Significance
Antibiotic development is frequently plagued by the rapid emergence of drug resistance. However, assessing the risk of resistance development in the preclinical stage is difficult. By building on multiplex automated genome engineering, we developed a method that enables precise mutagenesis of multiple, long genomic segments in multiple species without off-target modifications. Thereby, it enables the exploration of vast numbers of combinatorial genetic alterations in their native genomic context. This method is especially well-suited to screen the resistance profiles of antibiotic compounds. It allowed us to predict the evolution of resistance against antibiotics currently in clinical trials. We anticipate that it will be a useful tool to identify resistance-proof antibiotics at an early stage of drug development.
Keywords: directed evolution, antimicrobial resistance, high-throughput mutagenesis, multiplex automated genome engineering
Abstract
Antibiotic development is frequently plagued by the rapid emergence of drug resistance. However, assessing the risk of resistance development in the preclinical stage is difficult. Standard laboratory evolution approaches explore only a small fraction of the sequence space and fail to identify exceedingly rare resistance mutations and combinations thereof. Therefore, new rapid and exhaustive methods are needed to accurately assess the potential of resistance evolution and uncover the underlying mutational mechanisms. Here, we introduce directed evolution with random genomic mutations (DIvERGE), a method that allows an up to million-fold increase in mutation rate along the full lengths of multiple predefined loci in a range of bacterial species. In a single day, DIvERGE generated specific mutation combinations, yielding clinically significant resistance against trimethoprim and ciprofloxacin. Many of these mutations have remained previously undetected or provide resistance in a species-specific manner. These results indicate pathogen-specific resistance mechanisms and the necessity of future narrow-spectrum antibacterial treatments. In contrast to prior claims, we detected the rapid emergence of resistance against gepotidacin, a novel antibiotic currently in clinical trials. Based on these properties, DIvERGE could be applicable to identify less resistance-prone antibiotics at an early stage of drug development. Finally, we discuss potential future applications of DIvERGE in synthetic and evolutionary biology.
The emergence of drug resistance against existing antimicrobials is currently responsible for 700,000 worldwide deaths annually (1). However, many pharmaceutical companies have discontinued their antibiotic research programs. This is partly due to the rapid spread of multidrug-resistant bacteria, which makes the commercial success of new antimicrobial drugs unpredictable (2). Meanwhile, serious infection-causing Gram-negative bacteria are becoming resistant to all previously effective antibiotics on the market. The case of GSK2251052—a novel compound developed by GlaxoSmithKline (GSK)—highlights these problems. GSK2251052 inhibits bacterial leucyl-tRNA synthetase and possesses many beneficial properties of an antibiotic for treating human infections by Gram-negative pathogens. However, resistance against GSK2251052 by genomic mutations was identified in animal models and in patients during a phase 2b trial (3). As a consequence, GSK has discontinued clinical development of GSK2251052 (4).
In general, resistance can be mediated by chromosomal gene-resistance mutations or by broad host-range plasmids. The relative importance of these genetic mechanisms in resistance evolution depends on the antibiotic and the bacterial pathogens considered (5). For example, clinically significant levels of resistance against DNA gyrase and topoisomerase IV inhibitors are generally encoded by multiple resistance mutations in the pathogen genome. Resistance genes encoded by plasmids provide a much lower level of resistance against these antibiotics. Accordingly, this work studies chromosomal gene-resistance mutations with a focus on trimethoprim and DNA gyrase inhibitors.
In the early phase of drug development, researchers typically identify numerous lead molecules with antimicrobial activities. It is imperative to estimate the rate of resistance evolution at this early stage of development (3, 6). However, this is a complex problem for three main reasons: (i) the large diversity of molecular mechanisms contributing to resistance, (ii) the numerous pathogenic bacteria to be considered, and (iii) the large number of potential candidate molecules to be tested. Unfortunately, standard microbial protocols are slow, have a low coverage, and may fail to predict the frequency and molecular mechanisms of antibiotic resistance by genomic mutations (3, 7, 8). The two most widespread methods, fluctuation tests and serial passage experiments, generally rely on spontaneous mutational processes and therefore can explore only a small fraction of the sequence space (6, 9). This is especially problematic if a high level of resistance demands the simultaneous acquisition of multiple, rare mutations, many of which seemingly provide little benefit individually (10–12).
Recent genome-engineering technologies enable targeted mutagenesis of predefined DNA segments and therefore are promising alternative tools to explore resistance mutations in a systematic manner. However, current methods suffer from several limitations. The length of the targeted regions is generally limited (13–16), precise adjustment of the mutation rate is unsolved (13–15), or the mutational spectrum is highly biased (13–15, 17). Furthermore, throughput is often moderate due to constraints on maximum library size (18) and the maximum number of targeted regions (17, 19, 20). For a more detailed comparison of existing in vivo mutagenesis protocols, see Dataset S1. Limitations also hold for multicopy plasmid-based mutagenesis platforms, including error-prone PCR, as they distort native expression levels and fail to identify recessive mutations (21–25).
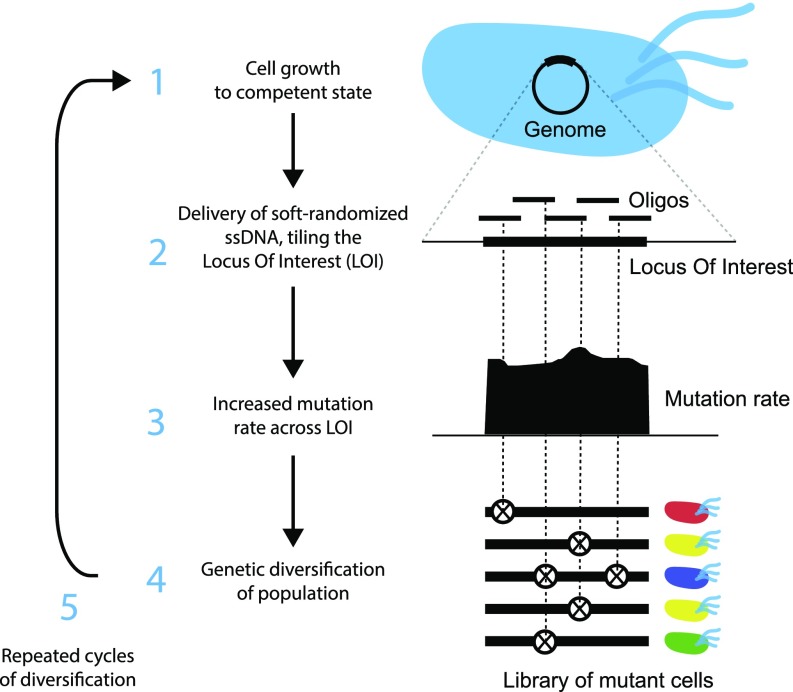
Multiplex automated genome engineering (MAGE) and related methods have been proposed to mutagenize the full length of individual genes (26–30). Existing approaches demand a large number of individual oligos even for targets of modest size, and therefore targeted mutagenesis of long genomic regions is tedious (Dataset S1). Here, we present directed evolution with random genomic mutations (DIvERGE), a method that addresses the aforementioned shortcomings of current in vivo mutagenesis techniques. DIvERGE enables mutagenesis of predefined long genomic regions (Fig. 1). Moreover, it (i) has broad, controllable mutagenesis spectra for each nucleotide position, (ii) allows very high mutation rates of the target sequences, (iii) enables multiple rounds of mutagenesis and selection, (iv) is applicable to a range of host species without the need for prior genomic modification, and is also (v) cost-effective.
Fig. 1.
Overview of DIvERGE mutagenesis. Randomized DNA oligo synthesis precisely controls the rate of mutations in partially overlapping oligos. These oligos fully cover the locus of interest and induce mutagenesis at their target after incorporation. Similarly to prior approaches (26, 28), DIvERGE proceeds via cell growth (1), oligo delivery and incorporation (2), and mutagenesis (3), leading to a highly elevated genetic diversity of the mutagenized population (4).
We show that this method is particularly well-suited to screen the resistance profiles of antibiotic compounds. Specifically, in a single day DIvERGE generated clinically significant resistance against trimethoprim and ciprofloxacin in multiple bacterial species. It also allowed us to predict the rise of high level of resistance (and the underlying mutational combinations) against gepotidacin, a novel antibiotic currently in clinical trials.
Results
Overview of DIvERGE.
DIvERGE is based on a unique single-strand (ss) DNA oligonucleotide (oligo) design strategy where long, continuous genomic segments are covered by the alignment of partially overlapping mutagenized DNA oligos (Fig. 1). Tuning of the nucleotide composition in each synthetic DNA oligo ensures that each possible mutation is represented in the synthesized oligo pool. These oligo pools are synthesized using a soft-randomization protocol (31). In a nutshell, soft-randomization introduces a small amount of nucleotide mixture at specific variable positions of the wild-type sequence. It thereby generates oligos with randomly positioned mutations. Moreover, soft-randomization enables the precise control of the rate and spectrum of mutations in the targeted segment (SI Appendix, Fig. S1).
The randomized oligos fully cover the locus of interest and induce mutagenesis at their target after incorporation. Indeed, prior works indicate that limiting the number of mismatches compared with the target sequence allows for an efficient genomic integration of DNA oligos (26, 32, 33) (SI Appendix, Fig. S2), while the overlapping design permits random and uniformly distributed mutagenesis along exceptionally long DNA segments. We note that application of soft-randomized oligos circumvents the need for cost-demanding, massively parallel oligonucleotide synthesis.
Similarly to prior approaches (26, 28), DIvERGE (Fig. 1) proceeds via cell growth (1), oligo delivery and incorporation (2), mutagenesis (3), leading to a highly elevated genetic diversity of the mutagenized population (4). In the following, we step-by-step demonstrate the potentials of the method and apply it to study antibiotic resistance.
Uniform and Adjustable Mutagenesis of Selected Genomic Targets.
We first tested whether a single randomized oligo pool can keep the mutation rate and mutational spectrum uniform along its sequence. To this end, 90-nt-long DNA oligos, complementary to a landing pad sequence, previously integrated into the genome of Escherichia coli K-12 MG1655 (34), were synthesized in a way that each nucleotide position was spiked with up to 20% of all three possible mismatching nucleotides. Spiking ratio is defined as the fraction of the mismatching nucleotides during oligo synthesis. For the estimated distribution of mutation numbers at each spiking ratio, see SI Appendix, Table S1.
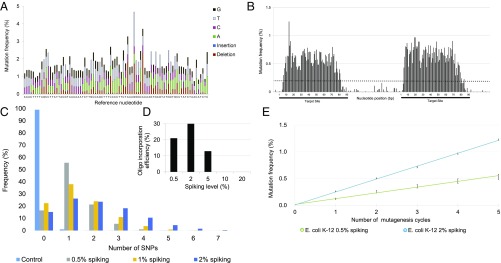
The nucleotide composition within the oligo pool was confirmed using Illumina high-throughput (HT) sequencing. Reassuringly, we achieved balanced mutational distribution across the entire length of the oligo (Fig. 2A). Moreover, the number of mutations within each oligo was precisely adjustable depending on the level of spiking (SI Appendix, Fig. S1 and Table S1).
Fig. 2.
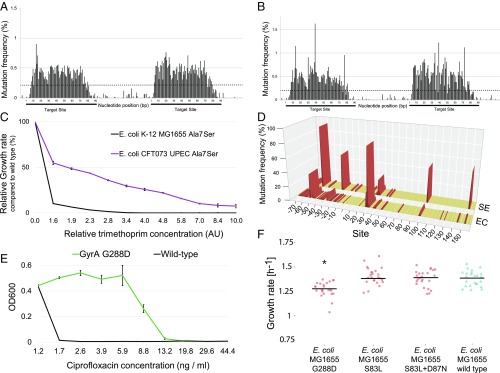
DIvERGE mutagenizes selected genomic targets. (A) Mutation frequency along a randomized 90-mer oligonucleotide. A 0.5% nucleoside-phosphoramidite spiking ratio was used to mutate each nucleotide position along the entire length of the sequence. (B) Genomic mutation frequency, measured as the background-normalized frequency of mutations at a given position, with DIvERGE in E. coli K-12 MG1655 after five cycles of mutagenesis. Two 90-bp genomic regions separated by an intermediate 70-bp region were targeted for mutagenesis (indicated as black lines beneath the x axis). A cut-off of 0.2% (dashed line) was used as a threshold to qualify diversified positions, all values below indicate detection limits of the employed sequencing technology (SI Appendix, SI Materials and Methods). (C) The frequency of SNP events per site, introduced by DIvERGE mutagenesis, as a function of the DNA-synthesis spiking ratio. Samples represent allelic composition after five mutagenesis cycles with the corresponding spiking ratio, while control represents mutagenesis with a nonspiked oligo. (D) Incorporation efficiencies of randomized oligos with increasing levels of the DNA-synthesis spiking ratio during DIvERGE in E. coli K-12 MG1655. Ten percent and 20% spiked oligo incorporations were below the detection threshold of 0.01 and therefore not visible on the graph. (E) Mutation frequency at the target site in consecutive DIvERGE cycles in E. coli K-12 MG1655. Oligo soft-randomization was applied at two spiking ratios (0.5% and 2%, respectively). Mutation frequency indicates background-normalized mutation frequency at a given position. Error bars represent SE based on diversified positions (n = 154).
Next, we examined the incorporation of these ssDNA oligo pools into the bacterial genome at the landing pad sequence. Iterative recombineering was performed utilizing pORTMAGE, an approach capable of achieving highly efficient allelic replacement without off-target effects (34). Using randomized oligos, we targeted two 90-bp-long regions within the landing pad and performed five DIvERGE cycles with each. The mutation frequency was then determined using HT sequencing of the targeted landing pad sequence, revealing that mutagenesis was extended to 87% of the length of the entire oligos (Fig. 2B). Consistent with prior works that characterized oligo incorporation in E. coli (33, 35, 36), we noted a sharp drop in mutation frequency at each oligo terminus (Fig. 2B).
Intermediate (2%) spiking ratios produced the highest mutation frequency at the target sequence. The average number of mutations within the target sequence increased with the spiking ratio (Fig. 2C). However, oligos with low similarities to their target sequence have a reduced capacity to integrate into the genome (26, 32) (SI Appendix, Fig. S2). Therefore, oligo pools with numerous mismatches may actually limit the efficiency of mutagenesis of the target sequence. Indeed, we observed a sharp decrease in the incorporation efficiency of oligo pools with a spiking ratio above 5% (Fig. 2D).
Reassuringly, no major bias in the mutational spectrum was detected along the target region: all individual mutation categories fell between 13.8% and 22.4% in the genomic pool (Table 1). This is a significant advantage over typical in vitro mutagenesis methods [such as error-prone PCR (37–39)], which tend to display significant mutational bias. Moreover, the extent of sequence diversity of the population was tunable by the number of DIvERGE cycles and the spiking ratio of the oligo pool (Fig. 2E).
Table 1.
Mutagenic spectrum of DIvERGE
| Oligonucleotide | Genomic | |||
| Mutational bias indicator | Frequency | SD | Frequency | SD |
| Transitions/Transversions (Ts/Tv) | 0.6 | 0.1 | 0.6 | 0.1 |
| A → G, T → C (%) | 13.8 | 0.8 | 13.8 | 0.4 |
| G → A, C → T (%) | 22.4 | 1.5 | 22.2 | 1.8 |
| A → T, T → A (%) | 15 | 1.1 | 17.1 | 1.4 |
| A → C, T → G (%) | 12.9 | 0.5 | 13.8 | 0.2 |
| G → C, C → G (%) | 14.2 | 0.4 | 14.8 | 1.1 |
| G → T, C → A (%) | 21.7 | 0.2 | 18.2 | 0.6 |
Spectrum of mutations in a randomized oligo after 0.5% spiked DNA synthesis (TETRM1_05) and the resulting spectrum of mutations at the genomic target after five DIvERGE cycles in E. coli K-12 MG1655. Frequencies and SD values were calculated from sequencing data.
Performing five DIvERGE cycles with medium-level randomization (i.e., 2% spiking ratio within the synthetic oligo sequence) resulted in an over 106-fold increase in mutation rate at the target region compared with the background level. This corresponds to an increase from 2.2 × 10−10 to 2.4 × 10−4 mutations per nucleotide per generation (SI Appendix, Table S6), as measured using HT sequencing of the landing pad sequence. Importantly, the mutation rate of nontargeted regions—measured at the intermediate region of the landing pad between the two target sequences—remained low (Fig. 2B). Overall, DIvERGE allowed efficient control of sequence diversification at a chosen target site by oligo randomization.
DIvERGE Can Mutagenize Extended Genomic Regions.
Next, we aimed to extend the genomic target region undergoing mutagenesis by designing and synthesizing multiple, overlapping, and randomized oligos that cover an entire gene and its promoter region. As a target, we chose the essential gene folA, encoding dihydrofolate reductase (FolA) and its promoter region. The selection of folA has clinical relevance, as dihydrofolate reductase is the target of the widely used antimicrobial drug trimethoprim (40). Prior studies demonstrated that under prolonged trimethoprim pressure, the evolution of resistance proceeds predominantly through mutations in folA (41, 42); therefore, mutagenesis of this gene produces an easily detectable phenotype. Because resistance-conferring mutations have been detected both in the regulatory (43) and in the protein-coding regions of folA (22) (Dataset S2), we diversified both regions in E. coli K-12 MG1655. To this aim, we used eight overlapping mutagenized oligos to cover folA (SI Appendix, SI Materials and Methods). Based on the observed incorporation pattern of mutations from individual oligos (Fig. 2B), 9-nt-long overlaps were designed between neighboring oligos to ensure an equally high probability of mutagenesis at all positions. Overall, one oligo targeted the 81-bp-long promoter region, while seven targeted the 480-bp-long protein-coding sequence (Dataset S3).
We generated folA variant libraries with single-point mutations in the target sequence. Single rounds of DIvERGE mutagenesis cycles were carried out separately with each of the eight oligos. The resulting mutants were then subjected to mild trimethoprim selection pressure and the genotypes of resistant clones were determined by sequencing (SI Appendix, SI Materials and Methods). Clones with more than one mutation were excluded from further analysis, thus focusing on the adaptive single-step mutational landscape (SI Appendix, Table S2 and Dataset S4). We found that 81% of the identified single-point mutations reside in the protein-coding region, primarily localized in the active site cavity and the NADPH binding site of dihydrofolate reductase (44), while the rest were in the promoter region. DIvERGE detected 17 previously described (22, 41–43, 45) mutations against trimethoprim, many of which have been found clinical isolates (Dataset S2), and unveiled seven novel ones. Analysis of a subset of these mutations individually confirmed their resistance-conferring phenotypes (SI Appendix, Fig. S4 and Table S2). These results are all of the more remarkable, as a single round of DIvERGE mutagenesis takes only ∼5 h to perform.
DIvERGE Promotes Exceptionally Rapid Evolution of High-Level Antibiotic Resistance.
Because high-level trimethoprim resistance generally demands multiple mutations in folA, we next subjected E. coli K-12 MG1655 to multiple rounds of DIvERGE mutagenesis. Five consecutive mutagenesis cycles were carried out, simultaneously targeting all nucleotide positions in the regulatory and protein-coding regions.
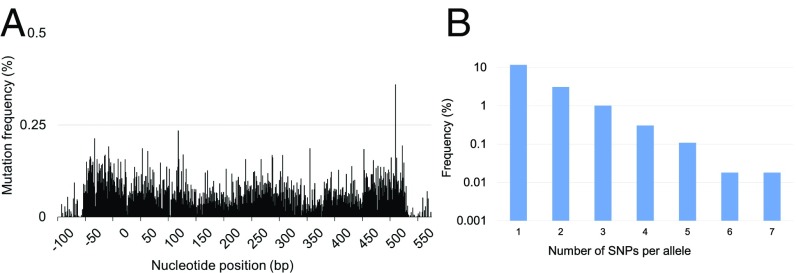
Iterative integration of the oligos resulted in randomization of the targeted regions (Fig. 3A, SI Appendix, Table S3, and Dataset S4) and successful generation of higher-order mutational combinations (Fig. 3B and Dataset S5). After only five DIvERGE cycles completed in a single day, we identified folA mutants with up to an 895-fold increase in trimethoprim resistance compared with that of the wild type (Table 2). Resistance was quantified by IC75, the trimethoprim concentration that inhibits growth by 75% compared with the drug-free condition.
Fig. 3.
DIvERGE mutagenesis along the full length of an antibiotic resistance gene. (A) Mutation frequency at the E. coli K-12 MG1655 folA locus after five cycles of DIvERGE mutagenesis. Positions 0 and 480 refer to the start and end of the protein-coding sequence of folA. Mutation frequency is defined as the background-normalized frequency of substitutions occurring at a given position. (B) Naïve library composition after five cycles of DIvERGE mutagenesis of the E. coli K-12 folA locus. The frequency of sequencing reads with the given number of SNPs within the target sequence.
Table 2.
Trimethoprim susceptibility of individually selected E. coli K-12 MG1655 folA mutants
| Strain ID | FolA regulatory mutation | FolA mutation | Trimethoprim IC75 (μg/mL) | IC75 (fold-change compared with wild type) |
| Escherichia coli K-12#1 | C-58T | A26T, L28R, P39R | 1,254 | 895.7 |
| Escherichia coli K-12#2 | C-58T, T-74A | P21P, L28R, N147D | 447.5 | 319.6 |
| Escherichia coli K-12#3 | C-58T | L28R | 610 | 492.8 |
| Escherichia coli K-12#4 | C-43T, C-58T | A26D, L28R, H45R | 794 | 567.1 |
IC75 (Trimethoprim) of the wild type equals 1.4 μg/mL. Data represent IC75 based on the average of three independent measurements.
With the multiround directed evolution of folA, higher-level mutational combinations were generated at high efficiency. Some of these variants displayed a more than 3,900-fold increase in their trimethoprim-resistance level compared with wild-type E. coli K-12 MG1655 (Table 3 and SI Appendix, Fig. S4).
Table 3.
Trimethoprim susceptibility of individually selected E. coli K-12 MG1655 folA mutants after two rounds of consecutive mutagenesis-selection cycles
| Strain ID | FolA regulatory mutation | FolA mutation (or same-sense DNA mutation) | IC75 (fold-change compared with wild type) |
| Escherichia coli K-12#8 | C-58A | A26S, L28R, W30C, (C132G) | >3,900 |
| Escherichia coli K-12#11 | C-58A | A26T, L28R, W30C | >3,900 |
| Escherichia coli K-12#12 | C-58A | A26T, L28R, (C132G), M88L | 2,140 |
| Escherichia coli K-12#23 | C-58A | A26T, L28R, (C132G) | 1,430 |
IC75 (trimethoprim) of the multiround DIvERGE generated folA variants in E. coli K-12 MG1655, with new mutation-combinations emerging compared with the parental variant (containing C-58A, W30C, and C132G, a same-sense mutation). Data represent IC75 based on the average of three independent measurements.
One may argue that the oligo set designed to target the wild-type folA may revert mutations that had accumulated at an earlier stage of laboratory evolution. To address this issue, we focused on a previously identified folA variant selected using mild trimethoprim stress with three mutations in E. coli K-12 MG1655. We carried out an additional five DIvERGE cycles on this mutant variant and sequenced the resulting library. Reassuringly, no substantial decrease in the level of nucleotide variation was observed in the folA variant, with the only exception being the nucleotides directly adjacent to the three preexisting mutations (SI Appendix, Fig. S3 and Datasets S4 and S7). Overall, DIvERGE generated a diverse set of trimethoprim-resistant variants, simultaneously retaining the three mutations introduced before the mutagenesis-selection cycles.
This indicates that there is no need for new oligo design and synthesis after each round of mutagenesis. Therefore, the mutagenesis cycles are rapid, uninterrupted, and can be potentially scaled up toward many parallel populations.
DIvERGE Minimizes Off-Target Effects.
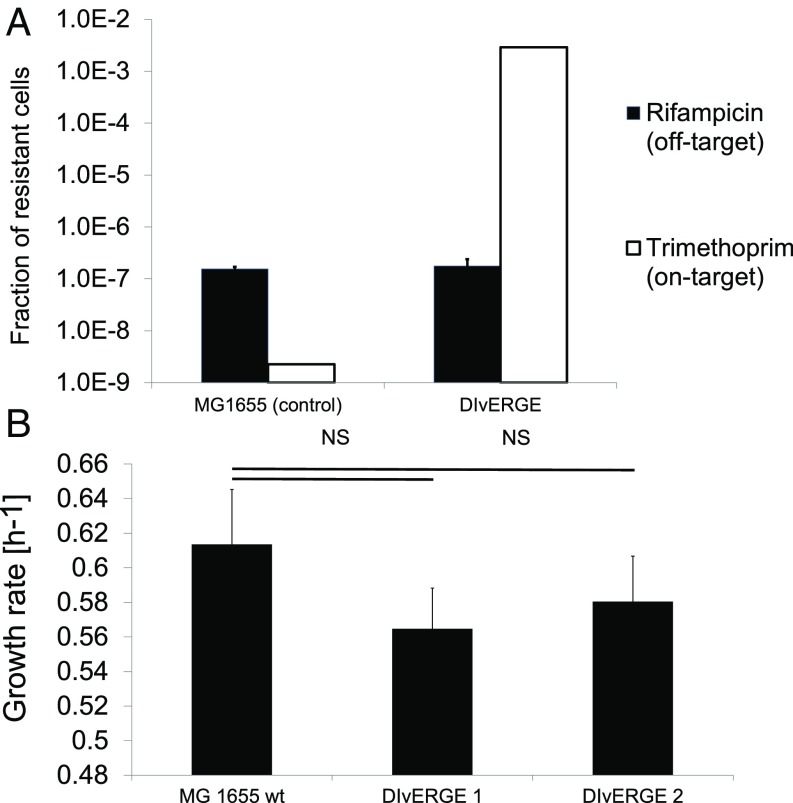
We next assessed the potential off-target mutagenesis of DIvERGE. The extent of off-target mutagenesis is particularly important because the accumulation of undesired, off-target mutations could interfere with the phenotypic effects of the engineered modifications. To quantify off-target mutagenesis, we measured the fraction of rifampicin-resistant cells in DIvERGE treated populations while simultaneously mutagenizing folA. Importantly, the corresponding resistance-conferring locus (rpoB) was not targeted by DIvERGE.
DIvERGE-treated populations showed no significant increase in the frequency of rifampicin-resistant cells after five cycles of mutagenesis compared with that of untreated wild-type populations (Fig. 4A). This confirms earlier results on the shortage of mutations in nontargeted regions of the landing pad sequence (Fig. 2B). Furthermore, no significant fitness decline was observed in DIvERGE-derived trimethoprim resistant clones (Fig. 4B), indicating a shortage of fitness-decreasing off-target mutations. Taking these data together, we find that DIvERGE selectively and efficiently targets predefined genomic loci, while minimizing off-target effects.
Fig. 4.
DIvERGE promotes the evolution of antibiotic resistance and simultaneously minimizes off-target effects. (A) The fraction of resistant cells in DIvERGE-mutagenized populations toward rifampicin (representing off-target) and trimethoprim (targeted by DIvERGE). E. coli K-12 MG1655 served as a wild-type control. Error bars denote the SEM for 12 biological replicates. (B) Fitness effect of DIvERGE mutagenesis. Growth rate measurements were performed on 3 μg/mL (DIvERGE 1) and 50 μg/mL (DIvERGE 2) trimethoprim-selected, DIvERGE-generated variants (n = 30 for each). The fitness of DIvERGE 1 and 2 are statistically equal to wild-type MG1655 fitness (MG1655 WT) (two-tailed t test, P = 0.23, and 0.49, respectively).
DIvERGE Is Applicable to Multiple Bacterial Species.
We next tested whether DIvERGE could be applicable to the mutagenesis of distant relatives of E. coli as well. We selected as models Salmonella enterica and the opportunistic pathogen Citrobacter freundii, all of which diverged from E. coli ∼100–200 million y ago (34).
To characterize the efficiency of mutagenesis with DIvERGE in a uniform manner across species, we integrated the same landing-pad sequence utilized for E. coli into the genomes of these bacterial species (34). We thereby characterized the incorporation of the oligos using the same oligo set and the same protocol as in E. coli (SI Appendix, SI Materials and Methods). Akin to what we observed in E. coli, the randomized oligos efficiently induced mutagenesis in both S. enterica and C. freundii (Fig. 5 A and B and SI Appendix, Fig. S5 and Table S4). Consistent with results in E. coli (Fig. 2C), the highest mutation frequency was achieved using oligos with 2% spiking ratios. As expected, mutation frequency correlated with the number of DIvERGE mutagenesis cycles (SI Appendix, Fig. S5). Overall, we achieved at least a 105-fold increase in mutation rate at the target sequence in both species (SI Appendix, Table S4).
Fig. 5.
DIvERGE is applicable to multiple hosts and discovers novel antibiotic resistance-conferring mutations. Elevation of mutation frequency, measured as the frequency of mutations occurring at a given position, after five consecutive DIvERGE cycles shows high locus specificity in (A) S. enterica and (B) C. freundii. Dashed line indicates cutoff as a qualification of diversified positions, all values below indicate detection limits of the employed sequencing technology (SI Appendix, SI Materials and Methods) (C) Trimethoprim susceptibility analysis of the strain-specific FolA A7S variant in E. coli K-12 MG1655 and in CFT073 UPEC. Data represent relative inhibitory concentration (AU) compared with the IC75 of the corresponding wild-type strain. Error bars are SDs based on three independent measurements. (D) DIvERGE revealed distinct mutational hot-spots during adaptation to 267-fold dosage above the wild-type minimum inhibitory concentration of trimethoprim in E. coli K-12 MG1655 (marked EC), and S. enterica (marked SE). Figure shows mutation frequency at the detected mutational hot-spots based on Single Molecule Real-Time (SMRT) sequencing. Promoter (DNA sites below 0) and protein coding regions (amino acid sites above 0) of folA are indicated. Mutational hot-spots above 0.5% mutation frequency are highlighted in red. (E) Resistance-conferring phenotype of the DIvERGE identified an E. coli GyrA G288D variant. OD600 indicates the optical density of the bacterial culture after 24 h incubation in the presence of the corresponding drug concentration. Error bars indicate SD for three parallel measurements. (F) Fitness of the antibiotic-resistant mutant and wild-type E. coli K-12 MG1655 strains. Using standard procedures (SI Appendix, SI Materials and Methods), fitness was estimated by growth rate in liquid Mueller Hinton II medium, 24 replicates per genotype. Growth curves were recorded by measuring optical density (OD600) every 7 min for 24 h at 37 °C. GyrA G288D mutant fitness is significantly lower than wild-type fitness (t test, *P < 4.91 × 10−5). By contrast, GyrA S83L mutant and S83L + D87N double-mutant fitness are statistically equal to wild-type fitness (t test, P = 0.83, and 0.67, respectively).
Differences in Mutational Effects Between Closely Related Pathogens.
We targeted folA and its promoter sequence in S. enterica and another clinically relevant strain, uropathogenic E. coli CFT073 (UPEC). Trimethoprim is frequently deployed against these pathogens in the clinic (46). The protocols employed were similar to that for E. coli K-12 MG1655. Species and strain-specific oligos were synthesized, resulting in eight overlapping oligos for each pathogen. A single round of DIvERGE generated folA variant libraries with single-point mutations in the target sequence. As for E. coli K-12 MG1655, ∼3,000 resistant clones were selected at mild trimethoprim dosage and were subsequently sequenced. Comparison of the adaptive mutations in E. coli K-12, E. coli CFT073, and S. enterica revealed novel resistance mutations (SI Appendix, Fig. S6 and Table S2 and Dataset S4). Despite the 99% sequence similarity of folA between E. coli K-12 and E. coli CFT073, the conferred relative resistance level by the same mutation frequently differed between the two strains (SI Appendix, Table S2). Most notably, the Ala7Ser mutation in the protein-coding region provided resistance in E. coli CFT073 only (Fig. 5C).
We next compared the mutational profiles of S. enterica and E. coli CFT073 using five rounds of DIvERGE. This resulted in the rapid generation of combinatorial folA mutants with trimethoprim resistance levels up to 3,873-times higher than that of the corresponding wild-type strains (SI Appendix, Table S5). It is worth noting that the corresponding trimethoprim dosages are comparable with the dosage regularly employed to treat urinary tract infections (46).
Sequence analysis of 1,000 resistant clones from each organism confirmed that high levels of resistance frequently required multiple mutations within the regulatory and protein-coding regions of folA (SI Appendix, Table S5 and Dataset S6). However, differences in mutational hot-spots were detected between the two species (Fig. 5D and SI Appendix, Fig. S6 and Table S5). Notably, a promoter position (C-61) and a C-terminal amino acid coding position (Phe-153) were mutated predominantly in S. enterica (Fig. 5D). Mutations at Phe-153 may act as compensatory mutations partially restoring catalysis, paired with active site mutations that lower catalytic activity (47, 48). In line with prior studies with E. coli (22, 42, 45), high levels of trimethoprim resistance mostly evolved via combinations of mutations in the promoter and the protein’s active site.
Taken together, these results indicate differences in mutational effects across related species.
DIvERGE Discovers Antibiotic Resistance Mutations Against a Fluoroquinolone.
Next, we addressed the question whether DIvERGE can mutagenize longer genomic regions with a comparable resolution. To this end, we mutagenized the gyrA locus in E. coli K-12 MG1655. GyrA encodes the A subunit of DNA gyrase, which is targeted by fluoroquinolones (49). GyrA is nearly five-times longer than folA mutagenized in the previous section.
We investigated resistance hot-spots toward ciprofloxacin, one of the most frequently used fluoroquinolones (49). After performing DIvERGE mutagenesis with 38 overlapping oligos covering the entire promoter and protein-coding regions, we selected 1,000 resistant mutants under mild ciprofloxacin stress [i.e., fivefold above the wild-type minimum inhibitory concentration (MIC)]. Sequence analysis revealed that all putative resistance mutations reside solely in the protein-coding region. Clinically occurring mutations at Ser-83 and Asp-87 and their combinations dominated the observed mutational landscape (10, 50–52). Remarkably, three frequently observed individual mutations at positions 288, 334, and 785 fall outside of the well-described quinolone resistance determining region of gyrA (51) (Dataset S6). To the best of our knowledge, this link of these positions to quinolone resistance in E. coli is unique. Reconstruction of the corresponding GyrA Gly288Asp (G288D) allele in a wild-type genetic background confirmed its resistant phenotype (Fig. 5E).
Future studies are needed to determine whether these positions are clinically relevant. There could be two reasons why this particular mutation has remained undetected so far. First, the level of ciprofloxacin resistance provided by G288D is relatively modest (Fig. 5E). Second, G288D may induce a fitness cost in the absence of antibiotic. To investigate the second possibility, we introduced G288D and two clinically known mutations (S83L, D87N) into wild-type E. coli, and measured the growth rates of the corresponding mutants in vitro (Fig. 5F). We found that G288D reduces growth rate by 8.3% compared with the wild type, while S83L and D87N have no detectable effects on fitness. These results indicate that future work should study resistance level and potential fitness costs of mutants simultaneously and in a HT manner.
Mutagenesis Along the Full Lengths of Multiple Genes.
Based on the results of the previous section, we sought to chart other putative resistance determinants along multiple target genes of ciprofloxacin. In Gram-negative bacteria (such as E. coli), ciprofloxacin’s primary drug target is the DNA gyrase complex (50). However, ciprofloxacin also has a lower binding affinity to the homologous DNA topoisomerase IV complex (53) as well. GyrA and GyrB constitute the gyrase complex, while ParC and ParE are involved in the topoisomerase IV complex.
To identify mutations that promote resistance, we mutagenized all four constituents along the full lengths of their corresponding protein-coding DNA regions. Accordingly, we performed a single round of DIvERGE mutagenesis in E. coli K-12 MG1655, using 130 oligos covering 9,503 base pairs (Dataset S3). The resulting mutants were then subjected to mild ciprofloxacin stress (i.e., at a dosage twofold higher than the wild-type MIC), and the genotypes of 3,000 resistant clones were determined (SI Appendix, SI Materials and Methods).
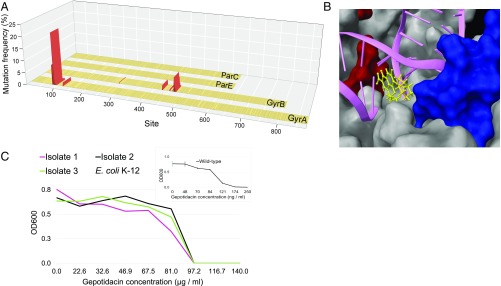
Sequence analysis indicated mutagenesis at multiple target loci and, as expected, the overwhelming majority of the identified alleles carried single mutations only (Fig. 6A and Dataset S6). Mutations were detected in gyrA and gyrB, but not in parC and parE (Fig. 6A). Most notably, the analysis revealed a 46-aa-long region of the GyrB protein mutated in 22.4% of the analyzed alleles (Dataset S6). Protein structure studies demonstrated that this protein region is in close proximity of GyrA (54, 55) in the gyrase complex and may interact with fluoroquinolones (Fig. 6B).
Fig. 6.
Mutagenesis along the full length of drug-targets discovers resistance-conferring mutations. (A) Map of DIvERGE-identified ciprofloxacin resistance-conferring mutations at the drug targets of fluoroquinolones in E. coli K-12 MG1655. Figure shows mutation frequencies at detected mutational hot-spots based on simultaneous Single Molecule Real-Time (SMRT) sequencing at the gyrA, gyrB, parC, and parE loci. Positions are marked with amino acid positions within the protein-coding region of the target loci. Mutational positions above a 0.5% mutation frequency cut-off are marked in red. (B) Ciprofloxacin resistance-determining regions of GyrA (red) and GyrB (blue), two subunits of the DNA gyrase complex (PDB ID code 5BS8). The figure also shows the interaction of the complex with a fluoroquinolone (yellow) and dsDNA (magenta). Mutated positions above the 0.5% mutation frequency cut-offs are indicated and qualified as mutational hot-spots. (C) Dose–response curves of three DIvERGE-generated gepotidacin resistant E. coli K-12 clones and that of the wild type, parental strain (Inset). OD600 indicates the optical density of the bacterial culture after 24 h incubation in the presence of the corresponding drug concentration. Measurements were performed in three replicates according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines. Error bars indicate SD.
Moreover, we note that the shortage of resistance-conferring mutations in ParC and ParE is in-line with prior observations (50). Mutations accounting for the first step of fluoroquinolone resistance development are generally in the primary drug target, and mutational effects in ParC and ParE rely heavily on specific epistatic interactions with mutations in GyrA (56). In a future work, DIvERGE can investigate the coevolution of the two protein complexes by performing repeated mutagenesis selection.
Predicting Resistance Against an Antibiotic Currently Under Clinical Trial.
Based on the above results, we anticipated that DIvERGE could forecast long-term clinical efficacy of multiple antibiotic candidates at an early stage of development. As the first step in this direction, we focused on gepotidacin (GSK2140944). Gepotidacin is an antibiotic candidate currently in clinical phase 2 trials. It selectively inhibits bacterial DNA gyrase and topoisomerase IV by an entirely novel mode-of-action, not observed in any other currently approved antibiotic (57). A recent study failed to recover resistant clones in Neisseria gonorrhoeae (58), but this result may reflect limitations of standard microbial assays for detection of high levels of resistance.
To investigate this problem in more detail, we first attempted to generate resistance mutants by exposing as many as 1010 wild-type E. coli cells to gepotidacin stress in a standard frequency of resistance assay (59). No resistant variants emerged after 72 h. In contrast, we subjected four potential target genes (gyrA, gyrB, parE, and parC) to a single round of DIvERGE mutagenesis. DIvERGE generated E. coli mutants displaying a 557-fold increase in resistance level compared with the corresponding wild type (Fig. 6C). Sequence analysis of three independently isolated clones indicated that a combination of two specific mutations is sufficient to reach such high levels of resistance. In particular, the subsequent introduction of the identified GyrA Asp82Asn and ParC Asp79Asn mutations into the genomes of E. coli CFT073 (UPEC), C. freundii ATCC 8090, and Klebsiella pneumoniae ATCC 10031 revealed that they greatly increase gepotidacin resistance levels in pathogenic strains as well (Table 4).
Table 4.
Gepotidacin resistance-causing effect of the GyrA Asp82Asn and ParC Asp79Asn mutation combination in human pathogenic bacteria
| Strain | Gepotidacin MIC (μg/mL) | MIC (fold-change compared with the corresponding wild type) |
| Escherichia coli O6:K2:H1 CFT073 (UPEC) | >150 | >750 |
| Citrobacter freundii ATCC 8090 | >150 | >330 |
| Klebsiella pneumoniae ATCC 10031 | 125 | 2,080 |
The MIC of gepotidacin against wild-type E. coli CFT073, C. freundii ATCC 8090, and K. pneumoniae ATCC 10031, equals 450, 200 and 60 ng/mL, respectively. MIC values were determined according to the EUCAST guidelines.
The above analyses indicate the utility of DIvERGE to explore rare combinations of resistance mutations. We anticipate that it will be a useful tool for rapid resistance-mutation screening of novel antimicrobial compounds at an early stage of drug development with the aim to identify candidate molecules that are less prone to resistance.
Discussion
In summary, DIvERGE offers a versatile solution for high-precision directed evolution: it allows directed mutagenesis along the full lengths of multiple loci in their native genomic context in multiple bacterial species. The properties of DIvERGE are as follows (Fig. 1).
First, and most importantly, DIvERGE can mutagenize the full lengths of multiple protein-coding genes (Fig. 6A). No constraint is placed on the target sequence by the availability of a protospacer adjacent motif, a strict requirement for CRISPR/Cas9-based mutagenesis techniques (13–15, 18). Second, DIvERGE allows the unbiased introduction of mutations at each targeted nucleotide position (Table 1). This is an advantage over techniques where mutational spectra are biased or limited to certain positions (13–15, 27, 29, 30). Third, the rate of mutagenesis can also be precisely adjusted (Fig. 2 C and E) and in certain cases can achieve an up to 106-fold increase compared with basal levels. This range exceeds that of several other in vivo methods (60). Fourth, DIvERGE can be performed iteratively using the same oligo pools, permitting multiple rounds of mutagenesis and selection. Fifth, as DIvERGE utilizes the pORTMAGE plasmids (34), it is applicable to multiple clinically relevant enterobacterial species. Sixth, as DIvERGE does not involve permanent inactivation of the endogenous mismatch-repair system (34), the targeted mutagenesis is coupled with the minimization of unwanted off-target mutations (Figs. 2B and 4A). Finally, DIvERGE is cost-effective, as it does not require a large set of predesigned oligos. It relies on a single set of soft-randomized oligos that can easily be manufactured at a modest cost.
In this work, we used DIvERGE to study antibiotic resistance. Standard microbial protocols, such as fluctuation tests and serial passage experiments, generally rely on spontaneous mutational processes. Therefore, it is possible that such experiments do not detect resistance because the underlying mutations (and combinations thereof) are too rare. As a good benchmark, mutation rate and population size should be high enough to test at least three mutations across 99.9% of nucleotide sites in the genome (6). This would require screening over 1011 wild-type E. coli cells in a standard fluctuation test (6). Alternatively, with an inoculum of 107 cells per population, one would have to propagate over 100 E. coli populations for 100 generations (∼40 d) in a serial passage experiment (6). Clearly, these two main approaches are tedious and slow. By allowing up to a million-fold increase in mutation rate at multiple predefined loci, DIvERGE allows deep scanning of resistance mutations. To reach the benchmark of at least three mutations across 99.9% of nucleotide sites at up to four defined loci, DIvERGE merely needs a population of around 1.4 × 107 E. coli cells, and a single day to perform five cycles. Therefore, DIvERGE is amenable to predicting resistance mutations in a rapid and parallel manner.
As a proof-of-concept, we demonstrated that DIvERGE identified several resistance mutations that had previously been detected in trimethoprim- and ciprofloxacin-resistant clinical isolates (Fig. 6A and Table 2). Within a single screen, DIvERGE not only detected the major known ciprofloxacin resistance hot-spots but also discovered resistance-conferring mutations at novel sites. DIvERGE also revealed major differences in the effects of resistance mutations across bacterial species (Fig. 5C and SI Appendix, Table S2). This is fully consistent with results of prior papers and underlines the importance of directly studying the resistance profiles of the relevant pathogens (61, 62). Finally, we focused on gepotidacin, an antibiotic currently under development. In contrast to previous claims on the shortage of high-level gepotidacin-resistance mutations (58), we demonstrated that a combination of two specific mutations yields an over 500-fold increase in resistance level in multiple bacterial species. This is all the more relevant, as a standard frequency of resistance assay failed to identify resistance mutants. Clearly, future studies are needed to determine whether these mutants are important in the clinical settings.
In the future, DIvERGE could contribute to the identification of the most promising, less resistance-prone hits from antimicrobial compound libraries. We anticipate that recombinases from other species (63, 64) and novel oligo chemistries (65) will facilitate implementation of DIvERGE across a set of host species, including yeasts and mammalian cells (66–70). DIvERGE may also be combined with microarray-based DNA synthesis (71), and thereby randomize up to megabase pairs of target DNA.
Currently, DIvERGE has some important limitations. First, the genes for targeted mutagenesis have to be defined in advance. If the loci involved in resistance are unknown, chemogenomics and related approaches are needed to identify the genes before a DIvERGE screen (72). Second, subsequent in vivo studies are needed to demonstrate the viability of the identified mutants in clinical settings. Third, DIvERGE cannot detect plasmid-mediated resistance (6, 8). For a broader analysis of horizontally transferrable resistance processes, protocols of functional metagenomics (73) and DIvERGE should be integrated in the future. For these reasons, we anticipate that DNA gyrase and topoisomerase IV inhibitors are ideal candidates for future DIvERGE studies. Of the antibiotics currently in clinical development, 24% target these complexes (74), endogenous mutations are the primary source of resistance, and the molecular targets of resistance are generally well-described.
Future Applications
DIvERGE can potentially address several other unmet needs in basic and applied research. Directed evolution of multiple genes in their native genomic context is cumbersome with other methods (14, 18). Improvement of complex traits demands coevolution of interacting amino acids that are coded at distinct loci and many of these mutations provide no benefit individually. For these reasons, directed evolution of protein complexes, multiple enzymatic steps of metabolic pathways, or large cellular subsystems have remained a formidable problem. DIvERGE offers a solution as it can explore combinatorial genotypes across the full lengths of multiple loci. We predict that DIvERGE will be applicable to optimize metabolic pathways in previously untapped species. For example, C. freundii is an efficient host for the production of valuable bioproducts (75), but optimization of metabolic pathways remains challenging in this organism. Other species, such as Pseudomonas putida, have great potential to serve as chassis for industrial biotechnology (76) but are thus far lacking efficient and targeted mutagenesis protocols.
Another potential application is optimization of synthetic DNA segments. Large de novo constructed DNA elements—from 10 kilobases up-to whole genomes—frequently suffer from suboptimal performance due to the unpredictability of their design guidelines (77). One solution is to design, build, and test mutant libraries of such constructs (78). DIvERGE can reduce the timeframe and costs to find an optimal genotype in such engineering endeavors.
Finally, DIvERGE could be employed to investigate some of the key issues in evolutionary biology. These issues include the evolution of mutational effects and epistasis between related species, the relative contribution of the promoter and protein-coding mutations to adaptation to novel conditions, and the extent of differences in the evolutionary routes toward antibiotic resistance in related pathogens.
Materials and Methods
Please see SI Appendix for the detailed description of materials and methods, including strains and oligonucleotides, DIvERGE oligonucleotides, DIvERGE cycling process, determination of mutation frequencies, selection of DIvERGE libraries, sequencing of folA target regions, nucleotide composition analysis in Landing Pad libraries and DIvERGE oligo pools, assessment of mutation profiles in folA libraries with Illumina sequencing, assessment of mutation profiles with Single Molecule Real-Time sequencing, high-throughput DIvERGE oligo sequencing, isolation of individual genotypes, fitness measurements, and antibiotic resistance measurements.
Supplementary Material
Acknowledgments
We thank Donald L. Court (National Cancer Institute) for providing Salmonella enterica LT2; Morten O. A. Sommer (Technical University of Denmark) for providing the Escherichia coli CFT073; Seqomics Ltd. for Illumina sequencing support; and Martin Krzywinski, Ave Tooming-Klunderud, Zoltán Farkas, Andrea Tóth, Zahid Kamal, Tamás Martinek, Ferenc Bogár, Tamás Kukli, Katalin László, and Mónika Pummer. The Pacific Biosciences sequencing service was provided by the Norwegian Sequencing Centre, a national technology platform hosted by the University of Oslo and supported by the “Functional Genomics” and “Infrastructure” programs of the Research Council of Norway and the Southeastern Regional Health Authorities. This work was supported by grants from the European Research Council H2020-ERC-2014-CoG 648364 - Resistance Evolution (to C.P.) and the Wellcome Trust (C.P. and B.P.); Economic Development and Innovation Operational Programme (GINOP) Grants (MolMedEx TUMORDNS) GINOP-2.3.2-15-2016-00020, GINOP (EVOMER) GINOP-2.3.2-15-2016-00014 (to C.P. and B.P.), GINOP-2.3.2-15-2016-00026 (iChamber) (to B.P.); the “Lendület” Program of the Hungarian Academy of Sciences (C.P. and B.P.); Hungarian Scientific Research Fund Grant OTKA PD 109572 (to B.C.); National Innovation Office of Hungary (NKFIH) Grant K120220 (to B.K.); and a PhD fellowship from the Boehringer Ingelheim Fonds (to Á.N.). I.N. and B.K. were supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences.
Footnotes
Conflict of interest statement: Á.N., B.C., B.K., and C.P. have filed a patent application toward the European Patent Office. I.N., B.B., B.M.V., and P.B. had consulting positions at SeqOmics Biotechnology Ltd. at the time the study was conceived. SeqOmics Biotechnology Ltd. was not directly involved in the design and execution of the experiments or in the writing of the manuscript.
This article is a PNAS Direct Submission.
Data deposition: All sequencing data reported in this paper have been deposited in NCBI’s Sequence Read Archive, https://www.ncbi.nlm.nih.gov/sra/SRP144255 (Accession no. SRP144255). Scripts used for analysis are available from http://group.szbk.u-szeged.hu/sysbiol/EvGEn/diverge-2018-script.html.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1801646115/-/DCSupplemental.
References
- 1.O’Neill J. 2016 The Review on Antimicrobial Resistance. Available at https://amr-review.org. Accessed September 7, 2017.
- 2.Fernandes P, Martens E. Antibiotics in late clinical development. Biochem Pharmacol. 2017;133:152–163. doi: 10.1016/j.bcp.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 3.O’Dwyer K, et al. Bacterial resistance to leucyl-tRNA synthetase inhibitor GSK2251052 develops during treatment of complicated urinary tract infections. Antimicrob Agents Chemother. 2015;59:289–298. doi: 10.1128/AAC.03774-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Silver LL. Antibacterial discovery: Problems and possibilities. In: Gualerzi CO, Brandi L, Fabretti A, Pon CL, editors. Antibiotics. Wiley-VCH; Weinheim, Germany: 2013. [DOI] [Google Scholar]
- 5.Hughes D, Andersson DI. Evolutionary consequences of drug resistance: Shared principles across diverse targets and organisms. Nat Rev Genet. 2015;16:459–471. doi: 10.1038/nrg3922. [DOI] [PubMed] [Google Scholar]
- 6.Bell G, MacLean C. The search for ‘evolution-proof’ antibiotics. Trends Microbiol. 2018;26:471–483. doi: 10.1016/j.tim.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 7.Martínez JL, Baquero F, Andersson DI. Predicting antibiotic resistance. Nat Rev Microbiol. 2007;5:958–965. doi: 10.1038/nrmicro1796. [DOI] [PubMed] [Google Scholar]
- 8.Martínez JL, Baquero F, Andersson DI. Beyond serial passages: New methods for predicting the emergence of resistance to novel antibiotics. Curr Opin Pharmacol. 2011;11:439–445. doi: 10.1016/j.coph.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 9.O’Neill AJ, Chopra I. Use of mutator strains for characterization of novel antimicrobial agents. Antimicrob Agents Chemother. 2001;45:1599–1600. doi: 10.1128/AAC.45.1.1599-1600.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marcusson LL, Frimodt-Møller N, Hughes D. Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 2009;5:e1000541. doi: 10.1371/journal.ppat.1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silver LL. Multi-targeting by monotherapeutic antibacterials. Nat Rev Drug Discov. 2007;6:41–55. doi: 10.1038/nrd2202. [DOI] [PubMed] [Google Scholar]
- 12.Sommer MOA, Munck C, Toft-Kehler RV, Andersson DI. Prediction of antibiotic resistance: Time for a new preclinical paradigm? Nat Rev Microbiol. 2017;15:689–696. doi: 10.1038/nrmicro.2017.75. [DOI] [PubMed] [Google Scholar]
- 13.Ma Y, et al. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13:1029–1035. doi: 10.1038/nmeth.4027. [DOI] [PubMed] [Google Scholar]
- 14.Hess GT, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13:1036–1042. doi: 10.1038/nmeth.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang L, et al. Engineering and optimising deaminase fusions for genome editing. Nat Commun. 2016;7:13330, and erratum (2017) 8:16169. doi: 10.1038/ncomms13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma L, et al. CRISPR-Cas9-mediated saturated mutagenesis screen predicts clinical drug resistance with improved accuracy. Proc Natl Acad Sci USA. 2017;114:11751–11756. doi: 10.1073/pnas.1708268114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Finney-Manchester SP, Maheshri N. Harnessing mutagenic homologous recombination for targeted mutagenesis in vivo by TaGTEAM. Nucleic Acids Res. 2013;41:e99. doi: 10.1093/nar/gkt150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garst AD, et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol. 2017;35:48–55. doi: 10.1038/nbt.3718. [DOI] [PubMed] [Google Scholar]
- 19.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soussy CJ, Wolfson JS, Ng EY, Hooper DC. Limitations of plasmid complementation test for determination of quinolone resistance due to changes in the gyrase A protein and identification of conditional quinolone resistance locus. Antimicrob Agents Chemother. 1993;37:2588–2592. doi: 10.1128/aac.37.12.2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Watson M, Liu J-W, Ollis D. Directed evolution of trimethoprim resistance in Escherichia coli. FEBS J. 2007;274:2661–2671. doi: 10.1111/j.1742-4658.2007.05801.x. [DOI] [PubMed] [Google Scholar]
- 23.Packer MS, Liu DR. Methods for the directed evolution of proteins. Nat Rev Genet. 2015;16:379–394. doi: 10.1038/nrg3927. [DOI] [PubMed] [Google Scholar]
- 24.Tee KL, Wong TS. Polishing the craft of genetic diversity creation in directed evolution. Biotechnol Adv. 2013;31:1707–1721. doi: 10.1016/j.biotechadv.2013.08.021. [DOI] [PubMed] [Google Scholar]
- 25.Wong TS, Roccatano D, Zacharias M, Schwaneberg U. A statistical analysis of random mutagenesis methods used for directed protein evolution. J Mol Biol. 2006;355:858–871. doi: 10.1016/j.jmb.2005.10.082. [DOI] [PubMed] [Google Scholar]
- 26.Wang HH, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelsic ED, et al. RNA structural determinants of optimal codons revealed by MAGE-seq. Cell Syst. 2016;3:563–571.e6. doi: 10.1016/j.cels.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallagher RR, Li Z, Lewis AO, Isaacs FJ. Rapid editing and evolution of bacterial genomes using libraries of synthetic DNA. Nat Protoc. 2014;9:2301–2316. doi: 10.1038/nprot.2014.082. [DOI] [PubMed] [Google Scholar]
- 29.Bonde MT, et al. Direct mutagenesis of thousands of genomic targets using microarray-derived oligonucleotides. ACS Synth Biol. 2015;4:17–22. doi: 10.1021/sb5001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordwald EM, Garst A, Gill RT, Kaar JL. Accelerated protein engineering for chemical biotechnology via homologous recombination. Curr Opin Biotechnol. 2013;24:1017–1022. doi: 10.1016/j.copbio.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 31.Hermes JD, Parekh SM, Blacklow SC, Köster H, Knowles JR. A reliable method for random mutagenesis: The generation of mutant libraries using spiked oligodeoxyribonucleotide primers. Gene. 1989;84:143–151. doi: 10.1016/0378-1119(89)90148-0. [DOI] [PubMed] [Google Scholar]
- 32.Wang HH, Church GM. Multiplexed genome engineering and genotyping methods applications for synthetic biology and metabolic engineering. Methods Enzymol. 2011;498:409–426. doi: 10.1016/B978-0-12-385120-8.00018-8. [DOI] [PubMed] [Google Scholar]
- 33.Wang HH, Xu G, Vonner AJ, Church G. Modified bases enable high-efficiency oligonucleotide-mediated allelic replacement via mismatch repair evasion. Nucleic Acids Res. 2011;39:7336–7347. doi: 10.1093/nar/gkr183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyerges Á, et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proc Natl Acad Sci USA. 2016;113:2502–2507. doi: 10.1073/pnas.1520040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li XT, Thomason LC, Sawitzke JA, Costantino N, Court DL. Bacterial DNA polymerases participate in oligonucleotide recombination. Mol Microbiol. 2013;88:906–920. doi: 10.1111/mmi.12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sawitzke JA, et al. Probing cellular processes with oligo-mediated recombination and using the knowledge gained to optimize recombineering. J Mol Biol. 2011;407:45–59. doi: 10.1016/j.jmb.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Copp JN, Hanson-Manful P, Ackerley DF, Patrick WM. Error-prone PCR and effective generation of gene variant libraries for directed evolution. In: Gillam E, Copp JN, Ackerley DF, editors. Directed Evolution Library Creation, Methods in Molecular Biology. Springer; New York: 2014. pp. 3–22. [DOI] [PubMed] [Google Scholar]
- 38.Rasila TS, Pajunen MI, Savilahti H. Critical evaluation of random mutagenesis by error-prone polymerase chain reaction protocols, Escherichia coli mutator strain, and hydroxylamine treatment. Anal Biochem. 2009;388:71–80. doi: 10.1016/j.ab.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 39.Agilent Technologies, Stratagene Products Division 2009 GeneMorph II EZClone Domain Mutagenesis Kit, Revision C.01. Available at https://www.chem-agilent.com/pdf/strata/200552.pdf. Accessed June 1, 2017.
- 40.Hitchings GH, Burchall JJ. Inhibition of folate biosynthesis and function as a basis for chemotherapy. Adv Enzymol Relat Areas Mol Biol. 1965;27:417–468. doi: 10.1002/9780470122723.ch9. [DOI] [PubMed] [Google Scholar]
- 41.Baym M, et al. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toprak E, et al. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2011;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flensburg J, Sköld O. Massive overproduction of dihydrofolate reductase in bacteria as a response to the use of trimethoprim. Eur J Biochem. 1987;162:473–476. doi: 10.1111/j.1432-1033.1987.tb10664.x. [DOI] [PubMed] [Google Scholar]
- 44.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: Crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 45.Palmer AC, et al. Delayed commitment to evolutionary fate in antibiotic resistance fitness landscapes. Nat Commun. 2015;6:7385. doi: 10.1038/ncomms8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wisell KT, Kahlmeter G, Giske CG. Trimethoprim and enterococci in urinary tract infections: New perspectives on an old issue. J Antimicrob Chemother. 2008;62:35–40. doi: 10.1093/jac/dkn147. [DOI] [PubMed] [Google Scholar]
- 47.Dion A, Linn CE, Bradrick TD, Georghiou S, Howell EE. How do mutations at phenylalanine-153 and isoleucine-155 partially suppress the effects of the aspartate-27–>serine mutation in Escherichia coli dihydrofolate reductase? Biochemistry. 1993;32:3479–3487. doi: 10.1021/bi00064a036. [DOI] [PubMed] [Google Scholar]
- 48.Brown KA, Howell EE, Kraut J. Long-range structural effects in a second-site revertant of a mutant dihydrofolate reductase. Proc Natl Acad Sci USA. 1993;90:11753–11756. doi: 10.1073/pnas.90.24.11753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hooper DC, Wolfson JS, Ng EY, Swartz MN. Mechanisms of action of and resistance to ciprofloxacin. Am J Med. 1987;82:12–20. [PubMed] [Google Scholar]
- 50.Hooper DC, Jacoby GA. Mechanisms of drug resistance: Quinolone resistance. Ann N Y Acad Sci. 2015;1354:12–31. doi: 10.1111/nyas.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Piddock LJV. Mechanisms of fluoroquinolone resistance: An update 1994-1998. Drugs. 1999;58:11–18. doi: 10.2165/00003495-199958002-00003. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida H, Bogaki M, Nakamura M, Nakamura S. Quinolone resistance-determining region in the DNA gyrase gyrA gene of Escherichia coli. Antimicrob Agents Chemother. 1990;34:1271–1272. doi: 10.1128/aac.34.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Khodursky AB, Zechiedrich EL, Cozzarelli NR. Topoisomerase IV is a target of quinolones in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:11801–11805. doi: 10.1073/pnas.92.25.11801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Heddle J, Maxwell A. Quinolone-binding pocket of DNA gyrase: Role of GyrB. Antimicrob Agents Chemother. 2002;46:1805–1815. doi: 10.1128/AAC.46.6.1805-1815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Laponogov I, et al. Structure of an ‘open’ clamp type II topoisomerase-DNA complex provides a mechanism for DNA capture and transport. Nucleic Acids Res. 2013;41:9911–9923. doi: 10.1093/nar/gkt749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moon DC, et al. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates. Int J Antimicrob Agents. 2010;35:76–79. doi: 10.1016/j.ijantimicag.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 57.Biedenbach DJ, et al. In vitro activity of gepotidacin, a novel triazaacenaphthylene bacterial topoisomerase inhibitor, against a broad spectrum of bacterial pathogens. Antimicrob Agents Chemother. 2016;60:1918–1923. doi: 10.1128/AAC.02820-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Farrell DJ, Sader HS, Rhomberg PR, Scangarella-Oman NE, Flamm RK. In vitro activity of gepotidacin (GSK2140944) against Neisseria gonorrhoeae. Antimicrob Agents Chemother. 2017;61:e02047-16. doi: 10.1128/AAC.02047-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Savage VJ, et al. Biological profiling of novel tricyclic inhibitors of bacterial DNA gyrase and topoisomerase IV. J Antimicrob Chemother. 2016;71:1905–1913. doi: 10.1093/jac/dkw061. [DOI] [PubMed] [Google Scholar]
- 60.Badran AH, Liu DR. Development of potent in vivo mutagenesis plasmids with broad mutational spectra. Nat Commun. 2015;6:8425. doi: 10.1038/ncomms9425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vogwill T, Kojadinovic M, MacLean RC. Epistasis between antibiotic resistance mutations and genetic background shape the fitness effect of resistance across species of Pseudomonas. Proc Biol Sci. 2016;283:20160151. doi: 10.1098/rspb.2016.0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vogwill T, Kojadinovic M, Furió V, MacLean RC. Testing the role of genetic background in parallel evolution using the comparative experimental evolution of antibiotic resistance. Mol Biol Evol. 2014;31:3314–3323. doi: 10.1093/molbev/msu262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Datta S, Costantino N, Zhou X, Court DL. Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci USA. 2008;105:1626–1631. doi: 10.1073/pnas.0709089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ricaurte DE, et al. A standardized workflow for surveying recombinases expands bacterial genome-editing capabilities. Microb Biotechnol. 2018;11:176–188. doi: 10.1111/1751-7915.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Ravesteyn TW, et al. LNA modification of single-stranded DNA oligonucleotides allows subtle gene modification in mismatch-repair-proficient cells. Proc Natl Acad Sci USA. 2016;113:4122–4127. doi: 10.1073/pnas.1513315113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brachman EE, Kmiec EB. Targeted nucleotide repair of cyc1 mutations in Saccharomyces cerevisiae directed by modified single-stranded DNA oligonucleotides. Genetics. 2003;163:527–538. doi: 10.1093/genetics/163.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dekker M, Brouwers C, te Riele H. Targeted gene modification in mismatch-repair-deficient embryonic stem cells by single-stranded DNA oligonucleotides. Nucleic Acids Res. 2003;31:e27. doi: 10.1093/nar/gng027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aarts M, Dekker M, de Vries S, van der Wal A, te Riele H. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res. 2006;34:e147. doi: 10.1093/nar/gkl896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.DiCarlo JE, et al. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013;2:741–749. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barbieri EM, Muir P, Akhuetie-Oni BO, Yellman CM, Isaacs FJ. Precise editing at DNA replication forks enables multiplex genome engineering in eukaryotes. Cell. 2017;171:1453–1467.e13. doi: 10.1016/j.cell.2017.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kosuri S, et al. Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nat Biotechnol. 2010;28:1295–1299. doi: 10.1038/nbt.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Girgis HS, Hottes AK, Tavazoie S. Genetic architecture of intrinsic antibiotic susceptibility. PLoS One. 2009;4:e5629. doi: 10.1371/journal.pone.0005629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Crofts TS, Gasparrini AJ, Dantas G. Next-generation approaches to understand and combat the antibiotic resistome. Nat Rev Microbiol. 2017;15:422–434. doi: 10.1038/nrmicro.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.The Pew Charitable Trusts 2017 Antibiotics Currently in Clinical Development. Available at www.pewtrusts.org/∼/media/assets/2017/12/antibiotics_currently_in_clinical_development_09_2017.pdf. Accessed June 8, 2017.
- 75.Jiang PX, et al. Pathway redesign for deoxyviolacein biosynthesis in Citrobacter freundii and characterization of this pigment. Appl Microbiol Biotechnol. 2012;94:1521–1532. doi: 10.1007/s00253-012-3960-0. [DOI] [PubMed] [Google Scholar]
- 76.Nikel PI, Martínez-García E, de Lorenzo V. Biotechnological domestication of pseudomonads using synthetic biology. Nat Rev Microbiol. 2014;12:368–379. doi: 10.1038/nrmicro3253. [DOI] [PubMed] [Google Scholar]
- 77.Raman S, Rogers JK, Taylor ND, Church GM. Evolution-guided optimization of biosynthetic pathways. Proc Natl Acad Sci USA. 2014;111:17803–17808. doi: 10.1073/pnas.1409523111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Esvelt KM, Wang HH. Genome-scale engineering for systems and synthetic biology. Mol Syst Biol. 2013;9:641. doi: 10.1038/msb.2012.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.