Significance
Seedlings of terrestrial flowering plants such as Arabidopsis display dramatically different morphologies depending on growing in the dark or light. Dark-grown seedlings exhibit long hypocotyls, apical hook formation, and closed small cotyledons with etioplast. In contrast, light represses hypocotyl elongation, unfolds apical hook, and promotes cotyledon development. How light-controlled seedling morphogenesis is regulated at the transcriptional level remains elusive. To date, three families of transcription factors—PIFs, EIN3/EIL1, and HY5/HYH—have been shown to mediate light responses in seedlings. This study systemically investigates their roles in regulating specific morphological aspects and provides both transcriptomic and genetic evidence that PIFs, EIN3/EIL1, and HY5 are master transcription factors for the proper establishment of seedling skotomorphogenesis and photomorphogenesis.
Keywords: light signal, ethylene signal, plant photobiology, PIFs, EIN3/EIL1
Abstract
Three families of transcription factors have been reported to play key roles in light control of Arabidopsis seedling morphogenesis. Among them, bHLH protein PIFs and plant-specific protein EIN3/EIN3-LIKE 1 (EIN3/EIL1) accumulate in the dark to maintain skotomorphogenesis. On the other hand, HY5 and HY5 HOMOLOG (HYH), two related bZIP proteins, are stabilized in light and promote photomorphogenic development. To systemically investigate the transcriptional regulation of light-controlled seedling morphogenesis, we generated HY5ox/pifQein3eil1, which contained mutations of EIN3/EIL1 and four PIF genes (pifQein3eil1) and overexpression of HY5. Our results show that dark-grown HY5ox/pifQein3eil1 seedlings display a photomorphogenesis highly similar to that of wild-type seedlings grown in continuous light, with remarkably enhanced photomorphogenic phenotypes compared with the pifQ mutants. Consistent with the genetic evidence, transcriptome analysis indicated that PIFs, EIN3/EIL1, and HY5 are dominant transcription factors in collectively mediating a wide range of light-caused genome-wide transcriptional changes. Moreover, PIFs and EIN3/EIL1 independently control the expression of light-regulated genes such as HLS1 to cooperatively regulate apical hook formation, hypocotyl elongation, and cotyledon opening and expansion. This study illustrates a comprehensive regulatory network of transcription activities that correspond to specific morphological aspects in seedling skotomorphogenesis and photomorphogenesis.
Light provides the energy source for photosynthesis and is a critical environmental regulator for plant growth. Terrestrial flowering plant seedlings grown in subterranean darkness adopt a skotomorphogenic developmental program, characterized by long hypocotyls, apical hook formation, and closed small cotyledons with etioplast (1, 2). Upon emerging from soil covering, light triggers a dramatic morphological and physiological transition from dark-grown skotomorphogenesis to light-grown photomorphogenesis, which is essential for seedling survival (2, 3). This transition involves hypocotyl elongation inhibition, apical hook unfolding, cotyledon opening and expanding, and etioplast-to-chloroplast transition (4–6).
Decades of research has identified positive and negative regulators of light-induced developmental transition. Positive regulators naturally include photoreceptors, among which the phytochromes (phyA to phyE in Arabidopsis) and cryptochromes are predominantly responsible for inducing photomorphogenesis in seedlings (1, 7, 8). Photoreceptors ultimately induce the light-controlled switch from skotomorphogenesis to photomorphogenesis via multiple downstream transcription factors. Two bZIP transcription factors—LONGATED HYPOCOTYL 5 (HY5) and HY5 HOMOLOG (HYH) proteins—have been genetically identified as positive regulators that promote light responses (9, 10). Loss of HY5 or HYH causes longer hypocotyl in the light (9, 10). In contrast to HY5, the bHLH transcription factors—PHYTOCHROME-INTERACTING FACTORs (PIFs), including PIF1, PIF3, PIF4, and PIF5—accumulate in the dark and maintain skotomorphogenesis (4, 11). With light exposure, photoactivated phytochromes are translocated into the nucleus, where they directly interact with, and trigger rapid degradation of, PIF proteins (12–15). The quadruple mutant of PIFs (pifQ) exhibits photomorphogenic phenotype in the dark (4, 11), similar to the mutant of COP1, a central repressor of photomorphogenesis (16, 17). COP1 is an E3 ubiquitin ligase and represses light signaling predominantly through the proteasome degradation system (18–20). HY5/HYH proteins are typical transcription factors that are targeted by COP1 for degradation in the dark (10, 18).
When grown under the soil, seedlings simultaneously confront darkness and mechanical pressure from the soil covering. In response to mechanical impedance, seedlings induce ethylene production to exaggerate apical hook formation and repress cotyledon development to reduce mechanical injuries (19, 21, 22). Ethylene is a gaseous hormone and its responses are mediated by the master transcription factors ETHYLENE-INSENSITIVE 3 (EIN3) and EIN3-LIKE 1 (EIL1) (23, 24). In the absence of ethylene, EIN3/EIL1 proteins are targeted by E3 ligase SCFEBF1/EBF2 complexes for degradation (25–27). Ethylene represses the action of EBF1/EBF2 to stabilize EIN3 and activate ethylene responses (28, 29). EIN3 proteins are highly accumulated in buried seedlings through both mechanical stress-evoked ethylene production and darkness (19, 21, 22). Previous studies suggest that EIN3 and PIFs regulate the expressions of their common target genes in either an additive or interdependent way (30). Recently, EIN3/EIL1 have been shown to promote seedling soil emergence by exaggerating apical hook formation (22), inhibiting cotyledon opening and expansion (19), and repressing chloroplast development (6).
Here, we present genetic and transcriptomic evidence to demonstrate that PIFs, EIN3/EIL1, and HY5 are master transcription factors for the proper establishment of seedling skoto- and photomorphogenesis. In addition, PIFs and EIN3/EIL1 are further shown to act independently in regulating the main processes of light-controlled seedling morphological development, including apical hook formation, hypocotyl elongation, and cotyledon opening and expansion.
Results
PIFs, EIN3/EIL1, and HY5 Are Essential for Establishment of Light-Controlled Seedling Morphogenesis.
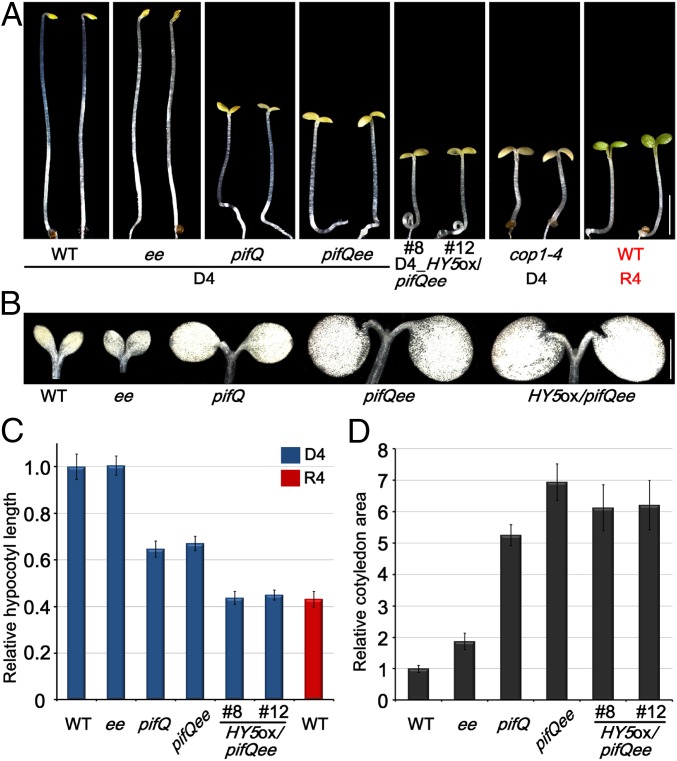
It is known that the proteins of four PIFs (PIF1, PIF3, PIF4, and PIF5) accumulate and act redundantly in maintaining skotomorphogenesis in the dark (4, 11), whereas HY5 protein is elevated in light to promote photomorphogenesis (9, 18). EIN3/EIL1 have been recently identified as a new class of regulators that suppress light-induced seedling developmental transition (5, 19). In 10-d-old dark-grown ein3eil1 mutant seedlings, apical hooks were unfolded and cotyledons were partially open, similar to those of pif1pif3 mutants (SI Appendix, Fig. S1), while the pif1pif3ein3eil1 quadruple mutants displayed an additive effect in cotyledon opening (SI Appendix, Fig. S1). As previously documented, the 4-d-old dark-grown pif1pif3pif4pif5 (pifQ) mutant showed partially constitutive photomorphogenesis, manifested by reduced hypocotyl elongation, unfolded apical hook, and largely opened and expanded cotyledons (Fig. 1A). To determine the relationship of EIN3/EIL1 and PIFs in repressing photomorphogenesis, we crossed pifQ and ein3eil1 mutants to generate the pifQein3eil1 (pifQee) sextuple mutant. The cotyledons of 4-d-old dark-grown pifQee seedlings were fully opened and expanded, similar to those of 4-d-old dark-grown cop1-4 mutant or continuous red light (R4)-grown wild-type (WT) seedlings (Fig. 1). Quantitative cotyledon areas and open angles indicate that PIFs and EIN3/EIL1 additively repressed cotyledon development in the dark (Fig. 1 B and D and SI Appendix, Fig. S2).
Fig. 1.
PIFs, EIN3/EIL1, and HY5 are the major transcription factors in directing light-controlled seedling morphogenesis. (A) Images of 4-d-old Col-0 (WT), ein3eil1 (ee), pifQ, pifQee, HY5ox/pifQee, and cop1-4 seedlings grown in the dark (D4) or in continuous red light (R4). (Scale bar, 2 mm). (B) Cotyledon photographs of 10-d-old dark-grown seedlings. (Scale bar, 0.5 mm.) (C) Hypocotyl lengths of 4-d-old seedlings grown in the dark (D4) or in R4. The WT value was set as 1, and the relative hypocotyl lengths were calculated. Error bars represent SD (n = 20). (D) Cotyledon areas of 10-d-old dark-grown etiolated seedlings. The WT value was set as 1, and the relative cotyledon areas were calculated. Error bars represent SD (n = 20).
Notably, mutations of EIN3/EIL1 did not further alter the hypocotyl lengths of pifQ, and the hypocotyls of pifQee were still much longer than those of light-grown WT (Fig. 1 A and C). HY5 has been shown to accumulate in light to repress hypocotyl elongation (9, 18). We therefore examined the protein levels of HY5 in the seedlings and found that HY5 proteins were stabilized in light-grown WT, but mutation of PIFs and EIN3/EIL1 in the dark did not change its abundance (SI Appendix, Fig. S3). To investigate whether HY5 regulates the hypocotyl lengths of pifQee, we constitutively overexpressed HY5 in dark-grown pifQee to reach a comparable level with light-grown WT (Fig. 1A). Although the developmental pattern of cotyledons was not altered by overexpressing HY5 (Fig. 1 A and B), the hypocotyl lengths of HY5ox/pifQee were further shortened (Fig. 1 A and C). Remarkably, the dark-grown HY5ox/pifQee showed photomorphogenic phenotypes most similar to those of dark-grown cop1-4 or light-grown WT (Fig. 1A). These pieces of genetic evidence indicate that PIFs, EIN3/EIL1, and HY5 are central transcription factors which function additively in establishing seedling morphogenesis responding to light.
Light-Directed Seedling Transcriptome Changes Are Largely Mediated by PIFs, EIN3/EIL1, and HY5.
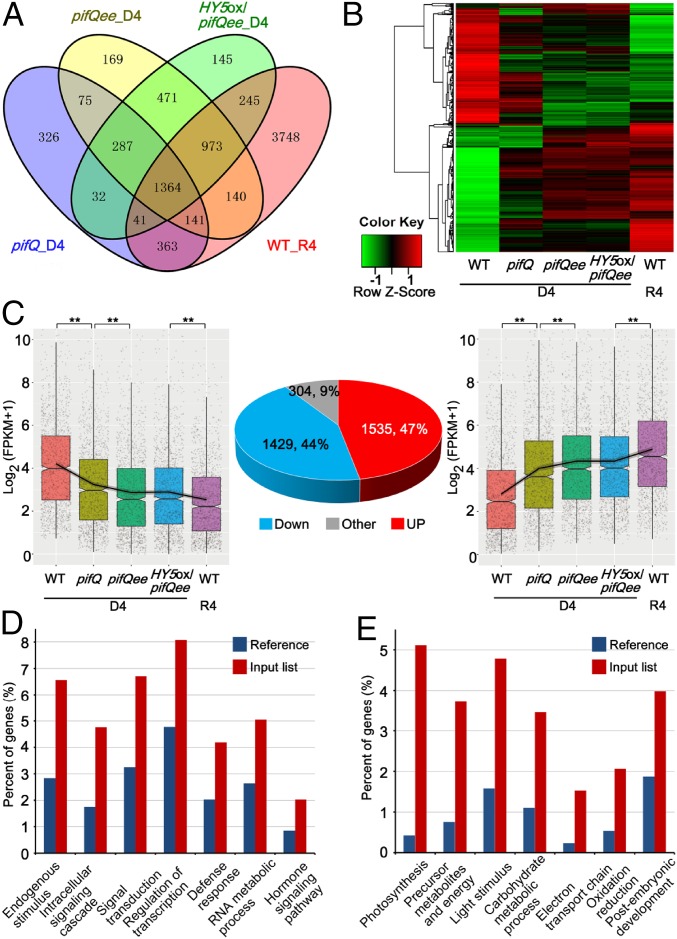
To understand how PIFs, EIN3/EIL1, and HY5 control light-responsive seedling morphogenesis, we further examined the transcriptomes in pifQ, pifQee, and HY5ox/pifQee in comparison with the light-grown profile by mRNA deep-sequencing analysis. Seedlings were grown in the dark (D4) or under R4 for 4 d and harvested for the transcriptomic analysis. Our results showed that about one-fourth (7,015 genes) of the whole genome in WT was statistically significantly twofold (SSTF) changed by red light (R4-WT versus D4-WT) (Fig. 2A). We then compared the gene expression profiles of WT with those of dark-grown pifQ, pifQee, and HY5ox/pifQee and identified 2,629 SSTF genes in pifQ; 3,620 SSTF genes in pifQee; and 3,558 SSTF genes in HY5ox/pifQee (Fig. 2A and SI Appendix, Fig. S4A). Moreover, 4,772 genes were shown to be significantly regulated at least in one mutant background and were identified as the PIFs/EIN3/HY5-regulated genes (Fig. 2A and SI Appendix, Fig. S4A). Among them, 71% (3,384 genes) were commonly regulated at least in two mutant backgrounds (Fig. 2A and SI Appendix, Fig. S4A). By comparing the R4- and PIFs/EIN3/HY5-regulated genes, we found that PIFs/EIN3/HY5-regulated genes in the dark were estimated to account for half of the light-directed transcriptome changes (Fig. 2A and SI Appendix, Fig. S4B). Together, the genome-wide gene expression analysis supports our genetic data demonstrating the essential roles of PIFs, EIN3/EIL1, and HY5 in regulating light-controlled seedling morphogenesis.
Fig. 2.
Light-induced genome-wide transcriptional changes are largely mediated by PIFs, EIN3/EIL1, and HY5. (A) Venn diagram showing overlaps among genes regulated by light, pifQ, pifQee, and HY5ox/pifQee. The seedlings were grown for 4 d in the dark (D4) or in R4. (B) Expression levels of the PIFs/EIN3/HY5- and light-coregulated 3,267 genes in different genotypes. The FPKM values of these genes were normalized based on the Z-score method. (C) Box plot representation of the PIFs/EIN3/HY5-mediated light down-regulated (Left) or light up-regulated (Right) gene FPKM values in different genotypes. **P < 0.01, Wilcoxon test. (D and E) Gene ontology analysis of the light down-regulated (D), or light up-regulated (E) genes mediated by the PIFs/EIN3/HY5 pathway.
PIFs, EIN3/EIL1, and HY5 Collectively Direct a Wide Range of Light-Regulated Biological Processes.
By comparing the SSTF genes regulated by PIFs/EIN3/HY5 and light, we identified 3,267 overlapped genes (SI Appendix, Fig. S4B and Dataset S1). After normalizing the expression levels [FPKM (fragments per kilobase of exon per million fragments mapped)] of the 3,267 genes in different samples based on the Z-score method, we found that about half of the 3,267 genes in D4-pifQ were redirected to the expressing patterns of R4-WT (Fig. 2B). Mutation of EIN3/EIL1 in pifQ remarkably exaggerated the expressing pattern changes (Fig. 2B), and most of the 3,267 genes in D4-HY5ox/pifQee were regulated in the same direction as those of R4-WT (Fig. 2B). We further analyzed the regulation pattern of these 3,267 genes and found that 91% (2,964 genes) were modulated in the same way by light and by PIFs/EIN3/HY5 (Fig. 2C). We referred to these 2,964 genes as members of the PIFs/EIN3/HY5-mediated light signaling pathway. The box plots of average FPKM values similarly showed that the expression of light-repressed genes was gradually inhibited, while the levels of light-activated genes were progressively increased in the seedlings of pifQ and pifQee (Fig. 2C). These analyses suggest that PIFs, EIN3/EIL1, and HY5 act collectively in directing light-induced transcriptome changes.
We then performed a gene ontology (GO) enrichment analysis to investigate the biological processes that PIFs/EIN3/HY5 mediate in light responses (Dataset S2). Our results revealed that among the light-repressed genes, PIFs/EIN3/HY5 most significantly regulated the following GO categories: response to endogenous stimulus (P < 10−11), intracellular signaling cascade (P < 10−11), signal transduction (P < 10−9), regulation of transcription (P < 10−6), defense response (P < 10−6), RNA metabolic process (P < 10−6), and hormone signaling pathway (P < 10−4) (Fig. 2D). In contrast, among the light-activated genes, the top enriched GO categories mediated by PIFs/EIN3/HY5 are photosynthesis (P < 10−48), precursor metabolites and energy (P < 10−19), response to light stimulus (P < 10−14), carbohydrate metabolic process (P < 10−11), electron transport chain (P < 10−10), oxidation reduction (P < 10−8), and postembryonic development (P < 10−6) (Fig. 2E). Thus, PIFs, EIN3/EIL1, and HY5 integrate both internal and external effects to mediate a wide range of biological processes in light responses.
PIFs and EIN3/EIL1 Independently Regulate the Expression of Common Downstream Genes.
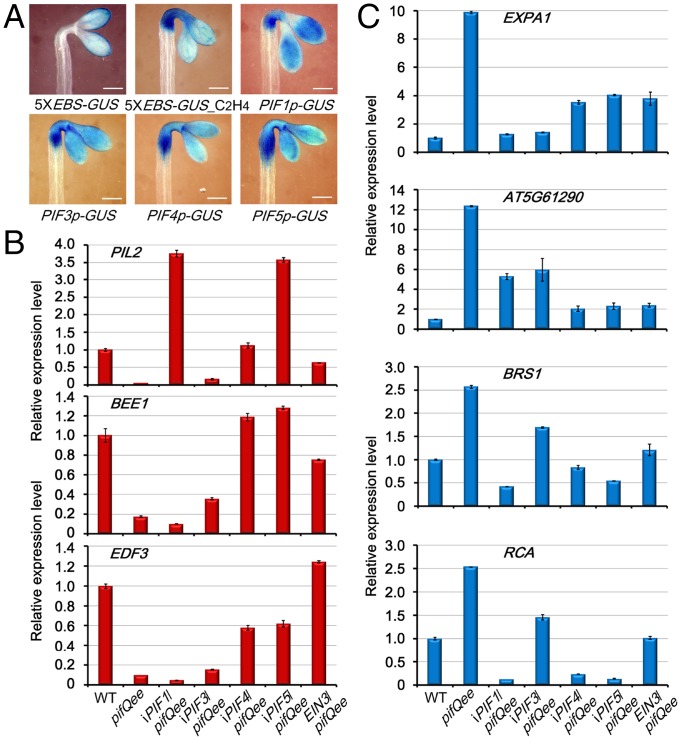
Given the essential roles of PIFs and EIN3/EIL1 in sustaining skotomorphogenesis, we next investigated how PIFs and EIN3/EIL1 regulate the expression of common downstream genes. We adopted a 5XEBS:GUS transgenic line in which the glucuronidase (GUS) reporter gene was driven by five tandem repeats of EIN3 binding sequence (EBS) to monitor the action of EIN3/EIL1 (31). GUS staining analysis showed that EIN3/EIL1 were active in the tissues of cotyledons, apical hook, and the upper hypocotyl, which was notably enhanced by ethylene treatment (Fig. 3A). Then we used the PIFs-promoter:GUS reporter lines in which the GUS gene was driven by each of the PIF promoters (32), and assayed the distribution of PIFs’ expression in etiolated seedlings. We found that all four PIF genes were highly expressed in the same tissues where EIN3/EIL1 function (Fig. 3A). These results suggest that EIN3/EIL1 and PIFs probably act in a similar spatial pattern.
Fig. 3.
PIFs and EIN3/EIL1 independently regulate the expression of light-responsive genes. (A) Representative images of GUS staining in 4-d-old dark-grown seedlings. 5XEBS-GUS transgenic lines harbor five tandem repeats of EBS fused with the GUS reporter gene. C2H4 indicates that the seedlings were treated with C2H4 gas for 4 h before staining. PIF1p- to PIF5p-GUS indicates that the GUS reporter gene is driven, respectively, by the promoter of PIF1 to PIF5 genes. (Scale bar, 0.2 mm.) (B and C) qRT-PCR analysis of the expression levels of light-repressed (B) or light-activated (C) genes in 4-d-old etiolated seedlings grown on half-strength Murashige and Skoog (1/2 MS) medium supplemented with 10 µM β-estradiol. iPIF1/pifQee to iPIF5/pifQee are the inducible transgenic plants in which the respective PIF1 to PIF5 genes driven by β-estradiol–induced pER8 promoters are transformed into pifQee background. EIN3/pifQee presents the transgenic plants with EIN3 native promoter-driven EIN3 gene in the pifQee background. The gene expression levels were normalized to that of two internal control genes (PP2A and SAND) and relative to the WT sample. Error bars indicate the SD of three technical repeats. All experiments were repeated independently three times.
Next, we expressed PIFs and EIN3 genes individually into the pifQee sextuple mutant and examined the expression levels of three pifQee down-regulated genes by qRT-PCR in these transgenic lines. Intriguingly, the transcriptions of all three highly repressed genes (PIL2, BEE1, and EDF3) in pifQee were remarkably activated by single PIFs or EIN3 to the comparable levels as seen in WT (Fig. 3B). Conversely, we also examined the expression of four pifQee up-regulated genes (EXPA1, AT5G61290, BRS1, and RCA), and found that the high expression of all four genes in pifQee was dramatically suppressed by introducing single PIFs or EIN3 (Fig. 3C). Thus, PIFs and EIN3/EIL1 could regulate downstream gene expressions in an independent and redundant manner.
PIFs and EIN3/EIL1 Additively Activate Transcription of HLS1 to Promote Apical Hook Formation in the Dark.
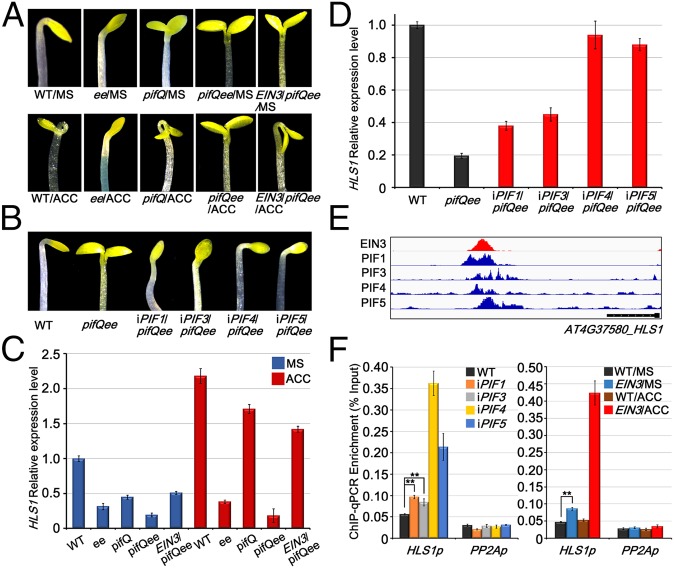
Apical hook formation is a characteristic feature of etiolated seedlings at the apex region. Because all four PIFs and EIN3 were expressed in apical hook (Fig. 3A), we wondered how PIFs and EIN3 may synergistically regulate this morphological trait. Previous studies have reported that, in response to ethylene, EIN3 activates HOOKLESS1 (HLS1) gene expression to promote apical hook formation (22, 33, 34). Agreeably, the angle of the apical hook was visibly reduced in ee (Fig. 4A). Treatment with aminocyclopropane carboxylic acid (ACC), the ethylene biosynthetic precursor, greatly enhanced the apical hook angles in WT, pifQ, and EIN3/pifQee, but not in lines that lacked EIN3/EIL1 (Fig. 4A). Mutation of PIFs showed dramatically reduced hook formation, and the apical hooks of pifQee seedlings were fully unfolded (Fig. 4A). We further expressed individual PIFs into pifQee and found that expression of each single PIF was sufficient to restore the hook formation to various degrees (Fig. 4B and SI Appendix, Fig. S5). This genetic evidence indicates that EIN3 or each of the PIFs alone is capable of promoting hook formation.
Fig. 4.
Apical hook formation is promoted by PIFs and EIN3/EIL1 through directly activating HLS1 transcription. (A and B) Apical hook images of 4-d-old etiolated seedlings grown on half-strength Murashige and Skoog (1/2 MS) medium without (MS) or with (ACC) 10 µM ACC supplementation (A) or on 1/2 MS medium supplemented with 10 µM β-estradiol (B). (C and D) qRT-PCR analysis of HLS1 gene expression levels in 4-d-old etiolated seedlings grown on 1/2 MS medium supplemented without or with 10 µM ACC (C) or on 1/2 MS medium supplemented with 10 µM β-estradiol (D). (E) Visualization of EIN3 and PIFs ChIP-seq data in the genomic regions encompassing the promoter region of HLS1 gene. (F) ChIP-qPCR analyzing the associations of PIFs and EIN3 with HLS1 promoter in 4-d-old etiolated seedlings grown on 1/2 MS medium supplemented with 10 µM β-estradiol (Left) and without or with 10 µM ACC (Right). iPIF1 to iPIF5 are the respective iPIF1/pifQee to iPIF5/pifQee transgenic plants, and EIN3 presents the EIN3/pifQee transgenic plants. WT and PP2A promoter (PP2Ap) were used as controls. **P < 0.01, Student’s t test. Error bars indicate the SD of three technical repeats. All experiments were repeated independently three times.
Since HLS1 is the main regulator of hook formation, with hls1 exhibiting fully unfolded apical hook (33), we next analyzed the regulation of HLS1 gene expression by EIN3 and PIFs. qRT-PCR results showed that mutation of either EIN3/EIL1 or PIFs notably reduced HLS1’s transcription, which was further declined in pifQee, suggesting that EIN3/EIL1 and PIFs function additively in activating the expression of HLS1 (Fig. 4C). In agreement with the hook formation phenotypes, the expression levels of HLS1 were partially restored by introducing EIN3 back into pifQee and were fully rescued to WT levels in ACC-treated pifQ or EIN3/pifQee (Fig. 4C). Furthermore, the highly inhibited HLS1 transcription in pifQee was reactivated by introducing single PIFs into it (Fig. 4D). Collectively, these data demonstrate that EIN3 and PIFs promote apical hook formation by activating HLS1 gene expression additively and independently of each other.
PIFs and EIN3/EIL1 Bind to the Promoter of HLS1 in an Independent Manner.
To further understand how PIFs and EIN3 regulate the gene expression of HLS1, we analyzed the genomic occupancy of PIFs and EIN3 based on previously reported chromatin immunoprecipitation sequencing (ChIP-seq) data (32, 35). The ChIP-seq results revealed the specific associations of EIN3 and PIFs with the promoter of HLS1 (Fig. 4E). Moreover, the binding peaks of EIN3 and PIFs in the HLS1 promoter region largely overlapped (Fig. 4E), suggesting that EIN3 and PIFs coassociated with the same regions. To investigate the interrelationship of PIFs and EIN3 in binding to the HLS1 promoter regions, we performed ChIP-qPCR assays using transgenic lines in which the individual PIFs or EIN3 was expressed in the pifQee background. Compared with the PP2A promoter region, the DNA fragments of the HLS1 promoter region were evidently enriched in immunoprecipitation by single PIFs or by EIN3 (Fig. 4F). In addition, the enrichment of HLS1 promoter fragments immunoprecipitated by EIN3 in the pifQee background was strongly enhanced by ACC treatment (Fig. 4F). Moreover, the relative enrichment of HLS1p by individual PIFs or EIN3 correlated with the expression levels of HLS1 and with the restoration of the apical hook in the corresponding transgenic lines (Fig. 4 A–D). Thus, these results imply that PIFs and EIN3 are each capable of binding to the promoter of HLS1 to activate HLS1’s transcription.
PIFs and EIN3 Cooperatively Regulate Hypocotyl Elongation and Cotyledon Development.
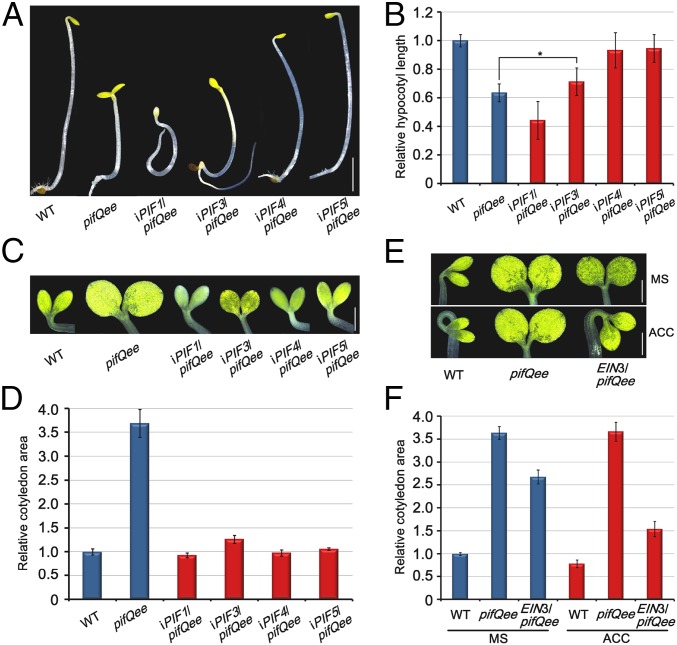
Decreasing rate of hypocotyl elongation is usually quantitatively correlated to the level of light responses in a plant seedling (18). We found that the shortened hypocotyl of pifQee could be reversed by expressing individual PIFs (PIF3, PIF4, or PIF5) in pifQee in the dark (Fig. 5 A and B). However, expressing EIN3 could not restore the hypocotyl elongation of pifQee (SI Appendix, Fig. S6). Instead, the hypocotyl lengths of EIN3/pifQee were further shortened by ACC application (SI Appendix, Fig. S6). In previous studies, we revealed that EIN3 concomitantly activates a PIF3-dependent promoting pathway and an ERF1-mediated repressing pathway, which antagonistically regulated hypocotyl elongation (36). Thus, it is likely that EIN3 activates the ERF1 pathway in pifQee to repress hypocotyl elongation.
Fig. 5.
PIFs and EIN3/EIL1 cooperatively regulate hypocotyl elongation, cotyledon opening, and cotyledon expansion. (A and B) Representative images (A) and hypocotyl lengths (B) of 4-d-old etiolated seedlings grown on half-strength Murashige and Skoog (1/2 MS) medium supplemented with 10 µM β-estradiol. (Scale bar, 2 mm.) The WT value was set as 1, and the relative hypocotyl lengths were calculated. Error bars represent SD (n = 20). *P < 0.05, Student’s t test. (C–F) Representative cotyledon images (C and E) and cotyledon areas (D and F) of 4-d-old etiolated seedlings grown on 1/2 MS medium supplemented with 10 µM β-estradiol (C and D) and without (MS) or with (ACC) 10 µM ACC (E and F). (Scale bar, 0.5 mm.) The WT value was set as 1, and the relative cotyledon areas were calculated. Error bars represent SD (n = 20).
Before emerging from the soil, cotyledon development of buried seedlings is strictly inhibited to reduce soil mechanical resistance. We investigated how PIFs and EIN3/EIL1 act in repressing cotyledon development. Remarkably, the results showed that individual PIFs alone could fully restore the etiolated cotyledon morphology, in contrast to the opened and expanded cotyledons of pifQee (Fig. 5 C and D). Expressing EIN3 in pifQee partially reduced the area of cotyledons, and ACC treatment further decreased the cotyledon expansion of EIN3/pifQee to a comparable level with WT (Fig. 5 E and F). Similar to the regulation of cotyledon expansion, a single PIF fully rescued the cotyledon-opening phenotype of pifQee (Fig. 4B and SI Appendix, Fig. S7). With ACC application, the cotyledons of EIN3/pifQee were also largely closed (Fig. 4A and SI Appendix, Fig. S7). Together, these genetic results indicate that PIFs and EIN3 repress cotyledon development in an independent way.
Discussion
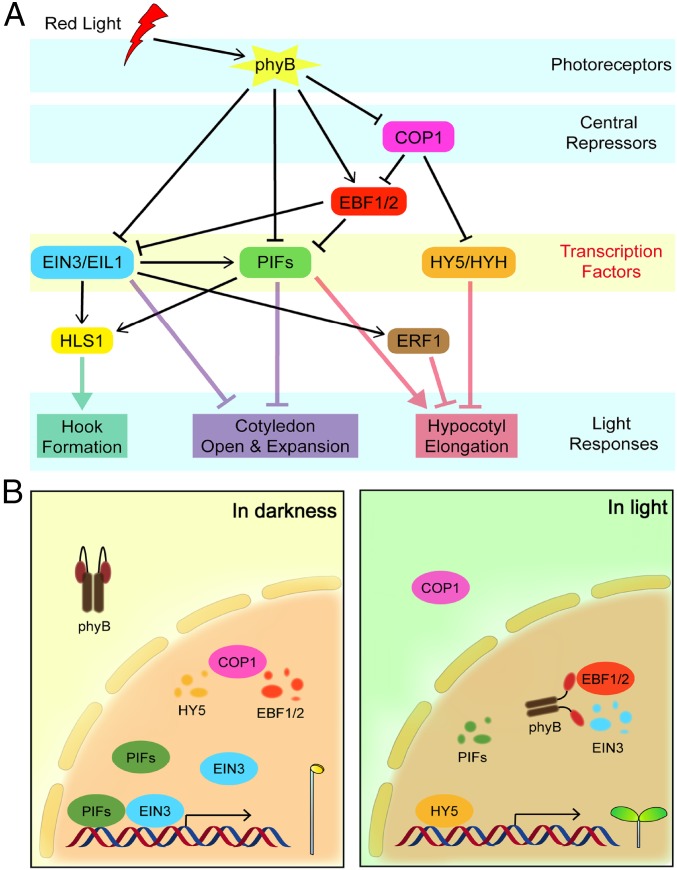
Most higher plants, such as Arabidopsis, exhibit distinct morphogenesis depending on the light environment. How light controls genomic gene expression to direct seedling morphogenesis has been extensively studied in the past decades (37). PIFs are well documented to accumulate and sustain seedlings’ skotomorphogenic state in darkness (4, 11, 38), whereas HY5 is accumulated in light and promotes light responses (9, 18, 39). EIN3/EIL1 have previously been reported to be indispensable for ethylene signaling transduction pathway (23, 24). In this study, we propose that the three families of transcription factors—PIFs, EIN3/EIL1, and HY5/HYH—function collectively in directing the transcriptional network underlying light-induced developmental transition (Fig. 6A). When grown in subterranean darkness, phyB is inactive and COP1 is activated to degrade HY5 and EBF1/EBF2. PIFs and EIN3/EIL1 proteins are highly accumulated and maintain skotomorphogenesis. Upon light exposure, photoactivated phyB directly binds to, and induces rapid degradation of, PIFs and EIN3/EIL1, thus removing the repression of PIFs and EIN3/EIL1 on light responses. At the same time, COP1 is repressed and HY5 proteins are stabilized to further promote light-induced photomorphogenesis (Fig. 6B).
Fig. 6.
Model of the transcriptional regulation of light-controlled seedling morphogenesis by PIFs, EIN3/EIL1, and HY5. (A) PIFs, EIN3/EIL1, and HY5 direct the transcriptional network underlying light responses in seedlings. PIFs and EIN3/EIL1 cooperatively repress light-induced morphological transition, including apical hook unfolding, cotyledon opening and expansion, and inhibition of hypocotyl elongation, whereas HY5 inhibits hypocotyl elongation to promote light responses. (B) Light oppositely regulates the protein levels of PIFs, EIN3/EIL1, and HY5 to initiate seedling morphological transition. When grown in the dark, photoreceptor phyB is inactive, and the central repressor COP1 is activated to control the protein abundance of downstream factors. PIFs and EIN3/EIL1 accumulate and function collectively in sustaining skotomorphogenesis. Upon light activation, phyB directly induces rapid degradation of PIFs and EIN3/EIL1, and indirectly stabilizes HY5 by repressing COP1’s action to promote the photomorphogenic development.
Skotomorphogenesis is a dark-adaptive strategy adopted by seedlings grown in subterranean darkness. The findings presented here indicate that maintenance of skotomorphogenic development requires the concerted action of PIFs and EIN3/EIL1. Remarkably, we demonstrate that overexpressing single PIFs or EIN3 alone can reverse the constitutive photomorphogenic phenotype of pifQee, including apical hook formation, hypocotyl elongation, and cotyledon development. The independent and redundant nature of PIFs and EIN3/EIL1 provides a molecular mechanism by which the skotomorphogenesis could be sustained when either darkness or mechanical stress exists alone. This idea may provide insight into how seedlings deal with the complex situations during soil emergence. For example, buried seedlings could grow in darkness with minimum mechanical stress in hollow soil or under dense canopy upon emergence. Skotomorphogenic pattern would be maintained by PIFs alone to allow seedlings to grow vigorously toward light. Upon breaking out, seedlings could be exposed to light, with soil block or stone retaining on the top. Mechanical stress-elevated EIN3 slows down apical hook unfolding and cotyledon development, enabling seedlings to push aside obstructions and fully emerge. With regard to the physiology, we have recently shown that EIN3 and PIF3 form an interdependent module in repressing chloroplast development (6). It is likely that seedlings have to rapidly initiate photoautotrophic growth upon light regardless of mechanical stress, due to the limited reserves in seeds. This hypothesis is consistent with the observations that dark-grown EIN3ox turned green normally under initial light irradiation, with apical hook folded and cotyledons closed and unexpanded (5).
Plant morphogenesis is profoundly influenced by signals from environmental factors as well as endogenous hormones. Collective studies have shown that PIFs, EIN3/EIL1, and HY5 serve as central integrators of environmental and hormonal signals in controlling seedling morphological development. Gibberellins (GAs) induce rapid degradation of DELLA proteins, which directly sequester the transcriptional activity of PIF3 and PIF4 (40, 41). Brassinosteroid-activated transcription factor BZR1 is found to interact and cooperatively promote hypocotyl cell elongation with PIF4 (42, 43), while GA and cytokinin have been reported to regulate the protein levels of HY5 (44, 45). During seedling soil emergence, EIN3/EIL1 are key transcription factors that precisely adjust seedling morphogenesis by converging both light and mechanical pressure signals (5, 19, 21). Thus, our studies provide genetic and molecular evidence and resources for elucidating the integration of environmental and hormonal signals.
Materials and Methods
The details and procedures of plant materials and growth conditions, immunoblot assays, ChIP analysis, RNA extraction, qRT-PCR analysis, transcriptome assays, and histochemical GUS staining assays are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Hongwei Guo for his valuable suggestions and Chang Xue for his assistance in generating the transgenic lines. This work was supported by grants from the National Science Foundation of China (31621001, 31770208, and 31570188), the National Key R&D Program of China (2016YFA0502900), and the NIH (GM-47850).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803861115/-/DCSupplemental.
References
- 1.Chen M, Chory J, Fankhauser C. Light signal transduction in higher plants. Annu Rev Genet. 2004;38:87–117. doi: 10.1146/annurev.genet.38.072902.092259. [DOI] [PubMed] [Google Scholar]
- 2.Huq E, et al. Phytochrome-interacting factor 1 is a critical bHLH regulator of chlorophyll biosynthesis. Science. 2004;305:1937–1941. doi: 10.1126/science.1099728. [DOI] [PubMed] [Google Scholar]
- 3.Zhong S, et al. EIN3/EIL1 cooperate with PIF1 to prevent photo-oxidation and to promote greening of Arabidopsis seedlings. Proc Natl Acad Sci USA. 2009;106:21431–21436. doi: 10.1073/pnas.0907670106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leivar P, et al. Multiple phytochrome-interacting bHLH transcription factors repress premature seedling photomorphogenesis in darkness. Curr Biol. 2008;18:1815–1823. doi: 10.1016/j.cub.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi H, et al. The red light receptor phytochrome B directly enhances substrate-E3 ligase interactions to attenuate ethylene responses. Dev Cell. 2016;39:597–610. doi: 10.1016/j.devcel.2016.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, et al. EIN3 and PIF3 form an interdependent module that represses chloroplast development in buried seedlings. Plant Cell. 2017;29:3051–3067. doi: 10.1105/tpc.17.00508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quail PH, et al. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- 8.Liu H, Liu B, Zhao C, Pepper M, Lin C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011;16:684–691. doi: 10.1016/j.tplants.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ang LH, et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 10.Holm M, Ma LG, Qu LJ, Deng XW. Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev. 2002;16:1247–1259. doi: 10.1101/gad.969702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shin J, et al. Phytochromes promote seedling light responses by inhibiting four negatively-acting phytochrome-interacting factors. Proc Natl Acad Sci USA. 2009;106:7660–7665. doi: 10.1073/pnas.0812219106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 13.Ni W, et al. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science. 2014;344:1160–1164. doi: 10.1126/science.1250778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dong J, et al. Light-dependent degradation of PIF3 by SCFEBF1/2 promotes a photomorphogenic response in Arabidopsis. Curr Biol. 2017;27:2420–2430.e6. doi: 10.1016/j.cub.2017.06.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, Paik I, Zhu L, Huq E. Illuminating progress in phytochrome-mediated light signaling pathways. Trends Plant Sci. 2015;20:641–650. doi: 10.1016/j.tplants.2015.06.010. [DOI] [PubMed] [Google Scholar]
- 16.Deng XW, et al. COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell. 1992;71:791–801. doi: 10.1016/0092-8674(92)90555-q. [DOI] [PubMed] [Google Scholar]
- 17.Lau OS, Deng XW. The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci. 2012;17:584–593. doi: 10.1016/j.tplants.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 18.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of i. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 19.Shi H, et al. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr Biol. 2016;26:139–149. doi: 10.1016/j.cub.2015.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoecker U. The activities of the E3 ubiquitin ligase COP1/SPA, a key repressor in light signaling. Curr Opin Plant Biol. 2017;37:63–69. doi: 10.1016/j.pbi.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 21.Zhong S, et al. Ethylene-orchestrated circuitry coordinates a seedling’s response to soil cover and etiolated growth. Proc Natl Acad Sci USA. 2014;111:3913–3920. doi: 10.1073/pnas.1402491111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shen X, Li Y, Pan Y, Zhong S. Activation of HLS1 by mechanical stress via ethylene-stabilized EIN3 is crucial for seedling soil emergence. Front Plant Sci. 2016;7:1571. doi: 10.3389/fpls.2016.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ecker JR. The ethylene signal transduction pathway in plants. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 24.Chao Q, et al. Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell. 1997;89:1133–1144. doi: 10.1016/s0092-8674(00)80300-1. [DOI] [PubMed] [Google Scholar]
- 25.Guo H, Ecker JR. Plant responses to ethylene gas are mediated by SCF(EBF1/EBF2)-dependent proteolysis of EIN3 transcription factor. Cell. 2003;115:667–677. doi: 10.1016/s0092-8674(03)00969-3. [DOI] [PubMed] [Google Scholar]
- 26.Potuschak T, et al. EIN3-dependent regulation of plant ethylene hormone signaling by two Arabidopsis F box proteins: EBF1 and EBF2. Cell. 2003;115:679–689. doi: 10.1016/s0092-8674(03)00968-1. [DOI] [PubMed] [Google Scholar]
- 27.Gagne JM, et al. Arabidopsis EIN3-binding F-box 1 and 2 form ubiquitin-protein ligases that repress ethylene action and promote growth by directing EIN3 degradation. Proc Natl Acad Sci USA. 2004;101:6803–6808. doi: 10.1073/pnas.0401698101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.An F, et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell. 2010;22:2384–2401. doi: 10.1105/tpc.110.076588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao H, et al. Processing and subcellular trafficking of ER-tethered EIN2 control response to ethylene gas. Science. 2012;338:390–393. doi: 10.1126/science.1225974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeong J, et al. Phytochrome and ethylene signaling integration in Arabidopsis occurs via the transcriptional regulation of genes co-targeted by PIFs and EIN3. Front Plant Sci. 2016;7:1055. doi: 10.3389/fpls.2016.01055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM. A link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell. 2005;17:2230–2242. doi: 10.1105/tpc.105.033365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y, et al. A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression-patterning of shared target genes in Arabidopsis. PLoS Genet. 2013;9:e1003244. doi: 10.1371/journal.pgen.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lehman A, Black R, Ecker JR. HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell. 1996;85:183–194. doi: 10.1016/s0092-8674(00)81095-8. [DOI] [PubMed] [Google Scholar]
- 34.An F, et al. Coordinated regulation of apical hook development by gibberellins and ethylene in etiolated Arabidopsis seedlings. Cell Res. 2012;22:915–927. doi: 10.1038/cr.2012.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang KN, et al. Temporal transcriptional response to ethylene gas drives growth hormone cross-regulation in Arabidopsis. eLife. 2013;2:e00675. doi: 10.7554/eLife.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhong S, et al. A molecular framework of light-controlled phytohormone action in Arabidopsis. Curr Biol. 2012;22:1530–1535. doi: 10.1016/j.cub.2012.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, et al. Genomic evidence for COP1 as a repressor of light-regulated gene expression and development in Arabidopsis. Plant Cell. 2002;14:2383–2398. doi: 10.1105/tpc.004416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leivar P, et al. Definition of early transcriptional circuitry involved in light-induced reversal of PIF-imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell. 2009;21:3535–3553. doi: 10.1105/tpc.109.070672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J, et al. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell. 2007;19:731–749. doi: 10.1105/tpc.106.047688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Lucas M, et al. A molecular framework for light and gibberellin control of cell elongation. Nature. 2008;451:480–484. doi: 10.1038/nature06520. [DOI] [PubMed] [Google Scholar]
- 41.Feng S, et al. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature. 2008;451:475–479. doi: 10.1038/nature06448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oh E, Zhu JY, Wang ZY. Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol. 2012;14:802–809. doi: 10.1038/ncb2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bai MY, et al. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandenbussche F, et al. HY5 is a point of convergence between cryptochrome and cytokinin signalling pathways in Arabidopsis thaliana. Plant J. 2007;49:428–441. doi: 10.1111/j.1365-313X.2006.02973.x. [DOI] [PubMed] [Google Scholar]
- 45.Alabadí D, et al. Gibberellins modulate light signaling pathways to prevent Arabidopsis seedling de-etiolation in darkness. Plant J. 2008;53:324–335. doi: 10.1111/j.1365-313X.2007.03346.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.