 | |||||
| Plasma | 17 M/41-42M | 151.40 | 12608 | 5.32 | 747.38 | 99.97% | 864 | |||||
| Amniotic fluid | 17 M/41-42M | 33.73 | 13152 | 1.05 | 256.42 | 98.93% | 862 | |||||
| Family B | ||||||||||||
| Mother | –SEA/αα | 20.78 | 12960 | 0.22 | 92.06 | 96.51% | 305 | 138 | 48 | 0 | 0 | 90 |
| Father | –SEA/αα | 23.54 | 12113 | 0.33 | 135.96 | 98.16% | 351 | |||||
| First child | –SEA/αα | 24.26 | 12341 | 0.37 | 151.11 | 98.57% | 338 | |||||
| Plasma | –SEA/–SEA | 100.41 | 13472 | 1.28 | 230.60 | 99.20% | 317 | |||||
| Amniotic fluid | –SEA/–SEA | 14.77 | 12639 | 0.30 | 129.93 | 89.03% | 315 | |||||
Dedup mean depth, mean depth after filtering of duplication reads; hap, haplotype; SEA, Southeast Asian.
Noninvasive prenatal diagnosis for family A
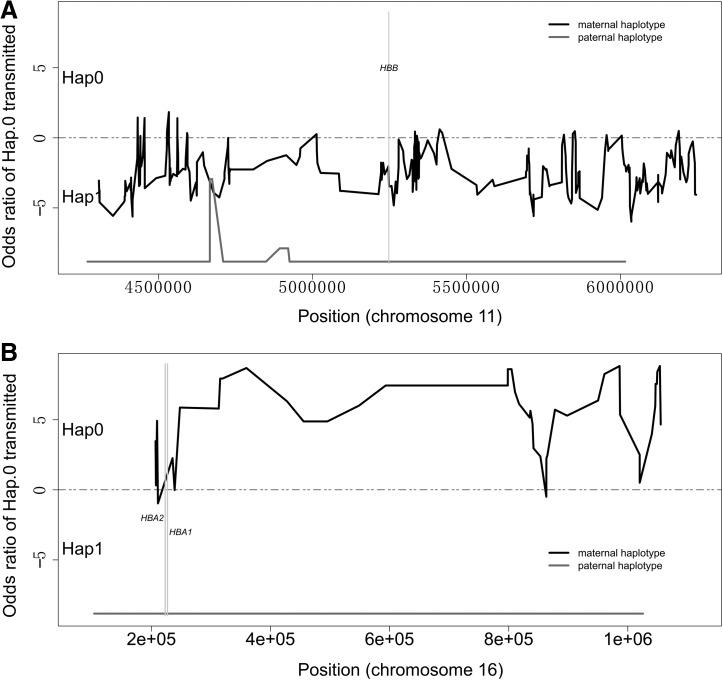
In family A, both parents were carriers of beta-thalassemia mutation, while their first child was normal in HBB gene (Fig. 1A). Thus, the father's haplotype 0 (F-hap0) and mother's haplotype 0 (M-hap0) were linked to the nonpathogenic allele. Father's haplotype 1 (F-hap1) was linked to the CD41-42(-TTCT) mutation in HBB, and the mother's haplotype 1 (M-hap1) was linked to the CD17 (A > T) mutation in HBB. The fraction of cell-free fetal DNA in the maternal plasma was estimated as 10.4%. A total of 857, 937, 834, and 864 SNPs were identified in the mother, the father, their first child, and the plasma analysis, respectively. Approximately 503 SNP markers were successfully phased and ready to be used for the noninvasive deduction of the fetal genotype in HBB (Table 1). About 223 SNPs were used in the paternal transmitted haplotype deduction. All of the SNPs suggested that the F-hap1, which was linked to CD41-42(-TTCT), was transmitted to the fetus. About 280 SNPs were used in maternal transmitted haplotype deduction, indicating that the CD17 (A > T)-linked M-hap1 was transmitted to the fetus (Fig. 2A). Thus, the fetal genotype is a compound heterozygote of CD17 (A > T) and CD41-42(-TTCT) mutations, and the baby will be affected by beta-thalassemia. This deduction was further confirmed by molecular diagnosis of the amniocentesis fluid. The detailed SNPs used for fetal genotype deduction are available in Supplementary Table S3.
FIG. 2.
(A) Noninvasive diagnosis results for family A. (B) Noninvasive diagnosis results for family B. hap0: the haplotype transmitted from the parents to the first child; hap1: the haplotype not inherited by the first child. The grey line represents the paternal haplotype prediction results, and the black lines the maternal haplotype prediction results. The grey line below zero indicates that the fetus inherited the hap1 from the father, and the black line above zero indicates that the fetus inherited the hap1 from the mother. Hap0, haplotype 0; Hap1, haplotype 1.
Noninvasive prenatal diagnosis for family B
In family B, the fraction of cell-free fetal DNA in the maternal plasma was estimated as 12.3%. A total of 305, 351, 338, and 317 SNP markers were detected in the mother, the father, their first child and the plasma data, respectively. Around 138 SNP markers were successfully phased and ready to be used for the noninvasive deduction of the fetal genotype in alpha-thalassemia mutation region (Table 2). As the mutations carried by the parents and the first child were the same (–SEA/αα), the origin of the normal allele was determined by the SNPs located within the breakpoint. A total of six SNPs located within the breakpoint were differentiated between the mother and the father, while their first child's genotypes in these loci were the same as the father's (Table 2), indicating that their first child inherits the normal allele from the father, the –SEA mutation from the mother. Thus, the F-hap0 and M-hap1 were linked to nonpathogenic allele, while F-hap1 and M-hap0 to the –SEA mutation. About 88 SNPs were used in paternal transmitted haplotype deduction, showing that the F-hap1 linked to the –SEA mutation was transmitted to the fetus. About 48 SNPs were used in maternal transmitted haplotype deduction demonstrating that the M-hap0 linked to the –SEA mutation was transmitted to the fetus (Fig. 2B). Thus, the fetus is homozygous of –SEA mutation and will be affected by the Hb Bart's syndrome. The detailed SNPs used for fetal genotype deduction are available in Supplementary Table S4.
Table 2.
The SNP Located Within the SEA Breakpoint in Family B
| Mother | Father | Proband | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Chromosome | Position | Reference allele | Alteration allele | Genotype | Allele depth | Genotype | Allele depth | Genotype | Allele depth |
| chr16 | 218735 | A | G | 1/1 | 0,85 | 0/0 | 116,0 | 0/0 | 131,1 |
| chr16 | 221057 | T | C | 1/1 | 0,64 | 0/0 | 101,0 | 0/0 | 102,0 |
| chr16 | 221126 | C | T | 1/1 | 0,64 | 0/0 | 86,0 | 0/0 | 98,1 |
| chr16 | 224619 | T | C | 1/1 | 1,97 | 0/0 | 115,0 | 0/0 | 152,0 |
| chr16 | 224647 | G | A | 1/1 | 0,94 | 0/0 | 111,0 | 0/0 | 170,0 |
| chr16 | 225653 | A | C | 1/1 | 3,97 | 0/0 | 137,1 | 0/0 | 136,0 |
The accuracy of fetal haplotype deduction
The capture sequencing performed with the amniocentesis fluid DNA of these two families showed that the SNPs of the amniocentesis fluid sample were 100% consistent to the deduced results (Supplementary Table S5).
Estimation of numbers of SNPs and sequence depth required for the accuracy of fetal genotype deduction
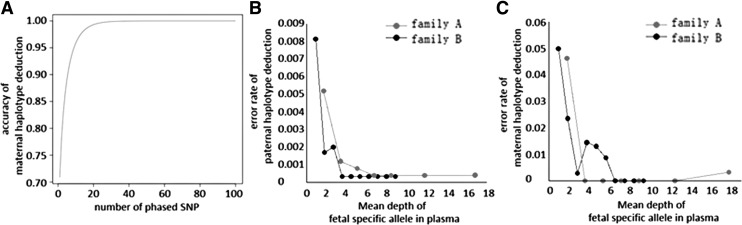
Paternal haplotype can easily be deduced by the existence of paternal-specific allele. However, maternal haplotype deduction is relatively more complicated. Therefore, only the accuracy of the maternal haplotype deduction with the number of informative SNPs was investigated. The simulation showed that the accuracy is increased with the numbers of available SNPs, ranging from 99% with 18 SNPs in haplotype deduction to 99.9% with 32 SNPs (Fig. 3A and Supplementary Table S6).
FIG. 3.
(A) Influence of numbers of SNPs on the accuracy of maternal haplotype deduction; (B) influence of the expected depth of fetal genome on the accuracy of paternal haplotype deduction; (C) influence of the expected depth of fetal genome on the accuracy of maternal haplotype deduction.
The analysis of sequence depth required to achieve accurate fetal haplotype deduction shows that when the expected depth for fetal genome reaches at least 10 × , the accuracy is higher than 99% (Fig. 3B, C). Thus, for family A, whose fetal DNA fraction is about 12.3%, a mean depth of 81 × is sufficient for its accurate deduction. And for family B, whose fetal DNA fraction is about 10.4%, a mean depth of 96 × is required.
Discussion
Main findings of this study
Given the high morbidity of alpha-thalassemia and beta-thalassemia in southern China, population screening and prenatal diagnosis are crucial to prevent the birth of affected children (Xiong et al., 2010). In this study, we demonstrated that haplotyping-assisted non-invasive prenatal diagnosis for alpha-thalassemia and beta-thalassemia can be achieved by capture sequencing of a small target region around the gene of interest for the detection of both SNV and large deletion-type mutations. One beta-thalassemia and one alpha-thalassemia case were successfully diagnosed by using our noninvasive prenatal diagnosis method with great consistency to the traditional invasive method using amniotic fluid.
Comparisons of previous studies
Generally, two kinds of methods were available for the noninvasive prenatal diagnosis of fetal thalassemia. One is to detect the presence of paternal pathogenic mutation in maternal plasma, which suggests that the fetus is exposed to a high risk of being affected. Xiong et al. reported one case of this type of noninvasive prenatal diagnosis using PCR and next-generation sequencing for the detection of paternal pathogenic mutation. Yan et al. (2011) and Ho et al. (2007) also reported exclusion of fetal homozygous alpha-thalassemia by the presence of father's SNP within the breakpoint. However, this method can only be used to exclude fetal risk, without knowing the fetus's exact genotype. The other is haplotype linkage-based noninvasive prenatal diagnosis, which could determine fetal genotypes with a high accuracy. Lam et al. reported noninvasive prenatal diagnosis of beta-thalassemia by the combined application of capture sequencing and haplotype analysis. However, the large target region in this report significantly increased the cost of sequencing, which may greatly prevent its clinical use. In addition, this method has not been applied to the detection of large deletion mutations yet.
In this study, we demonstrated that capture sequencing of highly heterozygous SNPs within the 1Mb flanking region of the gene of interest effectively shrinks the size of the target region, which can lead to a significant reduction of sequencing cost. In our study, the region used for alpha-thalassemia diagnosis is only 154 kb and for beta-thalassemia analysis only 287 kb. High heterozygous SNPs will increase the chance of getting enough informative SNPs. As the average recombination rate is about 1% per 1 Mb region, restricting the SNP within the 1 Mb flanking region of the gene of interest can reduce the probability of the occurrence of recombination events. Computer simulation suggested that 18 SNPs would be sufficient for an accurate maternal transmitted haplotype deduction, with the accuracy rate of 99%, and more SNPs are captured for analysis in our actual study. Moreover, the number of SNPs required for an accurate paternal transmitted haplotype deduction is even smaller given its less difficulty. Our study further suggested that capture sequencing and haplotype analysis could be used not only for noninvasive prenatal diagnosis of SNV or small INDEL but also for the detection of large deletions, including the SEA-type alpha-thalassemia deletion. Overall, capture sequencing of high heterozygous SNPs flanking the gene of interest is an effective way to reduce sequencing cost.
Limitations of the study
This newly developed method was validated with the successful diagnosis of one alpha-thalassemia and one beta-thalassemia case. Plasma samples were collected at 22 and 23 gestation weeks of the late second trimester, respectively. Non-invasive prenatal diagnosis (NIPD) in early gestation is very beneficial to clinical practice, as it can provide doctors and high-risk families more time to make decisions. Although fetal DNA fraction is variable among people, the overall fetal DNA fraction is lower in early gestation week. In this study, the fetal fraction was 12.3% in alpha-thalassemia case and 10.4% in beta-thalassemia case. As the accuracy is affected by fetal fraction, in the follow-up studies, more cases, especially samples collected in early gestation weeks, should be tested to thoroughly evaluate the method before its real clinical application.
Implications for clinical practice
Unlike the invasive prenatal diagnosis procedure, the noninvasive method in this study only requires 5–10 mL peripheral blood from the pregnant women and thereby greatly reduces traditional invasive procedures, which may be a great benefit to both pregnant women and hospitals.
Supplementary Material
Acknowledgments
We thank all the individuals who took part in the study. This work was supported by a grant from the National Natural Science Foundation of China (NSFC81360092) and by the Project of Technology of Guilin (20120121-1-12).
Author Disclosure Statement
No competing financial interests exist.
References
- Bianchi DW, Platt LD, Goldberg JD, et al. (2012) Genome-wide fetal aneuploidy detection by maternal plasma DNA sequencing. Obstet Gynecol 119:890–901 [DOI] [PubMed] [Google Scholar]
- Chiu RW, Lau TK, Leung TN, et al. (2002) Prenatal exclusion of β thalassaemia major by examination of maternal plasma. Lancet 360:998–1000 [DOI] [PubMed] [Google Scholar]
- Frampton GM, Fichtenholtz A, Otto GA, et al. (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbiati S, Brisci A, Lalatta F, et al. (2011) Full COLD-PCR protocol for noninvasive prenatal diagnosis of genetic diseases. Clin Chem 57:136–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge H, Huang X, Li X, et al. (2013) Noninvasive prenatal detection for pathogenic CNVs: the application in alpha-thalassemia. PLoS One 8:e67464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SS, Chong SS, Koay ES, et al. (2007) Microsatellite markers within –SEA breakpoints for prenatal diagnosis of HbBarts hydrops fetalis. Clin Chem 53:173–179 [DOI] [PubMed] [Google Scholar]
- Jin Y, Miao Z, Ge J, et al. (2014) Prenatal diagnosis of fetal chromosome aneuploidy by massively parallel genomic sequencing. Zhonghua Yi Xue Za Zhi 94:1788–1790 [PubMed] [Google Scholar]
- Kan YW, Nathan DG, Cividalii G, et al. (1974) Intrauterine diagnosis of thalassemia. Ann N Y Acad Sci 232:145–151 [DOI] [PubMed] [Google Scholar]
- Ko TM, Hsieh FJ, Hsu PM, et al. (1989) Prenatal diagnosis of Chinese homozygous alpha-thalassaemia 1 and haemoglobin H disease by analysis of alpha- and phi zeta-globin genes in chorionic villi and amniocytes. Prenat Diagn 9:715–725 [DOI] [PubMed] [Google Scholar]
- Lam KW, Jiang P, Liao GJ, et al. (2012) Noninvasive prenatal diagnosis of monogenic diseases by targeted massively parallel sequencing of maternal plasma: application to beta-thalassemia. Clin Chem 58:1467–1475 [DOI] [PubMed] [Google Scholar]
- Lim JH, Park SY, and Ryu HM. (2013) Non-invasive prenatal diagnosis of fetal trisomy 21 using cell-free fetal DNA in maternal blood. Obstet Gynecol Sci 56:58–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo YM, Chan KC, Sun H, et al. (2010) Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci Transl Med 2:61ra91. [DOI] [PubMed] [Google Scholar]
- Lo YM, Corbetta N, Chamberlain PF, et al. (1997) Presence of fetal DNA in maternal plasma and serum. Lancet 350:485–487 [DOI] [PubMed] [Google Scholar]
- Lun FM, Tsui NB, Chan KC, et al. (2008) Noninvasive prenatal diagnosis of monogenic diseases by digital size selection and relative mutation dosage on DNA in maternal plasma. Proc Natl Acad Sci U S A 105:19920–19925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv W, Wei X, Guo R, et al. (2015) Noninvasive prenatal testing for Wilson disease by use of circulating single-molecule amplification and resequencing technology (cSMART). Clin Chem 61:172–181 [DOI] [PubMed] [Google Scholar]
- Meng M, Li X, Ge H, et al. (2014) Noninvasive prenatal testing for autosomal recessive conditions by maternal plasma sequencing in a case of congenital deafness. Genet Med 16:972–976 [DOI] [PubMed] [Google Scholar]
- New MI, Tong YK, Yuen T, et al. (2014) Noninvasive prenatal diagnosis of congenital adrenal hyperplasia using cell-free fetal DNA in maternal plasma. J Clin Endocrinol Metab 99:E1022–E1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlandi F, Jakil C, Damiani G, et al. (1988) First trimester fetal blood sampling. Acta Eur Fertil 19:23–24 [PubMed] [Google Scholar]
- Papasavva TE, Lederer CW, Traeger-Synodinos , et al. (2013) A minimal set of SNPs for the noninvasive prenatal diagnosis of β-thalassaemia. Ann Hum Genet 77:115–124 [DOI] [PubMed] [Google Scholar]
- Tsang JC, Charoenkwan P, Chow KC, et al. (2007) Mass spectrometry-based detection of hemoglobin E mutation by allele-specific base extension reaction. Clin Chem 53:2205–2209 [DOI] [PubMed] [Google Scholar]
- Tungwiwat W, Fucharoen S, Fucharoen G, et al. (2006) Development and application of a real-time quantitative PCR for prenatal detection of fetal alpha(0)-thalassemia from maternal plasma. Ann N Y Acad Sci 1075:103–107 [DOI] [PubMed] [Google Scholar]
- Xiong F, Sun M, Zhang X, et al. (2010) Molecular epidemiological survey of haemoglobinopathies in the Guangxi Zhuang Autonomous Region of southern China. Clin Genet 78:139–148 [DOI] [PubMed] [Google Scholar]
- Xu Y, Li X, Ge HJ, et al. (2015) Haplotype-based approach for noninvasive prenatal tests of Duchenne muscular dystrophy using cell-free fetal DNA in maternal plasma. Genet Med 17:889–896 [DOI] [PubMed] [Google Scholar]
- Yan TZ, Mo QH, Cai R, et al. (2011) Reliable detection of paternal SNPs within deletion breakpoints for non-invasive prenatal exclusion of homozygous α-thalassemia in maternal plasma. PLoS One 6:e24779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Lim BC, Byeun J, et al. (2015) Noninvasive prenatal diagnosis of duchenne muscular dystrophy: comprehensive genetic diagnosis in carrier, proband, and fetus. Clin Chem 61:829–837 [DOI] [PubMed] [Google Scholar]
- You Y, Sun Y, Li X, et al. (2014) Integration of targeted sequencing and NIPT into clinical practice in a Chinese family with maple syrup urine disease. Genet Med 16:594–600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.