Significance
Stringent response is an important physiological process in microorganisms mediated by the alarmone molecules (p)ppGpp. RelA/SpoT homolog (RSH) proteins regulate the cellular alarmone concentration by their dual function as both synthetase and hydrolase. It has been known for decades that RSH proteins have an amino acid-binding ACT domain, with its role unknown. Here, we show that the ACT domain of Rel from Rhodobacter capsulatus indeed binds branched-chain amino acids (BCAAs) and that this binding increases its hydrolase activity. Thus, even though the stringent response is well known to be initiated by activation of alarmone synthetase activity via an interaction of Rel proteins with stalled ribosomes, there also exists another trigger involving the inhibition of (p)ppGpp hydrolysis via BCAA depletion.
Keywords: stringent response, RelA/SpoT homolog, branched-chain amino acids, Rhodobacter capsulatus, (p)ppGpp alarmone hydrolase
Abstract
When faced with amino acid starvation, prokaryotic cells induce a stringent response that modulates their physiology. The stringent response is manifested by production of signaling molecules guanosine 5′-diphosphate,3′-diphosphate (ppGpp) and guanosine 5′-triphosphate,3′-diphosphate (pppGpp) that are also called alarmones. In many species, alarmone levels are regulated by a multidomain bifunctional alarmone synthetase/hydrolase called Rel. In this enzyme, there is an ACT domain at the carboxyl region that has an unknown function; however, similar ACT domains are present in other enzymes that have roles in controlling amino acid metabolism. In many cases, these other ACT domains have been shown to allosterically regulate enzyme activity through the binding of amino acids. Here, we show that the ACT domain present in the Rhodobacter capsulatus Rel alarmone synthetase/hydrolase binds branched-chain amino acids valine and isoleucine. We further show that the binding of these amino acids stimulates alarmone hydrolase activity both in vitro and in vivo. Furthermore, we found that the ACT domain present in Rel proteins from many diverse species also binds branched-chain amino acids. These results indicate that the cellular concentration of amino acids can directly affect Rel alarmone synthetase/hydrolase activity, thus adding another layer of control to current models of cellular control of the stringent response.
Many species in the Bacteria and Archaea kingdoms induce a stringent response when faced with challenging environmental conditions. Stresses such as deprivation of amino acids and/or carbon sources, temperature changes, dehydration, and so forth, result in increased production of the alarmones guanosine 5′-diphosphate,3′-diphosphate (ppGpp) and guanosine 5′-triphosphate,3′-diphosphate (pppGpp). These alarmones regulate various downstream cellular functions at transcriptional, translational, and posttranslational levels (1). Physiological consequences of the stringent response include lowered stable RNA synthesis and increased transcription of amino acid biosynthesis genes.
Escherichia coli and other Betaproteobacteria and Gammaprotobacteria members produce ppGpp (or pppGpp) from GDP (or GTP) and ATP with the monofunctional alarmone synthetase, RelA. The stringent response triggered by ppGpp and pppGpp synthesis is dampened by SpoT, which has a hydrolase domain (HD) exhibiting 3′-pyrophosphatase activity that degrades ppGpp/pppGpp to GDP/GTP. RelA and SpoT are structurally very similar and, indeed, SpoT does have a RelA-like (p)ppGpp synthetase domain that exhibits weak synthetase activity. RelA also contains a SpoT-like HD, but this domain is apparently inactive. In these organisms, RelA is thus responsible for alarmone synthesis, whereas structurally related SpoT is involved in alarmone degradation (1).
In genera outside of Betaproteobacteria and Gammaprotobacteria, there is typically a single RelA/SpoT homolog protein often termed RSH (or Rel) with linked functional hydrolase and synthetase domains that, together, regulate alarmone degradation and synthesis, respectively (1–3). These opposing activities are tightly controlled, but details of this regulation remain unclear. Under normal growth conditions, the synthetase activity of Rel is thought to be self-inhibited; however, during times of amino acid starvation, Rel interacts with stalled ribosomes, which activates synthetase activity to produce (p)ppGpp. The regulation of hydrolase activity is less understood but may involve one or more downstream domains called the TGS and ACT domains. The TGS domain of SpoT has been shown to interact with an acyl carrier protein, so it is presumed to sense the status of fatty acid metabolism in E. coli (4). The function of the ACT domain is not as clear; however, recent cryo-EM structures of E. coli RelA show that this domain is involved in binding deacyl-tRNA as well as the ribosome (5–7). Interestingly, the ACT domain is present in several other enzymes where it allosterically regulates enzyme activity upon the binding of amino acids (8). In this study, we analyzed the role of the ACT domain in controlling the activity of the Rel protein in Rhodobacter capsulatus. We observed that the ACT domain indeed binds valine and isoleucine and that the binding of these amino acids increases (p)ppGpp hydrolase activity. We further show that ACT domains present in RSH proteins from a wide range of species also bind one or more branched-chain amino acids (BCAAs).
Results
Valine and Isoleucine Bind to RcRel C-Terminal Domain to Promote Its Hydrolase Activity.
We first addressed whether a Rel protein with bifunctional alarmone synthetase/alarmone hydrolase activity is capable of binding one or more BCAAs. For this analysis, we focused on the Rel protein that is present in R. capsulatus (RcRel coded by gene rcc03317). Previous genetic analysis has shown that RcRel is the only protein responsible for synthesizing the alarmones ppGpp and pppGpp in vivo in R. capsulatus (9). There is also no other gene identified as a RelA or SpoT gene in the annotated genome, nor are there any RelA/SpoT homologs identifiable by homology searches. These results indicate that rcc03317 most likely codes for a typical bifunctional Rel protein.
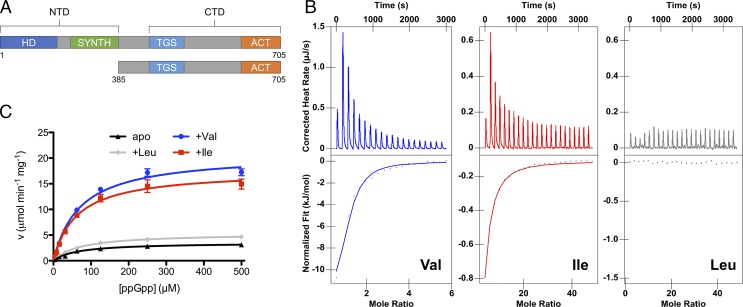
To access the possibility of binding BCAAs in vitro, we isolated both a full-length and an N-terminally truncated RcRel. Full-length RcRel exhibits poor solubility, which prevented in vitro binding studies; however, the C-terminal domain (CTD) region containing the TGS and ACT domains (RcRel385–705) did exhibit high solubility (Fig. 1A). Consequently, we obtained isothermal titration calorimetry (ITC) binding isotherms of the binding of seven different mixtures of amino acids to RcRel385–705, screening a total of 20 amino acids by this batch approach. As shown in the isotherms in SI Appendix, Fig. S1A, the BCAA group was the only one that exhibited binding to RcRel385–705. We further titrated each BCAA in this group to RcRel385–705, which shows that RcRel385–705 selectively binds valine and isoleucine, but not leucine (Fig. 1B). The highest binding affinity was to valine with a Kd = 12.6 µM, followed by isoleucine with a Kd =192 µM.
Fig. 1.
Valine and isoleucine bind to RcRel CTD to promote its hydrolase activity. NTD, N-terminal domain. (A) The domain structure of full-length RcRel and RcRel385–705 that was used in ITC experiments. (B) ITC binding profiles of RcRel385–705 (74 μM) titrated with BCAAs (5 mM Val, 25 mM Ile, or 25 mM Leu). (C) Initial rates of ppGpp hydrolysis plotted against ppGpp concentration for apo RcRel protein and RcRel with BCAAs. Initial rates were calculated based on the production of GDP. Data are represented as mean ± SEM (n = 3) and are fitted to the Michaelis–Menten equation in GraphPad Prism.
We also investigated the effect of valine and isoleucine on in vitro enzyme activities of full-length RcRel. We did not observe any synthesis of ppGpp or pppGpp with or without the presence of 1 mM valine or isoleucine, which is not surprising, given that bifunctional RSH proteins are known to exhibit poor, sometimes not observable, alarmone synthetase activity in vitro (10). On the other hand, purified RcRel did exhibit ppGpp hydrolase activity in the presence of divalent cations (SI Appendix, Table S1), with this activity increased by the addition of 1 mM valine or isoleucine, but not leucine, to the reaction mixture (Fig. 1C and SI Appendix, Table S2). Valine increased Vmax approximately sixfold, and isoleucine by approximately fivefold. Based on these results, we hypothesized that R. capsulatus adjusts the level of (p)ppGpp, in part, by controlling Rel hydrolase activity in response to intracellular changes in valine and/or isoleucine concentrations.
In Vivo Effect of BCAAs on Alarmone Levels.
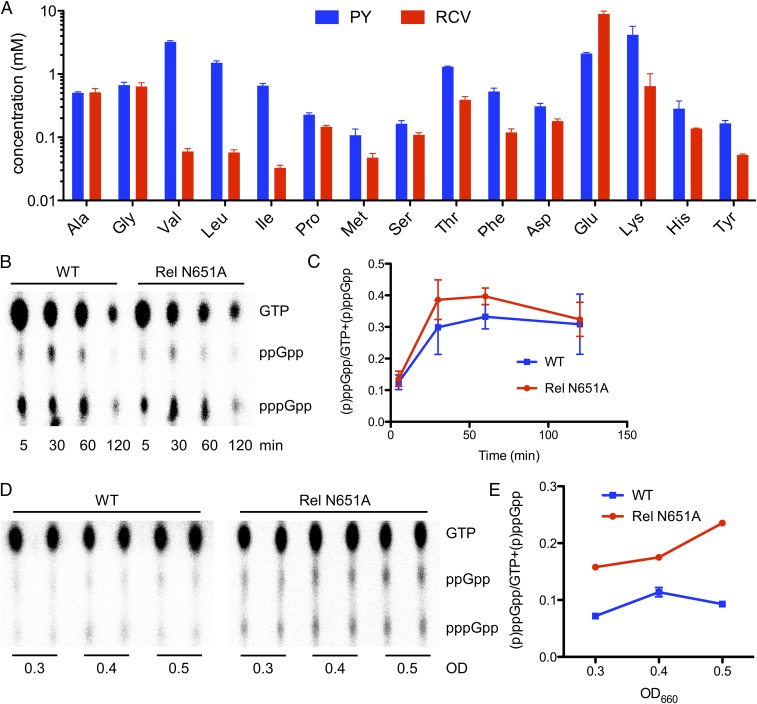
We evaluated the saturation status of valine and isoleucine to the ACT domain in vivo by determining amino acid concentrations in R. capsulatus cells grown in either rich-medium PY (peptone-yeast extract) or the defined minimum-medium RCV that contained malate as the single carbon and energy source. Results showed that in vivo concentrations of BCAAs were significantly different in cells grown in these two media, while other amino acids exhibited only minor changes in concentration (Fig. 2A and SI Appendix, Table S3). The biggest concentration difference was observed for valine, which exhibited a 54-fold lower level under nutrient-limiting versus nutrient-rich growth conditions (60 µM in RCV versus 3.2 mM in PY; SI Appendix, Table S3). This large nutrient-driven difference in BCAA concentration suggests that these cells are indeed capable of regulating alarmone hydrolase activity in response to changes in BCAA concentration.
Fig. 2.
In vivo effect of BCAAs on alarmone levels. (A) In vivo amino acid concentrations in WT R. capsulatus grown in rich PY medium versus minimal RCV medium as measured by GC-MS. Data are represented as mean ± SEM (n = 3). (B) In vivo alarmone levels in WT and Rel N651A mutant R. capsulatus strains at different time points after a down shift from rich medium to minimum medium as revealed by TLC of 32P-labeled cell extracts. (C) Quantification of alarmone levels shown in B using ImageQuant software. Data are represented as mean ± SEM (n = 2). (D) In vivo alarmone levels in WT and Rel N651A mutant R. capsulatus at different OD660, as revealed by TLC of 32P-labeled cell extracts. (E) Quantification of alarmone levels in D using ImageQuant software. Data are represented as mean ± SEM (n = 2).
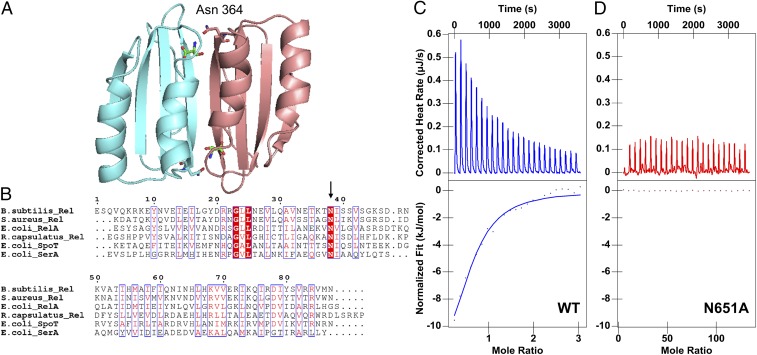
To further test the in vivo effect of BCAAs on alarmone levels, we generated a mutant RcRel that is not capable of binding BCAAs, which allowed us to address the role of Rel binding of BCAAs without depleting valine or isoleucine (depletion of BCAAs would trigger the stringent response by ribosomal stalling). For mutant construction, we used information from a previous crystallographic study of the ACT domain from the enzyme 3-phosphoglycerate dehydrogenase (SerA, PDB ID code 1PSD) that shows that Asn364 is bound to an amino acid ligand (11). Sequence alignment showed that the RcRel ACT domain also contains this conserved Asn (N651; Fig. 3 A and B). Consequently, we mutated N651 to Ala to test whether it has a role in Val/Ile binding. In vitro isothermal calorimetry binding studies with an isolated ACT domain containing the N651A substitution (RcRel385 N651A), shows that this mutant does not measurably bind valine or isoleucine (Fig. 3D). The effect of the N651A substitution on the ability of full-length RcRel to induce (p)ppGpp synthesis was tested in vivo by measuring (p)ppGpp levels in wild-type (WT) cells and in cells containing a RcRel N651A mutation. For this analysis, alarmone production was induced by shifting cells from rich PY growth medium, where amino acids are at a high concentration, to nutrient-limiting RCV medium that has no amino acids. As shown in Fig. 2 B and C, both strains exhibited an initial increase in (p)ppGpp levels, followed by a slow decrease. The observation that the Rel N651A mutant strain is capable of undergoing WT induction of alarmone synthesis indicates that this mutation does not affect Rel folding/stability. This result also demonstrates that disruption of BCAA binding to the ACT domain does not affect synthase activity. However, analysis of (p)ppGpp levels during sustained growth in minimum RCV medium shows that the Rel N651A strain had significantly higher steady-state amounts of (p)ppGpp (Fig. 2 D and E). These results are consistent with a model in which the WT cells have a Rel that is capable of binding BCAAs to the ACT domain, leading to an increase of alarmone hydrolase activity, which subsequently reduces (p)ppGpp levels. Conversely, the Rel N651A mutant strain has increased (p)ppGpp levels, presumably because Rel N651A is still capable of (p)ppGpp synthesis but also unable to bind BCAAs and, thus, does not increase alarmone hydrolase activity.
Fig. 3.
The conserved asparagine N651 is crucial for amino acid binding in RcRel. (A) ACT domain of E. coli 3-phosphoglycerate dehydrogenase (SerA, PDB ID code 1PSD), featuring the Asn364 interaction with ligand serine. (B) Multiple sequence alignment of ACT domains, with the red highlight representing fully conserved residues and the blue boxes outlining residues with similar physical and chemical properties. The arrow indicates the conserved Asn residues that were mutated to Ala. (C) ITC profile of titrating valine into WT RcRel385–705 (74 μM with 2.5 mM Val) or (D) RcRel385–705 N651A (56 μM with 50 mM Val).
Binding of BCAAs Occurs with Other RSHs.
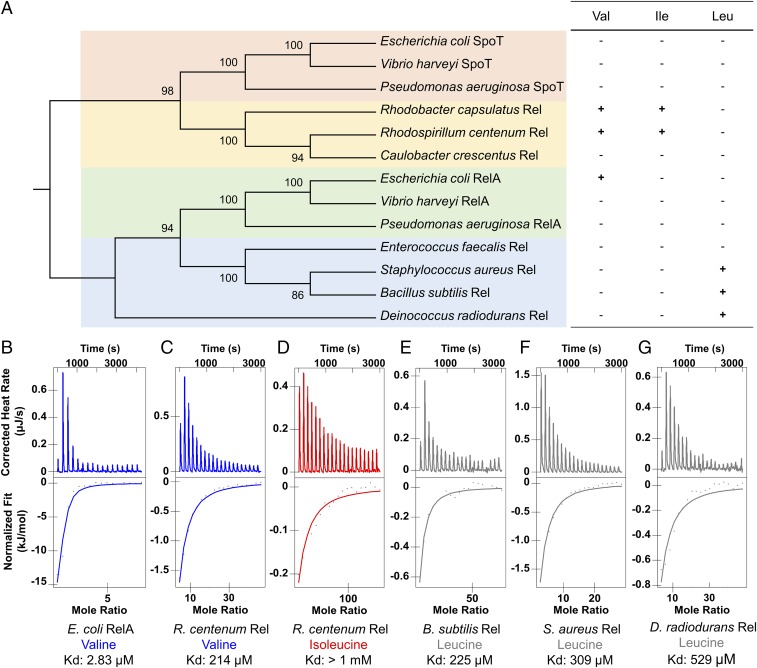
We further tested whether the binding of BCAAs in the ACT domain is widely spread in RSHs from other bacterial species. For this analysis, additional CTDs from 12 RSHs were purified and analyzed for the binding of BCAAs using ITC. The total 13 RSHs were from four groups: bifunctional SpoT from several gammaprotobacteria, bifunctional Rel from alphaprotobacteria, monofunctional synthetase RelA from gammaprotobacteria, and bifunctional Rel in Gram-positive species (Fig. 4A). The ITC results showed that CTDs from some, but not all, RSHs do bind BCAAs (Fig. 4 B–G). From the thermodynamic parameters obtained in the ITC experiments (SI Appendix, Table S4), it can be found that both enthalpy and entropy contribute to the binding between the CTD of RSH proteins and BCAAs. Only exception is Rhodospirillum centenum Rel binding with Val, which is an enthalpy-driven process. Interestingly, bifunctional Rels from the Gram-positive species Bacillus subtilis, Staphylococcus aureus, and Deinococcus radiodurans all bind leucine, instead of valine or isoleucine that is bound by bifunctional Rel in the alphaprotobacteria R. capsulatus and R. centenum (Fig. 4 C–G). One notable observation is that the Kd values of binding these ligands vary from low-micromolar to millimolar levels, with the strongest binding between EcRelA and valine (Fig. 4B). According to a previous study, the in vivo concentration of valine in E. coli cells grown in glucose medium is estimated to be 4 mM (12). With a Kd of 2.8 µM, it suggests that the domain is always saturated. However, EcRelA does not have a functional HD, so this binding might be an evolutionary relic, instead of having actual function. On the other hand, the Kd of BsRel for leucine is 225 µM (Fig. 4E), whereas the intracellular concentration of leucine in B. subtilis is reported to be ∼80 µM (13). These numbers suggest the possibility that B. subtilis may indeed use leucine concentration to regulate BsRel activity. Lastly, due to practical reasons, we only tested binding with BCAAs, so a failure to bind these amino acids does not rule out the possibility that the RSHs that do not bind Ile, Val, or Leu may actually bind other amino acids that subsequently affect alarmone synthesis or hydrolase activity.
Fig. 4.
Binding of BCAAs occurs with CTD present in other RSHs. (A, Left) The molecular phylogenetics of 13 RSHs were analyzed using the Maximum Likelihood method in MEGA7. The 13 RSHs were categorized into four groups: bifunctional SpoT from Gammaprotobacteria (pink), bifunctional Rel from Alphaprotobacteria (yellow), monofunctional synthetase RelA from Gammaprotobacteria (green), and bifunctional Rel in Gram-positive species (blue). (A, Right) “+” indicates binding revealed in ITC experiments. (B–G) CTDs of various RSHs titrated with BCAAs using ITC. (B) 20 μM E. coli RelA CTD with 1.5 mM Val. (C) 40 μM R. centenum Rel CTD with 15 mM Val. (D) 40 μM R. centenum Rel CTD with 50 mM Ile. (E) 40 μM B. subtilis Rel CTD with 25 mM Leu. (F) 108 μM S. aureus RelA CTD with 25 mM Leu. (G) 62 μM D. radiodurans RelA CTD with 25 mM Leu.
Discussion
It is an accepted model that RSH proteins sense stalled ribosomes caused by the accumulation of deacylated tRNA to activate its alarmone synthetase activity under amino acid starvation conditions (1). However, more attention has been given to the control of synthetase activity than to the control of hydrolase activity of RSH proteins. Indeed, cells need to balance the synthesis of (p)ppGpp with its hydrolysis to maintain optimal cellular concentrations of these alarmones. Our discovery that BCAAs activate the hydrolase activity of RcRel provides evidence that Rel proteins also sense the concentration of BCAAs to fine-tune their hydrolase activity to properly regulate cellular alarmone levels. It is intriguing that these cells appear to be monitoring BCAAs as an indication of nutrient level that, in turn, regulates (p)ppGpp levels. As discussed by Shivers and Sonenshein (14), BCAAs might be important because of their close relationships to various molecules crucial for cell growth and their role as precursors of branched-chain fatty acids that are the major component of the cell membrane.
Our work also shows that the regulation of RSH activity by BCAAs might be widely distributed in prokaryotes, with the caveat that Gram-positive bacteria appear to use Leu instead of Val or Ile, as appears to be the case in several Gram-negative bacteria (Fig. 4). Other than presumed differences in the BCAA binding pocket of RSH proteins, different BCAA regulatory systems used by Gram-positive and Gram-negative bacteria might contribute to the different ligand preferences for the RHS proteins. For example, in B. subtilis, the transcription factor CodY directly regulates the transcription of BCAA biosynthesis operons (14). Under nutrient-sufficient conditions, CodY binds BCAA promoters to repress the synthesis of enzymes used for BCAA biosynthesis. Interestingly, the binding of CodY to ilvB promoter is enhanced by isoleucine and valine, but not leucine (14). In E. coli, the leucine-responsive protein Lrp senses leucine to regulate BCAA biosynthesis and transport (15). In each of these cases, the BCAA that is sensed by the transcription factor involved in regulating BCAA production is different from the BCAA that is used to regulate its respective RSH.
In addition to direct BCAA regulation via Lrp and CodY, E. coli and B. subtilis are also known to regulate transcription of BCAA biosynthesis genes in response to alarmone levels (16–19). In E. coli, (p)ppGpp binds to RNAP and its cofactor DksA to selectively stimulate transcription activation of amino acid biosynthesis genes (19). In B. subtilis, this activation is more indirect, involving a mechanism that relies on a reduction in GTP concentration and de-repression of CodY (17). Thus, (p)ppGpp levels appear to form a secondary regulatory circuit used by cells to maintain cellular BCAA levels.
In conclusion, we find that valine and isoleucine bind to RcRel alarmone synthetase/hydrolase, resulting in the stimulation of hydrolase activity. We further show that binding of these BCAAs keeps cells from accumulating excess (p)ppGpp in vivo. This mechanism also appears to be used by many microorganisms, as shown by the binding of BCAAs to CTDs of different RSH proteins. Our finding thus adds another layer of regulation to the existing stringent response model. Due to its importance in microbial physiology, the stringent response has been a target for the development of antimicrobials (20, 21). Indeed, it has been proposed that inhibiting hydrolase activity of the RSH proteins could result in the accumulation of lethal amounts of (p)ppGpp (18). Our finding indicates that the ACT domain in RSH proteins could also be a good target for such a strategy.
Materials and Methods
Strains and Growth Conditions.
R. capsulatus strain SB1003 was used as the WT parental strain (22). R. capsulatus cells were routinely grown aerobically with shaking in PY medium or RCV medium (23) at 34 °C. E. coli cells were routinely grown in LB medium at 34 °C with E. coli BL21 (DE3) used to overexpress protein, while E. coli S17-1 λpir was used to introduce plasmid to R. capsulatus via conjugation. Antibiotics for plasmid selection were used at concentrations of 1 μg/mL gentamicin for R. capsulatus, whereas for E. coli, gentamicin and kanamycin were used at 10 μg/mL and 50 μg/mL, respectively.
Plasmid and Strain Construction.
Suicide plasmid pZJD29a was used to generate chromosomal Rel N651A mutation in R. capsulatus. The 1 kb centering the mutation site was amplified and ligated into XbaI/SacI sites on pZJD29a. Site-directed mutagenesis was performed to introduce mutation on the suicide plasmid. The mutation bearing plasmid was introduced to R. capsulatus SB1003 via E. coli S17-1 λpir. Double-recombinant event was selected as described before (24), and the successful generation of chromosomal mutation was confirmed by PCR amplification coupled with sequence analysis.
Commercially available plasmids pET28a and pET29a were used for protein overexpression in E. coli BL21 (DE3). Primers (SI Appendix, Table S5) with Strep tag sequence and restriction sites were used to amplify genes from genomic DNA with amplified fragments digested and ligated into NcoI/SacI (pET28a) or NdeI/SacI (pET29a) sites. N-terminal Strep tag (WSHPQFEK) was used for purification of CTD from 13 RSHs, whereas a C-terminal Strep tag was used for purification of full-length RcRel.
Protein Overexpression and Purification.
Plasmid bearing E. coli BL21 (DE3) strains were grown to OD600 0.4 with appropriate antibiotics before overnight induction of protein expression at 16 °C by the addition of 200 µM isopropyl β-D-1-thiogalactopyranoside. Cells were collected by centrifugation and resuspended in StrepTactin wash buffer (50 mM Tris pH 8.9, 1 M NaCl, 20% glycerol) at 4 °C. Cells were lysed by French Press, and the lysates were clarified by centrifugation at 27,000 × g (Sorvall SS-34 rotor) at 4 °C before loading onto a StrepTactin resin column (IBA Lifesciences). The column was washed with StrepTactin wash buffer, followed by protein elution with same buffer containing desthiobiotin (2.5 mM). The eluents were concentrated using spin concentrators and loaded onto a Superose 12 column equilibrated in the same StrepTactin wash buffer for size exclusion chromatography. In cases where proteins were isolated for ITC binding assays, ITC buffer (50 mM sodium phosphate pH 8.0, 1 M NaCl, 20% glycerol) was used for Superose 12 chromatography.
ITC.
Purified proteins were dialyzed in ITC buffer (50 mM sodium phosphate pH 8.0, 1 M NaCl, 20% glycerol) overnight with the final dialysate used as a buffer into which tested amino acids were dissolved. All solutions were filtered and degassed before ITC experimentation. Protein concentrations were determined using Bradford assay before titration. ITC experiments were carried out using a NanoITC (TA Instruments) at 25 °C. Different concentrations of protein (20 to 108 µM) were titrated with various concentrations of BCAAs (2.5 to 50 mM), with the samples stirred at 250 rpm. Data were analyzed by NanoAnalyze software (TA Instruments) using independent binding model with binding stoichiometry set at 1. Dilution heat from amino acids titrating to ITC buffer was subtracted.
Enzymatic Kinetics.
All enzymatic reactions were carried out at 25 °C. For hydrolase assays, full-length RcRel was incubated with various concentrations of ppGpp (TriLink Biotechnologies) for 1 min in 50 mM Tris pH 7.5, 150 mM NaCl, and 1 mM MnCl2. BCAAs were added at 1 mM. Reactions were stopped by heating at 75 °C for 5 min. Nucleotides in the reactions were separated by HPLC using previously described methods (25). The production of GDP from ppGpp hydrolysis was quantified using standard curve based on HPLC peak area, and kinetics data were analyzed with GraphPad Prism (GraphPad Software).
In Vivo Amino Acid Quantification.
R. capsulatus cells were grown to OD660 ∼0.3 in either PY medium or RCV medium before collecting by centrifugation at 4 °C. Cells were washed twice with ice-cold Tris-buffered saline (50 mM Tris pH 7.5, 150 mM NaCl) and then extracted using −20 °C 40:40:20 acetonitrile:methanol:water with 0.1 M formic acid. Internal standards of 2-aminoisobutyric acid and norvaline were added together with the extraction solvent. Clarified extracts were dried to complete dryness on a SpeedVac concentrator (Savant). The resulting films were resuspended in pyridine and derivatized with N-tert-butyldimethylsilyl-N-methyltrifluoroacetamide for 2 h at 120 °C. The derivatized amino acids were then separated and quantified by coupled GC-MS using a 30 m × 0.25 mm × 0.25 μm Zebron ZB-5MS column (26). Intracellular concentrations of amino acids were calculated using a coefficient of 1.2 × 109 cells⋅ mL−1⋅OD660 unit−1 and a volume of 6.4 × 10−16 L per cell.
In Vivo Alarmone Labeling.
Overnight cultures grown in either PY medium or RCV medium were diluted to OD660 0.05 and then further grown to 0.2 at 34 °C before the addition of 100 µCi/mL H332PO4 (PerkinElmer). For nutrient shift-down experiments, labeled cells grown in PY medium were collected by centrifugation at OD660 0.3 and washed with 4 °C RCV medium twice before finally being resuspended in RCV medium (supplemented with 100 µCi/mL H332PO4) at 34 °C. Cells were collected at desired time points with the labeled nucleotides, extracted with 8 M formic acid. Supernatant of the extracts were spotted on polyethyleneimine-cellulose plates (Merck Millipore) for TLC separation of nucleotides with 1.5 M KH2PO4 (pH 3.4). TLC plates were dried before exposed to phosphor screen (Molecular Dynamics). Radioactive spots were visualized using Typhoon imager (GE Healthcare Life Sciences) and quantified using ImageQuantTL software (GE Healthcare Life Sciences).
Phylogenetic Analysis.
Amino acid sequences of 13 RSHs were retrieved from the NCBI database and aligned using ClustalW in MEGA 7 (27). Maximum Likelihood phylogenetic analysis was also performed using MEGA 7.
Data Availability Statement.
No data sets were generated or analyzed in this study.
Supplementary Material
Acknowledgments
We thank Johnathan Karty (Indiana University Mass Spectrometry Facility) for help with coupled GC-MS analysis. Funding for this study was provided by the National Institutes of Health Award R01GM040941 (to C.E.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1803220115/-/DCSupplemental.
References
- 1.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson GC, Tenson T, Hauryliuk V. The RelA/SpoT homolog (RSH) superfamily: Distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One. 2011;6:e23479. doi: 10.1371/journal.pone.0023479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hogg T, Mechold U, Malke H, Cashel M, Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 4.Battesti A, Bouveret E. Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol Microbiol. 2006;62:1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x. [DOI] [PubMed] [Google Scholar]
- 5.Loveland AB, et al. Ribosome•RelA structures reveal the mechanism of stringent response activation. eLife. 2016;5:e17029. doi: 10.7554/eLife.17029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown A, Fernández IS, Gordiyenko Y, Ramakrishnan V. Ribosome-dependent activation of stringent control. Nature. 2016;534:277–280. doi: 10.1038/nature17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arenz S, et al. The stringent factor RelA adopts an open conformation on the ribosome to stimulate ppGpp synthesis. Nucleic Acids Res. 2016;44:6471–6481. doi: 10.1093/nar/gkw470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grant GA. The ACT domain: A small molecule binding domain and its role as a common regulatory element. J Biol Chem. 2006;281:33825–33829. doi: 10.1074/jbc.R600024200. [DOI] [PubMed] [Google Scholar]
- 9.Masuda S, Bauer CE. Null mutation of HvrA compensates for loss of an essential relA/spoT-like gene in Rhodobacter capsulatus. J Bacteriol. 2004;186:235–239. doi: 10.1128/JB.186.1.235-239.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mechold U, Cashel M, Steiner K, Gentry D, Malke H. Functional analysis of a relA/spoT gene homolog from Streptococcus equisimilis. J Bacteriol. 1996;178:1401–1411. doi: 10.1128/jb.178.5.1401-1411.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuller DJ, Grant GA, Banaszak LJ. The allosteric ligand site in the Vmax-type cooperative enzyme phosphoglycerate dehydrogenase. Nat Struct Biol. 1995;2:69–76. doi: 10.1038/nsb0195-69. [DOI] [PubMed] [Google Scholar]
- 12.Bennett BD, et al. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brinsmade SR, Kleijn RJ, Sauer U, Sonenshein AL. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J Bacteriol. 2010;192:6357–6368. doi: 10.1128/JB.00937-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shivers RP, Sonenshein AL. Activation of the Bacillus subtilis global regulator CodY by direct interaction with branched-chain amino acids. Mol Microbiol. 2004;53:599–611. doi: 10.1111/j.1365-2958.2004.04135.x. [DOI] [PubMed] [Google Scholar]
- 15.Cho BK, Barrett CL, Knight EM, Park YS, Palsson BO. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc Natl Acad Sci USA. 2008;105:19462–19467. doi: 10.1073/pnas.0807227105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Traxler MF, et al. The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008;68:1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geiger T, Wolz C. Intersection of the stringent response and the CodY regulon in low GC Gram-positive bacteria. Int J Med Microbiol. 2014;304:150–155. doi: 10.1016/j.ijmm.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 18.Gaca AO, Colomer-Winter C, Lemos JA. Many means to a common end: The intricacies of (p)ppGpp metabolism and its control of bacterial homeostasis. J Bacteriol. 2015;197:1146–1156. doi: 10.1128/JB.02577-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Srivatsan A, Wang JD. Control of bacterial transcription, translation and replication by (p)ppGpp. Curr Opin Microbiol. 2008;11:100–105. doi: 10.1016/j.mib.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Wexselblatt E, et al. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathog. 2012;8:e1002925. doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Syal K, et al. Synthetic (p)ppGpp analogue is an inhibitor of stringent response in mycobacteria. Antimicrob Agents Chemother. 2017;61:e00443-17. doi: 10.1128/AAC.00443-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen HC, Marrs B. Map of genes for carotenoid and bacteriochlorophyll biosynthesis in Rhodopseudomonas capsulata. J Bacteriol. 1976;126:619–629. doi: 10.1128/jb.126.2.619-629.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver PF, Wall JD, Gest H. Characterization of Rhodopseudomonas capsulata. Arch Microbiol. 1975;105:207–216. doi: 10.1007/BF00447139. [DOI] [PubMed] [Google Scholar]
- 24.Fang M, Bauer CE. The vitamin B12-dependent photoreceptor AerR relieves photosystem gene repression by extending the interaction of CrtJ with photosystem promoters. MBio. 2017;8:e00261-17. doi: 10.1128/mBio.00261-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebelein M, Merdes G, Berger MR. Nucleotide preparation from cells and determination of nucleotides by ion-pair high-performance liquid chromatography. J Chromatogr A. 1992;577:146–150. doi: 10.1016/0378-4347(92)80610-3. [DOI] [PubMed] [Google Scholar]
- 26.Sobolevsky TG, et al. Comparison of silylation and esterification/acylation procedures in GC-MS analysis of amino acids. J Sep Sci. 2003;26:1474–1478. [Google Scholar]
- 27.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data sets were generated or analyzed in this study.