Significance
Single-molecule live-cell imaging can answer many biological questions, but a significant obstacle is photobleaching of the dye molecules by dissolved oxygen. An excellent way to improve photostability is to reduce the oxygen content of the solutions using glucose oxidase, an oxygen scavenger. However, for live-cell experiments a critical downside of the scavengers is that ATP levels fall as mitochondria become oxygen-deprived. We show that certain metabolic supplements significantly slow the depletion of ATP by oxygen scavengers and substantially restore ATP-dependent functions like lipid synthesis and receptor endocytosis. This method could be valuable for scientific studies that need to control oxygen tension and intracellular ATP levels in live cells.
Keywords: Single-molecule live-cell imaging, metabolite, ATP, oxygen scavenger, fluorescence microscopy
Abstract
The stability of organic dyes against photobleaching is critical in single-molecule tracking and localization microscopy. Since oxygen accelerates photobleaching of most organic dyes, glucose oxidase is commonly used to slow dye photobleaching by depleting oxygen. As demonstrated here, pyranose-2-oxidase slows bleaching of Alexa647 dye by ∼20-fold. However, oxygen deprivation may pose severe problems for live cells by reducing mitochondrial oxidative phosphorylation and ATP production. We formulate a method to sustain intracellular ATP levels in the presence of oxygen scavengers. Supplementation with metabolic intermediates including glyceraldehyde, glutamine, and α-ketoisocaproate maintained the intracellular ATP level for at least 10 min by balancing between FADH2 and NADH despite reduced oxygen levels. Furthermore, those metabolites supported ATP-dependent synthesis of phosphatidylinositol 4,5-bisphosphate and internalization of PAR2 receptors. Our method is potentially relevant to other circumstances that involve acute drops of oxygen levels, such as ischemic damage in the brain or heart or tissues for transplantation.
This paper develops a strategy to sustain intracellular ATP levels by stimulation of specific metabolic pathways for single-molecule fluorescence imaging in live cells under low-oxygen conditions.
Single-molecule live-cell imaging is used increasingly in biophysical and cell biological studies. Prolonging the bleaching time of fluorescent dyes is crucial for single-molecule tracking and localization microscopy to follow intermolecular interactions or to monitor conformational changes of biomolecules. Photobleaching often limits single-molecule imaging. Fortunately, the dye bleaching time can be prolonged by lowering oxygen, since photobleaching is accelerated by oxygen (1). One practical approach uses an enzyme such as glucose oxidase to remove oxygen (1–3).
In live cells, the use of oxygen scavengers is limited by lowered ATP production. Therefore, we tried to find an optimal concentration of oxygen scavenger and energy metabolites to improve dye photostability while minimizing the loss of ATP in live cells. To preserve ATP levels, we considered ways to boost both glycolysis and oxidative phosphorylation in the TCA or Krebs cycle. Glyceraldehyde (GLA) can stimulate both processes to generate ATP. It is converted to glyceraldehye-3-phosphate, which goes into glycolysis, and to glycerol-3-phosphate (G-3-P), which produces the reduced form of flavin adenine dinucleotide (FADH2) to transfer electrons to complex II in the mitochondrial electron transport chain (ETC) (4). To stimulate the TCA cycle in mitochondria, we tried glutamine and α-ketoisocaproate (KIC) because those metabolites can be converted to α-ketoglutarate, an intermediate in the TCA cycle (5–7). To test whether ATP levels can be controlled by introducing such metabolites, we used an intracellular ATP biosensor, PercevalHR (8, 9). Further, to test for functional improvements, we measured ATP-dependent phosphatidylinositol 4,5-bisphosphate (PIP2) synthesis and ATP and PIP2-dependent internalization of a G protein-coupled receptor. With these tools and metabolites, we demonstrate that GLA, KIC, and glutamine prevent the loss of ATP, PIP2, and receptor endocytosis in the presence of the pyranose-2-oxidase (P2O) oxygen scavenger.
Results
An Oxygen Scavenger Slows Dye Bleaching.
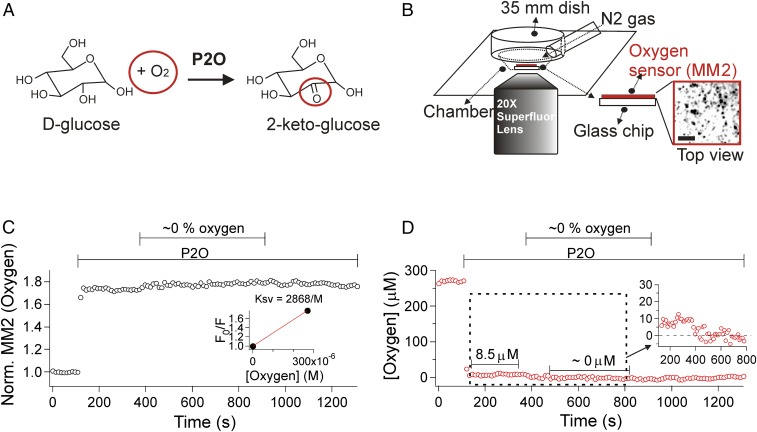
We wanted to measure the effect of an oxygen scavenger on the bleaching time of organic dyes in living cells. Traditionally, glucose oxidase has been used to deplete oxygen in microscopy, but it has the disadvantage of producing gluconic acid and a continuous drop of pH (10). We turned instead to another oxygen scavenger, P2O (11). It consumes O2 as it oxidizes d-glucose to 2-keto-d-glucose and H2O2 without generating acid equivalents (Fig. 1A and refs. 12 and 13). In our work, P2O was always paired with catalase to remove the hydrogen peroxide product (11). We measured the oxygen tension achieved using MM2 nanoparticles (14) as an oxygen sensor (Fig. 1B, Inset). They were affixed to a glass chip at the bottom of the chamber (at ∼5 mm depth). The fluorescence of the nanoparticles is normally quenched by collisions with molecular oxygen and brightens when the oxygen tension is lowered. The fluorescence was calibrated by covering the chamber when it contained 21% oxygen (air), adding oxygen scavengers, and delivering N2 gas at the upper fluid surface (Fig. 1B). As the oxygen tension is lowered by the scavenger, the fluorescence from MM2 is dequenched, and the signal brightens (Fig. 1C). The signal brightens even more as nitrogen replaces the air. The Fig. 1C, Inset is the two-point calibration curve for 21% and 0% oxygen. Applying the calibration shows that the oxygen concentration at the bottom of the chamber falls quickly from 270 µM (air) to 8.5 µM (0.7% partial pressure of oxygen) during addition of the oxygen scavengers (7.5 unit/mL P2O and 1,000 units/mL catalase) before the ambient air is replaced by nitrogen (Fig. 1D), much as reported for traditional glucose oxidase (11). In separate measurements, the enzyme consumed glucose at 0.2 mM/min (SI Appendix, Fig. S1), indicating that 20% of 10 mM glucose in the Ringer solution would be consumed during the first 10 min.
Fig. 1.
P2O reduces oxygen concentration in solution. (A) Reaction diagram of P2O with d-glucose and O2. (B) Configuration of the cell chamber system and the 20× superfluor objective lens used to collect photons from oxygen-sensitive nanoparticles (MM2) in epifluorescence mode. For calibration experiments, the chamber was covered by an inverted 35-mm dish with an inlet tube for gas flow. The oxygen scavengers were applied in the solution first, and then, where marked, nitrogen gas was switched on. (Inset ) The image of fluorescent particles in negative contrast. (Scale bar, 5 µm.) (C) Fluorescence of the MM2 oxygen sensor rises quickly with addition of oxygen scavengers (7.5 units/mL P2O and 1,000 units/mL catalase), and it rises slightly more with flow of nitrogen gas (∼0% oxygen), giving the calibration curve and the Ksv (Stern–Volmer constant, Inset). (D) The normalized intensity of MM2 fluorescence was converted to the oxygen concentration in D. By themselves, the oxygen scavengers reduced the oxygen concentration to 8.5 µM (see magnified inset).
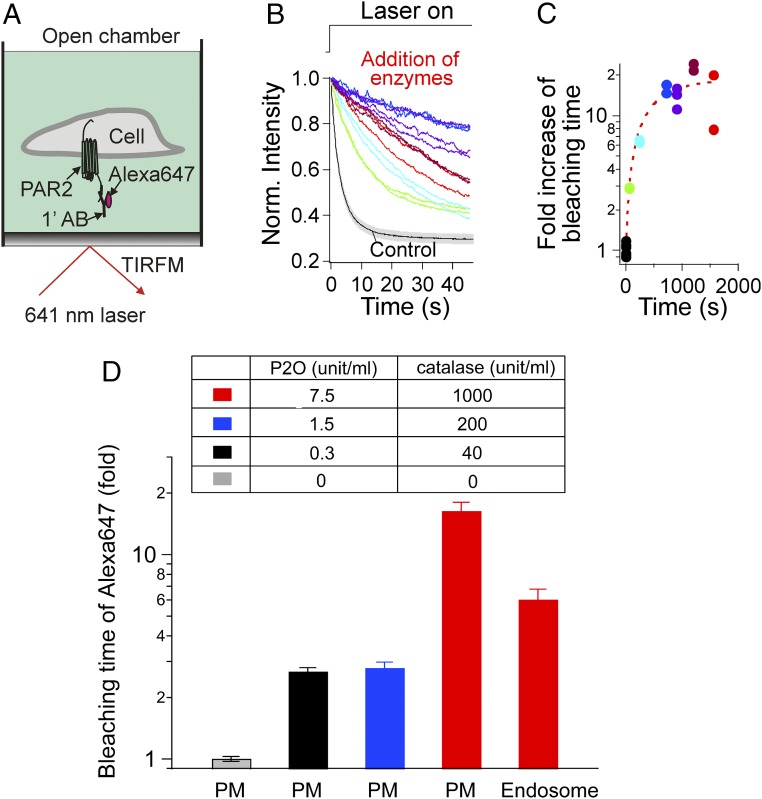
Next, we examined the bleaching rate of an organic dye with the oxygen scavengers. Protease-activated receptor 2 (PAR2) was transiently expressed in tsA201 cells and then labeled using Alexa647 dye-conjugated primary antibody targeted to the receptor extracellular N terminus. Cells were viewed in an open chamber (air above) by total internal reflection fluorescence (TIRF) microscopy with single-molecule sensitivity (Fig. 2A). When exposed to a 641-nm laser (100 mW), the intensity of Alexa647 fluorescence decayed as the dye was bleached (mean exponential time constant = 3.8 s; Fig. 2B, control). Addition of the P2O oxygen scavenger (always together with catalase) gradually prolonged the bleaching times of the fluorescent Alexa dye (Fig. 2B, colored lines). The protective effect developed fully by about 10 min (Fig. 2C). The maximum effect was a ∼20-fold prolongation of the bleaching time when using a high P2O activity (7.5 unit/mL), and approximately threefold when using a lower P2O activity (0.3 unit/mL) (Fig. 2D). In addition, we found that the bleaching time of endocytosed Alexa dyes is also prolonged by the oxygen scavengers. For this we used highly inclined and laminated optical sheet microscopy (15), a mode with signal-to-noise ratio comparable to that of the TIRF mode that can illuminate the inside of the cell with a thin sheet of laser light. The bleaching time was prolonged approximately sixfold in the presence of a high activity of oxygen scavengers in the bath (Fig. 2D and SI Appendix, Fig. S2). When we tried the same kind of experiment, asking whether oxygen scavengers slow the bleaching rate of GFP, the answer was negative (SI Appendix, Fig. S3). The literature reports that different fluorescent proteins show quite differing oxygen sensitivities (SI Appendix, Supplementary Discussion).
Fig. 2.
P2O slows the bleaching of Alexa647 dye. (A) Schematic diagram of a photobleaching experiment using single-molecule TIRF microscopy. PAR2 was transiently expressed in tsA201 cells before the experiments. Extracellular Alexa647-primary PAR2 antibody was used for labeling PAR2 at the plasma membrane (PM). (B) Laser power was increased from 10 to 100 mW to start bleaching. Bleaching curves are shown before (control, black line) and 100–1,700 s after addition of 7.5 units/mL P2O and 1,000 units/mL catalase (colored lines). These data were fitted with single-exponential decays to get time constants representing bleaching time. Background intensity was not removed. Different colors were used to emphasize the gradual improvement of bleaching time. The colors match the symbols at the different incubation times in C. (C) Temporal course of change of dye bleaching time in the presence of 7.5 units/mL P2O and 1,000 units/mL catalase. The points were fitted with a single-exponential curve to estimate how fast oxygen scavengers improve the bleaching time of the Alexa dye (red dashed line). (D) Summary of fold increase of Alexa647 dye bleaching time with different activities of added P2O and catalase. “PM” and “Endosome” indicate the locations of PAR2 receptors. To make PAR2 receptors localized in endosomes, PAR2 agonist AP was added for 1 h to trigger receptor endocytosis (SI Appendix, Fig. S2). Control (gray, PM, n = 20), 0.3 P2O and 40 catalase (black, PM, n = 8), 1.5 P2O and 200 catalase (blue, PM, n = 8), 7.5 P2O and 1,000 catalase (red, PM, n = 9), and 7.5 P2O and 1,000 catalase (red, Endosome, n = 9).
P2O Decreases Intracellular ATP.
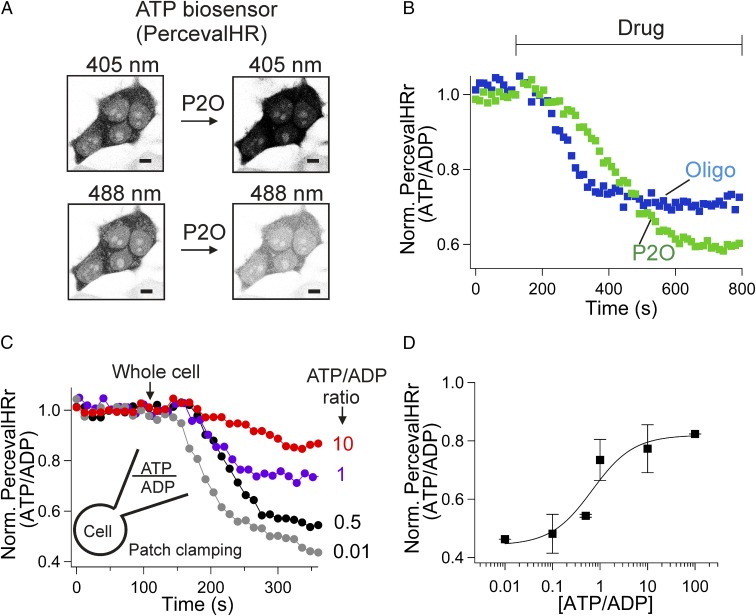
Although lowering oxygen successfully lengthens the bleaching time of the dye, it is also likely to reduce oxidative phosphorylation in mitochondria, causing a drop in ATP. We first tried to measure effects on mitochondrial membrane potential, which was not very informative. The initial changes were only small fluctuations of the membrane potential, but after 250 s an expected depolarization began to develop slowly (SI Appendix, Fig. S4 and Supplementary Discussion). We turned instead to monitoring intracellular ATP directly using a genetically expressed ATP biosensor, PercevalHR. This fluorescent biosensor detects the intracellular ATP-to-ADP ratio using competitive binding of ATP versus ADP to a single binding site on the probe (9). When the ATP-to-ADP ratio falls, the ratio of 530-nm emission during sequential excitation by 488- and 405-nm light (PercevalHRr = F488/F405) decreases. In confocal images of tsA201 cells, the expressed PercevalHR was distributed in the cytoplasm and the nucleus (Fig. 3A). Adding P2O initiated a strong reduction in PercevalHRr (Fig. 3B; see also pH calibration in SI Appendix, Fig. S5). As a control, oligomycin, which blocks mitochondrial ATP synthase, also reduced PercevalHRr, confirming that the biosensor responds to intracellular ATP reduction. We determined the lower limit of the probe ratio by dialyzing with patch pipettes containing different ratios of ATP and ADP (Fig. 3 C and D). The minimum ratio of PercevalHRr with the lowest ATP:ADP was ∼0.45 of the normoxic basal ratio without any drugs.
Fig. 3.
P2O decreases the ATP-to-ADP ratio. tsA201 cells were transfected with the ATP biosensor PercevalHR. (A) Confocal images (negative contrast) before and after treatment with high activity of P2O and catalase (P2O). (Scale bars, 5 µm.) (B) Both P2O and oligomycin decrease the ATP-to-ADP ratio. Oligomycin (n = 4), P2O (n = 6). The bar indicates time for application of drugs. Oligomycin (Oligo) and P2O (7.5 units/mL) containing catalase (1,000 units/mL) decreases the PercevalHR ratio (PercevalHRr). (C) Individual cells expressing the probe were internally dialyzed through the whole-cell pipette with solutions containing different ATP-to-ADP ratios at pH = 7.43 (Fig. 2C, Inset). The total ATP plus ADP was kept at ∼3 mM. At low ATP/ADP (0.01), PercevalHRr fell to 0.45, indicating the lower detection limit of the biosensor. (D) Dependence of PercevalHRr on the ATP-to ADP ratio. Each point is the mean of two or three independent results. The plotted data were fitted with the Hill equation (black line). The Hill coefficient was fixed at unity because the PercevalHR biosensor has only one nucleotide binding site.
ATP Depletion Slows PIP2 Synthesis.
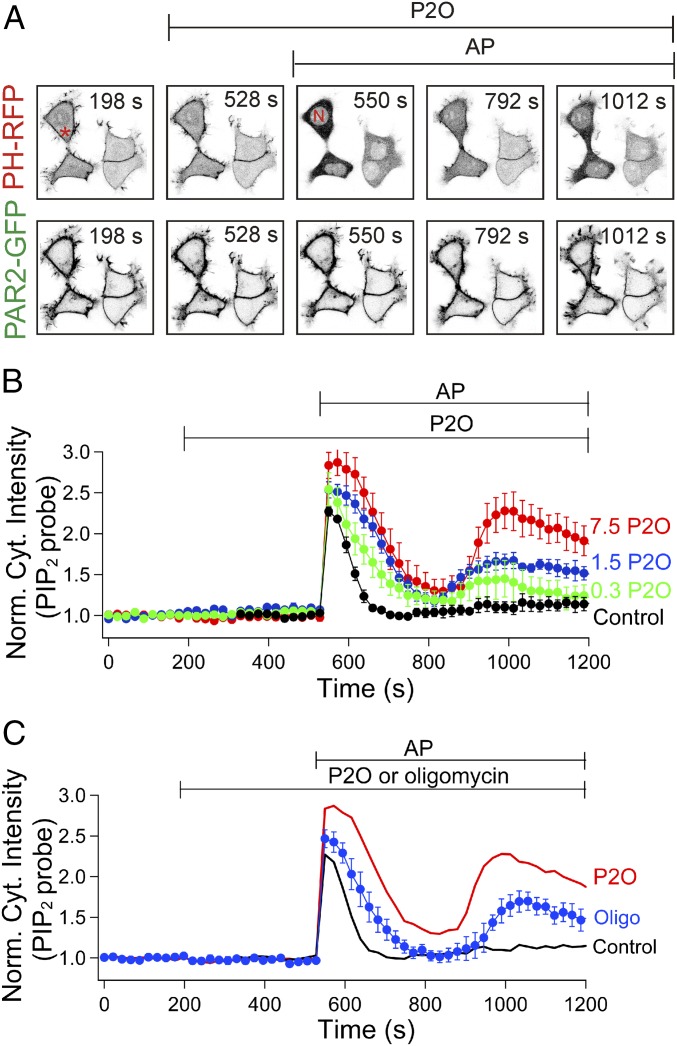
Does the P2O-induced ATP depletion perturb ATP-dependent cellular mechanisms? As a first test, we asked whether synthesis of PIP2, an ATP-dependent process, is depressed (16–18). The plasma membrane PIP2 level was monitored with a red fluorescent protein (RFP)-tagged PIP2-binding pleckstrin homology (PH) probe (PH-RFP probe) and perturbed by transiently activating phospholipase C (PLC). We coexpressed the PLC-coupled PAR2 together with the PH-RFP probe. PAR2 can deplete PIP2 quickly after agonist addition but then desensitizes within a minute, allowing ATP-dependent PIP2 restoration from phosphatidylinositol by lipid kinases (19). In the control experiment, the PH-RFP probe is initially localized at the plasma membrane (Fig. 4 A and B), briefly translocates to the cytosol (increased cytosolic PH-RFP intensity) after activation of PAR2 with the receptor agonist, activating peptide (AP), but then returns to the plasma membrane despite the continued presence of the agonist. Thus, as expected, PIP2 is initially broken down by PLC but recovers quickly after receptor desensitization. Adding P2O to lower the oxygen concentration compromised the recovery of PIP2 (Fig. 4B). Now the recovery became only partial and also biphasic, with a transient PIP2 increase after the initial phase of PIP2 depletion, followed by a second phase of PIP2 depletion (“hump” in the probe trace). Oligomycin similarly compromised the recovery (Fig. 4C), again with a partial biphasic response. We conclude that the oxygen scavenger does interfere with ATP-dependent PIP2 formation. The scavenger might also interfere with the ATP-dependent phosphorylation of PAR2 receptors by protein kinases that initiate receptor desensitization (19). ATP is needed for both pathways.
Fig. 4.
P2O induces PIP2 depletion during PAR2 desensitization. (A) Confocal images of PH-RFP (PIP2 probe) and PAR2-GFP. The cytosolic intensity of the probe was monitored from the cytosolic area denoted by a red asterisk and normalized with basal intensity of the probe without any drugs (Norm. cyt. intensity). After addition of 100 µM AP (PAR2 agonist), the PIP2 probe translocates from the plasma membrane to the cytoplasm but not the nucleus denoted by “N” at 550 s. Increasing intensity of PH-RFP in the cytoplasm indicates depletion of PIP2 at the plasma membrane. PAR2-GFP fluorescence indicates the localization of PAR2 at the plasma membrane. (B) Averaged PIP2 probe in progressively increased P2O and catalase activities. For example, 7.5 P2O indicates that 7.5 units/mL P2O in the presence of 1,000 units/mL catalase (Fig. 2D, inset table). 7.5 P2O (n = 4), 1.5 P2O (n = 3), 0.3 P2O (n = 4), and control (n = 8). (C) Averaged PIP2 probe with (7.5 P2O, red line) or without (control, black line) P2O or with oligomycin (Oligo, blue symbols, n = 6).
Metabolites Can Restore Oxygen-Scavenger–Induced ATP Depletion and PIP2 Reduction.
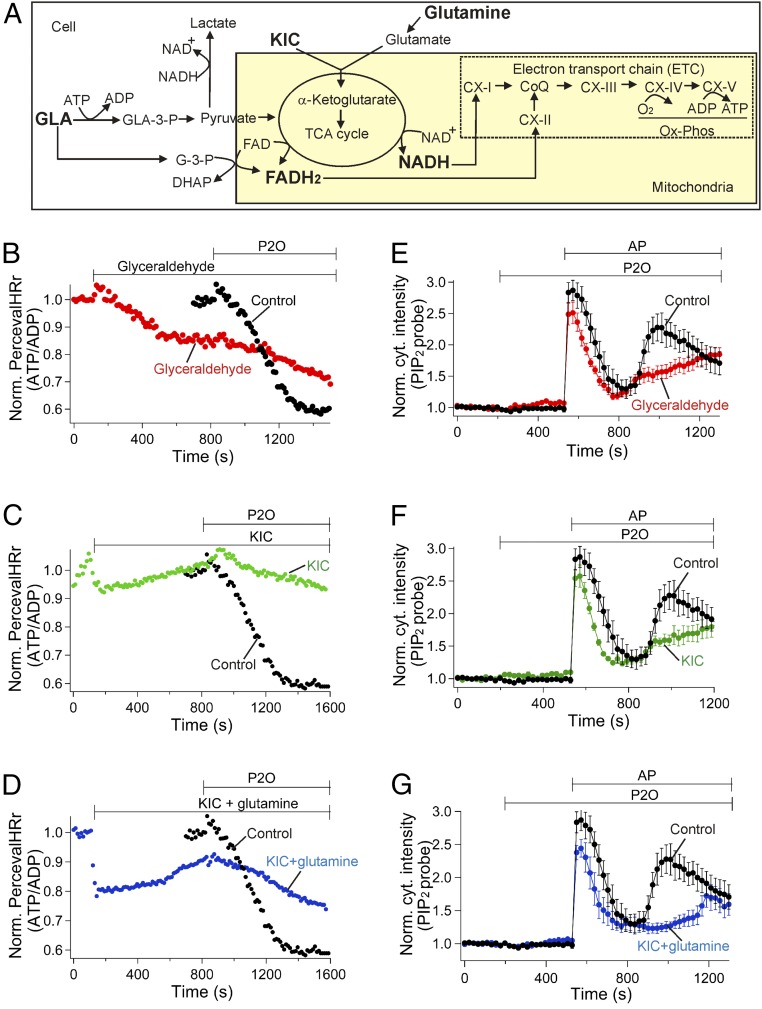
We developed protocols to maintain the ATP-to-ADP ratio during P2O treatment. Raising glucose from 10 mM to 25 mM turned out not to be a solution since a strong decrease of the ATP/ADP ratio still developed (SI Appendix, Fig. S6). This occurred even though the raised glucose did accelerate glycolysis, as evidenced by increased production of lactate (SI Appendix, Fig. S7). Therefore, we adopted a two-pronged strategy, illustrated in Fig. 5A, that included providing substrate for glycolysis while also directly activating the TCA cycle. Both pathways are coupled to the ETC and oxidative phosphorylation. The goal was to provide carbon substrates to generate reducing equivalents in the form of both NADH and FADH2 that donate electrons to the ETC. To feed the glycolytic pathway, we used GLA, which can be converted to glyceraldehyde-3-phosphate (GLA-3-P) by GLA kinase. GLA-3-P in turn enters the glycolytic pathway by a reaction linked to NADH production. In parallel, GLA produces G-3-P by a reaction linked to FADH2 production. The levels of NADH and FADH2 can be estimated by imaging NADH and FAD autofluorescence, respectively (SI Appendix, Fig. S8 A and B). The typical short-wavelength autofluorescence (excitation = 360 nm, emission = ∼450 nm) of live cells comes from NADH in mitochondria and in the cytosol (ref. 20 and Fig. 5A) compared with NADPH, which does not have strong regulation by glucose and metabolites (21). FAD autofluoresces at longer wavelength (excitation = 450 nm and emission = 530 nm), whereas FADH2 does not emit autofluorescence, so a decrease of the FAD signal corresponds to an increase of FADH2. Addition of GLA to the medium initially modestly decreased PercevalHRr (ATP/ADP ratio), probably due to consumption of cytosolic ATP by GLA kinase (Fig. 5B), with no intracellular pH (pHi) change (SI Appendix, Fig. S9). As hoped, the subsequent decrease of PercevalHRr by the oxygen scavenger was significantly slowed in the presence of GLA as GLA supplied both NADH and FADH2 (SI Appendix, Fig. S8 A and B).
Fig. 5.
Metabolites sustain the ATP level against reduction by P2O. (A) Schematic diagram of cellular metabolism of GLA, KIC, and glutamine. GLA can be converted to GLA-3-P and G-3-P. After many glycolytic steps, pyruvate is made from the GLA-3-P and enters into mitochondria to increase NADH in the TCA cycle. Mitochondrial complex I (CX-I) and complex II (CX-II) oxidize NADH and FADH2, respectively, to transport electrons into the ETC and, at the same time, a proton gradient develops at complexes I, III, and IV. Finally, to make ATP from ADP in complex V, complex IV consumes oxygen by using protons and electrons to produce water molecules. The G-3-P shuttle produces FADH2, which goes into the ETC. (B–D) tsA201 cells transfected with PercevalHR ATP biosensor and dark PAR2. (B) GLA slowed the reduction of the ATP-to-ADP ratio induced by P2O. Red symbols indicate the normalized PercevalHRr after pHi corrections in the presence of 10 mM GLA. (B–D) The black symbols (control) were obtained from green symbols in Fig. 3B. GLA (B, n = 8), KIC (C, n = 5), amd KIC + glutamine (D, n = 7). (E–G) GLA (E, n = 6), KIC (F, n = 6), and KIC plus glutamine (G, n = 5) restored the plasma membrane PIP2 resynthesis. Oxygen scavenger (7.5 units/mL P2O and 1,000 units/mL catalase) was applied for B–G. The control data in E–G were obtained from 7.5 P2O without additional metabolites in Fig. 4B.
To provide an intermediate metabolite of the TCA cycle (Fig. 5A), cells were treated with KIC, a precursor of α-ketoglutarate. There was a quick decrease of PercevalHRr (SI Appendix, Fig. S10A) upon adding KIC due to a quick pHi drop (SI Appendix, Fig. S10B). After correction for the pH effect using the calibration curve (SI Appendix, Fig. S5), a slight drop of the PercevalHRr with KIC still persisted, and NADH decreased (SI Appendix, Fig. S8A). Similarly, cell-permeable dimethyl α-ketoglutarate decreased NADH (SI Appendix, Fig. S8A) but increased FADH2 (SI Appendix, Fig. S8B). Nevertheless, KIC significantly improved the maintenance of PercevalHRr during P2O application (Fig. 5C) without increase of lactate production (SI Appendix, Fig. S7), suggesting that α-ketoglutarate produced from KIC in the TCA cycle helps to maintain ATP levels by an FADH2 gain sufficient to offset any NADH loss (Discussion).
We further tested the effect of glutamine since it also yields α-ketoglutarate via formation of glutamate (Fig. 5A). Indeed, the FADH2 level increased significantly with glutamine alone (SI Appendix, Fig. S8B), and again NADH declined (SI Appendix, Fig. S8A). The basal ATP-to-ADP ratio was minimally changed by glutamine (SI Appendix, Fig. S11). In the presence of P2O, glutamine sustained the ATP-to-ADP ratio but only for 5 min. Therefore, we tried a combination of KIC and glutamine (Fig. 5D). The combination did depress PercevalHRr somewhat more than with KIC alone (Fig. 5C), and NADH (SI Appendix, Fig. S8A) and pHi (SI Appendix, Fig. S10B) were decreased; however, a significant ATP-to-ADP ratio was sustained for at least 10 min in the presence of P2O (Fig. 5D). In summary, adding GLA, KIC, glutamine, or KIC plus glutamine aids the production of ATP during oxygen depletion perhaps by augmenting FADH2 (SI Appendix, Supplementary Discussion).
Added Metabolites Support ATP- and PIP2-Dependent Receptor Endocytosis.
We investigated whether the metabolic supplements also preserved ATP-dependent PIP2 resynthesis during P2O treatment. Using the PIP2-sensitive PH-RFP probe, we monitored the time course of PIP2 after depletion by PAR2 receptor stimulation. Again in the control, in the presence of P2O, the cytoplasmic PH-probe signal rose and partially recovered (as PIP2 was depleted and restored) after PAR2 stimulation, followed by a second phase of further depletion of PIP2 (hump) due to the low ATP (Fig. 5 E–G, control). The initial and especially the secondary depletion of the PIP2 with P2O were noticeably diminished by additions of the metabolites (Fig. 5 E–G, metabolites), showing that they improved the ATP supply enough to restore some PIP2 synthesis in living cells.
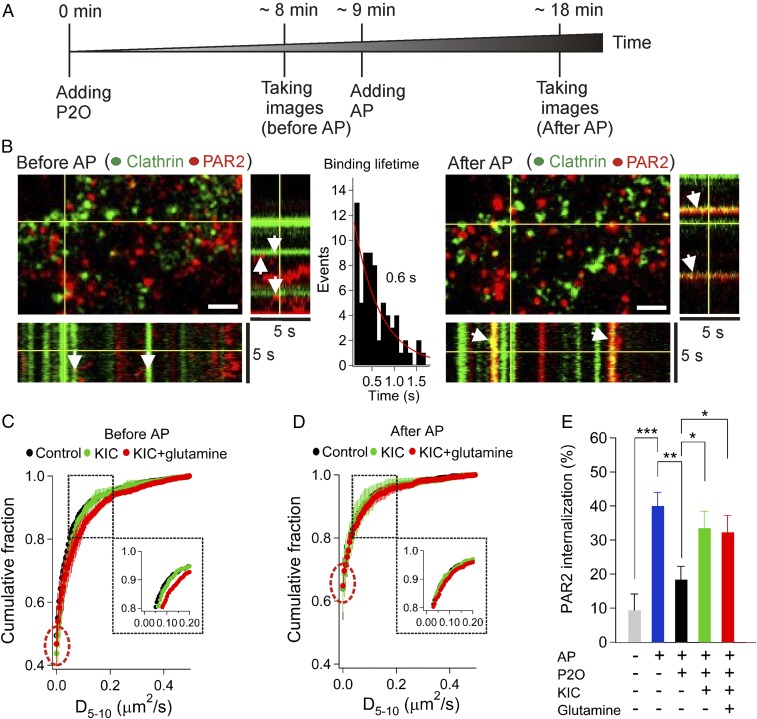
Our next goal was to test whether metabolites can affect molecular interactions. As an example, we monitored the interaction between PAR2 and clathrin using single-molecule TIRF microscopy. PAR2 uses at least two steps for its complete desensitization (22): (i) interactions with clathrin-coated pits to be captured in small vesicles and (ii) final endocytosis. As is well known, the formation of clathrin-coated pits can be down-regulated by PIP2 reduction (23). Thus, receptor–clathrin interaction can be negatively influenced by reduction of PIP2 induced by hypoxia. Our initial expectation was that metabolites would restore receptor–clathrin interactions at low oxygen conditions via recovery of PIP2.
Cells were pretreated with P2O to lengthen the photobleaching time of dyes in all cases (Fig. 6A). As expected, we could monitor the Alexa647-primary-antibody-tagged PAR2 with little photobleaching in the presence of P2O (Fig. 6B, before AP, see kymograph record). Clathrin-coated pits were themselves relatively immobile within a 5-s time scale as reported previously (24) but interacted transiently with and immobilized mobile receptors (∼0.6 s, Fig. 6B, Inset). After addition of the PAR2 agonist, AP, a significant portion of the receptors became colocalized with clathrin-coated pits for the duration of the recording (Fig. 6B, after AP, see kymograph). We next tested whether the interaction of receptors and clathrin-coated pits is affected by the metabolic supplements. To do this, we determined the distribution of receptor diffusion coefficients (D) during single-molecule tracking to distinguish mobile receptors from receptors immobilized by interaction with clathrin-coated pits. Before treatment with the agonist for PAR2, ∼60% of the PAR2 receptors were mobile (D > 0.005 µm2/s) and ∼40% of the receptors were immobile (D < 0.005 µm2/s). After addition of AP, a significant portion of the receptors became immobilized, as indicated by an increase of the fraction with low mobility (Fig. 6 C and D, red dotted circles). The immobilized fraction was not changed by supplementation with the different metabolites, suggesting that receptor–clathrin–coated-pit interactions may not be significantly influenced by the metabolites in the presence of oxygen scavengers.
Fig. 6.
Metabolites prevent reduction of PAR2 endocytosis by the oxygen scavenger. PAR2 is labeled with Alexa647-dye-conjugated primary antibody against the N terminus of the PAR2. Clathrin is labeled with cell-permeable SNAP cell-505 dye for intracellularly localized SNAP-tagged light chain of clathrin. (A) Protocol for treatment of P2O and AP, and timing for taking images. (B) Cells were incubated with 10 mM KIC and 2 mM glutamine before addition of P2O (7.5 units/mL) and catalase (1,000 units/mL). Line scan for 3D (x,y,t) projection before or after 100 µM AP treatment in the presence of the oxygen scavengers (see the protocol in A). White arrows indicate transient or long binding events. Most PAR2 molecules bind transiently to the clathrin-coated pits before addition of AP. (Scale bars in all figures, 2 µm.) (C and D) Receptor diffusion coefficient in the absence or presence of metabolites. Metabolites (10 min), oxygen scavengers (10 min), and 100 µM AP (10 min) were added sequentially into the nonperfused chamber. Two movies in the absence (C) and presence of AP (D) from same cells were taken for 5 s with a 100-ms time interval. The diffusion coefficient between frames 5 and 10 (D5–10) were estimated. All experiments were performed with oxygen scavenger (7.5 units/mL P2O and 1,000 units/mL catalase). Receptor molecules (200–500 ) were analyzed for the cumulative fraction for each condition and then the cumulative fraction was averaged from three replicate experiments. The bin size for the cumulative fraction was 0.005 µm2/s. The population of mobile receptors (∼0.1 µm2/s) with KIC plus glutamine exhibited a broader range of diffusion coefficient than did the control or KIC alone (dotted rectangular inset). (E) PAR2 internalization. KIC alone or KIC plus glutamine prevented reduction of PAR2 endocytosis by the oxygen scavenger (7.5 units/mL P2O and 1,000 units/mL catalase). For each condition (Materials and Methods and SI Appendix, Fig. S12), we analyzed at least 8–11 cells. Control (n = 8), AP (n = 11), AP+P2O (n = 10), AP+P2O+KIC (n = 11), and AP+P2O+KIC+glutamine (9). ***P < 0.0005, **P < 0.001, *P < 0.05.
Finally, we investigated endocytosis of PAR2 receptors, a downstream step after receptor–clathrin interaction. We defined endocytosis as the appearance of PAR2-labeled vesicles in the cytoplasm after addition of the PAR2 agonist (SI Appendix, Fig. S12). Consistent with a need for ATP and PIP2 in PAR2 endocytosis, the percentage of internalized PAR2 decreased during treatments with the oxygen scavenger (Fig. 6E) and further addition of KIC or KIC plus glutamine significantly restored the receptor endocytosis.
Discussion
Improving photostability of dyes has been a big hurdle in the single-molecule field. One approach to increase photostability has been to develop new dyes that have longer bleaching time and that generate more photons (25, 26). However, increasing bleaching time of dyes for solution-based experiments is fundamentally challenging because oxygen often accelerates dye photobleaching (SI Appendix, Supplementary Discussion).
To lessen photobleaching, experimenters have developed encapsulated dyes that are protected from oxygen molecules in solution (27). Quantum dots (28, 29) and polymer dots (30) overcome this issue as well. However, those approaches are limited for some biological experiments by the larger size of the fluorescent probes (29), that is, dye-encapsulated nanoparticles (∼30 nm), commercial quantum dots (∼15–20 nm), and polymer dots (∼25 nm). An alternative approach has been to use oxygen scavengers such as glucose oxidase and catalase, to prolong the bleaching time of dyes. Although this method is very useful for in vitro single-molecule assays to see protein–protein interaction and conformational dynamics inside proteins, a downside is that it decreases the ATP content in intact live cells since oxidative phosphorylation in mitochondria requires oxygen. Here, we wanted to lower the oxygen but minimize ATP depletion in live cells. As we demonstrated, P2O prolonged the bleaching time of Alexa647 dye by ∼20-fold while also significantly reducing the ATP level in live cells. Increasing glucose concentration or adding glutamine in the presence of oxygen scavengers did not keep up ATP levels in live cells. Increasing glycolysis and the TCA cycle by adding other metabolites such as GLA and KIC successfully boosted ATP production despite lowered oxygen. The supplements also supported ATP-dependent cellular processes, such as lipid phosphorylation and receptor endocytosis. Interestingly, according to our single-molecule live-cell imaging, receptor–clathrin interaction before endocytosis was not much affected by addition of the supplements although endocytosis was. We deduce that ATP- and PIP2-dependent pathways are engaged more in final steps of receptor endocytosis at the plasma membrane rather than in the earlier capture of receptors into clathrin-coated pits.
GLA increased NADH (SI Appendix, Fig. S8A) and FADH2 (SI Appendix, Fig. S8B). Thus, GLA stimulates oxidative phosphorylation in mitochondria via complex I (NADH) and complex II (FADH2) (Fig. 5A). An important consideration for supplementary metabolites is how readily they can enter cells. KIC can be transported into cells by the lactate transporter (31). Unlike KIC, which bears a negative charge, GLA is electrically neutral at physiological pH (∼7.0) and could therefore enter into cells by passive diffusion through the membrane. However, the low octanol/water partition coefficient of GLA (0.025) predicts a low passive membrane permeability coefficient of 0.0017 µm/s and a long equilibration time constant for tsA201 cells near 60 min (32), which suggests that permeation of GLA might be slow. However, GLA must actually load much faster since it changes FADH2 and NADH within 20 s as KIC does (SI Appendix, Fig. S6). Hence, GLA transporters are probably present in tsA201 cells. A GLA transporter has been suggested in insulin-secreting pancreatic β-cells (33, 34).
It has been questioned whether mitochondrial oxidative phosphorylation could be a major source of ATP during hypoxia (35). It is important to note that hypoxia is not anoxia. In normal conditions, the mitochondrial contribution to ATP production in transformed renal epithelial cells is 80%, and the glycolytic pathway is 20% (35). In hypoxia, the mitochondrial production is reduced but still can contribute up to 60% of the ATP by using glutamine to power the ETC (35). Glutamine has been suggested to enhance the TCA cycle via generation of α-ketoglutarate, a key intermediate metabolite of the TCA cycle. α-Ketoglutarate stimulates the ETC, which in turn continues to make some ATP even under lowered oxygen. In agreement with this concept, we confirmed that even with lowered oxygen (0.7%), glutamine can increase FADH2 production and maintain ATP levels for a while. However, probably due to a limited production of α-ketoglutarate, supplementation of glutamine together with KIC prolongs the beneficial effects further. The simplest way to increase ATP in cells is to add more glucose to the solution because additional glucose can increase glycolysis further. Indeed, high glucose increased lactate production (SI Appendix, Fig. S7). However, raising the glucose only marginally prevented lowering of the ATP/ADP ratio by P2O (SI Appendix, Fig. S6). This suggests that the contribution of the glycolytic pathway to keep the ATP level in hypoxia is still weaker than mitochondrial ATP production, as demonstrated by Fan et al. (35). Similarly, KIC did not increase lactate but boosted ATP levels, supporting again that ATP production via mitochondria is more efficient than glycolysis in hypoxia.
Taken together, we demonstrated (i) that an open-chamber system is suitable to deplete oxygen levels from 21 to 0.7%, (ii) that oxygen scavenging decreases oxygen, ATP production, and plasma membrane PIP2 synthesis, (iii) that the reduction of ATP and PIP2 synthesis in live cells can be compensated by adding metabolites (GLA, glutamine, and KIC), and (iv) that these supplements restored receptor-mediated endocytosis. Metabolic supplementation preserves physiological functions when using lowered oxygen for single-molecule imaging in live cells. Our method is potentially relevant to other cases of acute reduction of oxygen levels, such as ischemic damage in the brain or heart or handling tissues for transplantation.
Materials and Methods
Cell Culture.
tsA201 cells, derived from Human Embryonic Kidney 293 (HEK293) cells, were obtained from Sigma-Aldrich. They were cultured in DMEM containing glutamine (2 mM), sodium pyruvate, and glucose (25 mM) supplemented with 10% FBS and 1% penicillin and streptomycin and subcultured every 3–4 d using 0.05% trypsin containing EDTA. For confocal experiments, tsA201 cells were plated on small glass chips coated with poly-l-ornithine. All experiments were performed in normal Ringer solution at room temperature (22–24 °C). For some protocols, we used 37 °C CO2 incubator [i.e., labeling of SNAP dye for clathrin and tetramethylrhodamine, ethyl ester (TMRE) and JC-1 dyes for mitochondria]. All experiments were performed after incubation of the tsA201 cells in the normal external saline solution without any additional supplements such as glutamine and other amino acids for at least 30 min.
Reagents and Solutions.
dl-GLA, l-glutamine, ΚΙC acid sodium salt (K0629), cell-permeable α-ketoglutarate, P2O (P4234-250 unit), and catalase were purchased from Sigma-Aldrich. Alexa647-labeled primary antibody against PAR2 was from Santa Cruz Biotechnology (SAM11, 100 µg/2 mL). PAR2 AP (N-SLIGKT-C) was from United Biosystems Inc. BCECF-AM was purchased from Molecular probes. Cell-permeable SNAP-cell 505-star dye was from New England Biolabs Inc. Cell culture medium (DMEM) was from Invitrogen (11995). Normal external Ringer solution contained (in millimolar) 137.5 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 10 glucose, and 10 Hepes (pH adjusted to 7.3 with NaOH). The stocks of glutamine (200 mM) and KIC (1 M) were dissolved in water. The addition of 2 mM glutamine and 10 mM KIC did not change the pH of the normal external Ringer solution. GLA (20 mM) was freshly made up with the normal external Ringer solution before experiments and then it was diluted with 1:1 to make 10 mM for the experiment. The stocks of the P2O (81 mg/mL = ∼750 units/mL) and catalase (20 mg/mL = ∼100,000 units/mL) were made with T50 saline solution (50 mM NaCl and 10 mM Tris⋅HCl at pH 8.0). Usually, 1:100 diluted enzymes were used for the oxygen-scavenging system except for concentration-dependent experiments. The SNAP-cell 505-star dye (1 mM) and BCECF-AM (2 mM) were dissolved in DMSO. AP was dissolved as a 10 mM stock in distilled water. To label PAR2, the Alexa647-labeled PAR2 antibody was diluted (1:20) into the normal saline solution before experiments.
Transfecting cDNA.
tsA201 cells were transfected with different concentrations of cDNA constructs depending on the type of experiment. For normal confocal experiments, we used 0.5 µg cDNA except for the PH-RFP probe (0.3 µg). For single-molecule experiments, we used a ∼10-fold lower amount of plasmids. X-tremegene 9 DNA transfection reagent (10 µL; Roche Applied Science) and cDNA were added to Opti-MEM solution (∼87 µL) sequentially. For transfection, cells at ∼75% confluency in a 35-mm culture dish were washed with Opti-MEM and supplemented with 0.5 mL Opti-MEM solution. Then the mixture of the transfection reagent and cDNA (∼100 µL) was added into the Opti-MEM–containing culture dish. We incubated the cells for at least for 4 h and up to 6 h longer when needed. Finally, cells were treated with trypsin (0.05%)-EDTA to culture them. After washing off the trypsin with fresh medium twice, the cell pellet was resuspended. Cells were seeded onto small 5-mm glass chips or 25-mm coverslips coated with poly-l-orinithine (1 mg/mL) and then incubated for 1 d before the experiments. The PH-RFP probe (PIP2-sensitive PH domain of PLCδ1 tagged with RFP) was provided by C. S. Kearn, University of Washington, Seattle. GFP-tagged human PAR2 was obtained from N. W. Bunnett, Monash Institute of Pharmaceutical Sciences, Parkville, VIC, Australia. Unlabeled PAR2 (no GFP tag) was subcloned in pCDNA3.1 vector (19). No difference between unlabeled PAR2 and PAR2-GFP was detected with respect to PIP2 hydrolysis and desensitization kinetics (19).
Confocal Microscopy.
Confocal images were taken every 12 s for the PercevalHR probe and the TMRE dye and every 1 min for the JC-1 dye with a Zeiss 710 confocal microscope. Excitation and emission light was delivered and collected by a 40× oil immersion lens (1.3 N.A.). Agonist and drugs were applied by pipette into the imaging chamber without continuous perfusion.
For the PercevalHR biosensor, cells were excited by 488- and 405-nm lasers sequentially and emission was collected at ∼530 nm for calculation of the fluorescence ratio (PercevalHRr = F488/F405). The ATP-bound biosensor absorbs 488 nm more than 405 nm, whereas the ADP-bound biosensor absorbs 405 nm more than 488 nm. Both forms emit fluorescence at ∼530 nm. To remove cell-to-cell variation, plotted values of PercevalHRr were normalized to the initial resting value. The biosensor was present in the cytoplasm and nucleus. Since nuclear pores are permeable to ATP and ADP, we analyzed the fluorescence signals from the whole cell. Because the Venus fluorescent protein can be protonated as pH falls, the Perceval biosensor is pH-sensitive. Therefore, we checked whether the oxygen scavenger changes pHi. Compared with oligomycin or control, P2O did decrease pHi slightly (SI Appendix, Fig. S5B). We corrected the pH effect with the calibration curve in SI Appendix, Fig. S5C.
Mitochondrial membrane potential (MMP) was monitored from the whole-cell red/green fluorescence ratio of JC-1 dye expressed as a ratio (F561/F488). Red indicates hyperpolarized MMP and green indicates depolarized MMP. After loading with JC-1 dye (10 µg/mL) in normal external Ringer solution for 30 min in the 37 °C incubator, cells were washed twice with Ringer. The red dye form was excited by a 561-nm diode-pumped solid-state laser with emission collected from 591 to 700 nm. The green dye form was excited by 488-nm argon laser with emission collected from 518 to 540 nm. Additional measurements of MMP were made with TMRE. Like JC-1, the positively charged TMRE dye is sequestered into energized mitochondria depending on their membrane potential. Mitochondrial depolarization releases the dye from mitochondria. After loading TMRE (40 nM) in cell culture medium for 10 min in the 37 °C incubator, cells were washed with the culture medium and then the cells were transferred into normal saline Ringer solution. The TMRE fluorophore was excited at 561 nm.
For measuring internalization of PAR2, tsA201 cells transfected with PAR2-GFP were labeled with extracellular PAR2-primary antibody conjugated with Alexa647 dye (SAM11; Santa Cruz Inc.) for 15 min. The live cells were preincubated with metabolites for 10 min and then treated with oxygen scavengers (P2O plus catalase) before addition of PAR2 agonist. After treatment with the agonist AP for 1 h, the cells were fixed with 4% paraformaldehyde. Cytosolic and total intensities of the antibody were measured with Image J (NIH). Relative internalization of PAR2 was calculated from the ratio of the cytosolic intensity to the total intensity of the antibody in each cell after subtracting the average baseline fluorescence ratio before incubation with drugs.
For dialysis combined with confocal microscopy, cells were patch-clamped using an Axopatch amplifier controlled by the HEKA Pulse program. Whole-cell pipette resistance was 4–6 MΩ. The tsA201 cell was dialyzed with different ratios of ATP/ADP in the pipette. While breaking into the cell, the membrane potential was held at −40 mV and maintained for the entire experiment. The internal (pipette) solution for whole-cell clamping contained (in millimolar) 130 KCl, 10 NaCl, 1 MgCl2, and 10 Hepes (pH = 7.43) with appropriate amounts of ATP and ADP. The total adenosine nucleotides (ATP plus ADP) was fixed at ∼3 mM.
Single-Cell pHi Imaging.
The pHi was measured with BCECF, pH-sensitive fluorescent dye, without any other optical probes to avoid spectral overlap. Cells were preincubated with 2 µM BCECF-AM for 20 min at room temperature. The BCECF was excited at 450 and 495 nm using a Polychrome IV Monochromator (Till Photonics) and the emission at 530 nm was recorded at every 12 s with the MetaFluor system (Universal Imaging). The background signal, measured from a cell-free area, was subtracted before estimation of the emission ratio (R = F495/F450). The ratio (R) was converted into pHi values using the equation (36)
where Ra and Rb are the ratios at pH 5.0 and 9.0, respectively. Ra, Rb, and Kd were measured using 10 µM nigericin, a K+–H+ exchanger, in solutions containing (in millimolar) 130 KCl, 10 NaCl, 1 MgCl2, at different pH values using MES (pH 5.0), 2-(cyclohexylamino)ethanesulfonic acid (pH 9.0), and Hepes (pH 7.0) buffers. Ra, Rb, and Kd were 1.7, 6.3, and 97 nM, respectively. To calibrate the pH dependence of PercevalHR (SI Appendix, Fig. S3), the intracellular pH of resting cells was quickly stepped away from neutrality by exposure to NH4Cl or by treatment with different pH buffers in the presence of nigericin (10 µM) as previously reported (9).
Measurement of Glucose Consumption Rate.
Glucose in the bath solution was measured by the OneTouch UltaMini (Amazon). For calibration of the instrument, we used known concentrations of glucose dissolved in normal external Ringer solution without glucose.
Static Lactate Assay.
The assay was performed according to the manufacturer’s instructions (K607-100; BioVision). Briefly, 70% confluent cells were prepared in 24-well plates. After removal of culture medium, cells were gently rinsed with normal external Ringer solution twice. Then, they were incubated with the normal Ringer solution for at least 10 min to reach metabolic equilibrium before addition of 25 mM glucose or 10 mM KIC. After removal of the Ringer solution, 0.5 mL high-glucose or KIC solution was added gently into the wells and incubated for 10 min or 10 s. Samples (0.2 mL) were collected from each well and refrigerated until analysis by plate reader. Fluorescence was excited at ∼530 nm, collected at ∼590 nm, and calibrated as described by the manufacturer. We could not obtain clear data with oxygen scavengers because the scavenger enzymes generate a bright signal without any lactate addition when the lactate probe was added.
Imaging of NAD(P)H and FAD Autofluorescences.
Cells were placed onto small glass chips coated with poly-l-ornithine for autofluorescence imaging. The excitation and emission wavelengths for the NAD(P)H autofluorescence in live cells were 360 nm and >470 nm, respectively (6). The signal was normalized by the control value before addition of any chemicals. To measure FAD autofluorescence, the cells were excited at 450 nm and emission light was collected using a dual band-pass filter at 520 and 685 nm (37).
Oxygen Sensing with MM2 Nanoparticle.
Oxygen concentration was measured by oxygen-sensing nanoparticles (MM2, 74161; Ibidi). The 5- × 5-mm small glass chips were coated with 1 mg/mL poly-l-ornithine for 30–60 min and then washed with distilled water. We prepared 1,000 µg/mL MM2 stock solution in distilled water and then diluted to 10 µg/mL to make the final MM2 solution in a proper buffer solution. Then, the poly-l-ornithine–coated chips were incubated with the diluted MM2 solution for 1–2 h. The MM2-coated chips were washed with the same Ringer solution. The chip was transferred into the imaging chamber and excited at 405 nm, and emission was collected at >470 nm. The gain of the electron-multiplying CCD (EMCCD) camera was adjusted to 100, and 405-nm UV light from the Till monochrometer was limited to 50 ms to reduce potential photobleaching issue for the intensity measurement. These experiments were controlled by the Metafluor imaging program. For calibration of the oxygen-sensitive nanoparticles we used the Stern–Volmer equation:
| [1] |
| [2] |
where F0 is the fluorescence intensity at zero oxygen, F270 is fluorescence intensity at 270 µM oxygen, F is fluorescence intensity at a certain oxygen level, and Ksv is the Stern–Volmer constant. We assumed that the 21% (air) saturated solution has ∼270 µM oxygen at ∼25 °C according to Henry’s law. To get fluorescence intensity at zero oxygen (F0), ∼100% nitrogen gas was blown over the top of the solution containing oxygen scavengers (7.5 units/mL P2O and 1,000 units/mL catalase). The blowing angle was important to deplete oxygen in the chamber because molecular nitrogen is lighter than air. To restrict the nitrogen gas locally, the chamber was covered with a 35-mm dish with a small inlet. From the fluorescence data at the two different oxygen tensions (0 and 270 µM oxygen), we first estimated the Ksv using Eq. 1. Finally, we could convert the whole intensity trace of MM2 to oxygen concentration using Eq. 2.
Single-Molecule Live-Cell Imaging with TIRF Microscopy.
No.1 coverslips of 25-mm diameter were autoclaved and coated with poly-l-ornithine (1 mg/mL; Sigma-Aldrich). Longer coating times favor cell flattening. For single-molecule experiments, we usually coated for ∼30 min. Most of the background signal from organic molecules bleached quickly after exposure to the laser light. The Alexa647-labeled primary antibody against PAR2 receptors was applied for 5–15 min at room temperature. Shorter antibody incubation times reduced nonspecific binding of the primary antibody to glass surfaces. The remaining free antibody was washed out at least three times with normal saline-buffered solution. For the bleaching experiments shown in Fig. 2, cells transiently expressed PAR2-GFP (0.5 µg cDNA per plate). For two-color imaging in Fig. 6, cells transiently expressed clathrin light chain (CLC) with dual SNAP-tags at the N and C termini (SNAP-CLC-SNAP, 0.05 µg, Addgene no. 38012; ref. 38) and dark PAR2 (without GFP tag, 0.05–0.15 µg cDNA per plate). For SNAP tag labeling, we used cell-permeable SNAP-cell 505-star dye (New England Biolabs Inc.), incubating cells for 45 min with 2.5 µM dye in cell culture medium in the 37 °C CO2 incubator and then washing three times with the culture medium. The dye-labeled cells were treated 3–4 h later with Alexa647-PAR2 primary antibody for 5 min to mark PAR2 receptors. SNAP dye nonspecifically attached to the glass surface was quickly bleached by 488-nm laser compared with the dye tagged on the SNAP-CLC-SNAP protein in live cells. Single-molecule TIRF images were taken on a custom-built microscope based on a Nikon Eclipse TE2000 microscope base with an APO TIRF 100×/1.49 N.A. objective. A 641-nm laser was used for the illumination. Images were collected by an EMCCD ixon897 camera (Andor) and a custom controller written in LabVIEW as described previously (39). Dual-color imaging used two excitation lasers (488 and 641 nm) and two EMCCD cameras collecting emission at the same time. Multifluorescence solid latex beads (0.5 µm; Polysciences, Inc.) were utilized for correcting position differences in the two EMCCD cameras in ImageJ (NIH). To estimate diffusion coefficient, we used a MATLAB-based program to track single PAR2 molecules (40). The lifetime of transient binding in Fig. 6B, Inset was obtained from 70 events in ImageJ (NIH) and then fitted with a single-exponential curve to get the time constant in Igor Pro-6.02 (Wavemetrics).
Data Analysis.
All statistical significance tests were performed with Student t test in Microsoft Excel. All data averaged in the Microsoft Excel were plotted with SEM in Igor Pro-6.02 (Wavematrics).
Supplementary Material
Acknowledgments
We thank Lea M. Miller for technical assistance; Drs. Tianyi Yuan, Yongdeng Zhang, and Liangyi Chen for advice and use of their MATLAB program for the tracking of single receptors; and Drs. Liangyi Chen, James Hurley, and Linda Wordeman for reading our manuscript and giving us critical comments. Our work was supported by NIH Grants R01-DK080840 (to D.-S.K.), R01-GM083913 (to B.H.), R37-NS08174 (to B.H.), R01-GM079373 (to C.L.A.), P01-GM105537 (to C.L.A.), and 1S10RR026406 (to C.L.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1717724115/-/DCSupplemental.
References
- 1.Dempsey GT, Vaughan JC, Chen KH, Bates M, Zhuang X. Evaluation of fluorophores for optimal performance in localization-based super-resolution imaging. Nat Methods. 2011;8:1027–1036. doi: 10.1038/nmeth.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuman KC, Chadd EH, Liou GF, Bergman K, Block SM. Characterization of photodamage to Escherichia coli in optical traps. Biophys J. 1999;77:2856–2863. doi: 10.1016/S0006-3495(99)77117-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roy R, Hohng S, Ha T. A practical guide to single-molecule FRET. Nat Methods. 2008;5:507–516. doi: 10.1038/nmeth.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Voet D, Voet JG. Biochemistry. 3rd Ed. Vol 22. Wiley; New York: 2004. p. 802. [Google Scholar]
- 5.Gao Z, et al. Distinguishing features of leucine and alpha-ketoisocaproate sensing in pancreatic beta-cells. Endocrinology. 2003;144:1949–1957. doi: 10.1210/en.2002-0072. [DOI] [PubMed] [Google Scholar]
- 6.Jung SR, et al. Reduced cytochrome C is an essential regulator of sustained insulin secretion by pancreatic islets. J Biol Chem. 2011;286:17422–17434. doi: 10.1074/jbc.M110.202820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klysz D, et al. Glutamine-dependent α-ketoglutarate production regulates the balance between T helper 1 cell and regulatory T cell generation. Sci Signal. 2015;8:ra97. doi: 10.1126/scisignal.aab2610. [DOI] [PubMed] [Google Scholar]
- 8.Tantama M, Martínez-François JR, Mongeon R, Yellen G. Imaging energy status in live cells with a fluorescent biosensor of the intracellular ATP-to-ADP ratio. Nat Commun. 2013;4:2550. doi: 10.1038/ncomms3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tantama M, Yellen G. Imaging changes in the cytosolic ATP-to-ADP ratio. Methods Enzymol. 2014;547:355–371. doi: 10.1016/B978-0-12-801415-8.00017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shi X, Lim J, Ha T. Acidification of the oxygen scavenging system in single-molecule fluorescence studies: In situ sensing with a ratiometric dual-emission probe. Anal Chem. 2010;82:6132–6138. doi: 10.1021/ac1008749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aitken CE, Marshall RA, Puglisi JD. An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys J. 2008;94:1826–1835. doi: 10.1529/biophysj.107.117689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swoboda M, et al. Enzymatic oxygen scavenging for photostability without pH drop in single-molecule experiments. ACS Nano. 2012;6:6364–6369. doi: 10.1021/nn301895c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leitner C, Volc J, Haltrich D. Purification and characterization of pyranose oxidase from the white rot fungus Trametes multicolor. Appl Environ Microbiol. 2001;67:3636–3644. doi: 10.1128/AEM.67.8.3636-3644.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fercher A, Borisov SM, Zhdanov AV, Klimant I, Papkovsky DB. Intracellular O2 sensing probe based on cell-penetrating phosphorescent nanoparticles. ACS Nano. 2011;5:5499–5508. doi: 10.1021/nn200807g. [DOI] [PubMed] [Google Scholar]
- 15.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 16.Suh BC, Hille B. Recovery from muscarinic modulation of M current channels requires phosphatidylinositol 4,5-bisphosphate synthesis. Neuron. 2002;35:507–520. doi: 10.1016/s0896-6273(02)00790-0. [DOI] [PubMed] [Google Scholar]
- 17.Thore S, Wuttke A, Tengholm A. Rapid turnover of phosphatidylinositol-4,5-bisphosphate in insulin-secreting cells mediated by Ca2+ and the ATP-to-ADP ratio. Diabetes. 2007;56:818–826. doi: 10.2337/db06-0843. [DOI] [PubMed] [Google Scholar]
- 18.Balla T. Phosphoinositides: Tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93:1019–1137. doi: 10.1152/physrev.00028.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jung SR, et al. Contributions of protein kinases and β-arrestin to termination of protease-activated receptor 2 signaling. J Gen Physiol. 2016;147:255–271. doi: 10.1085/jgp.201511477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Titov DV, et al. Complementation of mitochondrial electron transport chain by manipulation of the NAD+/NADH ratio. Science. 2016;352:231–235. doi: 10.1126/science.aad4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tao R, et al. Genetically encoded fluorescent sensors reveal dynamic regulation of NADPH metabolism. Nat Methods. 2017;14:720–728. doi: 10.1038/nmeth.4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ricks TK, Trejo J. Phosphorylation of protease-activated receptor-2 differentially regulates desensitization and internalization. J Biol Chem. 2009;284:34444–34457. doi: 10.1074/jbc.M109.048942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoncu R, et al. Loss of endocytic clathrin-coated pits upon acute depletion of phosphatidylinositol 4,5-bisphosphate. Proc Natl Acad Sci USA. 2007;104:3793–3798. doi: 10.1073/pnas.0611733104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimm JB, et al. A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat Methods. 2017;14:987–994. doi: 10.1038/nmeth.4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stennett EM, Ciuba MA, Levitus M. Photophysical processes in single molecule organic fluorescent probes. Chem Soc Rev. 2014;43:1057–1075. doi: 10.1039/c3cs60211g. [DOI] [PubMed] [Google Scholar]
- 27.Yan L, et al. Bright and stable near-infrared pluronic-silica nanoparticles as a contrast agent for in vivo optical imaging. J Mater Chem B Mater Biol Med. 2016;4:5560–5566. doi: 10.1039/C6TB01234E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cai E, et al. Stable small quantum dots for synaptic receptor tracking on live neurons. Angew Chem Int Ed Engl. 2014;53:12484–12488. doi: 10.1002/anie.201405735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SH, et al. Super-resolution imaging of synaptic and extra-synaptic AMPA receptors with different-sized fluorescent probes. eLife. 2017;6:e27744. doi: 10.7554/eLife.27744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu J, Rong Y, Kuo CT, Zhou XH, Chiu DT. Recent advances in the development of highly luminescent semiconducting polymer dots and nanoparticles for biological imaging and medicine. Anal Chem. 2017;89:42–56. doi: 10.1021/acs.analchem.6b04672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin PM, et al. Identity of SMCT1 (SLC5A8) as a neuron-specific Na+-coupled transporter for active uptake of L-lactate and ketone bodies in the brain. J Neurochem. 2006;98:279–288. doi: 10.1111/j.1471-4159.2006.03878.x. [DOI] [PubMed] [Google Scholar]
- 32.Yu H, Dickson EJ, Jung SR, Koh DS, Hille B. High membrane permeability for melatonin. J Gen Physiol. 2016;147:63–76. doi: 10.1085/jgp.201511526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott AC, Trebilcock R, Yates AP, Best L. Stimulation of HIT-T15 insulinoma cells by glyceraldehyde does not require its metabolism. Eur J Biochem. 1993;213:359–365. doi: 10.1111/j.1432-1033.1993.tb17769.x. [DOI] [PubMed] [Google Scholar]
- 34.Davies J, Tomlinson S, Elliott AC, Best L. A possible role for glyceraldehyde transport in the stimulation of HIT-T15 insulinoma cells. Biochem J. 1994;304:295–299. doi: 10.1042/bj3040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fan J, et al. Glutamine-driven oxidative phosphorylation is a major ATP source in transformed mammalian cells in both normoxia and hypoxia. Mol Syst Biol. 2013;9:712. doi: 10.1038/msb.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung SR, Kim K, Hille B, Nguyen TD, Koh DS. Pattern of Ca2+ increase determines the type of secretory mechanism activated in dog pancreatic duct epithelial cells. J Physiol. 2006;576:163–178. doi: 10.1113/jphysiol.2006.114876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bartolomé F, Abramov AY. Measurement of mitochondrial NADH and FAD autofluorescence in live cells. Methods Mol Biol. 2015;1264:263–270. doi: 10.1007/978-1-4939-2257-4_23. [DOI] [PubMed] [Google Scholar]
- 38.Jones SA, Shim SH, He J, Zhuang X. Fast, three-dimensional super-resolution imaging of live cells. Nat Methods. 2011;8:499–508. doi: 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sarangapani KK, et al. Sister kinetochores are mechanically fused during meiosis I in yeast. Science. 2014;346:248–251. doi: 10.1126/science.1256729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yuan T, et al. Diacylglycerol guides the hopping of clathrin-coated pits along microtubules for exo-endocytosis coupling. Dev Cell. 2015;35:120–130. doi: 10.1016/j.devcel.2015.09.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.