Significance
Our study exploits time—the relatively unexplored fourth dimension of gene regulatory networks (GRNs)—to learn the temporal transcriptional logic underlying dynamic nitrogen (N) signaling in plants. We introduce several conceptual innovations to the analysis of time-series data in the area of predictive GRNs. Our resulting network now provides the “transcriptional logic” for transcription factor perturbations aimed at improving N-use efficiency, an important issue for global food production in marginal soils and for sustainable agriculture. More broadly, the combination of the time-based approaches we develop and deploy can be applied to uncover the temporal “transcriptional logic” for any response system in biology, agriculture, or medicine.
Keywords: systems biology, plant biology, nitrogen assimilation, transcriptional dynamics, network inference
Abstract
This study exploits time, the relatively unexplored fourth dimension of gene regulatory networks (GRNs), to learn the temporal transcriptional logic underlying dynamic nitrogen (N) signaling in plants. Our “just-in-time” analysis of time-series transcriptome data uncovered a temporal cascade of cis elements underlying dynamic N signaling. To infer transcription factor (TF)-target edges in a GRN, we applied a time-based machine learning method to 2,174 dynamic N-responsive genes. We experimentally determined a network precision cutoff, using TF-regulated genome-wide targets of three TF hubs (CRF4, SNZ, and CDF1), used to “prune” the network to 155 TFs and 608 targets. This network precision was reconfirmed using genome-wide TF-target regulation data for four additional TFs (TGA1, HHO5/6, and PHL1) not used in network pruning. These higher-confidence edges in the GRN were further filtered by independent TF-target binding data, used to calculate a TF “N-specificity” index. This refined GRN identifies the temporal relationship of known/validated regulators of N signaling (NLP7/8, TGA1/4, NAC4, HRS1, and LBD37/38/39) and 146 additional regulators. Six TFs—CRF4, SNZ, CDF1, HHO5/6, and PHL1—validated herein regulate a significant number of genes in the dynamic N response, targeting 54% of N-uptake/assimilation pathway genes. Phenotypically, inducible overexpression of CRF4 in planta regulates genes resulting in altered biomass, root development, and 15NO3− uptake, specifically under low-N conditions. This dynamic N-signaling GRN now provides the temporal “transcriptional logic” for 155 candidate TFs to improve nitrogen use efficiency with potential agricultural applications. Broadly, these time-based approaches can uncover the temporal transcriptional logic for any biological response system in biology, agriculture, or medicine.
Nitrogen (N)—a nutrient/signal—is a core component of fertilizer used in modern agriculture to alleviate worldwide hunger (1). However, this comes at environmental costs, through excess nitrogen run-off due to inefficient N-use efficiency by crops (2). Thus, improving plant N uptake, assimilation, and utilization is highly desirable. With this goal, studies have attempted to capture and model the N-regulatory networks controlling N uptake/assimilation (3–6). Validation studies have identified several transcription factors (TFs) (7–12) as key regulators of N signaling. However, we lack knowledge of the dynamics and temporal hierarchy of these known—and as yet unknown—TFs in controlling N signaling and N uptake/assimilation. A meta-analysis placed some known regulators within network modules (13). However, such correlation-based networks are unable to predict causality. By contrast, time-based machine learning approaches can predict the regulatory influence of TFs on their targets in the dataset and in out-of-sample data, the ultimate goal of systems biology (5, 14, 15).
In this study, we derived the temporal dynamics of N-regulatory networks by devising and combining several time-based approaches. First, our “just-in-time” (JIT) analysis uncovered a temporal cis-element cascade underlying dynamic N signaling. Second, we used a validated time-driven machine-learning approach, dynamic factor graph (DFG) (5, 14, 15), to infer TF–target interactions in 2,174 N-response genes in shoots. Third, we “pruned” the inferred TF-target edges in this gene regulatory network (GRN) using a precision cutoff threshold derived from experimentally regulated genome-wide targets of six regulators of N uptake/assimilation—CRF4, SNZ, CDF1, HHO5/6, and PHL1—validated herein. This pruned GRN predicts the influence of 155 TFs on 608 N-responsive genes. Fourth, to provide further support for the edges in the GRN, we used available TF-target binding data (DNA affinity purification sequencing, DAP-Seq) (16), also used to calculate a TF “N-specificity” index. This time-based GRN now reveals the temporal relationships of TFs previously validated in the N response [e.g., NLP7/8 (7, 17), TGA1/4 (8), NAC4 (9), HRS1 (10), and LBD37, 38, 39 (11)]. It also connects these known TFs with previously undescribed TFs in the N-response cascade, including ones we validated herein—CRF4, SNZ, CDF1, HHO5/6, and PHL1—to regulate a significant number of genes in the dynamic N response, including 54% of nitrate uptake/assimilation pathway genes. Finally, we show that perturbation of CRF4, the earliest N-responsive TF in this GRN, affects genes and processes that result in altered nitrate uptake, root development, and plant biomass, under low-N input conditions. Beyond these proof-of-principle examples, the pruned GRN of dynamic N signaling we derived now provides the temporal “transcriptional logic” for 155 candidate TFs for perturbations aimed at improving nitrogen use efficiency (NUE) with potential applications in agriculture. More broadly, these time-based approaches can be applied to uncover the temporal transcriptional logic for any biological response system in biology, agriculture, or medicine.
Results
A Fine-Scale Time-Course Transcriptome of Dynamic Nitrogen Signaling.
N nutrient signal elicits dynamic responses in plant metabolism and development (5, 13, 18–21). However, most prior transcriptome studies assayed only one or two time points following N treatment (3, 6, 13) or widely spaced time points not amenable to learning GRN causality (22). A previous study uncovered the very early (3–20 min) transcriptional response to nitrate treatment in Arabidopsis roots (5). Herein, we captured early-to-late transcriptome responses (5, 10, 15, 20, 30, 45, 60, 90, and 120 min) to an N supply (NO3− and NH4+) shown to elicit inorganic- and organic-N responses (4) (Methods). Genes responding to N as a function of time (NxTime genes) were identified using a cubic-spline model [false discovery rate (FDR) P < 0.01] (23) (SI Appendix, Supplementary Methods and Fig. S1 A and B). This analysis identified NxTime response genes in shoots (2,174 genes) and in roots (2,681 genes) [SI Appendix, Fig. S2C (shoots: green bars), Dataset S1, Table S1, and SI Appendix, Fig. S4B (roots: brown bars), Dataset S1, Table S2]. These NxTime gene sets are largely organ-specific but share 778 genes, including 54 TFs (SI Appendix, Fig. S1C). These include many known N-responsive genes (3–6, 13) and also 2,737 unique N-responsive genes (SI Appendix, Fig. S3) (24), due to increased sensitivity from RNA sequencing and 511 genes absent on microarrays (13). We also captured transient responses to N supply, including the well-known N-regulator TF, NLP7 (7, 17) (SI Appendix, Fig. S12). Our dataset captures dynamic effects of N signaling in shoots, on metabolism, RNA processing, photosynthesis (25), and circadian rhythm (4) (SI Appendix, Figs. S2 and S8–S10 and Dataset S1, Table S1).
Just-in-Time Analysis Uncovers a Temporal Cascade of Cis-Regulatory Elements and Biological Processes in Response to N Supply.
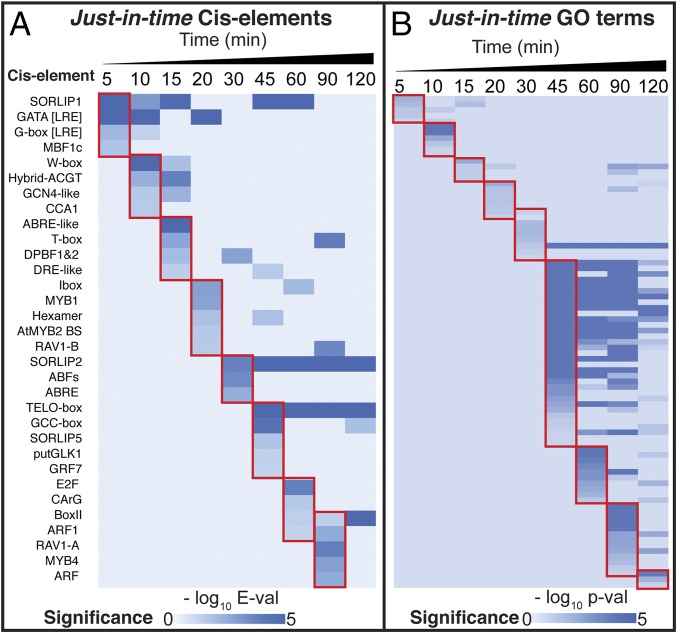
To uncover the regulatory cascade underlying dynamic N signaling, we implemented a JIT analysis (Methods). This JIT analysis bins NxTime genes, based on the first time point at which its mRNA levels are affected by N signaling (fold change ≥ 1.5) (SI Appendix, Fig. S2 B and C, blue bars, and Dataset S1, Table S1). We then identified overrepresented known cis motifs (16, 26, 27) in each JIT bin, using a hypergeometric distribution on a genome-wide promoter background (28). This analysis uncovered a temporal cascade of overrepresented cis-regulatory motifs (e-value < 0.05) in the promoters of genes first responding to N signaling at each JIT point (Fig. 1A). The set of enriched cis elements are different between the JIT sets of shoots (Fig. 1A) vs. roots (SI Appendix, Fig. S4C). The temporal enrichment of unique cis-element motifs in shoots is particularly noticeable at the 10-, 15-, and 20-min JIT points (Fig. 1A). Conversely, certain cis-element motifs—such as SORLIP2 and TELO-box—are overrepresented at consecutive JIT sets (Fig. 1A). This JIT analysis also uncovered a temporal cascade of enriched Gene Ontology (GO) terms enriched in each JIT gene set in shoots (FDR adjusted P < 0.01) (Fig. 1B and SI Appendix, Fig. S2D). The early JIT gene sets (5–15 min) are significantly enriched in genes related to N uptake/assimilation. Intermediate JIT gene sets (20–30 min) are enriched in energy generation. The later JIT gene sets (≥45 min) are enriched in genes for metabolic and developmental processes (SI Appendix, Figs. S2D and S8–S11). Overall, JIT cis-element and GO analysis implicates a cascade of associated TFs regulating largely nonoverlapping sets of genes at consecutive JIT time points in N signaling (Fig. 1). However, the current cis-motif datasets (16, 26, 27) are generalized for TF families and cannot associate individual TFs with specific target genes. We thus associated specific TFs with targets in the NxTime cascade by using a time-based network inference method described below.
Fig. 1.
JIT gene set analysis identifies a temporal cascade of N-response genes in shoots. Genes responding to NxTime by cubic-spline analysis (23) were binned into the first time point at which mean expression changes by ≥1.5 fold (Methods and Dataset S1, Table S1). (A) A cascade of unique cis-element motifs are significantly enriched in each JIT gene set (Methods). (B) The JIT gene sets have nonoverlapping sets of GO terms enriched at each time point (Methods and SI Appendix, Fig. S2D).
Assigning an N-Specificity Index to TFs in the Dynamic N-Response Cascade.
Our time course captures 172 TFs responding to N supply within 2 h (Dataset S1, Table S13). To identify TFs that play a specific role in N signaling, we computed an N-specificity index (Dataset S1, Table S12), based on available TF-target binding data (16). For each NxTime regulated TF with genome-wide binding data (40 TFs; Dataset S1, Table S12) we tested if the proportion of its genome-wide targets (16) in the NxTime shoot genes are significantly overrepresented, relative to the proportion of all of the TF-bound targets in the genome (Methods). This identified 19 TFs with a highly significant N-specificity score (P < 0.05) in shoots (Dataset S1, Table S12). These N-specific TFs include four validated regulators of the N response [NLP7 (7), TGA1/4 (8), and NAC4 (9)] and 15 additional TFs whose targets are enriched in N-signal-responsive genes in shoots (Dataset S1, Table S12). We note that this N-specificity calculation is limited to TFs with TF-target binding data for 529 TFs currently in the DAP-seq database (16). However, this N-specificity calculation may be applied to any TF with known genome-wide targets, as we show with SNZ and CDF1 (SI Appendix, Fig. S5B), as detailed below.
Inferring a Time-Derived GRN Driving the Temporal N Response in Shoots.
De novo network inference is a valuable approach to build GRNs (29–32). Because causality moves forward in time, fine-scale time-series transcriptome experiments are an especially valuable resource to infer GRNs that can predict out-of-sample target gene behavior, the ultimate goal of systems biology (5, 14, 33). Previously, we applied a time-based machine-learning method, DFG (15), to learn and predict causal relationships between TFs and their targets (5, 14). Briefly, DFG identifies the likely set of TFs driving target gene expression, by learning an f function that explains the target gene expression at each time point, based on the expression of the TFs at previous time points (15). Here, we used the DFG method to predict the influence of every TF on every gene in the shoot NxTime gene set, implementing rigorous hyperparameterization steps (Methods). The resultant DFG network provides a measure of the influence (i.e., TF-target edge score) of each of the 172 TFs on the 2,174 N-responsive genes in shoots (e.g., 374,000 predicted edges). However, a major challenge in de novo network inference is the high false-positive rate of TF-target predictions (30). We thus estimated confidence in the edges of our time-inferred DFG network by comparing the predicted TF-target edges in the GRN to experimentally validated TF targets (30). This method establishes the precision (i.e., proportion of predicted TF-target edges that are real) and recall (i.e., proportion of real TF-target edges that are predicted) of the GRN, which can then be used to prune the network to enrich for higher-confidence TF-target predictions (30). To implement this network pruning step, we first retained the top 10% of DFG predictions (Dataset S1, Table S19) and experimentally validated the genome-wide targets of seven TF hubs in this initial DFG network. This genome-wide TF-target validation step established the precision vs. recall for the larger GRN of 155 TFs (SI Appendix, Figs. S6 and S7), as detailed below.
Validation of TFs CRF4, SNZ, and CDF1 in the Time-Inferred GRN That Regulate N-Response and N-Uptake/Assimilation Pathway Genes.
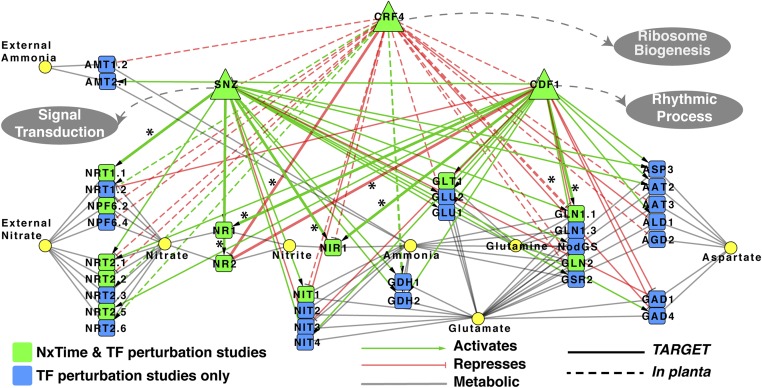
To implement genome-wide validation of the DFG-predicted GRN, CRF4 was selected for initial TF-target validation as it (i) is early N-responsive in both shoots and roots (SI Appendix, Fig. S15A), (ii) is a TF hub (422 out-edges) in the top 10% unpruned DFG shoot network (∼35,200 edges) (Dataset S1, Table S19), (iii) has a high N-specificity index (Dataset S1, Table S12), and (iv) is a TF in N signaling with potential links to the cytokinin pathway (34). We identified the genome-wide targets of CRF4 in an inducible overexpression transplanta line (CRF4-OX) (35) and also via TF perturbation in shoot cells using the TARGET (Transient Assay Reporting Genome-wide Effects of Transcription factors) assay (10, 36–38). These results confirm the early and central role that CRF4 plays in the dynamic N response. In planta CRF4-regulated targets are significantly overrepresented in NxTime genes in both shoots and roots, spanning early and later JIT NxTime points (SI Appendix, Fig. S15 B and D). The validated genome-wide targets of CRF4 in shoots include 16 downstream TFs responsive to NxTime (SI Appendix, Fig. S5A and Dataset S1, Table S5). We next selected two validated TF targets of CRF4—an “early” (SNZ, 10-min JIT) and “late” (CDF1, 45-min JIT) N responder—for TF-perturbation studies in shoot cells using the TARGET system (36) (SI Appendix, Fig. S5 A and B). These results revealed that the targets regulated by CRF4, SNZ, and CDF1 (Dataset S1, Tables S5–S7) (i) are significantly enriched in NxTime genes, (ii) support a high N-specificity index, and (iii) are enriched in GO terms related to nitrate assimilation/metabolism (for CRF4, SNZ, and CDF1), ribosome biogenesis (for CRF4), and rhythmic processes (for CDF1) (SI Appendix, Fig. S5B and Dataset S1, Tables S8–S10). Collectively, the targets of CRF4, SNZ, and CDF1 encompass (i) 54% of N-uptake/assimilation pathway (35/65) genes, (ii) 75% of the NxTime genes in the N-uptake/assimilation pathway (12/16), and (iii) 23 N-pathway genes that are not NxTime-responsive (Fig. 2). We note the cell-based TARGET system can identify direct targets based on TF regulation (Fig. 2, solid lines), because translation of mRNA from primary TF targets is blocked (Methods) (36). By contrast, in planta TF perturbations cannot distinguish direct vs. indirect regulated targets (Fig. 2, dashed lines).
Fig. 2.
Three TFs—CRF4, SNZ, and CDF1—regulate 53% of the N-uptake/assimilation pathway genes. A time-based machine learning approach DFG (5, 15) was used to infer TF-target influence in an N-response GRN in shoots (Methods). Validated genome-wide targets of three TFs in this GRN—CRF4, SNZ, and CDF1—are shown to regulate 53% (35/65) genes in the N-uptake/assimilation pathway (SI Appendix, Fig. S5B and Dataset S1, Tables S5–S7 and S15). TF edges to N-responsive genes (green nodes) that are predicted by the GRN and validated by TF perturbations are shown by asterisks and thicker edge width (Dataset S1, Table S14). Gray circles indicate other cellular processes validated to be regulated by these three TFs (Dataset S1, Tables S8–S10).
The GRN Is Pruned Using Genome-Wide TF-Target Validation Data to Identify Higher-Confidence Edge Predictions.
Next, to validate our edge predictions in the time-derived GRN, we used experimentally derived TF-target regulation data for CRF4, SNZ, and CDF1 (SI Appendix, Fig. S6). First, we tested the significance of the DFG TF-target edge rankings in our GRN by performing an area under precision recall (AUPR) curve analysis (30, 39, 40) (SI Appendix, Fig. S7). We compared the ranked TF-target edge predictions from the DFG-inferred network to a random ranking of TF-target edges (1,000 iterations). This analysis showed that the AUPR of the DFG-inferred network (0.24) is significantly better than the mean AUPR for random networks (0.14) (P < 0.001) (SI Appendix, Fig. S7C). Next, to identify higher-confidence edges in the GRN (39), we chose a cutoff point (precision = 0.345) before the AUPR curve flattens (SI Appendix, Fig. S7D). This precision cutoff point of 0.345 matches a TF-target edge score of 0.95554 in our GRN (SI Appendix, Fig. S7 A and D). Thus, only TF-target edges with an edge score ≥0.95554 were retained in our pruned DFG network (SI Appendix, Fig. S7A and Dataset S1, Table S3). This pruned GRN includes 85 validated targets out of the 245 predicted TF-target edges between CRF4, SNZ, and CDF1 and genes in the NxTime shoot set. These predicted and validated targets of CRF4, SNZ, and CDF1 include five key genes in N uptake/assimilation (NRT1.1, NR1 and NR2, NIR, and GLN1.1) (Fig. 2, edges denoted by asterisks), 10 genes involved in transcriptional/translation, and genes in the circadian clock (e.g., TIC) (Dataset S1, Table S14). As our pruned GRN was optimized to increase precision at the cost of low recall, it likely underestimates the influence of a given TF on GRN. For example, only 9/24 experimentally validated edges from these three TFs to the 12 N-responsive genes in the N-assimilation pathway (Fig. 2, green nodes) are in the pruned network (Fig. 2, edges with asterisks) (Dataset S1, Table S14).
Cross-Validation of the Pruned N-Signaling GRN Using TF-Target Regulation Data from Four Additional TFs.
The above pruned GRN at an average precision of ∼0.345 can now predict the influence of 155 N-responsive TFs on 608 NxTime genes in the dynamic N response in shoots (SI Appendix, Fig. S6 and Dataset S1, Table S3). To independently validate this precision rate, we identified the regulated TF targets of four additional TFs in the GRN—HHO5/6, PHL1, and TGA1—in shoot cells using the TARGET system (Dataset S1, Tables S22–S25). The precision for each of these four TFs in the GRN ranged from 0.17 to 0.45, for an overall average of 0.32 (SI Appendix, Figs. S7B and S16B). In total, 110/349 predicted TF targets in the pruned GRN were experimentally validated, including six genes involved in N uptake/reduction (SI Appendix, Fig. S16 A–C). These four TFs also influence a significant number of genes and processes in the NxTime gene set in shoots (SI Appendix, Fig. S5C and Dataset S1, Tables S27–S30). This independent TF validation proves that the initial network precision of 0.345 (SI Appendix, Fig. S6) used to prune the shoot N-response GRN extends beyond the three TFs used in the network inference pruning stage (e.g., CRF4, SNZ, and CDF1) and can be used to predict targets of 155 TFs in the N-response GRN with an overall precision of ∼0.33. We note that our precision cutoff of 0.33 (i.e., one in three predicted edges are likely to be true), is of a scale comparable to the maximum precision of 0.5 achieved using an ensemble approach of multiple network inference methods in microbes (30).
Independent TF-Target Binding Data Support Predicted Edges in the NxTime GRN.
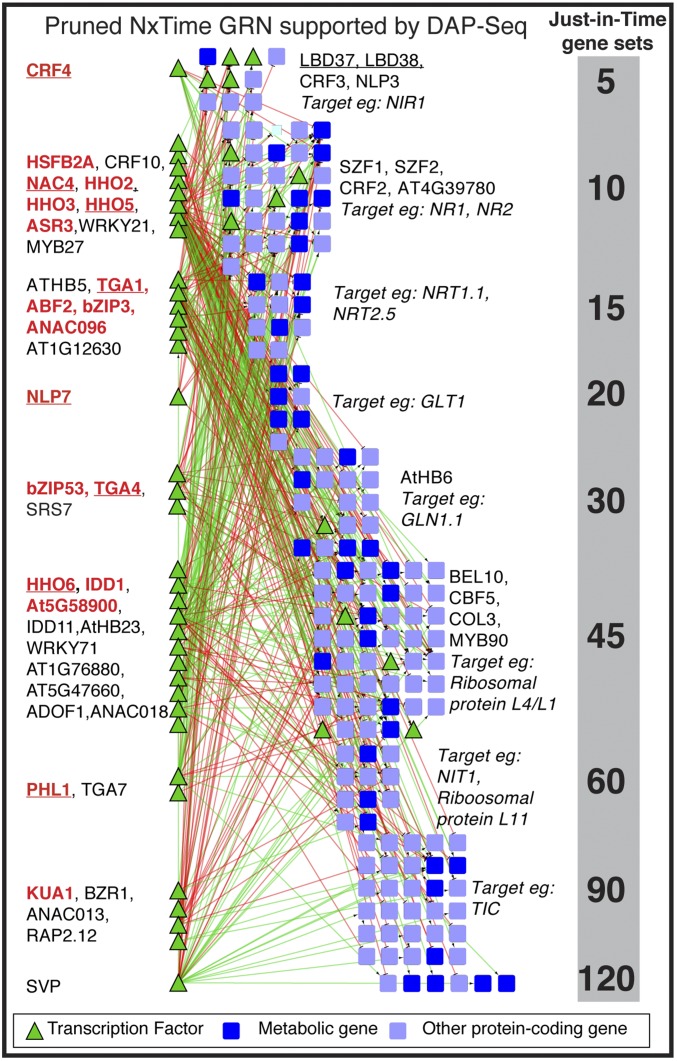
The TF-target edges in the pruned DFG network (Dataset S1, Table S3) identified as hubs (i.e., influential TFs) multiple known/validated regulators of N signaling (e.g., TGA1/4, NLP7/8, NAC4, HRS1, and LBD37/38/39) (7–11, 17), as well as 146 potential regulators, including six validated herein: CRF4, SNZ, CDF1, HHO5/6, and PHL1 (SI Appendix, Fig. S5 B and C and Dataset S1, Tables S5–S7 and S22–S24). To add further edge support, the DFG-predicted edges in the pruned GRN were queried using an independent source of TF-target binding data for the 40 NxTime TFs in shoots that are present in the DAP-Seq dataset (16) (Fig. 3). A TF-target edge in the pruned DFG network is supported by DAP-Seq TF-target data (16) only if that TF is shown to bind to the promoter of the target gene in the DAP-Seq assay (41) (Fig. 3 and Dataset S1, Table S4). We note that the actual DAP-Seq TF-DNA binding data (41) were used to establish TF-target binding, not the in silico cis-motif information (16). The 19 TFs in the pruned GRN that have DAP-seq data and high-N specificity include four known TFs in the N response (TGA1/4, NAC4, and NLP7) and four TFs (CRF4, HHO5/6, and PHL1) validated herein (Fig. 3, red underlined TFs).
Fig. 3.
A time-dependent GRN uncovers TFs in dynamic N signaling in shoot. A time-based machine learning approach DFG (5, 15) was used to infer TF-target influence in a GRN. Validated genome-wide targets of three TFs—CRF4, SNZ, and CDF1 (SI Appendix)—were used to prune the GRN for TF-target precision based on AUPR analysis (SI Appendix, Figs. S5–S7 and Dataset S1, Tables S5–S7). This TF-target precision was reconfirmed using data for four independent TFs—TGA1, HHO5/6, and PHL1 (SI Appendix, Fig. S7B and Dataset S1, Tables S22–S25). The TF-target edges supported by an independent source of TF-target binding data [DAP-SEq (16, 41)] capture regulation of 208 N-responsive target genes by 35 TFs (Dataset S1, Table S4). TFs with a significant N-specificity index are highlighted in red (Dataset S1, Table S12). Validated TF regulators of the N-response are underlined: NLP7 (7), TGA1/4 (8), NAC4 (9), LBD37, 38 (11), and CRF4, HHO5/6, and PHL1 (this study).
CRF4—the Earliest TF in the N-signaling GRN—Regulates N Uptake and N Use in Planta.
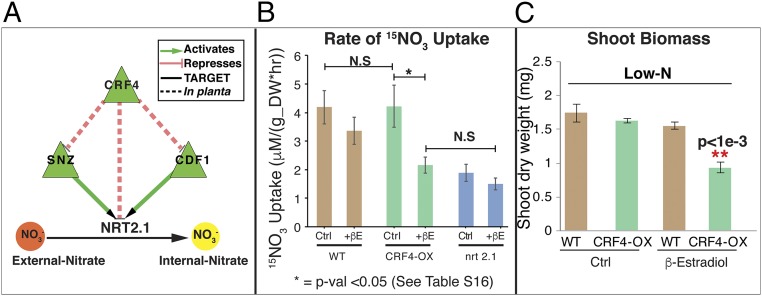
The pruned DFG network—refined by TF-target binding data—places CRF4 at the top of the N-signaling cascade (Fig. 3), based on its early response (5-min JIT) and its GRN connections (Dataset S1, Table S3). Indeed, our validation studies support the early and specific role of CRF4 in mediating the dynamic N-response GRN in planta. Inducible expression using a CRF4-OX transplanta line (35) (Methods) reveals that CRF4 controls a highly significant number of NxTime genes, spanning early and later JIT gene sets (SI Appendix, Fig. S15 B and D and Dataset S1, Tables S18 and S21). Impressively, CRF4 directly or indirectly regulates approximately one-third of the genes in the N-uptake/assimilation pathway (21/65), including seven N-uptake genes (Fig. 2). In planta CRF4 targets are also enriched in N-metabolic processes and translation (in shoots) and response to nitrate and root development (in roots) (SI Appendix, Fig. S15C). Moreover, these CRF4-mediated changes in gene regulation affect N uptake and use in planta (Fig. 4). CRF4-OX overexpression results in significantly lowered shoot biomass (P < 1e-5) (Fig. 4C), primary root length, and number of lateral roots, under low-N conditions (SI Appendix, Fig. S13 A and B), where the high-affinity N-transporter NRT2.1 is the major functional nitrate-uptake system (42). Further, repression of NRT2.1 in shoots and roots of CRF4-OX plants (Fig. 4A and SI Appendix, Fig. S13C and Dataset S1, Tables S5 and S20) leads to lower rates of nitrate uptake under low-N conditions in the CRF4-OX line (35) (Methods). Using 15NO3 tracer (43), 15NO3 uptake was significantly reduced in the induced CRF4-OX overexpression line, at levels comparable to the nrt2.1 mutant impaired in high-affinity nitrate uptake (44), compared with uninduced CRF4-line and wild-type controls, under low-N conditions (two-way ANOVA with Tukey honestly significant difference analysis) (Methods) (Fig. 4B and Dataset S1, Table S16). These results validate the important role CRF4 plays in regulating N uptake/use—acting either directly or indirectly through its downstream TFs, such as SNZ and CDF1 (Fig. 4A).
Fig. 4.
CRF4 overexpression represses high-affinity nitrate uptake and biomass in planta. (A) CRF4 overexpression via β-estradiol (+βE) induction (35) represses SNZ, CDF1, and NRT2.1 (SI Appendix, Fig. S13C and Dataset S1, Table S5). SNZ and CDF1 overexpression in shoot cells (36) induces NRT2.1 expression (Dataset S1, Tables S6 and S7). CRF4 overexpression in low-N (1 mM NO3) conditions significantly reduces (B) the rate of nitrate 15NO3− uptake (Dataset S1, Table S16) and (C) shoot biomass in planta (SI Appendix, Fig. S13A). N.S, not significant.
Discussion
N—a key nutrient/signal—regulates dynamic plant processes including circadian rhythm (4) and root foraging (5, 13, 18–20). However, the underlying temporal mechanisms are unknown. Our JIT analysis uncovered discrete waves of transcriptional responses to N signaling in shoots (Fig. 1). For example, we confirm and extend the role of N signaling as an input to the circadian clock in plants (4). N signaling regulates TFs in the circadian clock, inducing TOC1 and CDF1, and repressing ZTL within 20–45 min after N supply (SI Appendix, Fig. S10). Overall, the shoot NxTime gene set shows significant enrichment for genes with peak expression at predawn (45) (Dataset S1, Table S26).
A Fine-Scale Time Course and GRN Establishes the Temporal Hierarchy of N-Signaling Regulators.
Next, we used the DFG network inference method (5, 15) to derive GRNs that reveal the transcriptional logic underlying dynamic N signaling in shoots. The resulting N-response network pruned for precision (Dataset S1, Table S3) now places 155 N-responsive TFs in shoots in a temporal hierarchy (SI Appendix, Fig. S14) and predicts their likely temporal interactions. For example, the 12 TFs that respond earliest to the N signal in shoots (5-min JIT) (SI Appendix, Fig. S14) include TFs previously validated in the N response: LBD37/38/39 (11) and HRS1 (10), and an “early” TF validated herein, CRF4 (Fig. 3). We note that some of the earliest steps of N-signal transduction are also likely to occur via posttranslational modifications (18) or changes in TF localization, as shown for NLP7 (7). This pruned network is further supported by TF-target binding data and reveals a set of 15 TFs that are specific to the N response (Fig. 3, TFs in red) (Dataset S1, Table S12). This establishes the power of applying de novo GRN inference approaches to expression datasets, as shown for GRNs mediating environmental responses in rice (29) and drought responses in Arabidopsis (46).
CRF4 Regulates N Uptake and N Use in Planta.
Our time-based N-regulatory network revealed CRF4 as an early player in mediating the N-signaling response. Indeed, our genome-wide target studies and phenotypic analysis support the key role CRF4 plays in mediating nitrate uptake and use in planta (Fig. 4 and SI Appendix, Fig. S13). In response to N supply, CRF4 represses genes in the N-assimilation pathway, including the high-affinity nitrate transporter NRT2.1 (SI Appendix, Fig. S13C and Dataset S1, Tables S5 and S20), which is repressed under high-N conditions in wild type (44). Additionally, we validated two downstream TF targets of CRF4 and found that SNZ is largely an activator, while CDF1 activates or represses genes in the N-assimilation pathway (Fig. 2). CRF4 targets in shoots include ribosomal proteins, induced within 30–45 min of N supply (SI Appendix, Figs. S2D and S15C and Dataset S1, Table 5). In roots, CRF4 regulates nitrate uptake and root development processes, consistent with the in vivo phenotypes (Fig. 4, SI Appendix, Figs. S13 and S15C, and Dataset S1, Tables S20–S21). N-signaling is an additional role for CRF4, whose only previous described role was in the plant cold response (47). In addition, we discovered that 11/12 members of the CRF family (47) are N-responsive, including eight in shoots (CRF1–6, CRF10, and CRF11) and three (CRF3–4 and CRF11) in roots (Dataset S1, Tables S1 and S2). This highlights a potential role of the CRF family in linking the N response and cytokinin signaling (34). Our study also identifies multiple TFs that link nitrogen and phosphate responses (HHO5/6 and PHL1), as previously shown for HRS1 (10).
In addition to discovering TFs in the N-response network, the “transcriptional logic” of N signaling uncovered herein can also suggest the temporal mode-of-action for TFs and combinatorial TF experiments which will be valuable for the global goal of enhancing NUE. More broadly, our time-centric approach that uses fine-scale time-course data to fuel causal network inference can now be applied to understand any stimulus-driven GRN in any organism. Moreover, the analysis approaches we described—JIT and N-specificity index—can be used to uncover the regulatory structure and signal specificity in any time-series transcriptome datasets. When coupled with genome-wide TF-target binding data [e.g., ChIP-Seq and DAP-Seq (16)] and other layers of genome-wide dynamic interaction data [e.g., chromatin accessibility maps (27)], the approach employed in our time-based study can identify key molecular players, their hierarchy, and other emergent network properties in any complex transcriptional regulatory system in biology, agriculture, or medicine.
Methods
Plant Material and N Treatments.
Arabidopsis thaliana (Col-0) seeds were grown on N-free Murashige and Skoog (MS) media + 1 mM KNO3 for 2 wk in long-day conditions. Two hours after the start of the light period, plants were treated with (i) standard MS media (20 mM KNO3 + 20 mM NH4NO3) (4, 48) or (ii) 20 mM KCl. Triplicate shoot and root samples were harvested at 0, 5, 10, 15, 20, 30, 45, 60, 90, and 120 min after treatments.
CRF4-OX NUE Phenotyping.
Col-0 and CRF4-OX transplanta line (CS2104639) (35) were grown on 30 mM N [½ MS (48)] for 7 d and treated with (i) 10 μΜ β-estradiol or DMSO (solvent) and (ii) low N (1mM nitrate) or high N [30 mM ½ MS (48)] for 5 d. Shoot dry weight, root length, lateral root number, and lengths were assayed. Influx of 15NO3 was assayed as described previously on Col-0, CRF4-OX, and nrt2.1 mutant (44) plants.
Genome-Wide TF-Target Validation.
In planta genome-wide targets of CRF4 were identified by differential expression in Col-0 vs. CRF4-OX line (CS2104639) (35), 24 h after TF induction by 10 µM β-estradiol. The TARGET system (36) was used to identify the genome-wide targets of seven TFs (CRF4, SNZ, CDF1, TGA1, HHO5/6, or PHL1) (Dataset S1, Tables S5–S7 and S22–S25) as in ref. 37. Transformed shoot cells were treated sequentially with (i) 20 mM KNO3 + 20 mM NH4NO3 for 2 h, (ii) 35 μM cycloheximide for 20 min before, and (iii) TF-nuclear localization by 10 μM dexamethasone (37). Shoot cells overexpressing the TF or empty vector were collected in triplicate and transcriptomes profiled on the Illumina NextSEq 500 platform. Genes differentially expressed in response to TF overexpression were identified using DESeq2 package (FDR < 0.05).
Time-Course Transcriptome Profiling.
RNA was extracted from ∼100 mg shoot or root tissue and used for Illumina compatible library preparation. cDNA libraries were sequenced on Illumina HiSEq 2500 v4 platform (100 bp, PE). Gene expression values were determined after quality filtering, genome alignment, and quantile normalization. Differentially expressed genes (FDR < 0.05) were identified by fitting a cubic spline model (df = 5) (23).
GRN Inference and Network Pruning.
DFG (15) was used to infer interactions between 172 TFs and 2,174 NxTime genes in shoots (Dataset S1, Table S1). Experimentally determined TF-target relationships from CRF4, SNZ, and CDF1 were used to perform an AUPR analysis and identify a pruning threshold of precision = 0.345, which was independently confirmed with validated targets of four TFs in the GRN: TGA1, HHO5/6, and PHL1(SI Appendix, Figs. S6 and S7). Further support for predicted TF–target interactions was obtained from in vitro TF-promoter binding for 40 TFs that have binding data (16, 41) (Fig. 3).
JIT Analysis of Time-Series Transcriptome Data.
Each NxTime gene was assigned to the first time bin at which gene expression in N-treated samples is ≥1.5 fold of control (SI Appendix, Fig. S2B and Dataset S1, Tables S1 and S2). Each JIT gene set (SI Appendix, Fig. S2C, blue bars) was analyzed to identify overrepresentation of cis-regulatory motifs [FDR E-value < 0.05, Elefinder (28)] and such cis elements were hierarchically clustered (Fig. 1A). JIT gene sets were also analyzed to identify overrepresented GO terms (49) (Fig. 1B and SI Appendix, Fig. S2D).
Nitrogen-Specificity Index for TFs in the GRN.
For each of the 40 NxTime TFs with in vitro TF-target genome-wide binding data (16, 41) we retrieved genome-wide targets in shoot NxTime set. The TFs with a significantly higher proportion of targets in the NxTime set relative to their genome-wide distribution (one-tailed t test, P < 0.01) were accepted as being specific to the N signal (Dataset S1, Table S12).
Data Availability.
All raw sequence data from this project have been deposited into NCBI’s GEO database (accession no. GSE97500). All data and scripts used in this study are available in Data Dryad, dx.doi.org/10.5061/dryad.248g184.
Supplementary Material
Acknowledgments
This work was supported by NIH Grant R01-GM032877 (to G.M.C.), NSF Grant IOS-1339362 (to G.M.C.), NIH National Research Service Award GM095273 (to A.M.-C.), and NIH National Institute of General Medical Sciences Fellowship 1F32GM116347 (to M.D.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: All raw sequence data from this project have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE97500). All data and scripts used in this study are available at Data Dryad, dx.doi.org/10.5061/dryad.248g184.
See Commentary on page 6325.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1721487115/-/DCSupplemental.
References
- 1.Borlaug N. Feeding a hungry world. Science. 2007;318:359. doi: 10.1126/science.1151062. [DOI] [PubMed] [Google Scholar]
- 2.Zhang X, et al. Managing nitrogen for sustainable development. Nature. 2015;528:51–59. doi: 10.1038/nature15743. [DOI] [PubMed] [Google Scholar]
- 3.Wang R, Okamoto M, Xing X, Crawford NM. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003;132:556–567. doi: 10.1104/pp.103.021253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gutiérrez RA, et al. Systems approach identifies an organic nitrogen-responsive gene network that is regulated by the master clock control gene CCA1. Proc Natl Acad Sci USA. 2008;105:4939–4944. doi: 10.1073/pnas.0800211105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krouk G, Mirowski P, LeCun Y, Shasha DE, Coruzzi GM. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010;11:R123. doi: 10.1186/gb-2010-11-12-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang R, et al. Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiol. 2004;136:2512–2522. doi: 10.1104/pp.104.044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchive C, et al. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat Commun. 2013;4:1713. doi: 10.1038/ncomms2650. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez JM, et al. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014;80:1–13. doi: 10.1111/tpj.12618. [DOI] [PubMed] [Google Scholar]
- 9.Vidal EA, Álvarez JM, Gutiérrez RA. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1.1 nitrate transport function. Plant Signal Behav. 2014;9:e28501. doi: 10.4161/psb.28501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medici A, et al. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat Commun. 2015;6:6274. doi: 10.1038/ncomms7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan P, et al. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc Natl Acad Sci USA. 2017;114:2419–2424. doi: 10.1073/pnas.1615676114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Canales J, Moyano TC, Villarroel E, Gutiérrez RA. Systems analysis of transcriptome data provides new hypotheses about Arabidopsis root response to nitrate treatments. Front Plant Sci. 2014;5:22. doi: 10.3389/fpls.2014.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krouk G, Lingeman J, Colon AM, Coruzzi G, Shasha D. Gene regulatory networks in plants: Learning causality from time and perturbation. Genome Biol. 2013;14:123. doi: 10.1186/gb-2013-14-6-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mirowski P, LeCun Y. 2009. Dynamic factor graphs for time series modeling. Machine Learning and Knowledge Discovery in Databases, eds Buntine W, Grobelnik M, Mladenic D, Shawe-Taylor J (Springer, Berlin), Part 2, pp 128–143.
- 16.O’Malley RC, et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;166:1598. doi: 10.1016/j.cell.2016.08.063. [DOI] [PubMed] [Google Scholar]
- 17.Yan D, et al. NIN-like protein 8 is a master regulator of nitrate-promoted seed germination in Arabidopsis. Nat Commun. 2016;7:13179. doi: 10.1038/ncomms13179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medici A, Krouk G. The primary nitrate response: A multifaceted signalling pathway. J Exp Bot. 2014;65:5567–5576. doi: 10.1093/jxb/eru245. [DOI] [PubMed] [Google Scholar]
- 19.Ruffel S, et al. Nitrogen economics of root foraging: Transitive closure of the nitrate-cytokinin relay and distinct systemic signaling for N supply vs. demand. Proc Natl Acad Sci USA. 2011;108:18524–18529. doi: 10.1073/pnas.1108684108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, von Wirén N. Ammonium as a signal for physiological and morphological responses in plants. J Exp Bot. 2017;68:2581–2592. doi: 10.1093/jxb/erx086. [DOI] [PubMed] [Google Scholar]
- 21.Krapp A. Plant nitrogen assimilation and its regulation: A complex puzzle with missing pieces. Curr Opin Plant Biol. 2015;25:115–122. doi: 10.1016/j.pbi.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 22.Patterson K, et al. Distinct signalling pathways and transcriptome response signatures differentiate ammonium- and nitrate-supplied plants. Plant Cell Environ. 2010;33:1486–1501. doi: 10.1111/j.1365-3040.2010.02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poultney CS, et al. Sungear: Interactive visualization and functional analysis of genomic datasets. Bioinformatics. 2007;23:259–261. doi: 10.1093/bioinformatics/btl496. [DOI] [PubMed] [Google Scholar]
- 25.Bloom AJ. Photorespiration and nitrate assimilation: A major intersection between plant carbon and nitrogen. Photosynth Res. 2015;123:117–128. doi: 10.1007/s11120-014-0056-y. [DOI] [PubMed] [Google Scholar]
- 26.Yilmaz A, et al. AGRIS: The arabidopsis gene regulatory information server, an update. Nucleic Acids Res. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan AM, et al. Mapping and dynamics of regulatory DNA and transcription factor networks in A. thaliana. Cell Rep. 2014;8:2015–2030. doi: 10.1016/j.celrep.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Swaminathan K, Hudson ME. Rapid, organ-specific transcriptional responses to light regulate photomorphogenic development in dicot seedlings. Plant Physiol. 2011;156:2124–2140. doi: 10.1104/pp.111.179416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilkins O, et al. EGRINs (Environmental Gene Regulatory Influence Networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell. 2016;28:2365–2384. doi: 10.1105/tpc.16.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marbach D, et al. DREAM5 Consortium Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li Y, Pearl SA, Jackson SA. Gene networks in plant biology: Approaches in reconstruction and analysis. Trends Plant Sci. 2015;20:664–675. doi: 10.1016/j.tplants.2015.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Gaudinier A, Brady SM. Mapping transcriptional networks in plants: Data-driven discovery of novel biological mechanisms. Annu Rev Plant Biol. 2016;67:575–594. doi: 10.1146/annurev-arplant-043015-112205. [DOI] [PubMed] [Google Scholar]
- 33.Penfold CA, Buchanan-Wollaston V, Denby KJ, Wild DL. Nonparametric Bayesian inference for perturbed and orthologous gene regulatory networks. Bioinformatics. 2012;28:i233–i241. doi: 10.1093/bioinformatics/bts222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ruffel S, Poitout A, Krouk G, Coruzzi GM, Lacombe B. Long-distance nitrate signaling displays cytokinin dependent and independent branches. J Integr Plant Biol. 2016;58:226–229. doi: 10.1111/jipb.12453. [DOI] [PubMed] [Google Scholar]
- 35.Coego A, et al. TRANSPLANTA Consortium The TRANSPLANTA collection of Arabidopsis lines: A resource for functional analysis of transcription factors based on their conditional overexpression. Plant J. 2014;77:944–953. doi: 10.1111/tpj.12443. [DOI] [PubMed] [Google Scholar]
- 36.Bargmann BO, et al. TARGET: A transient transformation system for genome-wide transcription factor target discovery. Mol Plant. 2013;6:978–980. doi: 10.1093/mp/sst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Para A, et al. Hit-and-run transcriptional control by bZIP1 mediates rapid nutrient signaling in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:10371–10376. doi: 10.1073/pnas.1404657111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doidy J, et al. “Hit-and-Run” transcription: De novo transcription initiated by a transient bZIP1 “hit” persists after the “run”. BMC Genomics. 2016;17:92. doi: 10.1186/s12864-016-2410-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stolovitzky G, Prill RJ, Califano A. Lessons from the DREAM2 challenges. Ann N Y Acad Sci. 2009;1158:159–195. doi: 10.1111/j.1749-6632.2009.04497.x. [DOI] [PubMed] [Google Scholar]
- 40.Wang M, et al. LegumeGRN: A gene regulatory network prediction server for functional and comparative studies. PLoS One. 2013;8:e67434. doi: 10.1371/journal.pone.0067434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ecker JR. 2017 Base-pair resolution atlases of the plant cistrome and epicistrome. Available at neomorph.salk.edu/dap_web/pages/index.php. Accessed November 8, 2016.
- 42.Wang YY, Hsu PK, Tsay YF. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012;17:458–467. doi: 10.1016/j.tplants.2012.04.006. [DOI] [PubMed] [Google Scholar]
- 43.Muños S, et al. Transcript profiling in the chl1-5 mutant of Arabidopsis reveals a role of the nitrate transporter NRT1.1 in the regulation of another nitrate transporter, NRT2.1. Plant Cell. 2004;16:2433–2447. doi: 10.1105/tpc.104.024380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Filleur S, et al. An arabidopsis T-DNA mutant affected in Nrt2 genes is impaired in nitrate uptake. FEBS Lett. 2001;489:220–224. doi: 10.1016/s0014-5793(01)02096-8. [DOI] [PubMed] [Google Scholar]
- 45.Mockler TC, et al. The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol. 2007;72:353–363. doi: 10.1101/sqb.2007.72.006. [DOI] [PubMed] [Google Scholar]
- 46.Bechtold U, et al. Time-series transcriptomics reveals that AGAMOUS-LIKE22 affects primary metabolism and developmental processes in drought-stressed Arabidopsis. Plant Cell. 2016;28:345–366. doi: 10.1105/tpc.15.00910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zwack PJ, Compton MA, Adams CI, Rashotte AM. Cytokinin response factor 4 (CRF4) is induced by cold and involved in freezing tolerance. Plant Cell Rep. 2016;35:573–584. doi: 10.1007/s00299-015-1904-8. [DOI] [PubMed] [Google Scholar]
- 48.Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant. 1962;15:473–497. [Google Scholar]
- 49.Katari MS, et al. VirtualPlant: A software platform to support systems biology research. Plant Physiol. 2010;152:500–515. doi: 10.1104/pp.109.147025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence data from this project have been deposited into NCBI’s GEO database (accession no. GSE97500). All data and scripts used in this study are available in Data Dryad, dx.doi.org/10.5061/dryad.248g184.