Significance
The development of targeted therapies represents a major advance in cancer therapy, with its most prominent examples including agents targeting HER2 in breast cancer, EGFR and ALK in lung cancer, and BRAF in melanoma. However, a key challenge confronting such precision medicine approaches concerns the presence of subtypes of each of these three common, lethal solid tumors that lack identified molecular drivers, and are thus not amenable to targeted therapies. Our studies identify a role for pleckstrin homology domain-interacting protein (PHIP) in promoting the progression of “driver-negative” subtypes of these common solid tumors. In addition, they demonstrate a physical interaction between PHIP and an activating histone modification, thereby identifying PHIP as a rational target for the therapy of these solid tumor subtypes.
Keywords: PHIP, driver gene-negative, target, chromatin remodeling
Abstract
The identification and targeting of key molecular drivers of melanoma and breast and lung cancer have substantially improved their therapy. However, subtypes of each of these three common, lethal solid tumors lack identified molecular drivers, and are thus not amenable to targeted therapies. Here we show that pleckstrin homology domain-interacting protein (PHIP) promotes the progression of these “driver-negative” tumors. Suppression of PHIP expression significantly inhibited both tumor cell proliferation and invasion, coordinately suppressing phosphorylated AKT, cyclin D1, and talin1 expression in all three tumor types. Furthermore, PHIP’s targetable bromodomain is functional, as it specifically binds the histone modification H4K91ac. Analysis of TCGA profiling efforts revealed PHIP overexpression in triple-negative and basal-like breast cancer, as well as in the bronchioid subtype of nonsmall cell lung cancer. These results identify a role for PHIP in the progression of melanoma and breast and lung cancer subtypes lacking identified targeted therapies. The use of selective, anti-PHIP bromodomain inhibitors may thus yield a broad-based, molecularly targeted therapy against currently nontargetable tumors.
Improved understanding of the molecular basis of cancer has resulted in the development of targeted therapies for various solid tumors, which have been shown to prolong survival in the setting of metastatic disease. These developments have led to the approval by the US Food and Drug Administration of agents targeting HER2 in breast cancer (1), EGFR and ALK in nonsmall cell lung cancer (2, 3), and BRAF in melanoma (4–6). As a result, precision medicine using such targeted approaches can revolutionize current approaches to cancer classification and treatment. To determine the appropriate targeted therapy, tumors must be classified according to the major molecular drivers of the malignant phenotype.
However, a significant proportion of common solid tumor types currently lack clearly identified targetable molecular aberrations, highlighted by the triple-negative subset of breast cancers, and therefore cannot be effectively treated with currently available targeted approaches. As a result, the treatment of cancers without known molecular drivers has lagged significantly behind that of targetable tumors. Furthermore, such nontargetable tumors are often more treatment resistant than tumors displaying currently targetable molecular drivers. To date, genome-wide analyses of such tumors have not resulted in effective targetable interventions. Thus, identifying molecular factors that promote the progression of diverse solid tumors lacking known molecular drivers would represent a significant advance in our understanding of cancer progression and define targets for the therapy of such tumors. Previously, we identified a role for the pleckstrin homology domain-interacting protein (PHIP) in the progression of melanoma (7). Here we identify a more broad-based role for PHIP in driving the progression of three solid tumor subtypes, each lacking identified mutational drivers or molecular targets for precision therapy.
Results
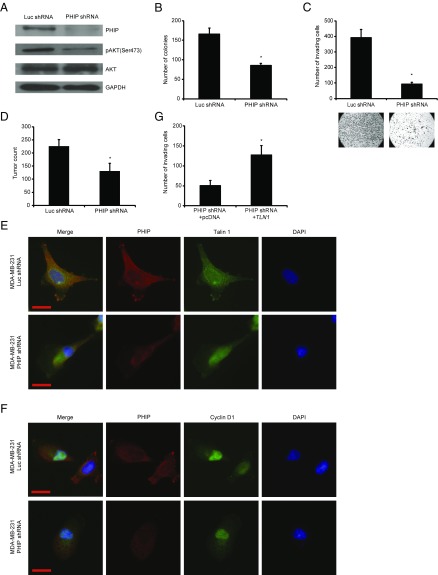
The effect of PHIP on the progression of currently nontargetable subsets of three solid tumors in which targeted therapeutics play an important role (breast and lung cancer and melanoma) was evaluated using shRNA-mediated gene silencing as well as PHIP cDNA overexpression. Initially, the effects of regulating PHIP expression were evaluated using lentiviral transduction of a well-characterized shRNA targeting human PHIP (7, 8) in the triple-negative MDA-MB-231 human breast cancer cell line. Suppression of PHIP expression (85% knockdown by TaqMan; Fig. 1A) produced significant reductions in MDA-MB-231 cell colony formation (Fig. 1B) compared with control clones expressing a shRNA targeting firefly luciferase (luc shRNA). In addition, anti-PHIP shRNA expression reduced the invasive capacity of MDA-MB-231 cells into Matrigel by 75% (Fig. 1C). Finally, shRNA-mediated targeting of PHIP resulted in 42% reduction in the metastatic potential of MDA-MB-231 cells after i.v. injection into nude mice (P < 0.0005; Fig. 1D).
Fig. 1.
Effects of stable shRNA-mediated suppression of PHIP in MDA-MB-231 cells. (A) Western analysis of expression of PHIP and other proteins in MDA-MB-231 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (B) Colony formation ability of MDA-MB-231 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P < 0.01). (C) Invasion into Matrigel of MDA-MB-231 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) with corresponding microphotographs (P < 0.001). (D) Quantification of total tumor counts on day 36 in the lungs of nude mice i.v. injected with MDA-MD-231 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P < 0.00001). (E and F) Qualitative immunofluorescence analysis of (E) PHIP and Talin 1 and (F) PHIP and Cyclin D1 in MDA-MB-231 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (Scale bar, 20 µm.) (G) Effects of overexpression of TLN1 cDNA or control plasmid on invasive capacity into Matrigel of MDA-MB-231 cells expressing anti-PHIP shRNA (127738) (P = 0.02).
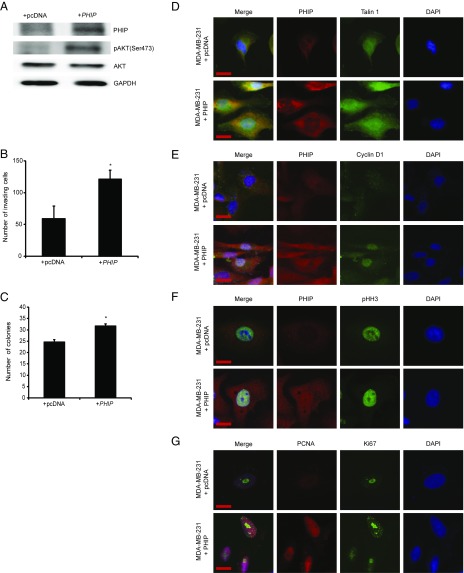
We then assessed the downstream pathways by which PHIP exerts its effects on human breast cancer progression and metastasis. Previously, we showed that suppression of PHIP expression was accompanied by down-regulation of phosphorylated AKT (pAKT Ser473) and Talin 1 (7). Accordingly, anti-PHIP shRNA-expressing MDA-MB-231 cells expressed reduced levels of PHIP and pAKT by Western analysis (Fig. 1A), as well as of PHIP and Talin 1 by immunofluorescence analyses (Fig. 1E and SI Appendix, Fig. S1 A and B). In addition, suppression of PHIP resulted in reduced expression of Cyclin D1 (Fig. 1F and SI Appendix, Fig. S1C), which plays an important role in breast cancer progression and is regulated by the PI3K/AKT pathway (9, 10). Transfection of a plasmid encoding TLN1 cDNA into MDA-MB-231 cells expressing anti-PHIP shRNA rescued the effects of PHIP suppression on invasion (Fig. 1G), indicating the requirement of Talin 1 for the proinvasive effects of PHIP. The effects of shRNA-based targeting of PHIP on expression of PHIP, pAKT, Talin 1, and Cyclin D1, as well as in reducing the invasive capacity and colony formation ability of MDA-MB-231 cells, were confirmed using a second shRNA targeting human PHIP mRNA (78% knockdown by TaqMan; SI Appendix, Fig. S1 D–K). Finally, overexpression of PHIP cDNA in MDA-MB-231 cells resulted in increased invasive and proliferative capacity, together with increased expression of pAKT, Talin 1, and Cyclin D1 (Fig. 2 A–E and SI Appendix, Fig. S2 A–C). In addition, as a recent study has shown an important role for PHIP in DNA replication (11), we observed significant overexpression of the proliferation markers PCNA, Ki67, and phospho-Histone H3 (pHH3) on PHIP overexpression (Fig. 2 F and G and SI Appendix, Fig. S2 D–F).
Fig. 2.
Effects of modulation of PHIP expression in MDA-MB-231 cells. (A) Western analysis of expression of PHIP and other proteins in MDA-MB-231 cells overexpressing PHIP cDNA or control plasmid. (B) Effects of overexpression of PHIP cDNA or control plasmid on invasive capacity into Matrigel of MDA-MB-231 cells (P < 0.05). (C) Effects of overexpression of PHIP cDNA or control plasmid on colony-formation ability of MDA-MB-231 cells (P < 0.05). (D–G) Qualitative immunofluorescence analysis of (D) PHIP and Talin 1, (E) PHIP and Cyclin D1, (F) PHIP and pHH3, and (G) PCNA and Ki67 in MDA-MB-231 cells overexpressing PHIP cDNA or control plasmid. (Scale bar, 20 µm.)
The effect of PHIP on human breast cancer progression was subsequently assessed in triple-negative MDA-MB-436 cells (12). Specifically, shRNA-mediated suppression of PHIP (83% knockdown by TaqMan; SI Appendix, Fig. S3A) resulted in significant suppression of MDA-MB-436 cell proliferation (60% reduction in colony formation, SI Appendix, Fig. S3B) and invasion (by 78%, SI Appendix, Fig. S3C), and was accompanied by down-regulation of pAKT (SI Appendix, Fig. S3A), Talin 1, and Cyclin D1 (SI Appendix, Fig. S3 D–H). Similar to MDA-MB-231 cells, transfection of a plasmid encoding TLN1 cDNA into MDA-MB-436 cells expressing anti-PHIP shRNA rescued the effects of PHIP suppression on invasion (SI Appendix, Fig. S3I). Taken together, these results demonstrate an important role for PHIP in the progression of triple-negative breast cancer, thus identifying a target for its therapy.
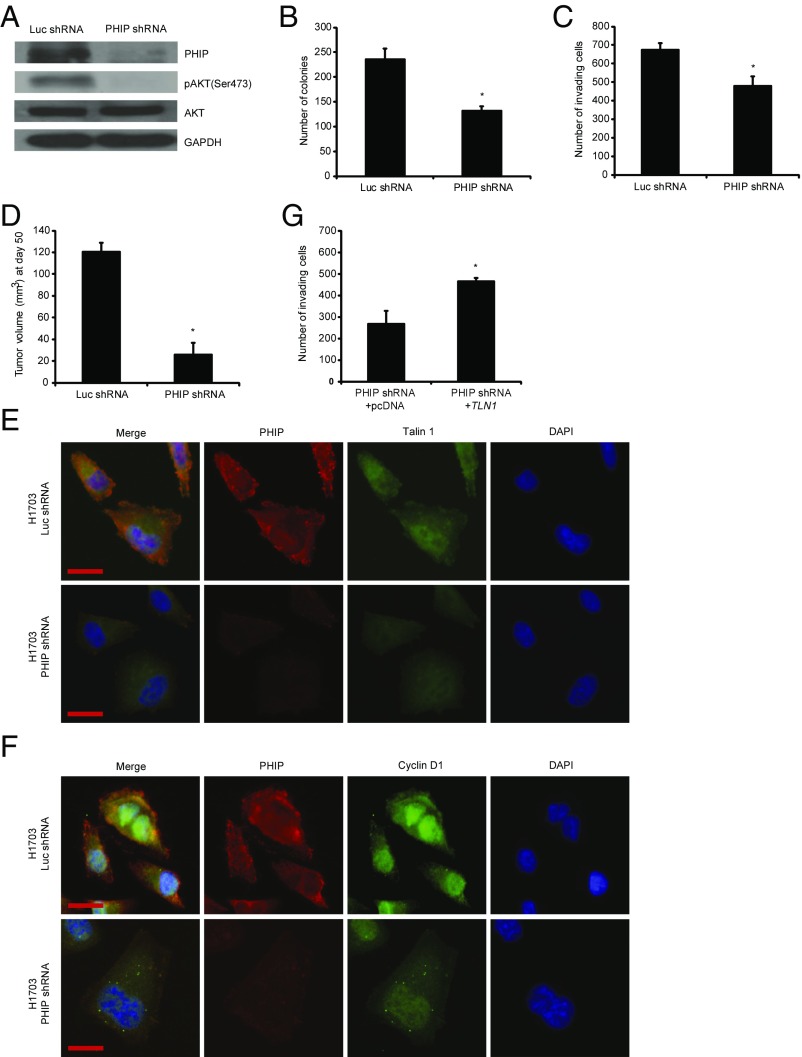
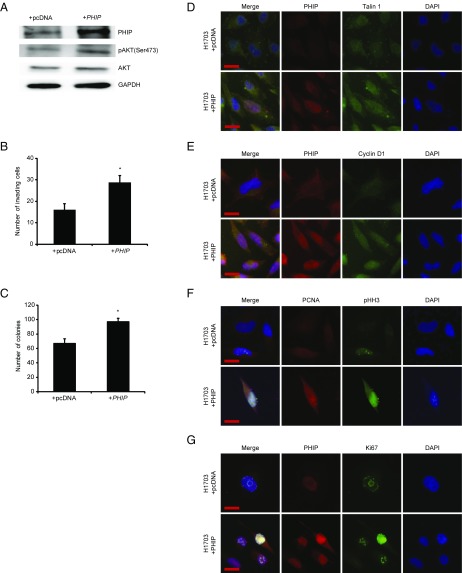
We then examined the role of PHIP in the progression of human nonsmall cell lung cancer, focusing on H1703 and Calu-3 cells, which lack molecular aberrations in EGFR, KRAS, PIK3CA, BRAF (13), and ALK (SI Appendix, Fig. S4A). Initially, shRNA-mediated PHIP knockdown (84% knockdown by TaqMan; Fig. 3A) in H1703 cells resulted in significant reduction in lung cancer proliferation (Fig. 3B) and invasion (Fig. 3C). Inoculation of H1703 cells expressing anti-PHIP shRNA s.c. produced profound (78%) reduction in the growth of lung cancer cells in vivo (P < 0.000005; Fig. 3D). PHIP suppression in H1703 cells was accompanied by down-regulation of pAKT (Fig. 3A), Talin 1, and Cyclin D1 (Fig. 3 E and F and SI Appendix, Fig. S4 B–D), similar to that observed in triple-negative breast cancer cells. The suppression of H1703 cell invasion by PHIP knockdown was rescued by TLN1 overexpression (Fig. 3G). In addition, the reduced expression of PHIP, accompanied by decreased pAKT, Talin 1, and Cyclin D1 expression, cell proliferation, and invasive capacity, was confirmed by targeting PHIP with a second shRNA (82.8% knockdown by TaqMan; SI Appendix, Fig. S5). Finally, overexpression of PHIP cDNA in H1703 cells resulted in increased invasive and proliferative capacity along with increased expression of pAKT, Talin 1, Cyclin D1, PCNA, Ki67, and pHH3 (Fig. 4 and SI Appendix, Fig. S4 E–J).
Fig. 3.
Effects of stable shRNA-mediated suppression of PHIP in H1703 cells. (A) Western analysis of expression of PHIP and other proteins in H1703 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (B) Colony formation ability of H1703 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P < 0.02). (C) Invasion into Matrigel of H1703 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P < 0.04). (D) Tumor volume 50 d after s.c. injection of H1703 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P = 0.000006). (E and F) Qualitative immunofluorescence analysis of (E) PHIP and Talin 1 and (F) PHIP and Cyclin D1 in H1703 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (Scale bar, 20 µm.) (G) Effects of overexpression of TLN1 cDNA or control plasmid on invasive capacity into Matrigel of H1703 cells expressing anti-PHIP shRNA (127738) (P < 0.001).
Fig. 4.
Effects of modulation of PHIP expression in H1703 cells. (A) Western analysis of expression of PHIP and other proteins in H1703 cells overexpressing PHIP cDNA or control plasmid. (B) Effects of overexpression of PHIP cDNA or control plasmid on invasive capacity into Matrigel of H1703 cells (P < 0.05). (C) Effects of overexpression of PHIP cDNA or control plasmid on colony formation ability of H1703 cells (P < 0.05). (D–G) Qualitative immunofluorescence analysis of (D) PHIP and Talin 1, (E) PHIP and Cyclin D1, (F) PCNA and pHH3, and (G) PHIP and Ki67 in H1703 cells overexpressing PHIP cDNA or control plasmid. (Scale bar, 20 µm.)
The effects of PHIP on promoting lung cancer progression were subsequently examined in Calu-3 cells. shRNA-mediated PHIP knockdown (85.4% knockdown by TaqMan; SI Appendix, Fig. S6A) was shown to result in significant (85%) reduction in Calu-3 cell colony formation (SI Appendix, Fig. S6B). As Calu-3 cells do not invade into Matrigel, the effects of PHIP suppression were assessed in a scratch assay, in which the PHIP shRNA-expressing cells closed the wound to a significantly lesser extent than control shRNA-expressing cells (P < 0.0005; SI Appendix, Fig. S6C). shRNA-mediated PHIP suppression also resulted in reduced expression of pAKT (SI Appendix, Fig. S6A), Talin 1, and Cyclin D1 (SI Appendix, Fig. S6 D–H).
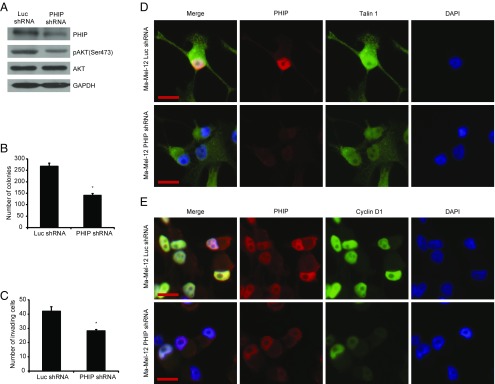
Finally, we assessed the role of PHIP in promoting the progression of human melanoma in early-passage, short-term melanoma cultures (Ma-Mel-12 and Ma-Mel-103b). Ma-Mel-12 cells have been characterized at the molecular level and found to harbor wild-type BRAF, NRAS, and NF1 [which defines the triple wild-type subtype defined by the recent TCGA analysis (14)]. Ma-Mel-12 cells also harbor wild-type PTEN and c-KIT, which represent additional mutational targets in melanoma. shRNA-mediated suppression of PHIP (85.6% knockdown by TaqMan; Fig. 5A) resulted in significant suppression of melanoma cell proliferation (Fig. 5B) and invasion (Fig. 5C), and was accompanied by down-regulation of pAKT (Fig. 5A), Talin 1, and Cyclin D1 (Fig. 5 D and E and SI Appendix, Fig. S7 A–C). In addition, the reduced expression of PHIP, accompanied by decreased pAKT, Talin 1, and Cyclin D1 expression, cell proliferation, and invasive capacity, was confirmed by targeting PHIP with a second shRNA (84.4% knockdown by TaqMan; SI Appendix, Fig. S7 D–K). The effects of PHIP on promoting melanoma progression were subsequently confirmed in another short-term melanoma culture, Ma-Mel-103b, which also lacks mutations in BRAF, NRAS, NF1, PTEN, and c-KIT (71.5% knockdown by TaqMan; SI Appendix, Fig. S8).
Fig. 5.
Effects of shRNA-mediated suppression of PHIP in Ma-Mel-12 cells. (A) Western analysis of expression of PHIP and other proteins in Ma-Mel-12 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (B) Colony formation ability of Ma-Mel-12 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P = 0.001). (C) Invasion into Matrigel of Ma-Mel-12 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738) (P = 0.005). (D and E) Qualitative immunofluorescence analysis of (D) PHIP and Talin 1 and (E) PHIP and Cyclin D1 in Ma-Mel-12 cells expressing anti-luc shRNA or anti-PHIP shRNA (127738). (Scale bar, 20 µm.)
Having established a role for PHIP in promoting the progression of “driver-negative” breast cancer, nonsmall cell lung cancer, and melanoma cell lines, we assessed PHIP expression levels in these malignancies using genome-wide analyses performed by TCGA. In the breast cancer analysis (15), PHIP transcript levels were significantly elevated in both basal-like tumors and triple-negative tumors (Fig. 6 A and B) compared with the other subtypes. In the analysis of lung adenocarcinomas (16), in the subset of tumors lacking mutations in EGFR, KRAS, and ALK, PHIP expression was significantly increased in the bronchioid and magnoid subtypes compared with the squamoid type, with a statistically significant increase in the bronchioid subtype (Fig. 6C). In the analysis of melanoma specimens (14), PHIP expression was not enriched in any particular molecular subtype, with its expression in the triple wild-type subtype not significantly different compared with all other subtypes (Fig. 6D). Thus, the PHIP gene is expressed in “driver-negative” tumors (and is enriched in the basal-like or triple-negative subtypes of breast cancer, and in the bronchioid subtype of lung cancer), justifying its role as a potential therapeutic target.
Fig. 6.
TCGA analysis of PHIP expression. (A–D) Box plots showing mean expression levels for PHIP in various molecular subtypes of human specimens from TCGA of (A) and (B) breast cancer, (C) lung cancer, and (D) melanoma.
Because PHIP promotes the progression of three currently nontargetable solid tumor subtypes, we assessed its potential druggability. The PHIP protein contains two bromodomains, which present an opportunity for small molecule targeting (17). Bromodomains are involved in chromatin remodeling by binding to acetylated lysine residues, enabling their interaction with specific histone modifications. However, whether the PHIP bromodomains are functional has not been conclusively demonstrated. Computational modeling has suggested the possible interaction of the second PHIP bromodomain with various acetylated lysine residues on histones (18). We therefore assessed the potential interaction of PHIP with one of these predicted modifications, H4K91ac.
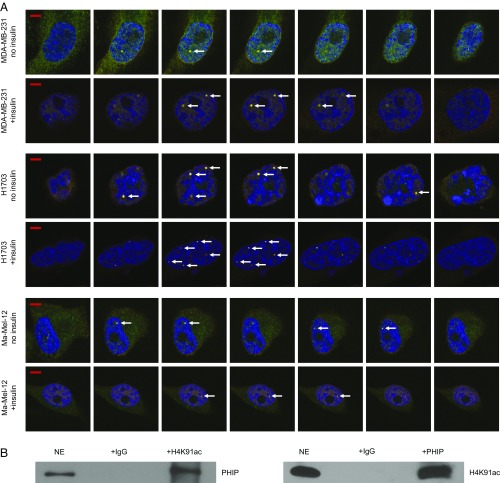
Initially, we determined the level of nuclear PHIP protein in our target tumor models. A cytoplasmic vs nuclear fractionation assay showed almost exclusive nuclear localization of PHIP in MDA-MB-231, H1703, and Ma-Mel-12 cells (Fig. 7A). Next, we assessed whether the PHIP protein has a spatial relationship with H4K91ac. Immunofluorescence analysis indicated a well-defined pattern of immunopositivity for both PHIP and H4K91ac staining in nuclei of MDA-MB-231, H1703, and Ma-Mel-12 cells, suggesting this colocalization (Fig. 7B). Intriguingly, areas of intense positivity, resembling distinct nuclear foci, were visible in all cell lines examined and were present in channels representing both PHIP and H4K91ac, suggesting this spatial relationship. We then assessed whether expression of PHIP and H4K91ac was coordinately regulated after a mitogenic stimulus. The intensity of nuclear immunopositivity was significantly increased for both proteins after treatment of MDA-MB-231, H1703, and Ma-Mel-12 cells with insulin (SI Appendix, Fig. S9 A–F). Further analysis using a laser-scanning microscope, with a resolution of 140 nm, indicated that these foci indeed occupy the same space when visualized over consecutive Z-stacks (Fig. 8A and Movies S1–S3). In addition, analysis of these consecutive Z-stacks in the confocal images revealed significant values for the Pearson’s colocalization coefficient between PHIP and H4K91ac in MDA-MB-231, H1703, and Ma-Mel-12 cells in both the presence or absence of insulin (SI Appendix, Fig. S9 G–L). Finally, a physical interaction between the two proteins was confirmed by coimmunoprecipitation of both PHIP and H4K91ac in nuclear extracts of H1703 cells. Both immunoprecipitated fractions were positive for their counterpart by Western analysis, indicating that PHIP binds to H4K91ac (Fig. 8B).
Fig. 7.
Colocalization of PHIP and H4K91ac. (A) Western analysis of PHIP and other proteins in nuclear (NE) versus cytoplasmic (CE) extracts of MDA-MB-231, H1703 and Ma-Mel-12 cells. Lamin B1 was used as a specific nuclear marker. (B) Qualitative immunofluorescence analysis showing colocalization of PHIP and H4K91ac (see arrows) in MDA-MB-231, H1703, and Ma-Mel-12 cells with or without insulin stimulation. (Scale bar, 20 µm.)
Fig. 8.
Colocalization and physical interaction of PHIP and H4K91ac. (A) Z-stacks of confocal images of PHIP and H4K91ac colocalization (see arrows) in MDA-MB-231, H1703, and Ma-Mel-12 cells with or without insulin stimulation. (Scale bar, 5 µm.) (B) Nuclear extracts from H1703 cells were subjected to the immunoprecipitation of endogenous H4K91ac using an anti-H4K91ac antibody or IgG control antibody followed by Western analysis of endogenous PHIP, using an anti-PHIP antibody (Left); nuclear extracts from H1703 cells were subjected to the immunoprecipitation of endogenous PHIP, using an anti-PHIP antibody or IgG control antibody followed by Western analysis of endogenous H4K91ac, using an anti-H4K91ac antibody (Right).
Discussion
The development of targeted therapies has revolutionized the treatment of many cancers. Such therapies require the identification of specific molecular drivers of the malignant phenotype that can be targeted by small molecules or monoclonal antibodies. This precision medicine approach has enabled the selection of drug therapies based on the molecular analysis of individual patient tumors. This restricts use of the therapeutic agent to those patients most likely to benefit from the targeted intervention, while sparing patients whose tumors lack these targets from the additional time, expense, and adverse events associated with treatments from which they are unlikely to benefit. Targeted therapies have been associated with both significantly increased response rates in matched patient populations and prolonged survival in multiple solid tumors, with breast and nonsmall cell lung cancer, as well as melanoma, emerging as the most prominent examples.
Despite these dramatic advances, significant challenges remain for the optimal development of targeted therapies for patients with cancer. First, the development of acquired drug resistance has emerged as a significant limitation of these approaches. In addition, many identified molecular targets are not currently druggable using available small molecules or monoclonal antibodies. One of the greatest challenges in the targeted therapy field remains the subset of patients whose tumors do not express the targetable molecular drivers. To date, no shared molecular targets for therapy have been identified for such tumor types. As a result, therapeutic approaches for these subgroups of patients have lagged significantly and continue to rely on conventional therapies.
In this study, we describe a broad-based role for PHIP in promoting the progression of three solid tumors (breast and nonsmall cell lung cancer as well as melanoma) in which targeted therapies have shown the greatest efficacy. We focused on subtypes of these three cancers that specifically lack identified targeted therapies, which are largely distinct in the three tumor types examined. The PHIP gene is expressed at high levels in each of these molecular subtypes, as evidenced by TCGA analyses of these malignancies. PHIP was specifically enriched in triple-negative or basal-like breast cancer. Interestingly, the PHIP locus has been recently identified as a possible breast cancer susceptibility gene (19). PHIP expression was also enriched in the bronchioid subtype of lung cancer lacking mutations in EGFR, KRAS, and ALK. This molecular subtype was previously shown to occur more commonly in females and to be associated with chemotherapy and radiation resistance, as well as poorer survival in late-stage disease (20). We show that regulation of PHIP expression, both through gene silencing and through PHIP cDNA overexpression, results in significant modulation of two hallmarks of the malignant phenotype; namely, tumor cell proliferation and invasion. These observations were supported by its coordinate regulation of a cascade of tumor cell proliferation and invasion markers (pAKT, Cyclin D1, and Talin 1, as well as Ki67, PCNA, and pHH3). Furthermore, significant suppression of cancer progression was observed in tumor-bearing mice by PHIP targeting in MDA-MB-231 and H1703 cells. In addition to the results presented here, prior studies have shown that PHIP regulates glycolysis and tumor angiogenesis, providing additional pathways by which it promotes tumor progression (8, 21, 22). Taken together, these results firmly establish a role for PHIP targeting in the setting of such “driver-negative” tumors.
However, PHIP targeting may be difficult to achieve, in part because of the lack of targetable enzymatic activity. As a result, we focused on PHIP’s bromodomains, given that the bromodomain-acetyl-lysine protein–protein interaction may be blocked by small molecule inhibitors (17). Previous studies of the BET protein family (including BRD2-4) indicating that the interaction between bromodomains and lysine residues on histones can be targeted by small molecules (17, 23, 24) have generated substantial interest in the druggability of bromodomain motifs (25). The PHIP bromodomains do not share significant sequence homology with that of BRD2-4 (18, 26), suggesting that novel small molecule inhibitors will need to be identified to most effectively target PHIP. Although a recent study has described a domain in the PHIP protein important for interacting with methylated histones (27), whether the PHIP bromodomains are functional has not been conclusively demonstrated. Our studies, using immunofluorescence, confocal microscopy, and coimmunoprecipitation, showed not only a colocalization but also a physical interaction between PHIP and H4K91ac, thereby indicating a role for the PHIP protein in chromatin remodeling. Our results are supported by a recent chromatin proteomic analysis that identified an association of PHIP with histone-marked genomic regions (28). In addition, an important role for PHIP in regulating site-specific initiation of DNA replication and cell cycle progression has been recently reported (11). DNA synthesis and histone modifications are highly coordinated to ensure the uninterrupted advance of the replication fork and assembly of the nascent DNA strands onto nucleosomes, a process that involves the deposition of histones H3 and H4 (29, 30). Interestingly, acetylation of H4K91 serves as a key modification associated with replication-coupled chromatin assembly (31).
Importantly, our studies suggest the potential therapeutic utility of pharmacological targeting of PHIP using small molecules inhibiting the PHIP bromodomain. Further supporting this hypothesis, a recent study has identified a small molecule specifically inhibiting one of the PHIP bromodomains (32). Additional refinements in medicinal chemistry will be required to develop an even more specific anti-PHIP small molecule inhibitor. Our studies clearly support the further development of such an approach.
In conclusion, these studies have identified a role for PHIP in promoting the progression of three common solid tumor types, each lacking an effective targeted therapy. In addition, they demonstrate a physical interaction between PHIP and a specific histone modification, thereby assigning a role to PHIP in chromatin remodeling. Last, our findings identify PHIP as a rational target for the therapy of each of these three important, now poorly treatable, “driver-negative” solid tumor subtypes lacking known targetable aberrations.
Materials and Methods
Additional technical details are provided in SI Appendix, SI Materials and Methods.
Cell Culture.
Ma-Mel-12 and Ma-Mel-103b short-term human melanoma cultures were provided by Dr. Schadendorf (Department of Dermatology, University of Duisburg-Essen, Essen, Germany). MDA-MB-231 and MDA-MB-436 human breast cancer cell lines and H1703 and Calu-3 human lung cancer cells were purchased from ATCC (Manassas, VA).
Generation of Stable Transformants.
All cells were generated by infection with a lentivirus containing anti-luc or anti-PHIP shRNA, as previously described (7).
Quantitative Real-Time PCR Analysis.
Gene expression was assessed as previously reported (7).
Invasion Assay.
The Matrigel assay for tumor invasion was performed as described (7).
Colony Formation Assay.
Five hundred to 2,000 cells were plated in a six-well plate and allowed to grow until visible colonies appeared. Then, they were stained with Crystal violet (Sigma) and counted.
Scratch Assay.
Calu-3 cells were plated in 10-cm dishes and a scratch was made by using a pipette tip on the confluent cells. Medium was aspirated to remove floating cells, and fresh medium was added to cells. At 24 h, several pictures were taken to evaluate the extent of wound closure. The percentage of area covered was quantified with ImageJ software.
Western Analysis.
Western analysis was performed as described (7). Protein extractions were carried out from adherent cells according to the manufacturer’s protocol (Santa Cruz Biotechnology). Antibodies to PHIP, ALK, Lamin B1, Actin, H4K91ac, pAKT (Ser473), total AKT, and GAPDH were used to detect the individual proteins (see SI Appendix, SI Materials and Methods for details).
Nuclear Fractionation and Coimmunoprecipitation.
All cells were stimulated with 5 µg/mL insulin for 30 min before nuclear and cytoplasmic fractionations and coimmunoprecipitation (described in SI Appendix, SI Materials and Methods).
Immunofluorescence and Confocal Imaging.
All cells were stimulated with 5 µg/mL insulin for 30 min before fixation. Quantification of protein expression using immunofluorescence was performed as previously described (7). Antibodies to PHIP, H4K91ac, Talin 1, pHH3, PCNA, Ki67, and Cyclin D1 were used to detect the individual proteins (see SI Appendix, SI Materials and Methods for details).
Animal Studies.
All animal care was in accordance with the NIH guide for the care and use of laboratory animals and with institutional protocol that was approved by the California Pacific Medical Center Research Institute. For experimental details, see SI Appendix, SI Materials and Methods.
Statistical Analysis.
All quantified data represent an average of at least triplicate samples or as indicated. Error bars represent standard error of the mean. Statistical significance was determined by the Student’s t test, Mann-Whitney test, or Kolmogorov-Smirnov test, and two-tailed P values <0.05 were considered significant. In all experiments, *P < 0.05 or otherwise indicated vs control.
Supplementary Material
Acknowledgments
This study was supported in part by US Public Health Service Grants R01CA114337 and R01CA175768 (to M.K.-S.), the Mary R. and Joseph R. Payden Foundation, the Kay Charitable Trust, and the E. A. Dickson Emeritus professorship of University California San Francisco (J.E.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1804779115/-/DCSupplemental.
References
- 1.Slamon DJ, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 2.Maemondo M, et al. North-East Japan Study Group Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N Engl J Med. 2010;362:2380–2388. doi: 10.1056/NEJMoa0909530. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–2394. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 4.Chapman PB, et al. BRIM-3 Study Group Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert C, et al. Improved overall survival in melanoma with combined dabrafenib and trametinib. N Engl J Med. 2015;372:30–39. doi: 10.1056/NEJMoa1412690. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 7.De Semir D, et al. Pleckstrin homology domain-interacting protein (PHIP) as a marker and mediator of melanoma metastasis. Proc Natl Acad Sci USA. 2012;109:7067–7072. doi: 10.1073/pnas.1119949109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bezrookove V, et al. Prognostic impact of PHIP copy number in melanoma: Linkage to ulceration. J Invest Dermatol. 2014;134:783–790. doi: 10.1038/jid.2013.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quintayo MA, et al. GSK3β and cyclin D1 expression predicts outcome in early breast cancer patients. Breast Cancer Res Treat. 2012;136:161–168. doi: 10.1007/s10549-012-2229-8. [DOI] [PubMed] [Google Scholar]
- 10.Rajput S, et al. Molecular targeting of Akt by thymoquinone promotes G(1) arrest through translation inhibition of cyclin D1 and induces apoptosis in breast cancer cells. Life Sci. 2013;93:783–790. doi: 10.1016/j.lfs.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat Commun. 2016;7:11748. doi: 10.1038/ncomms11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chavez KJ, Garimella SV, Lipkowitz S. Triple negative breast cancer cell lines: One tool in the search for better treatment of triple negative breast cancer. Breast Dis. 2010;32:35–48. doi: 10.3233/BD-2010-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soh J, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS One. 2009;4:e7464. doi: 10.1371/journal.pone.0007464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161:1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Network Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550, and erratum (2014) 514:262. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov. 2014;13:337–356. doi: 10.1038/nrd4286. [DOI] [PubMed] [Google Scholar]
- 18.Filippakopoulos P, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiao X, et al. NBCS Collaborators kConFab/AOCS Investigators PHIP–A novel candidate breast cancer susceptibility locus on 6q14.1. Oncotarget. 2017;8:102769–102782. doi: 10.18632/oncotarget.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hayes DN, et al. Gene expression profiling reveals reproducible human lung adenocarcinoma subtypes in multiple independent patient cohorts. J Clin Oncol. 2006;24:5079–5090. doi: 10.1200/JCO.2005.05.1748. [DOI] [PubMed] [Google Scholar]
- 21.Arbiser JL. PHIPing out: A genetic basis for tumor ulceration. J Invest Dermatol. 2014;134:600–602. doi: 10.1038/jid.2013.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arbiser JL, Bonner MY, Gilbert LC. Targeting the duality of cancer. NPJ Precis Oncol. 2017;1:23. doi: 10.1038/s41698-017-0026-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Filippakopoulos P, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller S, Filippakopoulos P, Knapp S. Bromodomains as therapeutic targets. Expert Rev Mol Med. 2011;13:e29. doi: 10.1017/S1462399411001992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidler LR, Brown N, Knapp S, Hoelder S. Druggability analysis and structural classification of bromodomain acetyl-lysine binding sites. J Med Chem. 2012;55:7346–7359. doi: 10.1021/jm300346w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Filippakopoulos P, Knapp S. The bromodomain interaction module. FEBS Lett. 2012;586:2692–2704. doi: 10.1016/j.febslet.2012.04.045. [DOI] [PubMed] [Google Scholar]
- 27.Morgan MAJ, et al. A cryptic Tudor domain links BRWD2/PHIP to COMPASS-mediated histone H3K4 methylation. Genes Dev. 2017;31:2003–2014. doi: 10.1101/gad.305201.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ji X, et al. Chromatin proteomic profiling reveals novel proteins associated with histone-marked genomic regions. Proc Natl Acad Sci USA. 2015;112:3841–3846. doi: 10.1073/pnas.1502971112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clemente-Ruiz M, Prado F. Chromatin assembly controls replication fork stability. EMBO Rep. 2009;10:790–796. doi: 10.1038/embor.2009.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prado F, Maya D. Regulation of replication fork advance and stability by nucleosome assembly. Genes (Basel) 2017;8:E49. doi: 10.3390/genes8020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ye J, et al. Histone H4 lysine 91 acetylation a core domain modification associated with chromatin assembly. Mol Cell. 2005;18:123–130. doi: 10.1016/j.molcel.2005.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cox OB, et al. A poised fragment library enables rapid synthetic expansion yielding the first reported inhibitors of PHIP(2), an atypical bromodomain. Chem Sci. 2016;7:2322–2330. doi: 10.1039/c5sc03115j. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.