Significance
The virulence of numerous bacterial pathogens is dependent on nanomachines that inject tens of proteins (effectors) into host cells. Effectors are often functionally redundant or act in an additive manner to usurp cellular processes, limiting their characterization via traditional top-down loss-of-function assays. To overcome this issue, we developed an innovative synthetic bottom-up platform to conduct gain-of-function screens. These screens identified previously missed Shigella effectors that inhibit induced epithelial cell death, an important arm of the host innate immune system triggered in response to invading pathogens. Our studies provide new mechanistic insight into the sophisticated strategies by which Shigella effectors interact with the host to enable this pathogen to establish a replicative niche within the cytosol of infected epithelial cells.
Keywords: type III secretion system, innate immunity, T3SS, pyroptosis, necrosis
Abstract
Over the course of an infection, many Gram-negative bacterial pathogens use complex nanomachines to directly inject tens to hundreds of proteins (effectors) into the cytosol of infected host cells. These effectors rewire processes to promote bacterial replication and spread. The roles of effectors in pathogenesis have traditionally been investigated by screening for phenotypes associated with their absence, a top-down approach that can be limited, as effectors often act in a functionally redundant or additive manner. Here we describe a synthetic Escherichia coli-based bottom-up platform to conduct gain-of-function screens for roles of individual Shigella effectors in pathogenesis. As proof of concept, we screened for Shigella effectors that limit cell death induced on cytosolic entry of bacteria into epithelial cells. Using this platform, in addition to OspC3, an effector known to inhibit cell death via pyroptosis, we have identified OspD2 and IpaH1.4 as cell death inhibitors. In contrast to almost all type III effectors, OspD2 does not target a host cell process, but rather regulates the activity of the Shigella type III secretion apparatus limiting the cytosolic delivery (translocation) of effectors during an infection. Remarkably, by limiting the translocation of a single effector, VirA, OspD2 controls the timing of epithelial cell death via calpain-mediated necrosis. Together, these studies provide insight into the intricate manner by which Shigella effectors interact to establish a productive intracytoplasmic replication niche before the death of infected epithelial cells.
Induced cell death is a major arm of the host innate immune response activated in response to recognition of invading bacterial pathogens. While the majority of studies in this area have focused on macrophages, infected epithelial cells, particularly those lining mucosal surfaces, behave similarly (1). Cell death results in the eradication of the niche that intracellular pathogens use for replication, as well as the release of alarmins and proinflammatory cytokines that recruit additional immune cells to sites of infection (2). In response, bacterial pathogens, particularly those that invade host cells, have evolved intricate means to manipulate cell death pathways to their own advantage (3); for example, Shigella species, professional intracytoplasmic pathogens, actively trigger cell death of macrophages while suppressing cytotoxicity of infected intestinal epithelial cells.
The causative agents of bacillary dysentery, Shigella are transmitted via a fecal-oral route. On reaching the colon, Shigella traverse the intestinal mucosa through microfold (M) cells after which they are engulfed by underlying resident macrophages. Once internalized, Shigella trigger rapid macrophage cell death via pyroptosis, primarily due to activation of canonical inflammasomes (4). This results in the release of viable Shigella at the basolateral surface of epithelial cells, which they preferentially invade. Within epithelial cells, Shigella inhibit cell death via both pyroptosis (5) and necrosis (6) to establish a replicative niche within the colonic epithelium.
The pathogenesis of Shigella, like many other Gram-negative bacteria, is dependent on a type III secretion system (T3SS), a syringe-like nanomachine that serves as a conduit to transfer proteins (effectors) directly from bacteria into the cytosol of targeted host cells (7). In addition to components of the translocon, the portion that forms a pore in the host cell membrane, T3SSs translocate tens to hundreds of effectors into host cells. In the case of Shigella, at least 30 secreted effectors have been identified, the majority of which are encoded on a large virulence plasmid (8, 9). The roles of effectors from Shigella and other pathogens have been traditionally studied via top-down approaches focused on screening for loss-of-function phenotypes associated with strains that no longer encode one or more effectors. However, this approach is limited when studying effectors that work in a functionally redundant or additive manner, a not too uncommon occurrence. For example, in the case of Shigella, one set of at least five effectors coordinates uptake into epithelial cells (10–13), while another comparably sized set inhibits the production of proinflammatory cytokines by blocking NF-κB activation (14–22).
Here we describe the development of a complementary bottom-up platform to conduct gain-of-function screens to identify roles of individual Shigella effectors in pathogenesis. This approach is an extension of a recombineering-based synthetic biology platform that we previously developed to introduce variants of the Shigella T3SS into laboratory strains of Escherichia coli (23, 24). The newest strain described herein, mT3.1_E. coli, not only invades epithelial cells at levels equivalent to wild-type (WT) Shigella, but also recognizes “added-back” Shigella effectors as secreted proteins. Using this platform, we find that the introduction of OspC3, IpaH1.4, or OspD2 into mT3.1_E. coli suppresses bacterial-triggered epithelial cell death. Notably, the absence of either of the latter two effectors was not observed to trigger excess cell death in a previous reciprocal top-down screen (5). Our follow-up studies demonstrate that in contrast to almost all characterized effectors, OspD2 does not target a host cell process, but rather regulates the activity of the Shigella type III secretion apparatus (T3SA), limiting effector translocation into host cells. Furthermore, we determined that OspD2 regulates Shigella-triggered cell death, not by inhibiting the delivery of bacterial PAMPS (pathogen-associated molecular patterns), but rather by restricting the translocation of another effector, VirA, into host cells, thus limiting cell death via calpain-mediated necrosis. These results highlight the complex means by which Shigella effectors interact to establish a replicative niche within the cytosol of infected epithelial cells.
Results
A Synthetic Bottom-Up Platform to Study Shigella Type III Secreted Effectors.
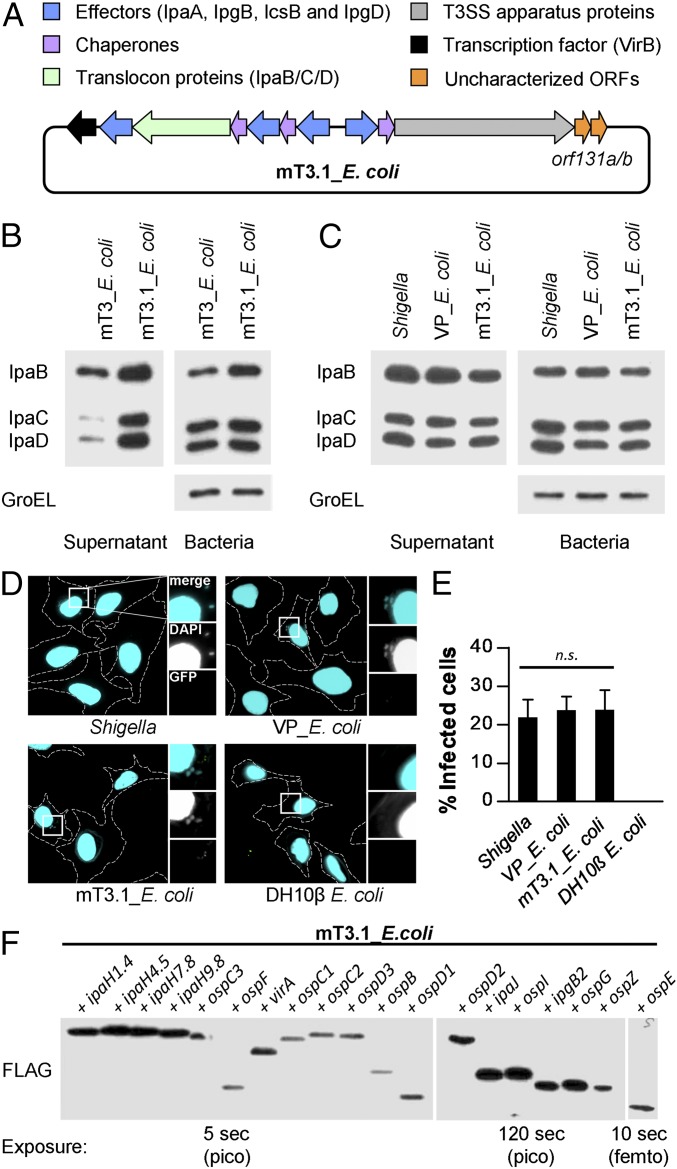
We recently developed a recombineering approach that we used to transfer a 31-kb region of the large Shigella virulence plasmid (VP) onto a smaller autonomously replicating plasmid (23, 24). The introduction into DH10β E. coli of this plasmid, which encodes all of the structural components of the Shigella T3SA plus a few embedded effectors, plus a second that carries VirB, a major T3SS transcriptional regulator, resulted in the generation of mT3_E. coli. Like WT Shigella, mT3_E. coli invade and enter the cytosol of epithelial cells (HeLa), albeit with lower efficiency (24). Here we extended the region of VP DNA introduced into DH10β E. coli to include two additional small, poorly characterized genes, orf131a and orf131b (Fig. 1A). This modification led to the development of mT3.1_E. coli, a strain that displays increased T3SA activity. In contrast to mT_E. coli, mT3.1_E. coli secretes components of the translocon (IpaB, IpaC, and IpaD) at levels equivalent to both WT Shigella and VP_E. coli, DH10β E. coli that carry the Shigella VP (Fig. 1 B and C). In addition, mT3.1_E. coli invade a similar percentage of HeLa cells as WT Shigella and VP_E. coli, as assessed using an inside/outside fluorescent microscopy assay that differentiates between internalized and extracellular bacteria (Fig. 1 D and E).
Fig. 1.
A synthetic bottom-up platform to study Shigella type III secreted effectors. (A) Schematic representations of the T3SS-encoding plasmid present in mT3.1_E. coli. (B and C) Secretion assays of designated strains. Each was grown under conditions that activate expression of its T3SS. A portion of the bacteria was removed and pelleted, and the remaining bacteria were resuspended in PBS plus Congo red, a condition that activates the machine. Proteins in the initial pellet and supernatant fractions collected at 30 min after the activation of secretion were separated by SDS/PAGE and transferred to nitrocellulose membranes. Membranes were immunoblotted with designated antibodies. GroEL, an unrelated cytosolic protein, served as a loading control. (D and E) HeLa cells were infected with each strain at an MOI of 100 for 30 min, followed by the addition of gentamicin to the media to kill extracellular bacteria. After 30 min, cells were labeled using an inside/outside microscopy assay to quantify intracellular (cyan) vs. extracellular (cyan/green) bacteria. (D) Representative images obtained with a 60× objective of labeled cells, with perimeters denoted by dashed lines. Enlarged images are of the boxed regions. (E) Quantification of infected cells. For each infection, 50 HeLa cells were counted. Data are expressed as the mean ± SD of three independent experiments. n.s., nonsignificant. (F) Secretion assays conducted as described in B and C. Membranes were immunoblotted with an antibody that recognizes the FLAG epitope. Multiple exposures were obtained to detect secretion of all effectors. femto, femtogram-level detection; pico, picogram-level detection. Blots in B, C, and F are representative of at least three experimental repeats.
We next investigated the breadth of Shigella effectors recognized as secreted proteins by mT3.1_E. coli. To directly compare levels of secreted effectors, we studied the behavior of FLAG-tagged variants. Furthermore, to avoid issues with the expression of effectors, which in Shigella are expressed and secreted in two waves (25), we studied effector variants expressed under the control of an isopropyl β-d-1-thiogalactopyranoside (IPTG)-inducible promoter. As shown in Fig. 1F, each effector tested was recognized as a secrete protein and, notably, found in the supernatant fractions at similar relative levels as previously observed when the same effector constructs were studied in WT Shigella (26). These observations suggested that mT3.1_E. coli could serve as a bottom-up platform to study Shigella effectors.
Activation of Noncanonical Inflammasomes by mT3.1_E. coli Is Suppressed by the Addition of OspD2, IpaH1.4, or OspC3.
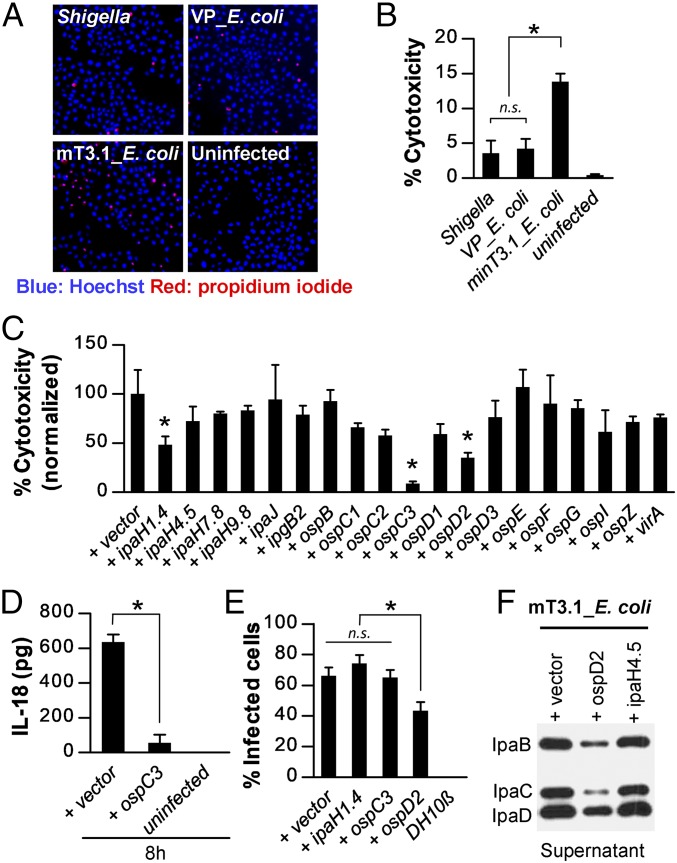
Given our prior observations that mT3_E. coli not only invade, but also escape into the cytosol of infected epithelial cells (24), we hypothesized that mT3.1_E. coli might serve to identify effectors involved in postentry steps of Shigella pathogenesis, including induced host cell death. To investigate this possibility, we compared the levels of cytotoxicity of epithelial cells infected with mT3.1_E. coli, WT Shigella, and VP_E. coli. To distinguish between live and dead cells, we used a fluorescent microscopy assay, in which all cell nuclei were visualized with Hoechst, but only those with a compromised plasma membrane with propidium iodide (PI). As shown in Fig. 2 A and B, HeLa cells infected with mT3.1_E. coli exhibited substantially more cytotoxicity than those infected with WT Shigella or VP_E. coli (13% vs. 4% vs. 4%), suggesting that mT3.1_E. coli trigger greater cytotoxicity due to differences in effector content rather than to inherent differences between Shigella and DH10β E. coli.
Fig. 2.
mT3.1_E. coli induces epithelial cell death via activation of noncanonical inflammasomes that are suppressed. (A and B) HeLa cells were infected with each strain at an MOI of 100 for 30 min, followed by the addition of gentamicin. After another 2 h, cells were stained with Hoechst (blue) and PI (red). (A) Representative images of stained cells obtained with 4× objective. (B) Quantification of percentage of PI-positive cells (percent cytotoxicity). (C) HeLa cells were infected with mT3.1_E. coli that express each designated effector at an MOI of 100 for 60 min, followed by the addition of gentamicin. After another 1.5 h, the cells were stained with Hoechst and PI. The percentage cytotoxicity normalized to vector control is shown. For each infection, at least 2,000 cells were imaged. *P < 0.05 compared with (+) vector control. (D) Quantification of IL-18 secreted by infected HeLa cells at 8 h postinvasion. (E) At 1 hpi of HeLa cells with each strain at an MOI of 100, the percentage of cells containing intracellular bacteria was determined by an inside/outside microscopy assay, as described in Fig. 1. Data are expressed as the mean ± SD of three experimental repeats. (F) Secretion assays of designated strains conducted as described in Fig. 1. Membranes were immunoblotted with designated antibodies. The blot shown is representative of three experimental repeats. All other data are expressed as the mean ± SD of at least two experimental repeats, each with two technical replicates (n ≥ 4). *P < 0.05 as indicated. n.s., nonsignificant.
We proceeded to screen for VP-encoded effectors that suppress mT3.1_E. coli triggered epithelial cell death. To increase the detection window for this screen, before adding gentamicin to kill extracellular bacteria, we infected epithelial cells for 60 min instead of 30 min, a change that resulted in a doubling of the percentages of both infected cells and dying cells (SI Appendix, Fig. S1). Our subsequent high-throughput and confirmatory screens identified three effectors—OspC3, IpaH1.4, and OspD2—that inhibited mT3.1_E. coli-triggered epithelial cell death (Fig. 2C and SI Appendix, Fig. S2). Of note, in a prior top down-screen, OspC3 was the sole effector identified to suppress epithelial cell death (5). Our observation that the addition of OspC3 suppresses cell death provided a proof of concept for our approach. In addition, the prior determination that OspC3 prevents Shigella triggered cell death via pyroptosis due to activation of noncanonical inflammasomes (5) suggested that mT3.1_E. coli trigger cell death via this pathway. Indeed, as shown in Fig. 2D, HeLa cells infected with mT3.1_E. coli but not OspC3-expressing mT3.1_E. coli secrete elevated levels of IL-18, a proinflammatory cytokine released by pyroptotic epithelial cells.
OspD2 Inhibits mT3.1_E. coli Host Cell Invasion by Inhibiting Activity of the Shigella T3SA.
We next focused our efforts on dissecting the role of OspD2, an effector about which essentially nothing was previously known, in regulating bacteria-triggered cell death. We first investigated whether OspD2 plays a role in mT3.1_E. coli host cell invasion. Using the inside/outside microscopy assay, we found that OspD2-expressing mT3.1_E. coli invade approximately 50% fewer HeLa cells than those expressing OspC3 or IpaH1.4 (Fig. 2E). The degree to which OspD2 blocks invasion was essentially equivalent to the degree to which it suppresses cell death. Thus, we hypothesized that OspD2 directly blocks either the translocation or the activity of mT3.1_E. coli effectors that promote host cell invasion. The former turned out to be true, as we observed that expression of OspD2 inhibits the secretion of IpaB, IpaC and IpaD, the three secreted components of the translocon (Fig. 2F). Notably, IpaH4.5-expressing mT3.1_E. coli did not exhibit a secretion defect, even though this effector is one of the most highly secreted effectors (Fig. 1F). These observations suggest that secretion inhibition is OspD2-specific.
OspD2 Regulates the Levels of Proteins Translocated into the Cytosol of Infected Cells.
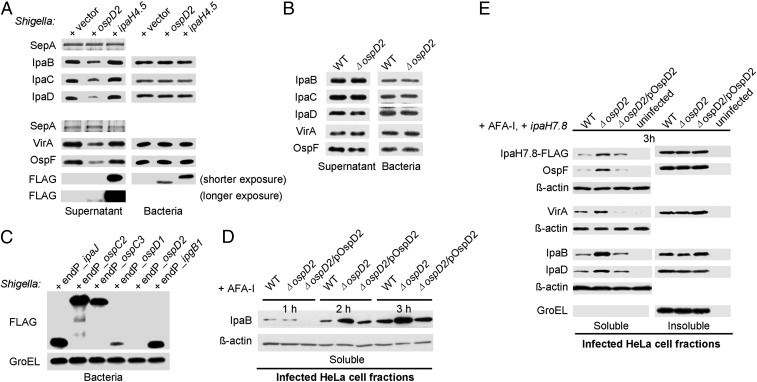
We next investigated whether OspD2 exhibits a similar regulatory role in the context of Shigella. As observed with mT3.1_E. coli, IPTG-induced expression of OspD2 resulted in decreased secretion of all three components of the translocon, IpaB, IpaC, and IpaD, as well as the two natively expressed effectors tested, OspF and VirA, but not the unrelated type V secreted protein, SepA (Fig. 3A). Consistent with this secretion inhibition, as observed with mT3.1_E. coli, IPTG-induced expression of OspD2 before contact with host cells inhibited epithelial cell invasion (SI Appendix, Fig. S3A). However, strains lacking OspD2 (ΔospD2 Shigella) secreted equivalent levels of IpaB, IpaC, IpaD, OspF, and VirA as WT Shigella (Fig. 3B), and ΔospD2 and WT Shigella invaded equivalent percentages of HeLa cells (SI Appendix, Fig. S3B). These seemingly conflicting observations suggested that at the onset of a Shigella infection, natively expressed OspD2 does not regulate activity of the Shigella T3SA.
Fig. 3.
OspD2 regulates the levels of proteins translocated into the cytosol of infected cells. (A and B) Secretion assays of Shigella as described in Fig. 1. Membranes were immunoblotted with the designated antibodies. The upper fragment of SDS/PAGE gels of supernatant fractions were silver-stained to visualize SepA, the loading control. (C) Immunoblots of FLAG-tagged effectors expressed under the control of their endogenous VirB-regulated promoters present in Shigella grown under conditions that induce T3SS expression, but not activation. (D and E) HeLa cells infected at an MOI of 30 with Shigella strains expressing Afa-1 at designated time points were treated with RIPA buffer to lyse mammalian, but not bacterial, cell membranes. The soluble and insoluble (bacterial-containing) fractions were separated and immunoblotted with designated antibodies. GroEL served as a fractionation and loading control for the insoluble fraction, and β-actin served as a loading control for the soluble fraction. Each immunoblot is representative of at least three experimental repeats.
Given recent evidence that the Shigella T3SA is inactivated after invasion of host cells (27), we reasoned that OspD2 might inhibit effector translocation of host cells postinvasion, perhaps after accumulating to elevated levels within the bacteria. Consistent with this hypothesis, natively expressed OspD2-FLAG was the only one of the six VirB-regulated effectors tested that we were unable to detect within Shigella grown under conditions that activate VirB, the transcription factor that coordinately regulates expression of multiple Shigella effectors and structural components of its T3SA (25) (Fig. 3C).
We next investigated whether OspD2 regulates T3SA activity after invasion of host cells by comparing the levels of proteins translocated into epithelial cells infected with ΔospD2 and WT Shigella. For these studies, to increase the detection of translocated proteins, HeLa cells were infected with Shigella expressing Afa-1, an adhesin that promotes Shigella attachment and uptake into host cells (28). As shown in Fig. 3D, starting as early as 2 h postinvasion (hpi), we observed significantly higher levels of IpaB in the cytosol of cells infected with ΔospD2 Shigella. Similarly, at 3 hpi, we observed increased levels of translocated natively expressed IpaB, IpaD, OspF, VirA, and IPTG-regulated IpaH7.8-FLAG in the cytosol of host cells infected with ΔospD2 Shigella (Fig. 3E). In each case, the hypertranslocation phenotype of ΔospD2 Shigella was complemented by the introduction of low-copy number plasmid expressing (untagged) OspD2 via its native promoter (Fig. 3E) or OspD2-FLAG via an IPTG-regulated promoter (SI Appendix, Fig. S4), the latter further supporting a role for OspD2 in the regulation of postinvasion T3SA activity. Notably, the levels of IpaB, IpaC, IpaD, OspF, and VirA present in the insoluble pellet fractions of WT and ΔospD2 Shigella infected cells, which contain intact bacteria, were similar, suggesting that OspD2 is not a transcriptional regulator, but rather directly controls the secretory activity of the T3SA.
OspD2 Regulates Timing of Epithelial Cell Death.
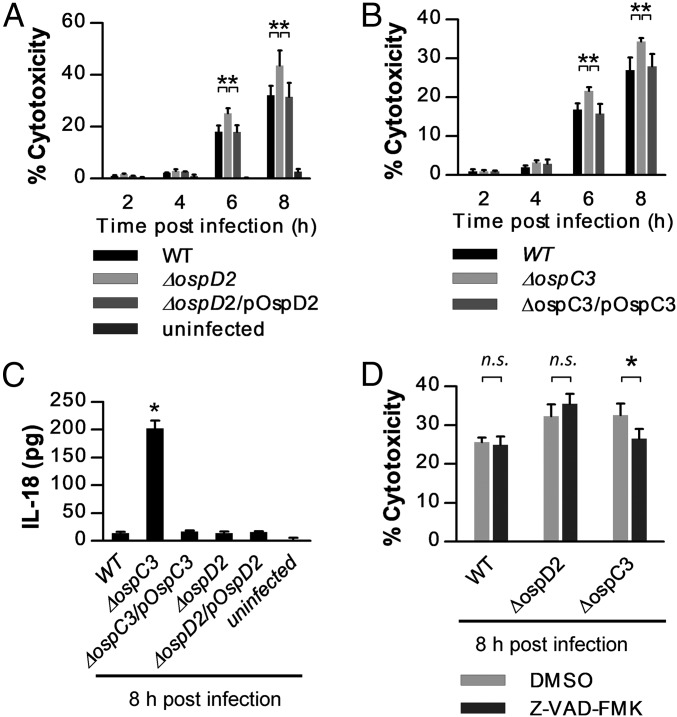
We next investigated whether, as in the context of mT3.1_E. coli, OspD2 modulates Shigella-triggered epithelial cell death. By 6 hpi, cells infected with ΔospD2 Shigella exhibited significantly more cytotoxicity than those infected with WT Shigella, a phenotype fully complemented by endogenously expressed (untagged) OspD2 (Fig. 4A). We also examined the consequences of overexpression of OspD2 postinvasion of host cells by inducing its expression with IPTG along with gentamicin, thus restricting OspD2 expression to live intracellular Shigella. When assayed at 8 h postinvasion, excess OspD2 expression markedly inhibits epithelial cell death (SI Appendix, Fig. S5A).
Fig. 4.
OspD2 and OspC3 regulate epithelial cell death via different pathways. HeLa cells were infected with each strain at an MOI of 100 for 20 min before the addition of gentamicin to media. (A, B, and D) At designated time points, cells were labeled with Hoechst and PI, and the percent cytotoxicity was quantified as described in Fig. 2. (C) Quantification of IL-18 secreted by infected HeLa cells at 8 h postinvasion. In D, Z-VAD-FMK, a pan-caspase inhibitor, or DMSO (vehicle control) was added to the cells at 3 h before the infection. All data are expressed as the mean ± SD of at least two experimental repeats, each with two technical replicates (n ≥ 4). *P < 0.05 as compared with WT or as indicated by brackets. n.s., nonsignificant.
Interestingly, under the same infection conditions, we observed similar levels of cytotoxicity of epithelial cells infected with ΔospC3 (Fig. 4B) and ΔospD2 Shigella, yet only cells infected with ΔospC3 released IL-18, a reporter of cell death via pyroptosis (Fig. 4C). Similarly, the addition of the pan-caspase inhibitor, Z-VAD-FMK, which inhibits cell death via both pyroptosis and apoptosis, suppressed ΔospC3 but not ΔospD2 Shigella triggered HeLa cell death (Fig. 4D). Together these observations suggested that OspD2 does not inhibit cell death via either pyroptosis or apoptosis.
OspD2 shares extensive homology (29% identify and 66% similarity) with EspL, an enteropathogenic E. coli (EPEC) type III effector (SI Appendix, Fig. S5B). EspL is a cysteine protease that limits EPEC-triggered epithelial cell death by targeting host cell proteins for degradation (29). To test whether OspD2 is also a cysteine protease, we generated OspD2_C79A, an OspD2 variant engineered to carry a mutation that would render it catalytically dead if it were a cysteine protease. Overexpression of OspD2 and OspD2_C79A postinvasion similarly reduce cell death (SI Appendix, Fig. S5A). Thus, it is highly unlikely that OspD2 is a cysteine protease that targets degradation of mammalian or bacterial proteins.
OspD2 Limits VirA-Triggered Calpain-Mediated Cell Death.
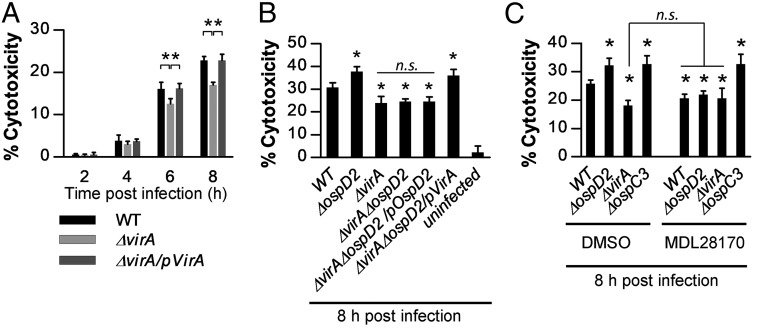
Based on the lytic appearance of the PI-stained epithelial cells infected with ΔospD2 and WT Shigella, we next investigated whether OspD2 delays epithelial cell death by limiting VirA-mediated necrosis (30). VirA, a type III secreted effector, is involved in multiple steps of Shigella pathogenesis. Initially, VirA works in concert with other effectors to mediate epithelial invasion and cell-to-cell spread. Later, after establishment of an infection, VirA-mediated calpain activation triggers epithelial cell death via necrosis (30). Under our experimental infection conditions, starting as early as 6 hpi, as reported previously, we observed that ΔvirA Shigella triggers substantially less cell death than WT Shigella (Fig. 5A). This phenotype is not due to decreased invasion or spread, as under these same infection conditions, we isolated equivalent numbers of ΔvirA and WT Shigella from infected HeLa cells at 2, 4, and 6 hpi (SI Appendix, Fig. S6).
Fig. 5.
OspD2 limits VirA-triggered calpain-mediated cell death. (A, B, and C) HeLa cells were infected with each strain at an MOI of 100 for 20 min before the addition of gentamicin to media. At designated time points, cells were labeled with Hoechst and PI, and the percent cytotoxicity was quantified as described in Fig. 2. In C, MDL28170, a calpain inhibitor, or DMSO (vehicle control) was added to the cells at 3 h before the infection. All data are expressed as the mean ± SD of two experimental repeats, each with at least two technical replicates (n ≥ 4). *P < 0.05 compared with WT or as indicated by the brackets. n.s., nonsignificant.
Next, to directly test whether OspD2 regulates epithelial cell death by limiting VirA translocation (Fig. 3E), we monitored levels of epithelial cell death triggered in response to WT, ΔvirA, ΔospD2, and ΔvirAΔospD2 Shigella. At 8 hpi, epithelial cells infected with the single deletion strains behaved as expected, with increased cytotoxicity observed with ΔospD2 and decreased cytotoxicity observed with ΔvirA Shigella. Remarkably, cells infected with ΔvirAΔospD2 Shigella exhibited levels of cytotoxicity essentially identical to those of cells infected with ΔvirA Shigella (Fig. 5B), suggesting the excess cell death observed in the absence of OspD2 is directly attributable to VirA translocation. The increased cytotoxicity of epithelial cells infected with ΔospD2 Shigella was similarly suppressed by the addition to the media of either of two well-established calpain inhibitors, ALLN and MDL28170 (Fig. 5C and SI Appendix, Fig. S7). Furthermore, the addition of MDL28170 had no effect on the cytotoxicity of cells infected with ΔvirA Shigella (Fig. 5C), supporting prior work establishing that in the absence of VirA, Shigella no longer induces calpain-mediated cell death (30). Finally, in the presence of MDL28170, HeLa cells infected with WT, ΔospD2, or ΔvirA Shigella exhibited equivalent levels of cytotoxicity (Fig. 5C), demonstrating that the removal of VirA or addition of a calpain inhibitor equivalently suppresses Shigella-triggered epithelial cell death. Together, these observations strongly suggest that by limiting VirA translocation into epithelial cells, OspD2 regulates the extent and timing of epithelial cell death via calpain-mediated necrosis.
Discussion
Induced inflammatory death of intestinal epithelial cells is emerging as a major arm of the host innate immune response activated in response to invading intestinal pathogens, including Shigella species. In response, Shigella, professional intracytoplasmic pathogens, have evolved intricate means of modulating host cell death signaling pathways to establish a productive replicative niche within the cytosol of intestinal epithelial cells. Here we describe the development of an innovative synthetic bottom-up platform to interrogate roles for Shigella effectors in specific steps in pathogenesis that circumvents issues with functional redundancy. Using this bottom-up platform, we have identified three effectors—OspC3, OspD2, and IpaH1.4—that suppress Shigella-triggered epithelial cell death, the latter two of which were previously missed by a reciprocal traditional top-down loss-of-function screen, thus demonstrating the strength of this platform. Our follow-up studies with OspD2 establish that it regulates the timing of epithelial cell death by modulating the activity of the Shigella T3SA, limiting the translocation of effectors into host cells during an infection.
OspD2 is the third effector from a bacterial pathogen that has been demonstrated to regulate translocation activity, the other two being Yersinia YopK and EPEC/EHEC EspZ (31, 32). After being translocated into host cells, YopK and EspZ likely block translocation through interactions with the translocon (31, 33). In contrast, our data suggest that OspD2 inhibits T3SA activity from within Shigella, as we observe inhibition of effector secretion into the media as well as translocation into host cells. Notably, unlike its homolog OspD1, a type III effector involved in regulating the activity of the transcription of second-wave Shigella effectors (34), we observe no evidence of similar activity for OspD2. One intriguing possibility is that OspD2 is an impassable substrate that over time accumulates and clogs the secretion apparatus. However, as shown in Fig. 1F, low levels of OspD2 are secreted into the media and, in previous studies, translocated into host cells (35), at least when under the control of an IPTG-regulated promoter. Additional studies are needed to assess the molecular mechanism by which OspD2 regulates T3SA activity.
Why would Shigella want to limit the translocation of effectors into epithelial cells? Our data demonstrate that by limiting VirA translocation, OspD2 delays calpain-mediated cell death via necrosis, thus enabling intracytoplasmic Shigella to replicate to higher titers before epithelial cell death occurs. Of note, however, our observations suggest that OspD2 acts to globally inhibit effector translocation. Might limiting the translocation of other effectors, such as those involved in inhibiting the production of proinflammatory cytokines or cell death via pyroptosis, have detrimental effects on Shigella survival? Interestingly, once inside epithelial cells, Shigella modify LPS, converting it to a hypoacylated state associated with reduced NF-κB and inflammasome activation (36), likely negating the need for effectors that inhibit these pathways at later time points. Thus, at this point the strategy likely turns to one of preserving the Shigella’s intracytoplasmic niche within the intestinal epithelium such that at a later point, when the massive intestinal inflammation, tissue destruction, and a profuse inflammatory diarrhea associated with Shigella infection have developed, the intracytoplasmic Shigella escape at high titers from dying epithelial cells into the intestinal lumen and spread to new hosts via a fecal-oral route.
Materials and Methods
Cell Lines.
HeLa cells (American Type Culture Collection; CCL2) were cultured as recommended in a 5% CO2 incubator at 37 °C. Bacterial strains, plasmids, and primers are summarized in SI Appendix, Tables S1–S3. Strain construction, growth, and infection conditions are described in SI Appendix, Materials and Methods.
Inside/Outside Microscopy.
As described previously (24), HeLa cells on coverslips were infected at a multiplicity of infection (MOI) of 100. At 1 h after the start of an infection, the coverslips were washed five times with PBS and then fixed with paraformaldehyde. Extracellular bacteria were labeled with rabbit anti-E. coli or anti-Shigella polyclonal antibodies, followed by labeling with anti-rabbit Alexa Fluor 488 secondary antibody. Cells were then permeabilized with 0.5% Triton X-100 and stained with DAPI. Bacteria labeled with blue, but not green, were defined as intracellular, and those labeled with both were defined as extracellular. HeLa cells containing intracellular bacteria were defined as infected. Additional details are provided in SI Appendix, Materials and Methods.
Cytotoxicity Assay.
HeLa cells were infected at an MOI of 100. At designated time points, cells were incubated with Hoechst and PI for 30 min before imaging. Stained cell nuclei were identified and quantified using CellProfiler 2.0 (37). Cell nuclei labeled with only blue were defined as live, and those labeled with both blue and red were defined as dead. Additional details are provided in SI Appendix, Materials and Methods.
Translocation Assays.
As reported previously (38), HeLa cells were infected at an MOI of 30 for 60 min before the addition of gentamicin to the media. At designated time points, cells were incubated in ice cold radioimmunoprecipitation assay (RIPA) buffer to lyse mammalian cells, but not bacterial cells. The pellet fraction, containing intact bacteria, was resuspended in Laemmli sample buffer while the supernatant was centrifuged a second time, and the resulting supernatant was designated the soluble fraction. Additional details are provided in SI Appendix, Materials and Methods.
Detailed information on all other methods, including secretion assays, quantification of secreted IL-18, gentamicin protection assays, alignment of protein sequences, and statistical analysis, are available in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank W. Spears, W. Polachek, and N. Ernst for helping with plasmid construction; L. Knodler, J. Coers, C. Gonzalez-Prieto, L. Goers, and J. Lynch for critically reading the manuscript; W. L. Picking, A. T. Maurelli, and M. B. Goldberg for sharing reagents/strains; and J. Boyd for assisting with high-content microscopy. This work was supported by National Institutes of Health Grant R01 AI064285 and a Brit d’Arebeloff Research Scholar award (to C.F.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801310115/-/DCSupplemental.
References
- 1.Sellin ME, Müller AA, Hardt W-D. Consequences of epithelial inflammasome activation by bacterial pathogens. J Mol Bio. 2017;430:193–206. doi: 10.1016/j.jmb.2017.03.031. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen I, Miao EA. Pyroptotic cell death defends against intracellular pathogens. Immunol Rev. 2015;265:130–142. doi: 10.1111/imr.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shin S, Brodsky IE. The inflammasome: Learning from bacterial evasion strategies. Semin Immunol. 2015;27:102–110. doi: 10.1016/j.smim.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Suzuki T, et al. Differential regulation of caspase-1 activation, pyroptosis, and autophagy via Ipaf and ASC in Shigella-infected macrophages. PLoS Pathog. 2007;3:e111. doi: 10.1371/journal.ppat.0030111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kobayashi T, et al. The Shigella OspC3 effector inhibits caspase-4, antagonizes inflammatory cell death, and promotes epithelial infection. Cell Host Microbe. 2013;13:570–583. doi: 10.1016/j.chom.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 6.Carneiro LA, et al. Shigella induces mitochondrial dysfunction and cell death in nonmyeloid cells. Cell Host Microbe. 2009;5:123–136. doi: 10.1016/j.chom.2008.12.011. [DOI] [PubMed] [Google Scholar]
- 7.Galán JE, Lara-Tejero M, Marlovits TC, Wagner S. Bacterial type III secretion systems: Specialized nanomachines for protein delivery into target cells. Annu Rev Microbiol. 2014;68:415–438. doi: 10.1146/annurev-micro-092412-155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ashida H, Toyotome T, Nagai T, Sasakawa C. Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol Microbiol. 2007;63:680–693. doi: 10.1111/j.1365-2958.2006.05547.x. [DOI] [PubMed] [Google Scholar]
- 9.Buchrieser C, et al. The virulence plasmid pWR100 and the repertoire of proteins secreted by the type III secretion apparatus of Shigella flexneri. Mol Microbiol. 2000;38:760–771. doi: 10.1046/j.1365-2958.2000.02179.x. [DOI] [PubMed] [Google Scholar]
- 10.Hachani A, et al. IpgB1 and IpgB2, two homologous effectors secreted via the Mxi-Spa type III secretion apparatus, cooperate to mediate polarized cell invasion and inflammatory potential of Shigella flexenri. Microbes Infect. 2008;10:260–268. doi: 10.1016/j.micinf.2007.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Niebuhr K, et al. IpgD, a protein secreted by the type III secretion machinery of Shigella flexneri, is chaperoned by IpgE and implicated in entry focus formation. Mol Microbiol. 2000;38:8–19. doi: 10.1046/j.1365-2958.2000.02041.x. [DOI] [PubMed] [Google Scholar]
- 12.Tran Van Nhieu G, Ben-Ze’ev A, Sansonetti PJ. Modulation of bacterial entry into epithelial cells by association between vinculin and the Shigella IpaA invasin. EMBO J. 1997;16:2717–2729. doi: 10.1093/emboj/16.10.2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Uchiya K, et al. Identification of a novel virulence gene, virA, on the large plasmid of Shigella, involved in invasion and intercellular spreading. Mol Microbiol. 1995;17:241–250. doi: 10.1111/j.1365-2958.1995.mmi_17020241.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhang L, et al. Cysteine methylation disrupts ubiquitin-chain sensing in NF-κB activation. Nature. 2011;481:204–208. doi: 10.1038/nature10690. [DOI] [PubMed] [Google Scholar]
- 15.Sanada T, et al. The Shigella flexneri effector OspI deamidates UBC13 to dampen the inflammatory response. Nature. 2012;483:623–626. doi: 10.1038/nature10894. [DOI] [PubMed] [Google Scholar]
- 16.Kim DW, et al. The Shigella flexneri effector OspG interferes with innate immune responses by targeting ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2005;102:14046–14051. doi: 10.1073/pnas.0504466102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ashida H, et al. A bacterial E3 ubiquitin ligase IpaH9.8 targets NEMO/IKKgamma to dampen the host NF-kappaB–mediated inflammatory response. Nat Cell Biol. 2010;12:66–73. doi: 10.1038/ncb2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arbibe L, et al. An injected bacterial effector targets chromatin access for transcription factor NF-kappaB to alter transcription of host genes involved in immune responses. Nat Immunol. 2007;8:47–56. doi: 10.1038/ni1423. [DOI] [PubMed] [Google Scholar]
- 19.Ashida H, Nakano H, Sasakawa C. Shigella IpaH0722 E3 ubiquitin ligase effector targets TRAF2 to inhibit PKC–NF-κB activity in invaded epithelial cells. PLoS Pathog. 2013;9:e1003409. doi: 10.1371/journal.ppat.1003409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, et al. Shigella flexneri T3SS effector IpaH4.5 modulates the host inflammatory response via interaction with NF-κB p65 protein. Cell Microbiol. 2013;15:474–485. doi: 10.1111/cmi.12052. [DOI] [PubMed] [Google Scholar]
- 21.Newton HJ, et al. The type III effectors NleE and NleB from enteropathogenic E. coli and OspZ from Shigella block nuclear translocation of NF-kappaB p65. PLoS Pathog. 2010;6:e1000898. doi: 10.1371/journal.ppat.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jong MF, Liu Z, Chen D, Alto NM. Shigella flexneri suppresses NF-κB activation by inhibiting linear ubiquitin chain ligation. Nat Microbiol. 2016;1:16084. doi: 10.1038/nmicrobiol.2016.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reeves AZ, et al. Engineering Escherichia coli into a protein delivery system for mammalian cells. ACS Synth Biol. 2015;4:644–654. doi: 10.1021/acssynbio.5b00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Du J, et al. The type III secretion system apparatus determines the intracellular niche of bacterial pathogens. Proc Natl Acad Sci USA. 2016;113:4794–4799. doi: 10.1073/pnas.1520699113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Le Gall T, et al. Analysis of virulence plasmid gene expression defines three classes of effectors in the type III secretion system of Shigella flexneri. Microbiology. 2005;151:951–962. doi: 10.1099/mic.0.27639-0. [DOI] [PubMed] [Google Scholar]
- 26.Schmitz AM, Morrison MF, Agunwamba AO, Nibert ML, Lesser CF. Protein interaction platforms: Visualization of interacting proteins in yeast. Nat Methods. 2009;6:500–502. doi: 10.1038/nmeth.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Campbell-Valois F-X, Sachse M, Sansonetti PJ, Parsot C. Escape of actively secreting Shigella flexneri from ATG8/LC3-positive vacuoles formed during cell-to-cell spread is facilitated by IcsB and VirA. MBio. 2015;6:e02567–14. doi: 10.1128/mBio.02567-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clerc P, Sansonetti PJ. Entry of Shigella flexneri into HeLa cells: Evidence for directed phagocytosis involving actin polymerization and myosin accumulation. Infect Immun. 1987;55:2681–2688. doi: 10.1128/iai.55.11.2681-2688.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pearson JS, et al. EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol. 2017;2:16258. doi: 10.1038/nmicrobiol.2016.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bergounioux J, et al. Calpain activation by the Shigella flexneri effector VirA regulates key steps in the formation and life of the bacterium’s epithelial niche. Cell Host Microbe. 2012;11:240–252. doi: 10.1016/j.chom.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 31.Berger CN, et al. EspZ of enteropathogenic and enterohemorrhagic Escherichia coli regulates type III secretion system protein translocation. MBio. 2012;3:e00317–12. doi: 10.1128/mBio.00317-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brodsky IE, et al. A Yersinia effector protein promotes virulence by preventing inflammasome recognition of the type III secretion system. Cell Host Microbe. 2010;7:376–387. doi: 10.1016/j.chom.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewoody R, Merritt PM, Houppert AS, Marketon MM. YopK regulates the Yersinia pestis type III secretion system from within host cells. Mol Microbiol. 2011;79:1445–1461. doi: 10.1111/j.1365-2958.2011.07534.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parsot C, et al. A secreted anti-activator, OspD1, and its chaperone, Spa15, are involved in the control of transcription by the type III secretion apparatus activity in Shigella flexneri. Mol Microbiol. 2005;56:1627–1635. doi: 10.1111/j.1365-2958.2005.04645.x. [DOI] [PubMed] [Google Scholar]
- 35.Costa SC, et al. A new means to identify type 3 secreted effectors: Functionally interchangeable class IB chaperones recognize a conserved sequence. MBio. 2012;3:e00243–11. doi: 10.1128/mBio.00243-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paciello I, et al. Intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation. Proc Natl Acad Sci USA. 2013;110:E4345–E4354. doi: 10.1073/pnas.1303641110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kamentsky L, et al. Improved structure, function and compatibility for CellProfiler: Modular high-throughput image analysis software. Bioinformatics. 2011;27:1179–1180. doi: 10.1093/bioinformatics/btr095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slagowski NL, Kramer RW, Morrison MF, LaBaer J, Lesser CF. A functional genomic yeast screen to identify pathogenic bacterial proteins. PLoS Pathog. 2008;4:e9. doi: 10.1371/journal.ppat.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.