Summary
Individuals with severe, undiagnosed developmental disorders (DDs) are enriched for damaging de novo mutations (DNMs) in developmentally important genes. We exome sequenced 4,293 families with individuals with DDs, and meta-analysed these data with another 3,287 individuals with similar disorders. We show that the most significant factors influencing the diagnostic yield of DNMs are the sex of the affected individual, the relatedness of their parents, whether close relatives are affected and parental ages. We identified 94 genes enriched for damaging DNMs, including 14 without previous compelling evidence. We have characterised the phenotypic diversity among these disorders. We estimate that 42% of our cohort carry pathogenic DNMs in coding sequences, and approximately half disrupt gene function, with the remainder resulting in altered-function. We estimate that developmental disorders caused by DNMs have an average birth prevalence of 1 in 213 to 1 in 448, depending on parental age. Given current global demographics, this equates to almost 400,000 children born per year.
Keywords: De novo mutation, Developmental Disease, Seizures, Intellectual Disability, PhenIcons, Average Faces, ANKRD11, ARID1B, KMT2A, DDX3X, ADNP, MED13L, DYRK1A, EP300, SCN2A, SETD5, KCNQ2, MECP2, SYNGAP1, ASXL3, SATB2, TCF4, CDK13, CREBBP, DYNC1H1, FOXP1, PPP2R5D, PURA, CTNNB1, KAT6A, SMARCA2, STXBP1, EHMT1, ITPR1, KAT6B, NSD1, SMC1A, TBL1XR1, CASK, CHD2, CHD4, HDAC8, USP9X, WDR45, AHDC1, CSNK2A1, GNAI1, GNAO1, HNRNPU, KANSL1, KIF1A, MEF2C, PACS1, SLC6A1, CNOT3, CTCF, EEF1A2, FOXG1, GATAD2B, GRIN2B, IQSEC2, POGZ, PUF60, SCN8A, TCF20, BCL11A, BRAF, CDKL5, NFIX, PTPN11, AUTS2, CHAMP1, CNKSR2, DNM1, KCNH1, NAA10, PPM1D, ZBTB18, ZMYND11, ASXL1, COL4A3BP, KCNQ3, MSL3, MYT1L, PDHA1, PPP2R1A, SMAD4, TRIO, WAC, CHD8, GABRB3, KDM5B, PTEN, QRICH1, SET, ZC4H2, ALG13, SCN1A, SUV420H1, SLC35A2
Approximately 2-5% of children are born with major congenital malformations and/or manifest severe neurodevelopmental disorders during childhood1,2. While diverse mechanisms can cause such developmental disorders, including gestational infection and maternal alcohol consumption, damaging genetic variation in developmentally important genes has a major contribution. Several recent studies have identified a substantial causal role for DNMs not present in either parent3–16. Despite the identification of many developmental disorders caused by DNMs, it is generally accepted that many more such disorders await discovery15, and the overall contribution of DNMs to developmental disorders is not known. Moreover, some pathogenic DNMs completely ablate the function of the encoded protein, whereas others alter the function of the encoded protein17; the relative contributions of these two mechanistic classes is also not known.
We recruited 4,293 individuals to the Deciphering Developmental Disorders (DDD) study15 via genetics services of the UK National Health Service and Republic of Ireland. Each of these individuals was referred with a severe undiagnosed developmental disorder and most were the only affected family member. Most (81%) individuals had been screened for large pathogenic deletions and duplications. We systematically phenotyped these individuals and sequenced the exomes of these individuals and their parents. Growth measurements, family history, and developmental milestones were collected, and detailed clinical phenotypes were captured using Human Phenotype Ontology (HPO) terms. Analyses of 1,133 of these trios were described previously15,18. We generated a high sensitivity set of 8,361 candidate DNMs in coding or splicing sequence (mean of 1.95 DNMs per proband), while removing systematic erroneous calls (Supplementary Table 1). This rate of candidate DNMs per proband is higher than other studies3–15, because we wish to maintain high sensitivity, and can address lower specificity via subsequent validation. 1,624 genes contained two or more DNMs in unrelated individuals.
Twenty-three percent of individuals had likely pathogenic protein-truncating or missense DNMs within the clinically curated set of genes robustly associated with dominant developmental disorders18. We investigated factors associated with whether an individual had a likely pathogenic DNM in these curated genes (Figure 1a, b, Supplementary Table 1). We observed that males had a lower chance of carrying a likely pathogenic DNM (P = 1.6 x 10-4; OR 0.75, 0.65 - 0.87 95% CI), as has also been observed in autism19. We also observed increased likelihood of having a pathogenic DNM with the extent of speech delay (P = 0.00115), but not other indicators of severity relative to the rest of the cohort. Individuals with other affected family members were less likely to have pathogenic DNMs (affected siblings: P = 7.3 x 10-18, affected parents: P = 5.7 x 10-9), and individuals who were from self-declared consanguineous unions were less likely to have a pathogenic DNM (P = 8.0 x 10-11). Furthermore, the total genomic extent of autozygosity (due to parental relatedness) was negatively correlated with the likelihood of having a pathogenic DNM (P = 1.7 x 10-7), for every log10 increase in autozygous length, the probability of having a pathogenic DNM dropped by 7.5%, likely due to increasing burden of recessive causation (Figure 1c). Nonetheless, 6% of individuals with autozygosity equivalent to a first cousin union or greater had a plausibly pathogenic DNM, underscoring the importance of considering de novo causation in all families.
Figure 1.
Association of phenotypes with presence of likely pathogenic de novo mutations (DNMs). Association of phenotypes with presence of likely pathogenic de novo mutations (DNMs). a, Odds ratios for binary phenotypes. Positive odds ratios are associated with increased risk of pathogenic DNMs when the phenotype is present. P-values are given for a Fisher’s Exact test. b, Beta coefficients from logistic regression of quantitative phenotypes versus presence of a pathogenic DNM. All phenotypes aside from length of autozygous regions were corrected for gender as a covariate. The developmental milestones (age to achieve first words, walk independently, sit independently and social smile) were log-scaled before regression. The growth parameters (height, birthweight and occipitofrontal circumference (OFC)) were evaluated as absolute distance from the median. c, Relationship between length of autozygous regions chance of having a pathogenic DNM. The regression line is plotted as the dark gray line. The 95% confidence interval for the regression is shaded gray. The autozygosity lengths expected under different degrees of consanguineous unions are shown as vertical dashed lines. n, number of individuals in each autozygosity group. d, Relationship between age of fathers at birth of child and number of high confidence DNMs. n, number of high confidence DNMs. e, Relationship between age of mothers at birth of child and number of high confidence DNMs. Error bars indicate 95% c.i. n, number of high confidence DNMs.
Paternal age has been shown to be the primary factor influencing the number of DNMs in a child20,21, and thus is expected to be a risk factor for pathogenic DNMs. Paternal age was only weakly associated with likelihood of having a pathogenic DNM (P = 0.016). However, focusing on the minority of DNMs that were truncating and missense variants in known DD-associated genes limits our power to detect such an effect. Analysing all 8,409 high confidence exonic and intronic autosomal DNMs confirmed a strong paternal age effect (P = 1.4 x 10-10, 1.53 DNMs/year, 1.07-2.01 95% CI), as well as highlighting a weaker, independent, maternal age effect (P = 0.0019, 0.86 DNMs/year, 0.32-1.40 95% CI, Figure 1d,e), as has recently been described in whole genome analyses22. These genome-wide estimates were scaled from exome-based estimates, of 0.0306 DNMs/year paternal effect and 0.0172 DNMs/year maternal effect.
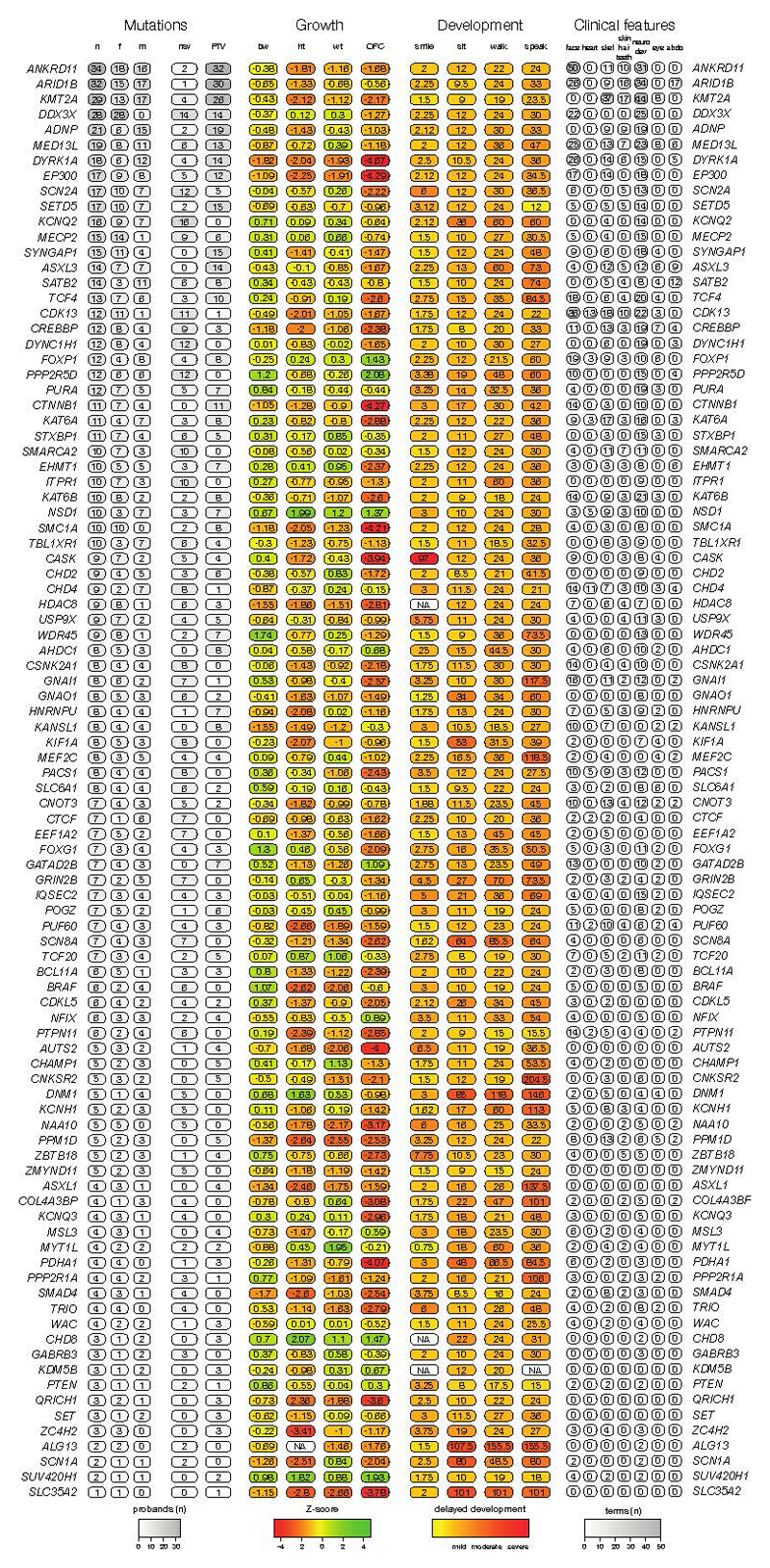
We identified genes significantly enriched for damaging DNMs by comparing the observed gene-wise DNM count to that expected under a null mutation model23, as described previously15. We combined this analysis with 4,224 published DNMs in 3,287 affected individuals from thirteen exome or genome sequencing studies (Supplementary Table 2)3–14 that exhibited a similar excess of DNMs in our curated set of DD-associated genes (Extended Data Figure 1). We found 93 genes with genome-wide significance (P < 5 × 10-7, Figure 2), 80 of which had prior evidence of DD-association (Supplementary Table 3). We have developed visual summaries of the phenotypes associated with each gene to facilitate clinical use. In addition, we created anonymised average face images from individuals with DNMs in genome-wide significant genes (Figure 2) from ordinary (2D) clinical photos using previously validated software24. These images highlight facial dysmorphologies specific to certain genes. After careful review by two experienced clinical geneticists, average face images for twelve genes were determined to be truly anonymised and of sufficient quality.To assess any increase in power to detect novel DD-associated genes, we excluded individuals with likely pathogenic variants in known DD-associated genes15, leaving 3,158 probands from our cohort, along with 2,955 probands from the meta-analysis studies. In this subset, fourteen genes for which no statistically-compelling prior evidence for DD causation was available achieved genome-wide significance: CDK13, CHD4, CNOT3, CSNK2A1, GNAI1, KCNQ3, MSL3, PPM1D, PUF60, QRICH1, SET, SUV420H1, TCF20, and ZBTB18 (P < 5 x 10-7, Table 1, Extended Data Figure 4). The clinical features associated with these newly confirmed disorders are summarised in Extended Data Figure 2, Extended Data Figure 3 and Supplementary Information. QRICH1 would not achieve genome-wide significance without excluding individuals with likely pathogenic variants in DD-associated genes. In addition to discovering novel DD-associated genes, we identified several new disorders linked to known DD-associated genes, but with different modes of inheritance or molecular mechanisms. We found USP9X and ZC4H2 had a genome-wide significant excess of DNMs in female probands, indicating these genes have X-linked dominant modes of inheritance in addition to previously reported X-linked recessive mode of inheritance in males25,26. In addition, we found truncating mutations in SMC1A were strongly associated with a novel seizure disorder (P = 6.5 x 10-19), while in-frame/missense mutations in SMC1A with dominant negative effects27 are a known cause of Cornelia de Lange Syndrome (CdLS). Individuals with truncating mutations in SMC1A lacked the characteristic facial dysmorphology of CdLS.
Figure 2.
Genes exceeding genome-wide significance. Manhattan plot of combined P-values across all tested genes. The red dashed line indicates the threshold for genome-wide significance (P < 7 x 10-7). Genes exceeding this threshold have HGNC symbols labelled. De-identified realistic average (‘composite’) faces were generated using previously validated software24 from clinical photos from individuals with DNMs in the same gene, and are shown here for the six most-significantly associated genes. Confirmation of de-identification was performed by careful review by two experienced clinical geneticists. Each face was generated from clinical photos of more than ten children.
Table 1.
Genes achieving genome-wide significant statistical evidence without previous compelling evidence for being developmental disorder genes. The numbers of unrelated individuals with independent de novo mutations (DNMs) are given for protein truncating variants (PTV) and missense variants. Counts of individuals in other cohorts are given in brackets if present. The P-value reported is the minimum P-value from the testing of the DDD dataset or the meta-analysis dataset. The subset providing the P-value is also listed. Mutations are considered clustered if the P-value from proximity clustering of DNMs is less than 0.01.
| Gene | Missense | PTV | P-value | Test | Clustering |
|---|---|---|---|---|---|
| CDK13 | 10 | 1 | 3.2 x 10-19 | DDD | Yes |
| GNAI1 | 7 (1) | 1 | 2.1 x 10-13 | DDD | No |
| CSNK2A1 | 7 | 0 | 1.4 x 10-12 | DDD | Yes |
| PPM1D | 0 | 5 (1) | 6.3 x 10-12 | Meta | No |
| CNOT3 | 5 | 2 (1) | 5.2 x 10-11 | DDD | Yes |
| MSL3 | 0 | 4 | 2.2 x 10-10 | DDD | No |
| KCNQ3 | 4 (3) | 0 | 3.4 x 10-10 | Meta | Yes |
| ZBTB18 | 1 (1) | 4 | 1.4 x 10-9 | DDD | No |
| PUF60 | 4 (1) | 3 | 2.6 x 10-9 | DDD | No |
| TCF20 | 1 | 5 | 2.7 x 10-9 | DDD | No |
| SUV420H1 | 0 (2) | 2 (3) | 2.9 x 10-9 | Meta | No |
| CHD4 | 8 (1) | 1 | 7.6 x 10-9 | DDD | No |
| SET | 0 | 3 | 1.2 x 10-7 | DDD | No |
| QRICH1 | 0 | 3 (1) | 3.6 x 10-7 | Meta | No |
We then explored two approaches for integrating phenotypic data into disease gene association: statistical assessment of Human Phenotype Ontology (HPO) term similarity between individuals sharing candidate DNMs in the same gene (as we described previously28) and phenotypic stratification based on specific clinical characteristics. Combining genetic evidence and HPO term similarity increased the significance of some known DD-associated genes. However, significance decreased for a larger number of genes causing severe DD but associated with non discriminative HPO terms (Extended Data Figure 5a). Although we did not incorporate categorical phenotypic similarity in the gene discovery analyses described above, the systematic acquisition of phenotypic data on affected individuals within DDD enabled aggregate representations to be created for each gene achieving genome-wide significance. We present these in the form of icon-based summaries of growth and developmental milestones (PhenIcons), heatmaps of the recurrently coded HPO terms and, where photos for at least ten children with mutations in the same gene were available, an anonymised average facial representation (Supplementary Information).
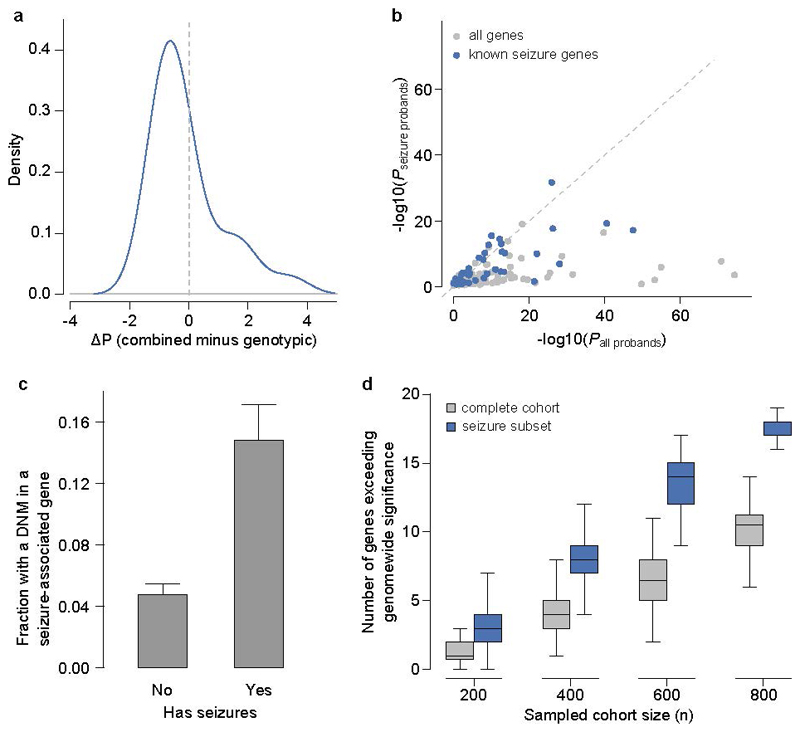
Twenty percent of individuals had HPO terms which indicated seizures and/or epilepsy. We compared analysis within this phenotypically stratified group with gene-wise analyses of the entire cohort, to see if it increased power to detect known seizure-associated genes (Extended Data Figure 5b). Fifteen seizure-associated genes were genome-wide significant in both the seizure-only and the entire-cohort analyses. Nine seizure-associated genes were genome-wide significant in the entire cohort but not in the seizure subset. Of the 285 individuals with truncating or missense DNMs in known seizure-associated genes, 56% of individuals had no coded terms related to seizures/epilepsy. These findings suggest that the power of increased sample size far outweighs specific phenotypic expressivity due to the shared genetic etiology between individuals with and without epilepsy in our cohort. Despite this, nearly three times as many individuals with seizures had a DNM in a seizure-associated gene compared to individuals without seizures (Extended Data Figure 5c). At matched sample sizes, more genes exceeded genomewide significance in seizure samples than in unstratified samples (Extended Data Figure 5d). This highlights the cost-benefit of recruiting a phenotypically more homogenous cohort.
The large number of genome-wide significant genes identified in the analyses above allows us to compare empirically different experimental strategies for novel gene discovery in a genetically heterogeneous cohort. We compared the power of exome and genome sequencing to detect genome-wide significant genes, assuming that budget and not samples are limiting, under different scenarios of cost ratios and sensitivity ratios (Extended Data Figure 6a). At current cost ratios (exome costs 30-40% of a genome) and with a plausible sensitivity differential (genome detects 5% more exonic variants than exome29) exome sequencing detects more than twice as many genome-wide significant genes. These empirical estimates were consistent with power simulations for identifying dominant loss-of-function genes (Extended Data Figure 6b). In summary, while genome sequencing gives greatest sensitivity to detect pathogenic variation in a single individual (or outside of the coding region), exome sequencing is more powerful for novel disease gene discovery (and, analogously, likely delivers lower cost per diagnosis currently).
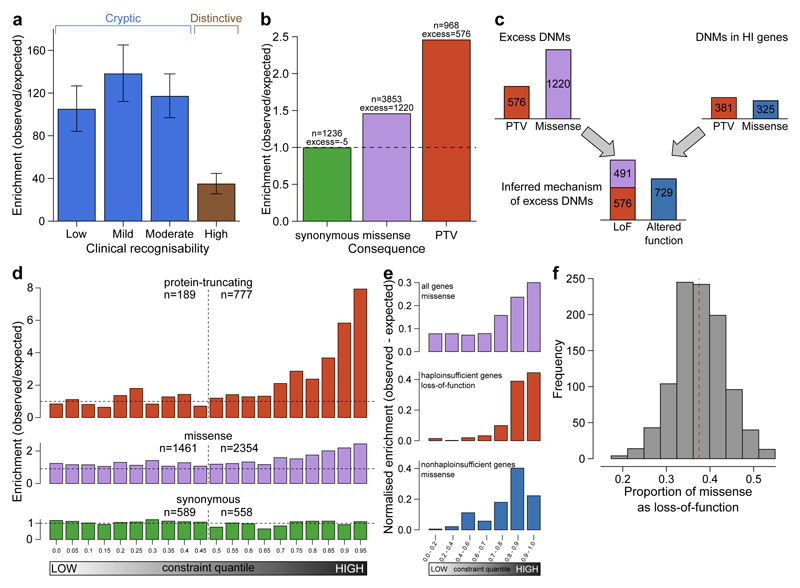
Our previous simulations suggested that analysis of a cohort of 4,293 DDD families ought to be able to detect approximately half of all haploinsufficient DD-associated genes at genome-wide significance15. Empirically, we have identified 47% (50/107) of haploinsufficient genes previously robustly associated with neurodevelopmental disorders18. We hypothesised that genetic testing prior to recruitment into our study may have depleted the cohort of the most clinically recognisable disorders. Indeed, we observed that the genes associated with the most clinically recognisable disorders were associated with a significant, three-fold lower enrichment of truncating DNMs than other DD-associated genes (~40-fold enrichment vs ~120-fold enrichment, Figure 3a). Removing these most recognisable disorders from the analysis, we identified 55% (42/76) of the remaining haploinsufficient DD-associated genes. The known DD-associated haploinsufficient genes that did not reach genome-wide significance were clearly enriched for those with lower mutability, which we would expect to lower power to detect in our analyses. We identified DD-associated genes (e.g. NRXN2) with high mutability, low clinical recognisability and yet no signal of enrichment for DNMs in our cohort, as assessed by ΔAIC (Extended Data Figure 7, Supplementary Table 4). Our analyses call into question whether these genes really are associated with haploinsufficient neurodevelopmental disorders and highlights the potential for well-powered gene discovery analyses to refute prior credence regarding disease gene associations or prior inferences regarding an underlying haploinsufficient mechanism.
Figure 3.
Excess of de novo mutations (DNMs). a, Enrichment ratios of observed to expected loss-of-function DNMs by clinical recognisability for dominant haploinsufficient neurodevelopmental genes as judged by two consultant clinical geneticists. Error bars indiciate 95% CI. b, Enrichment of DNMs by consequence normalised relative to the number of synonymous DNMs. c, Proportion of excess DNMs with loss-of-function or altered-function mechanisms. Proportions are derived from numbers of excess DNMs by consequence, and numbers of excess truncating and missense DNMs in dominant haploinsufficient genes. d, Enrichment ratios of observed to expected DNMs by pLI constraint quantile for loss-of-function, missense and synonymous DNMs. Counts of DNMs in each lower and upper half of the quantiles are provided. e, Normalised excess of observed to expected DNMs by pLI constraint quantile. This includes missense DNMs within all genes, loss-of-function including missense DNMs in dominant haploinsufficient genes and missense DNMs in dominant nonhaploinsufficient genes (genes with dominant negative or activating mechanisms). f, Proportion of excess missense DNMs with a loss-of-function mechanism. The red dashed line indicates the proportion in observed excess DNMs at the optimal goodness-of-fit. The histogram shows the frequencies of estimated proportions from 1000 permutations, assuming the observed proportion is correct.
We estimated the likely prevalence of pathogenic missense and truncating DNMs within our cohort by increasing the stringency of called DNMs until the observed synonymous DNMs equated that expected under the null mutation model (Extended Data Figure 8a), then quantifying the excess of observed missense and truncating DNMs across all genes (Figure 3b). We observed an excess of 576 truncating and 1,220 missense mutations, suggesting 41.8% (1,796/4,293) of the cohort has a pathogenic DNM. This estimate of the number of excess missense and truncating DNMs in our cohort is robust to varying the stringency of DNM calling (Extended Data Figure 8b). The vast majority of synonymous DNMs are likely to be benign, as evidenced by them being distributed uniformly (Figure 3d) among genes irrespective of their tolerance of truncating variation in the general population (as quantified by the probability of being LoF-intolerant (pLI) metric30). By contrast, missense and truncating DNMs are significantly enriched in genes with the highest probabilities of being intolerant of truncating variation (Figure 3d). The pLI-based distributions were similar to distributions which used functional constraint (Extended Data Figure 9)31. Only 51% (923/1,796) of these excess missense and truncating DNMs are located in DD-associated dominant genes, with the remainder likely to affect genes not yet associated with DDs. A much higher proportion of the excess truncating DNMs (71%) than missense DNMs (42%) affected known DD-associated genes. This suggests that whereas most haploinsufficient DD-associated genes have already been identified, many DD-associated genes characterised by pathogenic missense DNMs remain to be discovered.
Understanding the mechanism of action of a monogenic disorder is an important prerequisite for designing therapeutic strategies32. We sought to estimate the relative proportion of altered-function and loss-of-function mechanisms among the excess DNMs in our cohort, by assuming that the vast majority of truncating mutations operate by a loss-of-function mechanism and using two independent approaches to estimate the relative contribution of the two mechanisms among the excess missense DNMs (Methods). First, we used the observed ratio of truncating and missense DNMs within haploinsufficient DD-associated genes to estimate the proportion of the excess missense DNMs that likely act by loss-of-function (Figure 3c). This approach estimated that 59% (55 - 64% 95% CI) of excess missense and truncating DNMs operate by loss-of-function, and 41% by altered-function. Second, we took advantage of the different population genetic characteristics of known altered-function and loss-of-function DD-associated genes. Specifically, we observed that these two classes of DD-associated genes are differentially depleted of truncating variation in individuals without overt developmental disorders (pLI metric30). We modelled the observed pLI distribution of excess missense DNMs as a mixture of the pLI distributions of known altered-function and loss-of-function DD-associated genes (Figure 3e, f), and estimated that 63% (50 - 76% 95% CI) of excess missense DNMs likely act by altered-function mechanisms. Incorporating the truncating DNMs operating by a loss-of-function mechanism, this approach estimated that 57% (48 - 66% 95% CI) of excess missense and truncating DNMs operate by loss-of-function and 43% by altered-function.
We estimated the birth prevalence of monoallelic developmental disorders by using the germline mutation model to calculate the expected cumulative germline mutation rate of truncating DNMs in haploinsufficient DD-associated genes and scaling this upwards based on the composition of excess DNMs in the DDD cohort described above (see Methods), correcting for disorders that are under-represented in our cohort as a result of prior genetic testing (e.g. clinically-recognisable disorders and large pathogenic CNVs identified by prior chromosomal microarray analysis). This gives a mean prevalence estimate of 0.34% (0.31-0.37 95% CI), or 1 in 295 births. By factoring in the paternal and maternal age effects on the mutation rate (Figure 1) we modelled age-specific estimates of birth prevalence (Figure 4) that range from 1 in 448 (both mother and father aged 20) to 1 in 213 (both mother and father aged 45). Assuming a yearly global birth rate of 18.6 live births/1000 individuals, and a mean age when giving birth of 26.6 years, nearly 400,000 of the 140 million annual births will have a developmental disorder caused by a DNM.
Figure 4.
Prevalence of live births with developmental disorders caused by dominant de novo mutations (DNMs). The prevalence within the general population is provided as percentage for combinations of parental ages, extrapolated from the maternal and paternal rates of DNMs. Distributions of parental ages within the DDD cohort and the UK population are shown at the matching parental axis.
In summary, we have shown that de novo mutations account for approximately half of the genetic architecture of severe developmental disorders, and are split roughly equally between loss-of-function and altered-function. Whereas most haploinsufficient DD-associated genes have already been identified, currently many activating and dominant negative DD-associated genes have eluded discovery. This elusiveness likely results from these disorders being individually rarer, being caused by a relatively small number of missense mutations within each gene. It would be valuable to estimate the penetrance of de novo mutations in the genes we identified exceeding genome-wide significance, but we cannot formally assess penetrance with our data. Future evaluations could integrate depletion of damaging variation in large healthy populations with patterns of segregation in affected families. Discovery of the remaining dominant developmental disorders requires larger studies and novel, more powerful, analytical strategies for disease-gene association that leverage gene-specific patterns of population variation, specifically the observed depletion of damaging variation. The integration of accurate and complete quantitative and categorical phenotypic data into the analysis will improve the power to identify ultrarare DD with distinctive clinical presentations. We have estimated the mean birth prevalence of dominant monogenic developmental disorders to be around 1 in 295, which is greater than the combined impact of trisomies 13, 18 and 2133 and highlights the cumulative population morbidity and mortality imposed by these individually rare disorders.
Methods
Family recruitment
At 24 clinical genetics centers within the United Kingdom (UK) National Health Service and the Republic of Ireland, 4,293 patients with severe, undiagnosed developmental disorders and their parents (4,125 families) were recruited and systematically phenotyped. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South Research Ethics Committee and GEN/284/12, granted by the Republic of Ireland Research Ethics Committee). Families gave informed consent for participation.
Clinical data (growth measurements, family history, developmental milestones, etc.) were collected using a standard restricted-term questionnaire within DECIPHER34, and detailed developmental phenotypes for the individuals were entered using Human Phenotype Ontology (HPO) terms35. Saliva samples for the whole family and blood-extracted DNA samples for the probands were collected, processed and quality controlled as previously described15.
Exome sequencing
Genomic DNA (approximately 1 μg) was fragmented to an average size of 150 base-pairs (bp) and subjected to DNA library creation using established Illumina paired-end protocols. Adaptor-ligated libraries were amplified and indexed via polymerase chain reaction (PCR). A portion of each library was used to create an equimolar pool comprising eight indexed libraries. Each pool was hybridized to SureSelect ribonucleic acid (RNA) baits (Agilent Human All-Exon V3 Plus with custom ELID C0338371 and Agilent Human All-Exon V5 Plus with custom ELID C0338371) and sequence targets were captured and amplified in accordance with the manufacturer's recommendations. Enriched libraries were subjected to 75-base paired-end sequencing (Illumina HiSeq) following the manufacturer's instructions.
Alignment and calling single nucleotide variants, insertions and deletions
Mapping of short-read sequences for each sequencing lanelet was carried out using the Burrows-Wheeler Aligner (BWA; version 0.59)36 backtrack algorithm with the GRCh37 1000 Genomes Project phase 2 reference (also known as hs37d5). Sample-level BAM improvement was carried out using the Genome Analysis Toolkit (GATK; version 3.1.1)37 and SAMtools (version 0.1.19)38. This consisted of a realignment of reads around known and discovered indels followed by base quality score recalibration (BQSR), with both steps performed using GATK. Lastly, SAMtools calmd was applied and indexes were created.
Known indels for realignment were taken from the Mills Devine and 1000 Genomes Project Gold set and the 1000 Genomes Project phase low-coverage set, both part of the GATK resource bundle (version 2.2). Known variants for BQSR were taken from dbSNP 137, also part of the GATK resource bundle. Finally, single nucleotide variants (SNVs) and indels were called using the GATK HaplotypeCaller (version 3.2.2); this was run in multisample calling mode using the complete data set. GATK Variant Quality Score Recalibration (VQSR) was then computed on the whole data set and applied to the individual-sample variant calling format (VCF) files. DeNovoGear (version 0.54)39 was used to detect SNV, insertion and deletion de novo mutations (DNMs) from child and parental exome data (BAM files).
Variant annotation
Variants in the VCF were annotated with minor allele frequency (MAF) data from a variety of different sources. The MAF annotations used included data from four different populations of the 1000 Genomes Project40 (AMR, ASN, AFR and EUR), the UK10K cohort, the NHLBI GO Exome Sequencing Project (ESP), the Non-Finnish European (NFE) subset of the Exome Aggregation Consortium (ExAC) and an internal allele frequency generated using unaffected parents from the cohort.
Variants in the VCF were annotated with Ensembl Variant Effect Predictor (VEP)41 based on Ensembl gene build 76. The transcript with the most severe consequence was selected and all associated VEP annotations were based on the predicted effect of the variant on that particular transcript; where multiple transcripts shared the same most severe consequence, the canonical or longest was selected. We included an additional consequence for variants at the last base of an exon before an intron, where the final base is a guanine, since these variants appear to be as damaging as a splice donor variant28.
We categorized variants into three classes by VEP consequence:
protein-truncating variants (PTV): splice donor, splice acceptor, stop gained, frameshift, initiator codon, and conserved exon terminus variant.
missense variants: missense, stop lost, inframe deletion, inframe insertion, coding sequence, and protein altering variant.
silent variants: synonymous.
De novo mutation filtering
We filtered candidate DNM calls to reduce the false positive rate but maximize sensitivity, based on prior results from experimental validation by capillary sequencing of candidate DNMs15. Candidate DNMs were excluded if not called by GATK in the child, or called in either parent, or if they had a maximum MAF greater than 0.01. Candidate DNMs were excluded when the forward and reverse coverage differed between reference and alternative alleles, defined as P < 10-3 from a Fisher’s exact test of coverage from orientation by allele summed across the child and parents.
Candidate DNMs were also excluded if they met two of the three following three criteria: 1) an excess of parental alternative alleles within the cohort at the DNMs position, defined as P < 10-3 under a one-sided binomial test given an expected error rate of 0.002 and the cumulative parental depth; 2) an excess of alternative alleles within the cohort in DNMs in a gene, defined as P < 10-3 under a one-sided binomial test given an expected error rate of 0.002 and the cumulative depth, or 3) both parents had one or more reads supporting the alternative allele.
If, after filtering, more than one variant was observed in a given gene for a particular trio, only the variant with the highest predicted functional impact was kept (protein truncating > missense > silent).
De novo mutation validation
For candidate DNMs of interest, primers were designed to amplify 150-250 bp products centered around the site of interest. Default primer3 design settings were used with the following adjustments: GC clamp = 1, human mispriming library used. Site-specific primers were tailed with Illumina adapter sequences. PCR products were generated with JumpStart AccuTaq LA DNA polymerase (Sigma Aldrich), using 40 ng genomic DNA as template. Amplicons were tagged with Illumina PCR primers along with unique barcodes enabling multiplexing of 96 samples. Barcodes were incorporated using Kapa HiFi mastermix (Kapa Biosystems). Samples were pooled and sequenced down one lane of the Illumina MiSeq, using 250 bp paired end reads. An in-house analysis pipeline extracted the read count per site and classified inheritance status per variant using a maximum likelihood approach (see Supplementary Note).
Individuals with likely pathogenic variants
We previously screened 1,133 individuals for variants that contribute to their disorder15,18. All candidate variants in the 1,133 individuals were reviewed by consultant clinical geneticists for relevance to the individuals’ phenotypes. Most diagnosable pathogenic variants occurred de novo in dominant genes, but a small proportion also occurred in recessive genes or under other inheritance modes. DNMs within dominant DD-associated genes were very likely to be classified as the pathogenic variant for the individuals’ disorder. Due to the time required to review individuals and their candidate variants, we did not conduct a similar review in the remainder of the 4,293 individuals. Instead we defined likely pathogenic variants as candidate DNMs found in autosomal and X-linked dominant DD-associated genes, or candidate DNMs found in hemizygous DD-associated genes in males. 1,136 individuals in the 4,293 cohort had variants either previously classified as pathogenic15,18, or had a likely pathogenic DNM.
Gene-wise assessment of DNM significance
Gene-specific germline mutation rates for different functional classes were computed15,23 for the longest transcript in the union of transcripts overlapping the observed DNMs in that gene. We evaluated the gene-specific enrichment of PTV and missense DNMs by computing its statistical significance under a null hypothesis of the expected number of DNMs given the gene-specific mutation rate and the number of considered chromosomes23.
We also assessed clustering of missense DNMs within genes15, as expected for DNMs operating by activating or dominant negative mechanisms. We did this by calculating simulated dispersions of the observed number of DNMs within the gene. The probability of simulating a DNM at a specific codon was weighted by the trinucleotide sequence-context15,23. This allowed us to estimate the probability of the observed degree of clustering given the null model of random mutations.
Fisher’s method was used to combine the significance testing of missense + PTV DNM enrichment and missense DNM clustering. We defined a gene as significantly enriched for DNMs if the PTV enrichment P-value or the combined missense P-value less than 7 × 10-7, which represents a Bonferroni corrected P-value of 0.05 adjusted for 4 × 18500 tests (2 × consequence classes tested × protein coding genes).
Composite face generation
Families were given the option to have photographs of the affected individual(s) uploaded within DECIPHER34. Using images of individuals with DNMs in the same gene we generated de-identified realistic average faces (composite faces). Faces were detected using a discriminately trained deformable part model detector42. The annotation algorithm identified a set of 36 landmarks per detected face43 and was trained on a manually annotated dataset of 3100 images24. The average face mesh was created by the Delaunay triangulation of the average constellation of facial landmarks for all patients with a shared genetic disorder.
The averaging algorithm is sensitive to left-right facial asymmetries across multiple patients. For this purpose, we use a template constellation of landmarks based on the average constellations of 2000 healthy individuals24. For each patient, we align the constellation of landmarks to the template with respect to the points along the middle of the face and compute the Euclidean distances between each landmark and its corresponding pair on the template. The faces are mirrored such that the half of the face with the greater difference is always on the same side.
The dataset used for this work may contain multiple photos for one patient. To avoid biasing the average face mesh towards these individuals, we computed an average face for each patient and use these personal averages to compute the final average face. Finally, to avoid any image in the composite dominating from variance in illumination between images, we normalised the intensities of pixel values within the face to an average value across all faces in each average. The composite faces were assessed visually to confirm successful ablation of any individually identifiable features. Visual assessment of the composite photograph by two experienced clinical geneticists, alongside the individual patient photos, was performed for all 93 genome-wide significant DD-associated genes for which clinical photos were available for more than one patient, to remove potentially identifiable composite faces as well as quality control on the automated composite face generation process. Eighty-one composite faces were excluded leaving the twelve de-identified composite faces that are shown in Figure 2 and Extended Data Figure 3. Each of the twelve composite faces that passed de-identification and quality control was generated from photos of ten or more patients.
Assessing power of incorporating phenotypic information
We previously described a method to assess phenotypic similarity by HPO terms among groups of individuals sharing genetic defects in the same gene28. We examined whether incorporating this statistical test improved our ability to identify dominant genes at genome-wide significance. Per gene, we tested the phenotypic similarity of individuals with DNMs in the gene. We combined the phenotypic similarity P-value with the genotypic P-value per gene (the minimum P-value from the DDD-only and meta-analysis) using Fisher’s method. We examined the distribution of differences in P-value between tests without the phenotypic similarity P-value and tests that incorporated the phenotypic similarity P-value.
Many (854, 20%) of the DDD cohort experience seizures. We investigated whether testing within the subset of individuals with seizures improved our ability to find associations for seizure specific genes. A list of 102 seizure-associated genes was curated from three sources, a gene panel for Ohtahara syndrome, a currently used clinical gene panel for epilepsy and a panel derived from DD-associated genes18. The P-values from the seizure subset were compared to P-values from the complete cohort.
Assessing power of exome vs genome sequencing
We compared the expected power of exome sequencing versus genome sequencing to identify disease genes. Within the DDD cohort, 55 dominant DD-associated genes achieve genome-wide significance when testing for enrichment of DNMs within genes. We did not incorporate missense DNM clustering due to the large computational requirements for assessing clustering in many replicates.
We assumed a cost of 1,000 USD per individual for genome sequencing. We allowed the cost of exome sequencing to vary relative to genome sequencing, from 10-100%. We calculated the number of trios that could be sequenced under these scenarios. Estimates of the improved power of genome sequencing to detect DNMs in the coding sequence are around 1.05-fold29 and we increased the number of trios by 1.0–1.2-fold to allow this.
We sampled as many individuals from our cohort as the number of trios and counted which of the 55 DD-associated genes still achieved genome-wide significance for DNM enrichment. We ran 1000 simulations of each condition and obtained the mean number of genome-wide significant genes for each condition.
Associations with presence of likely pathogenic de novo mutations
We tested whether phenotypes were associated with the likelihood of having a likely pathogenic DNM. We analysed all collected phenotypes which could be coded in either a binary or quantitative format. Categorical phenotypes (e.g. sex coded as male or female) were tested by Fisher’s exact test while quantitative phenotypes (e.g. duration of gestation coded in weeks) were tested with logistic regression, using sex as a covariate.
We investigated whether having autozygous regions affected the likelihood of having a diagnostic DNM. Autozygous regions were determined from genotypes in every individual, to obtain the total length per individual. We fitted a logistic regression for the total length of autozygous regions on whether individuals had a likely pathogenic DNM. To illustrate the relationship between length of autozygosity and the occurrence of a likely pathogenic DNM, we grouped the individuals by length and plotted the proportion of individuals in each group with a DNM against the median length of the group.
The effects of parental age on the number of DNMs were assessed using 8,409 high confidence (posterior probability of DNM > 0.5) unphased coding and noncoding DNMs in 4,293 individuals. A Poisson multiple regression was fit on the number of DNMs in each individual with both maternal and paternal age at the child’s birth as covariates. The model was fit with the identity link and allowed for overdispersion. This model used exome-based DNMs, and the analysis was scaled to the whole genome by multiplying the coefficients by a factor of 50, based on ~2% of the genome being well covered in our data (exons + introns).
Excess of de novo mutations by consequence
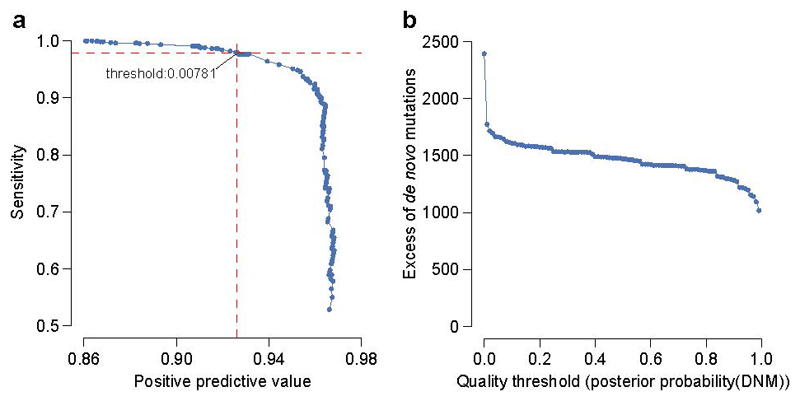
We identified the threshold for posterior probability of DNM at which the number of observed candidate synonymous DNMs equalled the number of expected synonymous DNMs. Candidate DNMs with scores below this threshold were excluded. We also examined the likely sensitivity and specificity of this threshold based on validation results for DNMs within a previous publication15 in which comprehensive experimental validation was performed on 1,133 trios that comprise a subset of the families analysed here.
The numbers of expected DNMs per gene were calculated per consequence from expected mutation rates per gene and the 2,407 male and 1,886 females in the cohort. We calculated the excess of DNMs for missense and PTVs as the ratio of numbers of observed DNMs versus expected DNMs, as well as the difference of observed DNMs minus expected DNMs.
Ascertainment bias within dominant neurodevelopmental genes
We identified 150 autosomal dominant haploinsufficient genes that affect neurodevelopment within our curated developmental disorder gene set. Genes affecting neurodevelopment were identified where the affected organs included the brain, or where HPO phenotypes linked to defects in the gene included either an abnormality of brain morphology (HP:0012443) or cognitive impairment (HP:0100543) term.
The 150 genes were classified for ease of clinical recognition of the syndrome from gene defects by two consultant clinical geneticists. Genes were rated from 1 (least recognisable) to 5 (most recognisable). Categories 1 and 2 contained 5 and 22 genes respectively, and so were combined in later analyses. The remaining categories had more than 33 genes per category. The ratio of observed loss-of-function DNMs to expected loss-of-function DNMs was calculated for each recognisability category, along with 95% confidence intervals from a Poisson distribution given observed counts.
We estimated the likelihood of obtaining the observed number of PTV DNMs under two models. Our first model assumed no haploinsufficiency, and mutation counts were expected to follow baseline mutation rates. Our second model assumed fully penetrant haploinsufficiency, and scaled the baseline PTV mutation expectations by the observed PTV enrichment in our known haploinsufficient neurodevelopmental genes, stratified by clinical recognisability into low (containing genes with our “low”, “mild” and “moderate” labels) and high categories. We calculated the likelihoods of both models per gene as the Poisson probability of obtaining the observed number of PTVs, given the expected mutation rates. We computed the Akaike’s Information Criterion for each model and ranked them by the difference between model 1 and model 2 (ΔAIC).
Proportion of de novo mutations with loss-of-function mechanism
The observed excess of missense/inframe indel DNMs is composed of a mixture of DNMs with loss-of-function mechanisms and DNMs with altered-function mechanisms. We found that the excess of PTV DNMs within dominant haploinsufficient DD-associated genes had a greater skew towards genes with high intolerance for loss-of-function variants than the excess of missense DNMs in dominant non-haploinsufficient genes. We binned genes by the probability of being loss-of-function intolerant30 constraint decile and calculated the observed excess of missense DNMs in each bin. We modelled this binned distribution as a two-component mixture with the components representing DNMs with a loss-of-function or function-altering mechanism. We identified the optimal mixing proportion for the loss-of-function and altered-function DNMs from the lowest goodness-of-fit (from a spline fitted to the sum-of-squares of the differences per decile) to missense/inframe indels in all genes across a range of mixtures.
The excess of DNMs with a loss-of-function mechanism was calculated as the excess of DNMs with a VEP loss-of-function consequence, plus the proportion of the excess of missense DNMs at the optimal mixing proportion.
We independently estimated the proportions of loss-of-function and altered-function. We counted PTV and missense/inframe indel DNMs within dominant haploinsufficient genes to estimate the proportion of excess DNMs with a loss-of-function mechanism, but which were classified as missense/inframe indel. We estimated the proportion of excess DNMs with a loss-of-function mechanism as the PTV excess plus the PTV excess multiplied by the proportion of loss-of-function classified as missense.
Prevalence of developmental disorders from dominant de novo mutations
We estimated the birth prevalence of monoallelic developmental disorders by using the germline mutation model. We calculated the expected cumulative germline mutation rate of truncating DNMs in 238 haploinsufficient DD-associated genes. We scaled this upwards based on the composition of excess DNMs in the DDD cohort using the ratio of excess DNMs (n=1816) to DNMs within dominant haploinsufficient DD-associated genes (n=412). Around 10% of DDs are caused by de novo CNVs44,45, which are underrepresented in our cohort as a result of prior genetic testing. If included, the excess DNM in our cohort would increase by 21%, therefore we scaled the prevalence estimate upwards by this factor.
Mothers aged 29.9 and fathers aged 29.5 have children with 77 DNMs per genome on average21. We calculated the mean number of DNMs expected under different combinations of parental ages, given our estimates of the extra DNMs per year from older mothers and fathers. We scaled the prevalence to different combinations of parental ages using the ratio of expected mutations at a given age combination to the number expected at the mean cohort parental ages.
To estimate the annual number of live births with developmental disorders caused by DNMs, we obtained country population sizes, birth rates, age at first birth46, and calculated global birth rate (18.58 live births/1000 individuals) and age at first birth (22.62 years), weighted by population size. We calculated the mean age when giving birth (26.57 years) given a total fertility rate of 2.45 children per mother47, and a mean interpregnancy interval of 29 months48. We calculated the number of live births given our estimate of DD prevalence caused by DNMs at this age (0.00288), the global population size (7.4 billion individuals) and the global birth rate.
Extended Data
Extended Data Figure 1.
Proportion of individuals with a de novo mutation (DNM) likely to be pathogenic. These only included individuals with protein altering or protein truncating DNMs in dominant or X-linked dominant developmental disorder (DD) associated genes, or males with DNMs in hemizygous DD-associated genes. The proportions given are for those individuals with any DNMs rather than the total number of individuals in each subset. Cohorts included in the DNM meta-analyses are shaded blue.
Extended Data Figure 2.
Phenotypic summary of genes without previous compelling evidence. Phenotypes are grouped by type. The first group indicates counts of individuals with DNMs per gene by sex (m: male, f: female), and by functional consequence (nsv: nonsynonymous variant, PTV: protein-truncating variant). The second group indicates mean values for growth parameters: birthweight (bw), height (ht), weight (wt), occipitofrontal circumference (OFC). Values are given as standard deviations from the healthy population mean derived from ALSPAC data. The third group indicates the mean age for achieving developmental milestones: age of first social smile, age of first sitting unassisted, age of first walking unassisted and age of first speaking. Values are given in months. The final group summarises Human Phenotype Ontology (HPO)-coded phenotypes per gene, as counts of HPO-terms within different clinical categories.
Extended Data Figure 3.
Phenotypic summary of individuals with de novo mutations in genes achieving genomewide significance. Phenotypes are grouped by type. The first group indicates counts of individuals with DNMs per gene by sex (m: male, f: female), and by functional consequence (nsv: nonsynonymous variant, PTV: protein-truncating variant). The second group indicates mean values for growth parameters: birthweight (bw), height (ht), weight (wt), occipitofrontal circumference (OFC). Values are given as standard deviations from the healthy population mean derived from ALSPAC data. The third group indicates the mean age for achieving developmental milestones: age of first social smile, age of first sitting unassisted, age of first walking unassisted and age of first speaking. Values are given in months. The final group summarises Human Phenotype Ontology (HPO)-coded phenotypes per gene, as counts of HPO-terms within different clinical categories.
Extended Data Figure 4.
Dispersion of de novo mutations and domains for each novel gene.a, CDK13, b, CHD4, c, CNOT3, d, CSNK2A1, e, GNAI1, f, KCNQ3, g, MSL3, h, PPM1D, I, PUF60, j, QRICH1, k, SET, l, SUV420H1, m, TCF20 and n, ZBTB18.
Extended Data Figure 5.
Effect of clustering by phenotype on the ability to identify genomewide significant genes.a, Comparison of P-values derived from genotypic information alone versus P-values that incorporate genotypic information and phenotypic similarity. b, Comparison of P-values from tests in the complete DDD cohort versus tests in the subset with seizures. Genes that were previously linked to seizures are shaded blue. c, Proportion of cohort with a de novo mutation (DNM) in a seizure-associated gene, stratified by whether seizure-affected status. Bars indicate 95% CI. d, Comparison of power to identify genomewide significant genes in probands with seizures, versus the unstratified cohort, at matched sample sizes.
Extended Data Figure 6.
Power of genome versus exome sequencing to discover dominant genes associated with developmental disorders. a, the number of genes exceeding genome-wide significance was estimated at three different fixed budgets (1 million (M) USD, 2M and 3M) and a range of relative sensitivities for genomes versus exomes to detect de novo mutations. The number of genes identifiable by exome sequencing are shaded blue, whereas the number of genes identifiable by genome sequencing are shaded green. The regions where exome sequencing costs 30-40% of genome sequencing are shaded with a grey background, which corresponds to the price differential in 2016. b, simulated estimates of power to detect loss-of-function genes in the genome at different cohort sizes, given fixed budgets.
Extended Data Figure 7.
Gene-wise significance of neurodevelopmental genes versus the expected number of mutations per gene. Points are shaded by clinical recognisability classification. Genes have been separated into two plots, one plot with genes for cryptic disorders with low, mild or moderate clinical recognisability, and one plot with genes for distinctive disorders with high clinical recognisability.
Extended Data Figure 8.
Stringency of de novo mutation (DNM) filtering. a, Sensitivity and specificity of DNM validations within sets filtered on varying thresholds of DNM quality (posterior probability of DNM). The analysed DNMs were restricted to sites identified within the earlier 1133 trios15, where all candidate DNMs underwent validation experiments. The labelled value is the quality threshold at which the number of candidate synonymous DNMs equals the number of expected synonymous mutations under a null germline mutation rate. b, Excess of missense and loss-of-function DNMs at varying DNM quality thresholds. The DNM excess is adjusted for the sensitivity and specificity at each threshold.
Extended Data Figure 9.
Enrichment of de novo mutations by consequence type, across RVIS functional constraint quantiles. A comparison of enrichment for RVIS values generated from ESP6500 data versus ExAC data is provided.
Extended Data Table 1.
Phenotypes tested for association with having a pathogenic de novo mutation.
| Category | Phenotype | Type | Value | 95% CI | P-value |
|---|---|---|---|---|---|
| Post-natal | abnormal cranial MRI | Odds ratio | 1.365 | 1.125 – 1.656 | 0.002 |
| feeding problems | Odds ratio | 1.176 | 1.01 – 1.369 | 0.039 | |
| neonatal intensive care | Odds ratio | 0.896 | 0.762 – 1.054 | 0.190 | |
| anticonvulsant drugs | Odds ratio | 0.582 | 0.246 – 1.377 | 0.270 | |
| Pre-natal | bleeding | Odds ratio | 0.892 | 0.714 – 1.114 | 0.346 |
| maternal illness | Odds ratio | 0.908 | 0.764 – 1.079 | 0.278 | |
| maternal diabetes | Odds ratio | 0.787 | 0.504 – 1.229 | 0.341 | |
| abnormal scan | Odds ratio | 0.839 | 0.692 – 1.017 | 0.078 | |
| assisted reproduction | Odds ratio | 0.868 | 0.554 – 1.36 | 0.584 | |
| increased nuchal translucency | Odds ratio | 1.432 | 0.903 – 2.271 | 0.126 | |
| Family history | consanguinity | Odds ratio | 0.234 | 0.138 – 0.397 | 8.0 x 10-11 |
| similar phenotype parents | Odds ratio | 0.295 | 0.184 – 0.474 | 5.7 x 10-9 | |
| similar phenotype relatives | Odds ratio | 0.553 | 0.402 – 0.761 | 1.5 x 10-4 | |
| similar phenotype siblings | Odds ratio | 0.311 | 0.23 – 0.421 | 7.3 x 10-18 | |
| only patient affected | Odds ratio | 2.478 | 2.001 – 3.068 | 3.9 x 10-19 | |
| X-linked inheritance | Odds ratio | 0.839 | 0.436 – 1.613 | 0.752 | |
| Multiple births | Beta | 0.043 | -0.058 – 0.144 | 0.403 | |
| History of pregnancy loss | Beta | -0.039 | -0.155 – 0.078 | 0.516 | |
| Developmental milestones | first words | Beta | 0.205 | 0.081 – 0.328 | 0.001 |
| walked independently | Beta | 0.125 | 0.016 – 0.235 | 0.025 | |
| sat independently | Beta | 0.050 | -0.069 – 0.17 | 0.408 | |
| social smile | Beta | 0.072 | -0.066 – 0.211 | 0.305 | |
| Growth | height | Beta | 0.008 | -0.111 – 0.126 | 0.897 |
| birthweight | Beta | -0.018 | -0.135 – 0.098 | 0.756 | |
| OFC | Beta | -0.094 | -0.215 – 0.026 | 0.125 | |
| weight | Beta | -0.331 | -1.278 – 0.615 | 0.493 | |
| Age | age at assessment | Beta | 0.116 | 0.015 – 0.217 | 0.025 |
| gestation | Beta | 0.079 | -0.033 – 0.19 | 0.167 | |
| father's age | Beta | 0.137 | 0.027 – 0.247 | 0.015 | |
| mother's age | Beta | 0.108 | -0.003 – 0.219 | 0.056 | |
| Other | phenotypic terms (n) | Beta | 0.104 | 0.004 – 0.203 | 0.041 |
| autozygosity length | Beta | -0.185 | -0.254 – -0.115 | 1.6 x 10-7 | |
| sex (male) | Odds ratio | 0.750 | 0.646 – 0.87 | 1.6 x 10-4 | |
Supplementary Material
Acknowledgments
We thank the families for their participation and patience. We are grateful to the Exome Aggregation Consortium for making their data available. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund (grant HICF-1009-003), a parallel funding partnership between the Wellcome Trust and the UK Department of Health, and the Wellcome Trust Sanger Institute (grant WT098051). The views expressed in this publication are those of the author(s) and not necessarily those of the Wellcome Trust or the UK Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South Research Ethics Committee and GEN/284/12, granted by the Republic of Ireland Research Ethics Committee). The research team acknowledges the support of the National Institutes for Health Research, through the Comprehensive Clinical Research Network. The authors wish to thank the Sanger Human Genome Informatics team, the Sample Management team, the Illumina High-Throughput team, the New Pipeline Group team, the DNA pipelines team and the Core Sequencing team for their support in generating and processing the data. D.R.F. is funded through an MRC Human Genetics Unit program grant to the University of Edinburgh. Finally we gratefully acknowledge the contribution of two esteemed DDD clinical collaborators, John Tolmie and Louise Brueton, who died in the course of the study.
Abbreviations
- PTV
Protein-Truncating Variant
- DNM
De Novo Mutation
- DD
Developmental Disorder
- DDD
Deciphering Developmental Disorders study
Footnotes
Code availability
Source code for filtering candidate DNMs, testing DNM enrichment, DNM clustering and phenotypic similarity can be found here: https://github.com/jeremymcrae/denovoFilter, https://github.com/jeremymcrae/mupit, https://github.com/jeremymcrae/denovonear, https://github.com/jeremymcrae/hpo_similarity
Author contributions
Patient recruitment and phenotyping: M. Ahmed, U.A., H.A., R.A., M. Balasubramanian, S. Banka, D. Baralle, A. Barnicoat, P.B., D. Baty, C. Bennett, J. Berg, B.B., M.B-G., E.B., M. Blyth, D. Bohanna, L. Bourdon, D. Bourn, L. Bradley, A. Brady, C. Brewer, K.B., D.J.B., J. Burn, N. Canham, B.C., K.C., D.C., A. Clarke, S. Clasper, J.C-S., V.C., A. Coates, T.C., A. Collins, M.N.C., F.C., N. Cooper, H.C., L.C., G.C., Y.C., M.D., T.D., R.D., S. Davies, J.D., C. Deshpande, G.D., A. Dixit, A. Dobbie, A. Donaldson, D. Donnai, D. Donnelly, C. Donnelly, A. Douglas, S. Douzgou, A. Duncan, J.E., S. Ellard, I.E., F.E., K.E., S. Everest, T.F., R.F., F.F., N.F., A. Fry, A. Fryer, C.G., L. Gaunt, N.G., R.G., H.G., J.G., D.G., A.G., P.G., L. Greenhalgh, R. Harrison, L. Harrison, V.H., R. Hawkins, S. Hellens, A.H., S. Hewitt, E.H., S. Holden, M. Holder, S. Holder, G.H., T.H., M. Humphreys, J.H., S.I., M.I., L.I., A.J., J.J., L.J., D. Johnson, E.J., D. Josifova, S.J., B. Kaemba, S.K., B. Kerr, H.K., U.K., E. Kinning, G.K., C.K., E. Kivuva, A.K., D. Kumar, V.A.K., K.L., W.L., A.L., C. Langman, M.L., D.L., C. Longman, G.L., S.A.L., A. Magee, E. Maher, A. Male, S. Mansour, K. Marks, K. Martin, U.M., E. McCann, V. McConnell, M.M., R.M., K. McKay, S. McKee, D.J.M., S. McNerlan, C.M., S. Mehta, K. Metcalfe, Z.M., E. Miles, S. Mohammed, T.M., D.M., S. Morgan, J.M., H. Mugalaasi, V. Murday, H. Murphy, S.N., A. Nemeth, L.N., R.N-E., A. Norman, R.O., C.O., K-R.O., S-M.P., M.J. Parker, C. Patel, J. Paterson, S. Payne, J. Phipps, D.T.P., C. Pottinger, J. Poulton, N.P., K.P., S. Price, A. Pridham, A. Procter, H.P., O.Q., N.R., J. Rankin, L. Raymond, D. Rice, L. Robert, E. Roberts, J. Roberts, P.R., G.R., A.R., E. Rosser, A. Saggar, S. Samant, J.S., R. Sandford, A. Sarkar, S. Schweiger, R. Scott, I. Scurr, A. Selby, A. Seller, C.S., N.S., S. Sharif, C.S-S., E. Shearing, D.S., E. Sheridan, I. Simonic, R. Singzon, Z.S., A. Smith, K.S., S. Smithson, L.S., M. Splitt, M. Squires, F.S., H.S., V. Straub, M. Suri, V. Sutton, E. Sweeney, K.T-B., C. Taylor, R.T., M. Tein, I.K.T., J.T., M. Tischkowitz, S.T., A.T., B.T., C. Turner, P.T., C. Tysoe, A.V., V.V., P. Vasudevan, J.V., E. Wakeling, S. Wallwark, J.W., A.W., D. Wellesley, M. Whiteford, S. Wilcox, D. Williams, N.W., L.W., G.W., C.W., M. Wright, L.Y., M.Y., H.V.F., D.R.F.
Sample and data processing: S. Clayton, T.W.F., E.P., D. Rajan, K.A., D.M.B., T.B., P.J., N.K., L.E.M., A.R.T., A.P.B., S. Brent, E.C., I.C., E.G., S.G., L. Hildyard, B.H., R.K., D.P., M.P., J. Randall, G.J.S., S. Widaa, E. Wilkinson
Validation experiments: J.F.M., E.P., D. Rajan, A. Sifrim, N.K., C.F.W.
Study design: M.J. Parker, H.V.F., C.F.W., D.R.F., J.C.B., M.E.H.
Method development and data analysis: J.F.M., S. Clayton, T.W.F., J.K., E.P., D. Rajan, A. Sifrim, S.A., N.A., M. Alvi, P.J., W.D.J., D. King, T.S., J.A., D.d.V., L. He, R.R., G.J.S., P. Vijayarangakannan, C.N., H.V.F., C.F.W., D.R.F., J.C.B., M.E.H.
Data interpretation: J.F.M., H.V.F., C.F.W., D.R.F., J.C.B., M.E.H.
Writing: J.F.M., C.F.W., D.R.F., M.E.H.
Experimental and analytical supervision: M.J. Parker, H.V.F., C.F.W., D.R.F., J.C.B., M.E.H.
Project Supervision: M.E.H.
Author information
Exome sequencing data are accessible via the European Genome-phenome Archive (EGA) under accession EGAS00001000775. Details of DD-associated genes are available at www.ebi.ac.uk/gene2phenotype. M.E.H. is a co-founder of, and holds shares in, Congenica Ltd, a genetics diagnostic company.
References
- 1.Sheridan E, et al. Risk factors for congenital anomaly in a multiethnic birth cohort: an analysis of the Born in Bradford study. Lancet. 2013;382:1350–9. doi: 10.1016/S0140-6736(13)61132-0. [DOI] [PubMed] [Google Scholar]
- 2.Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genomics Hum Genet. 2010;11:161–87. doi: 10.1146/annurev-genom-082509-141640. [DOI] [PubMed] [Google Scholar]
- 3.De Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. The New England Journal of Medicine. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 4.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epi4K Consortium & Epilepsy Phenome/Genome Project. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.EuroEPINOMICS-RES Consortium, Epilepsy Phenome/Genome Project & Epi4K Consortium. De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95:360–70. doi: 10.1016/j.ajhg.2014.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fromer M, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilissen C, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 9.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iossifov I, et al. De Novo Gene Disruptions in Children on the Autistic Spectrum. Neuron. 2012;74:285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Roak BJ, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012;485:1–7. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 13.Sanders SJ, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485:237–41. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–3. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Ligt J, Veltman JA, Vissers LELM. Point mutations as a source of de novo genetic disease. Current Opinion in Genetics & Development. 2013;23:257–263. doi: 10.1016/j.gde.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 17.Wilkie AO. The molecular basis of genetic dominance. J Med Genet. 1994;31:89–98. doi: 10.1136/jmg.31.2.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wright CF, et al. Genetic diagnosis of developmental disorders in the DDD study: a scalable analysis of genome-wide research data. The Lancet. 2014 doi: 10.1016/S0140-6736(14)61705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacquemont S, et al. A higher mutational burden in females supports a "female protective model" in neurodevelopmental disorders. Am J Hum Genet. 2014;94:415–25. doi: 10.1016/j.ajhg.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–5. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahbari R, et al. Timing, rates and spectra of human germline mutation. Nat Genet. 2016;48:126–33. doi: 10.1038/ng.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong WS, et al. New observations on maternal age effect on germline de novo mutations. Nat Commun. 2016;7:10486. doi: 10.1038/ncomms10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samocha KE, et al. A framework for the interpretation of de novo variation in human disease. Nature Genetics. 2014;46:944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferry Q, et al. Diagnostically relevant facial gestalt information from ordinary photos. eLife. 2014;3:e02020–e02020. doi: 10.7554/eLife.02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirata H, et al. ZC4H2 mutations are associated with arthrogryposis multiplex congenita and intellectual disability through impairment of central and peripheral synaptic plasticity. Am J Hum Genet. 2013;92:681–95. doi: 10.1016/j.ajhg.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Homan CC, et al. Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 2014;94:470–8. doi: 10.1016/j.ajhg.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu J, et al. SMC1A expression and mechanism of pathogenicity in probands with X-Linked Cornelia de Lange syndrome. Hum Mutat. 2009;30:1535–42. doi: 10.1002/humu.21095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akawi N, et al. Discovery of four recessive developmental disorders using probabilistic genotype and phenotype matching among 4,125 families. Nature Genetics. 2015;47:1363–1369. doi: 10.1038/ng.3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meynert AM, Ansari M, FitzPatrick DR, Taylor MS. Variant detection sensitivity and biases in whole genome and exome sequencing. BMC Bioinformatics. 2014;15:247. doi: 10.1186/1471-2105-15-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9:e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boycott KM, Vanstone MR, Bulman DE, Mackenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nature Reviews Genetics. 2013;14:681–91. doi: 10.1038/nrg3555. [DOI] [PubMed] [Google Scholar]
- 33.Springett A, et al. Congenital Anomaly Statistics 2011: England and Wales. 2013 [Google Scholar]
- 34.Bragin E, et al. DECIPHER: database for the interpretation of phenotype-linked plausibly pathogenic sequence and copy-number variation. Nucleic Acids Res. 2014;42:D993–D1000. doi: 10.1093/nar/gkt937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Köhler S, et al. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. American Journal of Human Genetics. 2009;85:457–464. doi: 10.1016/j.ajhg.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramu A, et al. DeNovoGear: de novo indel and point mutation discovery and phasing. Nature Methods. 2013;10:985–7. doi: 10.1038/nmeth.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abecasis GR, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McLaren W, et al. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–70. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Felzenszwalb PF, Girshick RB, McAllester D, Ramanan D. Object detection with discriminatively trained part-based models. IEEE transactions on pattern analysis and machine intelligence. 2010;32:1627–45. doi: 10.1109/TPAMI.2009.167. [DOI] [PubMed] [Google Scholar]
- 43.Xiong X, De la Torre F. Supervised Descent Method and Its Applications to Face Alignment. 2013 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); Portland, OR: IEEE; 2013. pp. 532–539. [Google Scholar]
- 44.Cooper GM, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46. doi: 10.1038/ng.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sagoo GS, et al. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med. 2009;11:139–46. doi: 10.1097/GIM.0b013e318194ee8f. [DOI] [PubMed] [Google Scholar]
- 46.Central Intelligence Agency. The World Factbook. 2016;2016 [Google Scholar]
- 47.The World Bank. Fertility rate, total (births per woman) World Development Indicators. 2016;2016 [Google Scholar]
- 48.Copen CE, Thoma ME, Kirmeyer S. National Vital Statistics Reports. Vol. 64. National Center for Health Statistics; Hyattsville, MD: 2015. Interpregnancy Intervals in the United States: Data From the Birth Certificate and the National Survey of Family Growth. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.