Significance
Cancer alters both the morphological and the biochemical properties of multiple cell types in a tissue. Generally, the morphology of epithelial cells is practical for routine disease diagnoses. Here, infrared spectroscopic imaging biochemically characterizes breast cancer, both epithelial cells and the tumor-associated microenvironment. Unfortunately, conventional spectral analyses are slow. Hence, we designed and built a laser confocal microscope that demonstrates a high signal-to-noise ratio for confident diagnoses. The instrument cuts down imaging time from days to minutes, making the technology feasible for research and clinical translation. Finally, automated human breast cancer biopsy imaging is reported in ∼1 hour, paving the way for routine research into the total tumor (epithelial plus microenvironment) properties and rapid, label-free diagnoses.
Keywords: spectroscopy, imaging, breast, cancer, pathology
Abstract
Histopathology based on spatial patterns of epithelial cells is the gold standard for clinical diagnoses and research in carcinomas; although known to be important, the tissue microenvironment is not readily used due to complex and subjective interpretation with existing tools. Here, we demonstrate accurate subtyping from molecular properties of epithelial cells using emerging high-definition Fourier transform infrared (HD FT-IR) spectroscopic imaging combined with machine learning algorithms. In addition to detecting four epithelial subtypes, we simultaneously delineate three stromal subtypes that characterize breast tumors. While FT-IR imaging data enable fully digital pathology with rich information content, the long spectral scanning times required for signal averaging and processing make the technology impractical for routine research or clinical use. Hence, we developed a confocal design in which refractive IR optics are designed to provide high-definition, rapid spatial scanning and discrete spectral tuning using a quantum cascade laser (QCL) source. This instrument provides simultaneously high resolving power (2-μm pixel size) and high signal-to-noise ratio (SNR) (>1,300), providing a speed increase of ∼50-fold for obtaining classified results compared with present imaging spectrometers. We demonstrate spectral fidelity and interinstrument operability of our developed instrument by accurate analysis of a 100-case breast tissue set that was analyzed in a day, considerably speeding research. Clinical breast biopsies typical of a patients’ caseload are analyzed in ∼1 hour. This study paves the way for comprehensive tumor-microenvironment analyses in feasible time periods, presenting a critical step in practical label-free molecular histopathology.
Histopathology is essential for both research and clinical management in a variety of diseases, including cancer. The diagnostic process relies on staining thin tissue sections, followed by a pathologist manually recognizing epithelial morphology and patterns using an optical microscope. While carcinomas originate in epithelial cells, it is now well understood that stromal (both cellular and extracellular) characteristics aid cancer progression (1, 2) and determine clinical outcomes (3, 4). Although holding tremendous potential for observing, understanding, and treating cancer, stromal changes are not routinely used for research or clinical diagnoses. This arises from a lack of practical technology to capture morphological and biochemical patterns; multiplex staining is time consuming and expensive, generally limited to proteins, and results are difficult to interpret. Computerized pattern recognition has been reported (4) to be effective in utilizing stromal signatures, but digital data have been limited to simple structural data or highly detailed but time-consuming molecular measurements (5–11). Methods based on optical vibrational spectroscopy (12–14) provide an avenue as they offer the same image scale and quality as traditional pathology, can be coupled to computational analysis (15), show contrast between different cell types and disease (16), and are sensitive to changes in both cells and extracellular material (17, 18). Moreover, label-free spectroscopic imaging does not perturb the tissue samples in any way and can be computed to resemble conventional stained (H&E or molecular) images, enabling integration into clinical or research workflows (12). Laboratory studies using tissue microarrays (TMAs) using Fourier transform infrared (FT-IR) imaging (19–21), for example, have provided extensive demonstration of this potential. Fundamental molecular vibrations provide strong signals across a spectral region (800–4,000 cm−1) 20-fold larger than the visible spectrum, making it efficient and ideally suited to thin sections commonly used in pathology.
Despite extensive histologic studies, there are no reports of tumor staging and tumor microenvironment use for (i) diagnostically relevant accuracies, and (ii) reasonable analysis times. These two factors are related. Data acquisition is slow compared with optical microscopy as thousands of spectral frequencies need to be acquired (22). A necessary trade-off to make imaging practical is to use larger pixel sizes, but large pixels result in biochemical averaging of the heterogeneous microenvironment reducing the chemical contrast. This spatial averaging over a 10-μm scale in turn necessitates exceptionally high signal-to-noise ratio (SNR) to resolve small differences, complex analysis models (23), and high computational overhead. The limits of pixel sizes are now established, with rigorous optical modeling fueling new instrumentation that provides so-called high-definition (HD) (24, 25) images, in which pixels are ∼25–100 times smaller than previously used. While understanding of optical design has been transformed, two unresolved issues persist: first, n-fold smaller pixel areas require n-fold more pixels scanned per specimen and n-fold less available light (SNR) per pixel; together, these imply ∼n2 larger scan time just to maintain SNR. Second, there is no evidence yet to show that HD imaging offers an analytical advantage beyond image quality. The smaller pixels may well need higher SNR, and subcellular heterogeneity may need yet more complex computations. Together, there appears to be no obvious pathway with current technology to clinically feasible, HD imaging that allows confident information extraction from both the tumor and microenvironment in real time.
This study focuses on three major advances: discovery of diagnostically useful tumor microenvironment classes along with precise detection of epithelial stages, development of a high-performance discrete-frequency IR (DFIR) microscope, and a demonstration of HD chemical characterization of human biopsies in clinically feasible times. First, we seek to develop confident tumor and microenvironment detection using HD FT-IR imaging. While there have been prior studies on breast tissue and its microenvironment using IR imaging (26–31), there are no reports of clinically feasible epithelial and microenvironment protocols. One challenge is to acquire consistent information that allows computerized recognition despite the tissue heterogeneity arising from small pixels. The underlying complexity is that the types and number of classes of microenvironment associated with disease are unknown. Hence, the first section of this study focuses on discovering a small number of characteristic microenvironment responses to cancer, potentially making diagnoses feasible. This fundamental understanding can broadly impact pathology as a step toward utilizing microenvironment information but must be translated to an approach with practical imaging times. Hence, second, we seek unique instrumentation to overcome current limitations. Recently, high-intensity, broadly tunable quantum cascade laser (QCL) sources have enabled rapid DFIR imaging (32–35), especially for problems in which the data dimensionality can be significantly reduced (36) by using a smaller set of frequencies (37–42). In current designs, however, laser coherence can degrade image quality and can subtly distort spectra. This distortion affects pattern recognition accuracy in an unpredictable manner due to its dependence on local structure and/or SNR. We systematically address these challenges by engineering an IR microscope. While providing unique analytical capabilities, this advance is useful for tumor staging and microenvironment analysis in breast cancer. We subsequently validate this confocal microscope design and ensure its capabilities in providing diffraction limited imaging with minimal noise and aberration. This enables accurate histological and pathological segmentation of tissue that was not previously possible. Next, we adapt the FT-IR epithelial subtyping and stromal differentiation using a discrete-frequency (DF) model to this instrument and assess its accuracy. Finally, in the third part of this report, we image tissue biopsy samples and demonstrate automated tumor-microenvironment classifications for breast cancer in clinically feasible time.
Results
Epithelial Subtyping and Microenvironment Detection of Breast Cancer.
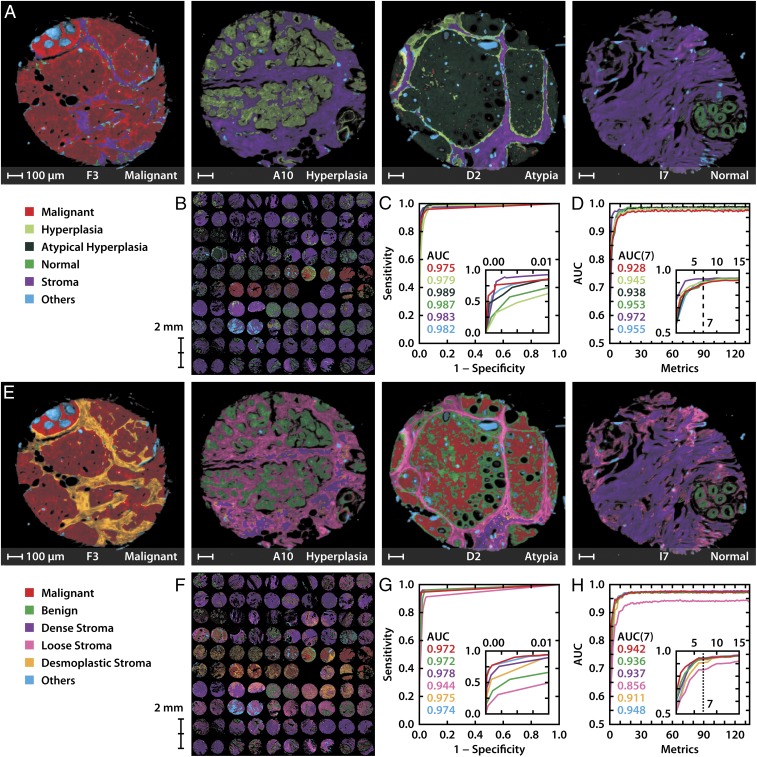
We developed a machine learning approach that provides detailed cell-level tumor diagnoses and discovers tumor-associated microenvironments. We first designed a study using TMAs with high disease diversity and accompanying heterogeneity of microenvironments. These samples were imaged with both FT-IR (unstained) and optical microscopy (stained), providing new spectral data and conventional histochemical diagnosis. Unperturbed by staining or any other intervention, broadband FT-IR spectroscopic imaging data provided 2,048 spectral data points per pixel and ∼106 pixels per patient sample. A board-certified pathologist carefully labeled cells and disease states for each sample with the help of stains, providing a ground truth for cellular and disease diagnoses. Both IR and pathology data were used to develop models and pattern recognition protocols, as described in Methods, to recognize different disease states within epithelium and stromal indicators of disease. The tissue sample is mainly divided into epithelial and the stromal compartments. The remaining cellular moieties such lymphocytes, red blood cells, secretions, mucin, and necrosis are grouped together in the “others” class. Our disease models directly relate to clinical pathology: as breast cancer is broadly characterized by four epithelial subtypes, namely, hyperplasia, atypical hyperplasia, invasive, and normal, we use the same. A typical diagnosis then consists of the four subtypes, the microenvironment (or collagen-dominated “stroma”) and an others class to account for the remaining cell types. The addition of the others class to the model increases the prediction accuracy with precise allocation of the stromal and epithelial regions. We term this the 6-class epithelial-focus (6E) model. The first question is whether the 6E model can accurately replicate clinical diagnoses. Going beyond conventional diagnoses, potential exists to develop interesting models for the microenvironment, but no clinical consensus on types or features of such changes exists. Stroma typically consists of dense, loose, and desmoplastic components that might contain cellular and acellular materials, including activated and inactivated stromal cells, inflammatory cells, fibroblasts, blood vessels, and extracellular matrix creating a diverse tumor microenvironment (43, 44). We assessed several simple to complex models that illustrate various aspects of tissue and disease; finally, we struck a balance between this complexity and the four epithelial subtypes. Here we report a parallel 6-class stromal (6S) model (dense, loose stroma and desmoplastic, other, and two epithelial classes—benign and malignant). Both the 6E and 6S models are described in detail in SI Appendix, Fig. S4. Together the 6E and 6S models will comprehensively characterize breast tissue. While the 6E model relates to conventional diagnoses, 6S provides distinctive insight.
For each of the two (6E, 6S) models, we developed numerical methods to classify each tissue pixel into one of the six classes. The process consists of acquiring full spectral data from a large number of diverse samples using TMAs, statistical formulation of spectral metrics that characterize tissues and can be interpreted biochemically. A sequential forward search approach is used to find the metrics that maximize the accuracy of prediction of the algorithm compared with pathologist marking, and quantitative enumeration of the results. The results are reviewed by a pathologist as well as quantitative statistical measures, as shown in Fig. 1 A and E, which compare automated tissue classifications for the 6E and 6S models. An entire TMA, with the 6E and 6S classifications shown in Fig. 1 B and F, respectively, can be seen in high resolution in SI Appendix, Figs. S1 and S2. A comparison of select samples with their corresponding H&E-stained images is shown in SI Appendix, Fig. S3. While spatially corresponding to H&E images, IR images provide greater capability and information. Unlike H&E images that are analyzed by a pathologist, first, IR analysis is all in silico and a prime example of fully digital pathology. Second, the microenvironment mapping is not manually possible. Third, the accuracy of diagnoses can be quantitatively assessed using receiver operating characteristic (ROC) curves as shown in Fig. 1 C and G for 6E and 6S models, respectively. The correlation between the pathologic state of the 6E model and stromal composition of the 6S model is apparent. For example, the malignant epithelium in Fig. 1E (F3) is accompanied by desmoplastic stroma while the hyperplasic biopsy (A10) is surrounded by loose and dense stroma. Stromal changes provide opportunities for differentiation of the clinically difficult and atypical cases from malignant, illustrating the value of chemical imaging in enabling precise diagnosis and decision-making. Finally, we sought to understand the prediction potential of the formulated spectral metrics. Fig. 1 D and H show that a quantitative measure of accuracy, namely the area under the curve (AUC) of the ROC curve, can be used to determine small number of metrics (∼10) that are sufficient for accurate segmentation. While more complex models for breast pathology or that of other tissues may be examined, this plot is generally representative of tissue classification as a function of spectral features, as shown in a number of studies (19, 45–47). The achievable classification accuracy is typically seen to saturate for a number of spectral frequencies (∼20 frequencies are used to build ∼10 metrics) that is much smaller than the number of spectral frequencies typically acquired in an FT-IR spectrum (∼2,000). This result facilitates the acquisition of only 12 frequencies needed and enables the use of emerging DF instruments for faster, yet accurate, digital pathology. For the optimal set of frequencies and their assignments, see SI Appendix, Tables S4 and S5.
Fig. 1.
Epithelial tumor classification and microenvironment models for breast cancer characterization using FT-IR imaging. Using the 6-class epithelial (6E) model, (A) classified images showing malignant, hyperplasia, atypia, and normal tissue samples, (B) 6E class image of the full tissue microarray (TMA), (C) the corresponding receiver operating characteristic (ROC) curve, and (D) area under the curve (AUC) of the ROC curve dependence on spectral metrics for the 6E model. The Inset shows that the accuracy saturates for a few metrics. The complementary 6-class stromal (6S) model on the same samples shows (E) disease-associated microenvironment changes, (F) 6S classification of the TMA, (G) the corresponding ROC curve, and (H) the impact of spectral metrics on AUC values for the 6S model.
We emphasize that low SNR does not permit accurate classification, as previously reported (48) and demonstrated with two illustrative examples in SI Appendix, Table S1. The apparent SNR for DF systems reported previously has ranged from ∼10 in early systems (49) to ∼100 using expensive cooled detectors in the latest state of the art (35). While large-format array detectors offer rapid imaging, the typical SNR cannot approach the levels reported here using a single-element detector. Even if laser stability or innovative designs allowed signal averaging to improve the SNR, the additional measurements will result in needing the same time as an FT-IR imaging experiment, providing no specific advantage to DFIR systems. A less discussed, but known issue, with using lasers is that coherence-induced speckle acts as a noise source since the underlying tissue structure changes from sample to sample. Speckle contributions may not appreciably manifest in images but do affect spectra. The variance introduced by speckle remains to be quantified using theory (25), but current estimates are of a few percent, which will not allow accurate histologic recognition. Thus, while it is possible to translate the FT-IR imaging results to DF instruments in principle, there are no reports yet of histopathological models with the complexity presented here being possible with speckle-free laser-based instruments using these few frequencies.
High-Performance IR Microscopy—a Confocal, Spectral-Spatial Scanning System.
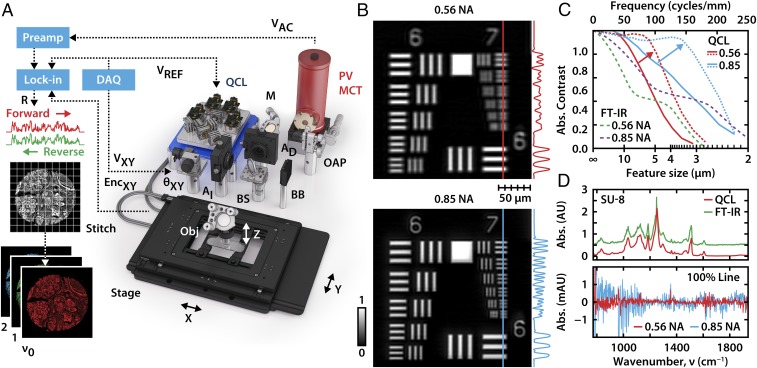
To address the need for a low-noise, speckle-insensitive, and diffraction-limited microscope, we developed a design precisely to address these shortcomings and enable translational studies. Our microscope, shown in Fig. 2A, consists of a multilaser unit spanning the mid-IR fingerprint region from 770 to 1,940 cm−1, interchangeable high numerical aperture (N.A.) optics with 0.56 N.A. and 0.85 N.A. image formation lenses, a cryogenically cooled, single-element mercury cadmium telluride (MCT) detector, and a high-speed linear motor microscopy stage. Each aspect of this design is optimized to overcome current drawbacks. First, we utilize the high brightness of the laser to provide confocal illumination using high-N.A. lenses and use apertures to limit the focal area. The confocal geometry not only provides spatial localization but also is effective in rejecting out-of-focus light as well as reduced speckle due to limited sample area illumination. Second, the use of a cooled single-element detector provides high SNR data due its superior noise characteristics compared with array detectors as well as the ability to lock into the modulated QCL pulsed signal. Third, real-time controls are integrated into the system to maintain high fidelity. Since the confocal geometry is effective in rejecting out-of-focus light, slight tilts in the sample can drastically affect the image’s focus while the high-speed stage is scanning the sample over large areas. We developed and integrated software controls that make axial adjustments to keep the sample in focus in real time, which also compensates for chromatic aberration and provides optimal SNR at every point. In addition to optimizing imaging performance, we also optimized spectral fidelity by resetting laser parameters at every wavenumber to the optimal repetition rates, pulse widths, and pointing alignment. The instrument can scan at up to 300 mm/s; for each pixel during the scan, axes encoders trigger detection electronics locked into the laser’s pulse frequency, allowing images to be acquired at a magnification limited only by the encoder resolution. The pixel size is optimally set at 1 μm, no larger than half the size of the point spread function (PSF) at the shortest tunable wavelength according to the Nyquist sampling criterion. The scanning is optimized for a “long direction” that limits acceleration–deceleration effects and achieve high pixel rates. Finally, the raw data undergo a multistep postprocess to obtain absorbance data from a background, integrate by aligning forward and backward stage sweeps using a nonrigid alignment algorithm that compensate for stage-induced aberrations, and interband alignment to account for instrument drift over time. If needed, the data can be deconvolved (50) against a simulated PSF at each wavenumber to further improve visualization.
Fig. 2.
Laser-based confocal IR microscopy. (A) The microscope consists of a quantum cascade laser (QCL) source, tunable for narrow-band emission across the mid-IR fingerprint region. A high-speed stage and focus unit raster scans the sample, while the detection is locked into the laser’s pulse rate and each pixel is triggered by the stage encoder counter. (B) Images of USAF 1951 resolution test targets show diffraction-limited performance for absorbance at 1,658 cm−1 with both the 0.56 N.A. objective (Top) at 2-μm pixel size and the 0.85 N.A. objective (Bottom) at 1-μm pixel size. (C) The optical contrast of each set of bars is plotted as a function of spatial frequency. The bars are no longer resolvable if the contrast drops below 26%, which corresponds to the Rayleigh criterion separation distance. The arrow indicates the deconvolution of the raw data (solid line) with the simulated PSF at the specified wavenumber to achieve a substantial resolution enhancement (dotted line). These results are compared with the simulated performance (dashed line) of a FT-IR instrument with optimized Schwarzschild objectives used in FT-IR imaging. (D) The unprocessed spectrum of a 5-μm layer of SU-8 epoxy acquired by our QCL instrument at 1 cm−1 resolution shows accurate spectral features compared with a reference FT-IR spectrometer. The 100% spectral profile lines show absorbance noise of ∼10−4 and 10−3 for the two objectives over most the fingerprint region.
Evaluation of Imaging Performance.
We sought to ensure spectral and spatial fidelity of the newly designed instrument. We first validated the spatial resolution against a USAF 1951 chrome-on-glass resolution target. The Rayleigh criterion is often used to determine the resolution of the instrument and signifies the minimum separation distance between two objects where they appear to remain distinct, corresponding to a contrast ratio of at least 26.4%. Accordingly, our system provides diffraction-limited resolution of 4 and 3 μm with the 0.56 and 0.85 N.A. objectives, as shown in Fig. 2B and seen in the absorbance contrast as a function of line spacing in Fig. 2C. After deconvolution, minor improvements in resolvability to 3 and 2 μm, respectively, are seen, but a drastic improvement in the contrast of structures up to 10 μm is observed, highlighting the potential to improve visualization. To our knowledge, the only comparable capability to this performance has previously been reported using synchrotrons (24). We further compared the experimental transfer functions with that of FT-IR instruments using optimized Schwarzschild objectives of the same N.A. using Code V optical design software. The underlying mechanism of image formation is apparent for a coherent source whose amplitude transfer function (ATF) has a flat frequency response followed by a sharp cutoff dependent on the aperture size of the pupil. As the beam coherence decreases, the transfer function will approach optical transfer function (OTF) behavior as seen with our measurements. Since the OTF of an instrument with an incoherent source is the autocorrelation of the ATF, the contrast gradually declines to zero at twice the ATF’s cutoff frequency. However, the higher cutoff frequency of these simulated incoherent systems falls below the Rayleigh criterion threshold where the features are indistinguishable. In FT-IR imaging instruments, the lower spatial frequencies are attenuated because the Schwarzschild objectives have an obscuration created by the secondary reflector and this presents in the simulations as an additional loss in contrast. Above the Rayleigh resolution, our confocal microscope outperforms and has substantially higher contrast levels compared with the current state-of-the-art IR imaging instruments. The spectral fidelity of the instrument was tested by measuring an epoxy photoresist, at 1 cm−1 resolution from 770 to 1,940 cm−1. The measurement accurately tracks the reference spectrum acquired with a FT-IR spectrometer, as indicated in the spectra in Fig. 2D. To provide the most representative noise characteristics, the spectra and SNR calculations are from data without deconvolution. Deconvolution provides substantial smoothing of the background as well as feature sharpening. Without postprocessing, we achieve noise levels on the order of 10−4 and 10−3 at 0.56 and 0.85 N.A., respectively. This data quality is not feasible in an FT-IR imaging system and has not been previously reported for an IR microscope.
High-Resolution and Rapid IR Imaging for Breast Tissue Segmentation.
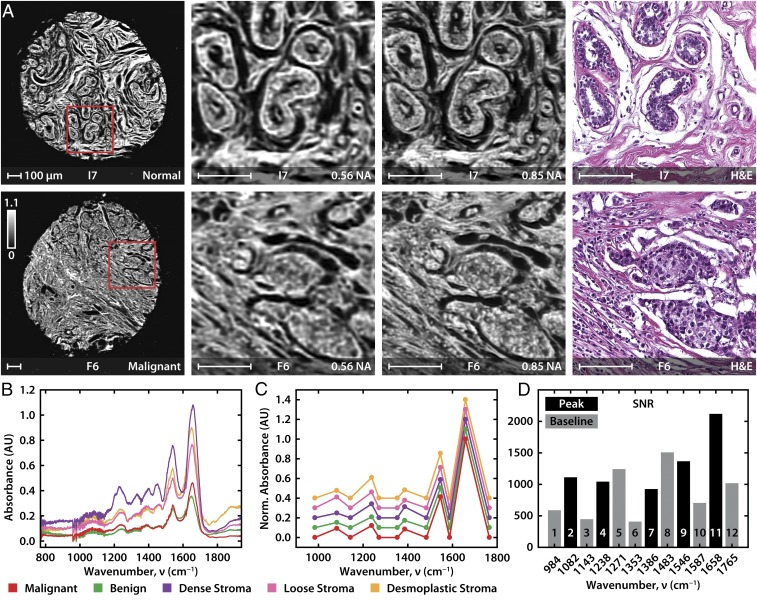
We imaged breast tissue previously analyzed by FT-IR imaging using the confocal microscope. Representative images and spectral data are shown in Fig. 3 A and B, respectively. In Fig. 3A, both the 0.56 and 0.85 N.A. objectives provide good quality data that match well with the conventional H&E images; the 0.85 N.A. images are sharper, as expected. Representative spectra from histologic units in breast tissue (Fig. 3B) match those acquired from FT-IR imaging and show the subtle spectral differences that permit accurate classification. We selected 12 DFs from the FT-IR 6E and 6S models (Fig. 3C) and also show differences in major classes as well as an average SNR of ∼1,000 for a single scan (Fig. 3D). This SNR enables high accuracy tissue classification with fewer frequencies that is only achieved in FT-IR imaging systems by extensive signal averaging and using mathematical noise rejection that needs hundreds of frequencies to estimate noise characteristics. To quantify the comparison between the FT-IR and confocal laser microscopes, scanning a full TMA shown here with a typical commercial FT-IR imaging system would require nearly 25,000 individual frames of the 128 × 128 array detector to cover the area and averaging each frame 32 times each (1.8 min of total acquisition) in a total acquisition time of ∼40 d. Due to the weak radiance from a FT-IR globar source, a large number of coadditions are required to achieve the required SNR for accurate tissue classification. To further increase the SNR, an additional processing time (30 s per frame) is needed using the most effective noise rejection algorithms available today (51), which adds another ∼10 d. The high SNR of our system allows us to avoid signal averaging and dispenses entirely with the need for numerical noise rejection. Thus, the microscope reported here permits imaging and analysis of the full TMA in feasible time (8 h), cutting the time to validate imaging results from months to 1 d and greatly accelerating the potential of IR imaging for biological research. This ∼50-fold reduction in time to acquire the needed quality data with the required set for obtaining a decision-ready image presents a practical comparison between conventional approaches and this instrument.
Fig. 3.
High-definition imaging of breast tissue. (A) A selection of one normal and one malignant tissue sample from a 20- × 20-mm TMA, acquired at 0.56 and 0.85 N.A. at 2 and 1 μm per pixel, respectively. The absorbance at 1,658 cm−1, indicative of the amide I vibrational mode, is shown in the image and enlarged subsections that are compared with an H&E-stained image of a serial section. (B) Point spectra from five tissue types at 1 cm−1 resolution; spectra are offset for clarity. (C) Normalized and baselined point spectra from five tissue types at DFs. (D) Average SNR for important spectral features.
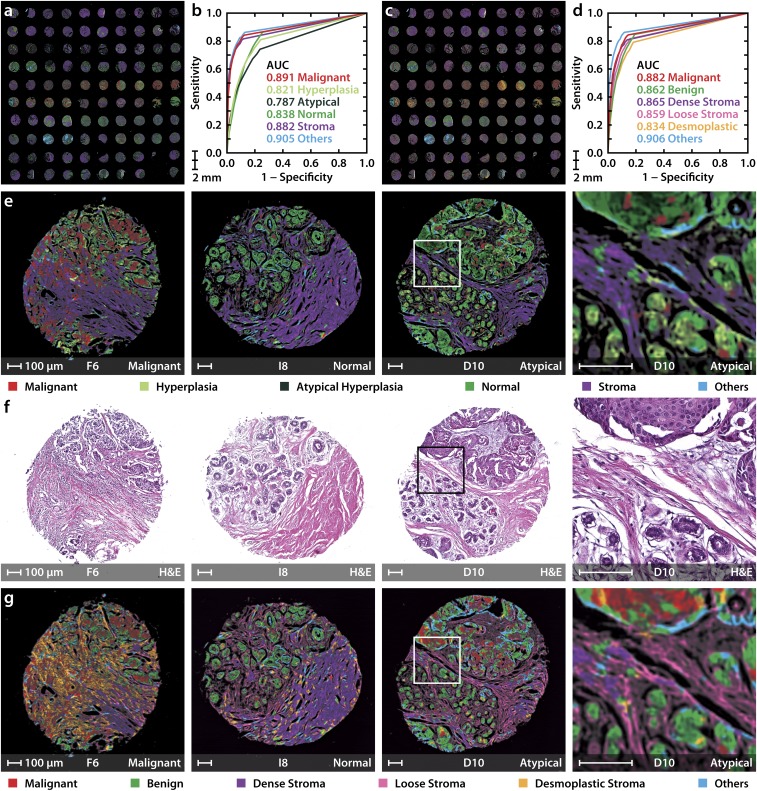
In addition to obtaining high SNR data, we demonstrate accurate classification of the TMA using the above discussed 6E and 6S models as shown in Fig. 4 A and C. The accuracy of both the 6E and 6S models is assessed by ROC curves, as shown in Fig. 4 B and D. The utility of IR imaging over conventional H&E images used in pathology can be seen in representative images (Fig. 4 E and G) from malignant to benign lesions. As opposed to H&E images that present only the structure and require pathologist decision-making, the classified IR images from the 6E model (Fig. 4E) provide ready visualizations of the diseased condition. This IR imaging approach offers an opportunity to be truly all digital, in recognition of disease and in quantification of the extent of disease, instead of manual staining, observation, and segmentation. While useful for visualizing routine samples, this digital approach is also an opportunity to leverage the recognition capabilities of computer algorithms for difficult cases or those of unclear pathology in limited tissue that is often available in modern biopsies. For example, epithelial hyperplasia with atypia (D10 in Fig. 4 E–G) is illustrative of a challenge facing pathologists. Although such cases are known to have specific histological features of malignancy (52), they are difficult to interpret and diagnose and are often confused with benign cases. In our approach, these samples are recognized as atypical hyperplasia in the 6E (epithelial) model. Moreover, the case is recognized as malignant epithelium surrounded by normal regions in the 6S (stromal) model. The 6S model also adds information that is not easily apparent in contemporary imaging (Fig. 4G). A desmoplastic reaction can be seen associated with malignant samples and normal stromal patterns can be seen in benign cases. Together, the 6E model shows efficacy for precise assessment of the diseased state, whereas, at times when the epithelial yield in biopsies is limited or inconclusive, the 6S model detects alterations in the surrounding stroma, offering unique avenues for precise and rapid detection of disease. This approach utilizing both the epithelial and stromal compartments enables analyses not possible without extensive, expensive, and time-consuming staining with multiple targets.
Fig. 4.
DF epithelial and stromal classification for rapid breast cancer diagnosis. (A) TMA classified using the 6-class epithelial (6E) model. (B) Receiver operating characteristic (ROC) curves represent the performance of each class in the 6E model. (C) TMA classified using the 6-class stromal (6S) differentiation model. (D) ROC curves for the 6S model. (E) 6E model classified images of three samples from the TMA with malignant, normal, and atypical hyperplasia states (Left to Right) along with their corresponding H&E-stained images in F. (G) Classification using the 6S model. A small region from the hyperplasia with atypia sample is also shown, along with its H&E stain, to demonstrate the spatial distribution of normal and malignant cells. The letter and numbers below each image correspond to the row and column of the TMA (A1 is the Top Left sample), respectively. All scale bars: 100 µm.
Clinical Translation to Biopsy and Surgical Specimens.
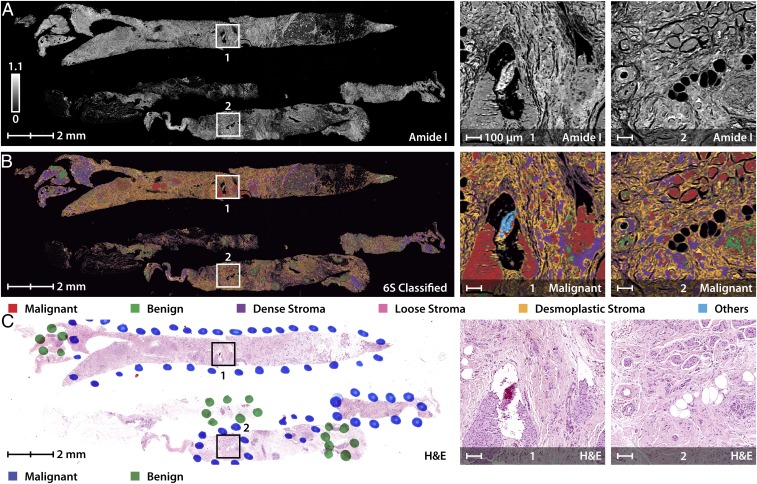
The impact of the three developments thus far—accurate HD histology, high-performance IR imaging, and fast histologic recognition—lies in a translation of the approach to typical samples in screening or operative care. We believe a primary utility of this technology will lie in rapidly triaging biopsies. Fig. 5 shows fast and accurate detection of tumor and characterization of the tumor microenvironment in needle biopsy sections. There are numerous implications of a clinically feasible (in both time and accuracy) imaging technology. With the microscope shown here, for example, biopsy results can now be provided on the day of biopsy itself, reducing wait times for patients. Combined with stainless staining techniques (13), both conventional and additional information can be provided to aid precise and accurate diagnoses. In atypical cases, additionally, both epithelial and stromal measurements (3) can be used to help pathologists make more confident decisions. This slide of biopsies shown in Fig. 5 can be imaged and classified in 3 h using the same 6-class technique discussed previously. We clearly see the spatial accuracy of the model wherein the cancerous regions are being classified with a majority of malignant epithelium surrounded by reactive collagen acting as further confirmation. Needle biopsies are the standard for breast cancer diagnosis and for many other solid tumors. Approximately 1.6 million biopsy samples are taken annually in the United States for breast cancer alone (53), with typical needle sizes of 14–22 gauge (54) (2- to 0.7-mm-diameter biopsy). There is an emphasis on returning biopsy results within days both to confirm the diagnosis in patients with breast cancer and to rapidly inform the large majority of biopsied women who do not have cancer.
Fig. 5.
Rapid triaging of malignant sections using stainless imaging of human breast biopsy samples in feasible times. (A) Image of needle biopsy sections using absorbance at 1,658 cm−1 with specific malignant regions enlarged for clarity. (B) The multispectral image was classified using a 6-class model separating cancerous and normal epithelial cells from various collagen-rich stromal types (6S). (C) Pathologist annotations to the H&E-stained image of a consecutive section demonstrate agreement with the IR-classified image.
Discussion and Conclusions
Here, we demonstrate histologic recognition by high-resolution imaging at a competitive level of morphological detail as optical microscopy along with further information regarding disease state and its progression; instrumentation for high-performance DFIR microscopy; and, enabling histologic imaging in clinically feasible times. This study forms a critical step toward practical IR imaging toward digital pathology with the three advances. Given that we use molecular composition in detection of disease, as opposed to morphology in conventional pathology, the capability of IR imaging opens the possibility of additional disease analysis. The first major result is that higher resolutions offer increased analytical sensitivity where comprehensive computational models, including those more complicated than presented here, can be used to then relate back to conventional spatial images. While the smaller pixels do indeed provide greater chemical localization, the subcellular sensitivity also implies that the heterogeneity in cellular responses may prevent accurate classification. Here, we are able to find specific metrics that allow differentiation of cell types and disease despite this limitation. We emphasize that the 6E and 6S models we use here do not reflect the full extent of information possible by IR imaging. We selected these models as a compromise between detail and clinical utility. Their successful implementation, however, implies that development of more complex models is possible in subcellular domains, including those from 3D spectral imaging (55). While only epithelial identification is considered here as this is the dominant site of origin in breast cancer, we note that cancer can arise in other cell types like mesenchymal cells (bone, cartilage, nerve), hematopoietic cells (lymphoma, leukemia), and germ cells (testicular cancer). The instrumentation and methods described here can also be used for other cases, but critical features of each disease will be different. Another possible outcome of this result is the association of subcellular changes with disease in both breast and other tissues that was not as definitively established using older technology (36). The sensitivity of both epithelial recognition and stromal engagement with cancer presents increased opportunities for clinical translation on one hand and a detailed view of the sample histology for research on the other. The quality of images provides confidence in spatial detail and can be further augmented by stainless-staining approaches as well as future tests based on stromal transformations. For research purposes, facile epithelial detection can aid in quantification of tumor volumes or identifying regions for further chemical analysis. In this regard, the stainless IR images preserve biochemistry for conventional analyses while providing information on epithelial as well as stromal transformations. While we have largely focused on the collagenous stroma, we anticipate that the 6S model’s demonstration will spur more detailed investigation into the microenvironment (4, 56) focusing on classification of other cell types as well.
While many optical scanning confocal designs are available for visible microscopy and several microscopy approaches available in the IR, enabling the use of a broadly tunable IR laser is not trivial. The large wavelength range in the mid-IR results in strong chromatic aberrations and makes design difficult using refractive optics; the use of reflective optics results in loss of light and weighting of collected signal on scattering due to the central obscuration in Schwarzschild objectives. Here, our optical design is combined with real-time control algorithms to reduce errors and achieve minimal-distortion images, demonstrating a performance that has not been previously seen for IR microscopy. While widefield configurations can typically provide greater speeds due to their multichannel advantage and increasingly larger sensor formats, area illumination using a laser typically suffers from a low SNR due to miniaturized detectors and presents speckle patterns across the image that can complicate spatial interpretation needed in pathology. The low noise in our single-element detector, ability to modulate the beam, and confocal geometry greatly reduce noise within each spectral band, allowing accurate classification models to be constructed from fewer number of DFs and without the need for extensive signal averaging or mathematical noise rejection. By providing enhanced spatial image quality, as shown by rigorous contrast analysis, the classified images allow for detailed tissue segmentation for tumor detection. The path to clinical translation is also enabled by the demonstration of HD histology and the instrument developed here. The key idea is that only a small number of spectral frequencies provide accurate and robust classification that allows us to overcome the slow data acquisition. The implementation of the presented approach on large biopsy sections emphasizes the potential of efficient clinical translation. Precise detection of the epithelial and stromal signatures in a few hours of tissue procurement can greatly complement and extend the capabilities of the current clinical practices. The combined instrument and epithelial–stromal segmentation opens possibilities for developing early detection automated algorithms, serve as a confirmation tool for diagnosis, especially facilitating the pathologists to focus on specific regions of interest. While focused on breast cancer, this work also paves the way for development of similar practical scanning for other tissues and disease types.
Methods
Sample Preparation.
A paraffin-embedded serial breast TMA (BR1003; US Biomax Inc.) consisting of a total of 101 cores of 1 mm diameter from 47 patients was obtained. Two sections were stained with H&E and smooth muscle actin and imaged with a light microscope. Corresponding 5-µm-thick adjacent unstained sections of the TMA were placed on a BaF2 salt plate for transmission FT-IR imaging and on a low-emissivity glass (MirrIR; Kevley Technologies) for reflective QCL-IR imaging. The sections were deparaffinized using a 16 h hexane bath.
The QCL confocal microscope was calibrated using a negative chrome on glass USAF 1951 test target (II-VI Max Levy) as well as a SU-8 photoresist target to evaluate spectral fidelity. The multispectral images were acquired in transflection configuration at 2-μm resolution with the 0.56 N.A. objective, at 1-μm resolution with the 0.85 objective, and at 12 distinct spectral frequencies from the fingerprint region. Point spectra were acquired at 1 cm−1 resolution.
FT-IR Imaging.
HD FT-IR imaging was conducted using a 680-IR spectrometer coupled to a 620-IR imaging microscope (Agilent Technologies) at 0.62 N.A. with a liquid-nitrogen–cooled MCT 128 × 128 focal plane array detector. Data were acquired over the 900–3,800 cm−1 spectral range and averaged over 32 scans per pixel. Afterward, the images were corrected against a background acquired in an empty space of BaF2 slide with 120 scans and Fourier transformed. The spectral resolution was 4 cm−1 with a pixel size of 1.1 µm. Resolutions Pro software was used for data collection and preliminary data processing. Each sample was imaged by raster scanning an ∼140- × 140-μm tile. Each of these tiles took ∼2 min for scanning and 30 s of processing. The individual spectroscopic image tiles were imported into ENVI + IDL 4.8 and mosaicked using in-house software. This was further processed using minimum noise fraction (MNF) for noise reduction. FT-IR images were manually labeled using correlation with the consecutive marked H&E-stained glass slide images under the supervision of a pathologist as ground truth for our analysis. A tissue mask based on intensity of amide I band was applied, to remove empty spaces and debris from further analysis.
QCL Confocal Microscope.
The QCL confocal microscope contains of a quantum cascade multilaser source (Block Engineering) that contains four individual tuner modules with beams combined into a single collinear output spanning the mid-IR fingerprint region from 770 to 1,940 cm−1. The general layout is illustrated in Fig. 1. The alignment of these tuners is assisted by a two-axis galvanometer pair (θXY) (6215H; Cambridge Technology). Imaging is performed by two interchangeable high-N.A. aspheric collimating lenses with 0.56 and 0.85 N.A. (LightPath Technologies) that focus the beam to a diffraction-limited spot. Complicated aberration corrected optics are not required as the instrument design compensates for many optical aberrations. When imaging via high-speed stage scanning (HLD117; Prior Scientific), all optics are illuminated with a zero-field angle where performance is optimal. Off-axis light rays due to scattering are rejected by the illumination (AI) and detection (AD) apertures limiting the focal area, thus increasing resolution and minimizing aberrations. The instrument also corrects for chromatic distortion using a calibration curve generated in optical design software (Code V; Synopsys). The instrument is also capable of running an autofocus subroutine per wavenumber by sweeping the axial position of the objective maximizing the signal.
The laser is split using a KBr beam splitter (BS) (Spectral Systems) with half discarded (BB) and the rest used to illuminate the sample in transflection mode. Light is absorbed by the sample and the remaining intensity is focused onto a cryogenically cooled, 0.5-mm active area, photovoltaic MCT detector with matched preamplification (MCT-13-0.5PV and MCT-1000PV; InfraRed Associates) using a 100-mm focal length parabolic mirror (OAP). After preamplification, the detector signal (VAC) is measured with a lock-in amplifier (MFLI; Zurich Instruments). A data acquisition card (PCIe-6361; National Instruments) generates the pulse reference frequency for the laser and the lock-in amplifier as well as the analog drive signal for the galvanometer pair. The demodulator samples are triggered by the stage encoder, and the magnitude (R) of the demodulator vector represents the pixel intensity. Since the stage velocity is not constant, this minimizes distortion since the image is formed as function of spatial distance rather than time. At 2-μm pixels, approximately half the size of the PSF and the lowest wavelength, with 50-nm encoder spacing, the counter outputs a single TTL pulse per 40 ticks (EncXY). The instrument can acquire images at any resolution as long as the pixel size is rounded to an integer multiple of the encoder spacing.
System performance is optimally stabilized by automatically adjusting several important parameters when tuning the laser, including repetition rate, pulse width, pointing angle, and detection sensitivity. These parameters are saved as a system configuration file for a subset of wavenumbers, typically the key classification bands. For hyperspectral scans where a predefined configuration is not available, a default set still provides acceptable performance for most wavenumbers. When a scan is first initiated, two subroutines are performed. First, the instrument finds the optimal focal point at various points across the scan field and calculates the best-fit plane that represents the substrate. The autofocus algorithm sweeps across z coordinates while monitoring the detector signal, which approximately follows a sinc-squared profile, until the global maximum is found. First, the operator must bring the sample near the focal point before the automation takes over the remaining fine adjustments. During the search, the signal is smoothed and the search window restricted to >5 µm to avoid converging onto errant local maxima. We autofocus only on bare substrate to avoid situations where the reflection off the sample surface exceeds the transflected signal resulting in an offset to the desired focal point. All system coordinates are then transformed by this matrix so its accuracy is critical. Small tilts can defocus the image by several microns for large specimens and create artifacts such as fades and ripples. The plane is shifted axially depending on the focal point of the objective at the specified wavenumber. Second, the system performs a test sweep calculating the minimum pixel dwell time as a function of stage velocity and setting the lock-in amplifier time constant to one-third this value. The instrument scans in both directions, creating a forward image and a backward image.
For each image, the background intensity is generated by surface fitting against empty regions of the image, predominantly along the edges. This compensates for slow power fluctuations as well as any residual sample tilt leftover from the mechanical correction. Forward and reverse images are then interlaced. These images have an offset due to system delays. When the pixel is triggered, assuming a constant time delay will still result in a spatial delay since velocity is constantly changing. The instrument records data throughout most of the acceleration period to maximize acquisition time. Any residual distortions are a function of velocity and signal delay. They are small, ∼0.02%, yet result in a misalignment gradually increasing to a several pixels over 10 s of millimeters. Therefore, aligning the odd and even rows in these forward and reverse images is a nonrigid image registration task. We estimate these local distortions and the displacement field that registers the reverse scan with the forward scan and the corrected images are warped and resampled using the original pixel grid (MATLAB; MathWorks). Next, to compensate for microscope drift over time arising from environmental changes, we register the adjacent wavenumber bands and aligning the entire stack using affine transforms (ImageJ; NIH). Last, the PSF of the optics at each wavenumber is simulated and used to deconvolve (50) the image according to the Tikhonov–Miller algorithm. The spectral images are rubber band baselined and visualized in ENVI + IDL (ITT Visual Information Solutions).
Supervised Classification.
First, regions of interest are identified by consulting the H&E-stained images annotated by a pathologist. These served as training data points for the algorithm to identify the spectral signatures associated with each class. These signatures were then used by the random forest classifier to map unknown pixels to the given classes (different histologic states). Random forest is an ensemble of decision trees in which a random subset of the training data (regions of interest) and random sampling of the features (spectral markers) is used to build a decision tree. Each decision tree votes for a particular class to be assigned to the sample at hand. The majority vote from all of the decision trees is used for the final class assignment. As random forest uses a parallel training algorithm, it is suitable for large datasets. The first set of spectral markers was manually determined by examining the individual cell spectra of all of the biological samples and also the average regions-of-interest spectra. The average spectra for different classes used in the model development is illustrated in SI Appendix, Fig. S4. It is evident from the average spectra that the absolute absorbance values are not that different across classes. Therefore, peak height and peak area ratios that can be mapped to known chemical features were used; 134 features (metrics) were defined, listed in SI Appendix, Table S2. Some of the most important features in both the models with their corresponding biochemical significance are listed in SI Appendix, Table S3. A supervised selection based on the classification error associated with each feature was also applied to the manually selected metrics to generate promising metrics candidates for a resilient approach in breast histopathology and future clinical applications. The number and type of histologic classes were identified based on breast histopathology. Breast cancer mainly comprises of four epithelial subtypes (hyperplasia, atypical hyperplasia, malignant, and normal) and three stromal compartments (dense, loose, and desmoplastic). Next, to ensure that both tumor and microenvironment features can be detected by the presented approach, an epithelial and stromal model were chosen. The epithelial model (6E) was composed of the four epithelial subtypes, all three stromal components combined into one as “stroma” and the remaining cellular entities (lymphocytes, red blood cells, necrosis, mucin, and secretions) merged into the “others” class. Similarly, the 6S model has three stromal components, malignant epithelium, benign epithelium (hyperplasia and normal combined together), and the “others” class. Both of these models are also described in SI Appendix, Fig. S4. The labeled pixels from the entire TMA were randomly split into half and assigned into the training and the validation sets. To assess the performance of the classifiers, prior distributions of different classes were varied to generate the ROC curves. This approach of ROC curve generation for a multiclass random forest classifier has been described in detail in our other work.
Finally, the top few features in the fingerprint region responsible for class differentiation were used to identify the frequencies for the DF measurement. A combination of these collected frequencies were then used for the classification of the DF-IR data.
Supplementary Material
Acknowledgments
We appreciate the technical assistance provided by Block Engineering. This work was supported in part by NIH Grant 2R01EB009745 (to R.B.), the Illinois Distinguished fellowship, and Beckman Graduate Student fellowship (to S.M.), and an NIH fellowship via the tissue microenvironment training program supported by Grant T32EB019944 (to S.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. T.J.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1719551115/-/DCSupplemental.
References
- 1.Hu M, et al. Distinct epigenetic changes in the stromal cells of breast cancers. Nat Genet. 2005;37:899–905. doi: 10.1038/ng1596. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Coussens LM. Accessories to the crime: Functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 3.Finak G, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 4.Beck AH, et al. 2011. Systematic analysis of breast cancer morphology uncovers stromal features associated with survival. Sci Transl Med 3108ra113.
- 5.Heijs B, et al. Histology-guided high-resolution matrix-assisted laser desorption ionization mass spectrometry imaging. Anal Chem. 2015;87:11978–11983. doi: 10.1021/acs.analchem.5b03610. [DOI] [PubMed] [Google Scholar]
- 6.Chaurand P, et al. Integrating histology and imaging mass spectrometry. Anal Chem. 2004;76:1145–1155. doi: 10.1021/ac0351264. [DOI] [PubMed] [Google Scholar]
- 7.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: Molecular snapshots of biochemical systems. Nat Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 8.Schwamborn K, Caprioli RM. Molecular imaging by mass spectrometry—looking beyond classical histology. Nat Rev Cancer. 2010;10:639–646. doi: 10.1038/nrc2917. [DOI] [PubMed] [Google Scholar]
- 9.Calligaris D, et al. Application of desorption electrospray ionization mass spectrometry imaging in breast cancer margin analysis. Proc Natl Acad Sci USA. 2014;111:15184–15189. doi: 10.1073/pnas.1408129111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baker MJ, Faulds K. Fundamental developments in clinical infrared and Raman spectroscopy. Chem Soc Rev. 2016;45:1792–1793. doi: 10.1039/c6cs90028c. [DOI] [PubMed] [Google Scholar]
- 11.Greenleaf JF, Bahn RC. Clinical imaging with transmissive ultrasonic computerized tomography. IEEE Trans Biomed Eng. 1981;28:177–185. doi: 10.1109/TBME.1981.324789. [DOI] [PubMed] [Google Scholar]
- 12.Orringer DA, et al. Rapid intraoperative histology of unprocessed surgical specimens via fibre-laser-based stimulated Raman scattering microscopy. Nat Biomed Eng. 2017;1:0027. doi: 10.1038/s41551-016-0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayerich D, et al. Stain-less staining for computed histopathology. Technology (Singap World Sci) 2015;3:27–31. doi: 10.1142/S2339547815200010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shemonski ND, et al. Computational high-resolution optical imaging of the living human retina. Nat Photonics. 2015;9:440–443. doi: 10.1038/NPHOTON.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baker MJ, et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat Protoc. 2014;9:1771–1791. doi: 10.1038/nprot.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bhargava R. Infrared spectroscopic imaging: The next generation. Appl Spectrosc. 2012;66:1091–1120. doi: 10.1366/12-06801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holton SE, Bergamaschi A, Katzenellenbogen BS, Bhargava R. Integration of molecular profiling and chemical imaging to elucidate fibroblast-microenvironment impact on cancer cell phenotype and endocrine resistance in breast cancer. PLoS One. 2014;9:e96878. doi: 10.1371/journal.pone.0096878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wald N, Bordry N, Foukas PG, Speiser DE, Goormaghtigh E. Identification of melanoma cells and lymphocyte subpopulations in lymph node metastases by FTIR imaging histopathology. Biochim Biophys Acta. 2016;1862:202–212. doi: 10.1016/j.bbadis.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez DC, Bhargava R, Hewitt SM, Levin IW. Infrared spectroscopic imaging for histopathologic recognition. Nat Biotechnol. 2005;23:469–474. doi: 10.1038/nbt1080. [DOI] [PubMed] [Google Scholar]
- 20.Benard A, et al. Infrared imaging in breast cancer: Automated tissue component recognition and spectral characterization of breast cancer cells as well as the tumor microenvironment. Analyst (Lond) 2014;139:1044–1056. doi: 10.1039/c3an01454a. [DOI] [PubMed] [Google Scholar]
- 21.Mayerich D, Walsh M, Schulmerich M, Bhargava R. Real-time interactive data mining for chemical imaging information: Application to automated histopathology. BMC Bioinformatics. 2013;14:156. doi: 10.1186/1471-2105-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suhalim JL, Boik JC, Tromberg BJ, Potma EO. The need for speed. J Biophotonics. 2012;5:387–395. doi: 10.1002/jbio.201200002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwak JT, et al. Improving prediction of prostate cancer recurrence using chemical imaging. Sci Rep. 2015;5:8758. doi: 10.1038/srep08758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nasse MJ, et al. High-resolution Fourier-transform infrared chemical imaging with multiple synchrotron beams. Nat Methods. 2011;8:413–416. doi: 10.1038/nmeth.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reddy RK, Walsh MJ, Schulmerich MV, Carney PS, Bhargava R. High-definition infrared spectroscopic imaging. Appl Spectrosc. 2013;67:93–105. doi: 10.1366/11-06568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian H, et al. Diagnosing benign and malignant lesions in breast tissue sections by using IR-microspectroscopy. Biochim Biophys Acta. 2006;1758:874–882. doi: 10.1016/j.bbamem.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 27.Verdonck M, et al. Breast cancer and melanoma cell line identification by FTIR imaging after formalin-fixation and paraffin-embedding. Analyst (Lond) 2013;138:4083–4091. doi: 10.1039/c3an00246b. [DOI] [PubMed] [Google Scholar]
- 28.Fabian H, Lasch P, Boese M, Haensch W. Infrared microspectroscopic imaging of benign breast tumor tissue sections. J Mol Struct. 2003;661–662:411–417. [Google Scholar]
- 29.Tian P, et al. Intraoperative diagnosis of benign and malignant breast tissues by Fourier transform infrared spectroscopy and support vector machine classification. Int J Clin Exp Med. 2015;8:972–981. [PMC free article] [PubMed] [Google Scholar]
- 30.Gao T, Feng J, Ci Y. Human breast carcinomal tissues display distinctive FTIR spectra: Implication for the histological characterization of carcinomas. Anal Cell Pathol. 1999;18:87–93. doi: 10.1155/1999/321357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kumar S, Desmedt C, Larsimont D, Sotiriou C, Goormaghtigh E. Change in the microenvironment of breast cancer studied by FTIR imaging. Analyst (Lond) 2013;138:4058–4065. doi: 10.1039/c3an00241a. [DOI] [PubMed] [Google Scholar]
- 32.Faist J, et al. Quantum cascade laser. Science. 1994;264:553–556. doi: 10.1126/science.264.5158.553. [DOI] [PubMed] [Google Scholar]
- 33.Bai Y, Slivken S, Kuboya S, Darvish SR, Razeghi M. Quantum cascade lasers that emit more light than heat. Nat Photonics. 2010;4:99–102. [Google Scholar]
- 34.Yao Y, Hoffman AJ, Gmachl CF. Mid-infrared quantum cascade lasers. Nat Photonics. 2012;6:432–439. [Google Scholar]
- 35.Yeh K, Kenkel S, Liu JN, Bhargava R. Fast infrared chemical imaging with a quantum cascade laser. Anal Chem. 2015;87:485–493. doi: 10.1021/ac5027513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bhargava R, Fernandez DC, Hewitt SM, Levin IW. High throughput assessment of cells and tissues: Bayesian classification of spectral metrics from infrared vibrational spectroscopic imaging data. Biochim Biophys Acta. 2006;1758:830–845. doi: 10.1016/j.bbamem.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 37.Kröger N, et al. Quantum cascade laser-based hyperspectral imaging of biological tissue. J Biomed Opt. 2014;19:111607. doi: 10.1117/1.JBO.19.11.111607. [DOI] [PubMed] [Google Scholar]
- 38.Bassan P, Weida MJ, Rowlette J, Gardner P. Large scale infrared imaging of tissue micro arrays (TMAs) using a tunable quantum cascade laser (QCL) based microscope. Analyst (Lond) 2014;139:3856–3859. doi: 10.1039/c4an00638k. [DOI] [PubMed] [Google Scholar]
- 39.Tiwari S, et al. Towards translation of discrete frequency infrared spectroscopic imaging for digital histopathology of clinical biopsy samples. Anal Chem. 2016;88:10183–10190. doi: 10.1021/acs.analchem.6b02754. [DOI] [PubMed] [Google Scholar]
- 40.Kröger-Lui N, et al. Rapid identification of goblet cells in unstained colon thin sections by means of quantum cascade laser-based infrared microspectroscopy. Analyst (Lond) 2015;140:2086–2092. doi: 10.1039/c4an02001d. [DOI] [PubMed] [Google Scholar]
- 41.Pilling MJ, Henderson A, Gardner P. Quantum cascade laser spectral histopathology: Breast cancer diagnostics using high throughput chemical imaging. Anal Chem. 2017;89:7348–7355. doi: 10.1021/acs.analchem.7b00426. [DOI] [PubMed] [Google Scholar]
- 42.Hughes C, et al. Introducing discrete frequency infrared technology for high-throughput biofluid screening. Sci Rep. 2016;6:20173. doi: 10.1038/srep20173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mao Y, Keller ET, Garfield DH, Shen K, Wang J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013;32:303–315. doi: 10.1007/s10555-012-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lloyd GR, Stone N. Method for identification of spectral targets in discrete frequency infrared spectroscopy for clinical diagnostics. Appl Spectrosc. 2015;69:1066–1073. doi: 10.1366/14-07677. [DOI] [PubMed] [Google Scholar]
- 46.Pounder FN, Reddy RK, Bhargava R. Development of a practical spatial-spectral analysis protocol for breast histopathology using Fourier transform infrared spectroscopic imaging. Faraday Discuss. 2016;187:43–68. doi: 10.1039/c5fd00199d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pilling MJ, et al. High-throughput quantum cascade laser (QCL) spectral histopathology: A practical approach towards clinical translation. Faraday Discuss. 2016;187:135–154. doi: 10.1039/c5fd00176e. [DOI] [PubMed] [Google Scholar]
- 48.Bhargava R. Towards a practical Fourier transform infrared chemical imaging protocol for cancer histopathology. Anal Bioanal Chem. 2007;389:1155–1169. doi: 10.1007/s00216-007-1511-9. [DOI] [PubMed] [Google Scholar]
- 49.Kole MR, Reddy RK, Schulmerich MV, Gelber MK, Bhargava R. Discrete frequency infrared microspectroscopy and imaging with a tunable quantum cascade laser. Anal Chem. 2012;84:10366–10372. doi: 10.1021/ac302513f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sage D, et al. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods. 2017;115:28–41. doi: 10.1016/j.ymeth.2016.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Leslie LS, et al. High definition infrared spectroscopic imaging for lymph node histopathology. PLoS One. 2015;10:e0127238. doi: 10.1371/journal.pone.0127238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Degnim AC, et al. Extent of atypical hyperplasia stratifies breast cancer risk in 2 independent cohorts of women. Cancer. 2016;122:2971–2978. doi: 10.1002/cncr.30153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elmore JG, et al. Diagnostic concordance among pathologists interpreting breast biopsy specimens. JAMA. 2015;313:1122–1132. doi: 10.1001/jama.2015.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang ML, et al. Comparison of the accuracy of US-guided biopsy of breast masses performed with 14-gauge, 16-gauge and 18-gauge automated cutting needle biopsy devices, and review of the literature. Eur Radiol. 2016;27:2928–2933. doi: 10.1007/s00330-016-4651-5. [DOI] [PubMed] [Google Scholar]
- 55.Martin MC, et al. 3D spectral imaging with synchrotron Fourier transform infrared spectro-microtomography. Nat Methods. 2013;10:861–864. doi: 10.1038/nmeth.2596. [DOI] [PubMed] [Google Scholar]
- 56.Ukkonen H, et al. Changes in the microenvironment of invading melanoma and carcinoma cells identified by FTIR imaging. Vib Spectrosc. 2015;79:24–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.